Abstract
Genetically encoded biological clocks are found broadly throughout life on Earth, where they generate circadian (about a day) rhythms that synchronize physiology and behavior with the daily light/dark cycle. Although the genetic networks that give rise to circadian timing are now fairly well established, our understanding of how the proteins that constitute the molecular ‘cogs’ of this biological clock regulate the intrinsic timing, or period, of circadian rhythms has lagged behind. New studies probing the biochemical and structural basis of clock protein function are beginning to reveal how assemblies of dedicated clock proteins form and evolve through post-translational regulation to generate circadian rhythms. This review will highlight some recent advances providing important insight into the molecular mechanisms of period control in mammalian clocks with an emphasis on structural analyses related to CK1-dependent control of PER stability.
Keywords: feedback loop, degradation, proteins, structure, dynamics, post-translational modifications
1.1. Introduction to the mammalian circadian clock
Circadian rhythms arise from genetically encoded molecular clocks that originate at the cellular level and operate with an intrinsic period of about a day (circa diem). The timekeeping encoded by these self-sustained biological clocks persists in constant darkness but responds acutely to changes in daily environmental cues, like light, to keep internal clocks aligned with the external environment [1]. Therefore, circadian rhythms are used to help organisms predict changes in their environment and temporally program regular changes in their behavior and physiology [2].
The circadian clock in mammals is driven by several interlocked transcription-translation feedback loops (TTFLs) [3]. The integration of these interlocked loops is a complicated process that is orchestrated by a core feedback loop in which the heterodimeric transcription factor complex, CLOCK:BMAL1, promotes the transcription of its own repressors, Cryptochrome and Period (CRY and PER) as well as other clock-controlled genes (Figure 1A). Notably, there is some redundancy in this system as paralogs of both PER (PER1–3) and CRY (CRY1–2) proteins participate in the core TTFL. In general, these proteins accumulate in the cytoplasm, interact with one another, and recruit a kinase that is essential for the clock, Casein Kinase 1 δ/ε (CK1δ/ε), eventually making their way into the nucleus as a large complex to repress CLOCK:BMAL1 transcriptional activity [4, 5].
Figure 1. Changes to the core molecular clock control mammalian circadian period.
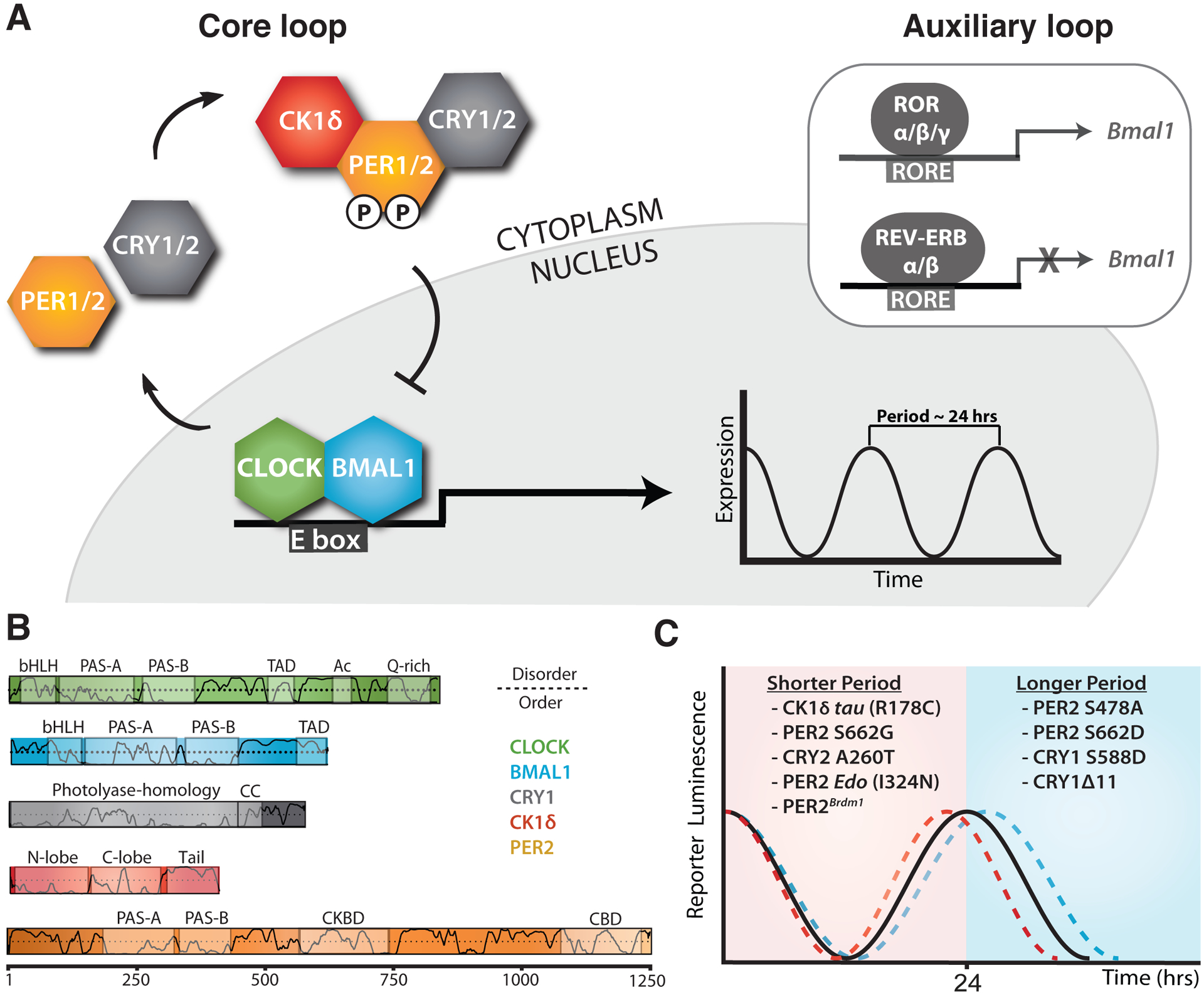
(A) Simplified schematic of the core mammalian feedback loop mediated by CLOCK:BMAL1 expression of CRY and PER genes with an auxiliary loop that consists of the retinoic acid receptor-related orphan receptor α (RORα) and the nuclear receptors REV-ERBα/β. (B) Functional domain architecture of core clock proteins with structured domains (boxes) and traces indicating the propensity for intrinsic disorder [92]: bHLH, basic helix-loop-helix; PAS, PER-ARNT-SIM; TAD, transactivation domain; Ac, acetyl-CoA binding, Q-rich, polyglutamine; CC, coiled-coil; CKBD, Casein Kinase 1-binding domain; and CBD, CRY-binding domain with residue numbering underneath. (C) Period effects from select mammalian clock alleles of core clock proteins.
Despite this relatively simple model for the core circadian feedback loop, there is growing evidence that different repressor complexes that exist throughout the evening may regulate CLOCK:BMAL1 in distinct ways [6, 7]. PER proteins are essential for the nucleation of large protein complexes that form early in the repressive phase [4] by acting as stoichiometrically-limiting factors that are temporally regulated through oscillations in expression [8, 9]. As a consequence, circadian rhythms can be disrupted by constitutively overexpressing PER proteins [8] or established de novo with tunable periods through inducible regulation of PER oscillations [10]. CK1δ/ε regulate PER abundance by controlling its degradation post-translationally [11–13]; accordingly, mutations in the kinases [14, 15] or their phosphorylation sites on PER2 [16, 17] can induce large (~4 hr) changes in circadian period, firmly establishing this regulatory mechanism as a central regulator of the mammalian circadian clock. CRY proteins bind directly to CLOCK:BMAL1 [18] and mediate the interaction of PER-CK1δ/ε complexes with CLOCK:BMAL1 leading to phosphorylation of the transcription factor and its release from DNA [6, 19] as well as acting as direct repressors of CLOCK:BMAL1 activity by sequestering the transcriptional activation domain (TAD) of BMAL1 from coactivators like CBP/p300 [20, 21].
Much remains to be elucidated about the assembly and activity of these core clock proteins, but insight into the molecular basis of circadian period determination is growing thanks to the integration of genetic studies from model organisms and humans with biochemical, structural and cell-based studies. High-resolution structures have now been determined for most of the globular domains of core clock proteins and some of their complexes [22], with a growing appreciation for the important role that flexible linkers and intrinsically disordered regions play in tuning clock protein function and clock timing (Figure 1B) [23]. We will focus here on recent advances in our understanding of the mechanisms of period control by some of the negative elements of the core feedback loop (Figure 1C), highlighting the nanoscale structural and dynamic properties of clock proteins that influence their functional roles as repressors within the core TTFL. For a review of mutations in human CRY1 [24, 25] and CRY2 [26] that influence circadian timing, please refer to [27].
1.2. Post-translational regulation of PER2 stability
Although transcriptional regulation by CLOCK:BMAL1 and downstream transcription factors is essential for generation of robust circadian rhythms, it is becoming increasingly clear that post-transcriptional and post-translational modifications of core clock components play an important role in both the generation of circadian rhythms and determination of its intrinsic period [28]. While many studies have identified roles for post-translational modification of CRY and PER proteins in clock timing, we will focus here on an in-depth analysis of the regulation of PER2 by CK1δ/ε, as it relies on an elaborate integration of post-translational modifications that ultimately determine the relative abundance of PER2 needed to maintain a ~24-hour period [29].
1.2.1. Structural organization of PER2
PER1 and PER2 serve as interaction hubs for both CK1δ/ε and cryptochromes with dedicated binding sites that maintain stable complexes with these core clock proteins throughout most of the repressive phase of the clock each day [4, 5] (Figure 2A), a feature that is notably absent in PER3 [30]. The flexibility of PER proteins likely contributes to their role as labile scaffolds for transcriptional regulators of the clock; PER1–3 are all predominantly intrinsically disordered, conferring a susceptibility to regulation by post-translational modifications and promiscuity for interaction partners [31–35] that is common among other intrinsically disordered proteins (IDPs) [36]. Two tandem PER-ARNT-SIM (PAS) domains present in the N-terminus of all three PER isoforms allow them to form homodimers and heterodimers (Figure 2A) [37–39] mediated by the β-sheet surface of the PAS-B domain (Figure 2B). Deletion of the core PAS-B motif in the Per2Brdm1 mutant leads to a loss of circadian rhythms, demonstrating that protein-protein interactions facilitated by this region are essential for clock function [40]. Notably, mutation of just a single residue, W419E, at the dimer interface in the mouse PER2 PAS-B domain potently disrupts formation of homodimers [37] and was recently shown to reduce phosphorylation by CK1δ [41]. Moreover, the PER2 earlydoors (Edo) mouse possesses a point mutation (I324N) in the interdomain linker connecting the PAS-A and B domains that reduces PER2 stability to shorten the circadian period [42]. Collectively, these findings demonstrate that dimerization via the PER PAS domains is critical for protein stability and clock timing, although more work is needed to understand the exact role that PAS domains play in orchestrating clock protein complexes and PER turnover.
Figure 2. PER2 stability is tightly regulated by post-translational modifications.
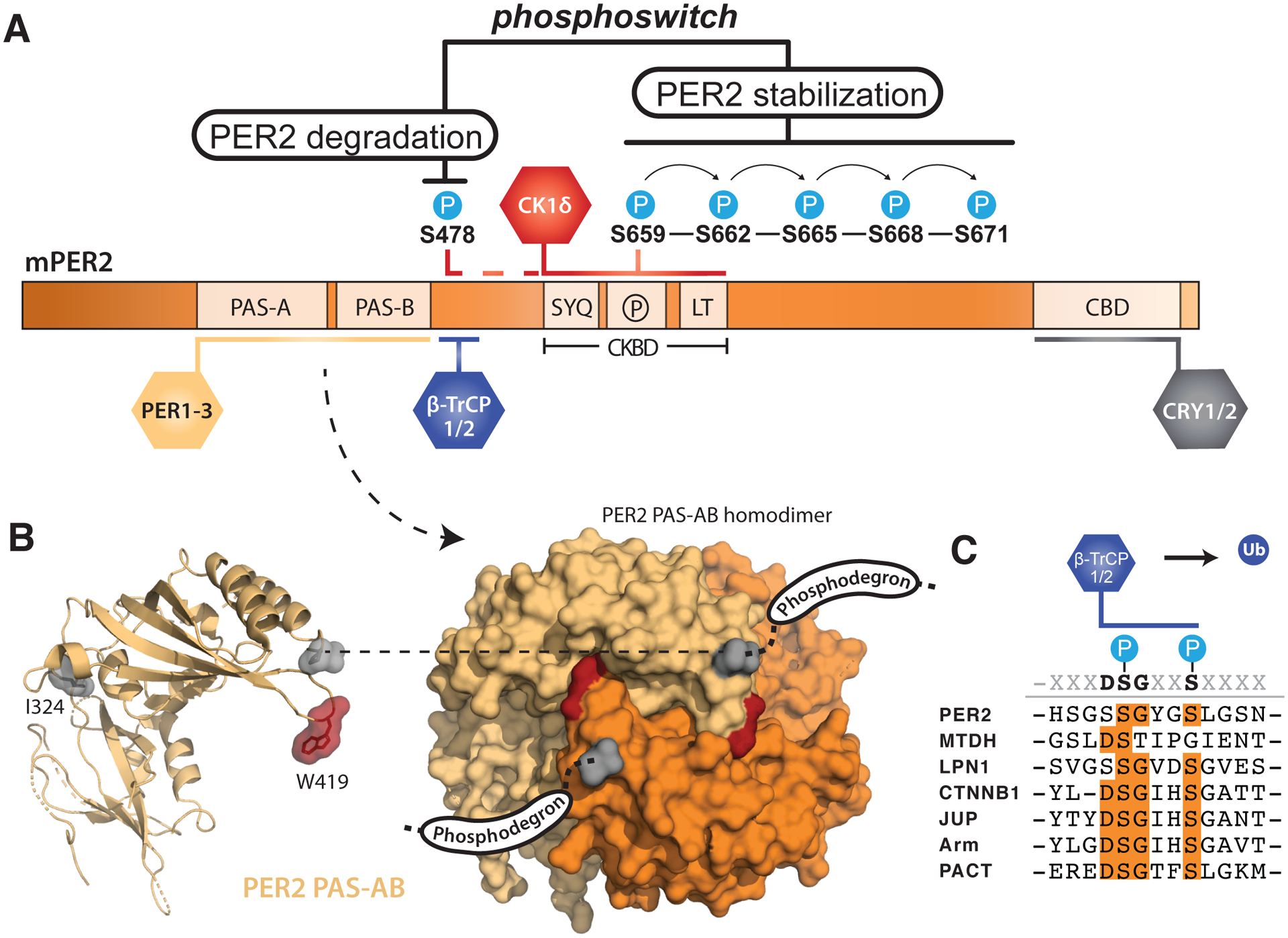
(A) Domain map of mouse PER2 depicting clock protein binding sites and the CK1δ-dependent phosphoswitch model. CK1δ-dependent phosphorylation of the FASP region (P) within the CKBD antagonizes activity at the upstream phosphodegron, which is used to recruit the E3 ubiquitin ligases, β-TrCP1/2, for subsequent proteasomal degradation of PER2. (B) Left, cartoon representation of the mouse PER2 PAS-AB domain monomer with the location of the Edo mutation (I324N) and Trp residue required for dimerization (W419) highlighted, PDB: 3GDI. Right, surface representation of the PER2 PAS-AB homodimer. The phosphodegron is located in a disordered region immediately downstream of the PAS-B domain; the C-terminal residue is depicted in surface mode (gray) to show how each respective phosphodegron is poised to protrude from the same face of the dimer. (C) Alignment of phosphodegrons within human proteins that are targeted by β-TrCP1/2.
1.2.2. Control of PER2 degradation by phosphodegrons
The turnover of PER2 is primarily mediated by CK1δ/ε-dependent phosphodegrons to intimately link kinase activity with PER2 stability. One well-studied phosphodegron site of mouse PER2 (S478) is located immediately downstream of the PAS-B domain. Dimerization of the PER2 PAS-AB domains positions the phosphodegron site (S478) of each monomer to protrude from the same face of the PAS-AB domain homodimer (Figure 2B) [37]. This phosphodegron largely conforms to the canonical β-TrCP recognition motif, DSGϕXS, where ϕ is a hydrophobic residue and the two conserved serines become phosphorylated to make the substrate competent for β-TrCP recognition (Figure 2C) [43]. CK1δ/ε phosphorylation of S478 in mouse PER2, the first of the two serines in the motif, is required for interaction with the E3 ubiquitin ligases β-TrCP1/2, leading to ubiquitination of PER2 and its proteasomal degradation [12, 44, 45]. However, this PAS-B phosphodegron is unique to PER2; PER1 utilizes a different CK1δ/ε-dependent phosphodegron N-terminal to the tandem PAS domains [13]. This N-terminal phosphodegron is also conserved in PER2 [44] and may play an auxiliary role in its turnover, as clock timing was only modestly impacted in the PER2 S478A transgenic mouse [46]. An interaction with the E3 ubiquitin ligase MDM2 also influences PER2 stability independently of CK1δ/ε activity [47], opening the door for a complex integration of signals to mediate PER2 degradation.
1.2.3. The Casein Kinase-Binding Domain stabilizes PER2 through a phosphoswitch
Although other kinases such as CK1α [48], CK2 [49], SIK3 [50] and Cdk5 [51] phosphorylate PER2, CK1δ/ε are the only kinases that stably associate with PER2 throughout the night, moving from the cytoplasm into the nucleus with the other core clock proteins [4, 5]. CK1δ and the related isoform CK1ε bind to the Casein Kinase-Binding Domain (CKBD) in PER2 via two conserved motifs that flank a serine-rich region [12, 30]. Notably, mutation of the first residue (S662G) in a series of five consecutive serines that are phosphorylated in human PER2 markedly decreases its stability and shortens circadian period by ~4 hours to manifest as Familial Advanced Sleep Phase Syndrome (FASPS) [16, 17]. Because CK1δ/ε-dependent phosphorylation of PER2 in this region links circadian timekeeping to this human sleep disorder, the serine-rich cluster in the CKBD has been named the FASP region.
Recent studies have begun to elucidate the molecular basis for CK1δ/ε activity in the FASP region of PER2 to understand how it exerts such powerful control over circadian period. Phosphorylation of the first serine in this cluster (S659 in mouse, S662 in humans) by CK1δ leads to the obligately sequential phosphorylation of downstream serines (Figure 2A) [52] Therefore, the human S662G FASPS allele eliminates the ability of CK1δ to prime its activity downstream, disrupting all phosphorylation in the FASP region. There is strong evidence that FASP phosphorylation plays a critical role in stabilizing PER2 protein [16], as the S662G mutation in human PER2 (or the analogous S659A mutation in mouse PER2) [53] leads to premature turnover of the protein and a dramatically shorter circadian period of ~20 hours in a transgenic mouse model, while use of a phosphomimetic mutation (S662D) in human PER2 that presumably leads to constitutive priming of sequential FASP phosphorylation confers a long period of ~25 hours in vivo [17]. Although it is not yet known how FASP phosphorylation contributes to regulation of PER2 stability, mutation of the priming serine that blocks phosphorylation of FASP downstream serines increases CK1δ/ε activity at the phosphodegron site S478 [54] to suggest that the phospho-FASP region could antagonize CK1δ/ε activity at the phosphodegron site S478. The opposing effects of FASP and phosphodegron phosphorylation likely involves cellular phosphatases like PP1 that contribute to CK1δ/ε-dependent regulation of circadian period through PER2 [55], although there could also be a direct mechanistic link between FASP phosphorylation and regulation of CK1δ/ε activity. In fact, the functional linkage of phosphorylation at the FASP region and phosphodegron by CK1δ/ε has been described as a phosphoswitch that introduces a phase-specific delay to PER2 degradation necessary for proper circadian timekeeping (Figure 2A) [56]. Interestingly, while introduction of the analogous priming site mutation in mouse PER1 (S714G) destabilized PER1 and led to a shorter circadian period, it also caused an advance in feeding rhythms not seen in the PER2 mutant [57], suggesting that further study of the regulation of PER turnover could help uncouple distinct functions of PER1 and PER2 in control of circadian period and clock outputs.
1.2.4. Other post-translational modifications influence PER2 stability
PER proteins are also subjected to a number of other post-translational modifications aside from phosphorylation. PER2 is O-GlcNacylated within the FASP region [58], modifying the priming serine along with two sites downstream. Both O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), the enzymes responsible for adding or hydrolyzing O-GlcNAc, respectively, are expressed or activated in a circadian manner, and factors that increase O-GlcNacylation also lead to a concomitant decrease in FASP phosphorylation that reduces PER2 protein levels [58, 59]. These results support a model for competition between O-GlcNacylation and phosphorylation at this key regulatory region, suggesting a mechanism by which glucose metabolism could modulate the circadian clock by antagonizing phosphorylation of PER2 in the stabilizing FASP region to represent a direct link between the circadian clock and metabolism as a “nutrition switch” [29, 58].
Acetylation also plays an important yet enigmatic role in PER2 regulation of the clock, first observed through manipulation of SIRT1, the nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase. Although PER2 becomes acetylated as it accumulates throughout the repressive phase of the circadian clock [60], the identity of the acetyltransferase(s) that modify PER2 is currently not known, nor is it known where PER2 is acetylated throughout the protein. Nonetheless, loss of SIRT1 results in elevated levels of acetylated PER2 in mouse liver to attenuate the robustness of circadian rhythms, while overexpression of SIRT1 facilitates PER2 degradation [60, 61]. Because acetylation and ubiquitination both target lysine residues, it is possible that these modifications compete for the same residues to directly control PER2 stability. However, there is some evidence that regulation of PER2 by acetylation could be more complicated, as acetylation at K680 on mouse PER2, located downstream of the serine cluster in the FASP region, is hyperacetylated following inhibition of SIRT1 and leads to a decrease in FASP phosphorylation [62], suggesting that an interplay between acetylation and phosphorylation of the FASP region could also control CK1δ/ε activity on PER2 to regulate the balance of the phosphoswitch.
1.3. A central role for CK1 in eukaryotic circadian period determination
The timing of eukaryotic circadian rhythms from green algae to humans is heavily influenced by CK1δ/ε and its related orthologs [15, 63]. Like other Ser/Thr kinases, CK1δ/ε has a typical two-lobed structure (Figure 3A), but little is known about the molecular mechanisms by which activity of the CK1 family is regulated. The activation loop is one key feature that distinguishes the CK1 family from other Ser/Thr kinases (Figure 3B) [64]. Unlike many other kinases, the kinase domain of CK1 family members is not regulated by activation loop phosphorylation; therefore, they are considered to be constitutively active [65]. The CK1 family acts as phosphate-directed kinases that preferentially recognize a D/E/pSxxS consensus motif, where a phosphorylated serine or a similar negative charge within the substrate templates activity at a serine located 3 or 4 residues downstream (Figure 3A) [66]. Interestingly, at least two functionally important CK1δ/ε-dependent phosphorylation sites on PER2, the phosphodegron and the FASP priming site, do not conform to this consensus motif and likely serve as slow, rate-limiting steps for PER2 regulation [52, 54, 67]. Therefore, understanding the molecular basis for kinase activity and substrate selectivity by CK1δ/ε has the potential to yield important insights into circadian timekeeping. In particular, a better understanding of the molecular mechanisms underpinning CK1δ/ε-dependent phosphorylation of PER will provide a framework for treating circadian disorders by targeting CK1δ/ε to modulate the clock [68].
Figure 3. Regulatory mechanisms control CK1δ activity in the mammalian clock.
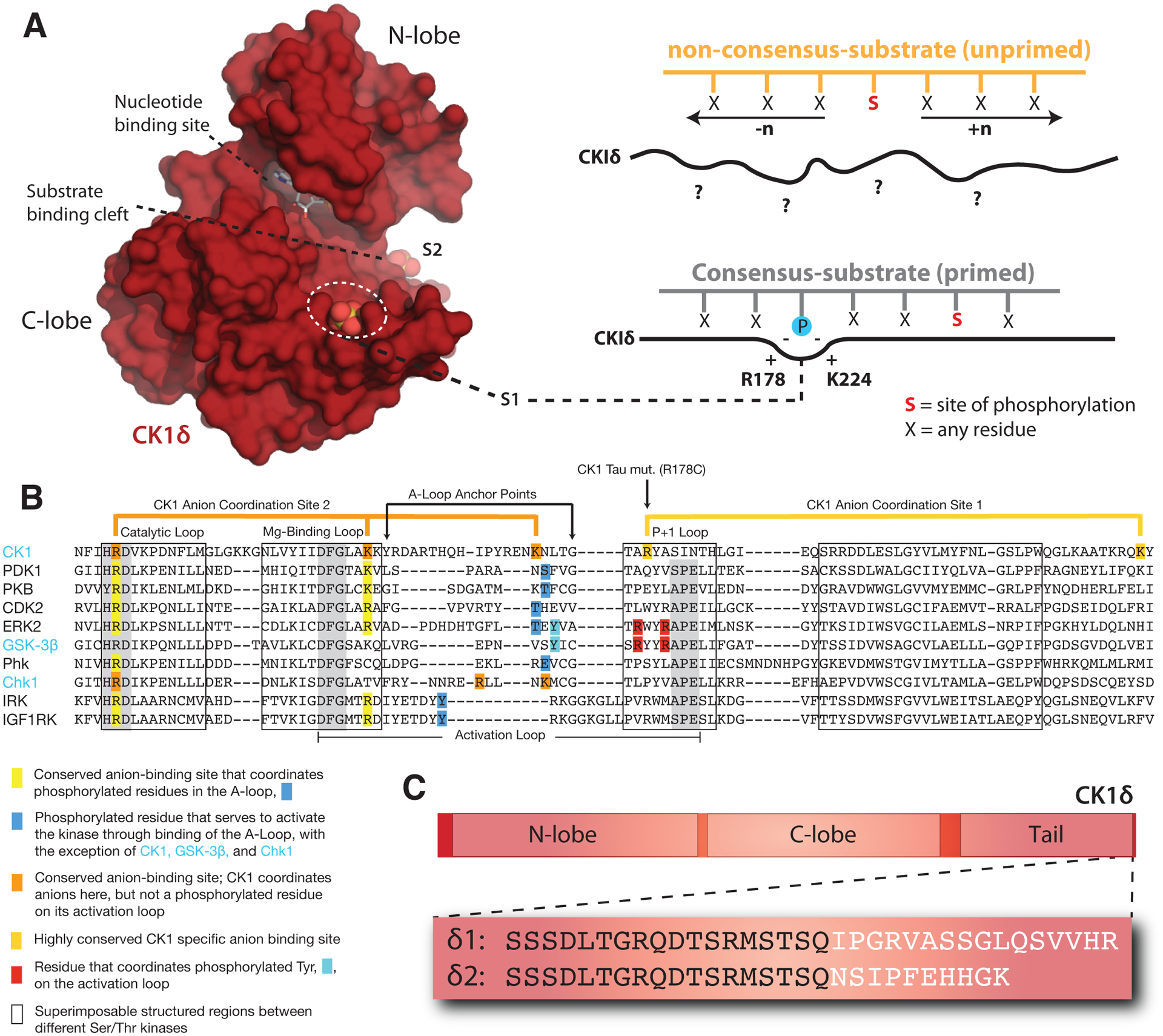
(A) Left, Surface representation of the CK1δ kinase domain, PDB: 5×17. The substrate binding cleft is flanked by two highly conserved anion binding sites, S1 and S2. Right, CK1 is thought to use S1 to bind phosphorylated (or ‘primed’) substrates, leading to phosphorylation of the CK1 consensus motif, pSxxS. CK1 also exhibits non-consensus activity on unprimed sites. (B) Alignment of the activation loop and nearby regions in representative Ser/Thr kinases. CK1 lacks the conserved APE motif in the P+1 loop involved in substrate recognition. (C) Differences between the alternatively spliced variants CK1δ1 and CK1δ2 at the C-terminus of the kinase.
1.3.1. Anion-dependent regulation of a structural switch in CK1δ
The kinase domain of CK1δ/ε contains several highly conserved anion binding sites located around the C-terminal lobe, including two that flank either side of the substrate binding cleft (Figure 3A, B) [69–71]. We recently showed that these anion binding sites regulate the overall kinase activity of CK1δ/ε, as well as influence the substrate specificity of the kinase at both consensus and non-consensus sites [54]. The significance of these highly conserved anion binding sites was initially suggested by the discovery of the first period-altering allele in mammals, the CK1ε tau allele that causes a dramatically shortened circadian period of ~20 hours [72]. The R178C substitution in the tau kinase was predicted to disrupt an anion-binding pocket near the substrate binding region to decrease CK1δ/ε activity [14]. While the tau mutant kinase did exhibit reduced activity on some generic kinase substrates (e.g., casein and phosvitin) as well as the FASP region of PER2, it led to a paradoxical gain of function at the PER2 phosphodegron that decreased stability of the protein [54, 73].
Crystal structures of the tau kinase domain recently revealed that disruption of the anion binding pocket at S1 in the mutant is linked to an allosteric structural switch in the activation loop that encodes a preference for the PER2 PAS-B phosphodegron site S478 [54]. Allostery is a common regulatory feature of protein kinases that allows for a switch-like, ultrasensitive regulation of their biological activity [74]. The activation loop and flanking regions distinguish CK1 from all other Ser/Thr kinases (Figure 3B) [75], containing residues involved in the coordination of anions at three conserved sites, S1-S3. Therefore, these sites likely play a role in the CK1 family-specific regulation of kinase activity, perhaps through binding of anionic, phosphorylated residues. Interestingly, the entire substrate binding cleft that allosterically links anion binding to substrate selectivity is 95% identical from humans to green algae [54], suggesting that the mechanisms discovered in mammalian CK1δ/ε may also regulate kinase activity and circadian period across other eukaryotic clocks.
1.3.2. Regulation of CK1δ/ε activity by its disordered C-terminal tail
The kinase activity of both CK1δ and CK1ε is inhibited by autophosphorylation of an intrinsically disordered inhibitory tail that follows the kinase domain to set these isoforms apart from other members of the CK1 family [76–79]. Because the full-length kinase autophosphorylates and slowly inactivates itself in vitro, most biochemical studies exploring the activity of CK1δ/ε on clock proteins utilize the truncated, constitutively active protein (CK1δ/ε ΔC) [52, 54, 70, 80], although new studies are finally beginning to explore the consequences of autophosphorylation in more detail [81, 82]. However, not much is known yet about how the phosphorylated tail interacts with the kinase domain to inhibit its activity; several autophosphorylation sites were previously identified on CK1ε at S323, T325, T334, T337, S368, S405, S407 and S408 using limited proteolysis and phosphatase treatment [78] or through Ser/Thr to Ala substitutions in vitro [79], although it is currently not known which (if any) of these sites are important for kinase regulation of the clock. One potential interface has been mapped between the kinase domain and autoinhibitory tail through crosslinking and mass spectrometry to suggest that the tail might dock some phosphorylated Ser/Thr residues close to the anion binding sites near the active site [83]. This study also provided evidence that the tail may be able to regulate substrate binding, and therefore control specificity of the kinase, by comparing the activity of CK1α, a tailless kinase, with CK1ε on two substrates, PER2 and Disheveled [83]. Understanding the role of tail autophosphorylation and its regulation of kinase activity is sure to shed light on control of circadian rhythms by CK1δ/ε.
Some sites within the C-terminal tail of CK1δ and/or CK1ε are known to be phosphorylated by other kinases, such as AMPK [84], PKA [85], Chk1 [86], PKCα [87], and cyclin-dependent kinases [88, 89]. PKA phosphorylates S370 in CK1δ to reduce its kinase activity; consistent with this, mutation of S370 to alanine increases CK1-dependent ectopic dorsal axis formation in Xenopus laevis [85]. Chk1 and PKCα also reduce CK1δ kinase activity through phosphorylation of overlapping sites at S328, T329, S331, S370, and T397 in the tail of rat CK1δ [86, 87]. Phosphorylation of CK1δ T347 influences its activity on PER2 in cells, and was found to be phosphorylated by proline-directed cyclin-dependent kinases rather than autophosphorylation [88]. CDK2 was also found to reduce the activity of rat CK1δ in vitro through phosphorylation of additional sites at T329, S331, T344, S356, S361, and T397 [89]. Unlike the other kinases listed here, phosphorylation of S389 on CK1ε by AMPK increases the apparent kinase activity on the PER2 phosphodegron in cells; consequently, activation of AMPK with metformin increased the degradation of PER2 [84]. Therefore, the phosphorylation of CK1δ and/or CK1ε tails by these other kinases therefore has the potential to link its regulation of PER2 and the circadian clock to metabolism, DNA damage response, and the cell cycle.
There is now strong evidence that the C-terminus of CK1δ plays a direct role in regulation of circadian period. Recently, tissue-specific methylation of CK1δ was shown to regulate alternative splicing of the kinase into two unique isoforms, δ1 and δ2, that differ only by the extreme C-terminal 15 residues (Figure 3C) [90]. Remarkably, expression of the canonical δ1 isoform decreases PER2 half-life and circadian period, while the slightly shorter δ2 isoform increases PER2 half-life and circadian period [90]. Further biochemical studies revealed that these two variants exhibit differential activity on the stabilizing priming site of the PER2 FASP region––the δ1 isoform has a lower activity than δ2, which also closely resembles the C-terminus of the ε isoform [52]. These data suggest that a very short region at the C-terminal end of the tail could play a major role in regulation of CK1δ and the PER2 phosphoswitch to control circadian period. This is bolstered by the discovery of a missense mutation in the same region of the CK1ε tail at S408N in humans that has been associated with protection from Delayed Sleep Phase Syndrome (DSPS) and Non-24-hr Sleep-Wake Syndrome (N-24) [91]. Further studies will help to reveal biochemical mechanisms behind regulation of kinase activity and substrate selectivity by the C-terminal tail of CK1δ and CK1ε to determine how they play into regulation of circadian rhythms.
1.4. Concluding remarks
While there is much more to be learned, data from human genetics and mammalian model systems are finally being integrated with biochemical and structural studies of the core clock components to provide clues to the molecular basis of the circadian clock. Recent advances in biophysical techniques, such as cryo-EM, will help to further enhance our understanding of how the molecular ‘cogs’ of the core clock come together to affect circadian period and influence human behavior and physiology.
Highlights.
Transcription-translation feedback loops underlie mammalian circadian rhythms
Biochemical studies reveal atomic details of clock proteins and their functions
Inherited changes to clock proteins can alter their structure and/or dynamics
Nanoscale studies of clock proteins reveal mechanisms of circadian period control
Acknowledgements
We would like to thank our collaborators Rajash Narasimamurthy, David Virshup (Duke-NUS Medical School) and Clarisse Ricci (UC San Diego) for fruitful conversations that have helped to frame our understanding of this system.
Funding
This work was supported by the National Institutes of Health (GM107069).
References
- 1.Aschoff J: Circadian Rhythms in Man. Science 1965, 148(3676):1427–1432. [DOI] [PubMed] [Google Scholar]
- 2.Bass J, Lazar MA: Circadian time signatures of fitness and disease. Science 2016, 354(6315):994–999. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi JS: Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 2017, 18(3):164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aryal RP, Kwak PB, Tamayo AG, Gebert M, Chiu PL, Walz T, Weitz CJ: Macromolecular Assemblies of the Mammalian Circadian Clock. Mol Cell 2017, 67(5):770–782 e776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM: Posttranslational mechanisms regulate the mammalian circadian clock. Cell 2001, 107(7):855–867. [DOI] [PubMed] [Google Scholar]
- 6.Chiou YY, Yang Y, Rashid N, Ye R, Selby CP, Sancar A: Mammalian Period represses and de-represses transcription by displacing CLOCK-BMAL1 from promoters in a Cryptochrome-dependent manner. Proc Natl Acad Sci U S A 2016, 113(41):E6072–E6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS: Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338(6105):349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo SH, Takahashi JS, Lee C: Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell 2009, 36(3):417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Chen R, Lee HM, Lee C: Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem 2011, 286(9):7033–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Alessandro M, Beesley S, Kim JK, Chen R, Abich E, Cheng W, Yi P, Takahashi JS, Lee C: A tunable artificial circadian clock in clock-defective mice. Nat Commun 2015, 6:8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akashi M, Tsuchiya Y, Yoshino T, Nishida E: Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol Cell Biol 2002, 22(6):1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM: Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol 2005, 25(7):2795–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirogane T, Jin J, Ang XL, Harper JW: SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem 2005, 280(29):26863–26872. [DOI] [PubMed] [Google Scholar]
- 14.Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS: Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 2000, 288(5465):483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH: Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 2005, 434(7033):640–644. [DOI] [PubMed] [Google Scholar]
- 16.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH: An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 2001, 291(5506):1040–1043. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptacek LJ: Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell 2007, 128(1):59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A: Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem 2011, 286(29):25891–25902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao X, Yang Y, Selby CP, Liu Z, Sancar A: Molecular mechanism of the repressive phase of the mammalian circadian clock. Proc Natl Acad Sci U S A 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gustafson CL, Parsley NC, Asimgil H, Lee HW, Ahlbach C, Michael AK, Xu H, Williams OL, Davis TL, Liu AC et al. : A Slow Conformational Switch in the BMAL1 Transactivation Domain Modulates Circadian Rhythms. Mol Cell 2017, 66(4):447–457 e447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Gustafson CL, Sammons PJ, Khan SK, Parsley NC, Ramanathan C, Lee HW, Liu AC, Partch CL: Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat Struct Mol Biol 2015, 22(6):476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Partch CL: Orchestration of Circadian Timing by Macromolecular Protein Assemblies. J Mol Biol 2020, 432(12):3426–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunlap JC, Loros JJ: Just-So Stories and Origin Myths: Phosphorylation and Structural Disorder in Circadian Clock Proteins. Mol Cell 2018, 69(2):165–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parico GCG, Perez I, Fribourgh JL, Hernandez BN, Lee HW, Partch CL: The human CRY1 tail controls circadian timing by regulating its association with CLOCK:BMAL1. Proc Natl Acad Sci U S A 2020, 117(45):27971–27979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patke A, Murphy PJ, Onat OE, Krieger AC, Ozcelik T, Campbell SS, Young MW: Mutation of the Human Circadian Clock Gene CRY1 in Familial Delayed Sleep Phase Disorder. Cell 2017, 169(2):203–215 e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirano A, Shi G, Jones CR, Lipzen A, Pennacchio LA, Xu Y, Hallows WC, McMahon T, Yamazaki M, Ptacek LJ et al. : A Cryptochrome 2 mutation yields advanced sleep phase in humans. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parico GCG, Partch CL: The tail of cryptochromes: an intrinsically disordered cog within the mammalian circadian clock. Cell Commun Signal 2020, 18(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crosby P, Partch CL: New insights into non-transcriptional regulation of mammalian core clock proteins. J Cell Sci 2020, 133(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirano A, Fu YH, Ptacek LJ: The intricate dance of post-translational modifications in the rhythm of life. Nat Struct Mol Biol 2016, 23(12):1053–1060. [DOI] [PubMed] [Google Scholar]
- 30.Lee C, Weaver DR, Reppert SM: Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock. Mol Cell Biol 2004, 24(2):584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, Rosbash M, Schibler U: PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 2005, 308(5722):693–696. [DOI] [PubMed] [Google Scholar]
- 32.Duong HA, Robles MS, Knutti D, Weitz CJ: A molecular mechanism for circadian clock negative feedback. Science 2011, 332(6036):1436–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duong HA, Weitz CJ: Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol 2014, 21(2):126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Padmanabhan K, Robles MS, Westerling T, Weitz CJ: Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 2012, 337(6094):599–602. [DOI] [PubMed] [Google Scholar]
- 35.Robles MS, Boyault C, Knutti D, Padmanabhan K, Weitz CJ: Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science 2010, 327(5964):463–466. [DOI] [PubMed] [Google Scholar]
- 36.Oldfield CJ, Dunker AK: Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem 2014, 83:553–584. [DOI] [PubMed] [Google Scholar]
- 37.Hennig S, Strauss HM, Vanselow K, Yildiz O, Schulze S, Arens J, Kramer A, Wolf E: Structural and functional analyses of PAS domain interactions of the clock proteins Drosophila PERIOD and mouse PERIOD2. PLoS Biol 2009, 7(4):e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kucera N, Schmalen I, Hennig S, Ollinger R, Strauss HM, Grudziecki A, Wieczorek C, Kramer A, Wolf E: Unwinding the differences of the mammalian PERIOD clock proteins from crystal structure to cellular function. Proc Natl Acad Sci U S A 2012, 109(9):3311–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yagita K, Yamaguchi S, Tamanini F, van Der Horst GT, Hoeijmakers JH, Yasui A, Loros JJ, Dunlap JC, Okamura H: Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev 2000, 14(11):1353–1363. [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A: The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 1999, 400(6740):169–173. [DOI] [PubMed] [Google Scholar]
- 41.Beesley S, Kim DW, D’Alessandro M, Jin Y, Lee K, Joo H, Young Y, Tomko RJ Jr., Faulkner J, Gamsby J et al. : Wake-sleep cycles are severely disrupted by diseases affecting cytoplasmic homeostasis. Proc Natl Acad Sci U S A 2020, 117(45):28402–28411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Militi S, Maywood ES, Sandate CR, Chesham JE, Barnard AR, Parsons MJ, Vibert JL, Joynson GM, Partch CL, Hastings MH et al. : Early doors (Edo) mutant mouse reveals the importance of period 2 (PER2) PAS domain structure for circadian pacemaking. Proc Natl Acad Sci U S A 2016, 113(10):2756–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP: Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell 2003, 11(6):1445–1456. [DOI] [PubMed] [Google Scholar]
- 44.Ohsaki K, Oishi K, Kozono Y, Nakayama K, Nakayama KI, Ishida N: The role of {beta}-TrCP1 and {beta}-TrCP2 in circadian rhythm generation by mediating degradation of clock protein PER2. J Biochem 2008, 144(5):609–618. [DOI] [PubMed] [Google Scholar]
- 45.Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, Herzel H, Kramer A: Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms 2007, 22(5):375–386. [DOI] [PubMed] [Google Scholar]
- 46.Masuda S, Narasimamurthy R, Yoshitane H, Kim JK, Fukada Y, Virshup DM: Mutation of a PER2 phosphodegron perturbs the circadian phosphoswitch. bioRxiv 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, Zou X, Gotoh T, Brown AM, Jiang L, Wisdom EL, Kim JK, Finkielstein CV: Distinct control of PERIOD2 degradation and circadian rhythms by the oncoprotein and ubiquitin ligase MDM2. Sci Signal 2018, 11(556). [DOI] [PubMed] [Google Scholar]
- 48.Hirota T, Lee JW, Lewis WG, Zhang EE, Breton G, Liu X, Garcia M, Peters EC, Etchegaray JP, Traver D et al. : High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIalpha as a clock regulatory kinase. PLoS Biol 2010, 8(12):e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oshima T, Niwa Y, Kuwata K, Srivastava A, Hyoda T, Tsuchiya Y, Kumagai M, Tsuyuguchi M, Tamaru T, Sugiyama A et al. : Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth. Sci Adv 2019, 5(1):eaau9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayasaka N, Hirano A, Miyoshi Y, Tokuda IT, Yoshitane H, Matsuda J, Fukada Y: Salt-inducible kinase 3 regulates the mammalian circadian clock by destabilizing PER2 protein. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brenna A, Olejniczak I, Chavan R, Ripperger JA, Langmesser S, Cameroni E, Hu Z, De Virgilio C, Dengjel J, Albrecht U: Cyclin-dependent kinase 5 (CDK5) regulates the circadian clock. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narasimamurthy R, Hunt SR, Lu Y, Fustin JM, Okamura H, Partch CL, Forger DB, Kim JK, Virshup DM: CK1delta/epsilon protein kinase primes the PER2 circadian phosphoswitch. Proc Natl Acad Sci U S A 2018, 115(23):5986–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A: Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev 2006, 20(19):2660–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Philpott JM, Narasimamurthy R, Ricci CG, Freeberg AM, Hunt SR, Yee LE, Pelofsky RS, Tripathi S, Virshup DM, Partch CL: Casein kinase 1 dynamics underlie substrate selectivity and the PER2 circadian phosphoswitch. Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee HM, Chen R, Kim H, Etchegaray JP, Weaver DR, Lee C: The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci U S A 2011, 108(39):16451–16456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou M, Kim JK, Eng GW, Forger DB, Virshup DM: A Period2 Phosphoswitch Regulates and Temperature Compensates Circadian Period. Mol Cell 2015, 60(1):77–88. [DOI] [PubMed] [Google Scholar]
- 57.Liu Z, Huang M, Wu X, Shi G, Xing L, Dong Z, Qu Z, Yan J, Yang L, Panda S et al. : PER1 phosphorylation specifies feeding rhythm in mice. Cell Rep 2014, 7(5):1509–1520. [DOI] [PubMed] [Google Scholar]
- 58.Kaasik K, Kivimae S, Allen JJ, Chalkley RJ, Huang Y, Baer K, Kissel H, Burlingame AL, Shokat KM, Ptacek LJ et al. : Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab 2013, 17(2):291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Durgan DJ, Pat BM, Laczy B, Bradley JA, Tsai JY, Grenett MH, Ratcliffe WF, Brewer RA, Nagendran J, Villegas-Montoya C et al. : O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem 2011, 286(52):44606–44619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U: SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 2008, 134(2):317–328. [DOI] [PubMed] [Google Scholar]
- 61.Foteinou PT, Venkataraman A, Francey LJ, Anafi RC, Hogenesch JB, Doyle FJ 3rd: Computational and experimental insights into the circadian effects of SIRT1. Proc Natl Acad Sci U S A 2018, 115(45):11643–11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levine DC, Hong H, Weidemann BJ, Ramsey KM, Affinati AH, Schmidt MS, Cedernaes J, Omura C, Braun R, Lee C et al. : NAD(+) Controls Circadian Reprogramming through PER2 Nuclear Translocation to Counter Aging. Mol Cell 2020, 78(5):835–849 e837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Ooijen G, Hindle M, Martin SF, Barrios-Llerena M, Sanchez F, Bouget FY, O’Neill JS, Le Bihan T, Millar AJ: Functional analysis of Casein Kinase 1 in a minimal circadian system. PLoS One 2013, 8(7):e70021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goldsmith EJ, Akella R, Min X, Zhou T, Humphreys JM: Substrate and docking interactions in serine/threonine protein kinases. Chem Rev 2007, 107(11):5065–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson LN, Noble ME, Owen DJ: Active and inactive protein kinases: structural basis for regulation. Cell 1996, 85(2):149–158. [DOI] [PubMed] [Google Scholar]
- 66.Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ: Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem 1990, 265(24):14264–14269. [PubMed] [Google Scholar]
- 67.Narasimamurthy R, Virshup DM: The phosphorylation switch that regulates ticking of the circadian clock. Mol Cell 2021, 81(6):1133–1146. [DOI] [PubMed] [Google Scholar]
- 68.Kim DW, Chang C, Chen X, Doran AC, Gaudreault F, Wager T, DeMarco GJ, Kim JK: Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy. Mol Syst Biol 2019, 15(7):e8838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Longenecker KL, Roach PJ, Hurley TD: Three-dimensional structure of mammalian casein kinase I: molecular basis for phosphate recognition. J Mol Biol 1996, 257(3):618–631. [DOI] [PubMed] [Google Scholar]
- 70.Shinohara Y, Koyama YM, Ukai-Tadenuma M, Hirokawa T, Kikuchi M, Yamada RG, Ukai H, Fujishima H, Umehara T, Tainaka K et al. : Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock. Mol Cell 2017, 67(5):783–798 e720. [DOI] [PubMed] [Google Scholar]
- 71.Zeringo NA, Murphy L, McCloskey EA, Rohal L, Bellizzi JJ 3rd: A monoclinic crystal form of casein kinase 1 delta. Acta Crystallogr Sect F Struct Biol Cryst Commun 2013, 69(Pt 10):1077–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ralph MR, Menaker M: A mutation of the circadian system in golden hamsters. Science 1988, 241(4870):1225–1227. [DOI] [PubMed] [Google Scholar]
- 73.Gallego M, Eide EJ, Woolf MF, Virshup DM, Forger DB: An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc Natl Acad Sci U S A 2006, 103(28):10618–10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kornev AP, Taylor SS: Dynamics-Driven Allostery in Protein Kinases. Trends Biochem Sci 2015, 40(11):628–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nolen B, Taylor S, Ghosh G: Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell 2004, 15(5):661–675. [DOI] [PubMed] [Google Scholar]
- 76.Graves PR, Roach PJ: Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J Biol Chem 1995, 270(37):21689–21694. [DOI] [PubMed] [Google Scholar]
- 77.Rivers A, Gietzen KF, Vielhaber E, Virshup DM: Regulation of casein kinase I epsilon and casein kinase I delta by an in vivo futile phosphorylation cycle. J Biol Chem 1998, 273(26):15980–15984. [DOI] [PubMed] [Google Scholar]
- 78.Cegielska A, Gietzen KF, Rivers A, Virshup DM: Autoinhibition of casein kinase I epsilon (CKI epsilon) is relieved by protein phosphatases and limited proteolysis. J Biol Chem 1998, 273(3):1357–1364. [DOI] [PubMed] [Google Scholar]
- 79.Gietzen KF, Virshup DM: Identification of inhibitory autophosphorylation sites in casein kinase I epsilon. J Biol Chem 1999, 274(45):32063–32070. [DOI] [PubMed] [Google Scholar]
- 80.Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, Kiuchi R, Ishida M, Ukai-Tadenuma M, Minami Y et al. : CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A 2009, 106(37):15744–15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo G, Wang K, Hu SS, Tian T, Liu P, Mori T, Chen P, Johnson CH, Qin X: Autokinase Activity of Casein Kinase 1 delta/epsilon Governs the Period of Mammalian Circadian Rhythms. J Biol Rhythms 2019, 34(5):482–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qin X, Mori T, Zhang Y, Johnson CH: PER2 Differentially Regulates Clock Phosphorylation versus Transcription by Reciprocal Switching of CK1epsilon Activity. J Biol Rhythms 2015, 30(3):206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dahlberg CL, Nguyen EZ, Goodlett D, Kimelman D: Interactions between Casein kinase Iepsilon (CKIepsilon) and two substrates from disparate signaling pathways reveal mechanisms for substrate-kinase specificity. PLoS One 2009, 4(3):e4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH: Activation of 5’-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem 2007, 282(29):20794–20798. [DOI] [PubMed] [Google Scholar]
- 85.Giamas G, Hirner H, Shoshiashvili L, Grothey A, Gessert S, Kuhl M, Henne-Bruns D, Vorgias CE, Knippschild U: Phosphorylation of CK1delta: identification of Ser370 as the major phosphorylation site targeted by PKA in vitro and in vivo. Biochem J 2007, 406(3):389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bischof J, Randoll SJ, Sussner N, Henne-Bruns D, Pinna LA, Knippschild U: CK1delta kinase activity is modulated by Chk1-mediated phosphorylation. PLoS One 2013, 8(7):e68803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meng Z, Bischof J, Ianes C, Henne-Bruns D, Xu P, Knippschild U: CK1delta kinase activity is modulated by protein kinase C alpha (PKCalpha)-mediated site-specific phosphorylation. Amino Acids 2016, 48(5):1185–1197. [DOI] [PubMed] [Google Scholar]
- 88.Eng GWL, Edison, Virshup DM: Site-specific phosphorylation of casein kinase 1 delta (CK1delta) regulates its activity towards the circadian regulator PER2. PLoS One 2017, 12(5):e0177834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ianes C, Xu P, Werz N, Meng Z, Henne-Bruns D, Bischof J, Knippschild U: CK1delta activity is modulated by CDK2/E- and CDK5/p35-mediated phosphorylation. Amino Acids 2016, 48(2):579–592. [DOI] [PubMed] [Google Scholar]
- 90.Fustin JM, Kojima R, Itoh K, Chang HY, Ye S, Zhuang B, Oji A, Gibo S, Narasimamurthy R, Virshup D et al. : Two Ck1delta transcripts regulated by m6A methylation code for two antagonistic kinases in the control of the circadian clock. Proc Natl Acad Sci U S A 2018, 115(23):5980–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takano A, Uchiyama M, Kajimura N, Mishima K, Inoue Y, Kamei Y, Kitajima T, Shibui K, Katoh M, Watanabe T et al. : A missense variation in human casein kinase I epsilon gene that induces functional alteration and shows an inverse association with circadian rhythm sleep disorders. Neuropsychopharmacology 2004, 29(10):1901–1909. [DOI] [PubMed] [Google Scholar]
- 92.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN: PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta 2010, 1804(4):996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]


