Abstract
We developed three type-specific PCR assays for the rapid and sensitive detection of Streptococcus suis serotype 1 (plus 14), serotype 2 (plus 1/2), and serotype 9 strains in tonsillar specimens from pigs. The PCR primers were based on the sequences of type-specific capsular genes of S. suis serotype 1, 2, and 9 strains. We recently characterized a major part of the capsular biosynthesis (cps) locus of S. suis serotype 2. Here we extended these studies and characterized major parts of the cps loci of S. suis serotypes 1 and 9. Type-specific genes were identified by cross-hybridization experiments between the individual cps genes and chromosomal DNAs from the 35 different serotypes. Four genes of S. suis serotype 1 specifically hybridized with serotype 1 and 14 strains only. Five genes of S. suis serotype 2 specifically hybridized with serotype 2 and 1/2 strains only, and two genes of S. suis serotype 9 specifically hybridized with serotype 9 strains. Until now rapid and sensitive diagnostic tests were available only for pathogenic strains of serotype 2 and highly pathogenic strains of serotype 1. The serotype-specific PCR assays can therefore be useful tools for the identification of serotype 1, 14, 2, 1/2, and 9 strains both for diagnostic purposes and in epidemiological and transmission studies. Therefore, these tests may facilitate control and eradication programs.
Streptococcus suis is an important cause of meningitis, septicemia, arthritis, and sudden death in young pigs (5, 32). It can, however, also cause meningitis in humans (2). Attempts to control the disease are still hampered by the lack of sufficient knowledge about the epidemiology of the disease and the lack of effective vaccines and sensitive diagnostics.
S. suis strains are usually identified and classified by their morphological, biochemical, and serological characteristics (11, 12, 31). Serological classification is based on the presence of specific antigenic polysaccharides. So far, 35 different serotypes have been described (7, 8, 10). In several European countries, S. suis serotype 2 is the most prevalent type isolated from diseased pigs, followed by serotypes 9 and 1. Serological typing methods for S. suis are very laborious and time-consuming and can be performed only with isolated colonies. The typing is carried out by different types of agglutination tests. In these tests, isolated and biochemically characterized S. suis cells are agglutinated with a panel of 35 specific sera.
The biosynthesis of capsular polysaccharide (cps) requires a complex pathway, and generally, the genes involved in this process are clustered in a single locus (14, 16, 22, 24). Moreover, in many gram-positive and gram-negative bacteria these cps loci show a common genetic organization. A region containing the serotype-specific glycosyltransferases, required for the successive linkage of the monosacchrides to the oligosaccharides, and the putative polysaccharide polymerase, required for the polymerization of the olisaccharide subunits, is preceded by a region presumed to encode proteins with common functions, such as regulation and transport of polysaccharide across the membrane (14, 16, 22, 24).
We recently isolated and characterized a major part of the cps2 locus of S. suis serotype 2 (27). The analyzed region comprised 13 open reading frames (ORFs), and we could assign tentative functions to most of the ORFs (27). Moreover, the organization of the cps2 locus of S. suis serotype 2 seemed to be in accord with the organization of the cps loci found in other bacteria (14, 16, 22, 24, 27).
In the present report we describe the isolation and molecular characterization of major parts of the cps loci of S. suis serotypes 1 and 9 as well as the identification of cps gene sequences specific for either serotype 1, serotype 2, or serotype 9. The specificities of these cps gene sequences were tested by hybridization of serotype 1, 2, or 9 cps sequences with chromosomal DNA of the reference strains of all known 35 serotypes of S. suis. On the basis of these data we could develop type-specific probes and type-specific PCR assays for serotype 9, serotype 2 (plus 1/2), and serotype 1 (plus 14). Compared with the serological serotyping methods, the PCR assays were rapid, reliable, and sensitive. Moreover, we showed that these PCR assays could be used directly with tonsillar specimens from infected or carrier animals so that isolation of single colonies can be omitted. Therefore, these tests will undoubtedly contribute to a more rapid and reliable diagnosis of S. suis and may facilitate control and eradication programs.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and serotyping.
The bacterial strains and plasmids used in this study are listed in Table 1. The S. suis reference strains were obtained from M. Gottschalk, Faculté de Médicine Vétérinaire, Université de Montréal, Saint-Hyacinthe, Québec, Canada. S. suis strains were grown in Todd-Hewitt broth (code CM189; Oxoid) and were plated on Columbia agar blood base (code CM331; Oxoid) containing 6% (vol/vol) horse blood. Escherichia coli strains were grown in Luria broth (20) and were plated on Luria broth containing 1.5% (wt/vol) agar. If required, 50 μg of ampicillin per ml was added to the plates. The S. suis strains were serotyped by the slide agglutination test with serotype-specific antibodies (31).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XL2 blue | Stratagene | |
| S. suis | ||
| 10 | Virulent serotype 2 strain | 32 |
| 3 | Serotype 2 | 28 |
| 735 | Reference serotype 2 strain | 28 |
| 6555 | Reference serotype 1 strain | 28 |
| 6388 | Serotype 1 | 28 |
| 6290 | Serotype 1 | 28 |
| 5673 | Serotype 1/2 | 28 |
| 5679 | Serotype 1/2 | 28 |
| 5209 | Reference serotype 1/2 strain | 28 |
| 5218 | Reference serotype 9 strain | 28 |
| 5973 | Serotype 9 | 28 |
| 6437 | Serotype 9 | 28 |
| Reference strains | Serotypes 1 to 34 | 7, 8, 10 |
| Plasmids | ||
| pKUN19 | Replication functions of pUC, Ampra | 17 |
| pCPS1-1 | pKUN19 containing 5-kb EcoRV fragment of cps operon of type 1 | This work (Fig. 1B) |
| pCPS1-2 | pKUN19 containing 2.2-kb HindIII fragment of cps operon of type 1 | This work (Fig. 1B) |
| pCPS9-1 | pKUN19 containing 1-kb HindIII-XbaI fragment of cps operon of serotype 9 | This work (Fig. 1C) |
| pCPS9-2 | pKUN19 containing 4.0-kb XbaI-XbaI fragment of cps operon of serotype 9 | This work (Fig. 1C) |
Ampr, ampicillin resistant.
DNA techniques.
Routine DNA manipulations and PCRs were performed as described by Sambrook et al. (23).
DNA sequence analysis.
DNA sequences were determined on a 373A DNA Sequencing System (Applied Biosystems, Warrington, United Kingdom). Samples were prepared by use of a ABI/PRISM dye terminator cycle sequencing ready reaction kit (Applied Biosystems). Custom-made sequencing primers were purchased from Life Technologies. Sequencing data were assembled and analyzed with the McMollyTetra program (Soft Gene GmbH). The BLAST program was used to search for protein sequences homologous to the deduced amino acid sequences (1).
Southern blotting and hybridization.
Chromosomal DNA was isolated as described by Sambrook et al. (23). DNA fragments were separated on 0.8% agarose gels and transferred to Zeta-Probe GT membranes (Bio-Rad) as described by Sambrook et al. (23). DNA probes were labelled with [α-32P]dCTP (3000 Ci mmol−1; Amersham) by use of a random primed labelling kit (Boehringer). The DNA on the blots was hybridized at 65°C with appropriate DNA probes, as recommended by the supplier of the Zeta-Probe membranes. After hybridization, the membranes were washed twice with a solution of 40 mM sodium phosphate (pH 7.2)–1 mM EDTA–5% sodium dodecyl sulfate (SDS) for 30 min at 65°C and twice with a solution of 40 mM sodium phosphate (pH 7.2)–1 mM EDTA–1% SDS for 30 min at 65°C.
PCR.
The primers used in the cps2J PCR correspond to positions 13791 to 13813 and 14465 to 14443 in the S. suis cps2 locus. The sequences of the primers were 5′-CAAACGCAAGGAATTACGGTATC-3′ and 5′-GAGTATCTAAAGAATGCCTATTG-3′, respectively. The primers used for the cps1I PCR correspond to positions 4398 to 4417 and 4839 to 4821 in the S. suis cps1 sequence. The sequences of the primers were 5′-GGCGGTCTAGCAGATGCTCG-3′ and 5′-GCGAACTGTTAGCAATGAC-3′, respectively. The primers used in the cps9H PCR correspond to positions 4106 to 4126 and 4494 to 4475 in the S. suis cps9 sequence. The sequences of the primers were 5′-GGCTACATATAATGGAAGCCC-3′ and 5′-CCGAAGTATCTGGGCTACTG-3′, respectively. The reaction mixtures (50 μl) contained 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, each of the four deoxynucleotide triphosphates at a concentration of 0.2 mM; each of the primers at a concentration of 1 μM, and 1 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer Applied Biosystems, Branchburg, N.J.). DNA amplification was carried out in a Perkin-Elmer 9600 thermal cycler, and the program consisted of incubation for 10 min at 95°C and 30 cycles of 1 min at 95°C, 2 min at 56°C, and 2 min at 72°C.
Preparation of tonsillar specimens.
Tonsillar specimens were prepared for PCR as described previously (33).
Nucleotide sequence accession numbers.
The nucleotide sequence data have been submitted to GenBank, in which they are listed under accession nos. AF155804 (serotype 1) and AF155805 (serotype 9).
RESULTS
Distribution of cps2 gene sequences among all known 35 S. suis serotypes.
On the basis of the genetic organization of the genes, we previously suggested that orf2X and cps2A to cps2K genes are part of one operon and that the proteins encoded by these genes are involved in polysaccharide biosynthesis (27). Orf2Y and Orf2Z are not part of this operon, and their role in polysaccharide biosynthesis is unclear. On the basis of sequence homology data, Orf2Y may be involved in regulation of the cps2 genes. Orf2Z is proposed to be unrelated to polysaccharide biosynthesis (27). To examine the homology between the cps2 genes and genes in the cps loci of other S. suis serotypes, we performed cross-hybridization experiments. DNA fragments of the individual cps2 genes were amplified by PCR, labelled with 32P, and hybridized to chromosomal DNAs of the reference strains of the 35 different S. suis serotypes. As a positive control we used a probe specific for S. suis 16S rRNA (28). The 16S rRNA probe hybridized with almost equal intensities with all serotypes tested (Table 2). However, none of the other genes were common in all serotypes (Table 2). Probes specific for the cps2A, cps2B, cps2C, and cps2D genes hybridized with most other serotypes. This indicates that the proteins encoded by these genes are not serotype specific but perform more common functions in biosynthesis of the capsular polysaccharide. This confirms previous data which showed that the cps2A to cps2D genes showed strong homology to the cps genes of Streptococcus pneumoniae involved in such common functions as regulation, chain length determination, and export (27). The cps2E gene hybridized with the DNAs of serotypes 1, 2, 14, 27, and 1/2. The cps2F, cps2G, cps2H, cps2I, and cps2J genes hybridized with the chromosomal DNAs of serotypes 2 and 1/2 only, indicating that these genes are specific for serotypes 2 and 1/2. The cps2G gene showed an additional weak hybridization signal with the DNA of serotype 34. In agglutination tests serotype 1/2 shows agglutination with sera specific for serotype 2 as well as with sera specific for serotype 1. This suggests that serotype 1/2 shares antigenic determinants with both serotypes 1 and 2. The cps2K gene hybridized with the DNAs of serotypes 1, 2, 14, and 1/2. Taken together, these hybridization data confirm earlier conclusions that in the cps2 locus the type-specific genes are preceded by genes with more common functions.
TABLE 2.
Hybridization of serotype 2 cps genes and neighboring sequences with chromosomal DNA of S. suis serotypes
| DNA probe | Hybridization with the following serotype:
|
||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 1/2 | |
| orf2Z | + | + | + | + | + | + | + | + | + | + | + | + | ± | + | + | + | + | + | + | − | + | − | + | + | + | − | + | + | + | + | + | − | − | − | + |
| orf2Y | + | + | + | + | + | + | + | + | + | + | + | + | ± | + | + | + | + | + | + | ± | + | ± | + | + | + | + | + | + | + | + | + | − | − | − | + |
| orf2X | + | + | + | + | + | + | + | + | + | + | + | + | ± | + | + | + | + | + | + | − | + | − | + | + | + | − | + | + | + | + | + | − | − | − | + |
| cps2A | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | + | + | + | − | + | + | + | + | + | − | − | − | + |
| cps2B | + | + | + | + | + | + | + | + | + | + | − | − | ± | + | − | − | ± | ± | ± | − | ± | − | + | + | + | − | − | − | + | ± | + | − | ± | − | + |
| cps2C | + | + | + | + | + | + | + | + | + | + | + | − | ± | + | − | ± | − | − | − | − | − | − | + | + | + | − | + | ± | − | − | + | − | ± | − | + |
| cps2D | + | + | + | + | + | + | + | + | + | + | + | ± | ± | + | − | ± | + | + | + | − | ± | − | + | + | + | − | + | + | + | ± | + | − | − | − | + |
| cps2E | + | + | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | + |
| cps2F | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| cps2G | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ± | + |
| cps2H | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| cps2I | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| cps2J | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| cps2K | + | + | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| 16S rRNA | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
Cloning of type-specific cps genes of serotypes 1 and 9.
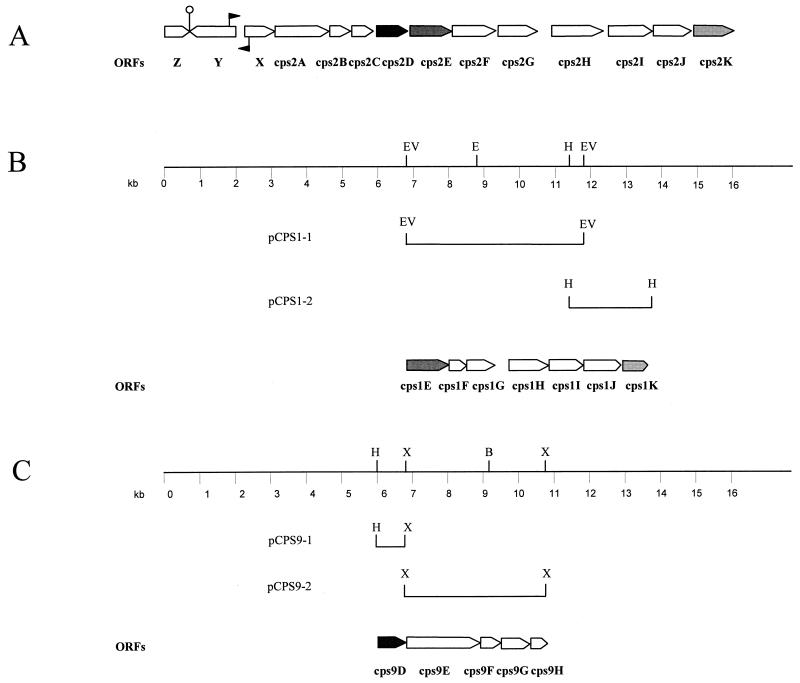
To clone the type-specific cps genes of S. suis serotype 1 we used the cps2E gene as a probe to identify chromosomal DNA fragments of serotype 1 containing homologous DNA sequences. A 5-kb EcoRV fragment was identified and was cloned in pKUN19. This yielded pCPS1-1 (Fig. 1B). This fragment was in turn used as a probe to identify an overlapping 2.2-kb HindIII fragment. pKUN19 containing the latter HindIII fragment was designated pCPS1-2 (Fig. 1B). The same strategy was followed to identify and clone the type-specific cps genes of serotype 9. In this case, we used the cps2D gene as a probe. A 0.8-kb HindIII-XbaI fragment was identified and cloned, yielding pCPS9-1 (Fig. 1C). This fragment was in turn used as a probe to identify a 4-kb XbaI fragment. pKUN19 containing this 4-kb XbaI fragment was designated pCPS9-2.
FIG. 1.
The cps2, cps1, and cps9 gene clusters of S. suis serotypes 2, 1, and 9. (A) Genetic organization of the cps2 gene cluster. The large arrows represent potential ORFs. Gene designations are indicated below the ORFs. Identically filled arrows represent ORFs which showed homology. The small closed arrows indicate the position of the potential promoter sequences.  , position of the potential transcription regulator sequence. (B) Physical map and genetic organization of the cps1 gene cluster. Restriction sites are as follows: E, EcoRI; EV, EcoRV; H, HindIII. The DNA fragments cloned in the various plasmids are indicated. The open arrows represent potential ORFs. As a result of the construction of the plasmids, cps1E is incomplete at its 5′ end and cps1K is incomplete at its 3′ end. (C) Physical map and genetic organization of the cps9 gene cluster. Restriction sites are as follows: B, BamHI; H, HindIII; X, XbaI. The DNA fragments cloned in the various plasmids are indicated. The open arrows represent potential ORFs. As a result of the construction of the plasmids, cps9D is incomplete at its 5′ end and cps9H is incomplete at its 3′ end.
, position of the potential transcription regulator sequence. (B) Physical map and genetic organization of the cps1 gene cluster. Restriction sites are as follows: E, EcoRI; EV, EcoRV; H, HindIII. The DNA fragments cloned in the various plasmids are indicated. The open arrows represent potential ORFs. As a result of the construction of the plasmids, cps1E is incomplete at its 5′ end and cps1K is incomplete at its 3′ end. (C) Physical map and genetic organization of the cps9 gene cluster. Restriction sites are as follows: B, BamHI; H, HindIII; X, XbaI. The DNA fragments cloned in the various plasmids are indicated. The open arrows represent potential ORFs. As a result of the construction of the plasmids, cps9D is incomplete at its 5′ end and cps9H is incomplete at its 3′ end.
Analysis of cloned cps1 and cps9 genes.
The complete nucleotide sequences of the inserts of pCPS1-1, pCPS1-2, pCPS9-1, and pCPS9-2 were determined. Examination of the cps1 sequence revealed the presence of five complete ORFs (Cps1F, Cps1G, Cps1H, Cps1I, and Cps1J) and two incomplete ORFs (Cps1E and Cps1K) (Fig. 1B). Examination of the cps9 sequence showed three complete ORFs (Cps9E, Cps9F, and Cps9G) and two incomplete ORFs (Cps9D and Cps9H) (Fig. 1C). All ORFs are preceded by a ribosome-binding site. In accord with data obtained for the cps2 genes of serotype 2, the majority of the serotype 1 and serotype 9 ORFs are very closely linked to each other. The only significant gap (718 bp) was found between Cps1G and Cps1H. No obvious promoter sequences or potential stem-loop structures could be found in the cps1 or cps9 sequences. This suggests that, as in serotype 2, the cps genes in serotype 1 and 9 form part of an operon.
An overview of the ORFs and their properties is shown in Table 3. As expected on the basis of the hybridization data (Table 2), Cps2E and Cps2K of S. suis serotype 2 showed significant homology to the proteins encoded by the cps1E and cps1K genes, respectively. Compared with Cps2E and Cps2K, the fragment cloned in pCPS1-1 lacked the coding region for the first 7 amino acids of the cps1E gene and the coding region for the last 58 amino acids of the cps1K gene. The proteins encoded by the cps1F, cps1G, cps1H, cps1I, and cps1J genes showed strong similarity with the Cps14F, Cps14G, Cps14H, Cps14J, and Cps14J proteins of Streptococcus pneumoniae serotype 14, respectively (15). Between Cps1G and Cps1H a gap of 718 bp was found. This region revealed three small putative ORFs. The three ORFs were present in three different reading frames and were not preceded by potential ribosome-binding sites, nor did they contain potential start sites. However, the three potential gene products encoded by this region showed some similarity with three successive regions of the C-terminal part of the EpsK protein of Streptococcus thermophilus (27% identity) (30). The region homologous to the first 82 amino acids of EpsK was lacking in S. suis serotype 1.
TABLE 3.
Properties of ORFs in the cps genes of S. suis serotypes 1 and 9 and similarities to gene products of other bacteria
| ORF | Nucleotide positions in sequence | Proposed function of gene product | Similar gene product (% identity) | Reference or accession no. |
|---|---|---|---|---|
| Cps1E | 1–1367 | Glucosyltransferase | Streptococcus suis Cps2E (86) | 27 |
| Cps1F | 1374–1823 | Unknown | Streptococcus pneumoniae Cps14F (83) | 15 |
| Cps1G | 1823–2317 | Glycosyltransferase | Streptococcus pneumoniae Cps14G (50) | 15 |
| Cps1H | 3036–4202 | CP polymerase | Streptococcus pneumoniae Cps14H (30) | 15 |
| Cps1I | 4195–5163 | Glycosyltransferase | Streptococcus agalactiae CpsH (45) | AB017355 |
| Streptococcus suis Cps2K (42) | 27 | |||
| Streptococcus pneumoniae Cps14J (38) | 14 | |||
| Streptococcus suis Cps2J (34) | 27 | |||
| Cps1J | 5172–6143 | Glycosyltransferase | Streptococcus suis Cps2J (47) | 27 |
| Cps1K | 6156–6992 | Glycosyltransferase | Streptococcus suis Cps2K (96) | 27 |
| Cps9D | 1–650 | Unknown | Streptococcus suis Cps2D (89) | 27 |
| Cps9E | 680–2506 | Glycosyltransferase | Staphylococcus aureus Cap1D (27) | 19 |
| Cps9F | 2679–3281 | Glycosyltransferase | Staphylococcus aureus Cap5M (52) | 18 |
| Cps9G | 3278–4087 | Unknown | Actinobacillus actinomycetemcomitans (43) | AB002668_4 |
| Cps9H | 4089–4520 | Glycosyltransferase | Yersinia enterolitica RfbB (28) | 35 |
| Streptococcus mutans RgpBc (28) | 34 |
As suggested by the hybridization data (Table 2) the Cps2D and Cps9D proteins showed significant homology (Table 3). Compared with Cps2D, the fragment cloned in pCPS9-1 lacked the coding region for the first 27 codons of the Cps9D protein. The protein encoded by the cps9E gene showed some similarity to the CapD protein of Staphylococcus aureus serotype 1 (19). The protein encoded by the cps9F gene showed some similarity to the CapM proteins of S. aureus serotypes 5 and 8 (18, 25, 26). The protein encoded by the cps9G gene showed some similarity to a protein of Actinobacillus actinomycetemcomitans (EMBL accession no. AB002668_4), whereas the protein encoded by the cps9H gene showed some similarity to the RfbB protein of Yersinia enterolitica (35) as well as to the RgpBc protein of Streptococcus mutans (34).
Serotype 1- and serotype 9-specific cps genes.
To determine whether the cloned fragments in pCPS1-1, pCPS1-2, pCPS9-1, and pCPS9-2 contained the type-specific genes for serotypes 1 and 9, respectively, cross-hybridization experiments were performed. DNA fragments of the individual cps1 and cps9 genes were amplified by PCR, labelled with 32P, and used to probe Southern blots of chromosomal DNA of the reference strains of the 35 different S. suis serotypes. The results are shown in Table 4. As expected, on the basis of the data obtained with the cps2E probe (Table 2), the cps1E probe hybridized with the chromosomal DNAs of S. suis serotypes 1, 2, 14, 27, and 1/2. The cps1K gene hybridized with the chromosomal DNAs of S. suis serotypes 1, 2, 14, and 1/2, whereas the cps1H probe hybridized with the chromosomal DNAs of various serotypes. However, the cps1F, cps1G, cps1I, and cps1J probes hybridized with the chromosomal DNAs of serotypes 1 and 14 only, indicating that these genes are specific for serotypes 1 and 14. The cps9E and cps9F probes hybridized with the chromosomal DNAs of various serotypes. However, the cps9G and cps9H probes hybridized with the chromosomal DNA of serotype 9 only, indicating that the cps9G and cps9H probes are specific for serotype 9.
TABLE 4.
Hybridization of serotype 1 and 9 cps genes with chromosomal DNA of S. suis serotypes
| DNA probe | Hybridization with the following serotype:
|
||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 1/2 | |
| cps1E | + | + | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | + |
| cps1F | + | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| cps1G | + | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| cps1H | + | − | + | + | + | − | + | − | + | + | + | ± | + | + | − | − | + | + | + | − | + | − | + | + | − | − | − | + | + | + | + | − | − | − | − |
| cps1I | + | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| cps1J | + | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| cps1K | + | + | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| cps9E | − | − | + | + | + | − | + | − | + | + | + | + | + | − | − | − | + | + | + | − | + | − | + | + | − | − | − | − | + | + | + | − | − | − | − |
| cps9F | − | − | − | − | − | − | − | − | + | + | ± | ± | − | − | − | − | − | − | − | − | ± | − | − | + | − | ± | − | ± | − | ± | − | − | ± | − | − |
| cps9G | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| cps9H | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 16S rRNA | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
Type-specific PCR.
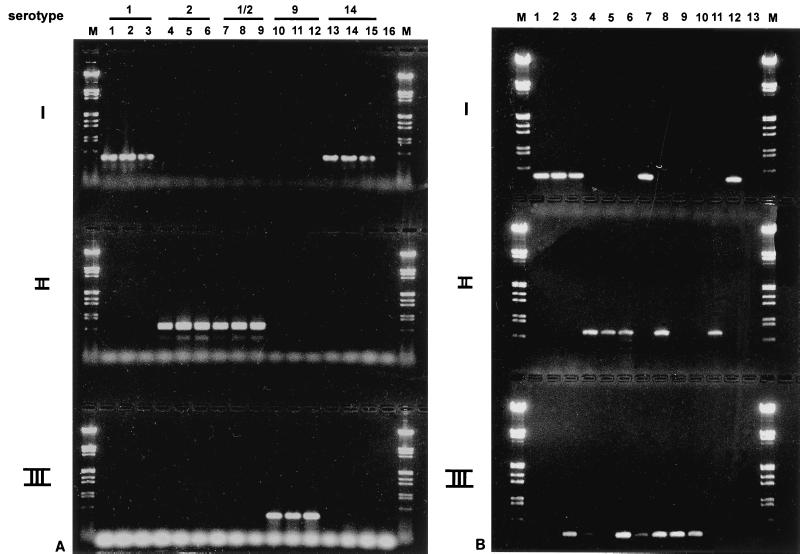
To test the diagnostic value of the type-specific cps probes further, several S. suis serotype 1, 2, 1/2, 9, and 14 strains, other than the reference strains, were used. In addition, we tested whether we could use PCR instead of hybridization for the serotyping of the S. suis strains. Three oligonucleotide primer sets within the cps2J, cps1I, and cps9H genes were chosen, and amplified fragments of 675, 440, and 390 bp, respectively, were expected. The results (Fig. 2A) show that, indeed, 675-bp fragments were amplified with serotype 2 and 1/2 strains with cps2J primers, 440-bp fragments were amplified with serotype 1 and 14 strains with cps1I primers, and 390-bp fragments were amplified with serotype 9 strains with cps9H primers.
FIG. 2.
(A) Ethidium bromide-stained agarose gel showing PCR products obtained with chromosomal DNA of S. suis strains belonging to serotypes 1, 2, 1/2, 9, and 14 and cps2J, cps1I, and cps2H primer sets. (I) cps1I primers; (II) cps2J primers; (III) cps9H primers. Lanes 1 to 3, serotype 1 strains; lanes 4 to 6, serotype 2 strains; lanes 7 to 9, serotype 1/2 strains; lanes 10 to 12, serotype 9 strains; lanes 13 to 15, serotype 14 strains; lane 16, negative control, no DNA, cps2J primers; lanes M, molecular size marker (bacteriophage lambda digested with EcoRI and HindIII). (B) Ethidium bromide-stained agarose gel showing PCR products obtained with tonsillar specimens collected from pigs carrying S. suis serotype 2, serotype 1, or serotype 9 strains and cps2J, cps1I, and cpsH primer sets. Bacterial DNA suitable for PCR was prepared by using the multiscreen methods described previously (21). (I) cps1I primers; (II) cps2J primers; (III) cps9H primers. Lanes 1 to 3, PCR products obtained with tonsillar specimens collected from pigs carrying S. suis serotype 1 strains (as detected by regular bacteriological examination); lanes 4 to 6, PCR products obtained with tonsillar specimens collected from pigs carrying S. suis serotype 2 strains; lanes 7 to 9, PCR products obtained with tonsillar specimens collected from pigs carrying S. suis serotype 9 strains; lanes 10 to 12, PCR products obtained with chromosomal DNA from serotype 9, 2, and 1 strains, respectively; lane 13, negative control, no DNA, cps2J primers; lanes M, molecular size marker (bacteriophage lambda digested with EcoRI and HindIII).
We next investigated whether these PCR assays could be used for the detection of S. suis serotype 2, 1/2, 1, 14, and 9 strains in clinical specimens. For that purpose, tonsils were collected from herds with a history of infection with either S. suis serotype 1, serotype 2, or serotype 9 strains. The results (Fig. 2B) show that specimens collected from animals known to be infected with S. suis serotype 1 strains were positive by the type 1-specific PCR (Fig. 2B, lanes 1 to 3). Moreover, specimens collected from animals known to be infected with either S. suis serotype 2 or serotype 9 strains were positive after the type 2- or 9-specific PCR assays, respectively (Fig. 2B, lanes 4 to 6 and 7 to 9, respectively. However, with some samples PCR-positive signals were obtained in two different type-specific PCR assays (Fig. 2B, lanes 3, 4, 6, 7, and 8). This suggested that these clinical samples contained S. suis strains of various serotypes which were not detected by regular bacteriological examination. To confirm this, these tonsillar specimens were next examined by using an extended bacteriological examination (33). To do this, the tonsillar specimens were plated and the colonies were hybridized with the type-specific probes. Hybridizing colonies were isolated and characterized by classical serotyping methods. The data showed that S. suis strains with the corresponding serotypes could be isolated from all samples that contained strains of more than one serotype, as identified by PCR. This confirmed the idea that the clinical samples contained S. suis strains of various serotypes. Moreover, these data show that the PCR assays are more sensitive than regular bacteriological examination.
DISCUSSION
In the present report we describe the identification and characterization of the cps genes specific for S. suis serotypes 1, 2, and 9. Previously, a major part of the cps2 locus of S. suis serotype 2 was cloned and characterized (27). Cross-hybridization experiments with the individual cps2, cps1 and cps9 genes as probes showed that the cps2F, cps2H, cps2I, and cps2J genes specifically hybridized with serotype 2 and 1/2 strains, whereas the cps1F, cps1G, cps1I, and cps1J genes specifically hybridized with serotype 1 and 14 strains. The cps9G and cps9H probes specifically hybridized with serotype 9 strains. On the basis of these data type-specific PCR assays were developed. These assays were highly specific and sensitive when they were used to test tonsillar specimens from pigs and more rapid and sensitive than regular bacteriological examination. Therefore, these assays may be important diagnostic tools for the detection of pigs carrying serotype 2, 1/2, 1, 14, and 9 strains and may facilitate control and eradication programs.
We previously developed a PCR method which can be used to detect pathogenic strains of S. suis serotype 2 as well as highly pathogenic strains of S. suis serotype 1 (33). However, besides S. suis serotype 1 and 2 strains, serotype 9 strains are frequently isolated from organs of diseased pigs. Until now a rapid and sensitive diagnostic test was not available for serotype 9 strains. Therefore, the serotype 9-specific probes as well as the serotype 9-specific PCR can be of great diagnostic value.
So far we have not found a probe which specifically hybridizes with serotypes 1 and 2. The cps1F, cps1G, and cps1I probes hybridized with serotype 1 as well as with serotype 14 strains. In coagglutination tests serotype 1 strains react with the anti-type 1 as well as with the anti-type 14 antisera (8). This suggests the presence of common epitopes between these serotypes. On the other hand, serotype 1 strains agglutinated only with anti-type 1 serum (8, 9), indicating that it should be possible to detect differences between those serotypes. Both serotype 1 and serotype 14 strains are highly pathogenic for young pigs (9, 29). This indicates that the type 1/14-specific PCR assays, which will detect both serotype 1 and serotype 14 strains, can be of great diagnostic value.
The cps2F, cps2G, cps2H, cps2I, and cps2J probes hybridized with serotypes 2 and 1/2 only. Serotype 34 showed a weak hybridizing signal with the cps2G probe. As shown in agglutination tests, serotype 1/2 strains react with sera directed against serotype 1 as well as with sera directed against serotype 2 strains (31). Therefore, serotype 1/2 shares antigens with both serotypes 1 and 2. This was supported by the recent observation of Charland and coworkers (4), who identified a monoclonal antibody against a capsular epitope shared by S. suis serotypes 1, 2, and 1/2. On the basis of the hybridization patterns of serotype 1/2 strains with the cps1- and cps2-specific probes, serotype 1/2 seemed to be more closely related to serotype 2 strains than to serotype 1 strains.
Putative functions for most of the cps1 gene products could be assigned by sequence homologies. The cps1E, cps1F, cps1G, cps1H, and cps1I genes revealed a striking similarity to the cps14E, cps14F, cps14G, cps14H, and cps14J genes of S. pneumoniae, respectively. Interestingly, S. pneumoniae serotype 14 is the serotype most commonly associated with pneumococcal infections in young children (3), whereas S. suis serotype 1 strains are most commonly isolated from piglets younger than 8 weeks (31). In S. pneumoniae the cps14E, cps14G, cps14I, and cps14J genes encode the glycosyltransferases required for the synthesis of the type 14 tetrameric repeating unit. This strongly suggests that the cps1E, cps1G, and cps1I genes similarly encode glycosyltransferases. In S. pneumoniae cps14E encodes a glycosyltransferase that links glucose to a lipid carrier (13). The structure of the S. suis serotype 1 capsule is unknown, but it is composed of glucose, galactose, N-acetylglucosamine, N-acetylgalactosamine, and sialic acid in a ratio of 1:2.4:1:1:1.4 (6). Therefore, a role of a cpsE-like glucosyltransferase activity can easily be envisaged. For polysaccharide biosynthesis in S. pneumoniae type 14, transfer of the second sugar of the repeating unit to the first lipid-linked sugar is performed by the gene products of cps14F and cps14G (15). Similar to Cps14F and Cps14G, the S. suis serotype 1 proteins Cps1F and Cps1G may act as one glycosyltransferase that performs the same reaction.
According to the homology found between the cps1H gene and the cps14H gene of S. pneumoniae (15), cps1H is expected to be encoded by a polysaccharide polymerase. The protein encoded by the cps1I gene showed some similarity to the Cps14J protein of S. pneumoniae (14) as well as to the Cps2K and Cps2J proteins of S. suis serotype 2 (27). The cps14J gene was shown to encode a β-1,4-galactosyltransferase activity, which is responsible for the addition of the fourth (i.e., last) sugar in the synthesis of the S. pneumoniae serotype 14 polysaccharide. In S. suis type 2 the proteins encoded by the cps2J and cps2K genes also showed homology to the Cps14J protein (27).
In the region between Cps1G and Cps1H three small putative ORFs were identified. Since the ORFs were present in three different reading frames and did not contain potential start sites, expression is not expected. However, the three potential gene products encoded by this region showed some similarity with three successive regions of the C-terminal part of the EpsK protein of Streptococcus thermophilus (27% identity) (30). The region homologous to the N-terminal 82 amino acids of the EpsK protein is lacking in S. suis serotype 1. The EpsK protein was suggested to play a role in the export of the exopolysaccharide by rendering the polymerized exopolysaccharide more hydrophobic through a lipid modification (30). These data could suggest that the sequences in the region between Cps1G and Cps1H originated from an epsK-like sequence. Hybridization experiments showed that this epsK-like region is also present in other serotype 1 strains as well as in serotype 14 strains (data not shown).
The structure as well as the composition of the serotype 9 capsular polysaccharide is unknown. Therefore, the function of most of the cloned serotype 9 genes remained unclear so far. On the basis of sequence homology data the cps9E, cps9F, and cps9H genes could be glycosyltransferases (18, 19, 24–26, 34). Moreover, the cps9G gene showed homology to a gene that is located in a region involved in polysaccharide biosynthesis but whose function is unknown (35).
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arends J P, Zanen H C. Meningitis caused by Streptococcus suis in humans. Rev Infect Dis. 1988;10:131–137. doi: 10.1093/clinids/10.1.131. [DOI] [PubMed] [Google Scholar]
- 3.Butler J C, Breiman R F, Lipman H B, Hofmann J, Facklam R R. Serotype distribution of Streptococcus pneumoniae infections among preschool children in the United States, 1978–1994: implications for development of a conjugate vaccine. J Infect Dis. 1995;171:885–889. doi: 10.1093/infdis/171.4.885. [DOI] [PubMed] [Google Scholar]
- 4.Charland N, Jacques M, Lacoutre S, Gottschalk M. Characterization and protective activity of a monoclonal antibody against a capsular epitope shared by Streptococcus suis serotypes 1, 2 and 1/2. Microbiology. 1997;143:3607–3614. doi: 10.1099/00221287-143-11-3607. [DOI] [PubMed] [Google Scholar]
- 5.Clifton-Hadley F A. Streptococcus suis type 2 infections. Br Vet J. 1983;139:1–5. doi: 10.1016/s0007-1935(17)30581-x. [DOI] [PubMed] [Google Scholar]
- 6.Elliott S D, Tai J Y. The type specific polysaccharide of Streptococcus suis. J Exp Med. 1978;148:1699–1704. doi: 10.1084/jem.148.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichsen J. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J Clin Microbiol. 1991;29:2590–2594. doi: 10.1128/jcm.29.11.2590-2594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottschalk M, Higgins R, Jacques M, Mittal K R, Henrichsen J. Description of 14 new capsular types of Streptococcus suis. J Clin Microbiol. 1989;27:2633–2636. doi: 10.1128/jcm.27.12.2633-2636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heath P J, Hunt B W, Duff J P. Streptococcus suis serotype 14 as a cause of pig disease in the UK. Vet Rec. 1996;2:450–451. [PubMed] [Google Scholar]
- 10.Higgins R, Gottschalk M, Boudreau M, Lebrun A, Henrichsen J. Description of six new capsular types (28 through 34) of Streptococcus suis. J Vet Diagn Invest. 1995;7:405–406. doi: 10.1177/104063879500700322. [DOI] [PubMed] [Google Scholar]
- 11.Hommez J, Devrieze L A, Henrichsen J, Castryck F. Identification and characterization of Streptococcus suis. Vet Microbiol. 1986;16:349–355. doi: 10.1016/0378-1135(86)90065-9. [DOI] [PubMed] [Google Scholar]
- 12.Killper-Balz R, Schleifer K H. Streptococcus suis sp. nov. nom. rev. Int J Syst Bacteriol. 1987;37:160–162. [Google Scholar]
- 13.Kolkman M A B, Morrison D A, van der Zeijst B A M, Nuijten P J M. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyl transferase gene cps14E. J Bacteriol. 1996;178:3736–3741. doi: 10.1128/jb.178.13.3736-3741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolkman M A B, Wakarchuk W, Nuijten P J M, van der Zeijst B A M. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 15.Kolkman M A B, van der Zeijst B A M, Nuijten P J M. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J Biol Chem. 1997;272:19502–19508. doi: 10.1074/jbc.272.31.19502. [DOI] [PubMed] [Google Scholar]
- 16.Kolkman M A B, van der Zeijst B A M, Nuijten P J M. Diversity of capsular polysaccharide synthesis gene clusters in Streptococcus pneumoniae. J Biochem. 1998;123:937–945. doi: 10.1093/oxfordjournals.jbchem.a022028. [DOI] [PubMed] [Google Scholar]
- 17.Konings R N H, Verhoeven E J M, Peeters B P H. pKUN vectors for the separate production of both DNA strands of recombinant plasmids. Methods Enzymol. 1987;153:12–34. doi: 10.1016/0076-6879(87)53045-2. [DOI] [PubMed] [Google Scholar]
- 18.Lee J C, Xu S, Albus A, Livolsi P J. Genetic analysis of type 5 capsular polysaccharide expression by Staphylococcus aureus. J Bacteriol. 1994;176:4883–4889. doi: 10.1128/jb.176.16.4883-4889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin W S, Cunneen T, Lee C Y. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol. 1994;176:7005–7016. doi: 10.1128/jb.176.22.7005-7016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 21.Reek F H, Smits M A, Kamp E M, Smith H E. Use of multiscreen plates for the preparation of bacterial DNA suitable for PCR. BioTechniques. 1995;19:282–285. [PubMed] [Google Scholar]
- 22.Roberts I S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Sau S, Bhasin N, Wann E R, Lee J C, Foster T J, Lee C Y. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology. 1997;143:2395–2405. doi: 10.1099/00221287-143-7-2395. [DOI] [PubMed] [Google Scholar]
- 25.Sau S, Lee C Y. Cloning of type 8 capsule genes and analysis of gene clusters for the production of different capsular polysaccharides in Staphylococcus aureus. J Bacteriol. 1996;178:2118–2126. doi: 10.1128/jb.178.7.2118-2126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sau S, Lee C Y. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol. 1997;179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith H E, Damman M, van der Velde J, Veenbergen V, Wagenaar F, Stockhofe-Zurwieden N, Smits M A. Identification and characterization of the complete cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun. 1999;67:1750–1756. doi: 10.1128/iai.67.4.1750-1756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith H E, Rijnsburger M, Stockhofe-Zurwieden N, Wisselink H J, Vecht U, Smits M A. Virulent strains of Streptococcus suis serotype 2 and highly virulent strains of Streptococcus suis serotype 1 can be recognized by a unique ribotype profile. J Clin Microbiol. 1997;35:1049–1053. doi: 10.1128/jcm.35.5.1049-1053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stockhofe-Zurwieden N, Vecht U, Wisselink H J, van Lieshout H, Smith H E. Comparative studies on the pathogenicity of different Streptococcus suis serotype 1 strains. In: Monetti P G, Vignola G, editors. Proceedings of the 14th International Pig Veterinary Society Congress, Bologna, Italy. 1996. p. 299. [Google Scholar]
- 30.Stringele F, Neeser J-R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vecht U, van Leengoed L A M G, Verheyen E R M. Streptococcus suis infections in pigs in The Netherlands (part one) Vet Q. 1985;7:315–321. doi: 10.1080/01652176.1985.9694005. [DOI] [PubMed] [Google Scholar]
- 32.Vecht U, Wisselink H J, van Dijk J E, Smith H E. Virulence of Streptococcus suis type 2 strains in newborn germfree pigs depends on phenotype. Infect Immun. 1992;60:550–556. doi: 10.1128/iai.60.2.550-556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisselink H J, Reek F H, Vecht U, Stockhofe-Zurwiede N, Smits M A, Smith H E. Detection of virulent strains of Streptococcus suis type 2 and highly virulent strains of Streptococcus suis type 1 in tonsillar specimens of pigs by PCR. Vet Microbiol. 1999;67:143–157. doi: 10.1016/s0378-1135(99)00036-x. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita Y, Tsukioka Y, Tomihisa K, Nakano Y, Koga T. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J Bacteriol. 1998;181:5803–5807. doi: 10.1128/jb.180.21.5803-5807.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Al-Hendy A, Toivanen P, Skurnik M. Genetic organization and sequence of the rfb gene cluster of Yersinia enterolitica serotype O:3: similarities to the dTDP-l-rhamnose biosynthesis pathway of Salmonella and to the bacterial polysaccharide transport systems. Mol Microbiol. 1993;9:309–321. doi: 10.1111/j.1365-2958.1993.tb01692.x. [DOI] [PubMed] [Google Scholar]