Abstract
Pituitary homeobox 3 (Pitx3) is required for the terminal differentiation of nigrostriatal dopaminergic neurons during neuronal development. However, whether Pitx3 contributes to the normal physiological function and cell-type identity of adult neurons remains unknown. To explore the role of Pitx3 in maintaining mature neurons, we selectively deleted Pitx3 in the mesodiencephalic dopaminergic (mdDA) neurons of Pitx3fl/fl/DATCreERT2 bigenic mice using a tamoxifen inducible CreERT2/loxp gene-targeting system. Pitx3fl/fl/DATCreERT2 mice developed age-dependent progressive motor deficits, concomitant with a rapid reduction of striatal dopamine (DA) content and a profound loss of mdDA neurons in the substantia nigra pars compacta (SNc) but not in the adjacent ventral tegmental area (VTA), recapitulating the canonical neuropathological features of Parkinson’s disease (PD). Mechanistic studies showed that Pitx3-deficiency significantly increased the number of cleaved caspase-3+ cells in SNc, which likely underwent neurodegeneration. Meanwhile, the vulnerability of SNc mdDA neurons was increased in Pitx3fl/fl/DATCreERT2 mice, as indicated by an early decline in glial cell line-derived neurotrophic factor (GDNF) and aldehyde dehydrogenase 1a1 (Aldh1a1) levels. Noticeably, somatic accumulation of α-synuclein (α-syn) was also significantly increased in the Pitx3-deficient neurons. Together, our data demonstrate that the loss of Pitx3 in fully differentiated mdDA neurons results in progressive neurodegeneration, indicating the importance of the Pitx3 gene in adult neuronal survival. Our findings also suggest that distinct Pitx3-dependent pathways exist in SNc and VTA mdDA neurons, correlating with the differential vulnerability of SNc and VTA mdDA neurons in the absence of Pitx3.
Subject terms: Neuroscience, Neurological disorders
Introduction
The main pathological characteristic of PD is a profound loss of mdDA neurons in SNc [1, 2]. However, the proximal neurons within VTA and retrorubral field (RRF) are less vulnerable to degeneration and are largely spared during the course of the disease [3]. Therefore, some inherent and different gene expression profiles of mdDA neuron subtypes impact their distinct vulnerability to PD pathology, i.e., a molecular code may be rooted in the neuroepithelium to specify the subsets of mdDA neurons [4].
During neuronal development, mdDA precursors are thought to originate from the floor plate of the midbrain [5]. Temporal and spatial studies suggest that SNc neurons are derived from a rostral neuronal population earlier, whereas VTA neurons from a caudal one later [6]. A cascade of developmental transcription factors has long been known to define the mdDA neuronal fate specification in the neuroepithelium, including Pitx3, Nurr1, Engrailed, and Lmx1b [7, 8]. Particularly, these molecules are not only involved in the early neuronal events but are also continuously expressed during neuronal maturation and even throughout adulthood [9]. However, how these transcription factors play a role in modulating the postnatal neurons is still largely unknown, a stark contrast to their well-documented involvement in the developmental stages [3]. Conventional knockout mice of Nurr1 die shortly after birth [10], whereas recent studies have shown that the haploinsufficient ones can survive until the late stages, given exhibiting mdDA neuron loss and locomotor deficits [11]. This Nurr1-related neuronal loss might be largely attributed to the intrinsic dysregulation of Bcl-2 and P53 genes, both critical for cell survival [12, 13]. Furthermore, the conditional deletion of Nurr1 in mature neurons results in a rapid reduction of striatal DA, a loss of mdDA neuronal markers, and neuronal degeneration [14, 15].
Pitx3, another transcription factor, has also gained particular interest, since its polymorphisms were genetically linked to PD cases [16, 17]. During mouse development, Pitx3 is expressed in the eye lens, skeletal muscles, and mdDA neurons [18, 19]. However, after birth, its expression is preserved only in mdDA neurons [18, 19], indicating that Pitx3 plays an important role in both development and maintenance of these neurons. Moreover, Aphakia (ak) mice, which are Pitx3-deficient, display SNc mdDA neuron preferential loss during embryonic and postnatal development [20], recapitulating Pitx3 is highly involved in early mdDA neuronal events. However, compared with Nurr1, the role of Pitx3 in mature mdDA neurons has not been studied. As an initial attempt to understand the role of Pitx3 in maintaining mature mdDA neurons, we established a Pitx3fl/fl/DATCreERT2 (Pitx3cKO) mouse model that postnatally knocks out the Pitx3 gene in the cells expressing the dopamine transporter (DAT) protein under tamoxifen (TAM) treatment. Our results showed that the conditional deletion of Pitx3 in adult mice caused striatal DA reduction and locomotor activity abnormalities. Moreover, a significant loss of SNc mdDA neurons was first noted at 9 months after TAM treatment in Pitx3cKO mice and was further aggravated at 15 months of age accompanied by a significant increase of cleaved caspase 3+ cells in SNc. In contrast to ak mice [20, 21], our mouse model showed that TH, DAT, and Nurr1, the hallmarks of mdDA neurons were less affected by Pitx3 within the adult VTA neurons during aging. However, GDNF, BDNF, and Aldh1a1, which are Pitx3-related genes, were all substantially reduced in both the SNc and VTA mdDA neurons at the early or/and advanced stages. Such distinct expression profiles of candidate genes within SNc and VTA may contribute to the differential vulnerabilities of neuronal subtypes to the loss of Pitx3. Interestingly, the somatic accumulation of α-syn was markedly increased in the mdDA neurons of 15-month-old Pitx3cKO mice. These data altogether establish the key role of Pitx3 in maintaining the normal physiological functions and preserving the specific molecular identities of postnatal mdDA neurons.
Methods
The generation of conditional knockout Pitx3 mouse model
The heterozygous mice Pitx3Flox/wt with C57BL/6J background were generated by ViewSolid Biotech Co., Ltd. (Beijing, China). To achieve the conditional knockout Pitx3 mouse model in the mdDA neuronal system, Pitx3fl/fl/DATCreERT2 mice were produced by breeding mice carrying an inducible Cre recombinase under the DAT promoter with the heterozygous mice Pitx3Flox/wt. The DATCreERT2 mice were kindly gifted by the Günther Schütz Group (German Cancer Research Center) [22], which were generated by recombining a construct containing an improved Cre recombinase fused to a modified ligand-binding domain of the estrogen receptor into a bacterial artificial chromosome containing the gene encoding DAT.
All experimental mice were maintained under specific-pathogen-free (SPF) conditions (temperature, 22°C ± 2°C; air exchange, per 20 min; 12 h/12 h light–dark cycle) with free access to food and water. Animal care and procedures were carried out in accordance with the Laboratory Animal Care Guidelines approved by the Institutional Animal Care Committee at Dalian Medical University. The protocol was approved by the Institutional Animal Care Committee at Dalian Medical University.
TAM (T-5648; Sigma-Aldrich) was dissolved in corn oil (S-5007, Sigma-Aldrich) and ethanol mixture with the ratio of 10:1. A fresh mixture was prepared by shaking overnight to dissolve TAM completely, and the mixture was then stored in a dark place at 4 °C. Inducible Cre recombinase was activated in 2-month-old transgenic mice by administering TAM as 2 mg/day via intraperitoneal injection for five consecutive days [23].
For the performance of genotyping, DATCreERT2 mice were identified by a PCR assay (2XEasyTaq PCR SuperMix, AS-111, Transgen Biotech) via tail biopsy. The sequence of the forward primer is AGA ACC TGA TGG ACA TGT TCA GG and the sequence of the reverse primer is CAG ACC AGG CCA GGT ATC TCT. The length of the target amplicon is 700 bp. PCR program: 95 °C for 45 s; 40X (95 °C for 15 s, 60 °C for 20 s, 72 °C for 30 s); 72 °C for 1 min; 4 °C for 2 min; hold at 4 °C. Pitx3Flox/wt mice were identified by a PCR assay (2XEasyTaq PCR SuperMix, AS-111, Transgen Biotech) via tail biopsy. The sequence of the forward primer is GTC AGT GGA TAG GAA AAG AGG C and the sequence of the reverse primer is TCA CTC TAC AGT GTG TAC CTG GTC. The PCR product size of wild-type allele is 167 bp and mutant allele is 201 bp. PCR program: 94 °C for 2 min; 32X (98 °C for 10 s, 60 °C for 30 s, 68 °C for 50 s); 68 °C for 5 min; hold at 16 °C.
Behavioral test
The open-field test was performed in a quiet testing room over 3 continuous days. To measure the locomotor activity, mice were placed into an Activity Monitor instrument (25 × 25 × 30 cm, Med Associates Inc., St. Albans, USA) equipped with computer-controlled photocells. Locomotor activity was automatically recorded for 40 min, and the total distance traveled and the number of rearings were calculated by the Med system.
The pole test was performed as described previously [14]. Mice were placed head-up on top of a vertical pole and allowed to descend freely to the bottom of the box with 1-day training before the test. On the test, the animals underwent three trials, and the time to orient downward (t-turn) and the total time to turn and descend the pole to the floor (t-total) were measured.
Rotarod assays were performed using the rotarod apparatus (Model 755, IITC Life Science). Both training and testing were performed at three trials per day for 3 consecutive days, as described previously [20]. At the start of the test, mice were stood on the rod and allowed to habituate for 1 min. After that, mice were trained to attain stable baseline levels of performance and climbing on the rod rotating at a constant speed of 4 rpm for 5 min. Subsequently, a protocol with an acceleration of 6 rpm/min was applied at a maximum speed of 40 rpm. The duration time for each mouse was then recorded.
Immunostaining
Mouse brains were collected at indicated time points after TAM injections. Mice were anesthetized by isoflurane with chloral hydrate perfused through the left ventricle with PBS. The brains were rapidly isolated and postfixed in ice-cold 4% paraformaldehyde and subsequently dehydrated for 24 h in 15% and 30% sucrose at 4 °C, as described previously [24].
For immunohistochemical (IHC) staining, rabbit (PV-9001, ZSGB-BIO Company, Beijing, China) or mouse (PV-9002, ZSGB-BIO Company, Beijing, China) two-step detection kit was used, as described previously [25]. A series of slides were incubated in Solution A for 10 min. After rinsing with PBS three times, blocking solution (5% normal goat serum, 0.2% Triton-X 100, and 0.05% NaN3 in PBS) was applied for 1 h at room temperature. The primary antibodies were used as follows: anti-TH (1:1000, AB152; Millipore, USA) and anti-α-syn (1:1000, 610786; BD Transduction Laboratories, USA). The use of Solutions B and C were performed according to manufacturer’s instructions and after subsequent exposure to diaminobenzidine (ZLI-9019, ZSGB-BIO Company, China) for 5 min. After rinsing with PBS, the sections were dehydrated through a graded ethanol series. Finally, IHC staining results were visualized directly by DP80 CCD brightfield microscopy (Olympus, Japan). The outlines of the SNc, VTA, dorsal striatum (CPu), and nucleus accumbens (NAc) were delimited according to anatomical landmarks [26].
For immunofluorescence staining (IFC), sections were incubated for 1 h in blocking solution (5% normal goat serum, 0.2% Triton-X 100, and 0.05% NaN3 in PBS). The primary antibodies were used as follows: anti-Nurr1 (1:100, M196; Santa Cruz Biotechnology, USA), anti-TH (1:1000, AB152; Millipore, USA), anti-DAT (1:2000, MAB369; Millipore, USA), anti-TH (1:2000, T1299; Sigma-Aldrich, USA), anti-TH (1:1000, TYH, Aves Labs, USA), anti-GDNF (1:100, SC-13147; Santa Cruz Biotechnology, USA), anti-BDNF (1:100, SC-65514; Santa Cruz Biotechnology, USA), anti-Aldh1a1 (1:1000, AF1351; Beyotime Biotechnology, China), anti-NeuN (1:1000, MAB377; Millipore, USA), anti-cleaved caspase-3 (1:1000, 9664; CST, USA), anti-Iba1 (1:1000, 019-19741, FUJIFILM Wako Pure Chemical Corporation, Japan), anti-GFAP (1:100, sc33673; Santa Cruz Biotechnology, USA), and anti-Pitx3 (provided by Dr. Marten P. Smidt’s lab at the University of Amsterdam, Netherlands). For the immunostaining of Nurr1, GDNF, and BDNF, the blocking steps were performed after antigen retrieval (citrate buffer was made with 3 g trisodium citrate and 0.4 g citrate diluted in 1 L distilled water, pH 6.0). Finally, section images were visualized and photographed directly with a confocal microscope (A1 confocal, Nikon Instruments [Shanghai]Co., Ltd.) and a DP80 CCD brightfield microscope (Olympus, Japan). The outlines of the SNc and VTA were delimited according to anatomical landmarks [27].
Image analysis
TH+ cells in the SNc and VTA were calculated in every three sections from −2.70 to −3.88 mm Bregma at 10× magnification by an observer who was blind to the genotype, and data were collected from 8 to 10 slices per animal. The IFC intensity and fiber density were analyzed using ImageJ software, and the data were collected from 2 to 3 slices per animal.
High-performance liquid chromatography
Mouse brains were rapidly dissected, and the whole striatum in the left brain was isolated. For high-performance liquid chromatography (HPLC) analysis (EICOM, HTEC-500, USA), the tissue specimen was first weighed and sonicated on ice. DA was then extracted from the tissue homogenate and measured by reverse-phase HPLC through the electrochemical detection method. Different DA standard concentrations were detected to plot a standard curve for data analysis, as described previously [28].
Statistical analysis
Data were expressed as means ± SEM and were analyzed using GraphPad Prism software (Version 7.0). Statistical comparisons were performed using non-parametric two-tailed Mann–Whitney test or Student’s two-tailed unpaired t-test as indicated in the figure legends, and p < 0.05 was considered significant. All experiments were repeated at least three times. No statistical methods were used to predetermine sample size, but our sample sizes are similar to those reported in previous publications.
Results
Selective deletion of Pitx3 in mature mdDA neurons
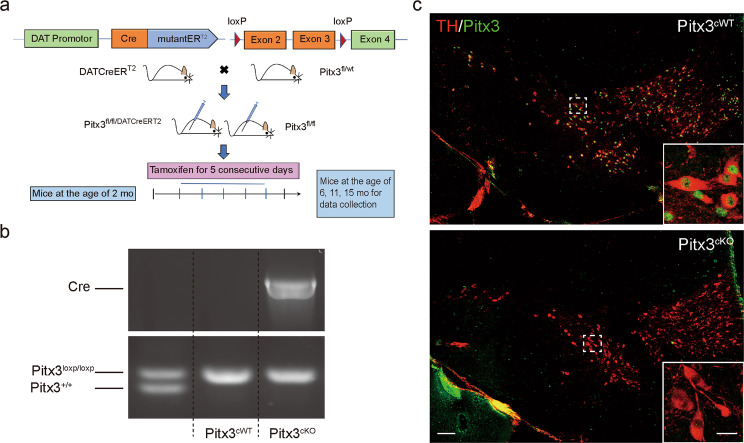
To investigate the role of Pitx3 in mature mdDA neurons, we first established a mouse model with selective Pitx3 deletion in mdDA neurons using a TAM-inducible CreERT2/loxp gene-targeting system (Fig. 1a). Cre-mediated recombination resulted in the removal of the second and third coding exons of Pitx3 (Fig. 1a). After breeding, homozygous Pitx3-floxed mice harboring either no copies (Pitx3cWT) or a single copy of the CreERT2/loxp gene (Pitx3cKO) were generated and characterized by conventional PCR analysis (Fig. 1b). At 2 months old, Pitx3cWT and Pitx3cKO mice were both injected intraperitoneally with TAM that provided a tightly spatial and temporal control of Pitx3 deletion. Tissues were first collected at 2 months after TAM administration (4 months of age), and the Pitx3 expression profile in mdDA neurons was detected by IFC staining (Fig. 1c). As expected, the expression levels of Pitx3 in the mdDA neurons of 4-month-old Pitx3cKO mice were not detectable, indicating the success of Pitx3 deletion. In all subsequent experiments, Pitx3cKO mice were treated with TAM at 2 months after birth, and the data were analyzed from three indicated time points (6, 11, and 15 months of age).
Fig. 1. Conditional knockout of Pitx3 in mdDA neurons.
a The basic strategy for the generation of TAM-inducible Cre/loxp-directed Pitx3 knockout mice. An experimental timeline for the administration of TAM and tissue collection is also presented. b PCR detection of Cre transgene (upper) and Pitx3 floxed allele (lower). c IFC analysis for Pitx3 expression in mdDA neurons was performed using an antibody against Pitx3 (green) together with TH (red) in 4-month-old Pitx3cWT and Pitx3cKO mice (scale bar: 100 μm; high-magnification, 25 μm).
Progressive mdDA neuronal loss and profound fiber pathology in Pitx3cKO mice
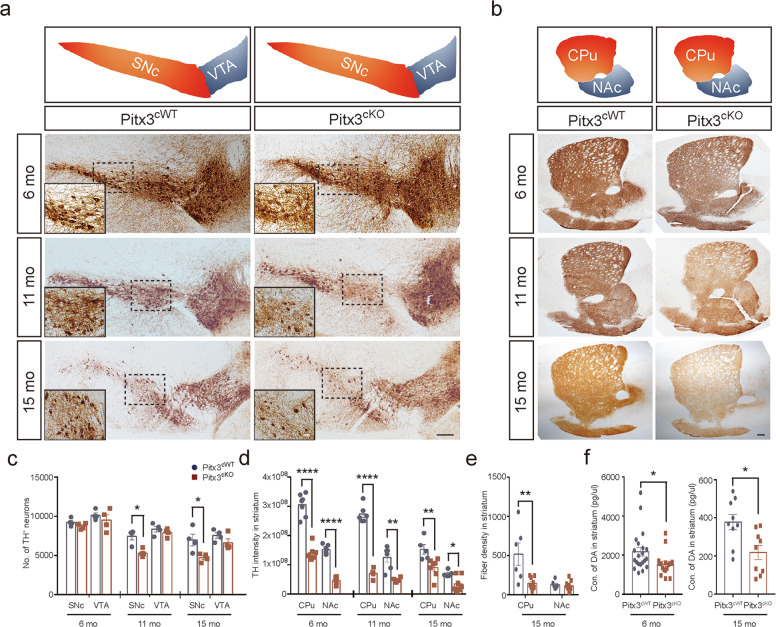
To further analyze the consequences of Pitx3 deletion in mature mdDA neurons, we examined the number of mdDA neurons in the SNc and VTA of Pitx3cKO mice at 6, 11, and 15 months of age following TAM administration. Notably, about 29% of SNc mdDA neurons were lost in 11-month-old Pitx3cKO mice, and the deficit was exaggerated at 15 months of age with around 32% neuronal loss in Pitx3cKO mice (Fig. 2a, c). However, VTA mdDA neurons were less affected by Pitx3-deficiency and remained intact during aging (Fig. 2a, c), resembling the neuropathological phenotype of ak mice [21]. Thus, our data re-emphasized that mdDA neurons consist of different subsets, each with a distinct vulnerability to neurodegeneration [4]. In addition to the neuron loss, Pitx3-deficiency also led to profound nerve fiber pathology. Our longitudinal data demonstrate that the striatal TH signals were progressively diminished in both Pitx3cWT and Pitx3cKO mice from 6 to 15 months of age (Fig. 2b, d). However, the lack of Pitx3 abnormally accelerated the decline at the early stages, i.e., IHC intensity of striatal TH was first noted to be dramatically decreased in 6-month-old Pitx3cKO mice, whereas no further marked reduction was identified from 11 to 15 months of age (Fig. 2b, d). On the other hand, TH+ fiber density showed a significant loss in the dorsal striatum of 15-month-old Pitx3cKO mice (Fig. 2b, e). The striatal DA levels were also dramatically reduced in Pitx3cKO mice, approximately a 50% reduction compared with Pitx3cWT mice at 15 months of age (Fig. 2f), suggesting that the disrupted dopaminergic innervation was closely associated with the alteration of DA content. Interestingly, as early as 6 months of age, the mean DA concentration was already reduced from 2174 pg/μL in Pitx3cWT to 1545 pg/μL in Pitx3cKO (Fig. 2f). However, the number of mdDA neurons was comparable between the two genotypes at this stage, suggesting that the perturbation of dopamine homeostasis may initially occur in axon terminals rather than in the soma.
Fig. 2. Neuropathological examination of the SNc-striatal pathway of Pitx3cKO mice.
a IHC staining of TH in the ventral midbrain sections from 6-, 11-, and 15-month-old Pitx3cWT and Pitx3cKO mice. SNc and VTA are highlighted with red and blue colors, respectively (scale bar: 100 μm; high-magnification, 50 μm). b IHC staining of TH in the striatal sections from 6-, 11-, and 15-month-old Pitx3cWT and Pitx3cKO mice. CPu and NAc are highlighted with red and blue colors, respectively (scale bar, 100 μm). c Quantification of TH+ neurons in the SNc and VTA from 6-, 11-, and 15-months-old Pitx3cWT and Pitx3cKO mice (N = 3-4 mice per genotype). Mann–Whitney test, *p = 0.0286 (11 months old for SNc); unpaired t-test, *p = 0.0299 (15 months old for SNc). d Quantification of TH IHC intensity in the striatum from 6-, 11-, and 15-months-old Pitx3cWT and Pitx3cKO mice (N = 4–7 mice per genotype). Unpaired t-test, ****p < 0.0001 (6 months old for CPu and NAc); ****p < 0.0001 (11 months for CPu); **p = 0.0053 (11 months for NAc); **p = 0.0095 (15 months for CPu); and *p = 0.0233 (15 months for NAc). e Quantification of TH+ fiber density in the striatum from 15-month-old Pitx3cWT and Pitx3cKO mice (N = 6–9 mice per genotype). Unpaired t-test, **p = 0.0078 (CPu). f Striatal DA levels in 6- and 15-month-old Pitx3cWT and Pitx3cKO mice (N = 9–21 mice per genotype). Mann–Whitney test, *p = 0.0148 (6 months old); and *p = 0.0102 (15 months old).
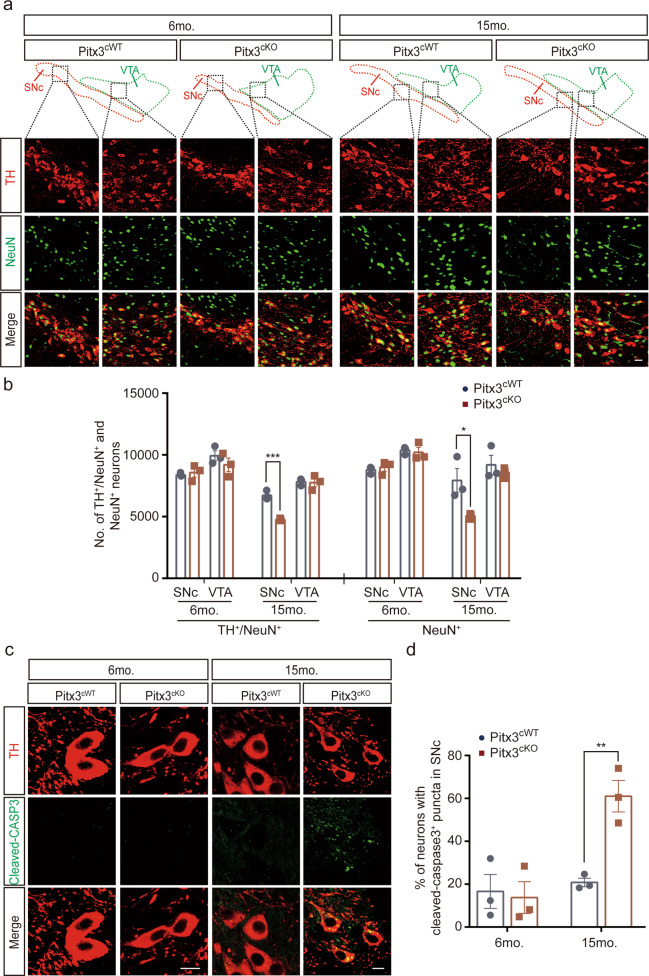
Since the downregulation of TH expressions was reported in ak mice previously [18], to make the identification of mdDA neurons more accurate, we also included another neuronal marker NeuN for neuron counting in the same SNc and VTA areas used for counting TH+ neurons [29]. Our results indicated that the number of NeuN+ cells was significantly lost, about 36% in the SNc area of 15-month-old Pitx3cKO mice (Fig. 3a, b), whereas the number of SNc NeuN+ neurons was comparable between two genotypes at 6 months of age (Fig. 3a, b), mirroring the DAB staining results of TH in the SNc area and further proving that the neuronal death indeed occurs at the advanced stage. In contrast to SNc, the NeuN+ cells in the VTA of 15-month-old Pitx3cKO mice were largely retained, and the two genotypes have comparable numbers of VTA NeuN+ cells (Fig. 3a, b). In addition, we examined the activation of caspase-3 in the SNc region, showing that more mdDA neurons in 15-month-old Pitx3cKO mice displayed concentrated cleaved-caspase3 puncta, compared to the age-matched Pitx3cWT mice (Fig. 3c, d), indicating that apoptosis may be promoted to trigger the neuronal death upon Pitx3-deficiency.
Fig. 3. Neurodegeneration in 15-month-old Pitx3cKO mice accompanied by the promoted apoptosis.
a IFC co-staining of TH and NeuN, and IFC staining of NeuN in the ventral midbrain sections from 6 and 15-month-old Pitx3cWT and Pitx3cKO mice. SNc and VTA were outlined, respectively (scale bar: 10 μm). b Quantification of TH+/NeuN+ and NeuN+ neurons in the SNc and VTA from 6- and 15-months-old Pitx3cWT and Pitx3cKO mice (N = 3 mice per genotype). unpaired t-test, ***p = 0.0008 (15 months old for TH+/NeuN+ co-staining); unpaired t-test, *p = 0.0424 (15 months old for NeuN+ staining). c IFC staining of cleaved-caspase3 in the SNc area from 6 and 15-month-old Pitx3cWT and Pitx3cKO mice (scale bar: 10 μm). d Quantification of the percentage of neurons with cleaved-caspase3+ puncta in SNc (N = 3 mice per genotype). unpaired t-test, **p = 0.0062.
Behavioral impairments in Pitx3cKO mice
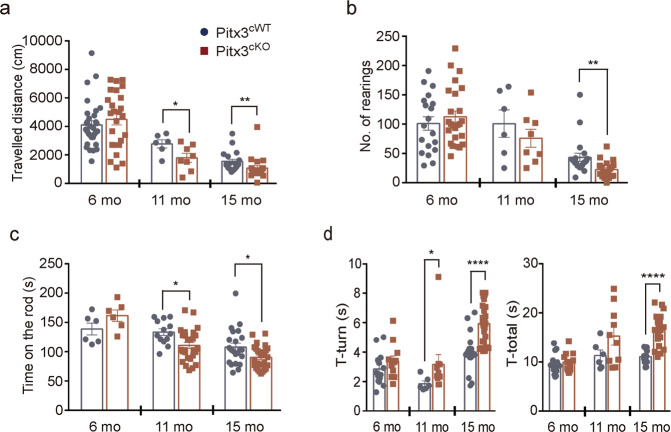
Consistent with the forementioned neuropathological changes, motor behaviors were abnormally altered in Pitx3cKO mice. The open-field test demonstrated that starting at 11 months of age, a significant difference in the distance traveled was identified between Pitx3cWT and Pitx3cKO mice (Fig. 4a). The deficit worsened in 15-month-old Pitx3cKO mice, together with a reduced number of rearing, indicating a progressive decline in locomotor activity with the progress of aging (Fig. 4a, b). The rotarod test indicated that young Pitx3cKO mice performed equally well with age-matched Pitx3cWT mice, whereas Pitx3cKO mice showed markedly less stay-time on the rotating rod at the advanced stages (Fig. 4c). Moreover, in the vertical pole test, a prolonged turning time was observed in 11- and 15-month-old Pitx3cKO mice, whereas a prolonged total task time was identified in 15-month-old Pitx3cKO mice (Fig. 4d). These observed motor activity abnormalities in Pitx3cKO mice correlate with the progressively diminished striatal DA levels during aging.
Fig. 4. Motor behavior in Pitx3cKO mice.
a Total traveled distance and b the number of rearings were performed in open-field tests (N = 6–31 mice per genotype). Unpaired t-test, *p = 0.0471 (total distance at 11 months of the age); Mann–Whitney test, **p = 0.0011 (total distance at 15 months old); and **p = 0.0011 (rearings at 15 months old). c The latency to fall from rotarod for 6-, 11-, and 15-month-old Pitx3cWT and Pitx3cKO mice (N = 6–27 mice per genotype). Unpaired t-test, *p = 0.0107 (11 months old); and *p = 0.0206 (15 months old). d The turning time and total task time for 6-, 11-, and 15-month-old Pitx3cWT and Pitx3cKO mice in pole tests (N = 6–27 mice per genotype). Unpaired t-test, ****p < 0.0001 (turning time at 15 months old); and ****p < 0.0001 (total time at 15 months old); Mann–Whitney test, *p = 0.016 (turning time at 11 months old).
Distinct cellular deficiency within SNc and VTA mdDA neurons of Pitx3cKO mice
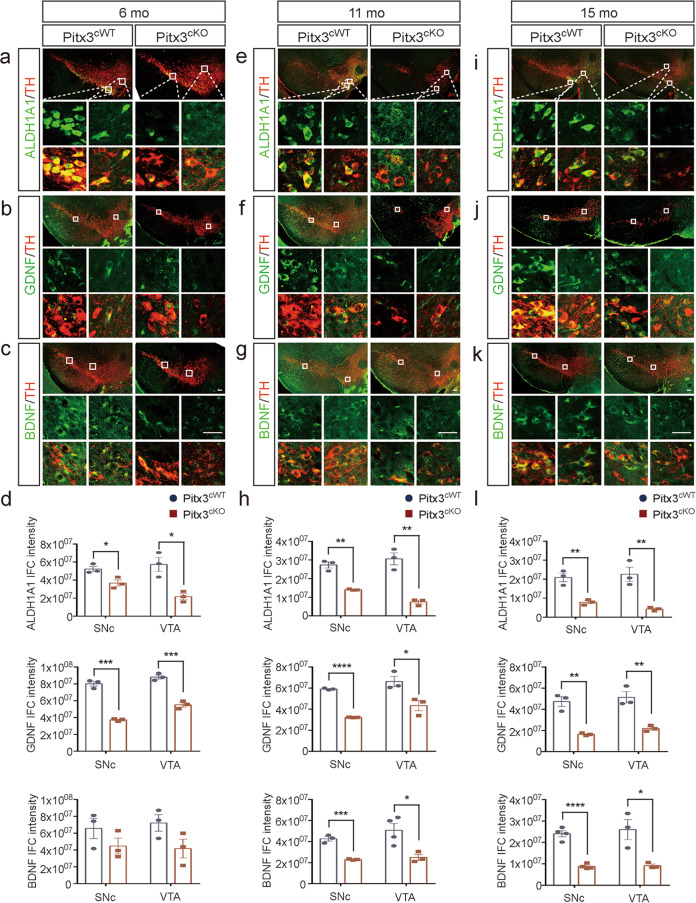
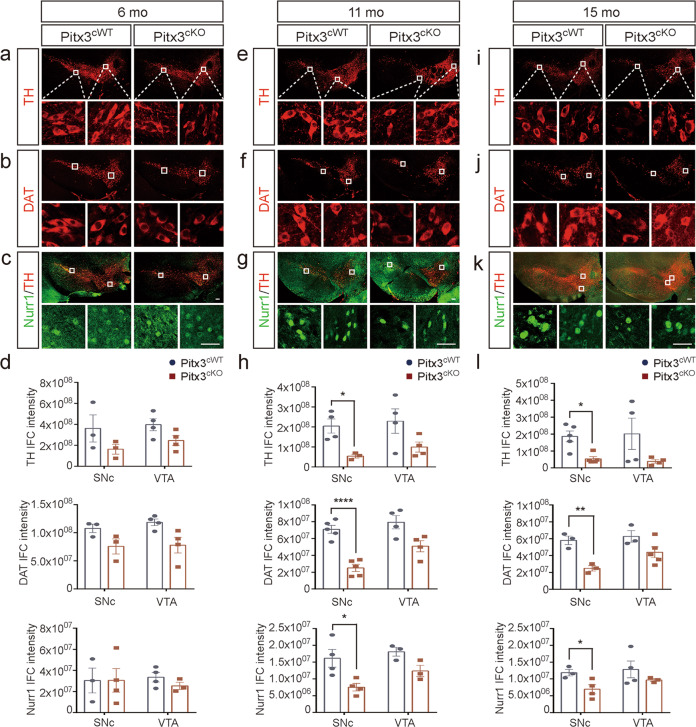
To further investigate cellular deficiency in different subtypes of mdDA neurons, a number of candidate genes were analyzed within SNc and VTA by immunostaining (Figs. 5 and 6). First, starting at 6 months of age Aldh1a1 and GDNF expressions were noted to decrease in both SNc and VTA mdDA neurons of Pitx3cKO mice (Fig. 5a, b) and showed a steady reduction in the later stages (Fig. 5e, f, i and j). We speculated that such a decrease may be possibly due to the disruption of their regulatory pathway due to Pitx3-deficiency. BDNF was significantly reduced in both SNc and VTA mdDA neurons of 11- and 15-month-old Pitx3cKO mice, but not at early stages (Fig. 5c, g and k), indicating that Aldh1a1 and GDNF may be more dependent on Pitx3 regulation than BDNF. On the other hand, a dramatic reduction in TH, DAT, and Nurr1 was observed in the SNc mdDA neurons of 11- and 15-month-old Pitx3cKO mice, which could be resulted from the profound neuronal loss (Fig. 6). In contrast to their expression in SNc, TH, DAT, and Nurr1 levels were not significantly altered in the spared VTA mdDA neurons between Pitx3cKO and Pitx3cWT mice at all stages (Fig. 6), indicating that these genes may be less regulated by Pitx3 in the adult VTA neurons. Altogether, the data suggest that distinct Pitx3-dependent pathways exist within SNc and VTA mdDA neurons, reflecting that Pitx3-deficiency specifically increases the vulnerability of SNc mdDA neurons, but not VTA, during adulthood. To better understand the distinct expression profiling of candidate genes within the mdDA neuronal subtypes, we summarized the above data in Supplementary Fig. 1.
Fig. 5. Expression analysis of Pitx3-related genes in Pitx3cKO mice.
Co-staining of Aldh1a1 and TH (a, e, i), GDNF and TH (b, f, j), and BDNF and TH (c, g, k) in the SNc and VTA from 6-, 11-, and 15-month-old Pitx3cWT and Pitx3cKO mice (scale bar: 100 μm; high-magnification, 25 μm). d, h, l Quantification of IFC intensity of Aldh1a1, GDNF, and BDNF in the SNc and VTA from 6-, 11-, and 15-month-old Pitx3cWT and Pitx3cKO mice (N = 3–4 mice per genotype). Unpaired t-test, *p = 0.0288 (Aldh1a1, 6 months old for SNc); ***p = 0.0001 (GDNF, 6 months old for SNc); *p = 0.0138 (Aldh1a1, 6 months old for VTA); ***p = 0.0007 (GDNF, 6 months old for VTA); **p = 0.0012 (Aldh1a1, 11 months old for SNc); ****p < 0.0001 (GDNF, 11 months old for SNc); ***p = 0.0002 (BDNF, 11 months old for SNc); **p = 0.0023 (Aldh1a1, 11 months old for VTA); *p = 0.0285 (GDNF, 11 months old for VTA); *p = 0.0225 (BDNF, 11 months old VTA); **p = 0.0052 (Aldh1a1, 15 months old for SNc); **p = 0.0026 (GDNF, 15 months old for SNc); ****p < 0.0001 (BDNF, 15 months old for SNc); **p = 0.0086 (Aldh1a1, 15 months old for VTA); **p = 0.0062 (GDNF, 15 months old for VTA); and *p = 0.0237 (BDNF, 15 months old for VTA).
Fig. 6. Expression analysis of mdDA neuronal markers in Pitx3cKO mice.
The IFC staining of TH (a, e, i), DAT (b, f, j) and the IFC co-staining of Nurr1 and TH (c, g, k) were presented in the SNc and VTA from 6-, 11-, and 15-month-old Pitx3cWT and Pitx3cKO mice (scale bar: 100 μm; high-magnification, 25 μm). d, h, l Quantification of TH, DAT, and Nurr1 IFC intensity in the SNc and VTA from 6-, 11-, and 15-month-old Pitx3cWT and Pitx3cKO mice (N = 3–5 mice per genotype). Unpaired t-test, *p = 0.0164 (TH, 11 months for SNc); ****p < 0.0001 (DAT, 11 months for SNc); *p = 0.0248 (Nurr1, 11 months for SNc); **p = 0.0051 (DAT, 15 months for SNc); and *p = 0.0395 (Nurr1, 15 months for SNc); Mann–Whitney test, *p = 0.0159 (TH, 15 months for SNc).
Aged Pitx3cKO mice developed progressive neuropathological abnormalities on α-syn, microgliosis, and astrocytosis
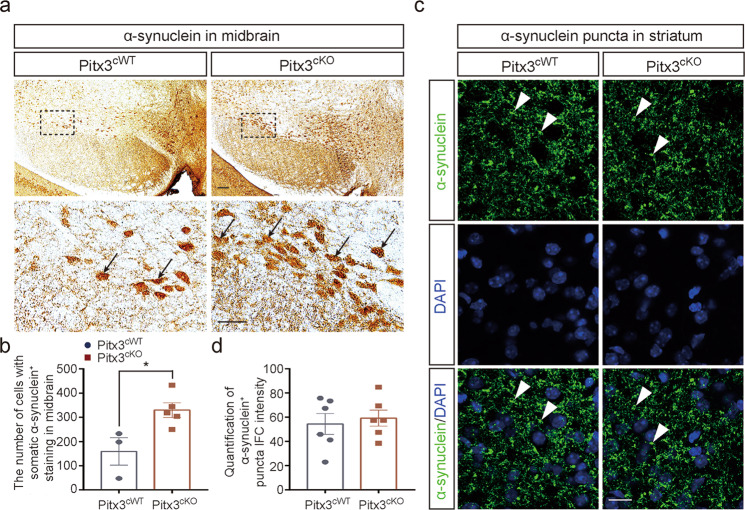
To further clarify the potential role of Pitx3 in PD-related pathology, we first examined the accumulation of endogenous α-syn by immunostaining. Increasing number of neurons with the somatic accumulation of α-syn were detected in 15-month-old Pitx3cKO mice (Fig. 7a, b). α-syn+ neurons within SNc and VTA were previously reported in wild-type mice [30, 31]. Such accumulation was exacerbated in our 15-month-old Pitx3cKO mouse model. Meanwhile, the data were closely correlated with the progression of neurodegeneration in Pitx3cKO mice at this advanced stage, suggesting that the increased somatic accumulation of α-syn may trigger the pathogenic cascades leading to cell death. By contrast, no significant difference of α-syn staining was observed in the striatum of 15-month-old Pitx3cWT and Pitx3cKO mice (Fig. 7c, d). In addition, no apparent α-syn staining was detected in soma of striatal neurons of 15-month-old Pitx3cWT and Pitx3cKO mice.
Fig. 7. The increasing number of neurons with somatic accumulation of α-syn in 15-month-old Pitx3cKO mice.
a IHC staining of α-syn in the ventral midbrain from 15-month-old Pitx3cWT and Pitx3cKO mice (scale bar: 100 μm; high-magnification, 50 μm). b Quantification of α-syn+ cells in the ventral midbrain from 15-month-old Pitx3cWT and Pitx3cKO mice (N = 3–4 mice per genotype). Mann–Whitney test, *p = 0.0357. c IFC staining of α-syn in the striatum from 15-month-old Pitx3cWT and Pitx3cKO mice (scale bar: 10 μm). d Quantification of α-syn+ puncta IFC intensity in the striatum from 15-month-old Pitx3cWT and Pitx3cKO mice (N = 6 mice per genotype).
Furthermore, we examined the brain sections of control and mutant animals for associated microgliosis and astrocytosis. The morphology of microglia was visualized by staining for ionized calcium-binding adapter molecule-1 (Iba1) [32]. The presence of reactive astrocytes was detected by staining for glial fibrillary acidic protein (GFAP) [33]. At 6 months of age, the reactive Iba1-positive cells were detected in both Pitx3cWT and Pitx3cKO mice, while more identified in Pitx3cKO mice, which reached statistical significance compared with Pitx3cWT mice. As expected, such phenotype aggravated in 15-month-old Pitx3cKO mice. The ratio of reactive Iba1-positive cells in Pitx3cKO mice increased to 30%, while around 8% in Pitx3cWT (Supplementary Fig. 2a, b). Interestingly, a similar augment of reactive GFAP-positive cells was observed at the age of 15 months. The ratio of reactive GFAP-positive cells in Pitx3cKO mice reached 48%, while around 30% in Pitx3cWT (Supplementary Fig. 2a, c). However, no apparent increase of reactive astrocytes was observed in 6-month-old Pitx3cKO mice compared to Pitx3cWT mice. Taken together, these findings demonstrate that a significant exacerbation of astrocytosis and microgliosis may contribute to neurodegeneration observed in the SNc mdDA neurons of 15-month-old Pitx3cKO mice.
Discussion
The mdDA neurons modulate many brain functions, including movement, emotion, and reward [7, 34]. Their degeneration and dysfunction contribute to several neurological disorders, including PD [35]. Extensive studies have been conducted to determine the regulatory pathways involved in cell fate specification and differentiation of mdDA neurons [36, 37], and several transcription factors have been well described. Remarkably, these molecules continue to be expressed in mdDA neurons throughout life, though at lower levels than during development [38]. In contrast to their comprehensive documentation at early stages, little information is available on how these transcription factors affect mdDA neurons at late stages. Pitx3, a transcription factor essential for the postmitotic development of mdDA neurons, has been identified to be severely reduced within PD brain tissues as well as in PD patients’ peripheral blood lymphocytes [8, 39]. Furthermore, several Pitx3 gene variants have been associated with sporadic PD [16, 40]. Thus, Pitx3 is critical not only for early differentiation but also for the maintenance of adult mdDA neurons. As an initial attempt to understand how Pitx3 contributes to the function of mature mdDA neurons, we established the Pitx3cKO mouse model and reported for the first time that conditional knockout of Pitx3 in fully differentiated mdDA neurons caused a rapid reduction of striatal DA, behavioral abnormalities, and progressive neuronal loss. Importantly, in our model, the striatal DA decline occurs long before mdDA neurons die, along with the distinct fiber pathology. These data suggest that perturbation on axonal terminals may be an early event in disease progression [41]. Taken together, the abnormalities caused by Pitx3-deficiency recapitulate the significant features of PD, suggesting that our mouse model can serve as a relevant PD model.
Our present studies provide a distinct, Pitx3-dependent expression profile in two subpopulations of mdDA neurons, namely SNc and VTA mdDA neurons, during aging. The data demonstrated that Pitx3 not only defines the molecular specification of neurons during early development but also maintains this characterization throughout adulthood. Pitx3 as a transcription factor is considered to modulate TH expression by binding to the 50 bp upstream area of the transcriptional start site [42]. During mdDA neuronal development, TH expression is initiated at the E11.5 stage, when ak mice show the comparable mdDA neuronal numbers with wild-type mice [21]. However, at E12.5 stage, the lateral TH+ neurons are largely lost in ak mice, and the TH+ signals are only detectable in a more dorsal field [21]. These studies indicate that Pitx3 is essential for the onset of TH expression in mdDA neurons during development [43]. However, what is the role of Pitx3 in TH regulation within adult mdDA neurons? In our Pitx3cKO mouse model, TH expression markedly diminished in SNc mdDA neurons at 9 months after TAM treatment but remained unaltered in the spared VTA mdDA neurons. Meanwhile, a moderate loss of mdDA neurons was also first noted at this stage. We thereby speculated that the severe neuronal loss within SNc may contribute to significant TH reduction. On the other hand, the unaltered TH levels within the VTA mdDA neurons of Pitx3cKO mice may be due to continuous Nurr1 expression. Nurr1, another regulator of TH [44], remained steadily expressed in the VTA mdDA neurons of 11- and 15-month-old Pitx3cKO mice, which may compensate for the on-site Pitx3 ablation and contribute to the unaltered TH expression within the VTA mdDA neurons at the advanced stages. Taken together, our study findings demonstrate that in adult mdDA neurons, Pitx3-deficiency may cause progressive TH reduction and significant neuronal loss in SNc. Conversely, in the spared VTA mdDA neurons, TH is steadily expressed even at the advanced stages, indicating that an alternative regulatory pathway in VTA may exist, possibly contributed by uninterrupted Nurr1 expression. We speculated that Nurr1 may play a vicarial role in justifying the late events during aging. Our data further indicate that the distinct vulnerability of mdDA neuronal subtypes is delicately controlled by the Pitx3-associated regulatory pathways during adulthood.
A follow-up question is why the mdDA neurons show progressive degeneration upon Pitx3-deficiency. Dramatically concentrated cleaved-caspase3 was characterized in Pitx3cKO mice, indicating that Pitx3-deficiency might promote the apoptosis of mdDA neurons. On the other hand, we have previously described that GDNF, BDNF, and Pitx3 are engaged in a feedforward regulatory pathway during development [45]. In our present study, the remarkable reduction of GDNF in SNc mdDA neurons appeared as early as 6 months in Pitx3cKO mice and continued to decrease into the advanced stages. As reported, GDNF is required for mdDA neuronal survival during development [46], we thereby speculate that such early GDNF deficit may contribute to late neuronal degeneration. Conversely, BDNF was steadily expressed in SNc mdDA neurons in both Pitx3cWT and Pitx3cKO mice at the early stages, but eventually decreased at the advanced stages, suggesting that a weaker association may exist between BDNF and Pitx3 during adulthood. Our model thus indicates that the feedforward interaction with GDNF, BDNF, and Pitx3 may not only protect mdDA neurons during embryogenesis but also promote neuronal survival during adulthood. Similarly, Aldh1a1, as a downstream target of Pitx3, is also reduced in 6-month-old Pitx3cKO mice. Aldh1a1 encodes for the retinoic acid-synthesizing enzyme and oxidizes the highly reactive DA catabolic intermediate dopamine, 3,4-dihydroxyphenylacetaldehyde (DOPAL), to prevent cytotoxicity [47]. Therefore, the decrease in Aldh1a1 may disrupt the DA metabolic equilibrium, resulting in mdDA neuronal dysfunction at the early stages. In addition, Aldh1a1-deficiency was reported to trigger α-syn aggregation in A53T transgenic mice [48, 49]. In our mouse model, the increasing number of neurons with the somatic accumulation of α-syn were characterized in 15-month-old Pitx3cKO mice, which was not previously recognized in the conventional Pitx3 knockout mouse lines. We thereby speculated that a severe loss of Aldh1a1 expression caused by Pitx3-deficiency may highly likely result in DOPAL formation, further increasing the somatic accumulation of α-syn [50, 51]. These data suggest that the reduction in GDNF and Aldh1a1 levels may cause neuronal dysfunction and disrupt DA metabolism within SNc mdDA neurons at the early stages of Pitx3cKO mice, possibly contributing to late neurodegeneration. On the other hand, the increased somatic accumulation of α-syn and reduced BDNF levels may promote the apoptosis and exacerbate the PD-related pathologies in Pitx3cKO mice at the advanced stages.
Taken together, the conditional knockout of Pitx3 in fully differentiated adult mdDA neurons results in progressive neurodegeneration, motor abnormalities, and promoted apoptosis. We propose that the early reduction in GDNF and Aldh1a1 levels may be associated with late neuronal loss via the disruption of both neuronal function and DA metabolism. In addition, the increased somatic accumulation of α-syn may exacerbate PD-related pathology at the advanced stages. Furthermore, Pitx3-deficiency triggers the differential expression of candidate genes within the distinct subtypes of mdDA neurons, indicating that distinct neuronal identities are delicately regulated through Pitx3-dependent pathways during adulthood. However, more detailed mechanistic studies are required to further elucidate how neuronal specification maintains in the subtypes of adult mdDA neurons.
Supplementary information
Acknowledgements
We are cordially thankful to Günther Schütz (German Cancer Research Center), David Engblom (Linköping University), Pierre Chambon (University of Strasbourg), Daniel Metzger (University of Strasbourg) for providing DAT-CreERT2 mouse strain, to Marten P. Smidt (University of Amsterdam) for providing Pitx3 antibody, and to all members in our laboratory for caring, help, and advice.
Author contributions
WY and LW designed the experiments. WY and CX contributed to imaging experiments and data analysis. WY, CX, and WYY contributed to behavior test and data analysis. WY, CX, SL, CH, and LW wrote and edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported in part by the National Key Research and Development Program of China (2016YFC1306600), the National Natural Science Foundation of China (NSFC81771521), Guangdong Provincial Key R & D Program 2018B030337001, and the intramural research program of National Institute on Aging, National Institutes of Health, USA (AG000943).
Data availability
All reagents will be available upon request.
Competing interests
The authors declare no competing interests.
Ethics declarations
Animal care and procedures were carried out in accordance with the Laboratory Animal Care Guidelines approved by the Institutional Animal Care Committee at Dalian Medical University. The protocol was approved by the Institutional Animal Care Committee at Dalian Medical University. This article does not contain any studies with human participants performed by any of the authors.
Footnotes
Edited by Professor Fabio Blandini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ying Wang, Xi Chen.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-021-04319-x.
References
- 1.Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/S0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 2.Shao Y, Le W. Recent advances and perspectives of metabolomics-based investigations in Parkinson’s disease. Mol Neurodegener. 2019;14:3. doi: 10.1186/s13024-018-0304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mesman S, Smidt MP. Acquisition of the midbrain dopaminergic neuronal identity. Int J Mol Sci. 2020;21:4638. doi: 10.3390/ijms21134638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakrabarty K, Von Oerthel L, Hellemons A, Clotman F, Espana A, Groot Koerkamp M, et al. Genome wide expression profiling of the mesodiencephalic region identifies novel factors involved in early and late dopaminergic development. Biol Open. 2012;1:693–704. doi: 10.1242/bio.20121230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smidt MP. Molecular programming of mesodiencephalic dopaminergic neuronal subsets. Front Neuroanat. 2017;11:59. doi: 10.3389/fnana.2017.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smits SM, von Oerthel L, Hoekstra EJ, Burbach JP, Smidt MP. Molecular marker differences relate to developmental position and subsets of mesodiencephalic dopaminergic neurons. PLoS ONE. 2013;8:e76037. doi: 10.1371/journal.pone.0076037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bissonette GB, Roesch MR. Development and function of the midbrain dopamine system: what we know and what we need to. Genes Brain Behav. 2016;15:62–73. doi: 10.1111/gbb.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Dani JA, Le W. The role of transcription factor Pitx3 in dopamine neuron development and Parkinson’s disease. Curr Top Med Chem. 2009;9:855–9. [PMC free article] [PubMed] [Google Scholar]
- 9.Asgrimsdottir ES, Arenas E. Midbrain dopaminergic neuron development at the single cell level: in vivo and in stem cells. Front Cell Dev Biol. 2020;8:463. doi: 10.3389/fcell.2020.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T, et al. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–50. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 11.Jiang C, Wan X, He Y, Pan T, Jankovic J, Le W. Age-dependent dopaminergic dysfunction in Nurr1 knockout mice. Exp Neurol. 2005;191:154–62. doi: 10.1016/j.expneurol.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 12.Imam SZ, Jankovic J, Ali SF, Skinner JT, Xie W, Conneely MO, et al. Nitric oxide mediates increased susceptibility to dopaminergic damage in Nurr1 heterozygous mice. FASEB J. 2005;19:1441–50. doi: 10.1096/fj.04-3362com. [DOI] [PubMed] [Google Scholar]
- 13.Zhang T, Wang P, Ren H, Fan J, Wang G. NGFI-B nuclear orphan receptor Nurr1 interacts with p53 and suppresses its transcriptional activity. Mol Cancer Res. 2009;7:1408–15. doi: 10.1158/1541-7786.MCR-08-0533. [DOI] [PubMed] [Google Scholar]
- 14.Kadkhodaei B, Alvarsson A, Schintu N, Ramskold D, Volakakis N, Joodmardi E, et al. Transcription factor Nurr1 maintains fiber integrity and nuclear-encoded mitochondrial gene expression in dopamine neurons. Proc Natl Acad Sci USA. 2013;110:2360–5. doi: 10.1073/pnas.1221077110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadkhodaei B, Ito T, Joodmardi E, Mattsson B, Rouillard C, Carta M, et al. Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J Neurosci. 2009;29:15923–32. doi: 10.1523/JNEUROSCI.3910-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le W, Nguyen D, Lin XW, Rawal P, Huang M, Ding YJ, et al. Transcription factor PITX3 gene in Parkinson’s disease. Neurobiol Aging. 2011;32:750–3. doi: 10.1016/j.neurobiolaging.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Jimenez-Jimenez FJ, Garcia-Martin E, Alonso-Navarro H, Agundez JA. PITX3 and risk for Parkinson’s disease: a systematic review and meta-analysis. Eur Neurol. 2014;71:49–56. doi: 10.1159/000353981. [DOI] [PubMed] [Google Scholar]
- 18.Smidt MP, van Schaick HS, Lanctot C, Tremblay JJ, Cox JJ, van der Kleij AA, et al. A homeodomain gene Ptx3 has highly restricted brain expression in mesencephalic dopaminergic neurons. Proc Natl Acad Sci USA. 1997;94:13305–10. doi: 10.1073/pnas.94.24.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smidt MP, Smits SM, Burbach JP. Homeobox gene Pitx3 and its role in the development of dopamine neurons of the substantia nigra. Cell Tissue Res. 2004;318:35–43. doi: 10.1007/s00441-004-0943-1. [DOI] [PubMed] [Google Scholar]
- 20.Le W, Zhang L, Xie W, Li S, Dani JA. Pitx3 deficiency produces decreased dopamine signaling and induces motor deficits in Pitx3(-/-) mice. Neurobiol Aging. 2015;36:3314–20. doi: 10.1016/j.neurobiolaging.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smidt MP, Smits SM, Bouwmeester H, Hamers FP, van der Linden AJ, Hellemons AJ, et al. Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development. 2004;131:1145–55. doi: 10.1242/dev.01022. [DOI] [PubMed] [Google Scholar]
- 22.Parkitna JR, Engblom D, Schutz G. Generation of Cre recombinase-expressing transgenic mice using bacterial artificial chromosomes. Methods Mol Biol. 2009;530:325–42. doi: 10.1007/978-1-59745-471-1_17. [DOI] [PubMed] [Google Scholar]
- 23.Wang P, Chen X, Wang Y, Jia C, Liu X, Wang Y, et al. Essential role for autophagy protein VMP1 in maintaining neuronal homeostasis and preventing axonal degeneration. Cell Death Dis. 2021;12:116. doi: 10.1038/s41419-021-03412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong J, Liu X, Wang Y, Cai H, Le W. Nurr1(Cd11bcre) conditional knockout mice display inflammatory injury to nigrostriatal dopaminergic neurons. Glia. 2020;68:2057–69. doi: 10.1002/glia.23826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merz H, Malisius R, Mannweiler S, Zhou R, Hartmann W, Orscheschek K, et al. ImmunoMax. A maximized immunohistochemical method for the retrieval and enhancement of hidden antigens. Lab Invest. 1995;73:149–56. [PubMed] [Google Scholar]
- 26.Bakker R, Tiesinga P, Kotter R. The scalable brain atlas: instant web-based access to public brain atlases and related content. Neuroinformatics. 2015;13:353–66. doi: 10.1007/s12021-014-9258-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y, Yuan Y, Halliday G, Rusznak Z, Watson C, Paxinos G. A cytoarchitectonic and chemoarchitectonic analysis of the dopamine cell groups in the substantia nigra, ventral tegmental area, and retrorubral field in the mouse. Brain Struct Funct. 2012;217:591–612. doi: 10.1007/s00429-011-0349-2. [DOI] [PubMed] [Google Scholar]
- 28.Xiao Q, Yang S, Le W. G2019S LRRK2 and aging confer susceptibility to proteasome inhibitor-induced neurotoxicity in nigrostriatal dopaminergic system. J Neural Transm. 2015;122:1645–57. doi: 10.1007/s00702-015-1438-9. [DOI] [PubMed] [Google Scholar]
- 29.Prasad K, Richfield EK. Number and nuclear morphology of TH+ and TH- neurons in the mouse ventral midbrain using epifluorescence stereology. Exp Neurol. 2010;225:328–40. doi: 10.1016/j.expneurol.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Taguchi K, Watanabe Y, Tsujimura A, Tanaka M. Expression of alpha-synuclein is regulated in a neuronal cell type-dependent manner. Anat Sci Int. 2019;94:11–22. doi: 10.1007/s12565-018-0464-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taguchi K, Watanabe Y, Tsujimura A, Tanaka M. Brain region-dependent differential expression of alpha-synuclein. J Comp Neurol. 2016;524:1236–58. doi: 10.1002/cne.23901. [DOI] [PubMed] [Google Scholar]
- 32.Yuan T, Manohar K, Latorre R, Orock A, Greenwood-Van Meerveld B. Inhibition of microglial activation in the amygdala reverses stress-induced abdominal pain in the male rat. Cell Mol Gastroenterol Hepatol. 2020;10:527–43. doi: 10.1016/j.jcmgh.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bardehle S, Kruger M, Buggenthin F, Schwausch J, Ninkovic J, Clevers H, et al. Live imaging of astrocyte responses to acute injury reveals selective juxtavascular proliferation. Nat Neurosci. 2013;16:580–6. doi: 10.1038/nn.3371. [DOI] [PubMed] [Google Scholar]
- 34.Van den Heuvel DM, Pasterkamp RJ. Getting connected in the dopamine system. Prog Neurobiol. 2008;85:75–93. doi: 10.1016/j.pneurobio.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q, Zhang B, Yue Z. Disentangling the molecular pathways of Parkinson’s disease using multiscale network modeling. Trends Neurosci. 2020;44:182–8. doi: 10.1016/j.tins.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smits SM, Burbach JP, Smidt MP. Developmental origin and fate of meso-diencephalic dopamine neurons. Prog Neurobiol. 2006;78:1–16. doi: 10.1016/j.pneurobio.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Mesman S, Kruse SJ, Smidt MP. Expression analyzes of early factors in midbrain differentiation programs. Gene Expr Patterns. 2018;27:8–15. doi: 10.1016/j.gep.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Veenvliet JV, Smidt MP. Molecular mechanisms of dopaminergic subset specification: fundamental aspects and clinical perspectives. Cell Mol Life Sci. 2014;71:4703–27. doi: 10.1007/s00018-014-1681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Wei L, Tao Q, Deng H, Ming M, Xu P, et al. Decreased NURR1 and PITX3 gene expression in Chinese patients with Parkinson’s disease. Eur J Neurol. 2012;19:870–5. doi: 10.1111/j.1468-1331.2011.03644.x. [DOI] [PubMed] [Google Scholar]
- 40.Anand D, Agrawal SA, Slavotinek A, Lachke SA. Mutation update of transcription factor genes FOXE3, HSF4, MAF, and PITX3 causing cataracts and other developmental ocular defects. Hum Mutat. 2018;39:471–94. doi: 10.1002/humu.23395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salvadores N, Sanhueza M, Manque P, Court FA. Axonal degeneration during aging and its functional role in neurodegenerative disorders. Front Neurosci. 2017;11:451. doi: 10.3389/fnins.2017.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cazorla P, Smidt MP, O’Malley KL, Burbach JP. A response element for the homeodomain transcription factor Ptx3 in the tyrosine hydroxylase gene promoter. J Neurochem. 2000;74:1829–37. doi: 10.1046/j.1471-4159.2000.0741829.x. [DOI] [PubMed] [Google Scholar]
- 43.Tekin I, Roskoski R, Jr, Carkaci-Salli N, Vrana KE. Complex molecular regulation of tyrosine hydroxylase. J Neural Transm. 2014;121:1451–81. doi: 10.1007/s00702-014-1238-7. [DOI] [PubMed] [Google Scholar]
- 44.Kim KS, Kim CH, Hwang DY, Seo H, Chung S, Hong SJ, et al. Orphan nuclear receptor Nurr1 directly transactivates the promoter activity of the tyrosine hydroxylase gene in a cell-specific manner. J Neurochem. 2003;85:622–34. doi: 10.1046/j.1471-4159.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- 45.Peng C, Aron L, Klein R, Li M, Wurst W, Prakash N, et al. Pitx3 is a critical mediator of GDNF-induced BDNF expression in nigrostriatal dopaminergic neurons. J Neurosci. 2011;31:12802–15. doi: 10.1523/JNEUROSCI.0898-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oo TF, Marchionini DM, Yarygina O, O’Leary PD, Hughes RA, Kholodilov N, et al. Brain-derived neurotrophic factor regulates early postnatal developmental cell death of dopamine neurons of the substantia nigra in vivo. Mol Cell Neurosci. 2009;41:440–7. doi: 10.1016/j.mcn.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchitti SA, Deitrich RA, Vasiliou V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharm Rev. 2007;59:125–50. doi: 10.1124/pr.59.2.1. [DOI] [PubMed] [Google Scholar]
- 48.Liu G, Yu J, Ding J, Xie C, Sun L, Rudenko I, et al. Aldehyde dehydrogenase 1 defines and protects a nigrostriatal dopaminergic neuron subpopulation. J Clin Invest. 2014;124:3032–46. doi: 10.1172/JCI72176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez-Zavala JS, Calleja LF, Moreno-Sanchez R, Yoval-Sanchez B. Role of aldehyde dehydrogenases in physiopathological processes. Chem Res Toxicol. 2019;32:405–20. doi: 10.1021/acs.chemrestox.8b00256. [DOI] [PubMed] [Google Scholar]
- 50.Burke WJ, Kumar VB, Pandey N, Panneton WM, Gan Q, Franko MW, et al. Aggregation of alpha-synuclein by DOPAL, the monoamine oxidase metabolite of dopamine. Acta Neuropathol. 2008;115:193–203. doi: 10.1007/s00401-007-0303-9. [DOI] [PubMed] [Google Scholar]
- 51.Cremades N, Cohen SI, Deas E, Abramov AY, Chen AY, Orte A, et al. Direct observation of the interconversion of normal and toxic forms of alpha-synuclein. Cell. 2012;149:1048–59. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All reagents will be available upon request.