Summary
Newts utilize their unique genes to restore missing parts by strategic regulation of conserved signaling pathways. Lack of genetic tools poses challenges to determine the function of such genes. Therefore, we used the Drosophila eye model to demonstrate the potential of 5 unique newt (Notophthalmus viridescens) gene(s), viropana1-viropana5 (vna1-vna5), which were ectopically expressed in L2 mutant and GMR-hid, GMR-GAL4 eye. L2 exhibits the loss of ventral half of early eye and head involution defective (hid) triggers cell-death during later eye development. Surprisingly, newt genes significantly restore missing photoreceptor cells both in L2 and GMR>hid background by upregulating cell-proliferation and blocking cell-death, regulating evolutionarily conserved Wingless (Wg)/Wnt signaling pathway and exhibit non-cell-autonomous rescues. Further, Wg/Wnt signaling acts downstream of newt genes. Our data highlights that unique newt proteins can regulate conserved pathways to trigger a robust restoration of missing photoreceptor cells in Drosophila eye model with weak restoration capability.
Subject areas: Biological sciences, Cell biology, Developmental biology
Graphical abstract
Highlights
-
•
Newt proteins regulate wingless/Wnt pathway to rescue eye mutant(s) in Drosophila
-
•
These proteins non-cell-autonomously rescue missing tissue in Drosophila eye
-
•
Promotes cell proliferation and downregulates cell death in Drosophila eye mutant(s)
-
•
These newt genes may have significant bearing on our understanding of regeneration
Biological sciences; Cell biology; Developmental biology
Introduction
A urodele amphibian, newts belong to the Salamandridae family (Weisrock et al., 2006). The newts are the only four-legged vertebrates that exhibit a remarkable capability to regenerate tissues like limbs, tail, heart, lens, spinal cord, brain, and retina throughout its lifetime (Mehta and Singh, 2019a; Sanchez Alvarado and Tsonis, 2006). This ability of the newt to regrow organs is because of their ability to reprogram and dedifferentiate the terminally differentiated cells to trigger regeneration response (Mehta and Singh, 2019a). Such an exceptional regeneration ability of newts has been attributed to unique gene(s) that may have evolved from forming the regeneration tool box (Bryant et al., 2017; Casco-Robles et al., 2018; Elewa et al., 2017; Evans et al., 2018; Keinath et al., 2015; Kumar et al., 2007; Matsunami et al., 2019; Mehta and Singh, 2019a; Nowoshilow et al., 2018; Sanor et al., 2020; Smith et al., 2009, 2019). Earlier, a newt gene Prod1, which encodes a transmembrane receptor, was found to be critical for maintaining proximodistal identity (pattern memory) during newt limb regeneration (da Silva et al., 2002; Echeverri and Tanaka, 2005; Kumar et al., 2007; Mehta and Singh, 2019a). Homologs of Prod1 are yet to be identified outside of salamanders (Garza-Garcia et al., 2009). Similarly, the transcript of a newt gene newtic1 was found to be significantly enriched in a subset of erythrocytes which formed an aggregate structure called erythrocyte clump. These newtic1 expressing erythrocytes promote limb regeneration (Casco-Robles et al., 2018). Surprisingly, our understanding of the underlying molecular genetic mechanism(s) responsible for promoting lifelong regeneration of missing structures and/or rescue of pattern defects in newts is far from complete. Notophthalmus viridescens (Newts) have enormous genome size (∼c × 1010 bases), a long reproductive cycle, and have limited genetic tools that makes it difficult to use this model to determine the underlying molecular mechanism(s) behind the function of its unique regeneration genes. Some strategies to determine their regeneration potential are to introduce these unique genes in genetically tractable model(s) that lack regeneration potential exhibited by newts such as mammals, Drosophila etc. Since the genetic machinery is conserved across the species, the Drosophila model has been successfully used for such cross species studies to model human disease and determine their underlying molecular genetic mechanism(s) (Hughes et al., 2012; Sarkar et al., 2016, 2018; Singh and Irvine, 2012; Tare et al., 2011).
Previously, a family of five protein members was identified in a comprehensive transcriptomic analysis from Notophthalmus viridescens (red-spotted newt). These proteins are expressed in adult tissues and are modulated during lens regeneration (Looso et al., 2013). The three-dimensional structure models of these newt proteins by ab initio methods suggested that these proteins could act as ion transporters and/or involved in signaling (Mehta et al., 2019). These unique newt genes were introduced in genetically tractable Drosophila melanogaster (fruit fly) model by transgenic approaches to test their regeneration potential. Such transgenic approaches were not feasible in N. viridescens, therefore, Drosophila served as a suitable system. The transcriptomic analysis was performed in the transgenic Drosophila that are expressing newt genes. Based on this study, graded expression of 2775 transcripts was reported in transgenic flies. In each transgenic fly one of the five newly identified newt genes was ectopically expressed using the GAL4/UAS binary target system (Brand and Perrimon, 1993; Mehta et al., 2019). Among 2775 transcripts, genes involved in the fundamental developmental processes like cell cycle, apoptosis, and immune response were highly enriched suggesting that these foreign genes from newt were able to modulate the expression of Drosophila gene(s) (Mehta et al., 2019; Mehta and Singh, 2019b).
Drosophila, a hemimetabolous insect, serves as a versatile model organism to study development and disease because of presence of fully sequenced and less redundant genome, presence of homologs or orthologs for human disease related genes, a shorter life cycle and economy to maintain fly cultures (Bier, 2005; Lenz et al., 2013; Singh and Irvine, 2012). Drosophila eye has been extensively used to understand the fundamental process of development and model neurodegenerative disorders (Gogia et al., 2020a; Kumar, 2018; Sarkar et al., 2016, 2018; Singh and Irvine, 2012; Tare et al., 2011; Yeates et al., 2019). The rich repository of genetic tools available in fly models, and the fact that eye is dispensable for survival, makes Drosophila eye suitable for screening genes from other animals that can promote growth and regeneration. The compound eye of the adult fly develops from a monolayer epithelium called the eye-antennal imaginal disc, which develops from an embryonic ectoderm (Cohen, 1993; Held, 2002). The imaginal disc, a sac-like structure present inside the larva, comprise of two different layers: the peripodial membrane (PM) and the disc proper (DP) (Kumar, 2020b). The DP develops into retina whereas the PM of the eye-antennal imaginal disc contributes to the adult head structures (Haynie and Bryant, 1986; Kango-Singh et al., 2003; Kumar, 2020b; Milner et al., 1983). In third instar larval eye imaginal disc, a wave of synchronous retinal differentiation moves anteriorly from the posterior margin of the eye disc and is referred to as Morphogenetic Furrow (MF) (Kumar, 2013, 2020a; Ready et al., 1976). The progression of MF transforms the undifferentiated cells into differentiated photoreceptor cells. The adult compound eye comprises of 800 unit eyes or ommatidia (Kumar, 2013, 2018, 2020a; Ready et al., 1976). The ommatidial cells in the compound eye are grouped together into two chiral forms, which are arranged in mirror image symmetry along the DorsoVentral (DV) midline called the equator. Ventral domain is the default state of the entire early eye imaginal primordium (Singh and Choi, 2003), and onset of the expression of the dorsal selector gene pannier (pnr), establishes the dorsoventral (DV) lineage in the eye (Maurel-Zaffran and Treisman, 2000; Singh and Choi, 2003; Tare et al., 2013). It has been reported that Lobe (L) is a cell survival gene required for ventral eye development and growth, and acts upstream to the Wg signaling pathway (Singh et al., 2006). During eye development L promotes cell survival by preventing ectopic induction of Wingless (Wg) and JNK-Signaling pathway in the ventral eye or in the entire early eye disc before the establishment of DV compartments (Singh et al., 2006, 2012a). L2 mutant exhibits a dominant, consistent phenotype of preferential loss of the ventral eye pattern from early larval eye imaginal disc to the adult eye (Singh et al., 2005). L inhibits the JNK signaling by downregulating Wg signaling and thereby prevent caspase-dependent and caspase-independent cell death (Singh et al., 2006). Thus, L plays an important role in cell survival during eye development.
The wg gene, a Drosophila homolog of Wnt, serves as a ligand, and encodes a secreted signaling protein that acts as a morphogen (Baker, 1987; Bejsovec, 2013; Swarup and Verheyen, 2012). The transcriptional effector of the Wg/β-catenin signaling is Drosophila T cell Factor (dTCF), which upon activation by Armadillo (Arm, human homolog β-catenin) leads to the transcription of Wg target genes. In the absence of ligand Wg, a destruction complex composed of Adenomatous polyposis coli (Apc), and Axin degrades Arm and thus prevents it to form a complex with dTCF. The pathway is activated by binding of Wg ligand to its co-receptors Frizzled (Fz) and Arrow (Arr). Upon activation, Fz binds to Disheveled (Dsh) and Arr, which binds to Axin and thus inactivating the destruction complex. This results in the translocation of Arm (β-catenin) from the cytosol to the nucleus where Arm binds to dTCF and activates transcription of Wg target genes (Bejsovec, 2006). During eye development, Wg plays different roles along the spatiotemporal axis. During early eye development, Wg is required for growth. In the eye-antennal disc, Wg acts as a negative regulator of eye development and inhibits morphogenetic furrow progression (Ma and Moses, 1995; Treisman and Rubin, 1995). In the early third instar eye-antennal imaginal disc, wg expresses on the ventral margin. Ectopic expression of Wg triggers cell death of photoreceptor cells causing eye defects (Baker, 1987; Cavodeassi et al., 1999; Singh, 2012; Singh et al., 2006). During pupal development, Wg is involved in programmed cell death to eliminate extra cells (Cordero et al., 2004; Lin et al., 2004; Swarup and Verheyen, 2012). Wg is also part of the dorsal eye gene hierarchy (Gogia et al., 2020a; Tare et al., 2013). . Previously it has been shown that Wg pathway was upregulated in the developing eye of L2 background. This perturbation in Wg pathway causes loss of ventral eye in 100% of flies of L2/+ background (Gogia et al., 2020a; Singh et al., 2006, 2011). Therefore, timing and level of wg expression in this region is crucial for formation of a normally patterned eye.
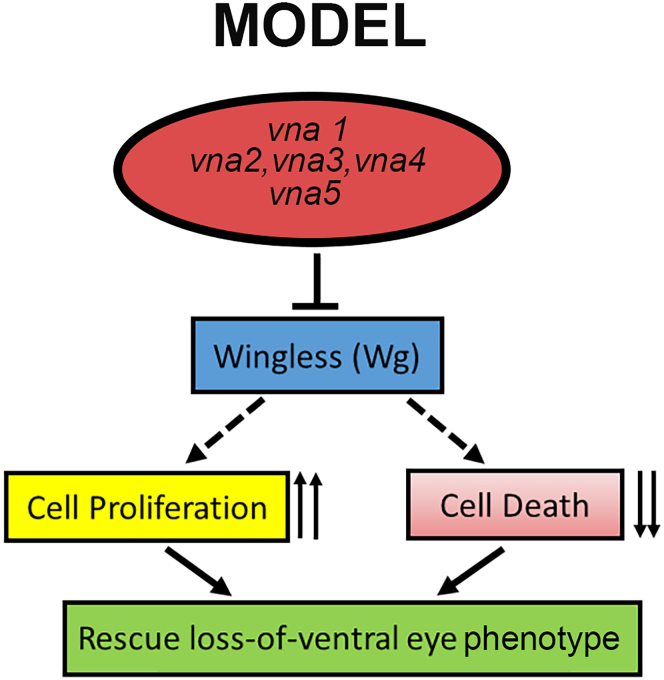
Here, we demonstrate the potential of five newly identified newt genes in significantly rescuing the missing photoreceptor cells in the ventral half of the eye of Drosophila L2 mutant eye. In addition, these genes can also rescue the reduced eye phenotype of GMR-hid, GMR-GAL4 flies, where activation of head involution defective (hid) triggers cell death. These two eye defect phenotypes represent the two different developmental time points. Our data suggests that these genes are effective in rescuing eye defects all along the developmental stages viz., early (L2 mutant) as well as later (GMR-hid,GMR-GAL4) time points of eye development. Ectopic expression of these genes restores eye mutant phenotypes by promoting cell proliferation, and downregulating cell death. Furthermore, these rescues by newt genes are non-cell-autonomous in nature. Finally, the misexpression of these newt genes in Drosophila downregulates Wg in L2 mutant background, resulting in significant rescue of missing photoreceptor cells in the developing Drosophila eye.
Results
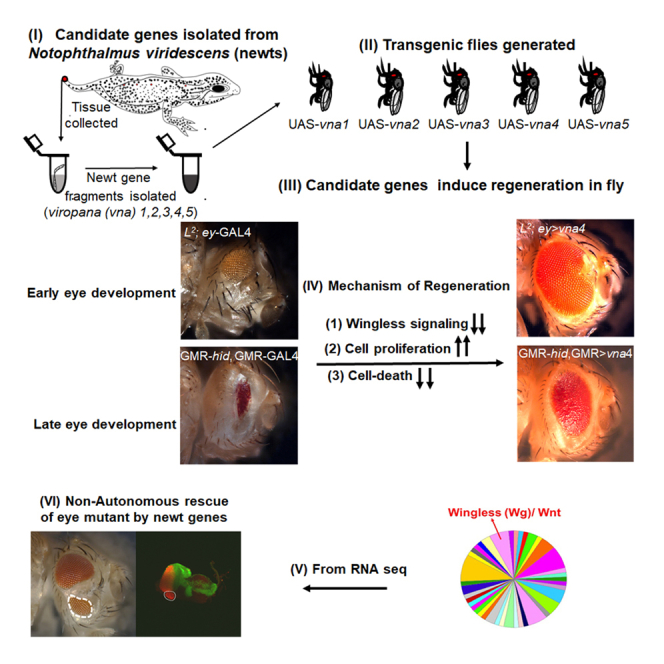
We employed Drosophila eye model to test the regeneration potential of the five unique newt genes, which have been named as: viropana1 (vna1), viropana2 (vna2), viropana3 (vna3), viropana4 (vna4), and viropana5 (vna5) (Mehta et al., 2021). All these genes consist of open reading frame (ORF), and additional 5′ and -3′ untranslated regions (UTR) (Figure S1) (Looso et al., 2013). They all share common signal peptides, which indicate that these proteins could be secreted (Petersen et al., 2011). Further, they might belong to the same family, which is defined by the common motif (L-x(1,3)-C-L-x(2)-[AL]-L-x(3)-[AL]-[AET]-x(2)-[LV]-x-[AS]-[ILV]-x-[DQ]-[LV]-[LV]-C-[AC]-[FIV]-x(3)-[DN]-[EP]-[AIV]-[EK]-x-K-[EN]-x-L) (Figure S1A). All five family members showed high similarity to the proteins from the other newt species- pleurodeles waltl) (Figure S1B). Previously, it has been reported that these five genes are highly expressed in the tail of the newt (Notophthalmus viridescens) (Looso et al., 2013). Therefore, in our study we isolated the total RNA from the newt tail, generated cDNA to amplify these newt genes, and used this cDNA to generate transgenic flies containing these five genes (Figure 1A). We employed GAL4/UAS system (Brand and Perrimon, 1993) to allow targeted misexpression of these newt transgenes in the fly tissues (Figure 1B). To confirm if targeted misexpression approach can translate newt proteins in Drosophila (Brand and Perrimon, 1993), we tested localization of the fusion protein. As there are no antibodies available against these newt proteins, we therefore used V5 epitope tagged transgenes to generate another set of transgenic flies (Wang et al., 2019). The V5 tag sequence of about 42 bp long is fused toward the 3′ end of the 501 bp long ORF. We tested all these transgenes using the antibody against V5 tag. Here we present the expression of one of the transgenes viz., vna4 using V5 tag (Figure S2). To detect if misexpression of these newt genes in Drosophila can generate any developmental defects, we used tubulin – GAL4 (tub –GAL4) to ubiquitously misexpress newt genes in flies in the wild-type background (Figure 1B) and found no defects.
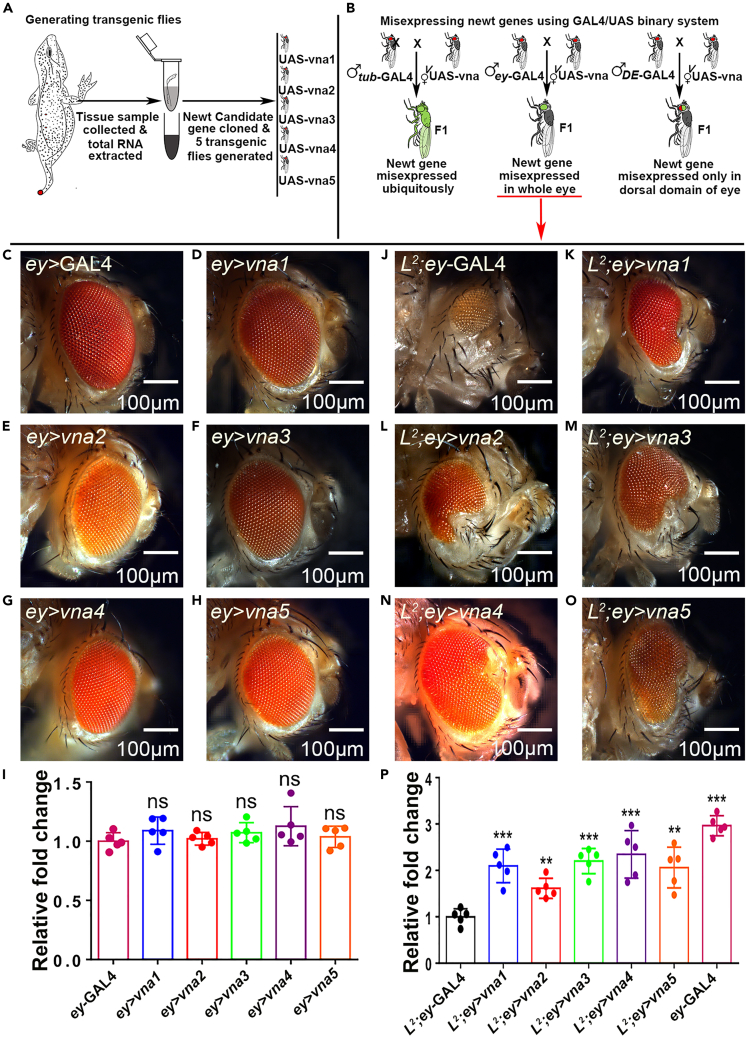
Figure 1.
Newt regeneration genes rescues L2 mutant’s loss-of-ventral eye phenotype
(A and B) Schematic representation of generating transgenic Drosophila carrying these newt (Notophthalmus viridescens) candidate genes (vna1, vna2, vna3, vna4, and vna5).
(C) Bright field image of a (C) wild type adult eye (ey-GAL4),
(D–H) adult eye phenotype when ey-GAL4 drives expression of newt transgenes (D) UAS-vna1 (ey>vna1), (E) UAS-vna2 (ey>vna2), (F) UAS-vna3 (ey>vna3), (G) UAS-vna4 (ey>vna4), and (H) UAS-vna5 (ey>vna5).
(I) The bar graph shows there is no significant (ns) change in eye size between control and where ey-GAL4 drives expression of newt transgenes (ey>vna1-vna5).
(J) Bright field image of a (J) L2 mutant (L2; ey-GAL4) adult eye where there is preferential loss-of- ventral eye phenotype,
(K–O) L2 mutant (L2; ey-GAL4) background where newt transgene (K) UAS-vna1 (L2; ey >vna1), (L) UAS-vna2 (L2; ey >vna2), (M) UAS-vna3 (L2; ey >vna3), (N) UAS-vna4 (L2;ey>vna4), and (O) UAS-vna5 (L2;ey>vna5) are expressed in the developing eye using ey-GAL4.
(P) L2 mutant adult eye is half the adult eye size of the ey-GAL4 which serves as a control. The bar graph clearly displays the significant increase in the adult eye size of L2-mutant background where unique newt genes were misexpressed compared to the adult eye size of the L2-mutant only (L2; ey-GAL4). Eye size was measured using ImageJ software tools (http://rsb.info.nih.gov/ij/). Sample size used for statistical analysis was five (n = 5). Statistical analysis was performed using the student’s t test for independent samples. Statistical significance was determined with 95% confidence (p < 0.05). Error bars show standard deviation (mean ± SD), and symbols above the error bar signify as: ns is non-significant p value, ∗ signifies p value < 0.05, ∗∗ signifies p value < 0.005, ∗∗∗ signifies p value < 0.0005, respectively. In this study ey is used as a driver to misexpress newt genes in the developing Drosophila eye. All the images are displayed in the same polarity as dorsal domain-towards top and ventral domain-towards bottom. Scale bar = 100 μm. See also Figures S1, S2, S3, and S4.
Newt genes can rescue Drosophila eye mutants
We used the Drosophila eye model to evaluate the function of unique newt genes as the eye is not required for the viability of the fly and it is easy to score the phenotypes in eye. We used ey-GAL4 driver (Hazelett et al., 1998) to misexpress these newt genes in the developing eye imaginal disc, which develops into an adult eye. The phenotypes of UAS-transgene alone were tested to rule out any contribution(s) in phenotype due to insertion of the transgene. In comparison to the control, ey-GAL4 (Figure 1C), misexpression of these transgenes ey-GAL4>UAS-vna1 (ey>vna1, Figure 1D), ey-GAL4>UAS-vna2 (ey>vna2, Figure 1E), ey-GAL4>UAS-vna3 (ey>vna3, Figure 1F), ey-GAL4>UAS-vna4 (ey>vna4, Figure 1G and ey-GAL4>UAS-vna5 (ey>vna5, Figure 1H), did not affect the eye size (Figure 1I). To investigate the regeneration potential of these genes, we used eye mutant(s) such as L2 mutant, which exhibits selective loss of ventral half of the eye (Figure 1J). L2 mutant exhibits loss-of-ventral-eye in 100% flies. Interestingly, targeted misexpression of the newt gene(s) in L2; ey-GAL4 background [(L2; ey>vna1; Figure 1K), (L2; ey>vna2; Figure 1L), (L2; ey>vna3; Figure 1M), (L2; ey>vna4; Figure 1N), (L2; ey>vna5; Figure 1O)] exhibit significant rescue of the loss-of-ventral eye phenotype (Figure 1P). For each cross, three independent cultures were established and two hundred flies for each set (triplicate, 600 flies) were counted to determine phenotype frequency. The rescue frequency was about 40.83% in vna1, 37.7% in vna2, 49% in vna3, 58.2% in vna4, and 38.7% in vna5 (Figure S3). Note that the strongest rescues (phenotype strength as well as rescue frequency %) were observed with vna4 transgene (Figures 1P and S3; Table S1).
Because the L2 eye mutant represents developmental defects in the genetic machinery involved in early eye development, it raises a question whether the regeneration potential of these newt genes is restricted to the early time window or even later stages of eye development. The GMR- GAL4 driver was used to drive expression of transgenes in the differentiating retinal neurons of the larval eye imaginal disc, which continues all along to the adult eye (Moses and Rubin, 1991). Gain-of-function of head involution defective (hid), triggers cell death (Bergmann et al., 1998; Grether et al., 1995; Hay et al., 1995; White et al., 1994). Misexpression of hid using GMR-GAL4 (GMR>hid) results in a “No-eye” or highly reduced eye phenotype (Figure S4B). However, misexpression of newt transgenes along with hid in the eye (GMR-GAL4>hid+vna4) results in a significant rescue of the “No-eye” phenotype (Figure S4C). Thus, confirming that the restoration potential of newt genes is not restricted to the early time window but can even extend to the later stages of eye development.
Ectopic expression of newt genes exhibits non-cell-autonomous rescue
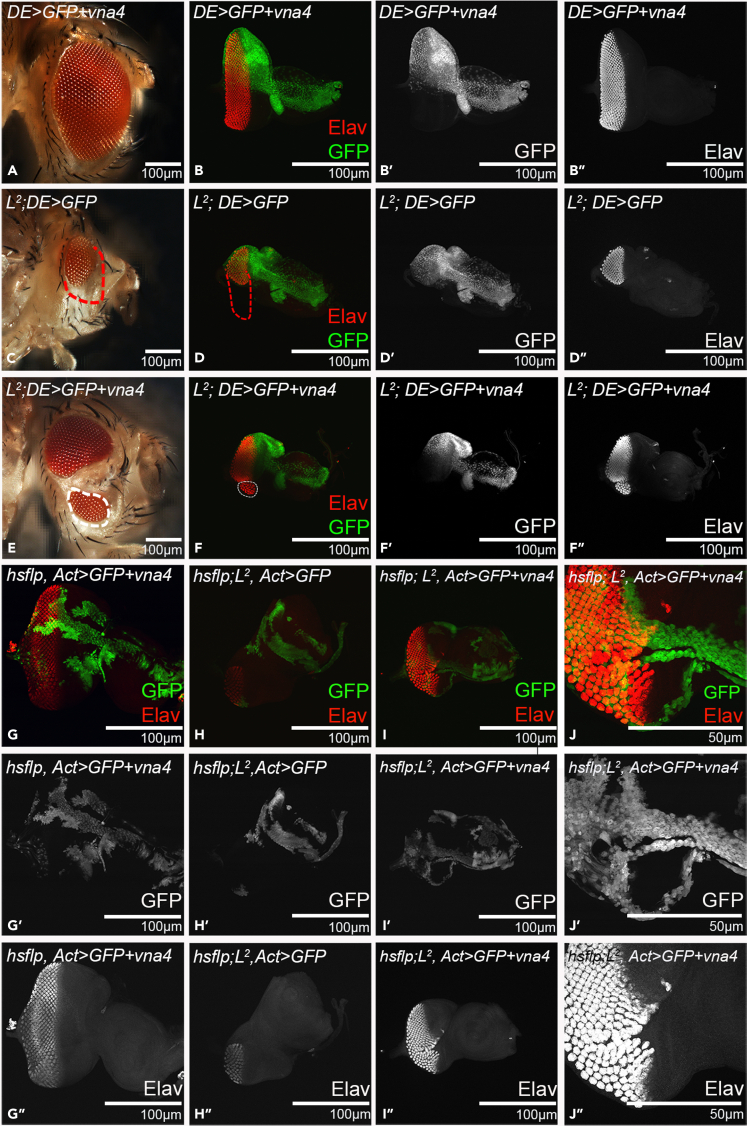
All 5 unique newt genes have signal peptides (Looso et al., 2013), so it is possible that these genes may have non-cell-autonomous effects. To determine any such possibility, we misexpressed these genes within a subset of retinal neuron population in the developing eye field of L2 mutant and assay their regeneration effect. Misexpression of vna4 using dorsal-eye - GAL4 (DE - GAL4) (Figure 2) directs its expression in the dorsal half of the developing eye (Morrison and Halder, 2010) (Figures 2A and 2E). The rationale of this experiment was to misexpress vna4 in the dorsal half of the eye and test its effect on an eye mutant, which results in loss of the ventral half of the developing eye. Misexpression of both GFP (reporter) and newt vna4 in the dorsal domain of the developing eye (+/+; DE>GFP+vna4) (Figures 2A, 2B, 2B′, and 2B″), exhibits a near wild-type eye. As a negative control, we tested GFP expression alone in L2-mutant background (L2/+; DE>GFP) (Figures 2C, 2D, 2D′, and 2D″). We found that GFP expression was restricted to the dorsal half as seen in the wild-type control (Figures 2B, 2B′, and 2B″). Furthermore, we did not see any rescue of L2 mutant phenotype of ventral eye loss (marked by red dotted boundary, Figures 2D, 2D′, and 2D″). Misexpression of both GFP+vna4 in the dorsal half of the L2 mutant eye (L2/+; DE>GFP+vna4) exhibits a significant rescue of the loss-of-ventral eye phenotype as seen in the adult eye, and the eye imaginal disc (marked by white-dotted boundary, Figures 2E, 2F, 2F′, and 2F″). Even though the targeted misexpression of vna4 is restricted to the dorsal half of the developing eye, it was able to rescue the loss-of-ventral eye phenotype suggesting that newt genes can show non-cell-autonomous rescue of Drosophila eye mutants.
Figure 2.
Misexpression of newt genes exhibits non-cell-autonomous rescue
(A, B, B′, and B″) Misexpression of vna4 in the dorsal half using DE-GAL4 driver (A) Adult eye and (B, B′, B″) developing third instar larval eye imaginal disc Note that Green fluorescence protein (GFP) (green) reporter marks the DE-GAL4 driver domain and proneural marker Embryonic lethal abnormal visual system (Elav) (red) marks the neuronal fate. (B′) is a single channel confocal image showing GFP expression, and (B″) is a single channel confocal image showing Elav expression.
(C, D, D′, and D″) L2 mutant eye where there is no misexpression of newt genes (L2/+; DE>GFP) (as negative control). (C) Adult eye bright field image and (D, D′, D″) third instar eye disc of L2 mutant (L2/+; DE>GFP) exhibiting loss-of-ventral eye phenotype and red dotted line marks the boundary of eye. GFP reporter (green) marks domain of DE-GAL4 expression domain in L2 mutant background.
(E, F, F′, and F″) Misexpression of vna4 in dorsal eye domain of L2 mutant (L2/+; DE>GFP; vna4) exhibits non-cell autonomous rescue of loss-of-ventral eye phenotype in (E) Adult eye (white dotted boundary) and (F, F′, F″) third instar eye disc. White dotted boundary marks the significantly rescued loss-of-ventral eye phenotype.
(G–J) Genetic mosaic “Flp out” somatic clones of (G, G′, and G″) newt gene vna4 in the developing eye disc wild-type, (H, H′, and H″) GFP in L2 background (L2/+; hsflp>GFP, served as the negative control) and (I, I′,I″, J, J′, and J″) newt gene vna4 clones in L2 mutant (L2/+; hsflp>GFP;vna4) background. All clones are marked by GFP reporter (green). Note that misexpressing vna4 rescue missing photoreceptor cells in a non-cell-autonomous manner, as photoreceptor cells rescue marked by Elav (in red) extends outside the boundary of the clone marked by GFP (in green). (J, J′, and J″) exhibits the magnified view of the vna4 misexpressing clone (green) in L2/+; hsflp>GFP; vna4 background. All the images are displayed in the same polarity as dorsal domain-towards top, and ventral domain-towards bottom. Scale bar = 100μm,
To rule out any domain specific restraint, or contribution of a DE-GAL4 background, we employed a genetic mosaic approach. We generated gain-of-function clones using Flp out approach (Xu and Rubin, 1993), which result in clones of cells that misexpress high levels of the vna4 transgene and these clones are marked by the GFP reporter (Figures 2G, 2H, 2I, and 2J). We found that the misexpression of these vna transgenes rescues the loss-of-ventral eye phenotypes; however, these rescues extend beyond the boundary of the clone. This data validates our prior observation that these newt genes exhibit non-cell-autonomous rescue due to their secretory nature. Next, we wanted to investigate the mechanism by which these newt genes can rescue loss-of-ventral eye phenotype.
Newt genes promote cell proliferation and downregulate cell death
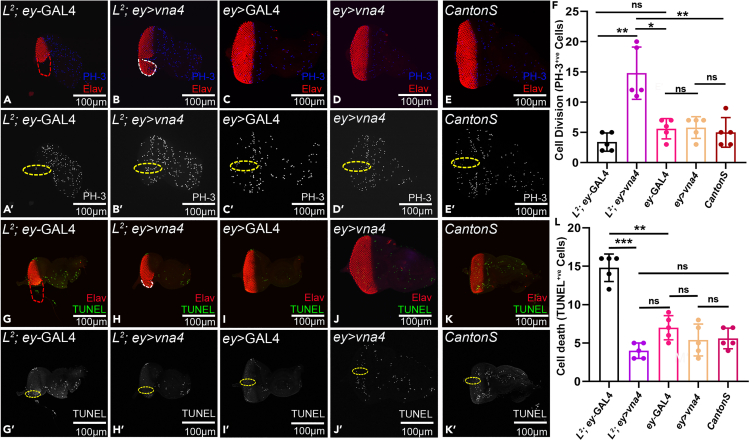
To test the possibility of upregulation of cell proliferation resulting in the rescue of the missing tissue in the L2 mutant background, we compared number of dividing cells in the zone of interest between the L2 mutant, and the L2 mutant background where the newt gene was misexpressed (Figures 3A–3F). The proliferating cells are marked by anti-phospho-histone H3 (PH3) marker (Figures 3A–3E, 3A′–3E′, S5A–S5H, and S5A′–S5H′). Phosphorylation of a highly conserved Serine residue (Ser-10) in the histone H3 tail is considered a crucial event for the onset of mitosis and represents completion of cell cycle (Crosio et al., 2002; Kim et al., 2017). Our results show robust increase in the number of dividing cells when vna4 is misexpressed in the L2-mutant background (L2; ey > vna4) (shown in white dotted boundary Figure 3B) with respect to the L2-mutant (L2; ey-GAL4) only (Figure 3F). Similar trend in the cell proliferation rate were observed with other vna transgenes (Figures S5A–S5I). The L2 mutant background, which served as control, did not show any rescue of the loss-of-ventral eye phenotype due to lack of cell proliferation as evident from PH3 staining (shown in red dotted boundary, Figure 3A). In addition, newt gene when misexpressed in the wild-type background (ey > vna4) (Figures 3D, 3F, S5G, and S5I) did not show significant increase in cell proliferation compared to its control (ey-GAL4) as well as wild-type fly (Canton-S) (Figures 3C, 3E, 3F, S5F, S5H, and S5I).We calculated dividing cells in GMR-hid, GMR-GAL4 alone background, and as expected found upregulation in cell proliferation when newt gene is misexpressed along with GMR-hid, GMR-GAL4 (GMR-hid, GMR-GAL4>vna4) (Figures S6B and S6C) compared to the GMR-hid, GMR-GAL4 background in which newt gene has not been misexpressed (Figures S6A and S6C). These results clearly show that the newt gene(s) can induce cell proliferation to restore the missing structures in L2-mutant and GMR-hid, GMR-GAL4 eye disc. These missing structures are t photoreceptor cells in our study .
Figure 3.
Newt genes promote cell proliferation and downregulate cell death to promote rescue of L2 mutant phenotype of loss-of-ventral eye
(A–E) PH-3 staining (blue), as a marker to calculate cell proliferation in the developing eye imaginal disc of (A) L2 mutant (red dotted boundary marking eye boundary), (B) L2 mutant background where vna4 gene is misexpressed (L2; ey>vna4), (C) ey-GAL4 driver, (D) misexpression of newt gene in ey-GAL4 domain (ey>vna4) (E) and wild type (CantonS). (A) Red dotted boundary represents the loss-of-ventral-eye area, and (B) white dotted boundary represents the rescue of loss-of-ventral eye phenotype where vna4 gene is misexpressed. (A′–E′) single channel confocal images demonstrating PH-3 stained nuclei. The region of interest used to count the number of dividing cells is marked in yellow dotted boundaries.
(F) Bar graph shows comparative increase in the rate of cell division. There is no significant change in cell proliferation rate among the (A and A′) L2; ey-GAL4, (C and C′) ey-GAL4, (D and D′) ey>vna4, (E and E′) Canton-S (wild-type) eye disc. (A, A′, B, B′, and F) However, there is robust increase in the cell proliferation rate in (B and B′) L2; ey>vna4 background in comparison to (A and A′) L2 mutant (L2; ey-GAL4).
(G–K) Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining serves as a marker for cell death assay. TUNEL staining (shown in green) marks the dying cells nuclei in (G and G′) L2; ey-GAL4, (H and H′) L2; ey>vna4, (I and I′) ey-GAL4, (J and J′) ey>vna4, (K) and wild type (Canton-S) eye imaginal disc. (G) Red dotted boundary represents the loss-of-ventral eye region, and (H) white dotted boundary represents the rescued loss-of-ventral eye region. (G′–K′) The TUNEL positive cells are counted within the zone of interest marked within the yellow dotted boundary as shown in single filter confocal images.
(L) Bar graph shows decrease in the cell death when a newt gene is misexpressed in the L2-mutant background compared to only L2-mutant. The downregulation in cell death is comparable to the cell death occurring in the normal wild- type background. Statistical analysis was performed using the student’s t-test for independent samples. Sample size used for the calculation was five in number (n = 5). Statistical significance was determined with 95% confidence (p < 0.05). All bar graphs show change in cell proliferation and cell death as average between 5 samples. Error bars show standard deviation (mean ± SD), and symbols above the error bar ∗ signify p value < 0.05, ∗∗ signify p value < 0.005, ∗∗∗ signify p value < 0.0005, and ns signify p value >0.05 respectively. All the images are displayed in the same polarity as dorsa domain-towards top, and ventral domain-towards bottom. Scale bar = 100 μm. See also Figures S5, S6, S7, S8, and S9.
During newt lens regeneration, both cell proliferation and apoptosis are observed (Tsonis et al., 2004). It has been shown that the loss-of-ventral-eye phenotype observed in L2 mutant is because of induction of both caspase-dependent and caspase-independent cell death (Singh et al., 2006). Therefore, we did TUNEL (Terminal deoxynucleotidyl transferase dUTP nick end labeling) assay to test if newt genes can block or downregulate cell death to rescue L2 mutant phenotype (Figures 3G–3L and S5J–S5R). TUNEL assay is based on the detection of fragmented DNA ends, which are characteristic of apoptotic cells (Sarkissian et al., 2014; White et al., 2001). The number of dying cells are compared between the L2 mutant, and the L2 mutant background where the newt gene was misexpressed. We found a significant reduction in the number of dying cells when newt gene (vna4) is misexpressed in the L2 mutant background (L2; ey > vna4) (Figures 3H, 3H′, and 3L) in comparison to the L2-mutant alone (L2; ey-GAL4) (Figures 3G, 3G′, and 3L). Similarly, ey-GAL4 (Figures 3I, 3I′, and 3L) exhibits signifiant downregulation in the rate of cell death with respect to the L2 mutant (L2; ey-GAL4) (Figures 3G, 3G′, and 3L). The value is significantly lower in comparison to the L2 mutant (L2; ey-GAL4), and almost equivalent to the L2 mutant where the newt gene is misexpressed (L2; ey > vna4) (Figure 3L). This suggests that newt gene(s) upon misexpression can rescue the L2 mutant’s loss-of-ventral eye phenotype by preventing excessive cell death of photoreceptor cells (marked in white dotted boundary, Figures 3H and 3H′). We found a similar trend of reduction in cell death when other members of this newt gene family were misexpressed (Figures S5J–S5R). However, when newt gene is misexpressed in the wild type background (ey>vna4) (Figures 3J, 3L, S5P, and S5R) no significant change in cell death was observed in comparison to its control (ey-GAL4) as well as wild type fly (CantonS) (Figures 3I, 3K, 3L, S5O, S5Q, and S5R). TUNEL assay was also performed in GMR-hid, GMR-GAL4 eye imaginal discs, and dying cells nuclei were quantified. Our results report that misexpressing newt genes in GMR-hid, GMR-GAL4 (GMR-hid, GMR-GAL4>vna4) robustly downregulates cell death (Figures S6E and S6F) in comparison to GMR-hid, GMR-GAL4 strain alone (Figures S6D and S6F). We also tested the role of cell death using Dcp1 (Death caspase-1) staining (Figure S7). Antibody against cleaved Dcp1 marks the dying cells in Drosophila (Sarkissian et al., 2014). Dcp1 staining exhibits reduction in the number of dying cells when newt genes are misexpressed in L2 mutant background (Figures S7C–S7G), in comparison to the L2 mutant alone (Figure S7B). These results suggest that newt genes upregulate cell proliferation and downregulate cell death to significantly rescue the L2 mutant loss-of-ventral eye phenotype, and eye loss caused by misexpression of hid.
We further validated the role of cell death by blocking caspase-dependent cell death using baculovirus p35 misexpression (Hay et al., 1994). Even though misexpression of p35 in L2 mutant background (L2/+; ey > p35) exhibits rescue of loss-of-ventral eye phenotype, the phenotype strength (relative eye size ratio of 1.42 ± 0.29 and rescue frequency of 22.7%) of L2/+; ey > p35 was significantly weaker than phenotype strength (relative eye size ratio = 2.34 ± 0.51 and rescue frequency of 58.2%) of L2/+; ey > vna4 (Figures S8C, S8D, S8F, and S8G). This data suggests that the newt genes can upregulate cell proliferation and also block cell death, but cell proliferation may be playing significant role in rescuing the missing tissues in the L2 mutant eye. To validate this hypothesis, we compared rate of cell proliferation and cell death between L2 mutant (L2; ey-GAL4), L2/+; ey > p35, and L2; ey > vna4 (Figure S9). As expected, there was a significant downregulation in cell death (Figures S9A, S9A′, S9B, S9B′, and S9D), and non-significant change in cell proliferation (Figures S9E, S9E′, S9F, S9F′, and S9H) between L2; ey-GAL4 and L2/+; ey > p35. However, an opposite trend is observed when comparing L2/+; ey > p35 and L2; ey > vna4. There is a non-significant change in cell death (Figures S9B, S9B′, S9C, S9C′, and S9D), but highly significant change in cell proliferation (Figures S9F, S9F′, S9G, S9G′, and S9H). Thus, it is clearly demonstrated that newt genes favor cell proliferation more than cell death in order to restore the missing structures. Therefore, it becomes warranted to understand what signaling pathway is triggered by these newt genes to promote the rescue.
Newt genes downregulate wg expression in developing eye
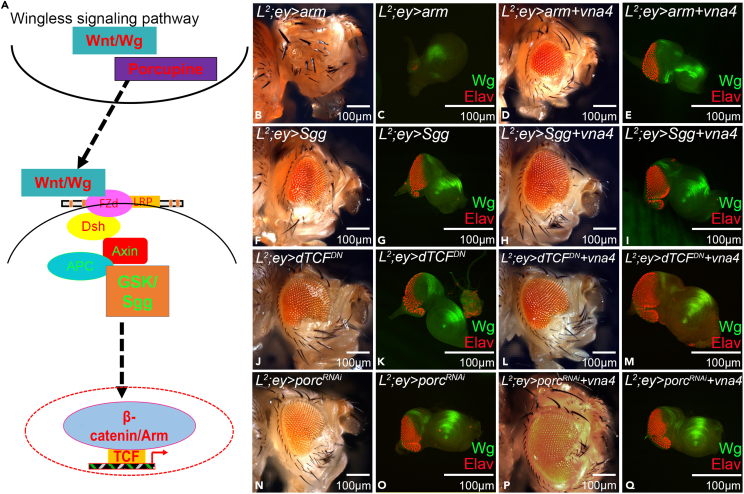
We screened for the signaling pathway(s) that are involved in the rescue of L2 mutant loss-of-ventral eye phenotype. We identified members of evolutionarily conserved Wg/Wnt signaling pathway in our high throughput screen (RNA sequencing) where newt genes were misexpressed in the fly tissue (Figure S10). It is reported that L2 mutant phenotype can be rescued by downregulating wg (Singh et al., 2006). To determine the involvement of wg in promoting rescue of L2 mutant phenotype by unique newt genes, we examined expression levels of Wg in larval eye-antennal imaginal disc (Figures 4A–4C, 4A′–4C′, and 4D). Wg expression is seen at the antero-lateral margins of the developing eye imaginal disc (Baker, 1987; Cavodeassi et al., 1999; Singh et al., 2006). In L2 mutant, robust ectopic expression of Wg is seen at the ventral eye margins (Figures 4B and 4B′) (Singh et al., 2006, 2011). Misexpression of newt gene (vna4) in L2 mutant background downregulates the Wg expression at the ventral margin of eye-antennal imaginal disc (Figures 4C and 4C′), as evident from the signal intensity plots (integrative density) in various backgrounds (Figure 4D). The semi-quantitative Western blot, showed that Wg levels increased 3 folds in L2 mutant as compared to ey-GAL4 control whereas misexpressing vna4 in the L2 mutant background decreased the Wg expression by 1.6-fold as compared to L2 mutant (L2; ey-GAL4) (Figures 4E and 4F). Similar trends were seen with other newt genes (vna1, vna2, vna3, and vna5) when misexpressed in L2 mutant background (Figures S11C–S11F, S11C′–S11F′, and S11G). Thus, the newt genes can downregulate wg expression in Drosophila L2 mutant to partially rescue ventral eye-loss-phenotype.
Figure 4.
Newt genes downregulate Wg expression to rescue loss-of-ventral eye phenotype.
(A–D) Immunohistochemistry showing Wg modulation. (A–C) Eye disc showing Wg staining in green, and Elav in red. (A′–C′) Single channel confocal images showing Wg expression. The green window is the area of interest that is utilized to calculate (D and D′) integrative density (intensity plot). The bar graph represents the Wg intensity at the ventral margin (within the green box) in (A and A′) wild type fly (ey-GAL4), (B and B′) L2-mutant fly, (C and C′) Misexpression of newt gene (vna4) in the L2 mutation background. Note that vna4 misexpression downregulates Wg expression.
(E and F) Western blot (WB) to semiquantitative modulation of Wg expression levels. (E) The blot shows the expression of Wg protein in the Drosophila eye. The samples were loaded in the following sequence: Lane 1- ey-GAL4, Lane 2- L2; ey-GAL4, Lane 3- L2; ey>vna4. Alpha tubulin is used as a loading control. Molecular weight of Wg is 46kD, and alpha tubulin is 55 kD. The samples were treated with anti-Wg antibody, and anti-α tubulin antibody. (F) Bar graph representing as a relative Wg level, is a measure of signal intensity of the bands, which clearly demonstrates that newt genes when misexpressed in the L2-mutant background downregulates Wg levels. Statistical analysis was performed using the student’s t-test for independent samples. Statistical significance was determined with 95% confidence (p < 0.05). All bar graphs show integrative density as a scale to measure Wg intensity for each sample represented as the average between 5. Error bars show standard deviation (mean ± SD), and symbols above the error bar ∗ signify p value < 0.05, and ∗∗ signify p value < 0.005 respectively. All the images are displayed in the same polarity as dorsal domain-towards top, and ventral domain-towards bottom. Scale bar = 100 μm. See also Figures S10 and S11.
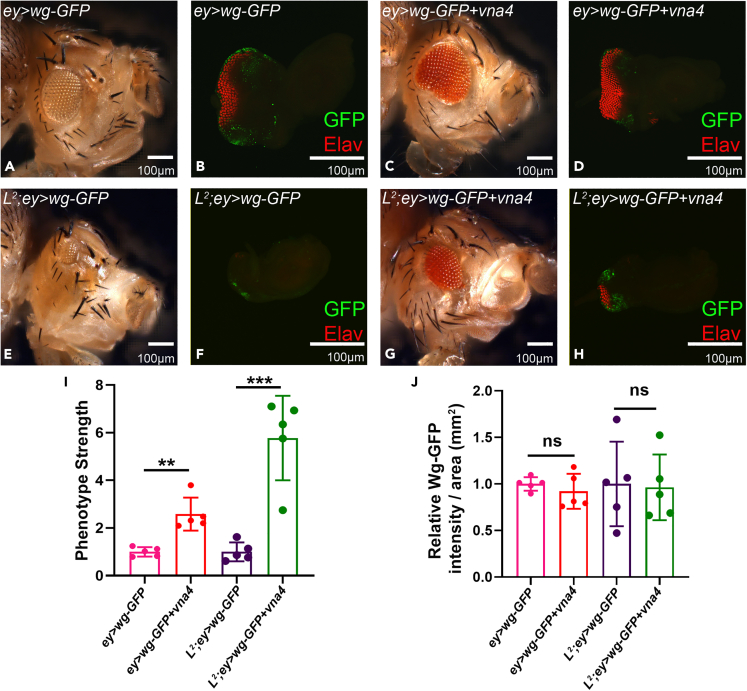
To further test our hypothesis, we misexpressed wg (the target identified in our screen) along with vna4 transgene in the L2 mutant background. As control, we misexpressed wg in the developing eye (ey>wg), which result in reduced eye phenotype (Figures 5A and 5B) (Lee and Treisman, 2001; Singh et al., 2012a). Misexpressing vna4 in the wild-type background where wg is ectopically induced (+/+; ey>wg+vna4), rescues the reduced eye phenotype (Figures 5C and 5D) at a frequency of 16.8% (Figure S12; Table S1). Misexpression of wg in the L2/+ mutants eye background (L2/+; ey>wg) completely eliminate the eye field as seen in the eye imaginal disc and the adult eye (Figures 5E and 5F) whereas, misexpressing vna4 along with wg in the L2/+ mutant eye disc (L2/+; ey>wg; vna4) partially rescues the loss-of-ventral eye phenotype (Figures 5G and 5H) in 21% flies (Figure S12; Table S1). The phenotype strength shows the significant rescue of the eye loss phenotype in both wild-type (+/+; ey>wg+vna4; Figures 5C and 5D) as well as L2 (L2/+; ey>wg; vna4; Figures 5G and 5H) background(s) where wg was ectopically co-expressed (Figure 5I). The changes in the ectopically overexpressed wg-GFP levels were non-significant, suggesting that newt genes can regulate endogenous Wg levels to promote rescue (Figure 4) and not the ectopically overexpressed Wg levels (Figure 5J).
Figure 5.
Newt gene regulates Wg both in the wild type, and in the L2-mutant background
(A–H) Misexpression of wg in the Drosophila developing eye. (A, B) Wild type background (+/+; ey>wg-GFP). (C and D) Newt gene and Wg are misexpressed in the wild type background (+/+; ey>wg-GFP; vna4) exhibits increase in the eye size compared to (+/+; ey>wg-GFP) as seen in (A and C) bright field adult eye picture, and (B and D) third instar eye-antennal imaginal disc confocal image. (E, F) Misexpression of wg in the L2–mutant background (L2/+; ey>wg-GFP) increases the severity of small-eye phenotype of L2–mutant. (G and H) Misexpression of the newt gene and wg in the L2-mutant background (L2/+; ey>wg-GFP; vna4) exhibits a significant rescue of the reduced eye phenotype as seen in (E and G) bright field adult eye picture, and (F and H) third instar eye-antennal imaginal disc confocal image. The GFP reporter (green) marks ectopic misexpression of wg in the developing eye, Elav a proneural marker is shown in red.
(I) The bar graph (I) represents the relative fold change in phenotype strength (eye size) compared between ey>wg-GFP and ey>wg-GFP; vna4, and between L2/+; ey>wg-GFP and L2/+; ey>wg-GFP; vna4.
(J) Similarly bar graph (J) represents relative fold change in the ratio of Wg -GFP intensity per area of GFP expression between respective samples. Statistical analysis was performed using the student’s t-test for independent samples. Statistical significance was determined with 95% confidence (p < 0.05). All bar graphs show values as the average between5 samples. Error bars show standard deviation (mean ± SD), and symbols above the error bar signify as: ns is non-significant p value, ∗∗ signifies p value < 0.005, ∗∗∗ signifies p value < 0.0005 respectively. All the images are displayed in the same polarity as dorsal domain-towards top, and ventral domain-towards the bottom. Scale bar = 100 μm. See also Figure S12.
Newt genes negatively regulate Wg/Wnt signaling
To investigate if newt genes regulate expression of wg alone or they regulate the Wg signaling pathway, we tested the effector and inhibitor of the Wg-signaling pathway. The rationale was to modulate the levels of Wg signaling and sample its effect on the L2 mutant phenotype as well as the L2 mutant background where vna4 is misexpressed (L2/+; ey> vna4) (Figure 6). Increasing the levels of Arm (a vertebrate homolog of β-Catenin, an effector of Wg signaling) in the wild-type eye (ey>arm) results in small eyes (Figures S13A and S13B) and in the L2/+ background, (L2/+; ey>arm) it results in elimination of the entire eye field (Figures 6B and 6C). Misexpressing vna4 along with arm in the L2/+ mutant background (L2/+; ey>arm; vna4) partially rescues the loss-of-ventral eye phenotype (Figures 6D and 6E) and the rescue frequency is 22.8% (Figure S14A; Table S1). Similarly, misexpressing vna4 and arm in the wild-type background (+/+; ey>arm; vna4) partially rescues the small eye phenotype defect caused by ey>arm phenotype (Figures S13C and S13D), and the rescue frequency is 21% (Figure S14A; Table S1). This data suggests that newt gene can modulate the Wg signaling pathway components to promote rescue of reduced eye phenotype both in L2-mutant as well as the wild-type background(s). However, the rescue frequency obtained by misexpressing newt gene in addition to ectopically misexpressing arm (L2/+; ey>arm; vna4, 22.8%) and/or wg (L2/+; ey>wg; vna4, 21%) (Figures S12 and S14A; Table S1) is lower than the rescue frequency obtained when only newt gene is misexpressed in the L2-mutant background (L2/+; ey>arm; vna4, 58.2%). This could be because of severity of loss-of-ventral eye phenotype caused by the combined effect of L2 mutation and gain-of-function of wg, or arm.
Figure 6.
Modulating positive and negative regulators of Wg signaling pathway affects the L mutant phenotype
(A) Schematic presentation of the Wg signaling pathway showing various members of the canonical pathway. The positive regulators are in red, and negative regulators of the Wg pathway are in green.
(B–E) Activating Wg signaling in the L2 mutant eye background by misexpression of (B and C) arm alone (L2/+; ey>arm), (D and E) arm and vna4 (L2/+; ey>arm; vna4). Note that vna4 misexpression with arm (L2/+; ey>arm; vna4) in the eye rescues the L2/+; ey>arm reduced eye phenotype.
(F–M) Blocking Wg signaling in L2 mutant eye background by misexpression of (F and G) sgg alone (L2/+; ey>sgg), (J and K) dTCFDN alone (L2/+; ey> dTCFDN) results in rescue of L2 loss-of-ventral eye phenotype. Similarly, in L2 mutant background misexpression of vna4 along with (H and I) sgg (L2/+; ey>sgg; vna4) and (L and M) dTCFDN (L2/+; ey> dTCFDN; vna4) significantly rescue the partial loss-of-ventral eye phenotype.
(N–Q) Blocking transport of Wg morphogen in L2 mutant eye background by misexpression of (N and O) porcRNAi alone (L2/+; ey> porcRNAi), (P and Q) porcRNAi and vna4 (L2/+; ey> porcRNAi; vna4). Note that L2/+; ey> porcRNAi exhibits weak rescue of loss-of-ventral eye phenotype whereas vna4 misexpression with porcRNAi (L2/+; ey> porcRNAi; vna4) enhances the phenotype strength. All the images are displayed in same polarity as dorsal domain-towards the top, and ventral domain-towards the bottom. Scale bar = 100 μm. See also Figures S13 and S14.
Blocking Wg signaling by misexpressing shaggy (sgg), an inhibitor of Wg signaling, in the eye (ey>sgg, Figures S13E and S13F) (Hazelett et al., 1998; Singh et al., 2002, 2006) or along with vna4 in the wild-type background (+/+; ey>sgg; vna4, Figures S13G and S13H) does not affect the eye size. Misexpression of sgg in the L2/+ mutants (L2/+; ey>sgg), significantly rescues the loss-of-ventral eye phenotype in 27.3% of flies (Figures 6F, 6G, and S14B; Table S1) (Singh et al., 2006). Similarly, misexpressing vna4 and sgg in the L2/+ mutant eye (L2/+; ey>sgg; vna4) rescues the loss-of-ventral eye phenotype (Figures 6H and 6I) and rescue frequency increased dramatically from 27.3% to 71.5% (Figure S14B; Table S1). The transcription factor dTCF is the downstream target of Wg signaling, and is downregulated by misexpression of the N-terminal deleted dominant-negative form of TCF (dTCFDN) (van de Wetering et al., 1997). Misexpression of dTCFDN alone in the eye (ey>dTCFDN, Figures S13I and S13J) or dTCFDN with vna4 (+/+; ey> dTCFDN; vna4, Figures S13K and S13L) does not affect the eye size. Misexpressing dTCFDN in the eye of L2/+ mutants (L2/+; ey>dTCFDN) significantly rescues the loss-of-ventral eye phenotype in 21.8% of flies (Figures 6J, 6K, and S14B; Table S1) (Singh et al., 2006). Similarly, misexpressing vna4 and dTCFDN in the eye of L2 mutants (L2/+; ey> dTCFDN; vna4) rescues the loss-of-ventral eye phenotype (Figures 6L and 6M). As expected, rescue frequency again dramatically increased from 21.8% to 70.6% (Figure S14B; Table S1).
Our results demonstrate that misexpression of newt genes when Wg signaling is downregulated (using inhibitors of Wg) in L2 mutant background (L2/+; ey> sgg; vna4, 71.5%) or (L2/+; ey> dTCFDN; vna4, 70.6%) significantly increased the rescue frequency of the L2 mutant phenotype. The rescue frequency obtained is greater than either misexpressing newt gene (L2/+; ey>vna4, 58.2%) alone or only misexpressing negative regulators (L2/+; ey> sgg, 27.3%) or (L2/+; ey> dTCFDN, 21.8%). The converse phenotypes were seen when Wg signaling was upregulated. This strongly suggests that newt gene(s) interact with the Wg signaling pathway to promote rescue of L2 mutant phenotype.
Porcupine (Porc) is involved in post-translational modification of Wg nascent protein in the producing cell and thus facilitates its transport outside to the receiving cell. Wg ligand can bind to its receptor on the receiving cell (Manoukian et al., 1995; Swarup and Verheyen, 2012) thus modulating downstream pathway components (Figure 6A). We blocked Wg transport from the producing cell using porcRNAi and sampled its effect on L2 mutant phenotype, and L2 mutant where vna4 is misexpressed. Misexpression of porcRNAi alone (ey> porcRNAi) (Figures S13M and S13N), and along with vna4 in the wild-type background (+/+; ey> porcRNAi; vna4) (Figures S13O and S13P) does not affect the eye size. Misexpressing porcRNAi in the L2 mutant eye (L2/+; ey> porcRNAi) significantly rescues the loss-of-ventral eye phenotype in 20.8% of flies (Figures 6N, 6O, and S14B; Table S1). Similarly, vna4 misexpressed in the eye of L2 mutants along with porcRNAi (L2/+; ey> porcRNAi; vna4) rescued the loss-of-ventral eye phenotype (Figures 6P and 6Q). The rescue frequency increased dramatically from 20.8% to 68.1% (Figure S14B; Table S1). Thus, confirming that newt genes can modulate levels of Wg signaling to promote restoration response in L2 eye mutants.
Discussion
Notophthalmus viridescens have an enormous genome size (∼ c × 1010 bases), a long reproductive cycle, and have limited genetic tools that makes it difficult to use this model to ascertain the molecular mechanism(s) behind the function of its unique genes (Mehta and Singh, 2019a). One of the strategies can be to introduce these genes into models with array of genetic tools and where regeneration potential is lower as seen in models like Drosophila, Bombyx mori etc (Gopinathan et al., 1998; Kango-Singh et al., 2001; Mehta and Singh, 2019a; Singh et al., 2007). The rationale is to test the efficacy of these newt genes in triggering restoration response in the model systems with lower restoration potential. We and others have introduced human, plant and other vertebrate genes by transgenic approaches in Drosophila (Deshpande et al., 2019; Gogia et al., 2020b; Hughes et al., 2012; Sarkar et al., 2018; Tare et al., 2011). These transgenic flies have been utilized for targeted expression of foreign genes in flies using a targeted misexpression approach (Brand and Perrimon, 1993). The successful misexpression of the unique newt genes (vertebrate genes) in Drosophila using the same targeted misexpression approach has been reported earlier (Mehta et al., 2019; Mehta and Singh, 2019b).
To test the regeneration potential of these newt genes, we took a distinctive approach of using Drosophila model because the signaling pathways, which are involved in regeneration and/or tissue growth, patterning and development are evolutionarily conserved across the species (Kango-Singh and Singh, 2009; Mehta and Singh, 2019a; Wang and Hu, 2020). Even though humans (Homo sapiens), which evolved approx. 200,000 years ago (Stringer, 2016) and are 541 million years apart from Drosophila, still share 75% of genes with flies (Pandey and Nichols, 2011). Despite the gap of 250 million years between newts and Drosophila, our studies suggest that the proteins encoded by unique newt genes are functional in Drosophila (Figures 1 and S2). Therefore, it is plausible that the pathways that newt genes can modulate in Drosophila might share parallels with their mechanism of action in newts. We found that the five newly identified genes from newt, can (i) rescue L mutant eye phenotype (early eye mutant) as well as GMR-hid GMR-GAL4 (late retinal degeneration) phenotype, (ii) promote cell proliferation, and downregulate cell death to restore the loss-of-ventral-eye phenotype, and (iii) downregulate Wg signaling (Figure 7). Our studies clearly demonstrate that the potential of these newt genes in triggering rescue of missing photoreceptor cells in Drosophila eye model exists along the temporal axis from early eye development (L2 mutant) to the later stages of eye development (GMR-hid GMR-GAL4).
Figure 7.
Model of newt genes function during development
These newt genes (vna1-vna5) can negatively regulate evolutionarily conserved Wg/Wnt signaling pathway in the developing Drosophila eye, promote cell proliferation, and block cell death, which results in the rescue of L2 mutant’s loss-of-ventral eye phenotype. This potential to restore missing structures by these newt genes extend from early eye development to late larval eye and adult eye.
Newt genes promote cell proliferation to restore Drosophila eye mutant phenotype
Regeneration involves the formation of a blastema, a proliferating zone of undifferentiated cells to restore the missing structures (Brockes and Kintner, 1986; Mehta and Singh, 2019a; Sanchez Alvarado and Tsonis, 2006). Furthermore, a network of newt gene(s) triggers proliferation response to promote lens regeneration (Eguchi and Shingai, 1971; Grogg et al., 2005; Looso et al., 2013; Madhavan et al., 2006). A similar proliferating response was observed when newt genes were misexpressed in the Drosophila L2 mutant eye (Figures 3B and S5), and in GMR-hid, GMR-GAL4 eye discs (Figures S6A–S6C). L2 mutant flies exhibit a loss-of-ventral eye phenotype, and misexpressing newt genes robustly increased the number of proliferating cells (Figures 3, 7, and S5), which clearly mimics the classic mechanism of epimorphic regeneration in the newt lens. Similarly, GMR-hid, GMR-GAL4>vna4 eye discs showed significant increase in the number of dividing cells (Figure S6C).
In our study, we found that newt genes can also modulate cell death in Drosophila developing eyes. When newt genes were misexpressed in the L2 mutant background (L2; ey>newt gene), the rate of cell death was lower as compared to the L2 mutant (L2; ey-GAL4) eye (Figures 3H, 3L, S5K–N, and S5R), but minimally higher compared to the control (ey-GAL4) (Figures 3I, 3L, S5O, and S5R). A minimal rate of apoptosis is observed during the newt lens regeneration as well (Tsonis et al., 2004). Newt genes can also robustly downregulate cell death caused by the overexpression of hid (GMR-hid, GMR-GAL4>vna4) (Figures S6D–S6F). Moreover, it was interesting to note that newt genes do not upregulate cell proliferation or downregulate cell death in the normal Drosophila eye (ey>vna4) (Figures 3D, 3J, 3F, and 3L). There is a strong possibility that these newt genes might be hijacking the same evolutionarily conserved signaling pathway(s) in Drosophila to regulate cell proliferation, and cell death that might also be involved in promoting regeneration in newts.
Newt genes tightly regulate Wg signaling pathway in Drosophila
We screened for the various signaling pathways using the candidate approach as well as compared them to our transcriptomic analysis (Mehta et al., 2019). Among the various genetic modifiers of the L2 mutant phenotype, we found wg as one of the possible candidates. Wg, a member of the evolutionarily conserved Wg/Wnt pathway, has been previously reported to promote regeneration, growth, development etc. (Harris et al., 2016; Schubiger et al., 2010; Singh et al., 2012b, 2018; Smith-Bolton et al., 2009; Yokoyama et al., 2007). Misexpression of newt genes in Drosophila wild-type background modulate both positive regulators of Wg/Wnt signaling like frizzled (fz), and microtubule star, (mts) and negative regulators like Casein Kinase IIβ (CKIIβ), and sinaH (Figure S10). This modulation in the levels of both inhibitors as well as effectors can be a homeostatic reaction shown by the Drosophila to normalize expression levels of Wg/Wnt signaling pathway. As a result, changes in the eye growth after misexpressing newt genes in the wild-type was non-significant when compared to the wild-type control (ey-GAL4) (Figures 1, 3, and S5). We did not observe any lethality, developmental defects, dysregulation in cell proliferation or cell death etc.
Interestingly, when newt genes were misexpressed in L2 mutant background, there was significant downregulation of Wg/Wnt expression (Figures 4 and S11), which resulted in rescue of the loss-of-ventral eye phenotype. It is known that overexpressing Wnt, and its receptor Fz promote lens regeneration in newts, whereas blocking Wnt pathway by expressing Dkk1 inhibits lens induction (Hayashi et al., 2006). Therefore, our study may have parallels with the regeneration mechanism in newts. Furthermore, we found that when Wg pathway components are modulated in Drosophila along with the newt genes, it not only rescues the L2 mutant loss-of-ventral eye phenotype but can also rescue the eye-loss caused by the activation of the Wg signaling pathway in the wild-type background. Hence, our data strongly suggest that the newt genes can modulate Wg signaling pathway in loss-of- eye background. Interestingly, these newt genes did not show any phenotype when they were misexpressed alone suggesting that restoration response is triggered only to the cues generated because of the tissue damage.
Newt proteins exhibit non-cell-autonomous response
Genetic mosaic experimental approach resulted in the generation of clone(s) of retinal neurons expressing high levels of newt genes within the L2 mutant eye disc. These experiments showed that clones expressing high levels of newt genes can rescue the L2 mutant phenotype; however, the rescue was not limited to the clone itself and spreads into the neighboring cells where newt genes were not misexpressed. This non-cell-autonomous rescue of the missing tissue is possible only if the newt proteins are getting secreted from one cell to another and/or newt proteins are acting on the signaling molecules. These newt proteins have signal peptides (Looso et al., 2013), which can facilitate its transport from one cell to another. Similarly, we have reported that these genes interact with Wg (Figure 4), which is a morphogen that can have a long-distance effect in the Drosophila eye (Zecca et al., 1996).
It will be interesting to investigate, (a) if newt genes can regulate Wg signaling pathway to promote regeneration in other tissues of Drosophila e.g., wing, leg etc., and in the background of other congenital mutations (Schubiger et al., 2010; Smith-Bolton et al., 2009), (b) if these proteins can rescue/regenerate missing tissues/organs in other animals with low regenerative potentials such as mammals because pathways that have roles in regeneration, growth, development, and cancer are evolutionarily conserved throughout the animal kingdom (Mehta and Singh, 2019a). For example, the Wg/Wnt signaling pathway, is one such evolutionarily conserved pathway that apart from promoting regeneration in hydra, planaria, newts, zebrafish, etc. has also been found to promote cell proliferation during regeneration of mammalian muscles, liver, and bone (Polesskaya et al., 2003; Sodhi et al., 2005; Zhong et al., 2006). Similarly, its dysregulation has been found to cause cancer in mammals (Nusse and Varmus, 1982; Taipale and Beachy, 2001). Interestingly newts are resistant to carcinogens (Tsonis and Eguchi, 1981), which could be because of its potential to regulate evolutionarily conserved pathways like Wnt, therefore in future it will also be interesting to investigate (c) potential of these newt genes to regulate overgrowth/cancer in Drosophila, and in mammals.
Limitations of study
We found that misexpressing these five newt genes (vna1, vna2, vna3, vna4, vna5) in the Drosophila loss-of-eye mutant background could not completely restore the eye size in the flies as observed in newts. In newts, there is complete regeneration of the appendage(s). It is possible that there may be many more players in the regeneration tool kits of newts or in future combining all these five genes in one transgenic fly is warranted to check if we can fully restore the missing eye. There could also be specific domains of these proteins that might be critical for their function, which need to be identified and characterized to investigate the course of their action in detail. Future studies using the established regeneration system in Drosophila could help provide more details into the function of these genes. In addition, we have not been able to determine the regeneration potential of these genes in newt species, because of absence of genetic tools in the newt model. Furthermore, our data using the dorsal-eye-Gal4 demonstrated that misexpression of these regeneration tools genes in the dorsal eye can rescue loss-of-eye phenotype non-autonomously even in the ventral half. However, because of challenges of studying V5 tag expression in the tissue by immunohistochemistry, we could not validate our observation that these newt genes can direct non-autonomous rescue of eye phenotype. Previously it has been suggested that highly regenerative animal models like newts might be employing unique regeneration tool kit genes, and/or unique regeneration enhancers that can modulate the evolutionarily conserved signaling pathways to promote restoration (regenerative) response (Casco-Robles et al., 2018; Looso et al., 2013; Mehta and Singh, 2019a; Rodriguez and Kang, 2020; Thompson et al., 2020; Wang and Hu, 2020). In absence of such genes, the low regeneration animal models like Drosophila, mammals etc. or the animals that lack regeneration potential (Kango-Singh et al., 2001; Singh et al., 2007) could not exhibit robust response to restore missing structures even though the Wnt/Wg signaling pathway members are present (Mehta and Singh, 2019a). Thus, introduction of these genes into flies can coax the genetic machinery to promote restoration response. Such studies will have significant bearings on our efforts to understand and trigger regenerative response in humans upon injury.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti Phospho histone-3 antibody | Cell Signaling | 9706 |
| Rabbit anti- Drosophila caspase-1 (DCP-1) | Cell Signaling | 9578 |
| Rat anti-Elav | Developmental Studies Hybridoma Bank | 7E8A10 |
| mouse anti-Wg | Developmental Studies Hybridoma Bank | 4D4 |
| goat anti-rat IgG conjugated with Cy5 | Jackson Laboratories | 112-175-143 |
| donkey anti-rat IgG conjugated to FITC | Jackson Laboratories | 712-095-153 |
| donkey anti-rat IgG conjugated to Cy5 | Jackson Laboratories | 712-606-153 |
| donkey anti-mouse IgG conjugated to Cy3 | Jackson Laboratories | 715-166-151 |
| donkey anti-goat IgG conjugated to Cy3 | Jackson Laboratories | 705-165-147 |
| goat anti-rabbit IgG conjugated with Cy5 | Jackson Laboratories | 111-175-144 |
| Chemicals | ||
| RNaseZap | Thermofisher scientific | AM9780 |
| TRIzol Reagent | Thermo Fisher | 15596926 |
| Chloroform | Sigma | 319988/W205702 |
| Vectashield | Vector labs | H1000 |
| ethyl-3-aminobenzoate methanesulfonic acid | Sigma | MS222 |
| Paraformaldehyde | Electron Microscopy Sciences | 15710 |
| Triton X-100 | Sigma | T8787 |
| Sodium citrate | Thermofisher Scientific | S279 |
| Tris Base | Sigma | T1503-500G |
| Sodium dodecyl sulfate | Sigma | L3771-100G |
| 10X Tris/Glycine/SDS buffer | Biorad | 1610732 |
| 10X Tris/Glycine buffer | Biorad | 1610734 |
| 20X TBS Tween-20 buffer | Thermofisher Scientific | 28360 |
| BSA | Fisher Bioreagents | 9048468 |
| Ammonium Persulfate | Fisher Bioreagents | BP179-25 |
| TEMED | Sigma | T9281-100ml |
| 2X Sample buffer | Sigma | S3401 |
| Phenylmethanesulfonyl fluoride | Sigma | P7626-5G |
| Glacial acetic acid | VWR | 64197 |
| 40%Acrylamide /bisacrylamide (29:1) | Fisher Bioreagents | BP-1408-1 |
| Precision plus Protein Kaleidoscope Prestained | Biorad | 1610395 |
| Ponceau S | Sigma | P7170 |
| Commercial assays | ||
| Super Signal West Dura Extended Duration Substrate | Thermo Scientific | 34076 |
| Pierce BCA protein assay kit | Thermo Scientific | 23225 |
| RNA clean and ConcentratorTM | Zymo research | R1080 |
| TUNEL dilution buffer | Roche Diagnostics | 11966006001 |
| Deposited data | ||
| viropana 1 (vna1) | This Manuscript | https://doi.org/10.26890/ddl.dbitmbv |
| viropana 2 (vna2) | This Manuscript | https://doi.org/10.26890/ddl.dbitmbv |
| viropana 3 (vna3) | This Manuscript | https://doi.org/10.26890/ddl.dbitmbv |
| viropana 4 (vna4) | This Manuscript | https://doi.org/10.26890/ddl.dbitmbv |
| viropana 5 (vna5) | This Manuscript | https://doi.org/10.26890/ddl.dbitmbv |
| RNA sequencing data | Gene Expression Omnibus (GEO) | GSE134348 |
| Software and algorithms | ||
| Image Studio Lite ver. 5.2 | LI-COR | https://www.licor.com/bio/image-studio-lite/d5 |
| VisionWorks Acquisition and analysis software | UVP | https://www.medicalexpo.com/prod/uvp/product-99853-693587.html |
| ImageJ software | NIH | http://rsb.info.nih.gov/ij/ |
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for the resources should be directed to and will be fulfilled by the lead contact, Dr. Amit Singh; Email ID: asingh1@udayton.edu, Telephone Number: +1 9372292894.
Materials availability
viropana1 (vna1), viropana2 (vna2), viropana3 (vna3), viropana4 (vna4), viropana5 (vna5) are the list of transgenic flies associated with this manuscript.
Experiment model and subject details
Animals
Handling and operations of Notophthalmus viridescens (newts) have been described previously (Mehta et al., 2019; Sousounis et al., 2013). For our studies, Notophthalmus viridescens (newts) were purchased from Charles Sullivan Inc. newt farm. Adult newts (gender unknown) of approximately 8-12 years of age were used for the experiment. Newts were anesthetized in 0.1% (w/v) ethyl-3-aminobenzoate methanesulfonic acid (MS222; Sigma) dissolved in phosphate buffered saline.
Ethics statement
All procedures involving animals were approved by the University of Dayton Institutional Animal Care and Use Committee (IACUC; Protocol ID: 011-02). The surgical procedures were performed by using a forceps to hold the tail, and surgical scissors to cut it. The operations were performed on an anesthetized newts, and appropriate procedures were undertaken to alleviate pain and distress while working with newts.
Drosophila stocks
In this study, we used transgenic flies UAS-vna1, UAS-vna2, UAS-vna3, UAS-vna4, UAS-vna5, and UAS-vna1-5-V5). Other fly stocks used in this study are described in Flybase (http://flybase.bio.indiana.edu). We used Tub-GAL4/SM6-TM6bTb (S-T) (Lee and Luo, 1999), L2/CyO; ey-GAL4/ey-GAL4, L2/C4; ey-GAL4/ey-GAL4, UAS-GFP (Singh et al., 2005, 2006; Singh and Choi, 2003). DE-GAL4/CyO; UAS-GFP/Tb (Morrison and Halder, 2010), UAS-wg-GFP (Azpiazu and Morata, 1998), UAS-arm (Zecca et al., 1996), UAS-sgg (Hazelett et al., 1998), UAS-dTCFDN (van de Wetering et al., 1997), UAS-p35 (Hay et al., 1994). Both male and female flies were used for our experiments.
Method details
Tissue harvest and RNA extraction
It is recommended to clean working space (RNA isolation bench), and all the equipment’s (e.g., scalpels, forceps, pipettes) with RNase decontamination solution e.g. use RNaseZap® (Thermo Fisher Scientific, Cat No. AM9780). Spray RNaseZap® Solution onto the surface to be decontaminated and rinse it off with RNase-free water.
Sample collection and storage
The tissue sample was collected and stored in RNAlater (Thermo Fisher Scientific, Cat. No. # AM7024) solution. The samples were then stored at 4°C. For long term storage we collected tissue in an empty RNase free tubes and store them at −70°C. Before starting the procedure to isolate RNA, the tubes were briefly centrifuged and RNAlater solution was completely removed. Then we added 500 μl of TRIzol Reagent (Thermo Fisher, Cat. No. # 15596926) to the tubes. TRIzol Reagent is used to isolate good quality RNA from tissue samples. TRIzol is a monophasic solution, primarily consisting of phenol and guanidine isothiocyanate along with other proprietary components, which disrupt samples, stabilize nucleic acids, and are compatible with downstream analysis. Tissue was homogenized in TRIzol using a 150 handheld homogenizer Motor (Thermo Fisher Scientific, Cat. No. # 15-340-167). The solution was incubated for 30 minutes in ice followed by 10 minutes of Centrifugation at 4°C and supernatant was transferred to clean tube. Chloroform (Sigma Catalog #: 319988/W205702) was added to the supernatant, and homogenate was allowed to separate into a clear upper aqueous layer. 200 μl of the aqueous phase was transferred to RNA clean and ConcentratorTM (Zymo research, Cat. No. R1080) columns, and the recommended protocol was followed, i.e., solution was passed through the RNA binding buffer, which binds RNA to the desired columns (Mehta and Singh, 2017). Then RNA wash buffer was added to remove all the impurities from the column. Finally, RNA was eluted in 20 μl of molecular grade water (DNase/RNase free) and collected in a separate tube. The molecular grade water serves as the elution buffer as it releases the RNA from the column.
RNA quantitation, and storage
RNA quantitation is an important and necessary step prior to most RNA analysis methods. Quality of RNA as well as concentration of RNA was determined by calculating absorbance at 260 nm (A260) and 280 nm (A280) wavelengths using Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). Good quality samples had A260/A280 ratio greater than 2 and a peak at 260 nm.
Sample preparation to clone newt genes
Total RNA was extracted from the newt tail as stated above. The quality and quantity of RNA was determined using Agilent RNA 6000 nano LabChip (Agilent 2100 Bioanalyzer). Approximately 200ng of total RNA with a RIN >9 was used for the cDNA synthesis using ImProm-II Reverse Transcription System (Promega) and random-primer hexamers. All PCR reactions were performed using PlatinumTaq DNA polymerase (Invitrogen). The primers used are:
Vna1Fw 5′-AAAGGATCCatgaagatctctctagctttcc-3′,
Vna1Rev 5′-AAATCTAGActaagaacagctgcgacaagtg-3′
Vna2Fw 5′-AAAGGATCCatgaagatctctctagctttcc-3′
Vna2Rev 5′-AAATCTAGActaagaacagctgcgataagtgg-3′
Vna3Fw 5′-AAAGGATCCatgaagatctctctagctttcc-3′
Vna3Rev 5′-AAATCTAGActagtcgactagaaggcctgc-3′
Vna4Fw 5′-AAACTCGAGatgaagattgctctcgttttcc-3′
Vna4Rev 5′-AAATCTAGAttagcacctaccgccaggcag-3′
Vna5Fw 5′-AAACTCGAGATGAAGATCGCTCTCGTTTTC-3′
Vna5Rev 5′-AAATCTAGActactgcttccacacttgccaaa-3′
The underlined sequences introduced XbaI and BamHI sites at the ends of vna1, vna2 and vna3 and XbaI and XhoI at the ends of vna4 and vna5 to facilitate the cloning in pUAST-attB plasmid. The fragments were cloned in pDrive (QIAGEN) followed by which the sequence for each gene was confirmed using the primer M13 forward −21 (5′-GTAAAACGACGGCCAGT-3′). Thereafter, the fragments vna1 (498bp), vna2 (402bp) or vna3 (426pb) were delivered from pDrive using the enzymes XbaI/BamHI and cloned into the sites XbaI/BglII downstream of 5XUAS-hsp70 sequence in pUAST-attB plasmid. The fragments vna4 (501bp) and vna5 (426bp) were delivered from pDrive using the enzymes XhoI/XbaI and cloned into the same sites in pUAST-attB plasmid. For the only purpose to validate the successful insertion of newt genes in Drosophila by targeted misexpression approach five more transgenic flies like ‘UAS-vna1-5-V5’ with 14 a.a. long V5-epitope sequence tagged towards the 3′ end of the vna ORF were generated by GenScript (https://www.genscript.com/). The complete gene fragment (5′-newt gene-V5-3′) was synthesized and cloned into the same sites as stated above in pUAST-attB plasmid. The integration and expression of transgene was verified by the western blots using anti-V5 antibody.
Generating transgenic flies
The design and generation of transgenic flies (UAS-vna1, UAS-vna2, UAS-vna3, UAS-vna4, UAS-vna5, UAS-vna1-V5, UAS-vna2-V5, UAS-vna3-V5, UAS-vna4-V5, UAS-vna5-V5) used in this study have been described (Mehta et al., 2019). Briefly, transgenic flies were generated using microinjection-based φC31 integrase mRNA-mediated method by genetivision (https://www.genetivision.com/). The pUAST-attB plasmid containing both the transgene and donor sequence (attB) was co-injected along with φC31 integrase mRNA into attP-containing recipient embryos, resulting in site-specific insertion of the transgene at Chromosome III (Fish et al., 2007; Groth et al., 2004).
RNA sequencing
Tissue collection
The third instar larvae were selected and were stored in RNAlater (Thermo Fisher, Cat. No. # AM7024) solution. The samples were then stored at 4°C. For long term storage we isolated larvae into empty RNase free tubes and store them at −70°C. The third instar larvae were selected and a high-quality RNA was isolated using the same protocol as stated above. High quality total RNA was shipped in dry ice to our collaborator for next generation RNA sequencing.
Analysis
Illumina reads were mapped to the Drosophila genome dm6 using TopHat splice-aware aligner (Kim et al., 2013). Expectation-Maximization (EM) approach was used to estimate transcript abundance (Jiang and Wong, 2009) . Reads per kilobase per million mapped reads (RPKM) approach was applied for within sample normalization (Mortazavi et al., 2008). Between sample normalization and differentially expressed test were performed by BioConductor DESeq package (v 1.20), which allows analysis of non-replicate experiments (Anders and Huber, 2010). The significant criteria were (1) detected transcript in at least one sample (RPKM>1), (2) fold change over 2 and (3) adjusted p-value less than 0.05. Gene ontology enrichment analysis and visualization tool (GOrilla) (Eden et al., 2009) and protein analysis through evolutionary relationships (PANTHER) 14.1 version tool (Mi et al., 2019) was used to identify enriched gene ontology (GO) terms, and pinpoint the signaling pathways that are getting differentially regulated.
Targeted misexpression studies
We used the GAL4/UAS system for targeted misexpression studies (Brand and Perrimon, 1993). All GAL4/UAS crosses were performed at 25°C (Singh et al., 2005; Singh and Choi, 2003). In order to misexpress the UAS-transgenes, we used tub –GAL4 (Lee and Luo, 1999) to ubiquitously misexpress genes, ey-GAL4 (Hazelett et al., 1998) to misexpress transgene in the developing eye, and DE-GAL4 (Morrison and Halder, 2010) that drives transgene in the dorsal domain of the developing eye.
Genetic mosaic analysis
Clones were generated using the FLP/FRT system of mitotic recombination (Xu and Rubin, 1993). To generate clones of vna4 in L2-mutant eye, yw hsFLP; Act>y+>GAL4, UAS-GFP/CyO females were crossed to L2/CyO; UAS-vna4/Tb males. vna4 is expressed in all the GFP+ cells of eye-antennal imaginal disc.
Immunohistochemistry
Dissection and fixation
Eye-antennal imaginal discs were dissected from wandering third instar larvae and stained following the standard protocol (Singh et al., 2006; Tare et al., 2016). The eye imaginal discs were dissected with the brain structures in cold 1X solution of Phosphate Buffered Saline (PBS) to maintain the tissue integrity. They were fixed for 20 minutes at room temperature using 4% paraformaldehyde [50μL 16% paraformaldehyde (Electron Microscopy Sciences, Catalog no. 15710) + 150μL 1X PBS]. Following fixation, the tissues were washed thrice for 10 minutes using 1X PBS-T [0.2% Triton X-100 (Sigma, Catalog no. T8787) in 1X PBS] to permeabilize the tissue.
Immunostaining
To visualize the protein of interest, specific primary antibodies were used. The primary antibodies used were mouse anti-phospho histone-3 (1:200) (Cell Signaling, Catalog no. 9706); Rabbit anti- Drosophila caspase-1 (DCP-1) (1:100) (Cell Signaling, Catalog no. 9578); rat anti-Elav (1:100) (Developmental Studies Hybridoma Bank, Catalog no. 7E8A10); mouse anti-Wg (1:100) (Developmental Studies Hybridoma Bank, Catalog no. 4D4), and goat anti-Hth (a gift from H. Sun and R. Mann). The working dilutions (as specified above) for the antibodies were prepared with 1X PBS-T-NDS (Normal Donkey Serum). Normal Donkey Serum is used as a blocking agent and helps reduce non-specific binding. The tissues were incubated overnight in the primary antibody at 4°C. The following day, the tissues were washed thrice for 10 minutes with 1X PBS-T and then incubated with the appropriate secondary antibody for two hours in the dark at room temperature. Secondary antibodies (Jackson Laboratories) used were goat anti-rat IgG conjugated with Cy5 (1:200) (Catalog no.112-175-143); donkey anti-rat IgG conjugated to FITC (1:200) (Catalog no. 712-095-153); donkey anti-rat IgG conjugated to Cy5 (1:200) (Catalog no. 712-606-153), donkey anti-mouse IgG conjugated to Cy3 (1:200) (Catalog no. 715-166-151); donkey anti-goat IgG conjugated to Cy3 (1:200) (Catalog no. 705-165-147), and goat anti-rabbit IgG conjugated with Cy5 (1:200) (Catalog no. 111-175-144). Like primary antibodies, secondary antibodies were also diluted in 1X PBS-T-NDS. The tissues were washed thrice with 1X PBST.
Confocal microscopy and analysis
The washed tissues are further dissected on a slide to remove all extra tissues leaving only eye antennal imaginal discs. The discs are mounted in Vectashield (Vector labs, Catalog no.H1000), sealed carefully and stored at −20°C till imaging is done. The slides are imaged using Olympus Fluoview 1000 or Olympus Fluoview 3000 microscope. All images were captured at 20X unless specified. For all genotypes, a minimum of 5 images were captured. Images were analyzed using Adobe Photoshop and ImageJ software. For quantification using ImageJ software, ROI tool was used to select different regions and measurements were done. Any graphs were plotted in GraphPad Prism.
Western blotting
Sample preparation
Protein samples were prepared from the adult fly head (n = 25) by using a standardized protocol (Gogia et al., 2017, 2020b). To each collected sample, 50μl of Laemmli 2X concentrate sample buffer and 3μl of phenylmethanesulfonyl fluoride were added. The samples were then macerated by sterilized pestle and then boiled for 5-10 minutes and snap-chilled on ice for 10 minutes. The samples were then centrifuged at 13,000 rpm for 20 minutes and supernatant was transferred to the fresh tube. The total protein concentration was measured by BCA protein assay (Thermo Scientific, Catalogue no. 23225). The protein concentration is proportional to the color change from green to purple that can be measured by spectrophotometry at absorbance of 562 nm wavelength. The protein samples were then normalized by calculating the required volume of protein for a total concentration of 50μg per well.
SDS-PAGE gel electrophoresis
For casting 10% SDS-PAGE gel, the gel apparatus was set up. First 4X separating gel was prepared by mixing 40% acrylamide/bisacrylamide, 4X separating buffer, 10% freshly prepared ammonium persulfate and tetramethylethylenediamine. This separating gel mixture was poured between two plates up to 70% of the total size of the glass plate and around 750 μl of 70%ethanol was poured on top of it to avoid air bubbles. After the polymerization of the gel, 70% ethanol was drained out. The 4X stacking gel was prepared by mixing 40% acrylamide/ bisacrylamide, 4X stacking buffer, 10% freshly prepared ammonium persulfate and tetramethylethylenediamine. The stacking gel was poured on top of the polymerized separating gel and the comb was inserted carefully by avoiding air bubbles. After the gel was polymerized, the gel cassette was immersed in 1X tris glycine SDS-buffer in the gel tank. The comb was removed, and the normalized protein samples were loaded along with a molecular weight marker (Precision plus protein standard kaleidoscope) in the well. The proteins were separated by running the gel at 70V for 2.5-3 hours.
Protein transfer
Once the gel electrophoresis was done, the gel was removed from the gel cassette, and it was equilibrated in a cold 1X tris/glycine buffer. Nitrocellulose membrane was equilibrated in methanol for 5-10 minutes. Transfer sandwich was prepared by keeping the membrane onto the gel without trapping any air bubbles. The sandwich was then placed in a transfer apparatus and filled with a small ice pack and pre-cooled 1X transfer buffer. The transfer process was performed in a cold room at 25V overnight.
Detection of protein and quantification
Once the protein transfer is done, the membrane was then removed from the transfer apparatus and washed three times with autoclaved water and then treated with 1% glacial acetic acid for 5 minutes. The membrane was then stained with Ponceau-S to visualize the total proteins in the sample. The membrane was destained by 1% glacial acetic acid and then washed with autoclaved water. The membrane was then washed with 1X TBST three times. Then the membrane was blocked with 5% bovine serum albumin (BSA) for 1 hour at room temperature. Primary antibodies used were anti-mouse Wg (1:1000) (DSHB, 4D4); anti-rabbit V5 (1∶15,000) (SIGMA, Cat. # V8137); and anti-mouse α-Tubulin (1:12000) (SIGMA, Cat. # T5168) overnight at 4°C. Then the membrane was washed three times with 1X TBST. Then secondary antibodies used were horseradish peroxidase conjugated goat anti-mouse IgG (1:5000) (Santa Cruz Biotechnology, Cat. Sc-2005) and horseradish peroxidase conjugated goat anti–rabbit IgG (1:5000) for half an hour. The signal was detected using the Super Signal West Dura Extended Duration Substrate kit (Thermo Fisher Scientific, Cat. #34076). Images were captured using the UVP Imaging System. Intensity of Wg protein was quantified by LI-COR Image Studio lite ver. 5.2 free software and graph was plotted in GraphPad Prism.
TUNEL assay for detection of cell death
Dissection and immunostaining before TUNEL labeling
Apoptosis was detected by using Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay (Tare et al., 2011; White et al., 2001). TUNEL assay is based on the detection of fragmented DNA ends, which are characteristic of apoptotic cells. A deoxynucleotidyl transferase (TdT) is used to add fluorescently labeled nucleotides to free 3′-OH DNA ends in a template-independent manner. The modified nucleotides can then be detected by fluorescence microscopy. Eye-antennal discs are processed following standard immunohistochemistry protocol described above (Immunohistochemistry section).
TUNEL assay
Following secondary antibody staining, tissues were washed thrice with 1X PBS-T for 10 minutes. Samples were then incubated in 0.1 M sodium citrate (Fisher Scientific) and 10% TritonX-100 (495μL of 0.1M Na-Citrate + 5μL of 10% TritonX-100 per sample tube) for 30 minutes in dark at 65°C. This step was performed to increase permeabilization and improve signal. Samples were then washed thrice with 1X PBS-T for 10 minutes. Next, the samples were incubated in the TUNEL dilution buffer (Roche Diagnostics, Catalog no. 11966006001) for 10 minutes at room temperature in the dark. Finally for the labelling step, samples were incubated for 2 hours at 37°C in 55μL TUNEL labelling solution [25μL Labeling Solution (Roche, Catalog no. 12156792910) + 25μL Dilution Buffer+5μL enzyme solution]. Samples were then washed thrice with 1X PBS-T for 10 minutes.
Confocal microscopy and analysis
The washed tissues are mounted onto a glass slide with Vectashield (Vector labs, Catalog no.H1000). The slides are sealed carefully and stored at −20°C till imaging is done. The slides are imaged using Olympus Fluoview 1000 or Olympus Fluoview 3000 microscope. All images were captured at 20X unless specified. For all genotypes, a minimum of 5 images were captured. Images were analyzed using Adobe Photoshop and ImageJ software. For quantification using ImageJ software, ROI tool was used to select different regions and measurements were done. Any graphs were plotted in GraphPad Prism.
Bright field imaging
Adult flies were selected and were transferred to −20°C for two hours, followed by removal of legs and wings. The legs and wings were removed in order to get a clear view of the compound eye. The bright field pictures of adult heads were taken using a Zeiss Apotome Imager Z1 (Sarkar et al., 2018; Singh et al., 2019; Wittkorn et al., 2015). Eye size was measured using ImageJ software tools (http://rsb.info.nih.gov/ij/). To normalize the eye size values and neglect any variations caused by overall different head size we took ratio of eye size to the head size for each individual picture. Then relative ratio was calculated for all the treatments, and the value reported is the relative fold change for each experimental treatment compared to the control.
Quantification and statistical analysis
Statistical analysis was performed using student’s t-test for independent samples. Sample size used for the calculation was five in number (n = 5), otherwise specified (Mehta et al., 2019; Raj et al., 2020; Yeates et al., 2020). Statistical significance was determined with 95% confidence (p < 0.05). Error bars show standard deviation (mean ± SD), and symbols above the error bar signify as: ns is non-significant p value, ∗ signifies p value < 0.05, ∗∗ signifies p value < 0.005, ∗∗∗ signifies p value < 0.0005 respectively.
Acknowledgments
We thank Bloomington Drosophila Stock Center (BDSC) for Drosophila strains, and the Developmental Studies Hybridoma Bank (DSHB) for antibodies. We would like to thank Katia Del Rio-Tsonis, Labib Rouhana, Madhuri Kango-Singh, Meghana Tare, Neha Gogia, Shilpi Verghese, Pothitos Pitychoutis, and Deepika K Sodhi for critical comments on the manuscript. We also thank Justin Kumar, Y. Henry Sun, and Kyung Ok Cho for the gift of fly strains and antibodies. Confocal microscopy was supported by the core facility at University of Dayton. A.S. is supported by 1R15GM124654-01, 1RO1EY032959-01 from NIH, Schuellein Chair Endowment Fund and STEM Catalyst Grant from the University of Dayton.
Author contributions
A. S. Mehta, P. A. Tsonis, A. Singh designed the study. P. Deshpande, A. V. Chimata, A. S. Mehta performed the experiments. A. Singh contributed the resources. A. Singh, A.S. Mehta., A V. Chimata, P. Deshpande analyzed the data. A. S.Mehta and A. Singh wrote the manuscript with input from all authors. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: October 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103166.
Supplemental information
Data and code availability
-
•
The RNA sequencing raw data is deposited in the public repository: Gene Expression Omnibus (GEO) repository, GEO accession number is GSE134348. The processed data (Excel sheets that contain list of genes that were significantly regulated) can be found in an institutional repository of the University of Dayton (eCommons) and are publicly accessible https://doi.org/10.26890/ddlla1a541sgr. The DNA sequence for viropana1 (vna1), viropana2 (vna2), viropana3 (vna3), viropana4 (vna4), viropana5 (vna5) is available at https://doi.org/10.26890/ddl.dbitmbv
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu N., Morata G. Functional and regulatory interactions between Hox and extradenticle genes. Genes Dev. 1998;12:261–273. doi: 10.1101/gad.12.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.E. Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: the spatial distribution of a transcript in embryos. EMBO J. 1987;6:1765–1773. doi: 10.1002/j.1460-2075.1987.tb02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejsovec A. Flying at the head of the pack: Wnt biology in Drosophila. Oncogene. 2006;25:7442–7449. doi: 10.1038/sj.onc.1210051. [DOI] [PubMed] [Google Scholar]
- Bejsovec A. Wingless/Wnt signaling in Drosophila: the pattern and the pathway. Mol. Reprod. Dev. 2013;80:882–894. doi: 10.1002/mrd.22228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann A., Agapite J., McCall K., Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brockes J.P., Kintner C.R. Glial growth factor and nerve-dependent proliferation in the regeneration blastema of urodele amphibians. Cell. 1986;45:301–306. doi: 10.1016/0092-8674(86)90394-6. [DOI] [PubMed] [Google Scholar]
- Bryant D.M., Johnson K., DiTommaso T., Tickle T., Couger M.B., Payzin-Dogru D., Lee T.J., Leigh N.D., Kuo T.H., Davis F.G. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017;18:762–776. doi: 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casco-Robles R.M., Watanabe A., Eto K., Takeshima K., Obata S., Kinoshita T., Ariizumi T., Nakatani K., Nakada T., Tsonis P.A. Novel erythrocyte clumps revealed by an orphan gene Newtic1 in circulating blood and regenerating limbs of the adult newt. Sci. Rep. 2018;8:7455. doi: 10.1038/s41598-018-25867-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavodeassi F., Diez Del Corral R., Campuzano S., Dominguez M. Compartments and organising boundaries in the Drosophila eye: the role of the homeodomain Iroquois proteins. Development. 1999;126:4933–4942. doi: 10.1242/dev.126.22.4933. [DOI] [PubMed] [Google Scholar]
- Cohen S.M. 1993. Imaginal Disc Development. Paper presented at: Bate, Martinez Arias, 1993. [Google Scholar]
- Cordero J., Jassim O., Bao S., Cagan R. A role for wingless in an early pupal cell death event that contributes to patterning the Drosophila eye. Mech. Dev. 2004;121:1523–1530. doi: 10.1016/j.mod.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Crosio C., Fimia G.M., Loury R., Kimura M., Okano Y., Zhou H., Sen S., Allis C.D., Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S.M., Gates P.B., Brockes J.P. The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev. Cell. 2002;3:547–555. doi: 10.1016/s1534-5807(02)00288-5. [DOI] [PubMed] [Google Scholar]
- Deshpande P., Gogia N., Singh A. Exploring the efficacy of natural products in alleviating Alzheimer's disease. Neural Regen. Res. 2019;14:1321–1329. doi: 10.4103/1673-5374.253509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri K., Tanaka E.M. Proximodistal patterning during limb regeneration. Dev. Biol. 2005;279:391–401. doi: 10.1016/j.ydbio.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi G., Shingai R. Cellular analysis on localization of lens forming potency in newt iris epithelium. Dev. Growth Differ. 1971;13:337–350. doi: 10.1111/j.1440-169x.1971.00337.x. [DOI] [PubMed] [Google Scholar]
- Elewa A., Wang H., Talavera-López C., Joven A., Brito G., Kumar A., Hameed L.S., Penrad-Mobayed M., Yao Z., Zamani N. Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration. Nat. Commun. 2017;8:2286. doi: 10.1038/s41467-017-01964-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Johnson A.D., Loose M. Virtual genome walking across the 32 Gb Ambystoma mexicanum genome; assembling gene models and intronic sequence. Sci. Rep. 2018;8:618. doi: 10.1038/s41598-017-19128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish M.P., Groth A.C., Calos M.P., Nusse R. Creating transgenic Drosophila by microinjecting the site-specific phiC31 integrase mRNA and a transgene-containing donor plasmid. Nat. Protoc. 2007;2:2325–2331. doi: 10.1038/nprot.2007.328. [DOI] [PubMed] [Google Scholar]
- Garza-Garcia A., Harris R., Esposito D., Gates P.B., Driscoll P.C. Solution structure and phylogenetics of Prod1, a member of the three-finger protein superfamily implicated in salamander limb regeneration. PLoS One. 2009;4:e7123. doi: 10.1371/journal.pone.0007123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogia N., Puli O.R., Raj A., Singh A. In: Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye. Singh A., Kango-Singh M., editors. Springer; 2020. Generation of third dimension: axial patterning in the developing Drosophila eye; pp. 53–95. [Google Scholar]
- Gogia N., Sarkar A., Mehta A.S., Ramesh N., Deshpande P., Kango-Singh M., Pandey U.B., Singh A. Inactivation of Hippo and cJun-N-terminal kinase (JNK) signaling mitigate FUS mediated neurodegeneration in vivo. Neurobiol. Dis. 2020;140:104837. doi: 10.1016/j.nbd.2020.104837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogia N., Sarkar A., Singh A. An undergraduate cell biology lab: Western Blotting to detect proteins from Drosophila eye. Dros Inf. Serv. 2017;100:218–225. [Google Scholar]
- Gopinathan K.P., Joy O., Singh A. In: Genome Analysis in Eukaryotes: Developmental and Evolutionary Aspects. Chatterjee R.N., Sanchez L., editors. Springer-Verlag; 1998. Developmental aspects of mulberry and nonmulberry silkworm species: a comparative study; pp. 59–91. [Google Scholar]
- Grether M.E., Abrams J.M., Agapite J., White K., Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Grogg M.W., Call M.K., Okamoto M., Vergara M.N., Del Rio-Tsonis K., Tsonis P.A. BMP inhibition-driven regulation of six-3 underlies induction of newt lens regeneration. Nature. 2005;438:858–862. doi: 10.1038/nature04175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A.C., Fish M., Nusse R., Calos M.P. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.E., Setiawan L., Saul J., Hariharan I.K. Localized epigenetic silencing of a damage-activated WNT enhancer limits regeneration in mature Drosophila imaginal discs. eLife. 2016;5:e11588. doi: 10.7554/eLife.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B.A., Wassarman D.A., Rubin G.M. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Hay B.A., Wolff T., Rubin G.M. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Mizuno N., Takada R., Takada S., Kondoh H. Determinative role of Wnt signals in dorsal iris-derived lens regeneration in newt eye. Mech. Dev. 2006;123:793–800. doi: 10.1016/j.mod.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Haynie J.L., Bryant P.J. Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. J. Exp. Zool. 1986;237:293–308. doi: 10.1002/jez.1402370302. [DOI] [PubMed] [Google Scholar]
- Hazelett D., Bourouis M., Walldorf U., Treisman J. Decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development. 1998;125:3741–3751. doi: 10.1242/dev.125.18.3741. [DOI] [PubMed] [Google Scholar]
- Held L.I.J. In: Imaginal Disc. Held L.I., editor. Cambridge University Press; 2002. The eye disc; pp. 197–236. [Google Scholar]
- Hughes T.T., Allen A.L., Bardin J.E., Christian M.N., Daimon K., Dozier K.D., Hansen C.L., Holcomb L.M., Ahlander J. Drosophila as a genetic model for studying pathogenic human viruses. Virology. 2012;423:1–5. doi: 10.1016/j.virol.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Wong W., H. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics (Oxford, England) 2009;25(8):1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kango-Singh M., Singh A. Regulation of organ size: insights from the Drosophila Hippo signaling pathway. Dev. Dyn. 2009;238:1627–1637. doi: 10.1002/dvdy.21996. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M., Singh A., Gopinathan K.P. The wings of Bombyx mori develop from larval discs exhibiting an early differentiated state: a preliminary report. J. Biosci. 2001;26:167–177. doi: 10.1007/BF02703641. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M., Singh A., Henry Sun Y. Eyeless collaborates with Hedgehog and Decapentaplegic signaling in Drosophila eye induction. Dev. Biol. 2003;256:49–60. doi: 10.1016/s0012-1606(02)00123-9. [DOI] [PubMed] [Google Scholar]
- Keinath M.C., Timoshevskiy V.A., Timoshevskaya N.Y., Tsonis P.A., Voss S.R., Smith J.J. Initial characterization of the large genome of the salamander Ambystoma mexicanum using shotgun and laser capture chromosome sequencing. Sci. Rep. 2015;5:16413. doi: 10.1038/srep16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-Y., Jeong H.S., Chung T., Kim M., Lee J.H., Jung W.H., Koo J.S. The value of phosphohistone H3 as a proliferation marker for evaluating invasive breast cancers: a comparative study with Ki67. Oncotarget. 2017;8:65064–65076. doi: 10.18632/oncotarget.17775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Godwin J.W., Gates P.B., Garza-Garcia A.A., Brockes J.P. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science. 2007;318:772–777. doi: 10.1126/science.1147710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J.P. In: Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye. Singh A., Kango-Singh M., editors. Springer; 2013. Catching the next wave: patterning of the Drosophila eye by the morphogenetic furrow; pp. 75–97. [Google Scholar]
- Kumar J.P. The fly eye: through the looking glass. Dev. Dyn. 2018;247:111–123. doi: 10.1002/dvdy.24585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J.P. In: Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye. Singh A., Kango-Singh M., editors. Springer; 2020. Catching the next wave: patterning of the Drosophila eye by the morphogenetic furrow; pp. 97–120. [Google Scholar]
- Kumar J.P. In: Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye. Singh A., Kango-Singh M., editors. Springer; 2020. Ghost in the machine: the peripodal membrane; pp. 121–142. [Google Scholar]
- Lee J.D., Treisman J.E. The role of Wingless signaling in establishing the anteroposterior and dorsoventral axes of the eye disc. Development. 2001;128:1519–1529. doi: 10.1242/dev.128.9.1519. [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lenz S., Karsten P., Schulz J.B., Voigt A. Drosophila as a screening tool to study human neurodegenerative diseases. J. Neurochem. 2013;127:453–460. doi: 10.1111/jnc.12446. [DOI] [PubMed] [Google Scholar]
- Lin H.V., Rogulja A., Cadigan K.M. Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development. 2004;131:2409–2418. doi: 10.1242/dev.01104. [DOI] [PubMed] [Google Scholar]
- Looso M., Preussner J., Sousounis K., Bruckskotten M., Michel C.S., Lignelli E., Reinhardt R., Höffner S., Krüger M., Tsonis P.A. A de novo assembly of the newt transcriptome combined with proteomic validation identifies new protein families expressed during tissue regeneration. Genome Biol. 2013;14:R16. doi: 10.1186/gb-2013-14-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Moses K. Wingless and patched are negative regulators of the morphogenetic furrow and can affect tissue polarity in the developing Drosophila compound eye. Development. 1995;121:2279–2289. doi: 10.1242/dev.121.8.2279. [DOI] [PubMed] [Google Scholar]
- Madhavan M., Haynes T.L., Frisch N.C., Call M.K., Minich C.M., Tsonis P.A., Del Rio-Tsonis K. The role of Pax-6 in lens regeneration. Proc. Natl. Acad. Sci. 2006;103:14848. doi: 10.1073/pnas.0601949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoukian A.S., Yoffe K.B., Wilder E.L., Perrimon N. The porcupine gene is required for wingless autoregulation in Drosophila. Development. 1995;121:4037–4044. doi: 10.1242/dev.121.12.4037. [DOI] [PubMed] [Google Scholar]
- Matsunami M., Suzuki M., Haramoto Y., Fukui A., Inoue T., Yamaguchi K., Uchiyama I., Mori K., Tashiro K., Ito Y. A comprehensive reference transcriptome resource for the Iberian ribbed newt Pleurodeles waltl, an emerging model for developmental and regeneration biology. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes. 2019;26:217–229. doi: 10.1093/dnares/dsz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel-Zaffran C., Treisman J.E. Pannier acts upstream of wingless to direct dorsal eye disc development in Drosophila. Development. 2000;127:1007–1016. doi: 10.1242/dev.127.5.1007. [DOI] [PubMed] [Google Scholar]
- Mehta A., Singh A. Real time quantitative PCR to demonstrate gene expression in an undergraduate lab. Dros Inf. Serv. 2017;100:5. [Google Scholar]
- Mehta A.S., Luz-Madrigal A., Li J.-L., Tsonis P.A., Singh A. Comparative transcriptomic analysis and structure prediction of novel Newt proteins. PLoS One. 2019;14:e0220416. doi: 10.1371/journal.pone.0220416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A.S., Singh A. Insights into regeneration tool box: an animal model approach. Dev. Biol. 2019;453:111–129. doi: 10.1016/j.ydbio.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A.S., Singh A. University of Dayton; 2019. Regeneration Data: RNA Sequences [Internet] eCommons. [Google Scholar]
- Mehta A.S., Tsonis P.A., Singh A. University of Dayton; 2021. Components of a Regeneration Toolkit: Notophthalmus viridescens. eCommons. [Google Scholar]
- Mi H., Muruganujan A., Huang X., Ebert D., Mills C., Guo X., Thomas P.D. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0) Nat. Protoc. 2019;14:703–721. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner M.J., Bleasby A.J., Pyott A. The role of the peripodial membrane in the morphogenesis of the eye-antennal disc of Drosophila melanogaster. Wilehm Roux Arch. Dev. Biol. 1983;192:164–170. doi: 10.1007/BF00848686. [DOI] [PubMed] [Google Scholar]
- Morrison C.M., Halder G. Characterization of a dorsal-eye Gal4 line in Drosophila. Genesis. 2010;48:3–7. doi: 10.1002/dvg.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B., A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Moses K., Rubin G.M. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5:583–593. doi: 10.1101/gad.5.4.583. [DOI] [PubMed] [Google Scholar]
- Nowoshilow S., Schloissnig S., Fei J.-F., Dahl A., Pang A.W.C., Pippel M., Winkler S., Hastie A.R., Young G., Roscito J.G. The axolotl genome and the evolution of key tissue formation regulators. Nature. 2018;554:50–55. doi: 10.1038/nature25458. [DOI] [PubMed] [Google Scholar]
- Nusse R., Varmus H.E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Pandey U.B., Nichols C.D. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 2011;63:411–436. doi: 10.1124/pr.110.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Polesskaya A., Seale P., Rudnicki M.A. Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell. 2003;113:841–852. doi: 10.1016/s0092-8674(03)00437-9. [DOI] [PubMed] [Google Scholar]
- Raj A., Chimata A.V., Singh A. Motif 1 binding protein suppresses wingless to promote eye fate in Drosophila. Sci. Rep. 2020;10:17221. doi: 10.1038/s41598-020-73891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready D.F., Hanson T.E., Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Rodriguez A.M., Kang J. Regeneration enhancers: starting a journey to unravel regulatory events in tissue regeneration. Semin. Cell Dev. Biol. 2020;97:47–54. doi: 10.1016/j.semcdb.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Alvarado A., Tsonis P.A. Bridging the regeneration gap: genetic insights from diverse animal models. Nat. Rev. Genet. 2006;7:873–884. doi: 10.1038/nrg1923. [DOI] [PubMed] [Google Scholar]
- Sanor L.D., Flowers G.P., Crews C.M. Multiplex CRISPR/Cas screen in regenerating haploid limbs of chimeric Axolotls. eLife. 2020;9:e48511. doi: 10.7554/eLife.48511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Gogia N., Glenn N., Singh A., Jones G., Powers N., Srivastava A., Kango-Singh M., Singh A. A soy protein Lunasin can ameliorate amyloid-beta 42 mediated neurodegeneration in Drosophila eye. Sci. Rep. 2018;8:13545. doi: 10.1038/s41598-018-31787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Irwin M., Singh A., Riccetti M., Singh A. Alzheimer's disease: the silver tsunami of the 21(st) century. Neural Regen. Res. 2016;11:693–697. doi: 10.4103/1673-5374.182680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkissian T., Timmons A., Arya R., Abdelwahid E., White K. Detecting apoptosis in Drosophila tissues and cells. Methods. 2014;68:89–96. doi: 10.1016/j.ymeth.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger M., Sustar A., Schubiger G. Regeneration and transdetermination: the role of wingless and its regulation. Dev. Biol. 2010;347:315–324. doi: 10.1016/j.ydbio.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. Neurodegeneration-a means to an end. J. Cell Sci. Ther. 2012;3 doi: 10.4172/2157-7013.1000e107. [DOI] [Google Scholar]
- Singh A., Chan J., Chern J.J., Choi K.W. Genetic interaction of Lobe with its modifiers in dorsoventral patterning and growth of the Drosophila eye. Genetics. 2005;171:169–183. doi: 10.1534/genetics.105.044180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Choi K.-W. Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development. 2003;130:6351. doi: 10.1242/dev.00864. [DOI] [PubMed] [Google Scholar]
- Singh A., Gogia N., Chang C.Y., Sun Y.H. Proximal fate marker homothorax marks the lateral extension of stalk-eyed fly Cyrtodopsis whitei. Genesis. 2019;57:e23309. doi: 10.1002/dvg.23309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Irvine K.D. Drosophila as a model for understanding development and disease. Dev. Dyn. 2012;241:1–2. doi: 10.1002/dvdy.23712. [DOI] [PubMed] [Google Scholar]
- Singh A., Kango-Singh M., Parthasarathy R., Gopinathan K.P. Larval legs of mulberry silkworm Bombyx mori are prototypes for the adult legs. Genesis. 2007;45:169–176. doi: 10.1002/dvg.20280. [DOI] [PubMed] [Google Scholar]
- Singh A., Kango-Singh M., Sun Y.H. Eye suppression, a novel function of teashirt, requires Wingless signaling. Development. 2002;129:4271–4280. doi: 10.1242/dev.129.18.4271. [DOI] [PubMed] [Google Scholar]
- Singh A., Shi X., Choi K.W. Lobe and Serrate are required for cell survival during early eye development in Drosophila. Development. 2006;133:4771–4781. doi: 10.1242/dev.02686. [DOI] [PubMed] [Google Scholar]
- Singh A., Tare M., Kango-Singh M., Son W.-S., Cho K.-O., Choi K.-W. Opposing interactions between homothorax and Lobe define the ventral eye margin of Drosophila eye. Dev. Biol. 2011;359:199–208. doi: 10.1016/j.ydbio.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Tare M., Puli O.R., Kango-Singh M. A glimpse into dorso-ventral patterning of the Drosophila eye. Dev. Dyn. 2012;241:69–84. doi: 10.1002/dvdy.22764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.N., Doyle M.J., Weaver C.V., Koyano-Nakagawa N., Garry D.J. Hedgehog and Wnt coordinate signaling in myogenic progenitors and regulate limb regeneration. Dev. Biol. 2012;371:23–34. doi: 10.1016/j.ydbio.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.N., Weaver C.V., Garry M.G., Garry D.J. Hedgehog and Wnt signaling pathways regulate tail regeneration. Stem Cells Dev. 2018;27:1426–1437. doi: 10.1089/scd.2018.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Bolton R.K., Worley M.I., Kanda H., Hariharan I.K. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev. Cell. 2009;16:797–809. doi: 10.1016/j.devcel.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.J., Putta S., Zhu W., Pao G.M., Verma I.M., Hunter T., Bryant S.V., Gardiner D.M., Harkins T.T., Voss S.R. Genic regions of a large salamander genome contain long introns and novel genes. BMC Genomics. 2009;10:19. doi: 10.1186/1471-2164-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.J., Timoshevskaya N., Timoshevskiy V.A., Keinath M.C., Hardy D., Voss S.R. A chromosome-scale assembly of the axolotl genome. Genome Res. 2019;29:317–324. doi: 10.1101/gr.241901.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi D., Micsenyi A., Bowen W.C., Monga D.K., Talavera J.C., Monga S.P. Morpholino oligonucleotide-triggered beta-catenin knockdown compromises normal liver regeneration. J. Hepatol. 2005;43:132–141. doi: 10.1016/j.jhep.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Sousounis K., Looso M., Maki N., Ivester C.J., Braun T., Tsonis P.A. Transcriptome analysis of newt lens regeneration reveals distinct gradients in gene expression patterns. PLoS One. 2013;8:e61445. doi: 10.1371/journal.pone.0061445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C. The origin and evolution of Homo sapiens. Philos. Trans. R. Soc. Lond. Ser. B, Biol. Sci. 2016;371:20150237. doi: 10.1098/rstb.2015.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S., Verheyen E.M. Wnt/Wingless signaling in Drosophila. Cold Spring Harb. Perspect. Biol. 2012;4:a007930. doi: 10.1101/cshperspect.a007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J., Beachy P.A. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- Tare M., Modi R.M., Nainaparampil J.J., Puli O.R., Bedi S., Fernandez-Funez P., Kango-Singh M., Singh A. Activation of JNK signaling mediates amyloid-ss-dependent cell death. PLoS One. 2011;6:e24361. doi: 10.1371/journal.pone.0024361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tare M., Puli O.R., Singh A. In: Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye. Singh A., Kango-Singh M., editors. Springer; 2013. Molecular genetic mechanisms of axial patterning: mechanistic insights into generation of axes in the developing eye; pp. 37–75. [Google Scholar]
- Tare M., Sarkar A., Bedi S., Kango-Singh M., Singh A. Cullin-4 regulates Wingless and JNK signaling-mediated cell death in the Drosophila eye. Cell Death Dis. 2016;7:e2566. doi: 10.1038/cddis.2016.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Ou J., Lee N., Shin K., Cigliola V., Song L., Crawford G.E., Kang J., Poss K.D. Identification and requirements of enhancers that direct gene expression during zebrafish fin regeneration. Development. 2020;147:dev191262. doi: 10.1242/dev.191262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J.E., Rubin G.M. Wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development. 1995;121:3519–3527. doi: 10.1242/dev.121.11.3519. [DOI] [PubMed] [Google Scholar]
- Tsonis P.A., Eguchi G. Carcinogens on regeneration. Effects of N-methyl-N'-nitro-N-nitrosoguanidine and 4-nitroquinoline-1-oxide on limb regeneration in adult newts. Differentiation. 1981;20:52–60. doi: 10.1111/j.1432-0436.1981.tb01155.x. [DOI] [PubMed] [Google Scholar]
- Tsonis P.A., Madhavan M., Tancous E.E., Del Rio-Tsonis K. A newt's eye view of lens regeneration. Int. J. Dev. Biol. 2004;48:975–980. doi: 10.1387/ijdb.041867pt. [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Cavallo R., Dooijes D., van Beest M., van Es J., Loureiro J., Ypma A., Hursh D., Jones T., Bejsovec A. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang X., Zhang K., Wang Q., Cao X., Wang Z., Zhang S., Li A., Liu K., Fang Y. Rapid depletion of ESCRT protein Vps4 underlies injury-induced autophagic impediment and Wallerian degeneration. Sci. Adv. 2019;5:eaav4971. doi: 10.1126/sciadv.aav4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Hu C.K. Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science. 2020;369:eaaz3090. doi: 10.1126/science.aaz3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisrock D.W., Papenfuss T.J., Macey J.R., Litvinchuk S.N., Polymeni R., Ugurtas I.H., Zhao E., Jowkar H., Larson A. A molecular assessment of phylogenetic relationships and lineage accumulation rates within the family Salamandridae (Amphibia, Caudata) Mol. Phylogenet. Evol. 2006;41:368–383. doi: 10.1016/j.ympev.2006.05.008. [DOI] [PubMed] [Google Scholar]
- White K., Grether M.E., Abrams J.M., Young L., Farrell K., Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- White K., Lisi S., Kurada P., Franc N., Bangs P. Methods in Cell Biology. Academic Press; 2001. Methods for studying apoptosis and phagocytosis of apoptotic cells in Drosophila tissues and cell lines; pp. 321–338. [DOI] [PubMed] [Google Scholar]
- Wittkorn E., Sarkar A., Garcia K., Kango-Singh M., Singh A. The Hippo pathway effector Yki downregulates Wg signaling to promote retinal differentiation in the Drosophila eye. Development. 2015;142:2002–2013. doi: 10.1242/dev.117358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Rubin G.M. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yeates C.J., Sarkar A., Deshpande P., Kango-Singh M., Singh A. A two-clone approach to study signaling interactions among neuronal cells in a pre-clinical Alzheimer's disease model. iScience. 2020;23:101823. doi: 10.1016/j.isci.2020.101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates C.J., Sarkar A., Kango-Singh M., Singh A. In: Insights into Human Neurodegeneration: Lessons Learnt from Drosophila. Mutsuddi M., Mukherjee A., editors. Springer; 2019. Unravelling Alzhiemer's disease using Drosophila; pp. 251–277. [Google Scholar]
- Yokoyama H., Ogino H., Stoick-Cooper C.L., Grainger R.M., Moon R.T. Wnt/beta-catenin signaling has an essential role in the initiation of limb regeneration. Dev. Biol. 2007;306:170–178. doi: 10.1016/j.ydbio.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M., Basler K., Struhl G. Direct and long-range action of a wingless morphogen gradient. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]
- Zhong N., Gersch R.P., Hadjiargyrou M. Wnt signaling activation during bone regeneration and the role of Dishevelled in chondrocyte proliferation and differentiation. Bone. 2006;39:5–16. doi: 10.1016/j.bone.2005.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The RNA sequencing raw data is deposited in the public repository: Gene Expression Omnibus (GEO) repository, GEO accession number is GSE134348. The processed data (Excel sheets that contain list of genes that were significantly regulated) can be found in an institutional repository of the University of Dayton (eCommons) and are publicly accessible https://doi.org/10.26890/ddlla1a541sgr. The DNA sequence for viropana1 (vna1), viropana2 (vna2), viropana3 (vna3), viropana4 (vna4), viropana5 (vna5) is available at https://doi.org/10.26890/ddl.dbitmbv
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.