Summary
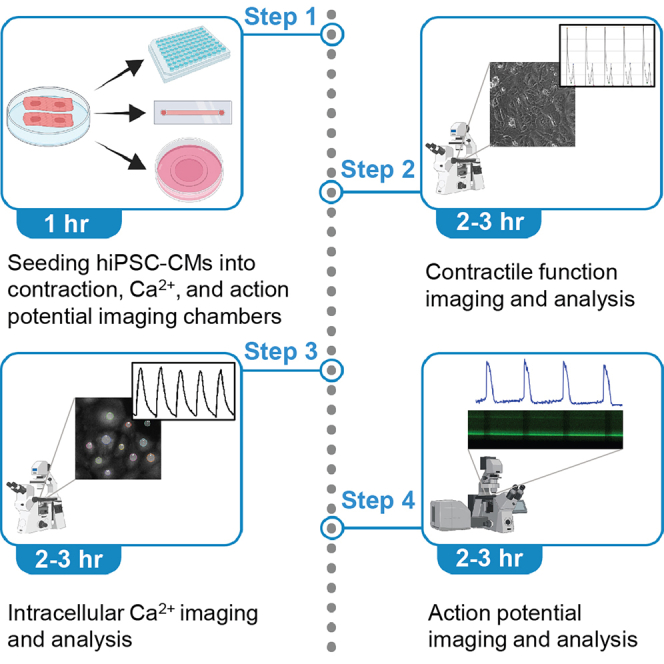
Multiple strategies have been developed to efficiently differentiate human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs). Here, we describe a protocol for measuring three key functional parameters of hiPSC-CMs, including contractile function, calcium (Ca2+) handling, and action potential.
For complete details on the use and execution of this protocol, please refer to Zhang et al. (2021).
Subject areas: Cell Biology, Cell culture, Microscopy, Stem Cells
Graphical abstract
Highlights
-
•
Automated measurement of hiPSC-CM contractile function
-
•
Single-cell calcium imaging of hiPSC-CMs loaded with Fura-2 AM
-
•
Imaging-based action potential measurement of hiPSC-CMs
Multiple strategies have been developed to efficiently differentiate human-induced pluripotent stem-derived cardiomyocytes (hiPSC-CMs). Here, we describe a protocol for measuring three key functional parameters of hiPSC-CMs, including contractile function, calcium (Ca2+) handling, and action potential.
Before you begin
Human-induced pluripotent stem cells (hiPSCs) can be requested from Stanford Cardiovascular Institute iPSC Biobank (https://med.stanford.edu/scvibiobank.html) or obtained from various commercial sources. For maintenance of hiPSCs, differentiation of hiPSCs into cardiomyocytes (hiPSC-CMs), expansion and cryopreservation of hiPSC-CMs, please refer to other STAR protocols (Lin and Zou, 2020; Lyra-Leite et al., 2021; Maas et al., 2021). Here, we used a small molecule-based cardiomyocyte differentiation method as described in Zhang et al. (2021).
Preparation of customized cell chambers for culturing hiPSC-CMs for Ca2+ imaging
Timing: 10 min
Note: The following steps should be conducted in a biosafety cabinet.
-
1.
Pour 10 mL of 75% ethanol into a petri dish.
-
2.
Sterilize glass coverslips by submerging them in 75% ethanol for 5 min.
-
3.
Air dry coverslips.
-
4.
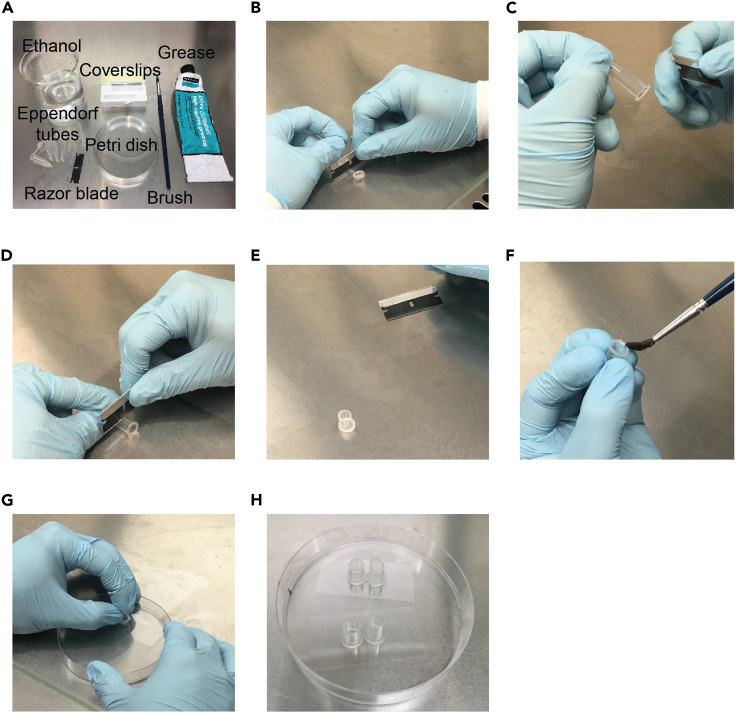
Cut off the lid and bottom half of 0.5 mL Eppendorf tubes using a razor blade (Figures 1A–1E).
-
5.
Carefully paint a thin layer of grease around the ring (not the cut side) of the Eppendorf tube (Figure 1F).
CRITICAL: Do not put too much grease.
-
6.
Put the tube on top of the glass coverslip and gently press the top of the tube to form a seal (Figure 1G).
Note: We recommend putting two tubes on one coverslip.
CRITICAL: Ensure that the tube does not slide on the coverslip when pressing, as this may leave a grease smear on the coverslip and affect cell attachment.
-
7.
Place two coverslips in a petri dish (Figure 1H).
-
8.
Leave the customized cell chambers under UV exposure in a biosafety cabinet for 1 h.
-
9.
After 1 h, seal the petri dish using parafilm for further use.
Figure 1.
Customized cell chambers for Ca2+ imaging
(A) Required materials for making customized cell chambers.
(B–G) The steps of making customized chambers. The small tubes are made by cutting off the lid and bottom half of 0.5 mL Eppendorf tubes using a razor blade. The grease is used for attaching tubes on coverslips. The paint brush is used for painting grease on the bottom side (not the cut side) of tubes.
(H) Example of customized cell chambers. Tubes with grease are put at the center of the coverslip. By gentle pressing the top of tubes, tubes are attached to the coverslips with a good seal.
Preparation of Matrigel-coated plates/coverslips/dishes
Timing: 30 min
-
10.
Thaw a bottle of Growth Factor Reduced (GFR) Matrigel (10 mL) for 18 h at 4°C.
Note: The following steps should be conducted in a biosafety cabinet.
-
11.
Aliquot 500 μL of Matrigel in sterile 1.5 mL Eppendorf tubes and store aliquoted Matrigel in a −20°C freezer for further use.
Note: Matrigel should be kept on ice at all times during the procedure to avoid solidification.
-
12.
Pipette 200 mL of ice-cold DMEM/F12 in a sterile plastic bottle and keep the bottle on ice.
-
13.
Take out one aliquot of Matrigel and leave it on ice to thaw.
-
14.
Add 500 μL of Matrigel in ice-cold DMEM/F12 medium at a concentration of 1:400 dilution (v/v).
-
15.
Rinse the Eppendorf tube with 1 mL of DMEM/F12 to collect the remaining Matrigel in the Eppendorf tube and add it to the bottle.
Note: Diluted Matrigel medium (DMEM/F12/Matrigel) can be stored in a 4°C fridge. We recommend using up diluted Matrigel medium within one week.
-
16.
Add 100 μL of DMEM/F12/Matrigel to each well of 96-well plates that will be used for seeding hiPSC-CMs for contraction measurement.
-
17.
Add 100 μL of DMEM/F12/Matrigel to each customized cell chamber that will be used for seeding hiPSC-CMs for Ca2+ imaging.
-
18.
Add 500 μL of DMEM/F12/Matrigel to each 35 mm glass-bottom cell culture dish that will be used for seeding hiPSC-CMs for action potential measurement.
-
19.
Transfer coated plates/coverslips/dishes to a humidified cell culture incubator at 37°C for 18 h before use.
Key resources table
| REGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| RPMI 1640 Medium | Life Technologies | 11875-119 |
| B27 supplements | Life Technologies | 17504-044 |
| Growth Factor Reduced (GFR) Basement Membrane Matrix, Phenol Red-Free, LDEV-Free | Corning | 356231 |
| DMEM/F12 | Gibco/Life Technologies | 12634028 |
| Accutase | Sigma-Aldrich | A6964 |
| KnockOut Serum Replacement | Thermo Fisher Scientific | 10828-028 |
| Pluronic™ F-127 (20% Solution in DMSO) | Thermo Fisher Scientific | P3000MP |
| Fura-2, AM, cell permeant | Thermo Fisher Scientific | F1221 |
| DPBS, no calcium, no magnesium | Fisher Scientific | 14-190-250 |
| NaCl | Sigma-Aldrich | S9888 |
| KCl | Sigma-Aldrich | 529552 |
| CaCl2 1 M in H2O | Sigma-Aldrich | 21115 |
| Glucose | Sigma-Aldrich | G8270 |
| MgCl2 | Sigma-Aldrich | M8266 |
| HEPES | Sigma-Aldrich | H3375 |
| NaOH 10 M in H2O | Sigma-Aldrich | 72068 |
| Trypan blue stain | Life Technologies | T10282 |
| Experimental models: Cell lines | ||
| Human induced pluripotent stem cell lines | Stanford Cardiovascular Institute Biobank | N/A |
| Bacterial and virus strains | ||
| ASAP2 lentivirus | ASAP2 plasmid is from Michael Lin’s Lab | N/A |
| Other | ||
| Polystyrene Storage Bottle, 250 mL | VWR | 8390 |
| 6-Well culture plate | E & K Scientific | EK-27160 |
| 96-Well flat clear bottom black polystyrene TC-treated microplates | Corning | 3603 |
| 35 mm Glass bottom dish with 20 mm micro-well #1.5 cover glass | Cellvis | D35-20-1.5-N |
| Rectangular glass coverslips | Warner Instruments | 64-0709 |
| 100 mm × 20 mm Petri dish, sterile | Sigma-Aldrich | P5606 |
| Dow Corning High Vacuum Grease | Sigma-Aldrich | Z273554-1EA |
| Fisherbrand Razor Blades | Fisher Scientific | 12-640 |
| Nikon Immersion Oil (Type F) 30cc PFS | Morrell Instrument Company | MXA22168 |
| Conical tubes, 15 mL | Corning Falcon | 352097 |
| LoBind micro-centrifuge tubes | Fisher Scientific | 13-698-794 |
| 5 mL Pipettes sterile, individually wrapped | Fisher Basix | 14-955-232 |
| 10 mL Pipettes sterile, individually wrapped | Fisher Basix | 14-955-234 |
| 2 mL Aspiration pipettes sterile, individually wrapped | Corning Falcon | 357558 |
| Reservoir, 10 mL sterile | MTC Bio | P8010-5S |
| Tube micro centrifuge, 0.5 mL | Dynalon Labware | 299265 |
| Nalgene™ Rapid-Flow™ Sterile Single Use Vacuum Filter Units | Fisher Scientific | 09-741-04 |
| Paint brush | Amazon | N/A |
| Narrow bath chamber with field stimulation | Warner Instruments | RC-27NE2 |
| Magnetic heated platforms | Warner Instruments | PM-6 |
| Class II, type A2 or better biosafety cabinet | Thermo Fisher Scientific | 1300 Series Class II |
| CO2 laboratory incubator | Laboratory Equipment Company | MCO-170AICUVL-PA |
| Centrifuge | VWR | Centrifuge 5810 R |
| pH meter | Hanna Instruments | HI2002-01 |
| Lab companion stirring hot plate, ceramic top | Cole-Parmer | 8657808 |
| Magnetic stirring bars | Fisher Scientific | 14-513-68 |
| Aspirator vacuum | N/A | N/A |
| Countess® Cell Counting Chamber Slides | Thermo Fisher Scientific | C10312 |
| Luna-FL automated fluorescence cell counter | WISBIOMED | LB-L20001 |
| Sony SI8000 Cell Motion Imaging System | Sony Biotechnology | N/A |
| Lambda DG 4 | Sutter Instrument Company | N/A |
| Zeiss LSM 710 confocal microscope | Zeiss | N/A |
| Software and algorithms | ||
| GraphPad Prism version 7.0 | GraphPad Software | N/A |
| ImageJ | NIH | N/A |
| Sony SI8000 cardiac analysis software | Sony Biotechnology | N/A |
Materials and equipment
-
•
hiPSC-CM culture medium: thaw a bottle of 50x B27 supplements (10 mL) and add B27 supplements to a bottle of RPMI 1640 medium (500 mL) (RPMI/B27).
Note: We recommend aliquoting 50x B27 supplements (stored in −20°C and used before the expiration date) and preparing RPMI/B27 weekly according to the required amount.
-
•
hiPSC-CM seeding medium: thaw a bottle of KnockOut serum replacement (KOSR) (500 mL) and aliquot the KOSR in 50 mL Falcon tubes (20 mL/tube and stored in −20°C) and prepare 10% KOSR RPMI/B27 by adding 20 mL of KOSR into 180 mL of RPMI/B27.
Note: This is the seeding medium that can be stored at 4°C for up to a month.
-
•
hiPSC-CM lentivirus infection medium: thaw a vial of the accelerated sensor of action potentials 2 (ASAP2) lentivirus (1.0 × 108 TU/mL) on ice, then add 1 μL of the virus into 1 mL of hiPSC-CM culture medium.
Note: Keep the virus on ice at all times. If the virus titer is different, the dilution should be adjusted according to the virus titer. The infection medium should be prepared on the day of use.
-
•
10× Tyrode’s stock solution: Dissolve the chemicals (see the table below) in 500 mL of distilled water to make 10× Tyrode’s stock solution with the defined concentrations.
| Reagent | Final concentration (mM) | Amount (g) |
|---|---|---|
| NaCl | 1350 | 39.45 |
| KCl | 54 | 2.01 |
| MgCl2 | 10 | 0.48 |
| HEPES | 100 | 11.92 |
| Distilled water | - | To final volume of 500 mL |
Note: The stock solution can be stored at 4°C for up to one year.
-
•
Tyrode’s solution: add 5 mL of 10× Tyrode’s stock solution, 90 μL of 1 M CaCl2, and 0.045 g of glucose in 40 mL of distilled water in a beaker (see the table below). Adjust the pH of the solution to 7.4 with NaOH. Add additional distilled water to bring the solution to a final volume of 50 mL. Consequently, Tyrode’s solution contains NaCl (135 mM), KCl (5.4 mM), MgCl2 (1 mM), CaCl2 (1.8 mM), glucose (5 mM), and HEPES (10 mM).
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× Tyrode’s stock solution | 1× | 5 mL |
| 1 M CaCl2 | 1.8 mM | 90 μL |
| Glucose | 5 mM | 0.045 g |
| Distilled water | - | To final volume of 50 mL |
Note: As Tyrode’s solution contains glucose that can be easily contaminated, we do not recommend storing it for a long time. Tyrode’s solution should be prepared in fresh on the day of the experiment.
-
•
Fura-2 AM indicator: Add 125 μL of Pluronic™ F-127 (20% solution in DMSO) into one vial of Fura-2 AM (50 μg) to make 0.4 mM stock of Fura-2 AM indicator solution. Aliquot 5 μL of Fura-2 AM indicator in a 1.5 mL Eppendorf tube, store it at −20°C, and protect it from light.
Note: Fura-2 AM indicator may take a few minutes to fully dissolve. Aliquoted Fura-2 AM can be stored up to one year.
-
•
Fura-2 AM loading solution: Take out one aliquot (5 μL) of Fura-2 indicator solution and add 995 μL of Tyrode’s solution to the Eppendorf tube to make the loading solution with 2 μM Fura-2 AM and 0.1% F-127.
Note: Loading solution should be prepared in fresh on the day of the experiment and protected from light.
Alternatives: For contraction measurement, this protocol uses Sony SI8000 Cell Motion Imaging System for imaging and analyses. Other microscopes equipped with a high-speed camera can also be used for recording hiPSC-CM contraction motion. For analyses, open-source motion tracking software is also available (Huebsch et al., 2015).
Alternatives: For Ca2+ imaging, this protocol uses Lambda DG 4 ultra-high-speed wavelength switching illumination system to excite Fura-2 AM. Others systems such as IonOptix can also be used for Fura-2 AM imaging.
Alternatives: For action potential measurement, this protocol uses ASAP2 lentivirus to record membrane potential (Chamberland et al., 2017; Chour et al., 2021; Zhang et al., 2019, 2021). Other probes such as FluoVolt can also be used for action potential measurement. This protocol uses Zeiss LSM 710 confocal microscope for action potential imaging. Other confocal microscopes such as Nikon AX/AXR can also be used for imaging.
Step-by-step method details
Enzymatic dissociation of hiPSC-CMs
Timing: 20 min
Day 23 hiPSC-CMs cultured in 6-well plates can be dissociated by enzymes for re-plating into desired imaging plates/coverslips/dishes at a defined cell density.
Note: hiPSC-CMs with the purity over 95% is necessary for the reproducible measurement.
-
1.
Aspirate the medium, wash hiPSC-CMs with DPBS twice, and incubate cells with Accutase (1 mL/well) for 6 min at 37°C.
Note: The incubation time may vary for different cell lines or culture conditions. We recommend observing cells under a microscope regularly to ensure that cells are not over-digested. A gentle tap on one side of the plate helps to agitate cells and improve the dissociation efficiency. Prolonged incubation of cells with enzymes may reduce cell viability.
-
2.
After incubation for 6 min, gently shake the plate and pipette the solution using a 1 mL pipette several times to fully dissociate the monolayer into single cells.
CRITICAL: hiPSC-CMs should be dissociated into single cells to ensure accurate cell count.
-
3.
Transfer the dissociated cell suspension into a 15 mL Falcon tube containing 8 mL of seeding medium.
-
4.
Add 1 mL of seeding medium to the well and wash off the remaining cells from the well; then transfer the cell suspension to the Falcon tube to give a final volume of 10 mL.
Note: If a large number of cells are required, multiple wells of hiPSC-CMs can be dissociated together and the cell suspension can be collected in a 50 mL Falcon tube.
-
5.
Centrifuge the cell suspension at 300 × g for 3 min at room temperature (22°C).
-
6.
Aspirate the supernatant and re-suspend cells in 1 mL of 37°C pre-warmed seeding medium.
Note: If multiple wells of hiPSC-CMs are dissociated, we recommend re-suspending the cell pellet according to the rule of 1 mL of seeding medium per well of 6-well plate.
-
7.
Mix 20 μL of cell suspension with 1:1 Trypan blue in a 0.5 mL Eppendorf tube.
-
8.
Transfer 15 μL of stained cell suspension in a Countess® Cell Counting Chamber Slide (each slide has a duplicate) and count cell number by the Luna-FL automated fluorescence cell counter. The cell suspension is ready for re-plating.
Seeding cells for imaging
Timing: 30 min (15 min every other day)
-
9.
Remove the coating Matrigel from each well/dish/customized chamber.
-
10.
Adjust the cell suspension to the density of 500,000 cells/mL in the seeding medium, transfer the cell suspension into a 10 mL reservoir, and add 100 μL of the cell suspension to each well of the 96-well plate.
CRITICAL: To re-plate cells in a 96-well plate, a multi-channel pipette should be used. To avoid cells from settling down at the bottom of the reservoir, we recommend pipetting up and down the cell suspension several times before re-plating.
-
11.
Add 50 μL of cell suspension and 50 μL of seeding medium to each customized cell chamber on the coverslip for Ca2+ imaging.
-
12.
Add 200 μL of cell suspension and 300 μL of seeding medium to each 35 mm glass-bottom dish for action potential measurement.
-
13.
After 24 h, aspirate the seeding medium.
Note: We recommend putting a 1 mL non-filtered tip over the 2 mL aspiration pipette and ensure that the tip does not touch cells.
-
14.
Add 100 μL of hiPSC-CM culture medium to each well in a 96-well plate or customized cell chamber. Replace the medium every other day until recording.
-
15.
Add 500 μL of the hiPSC-CM culture medium to 35 mm glass-bottom dishes. Replace the medium every other day until recording.
Automated recording of hiPSC-CM contraction motion
Timing: 1 h/96-well plate
-
16.
Turn on the microscope, camera, CO2 supply, and heating unit 10 min before use and set the heating unit temperature to 37°C.
-
17.
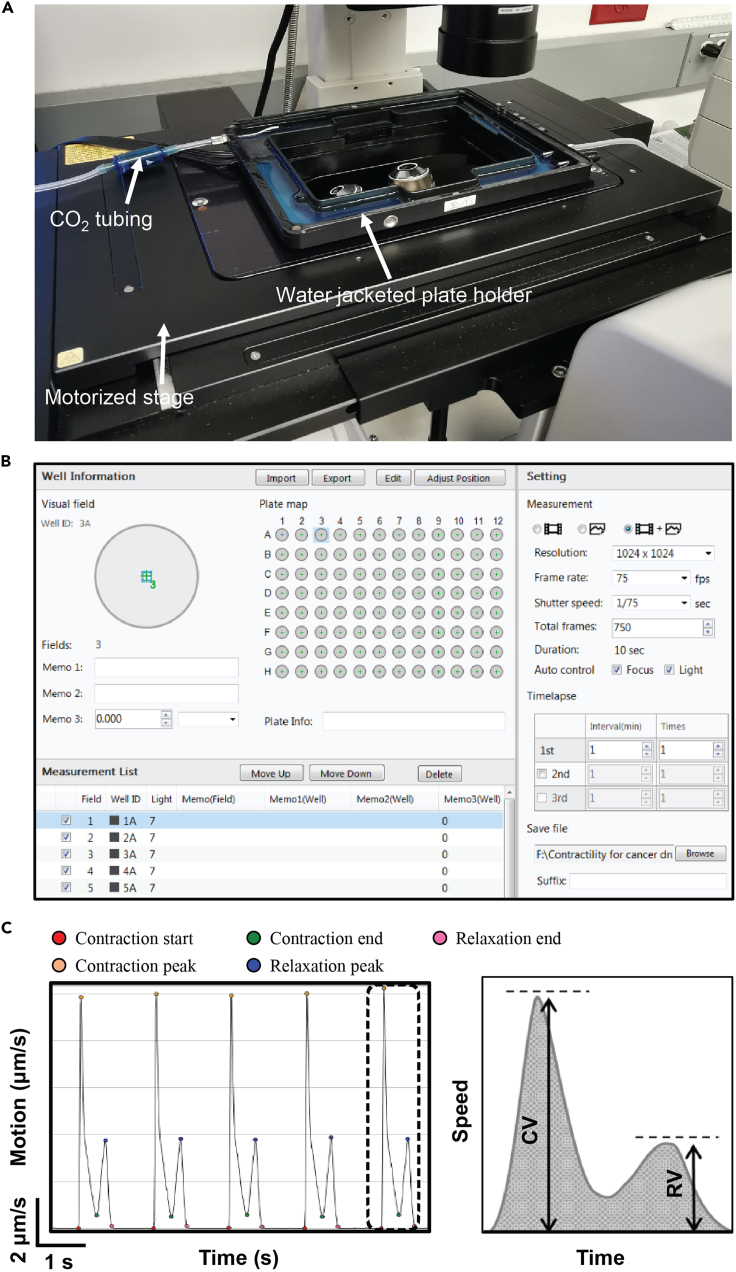
Add the desired amount of water to the water jacket of the plate holder (Figure 2A).
-
18.
Put the 96-well plate on the plate holder and allow cells to stabilize for 5 min before recording.
-
19.
In the software interface, activate the auto-focus and auto-brightness functions. Adjust the camera frame rate to 75 frames per second (fps) and the video/image format to 1024 × 1024 pixels (Figure 2B). The total recording time is 10 s. Live-cell imaging is performed by an inverted Nikon Eclipse Ti microscope (USA) equipped with a 10× objective (NA: 0.3).
Note: To avoid the biased selection of a region of interest (ROI), the center of each well is imaged. During recording, ensure the vibration of the microscope table is minimal.
-
20.
Open the Sony SI8000 cardiac analysis software; the motion detection and analyses are conducted automatically (Figure 2C).
Note: It is essential to detect contraction start, contraction peak, contraction end, relaxation peak, and relaxation end for each beating cycle to complete analyses.
Figure 2.
Measurement of contractile function
(A) Sony SI8000 Cell Motion Imaging System (hardware).
(B) The interface of Sony recording software.
(C) Representative contraction-relaxation traces of hiPSC-CMs and schematic diagrams showing the measurement of contraction velocity (CV) and relaxation velocity (RV).
Single-cell Ca2+ imaging
Timing: 2–3 h
-
21.
Prepare Tyrode’s solution as described in the materials and equipment section.
-
22.
Prepare the Fura-2 AM loading solution as described in the materials and equipment section.
-
23.
Aspirate the culture medium in the customized cell chambers and wash the hiPSC-CMs twice with 100 μL of Tyrode’s solution.
-
24.
Load 100 μL of Fura-2 AM loading solution to hiPSC-CMs in the customized cell chambers.
-
25.
Incubate the hiPSC-CMs at room temperature for 10 min.
-
26.
Remove Fura-2 AM loading solution from the hiPSC-CMs and wash twice with 100 μL of Tyrode’s solution.
-
27.
Incubate the hiPSC-CMs in Tyrode’s solution for another 5 min at room temperature to allow de-esterification of Fura-2 AM.
-
28.
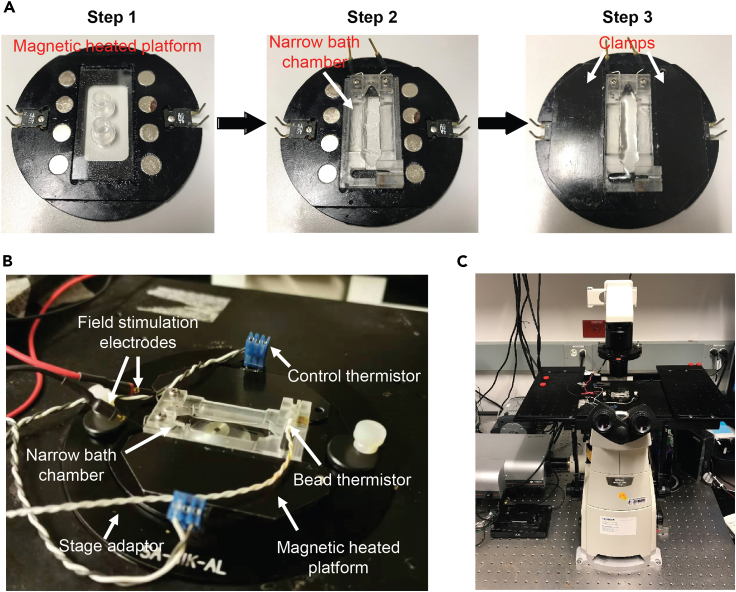
Put the coverslips in the magnetic heated platform (Figure 3A, step 1).
-
29.
Remove tubes on the coverslip and put the narrow bath chamber with grease at the bottom on top of the coverslip (Figure 3A, step 2).
-
30.
Secure the bath chamber in place using the magnetic clamps and ensure there is no leak (Figure 3A, step 3).
-
31.
Add 1 mL of Tyrode’s solution into the chamber (Figure 3A, step 3).
-
32.
Add a drop of immersion oil on the objective.
-
33.
Place the magnetic heated platform in a stage adapter on the stage of an inverted epifluorescence microscope (Nikon Eclipse Ti-S, USA) equipped with a 40× oil immersion objective (0.95 NA) (Figures 3B and 3C).
-
34.
Connect the temperature controller wires with the bath chamber.
-
35.
Put the temperature sensor in the bath chamber.
Note: Secure the sensor in place during the recording.
-
36.
Set the temperature controller at 37°C.
-
37.
Connect the field stimulation electrodes with a stimulator.
-
38.
Set the field stimulation at 0.5 Hz with a pulse duration of 20 ms.
-
39.
Fura-2 AM loaded cells are excited with Lambda DG 4 ultra-high-speed wavelength switching light source (Sutter Instrument, USA) at both 340 and 380 nm at fast-switching mode and the emission fluorescence signals are collected at 510 nm with the iXon Ultra 897 EMCCD camera (Andor Technology, UK). Find a region of interest (ROI) under the eye pieces first and then switch to the live view by camera.
Note: The light source switching mode can be set as indicated in the operation manual of Lambda DG 4. Ensure cells are focused under the live view by camera.
-
40.
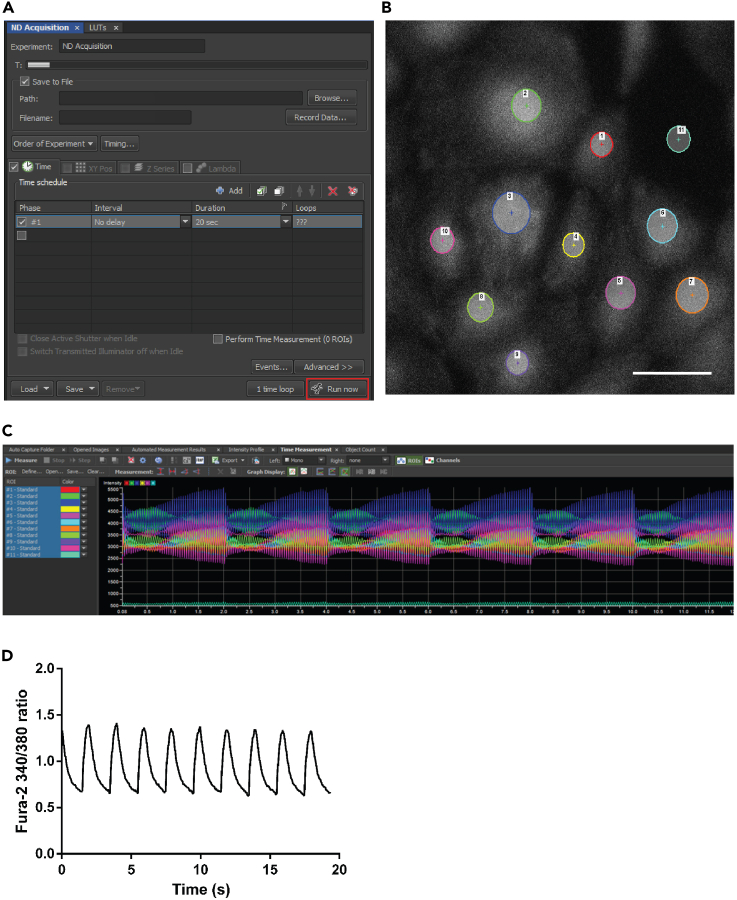
Click “Run now” to start recording under the ND Acquisition mode in NIS Elements AR software (Figure 4A). Exposure time for each wavelength is 20 ms and the total recording time is 20 s.
-
41.
The photons are detected by the camera and converted to arbitrary intensity values. Changes in fluorescence intensity of individual hiPSC-CMs over time are recorded by the NIS Elements AR software, which permits the recording of multiple cells.
-
42.
After recording, using the ROI selection tool to select individual hiPSC-CM in ROI as well as an area without any cells for background subtraction (Figure 4B).
-
43.
Measure the fluorescence intensity using the time measurement function (Figure 4C).
-
44.
Export the data to Excel for further analyses.
-
45.
Analyze the data using an open-source Excel-based program as previously published (Greensmith, 2014) (Figure 4D).
Figure 3.
Equipment used for Ca2+ imaging
(A) Steps of assembling coverslips, bath chamber, and the magnetic platform. Step 1: put the coverslip with tubes in the slot of magnetic heated platform; Step 2: remove tubes and put the bath chamber on top of the coverslip; and Step 3: clamp the bath chamber in place by magnetic clamps and add 1 mL of Tyrode’s solution.
(B) Picture of the bath chamber that has been connected with the heating and stimulation components.
(C) Picture of the microscope used for Ca2+ imaging.
Figure 4.
Software interface of Ca2+ transient recording and the first-step analysis
(A) The picture of the acquisition mode interface.
(B) Example of the region of interest showing the cell selection. Scale bar is 100 μm.
(C) Example of the time measurement of Ca2+ imaging, the color code corresponds to the cells shown in panel B.
(D) Representative trace of Ca2+ transients.
Fluorescence-based single-cell action potential imaging
Timing: 2–3 h
-
46.
Three days before imaging, prepare the lentivirus infection medium as described in materials and equipment and add 300 μL to each dish with the multiplicity of infection (MOI) of three.
Note: Use the minimum required volume for infection to improve efficiency.
-
47.
After 24 h, aspirate the infection medium.
-
48.
Add 500 μL of hiPSC-CM culture medium to each dish until imaging.
-
49.
On the day of imaging, turn on the confocal microscope and incubation system.
CRITICAL: The incubation system is essential for live-cell imaging. The temperature of the imaging chamber is kept at 37°C and is humidified to avoid evaporation of solution during imaging.
-
50.
Remove the medium and wash cells twice with 1 mL of Tyrode’s solution.
-
51.
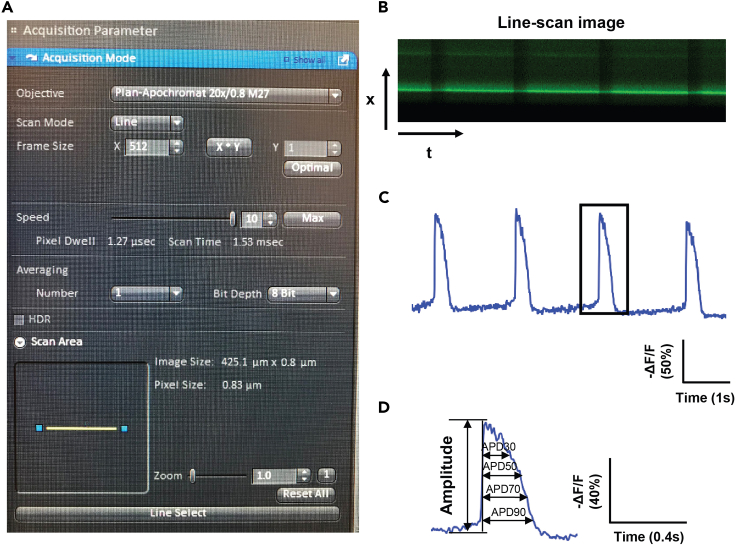
Remove the washing solution, add 1 mL of Tyrode’s solution to the dish, and place the dish onto a holding adapter to allow cells to stabilize for 5 min before imaging. Live-cell imaging is performed by an inverted Zeiss LSM 710 confocal microscope equipped with a 20× objective (NA: 0.8). The line-scan mode is used for measuring the action potential of single cells (Figures 5A and 5B).
CRITICAL: It is essential to allow cells to adapt to Tyrode’s solution before imaging. Cells usually start to show changes in fluorescence (changes in membrane potential) within 1 min after switching to Tyrode’s solution based on our experience.
-
52.
Live cell imaging of single cells is conducted in a line scan mode with an excitation wavelength of 488 nm and the emission is filtered with a band-pass filter (493–630 nm). The scanning line is selected across the cell membrane with the maximal scanning speed. The image size is 512 × 1 pixels with the total recording time of 8 s. The recording is conducted using the Zen software (USA) and images are saved as LSM files.
-
53.
For analyses, line-scan images are opened in Image J (USA). The fluorescence intensity of images is inverted using the Invert function under the Edit in ImageJ. The entire image is selected (Ctrl+A) to plot the profile of images (Ctrl+Alt+K). The amplitude, action potential duration (APD) 30, 50, 70, and 90 can be determined accordingly (Figures 5C and 5D).
Note: The cycle of scanning can be adjusted according to the experimental condition. If hiPSC-CMs beat slow, more recording time is required to allow recording multiple cycles of action potential.
Figure 5.
Imaging-based fluorescence measurement of the action potential
(A) The acquisition interface of the line-scan mode.
(B) Representative line-scan image of the action potential.
(C) Representative fluorescence signals of action potential trace.
(D) Example of single action potential trace (shown in the box in panel C) showing the measurement of amplitude and action potential duration (APD).
Expected outcomes
Here we introduce a protocol to measure hiPSC-CM contraction, Ca2+ handling, and action potential. Our previous studies have applied these approaches to investigate inherited cardiomyopathies and drug-induced cardiotoxicity (Kitani et al., 2019; Lan et al., 2013; Lee et al., 2019; Ma et al., 2018; Seeger et al., 2019; Sharma et al., 2018; Wu et al., 2019). This protocol provides an imaging-based assessment of hiPSC-CM functions, which does not require extensive technical training. When following the protocol, individuals who have cell culture and microscope operation experience will usually succeed in the recording in the first attempt. For the automated contraction measurement, over 100 experimental conditions with triplicates for each condition can be screened within a few hours. Moreover, this approach provides an opportunity for experimenters to repeatedly monitor the contractile function of the same ROI at different time points.
We have measured the stimulated Ca2+ transients in hiPSC-CMs by Ca2+ imaging. The arbitrary values of fluorescence intensity are measured by NIS Elements AR software and exported to an Excel sheet. After analyzing with the open-source Excel-based program (Greensmith, 2014), we can obtain the F340/F380 ratio over time for each cell. A representative raw trace of stimulated Ca2+ transients is shown in Figure 4D. In addition, we can obtain the values of three parameters (diastolic Ca2+, amplitude, and Ca2+ decay tau of stimulated Ca2+ transients) based on the Excel-based program. According to our experience, up to 1000 cells can be recorded per coverslip in one hour if necessary.
Optical imaging of hiPSC-CM action potential provides an accessible method to examine the electrophysiology of hiPSC-CMs. Although the single-cell patch-clamp is regarded as the gold-standard approach to determine the electrophysiological function of hiPSC-CMs, it requires highly specified equipment that is not always accessible. Moreover, as it requires extensive training for new users to master the skills, the labor-intensive nature of the patch-clamp hinders its application for high throughput studies. As an alternative, here we introduce an imaging-based fluorescence measurement of action potential using a highly sensitive and fast voltage sensor. This approach only requires minimal training in operating the confocal microscope. The brightness of fluorescence signals represents the resting membrane potential. When hiPSC-CMs depolarize, conformational changes in the ASAP2 lead to a reduction in the fluorescence signals. The line scan mode is normally utilized, and the detector gain is set at 600 with a digital gain of three. The scanning line is across the cell membrane and each line comprises 512 pixels. After a sequential scanning, a two-dimensional (2D) image of 512 × 2000 lines can be generated and stored for offline analyses. The fluorescence-based imaging approach allows experimenters to record over 100 cells per hour, which significantly improves the scale of throughput.
Quantification and statistical analyses
Statistical analyses and graph generation can be done using GraphPad Prism 9 (Graphpad, USA).
Limitations
Motion-based hiPSC-CM contraction measurement provides an easy and accessible approach to determine the hiPSC-CM contractile function. However, the major disadvantage of this approach is that it records the motion velocity when hiPSC-CMs contract and relax, so it does not report contractile force directly. Another disadvantage of this approach is that if hiPSC-CMs do not form a syncytium and contract coherently, the camera could capture multiple contraction peaks, which may make the analysis difficult.
Fura-2 AM, a ratiometric Ca2+ indicator, has been widely used because of advantages such as allowing the quantitative determination of the cytosolic Ca2+ level and being unaffected by uneven dye distribution and photo-bleaching. However, UV-excitation may lead to high background noise and bring photo-toxicity due to the high energy. Dual excitation dye requires fast filter switching, which is not suitable for confocal microscopy. In addition, like other Ca2+ dyes, Fura-2 AM may compartmentalize into intracellular compartments, making it challenging to indicate the cytosolic Ca2+ level. Either loading at lower temperatures or using a dextran-conjugated dye can be used to minimize this problem.
ASAP2 is a fast and sensitive voltage indicator (Chamberland et al., 2017). However, given the difficulty of transfection of hiPSC-CMs, the construct of the ASAP2 viral vector is required to achieve the desired efficiency of ASAP2 transduction in hiPSC-CMs. Another disadvantage of fluorescence-based imaging is the measurement of relative changes in membrane potential. In other words, this approach cannot provide the absolute recording of maximal diastolic membrane potential.
Troubleshooting
Problem 1
There are a lot of dead hiPSC-CMs floating 24 h after re-plating (step 11).
Potential solution
Over-digestion or under-digestion (leading to a forceful blow to attached cells) usually reduces cell viability. Continuous monitoring of cells during dissociation every minute under a microscope may be necessary. Adding 10 μM Y27632 in the hiPSC-CM seeding medium could improve cell viability. Another reason is the low attachment of hiPSC-CMs, and it may be necessary to use newly prepared Matrigel or increase the dilution to 1:200.
Problem 2
No monolayer is formed for contraction measurement (step 11).
Potential solution
We typically observe a monolayer when seeding 50,000 hiPSC-CMs per well in a 96-well plate with over 90% viability and attachment 1-week after re-plating. If cell viability is low, more cells could be seeded per well.
Problem 3
If hiPSC-CMs are clustered in the center on coverslips/dishes, is this a problem (step 13)?
Potential solution
The hiPSC-CMs need to be equally distributed over coverslips/dishes to allow single-cell recording. After seeding cells, using a pipette to give a gentle blow may help equalize the distribution. Do not swirl the dish, as this may lead to cell aggregation in the center.
Problem 4
There are small spikes on the contraction-relaxation graphs (step 21).
Potential solution
These small spikes could be caused by the vibration of the microscope table, or floating dead cells moving around during recording. If this “noise” does not affect the detection of contraction start, contraction peak, contraction end, relaxation peak, and relaxation end, it can be ignored. Ensure the microscope table is stable during recording. Change the medium the day before recording can help remove the cell debris, which may solve the problem.
Problem 5
No Ca2+ transients are observed and recorded in Ca2+ imaging (step 40).
Potential solution
Several points may cause this problem. First, make sure that hiPSC-CMs are in good conditions and beat normally before the start of Ca2+ imaging. Second, check whether Tyrode's stock solution has bacterial contamination. If there is contamination, filter the solution using a Rapid-Flow sterile filter unit with a pore size of 0.2 μm. Finally, the pH of the freshly prepared Tyrode's solution should be accurately adjusted.
Problem 6
Motion artifact during Ca2+ or action potential recording (steps 41 and 53).
Potential solution
When single-cells are recorded, we hardly detect the motion artifact. Avoid the regions where monolayers are formed.
Problem 7
No spontaneous electrical signals (step 52).
Potential solution
Ensure hiPSC-CMs are in good condition. After switching to Tyrode’s solution, give hiPSC-CMs more time to adapt to the new solution.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Joseph C. Wu, Email: joewu@stanford.edu.
Materials availability
hiPSC lines used in this study are available upon request from the lead contact, Joseph C. Wu, Email: joewu@stanford.edu.
Acknowledgments
This work was supported by NIH R01 HL123968, R01 HL141371, R01 HL145676, R01 HL150693, and Leducq 18CVD05 (J.C.W.).
Author contributions
J.Z.Z. wrote the manuscript, performed the experiments, and analyzed the data. S.R.Z., C.T., P.P., and M.Z. co-wrote the manuscript. J.Z.Z. and S.R.Z developed the full protocol. J.C.W. supervised the experimental studies, reviewed the manuscript, and provided funding support.
Declaration of interests
J.C.W. is a cofounder of Khloris Biosciences but has no competing interests, as the work presented here is completely independent. The other authors declare no competing interests.
Contributor Information
Joe Z. Zhang, Email: joezhang@szbl.ac.cn.
Joseph C. Wu, Email: joewu@stanford.edu.
Data and code availability
The published article includes all datasets generated or analyzed during this study. No new data have been generated for this protocol.
References
- Chamberland S., Yang H.H., Pan M.M., Evans S.W., Guan S., Chavarha M., Yang Y., Salesse C., Wu H., Wu J.C. Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. Elife. 2017;6:e25690. doi: 10.7554/eLife.25690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chour T., Tian L., Lau E., Thomas D., Itzhaki I., Malak O., Zhang J.Z., Qin X., Wardak M., Liu Y. Method for selective ablation of undifferentiated human pluripotent stem cell populations for cell-based therapies. JCI insight. 2021;6:10. doi: 10.1172/jci.insight.142000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greensmith D.J. Ca analysis: an Excel-based program for the analysis of intracellular calcium transients including multiple, simultaneous regression analysis. Comput. Methods Programs Biomed. 2014;113:241–250. doi: 10.1016/j.cmpb.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N., Loskill P., Mandegar M.A., Marks N.C., Sheehan A.S., Ma Z., Mathur A., Nguyen T.N., Yoo J.C., Judge L.M. Automated video-based analysis of contractility and calcium flux in human-induced pluripotent stem cell-derived cardiomyocytes cultured over different spatial scales. Tissue Eng. Part C Methods. 2015;21:467–479. doi: 10.1089/ten.tec.2014.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani T., Ong S.G., Lam C.K., Rhee J.W., Zhang J.Z., Oikonomopoulos A., Ma N., Tian L., Lee J., Telli M.L. Human-induced pluripotent stem cell model of trastuzumab-induced cardiac dysfunction in patients with breast cancer. Circulation. 2019;139:2451–2465. doi: 10.1161/CIRCULATIONAHA.118.037357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Lee A.S., Liang P., Sanchez-Freire V., Nguyen P.K., Wang L., Han L., Yen M., Wang Y., Sun N. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Termglinchan V., Diecke S., Itzhaki I., Lam C.K., Garg P., Lau E., Greenhaw M., Seeger T., Wu H. Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature. 2019;572:335–340. doi: 10.1038/s41586-019-1406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Zou J. Differentiation of cardiomyocytes from human pluripotent stem cells in fully chemically defined conditions. STAR Protoc. 2020;1:100015. doi: 10.1016/j.xpro.2020.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyra-Leite D.M., Fonoudi H., Gharib M., Burridge P.W. An updated protocol for the cost-effective and weekend-free culture of human induced pluripotent stem cells. STAR Protoc. 2021;2:100213. doi: 10.1016/j.xpro.2020.100213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Zhang J.Z., Itzhaki I., Zhang S.L., Chen H., Haddad F., Kitani T., Wilson K.D., Tian L., Shrestha R. Determining the pathogenicity of a genomic variant of uncertain significance using CRISPR/Cas9 and human-induced pluripotent stem cells. Circulation. 2018;138:2666–2681. doi: 10.1161/CIRCULATIONAHA.117.032273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas R.G.C., Lee S., Harakalova M., Snijders Blok C.J.B., Goodyer W.R., Hjortnaes J., Doevendans P.A.F.M., Van Laake L.W., van der Velden J., Asselbergs F.W. Massive expansion and cryopreservation of functional human induced pluripotent stem cell-derived cardiomyocytes. STAR Protoc. 2021;2:100334. doi: 10.1016/j.xpro.2021.100334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger T., Shrestha R., Lam C.K., Chen C., McKeithan W.L., Lau E., Wnorowski A., McMullen G., Greenhaw M., Lee J. A premature termination codon mutation in MYBPC3 causes hypertrophic cardiomyopathy via chronic activation of nonsense-mediated decay. Circulation. 2019;139:799–811. doi: 10.1161/CIRCULATIONAHA.118.034624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., McKeithan W.L., Serrano R., Kitani T., Burridge P.W., Del Álamo J.C., Mercola M., Wu J.C. Use of human induced pluripotent stem cell-derived cardiomyocytes to assess drug cardiotoxicity. Nat. Protoc. 2018;13:3018–3041. doi: 10.1038/s41596-018-0076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Yang H., Rhee J.W., Zhang J.Z., Lam C.K., Sallam K., Chang A.C.Y., Ma N., Lee J., Zhang H. Modelling diastolic dysfunction in induced pluripotent stem cell-derived cardiomyocytes from hypertrophic cardiomyopathy patients. Eur. Heart J. 2019;40:3685–3695. doi: 10.1093/eurheartj/ehz326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Belbachir N., Zhang T., Liu Y., Shrestha R., Wu J.C. Effects of cryopreservation on human induced pluripotent stem cell-derived cardiomyocytes for assessing drug safety response profiles. Stem Cell Reports. 2021;16:168–181. doi: 10.1016/j.stemcr.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Termglinchan V., Shao N.Y., Itzhaki I., Liu C., Ma N., Tian L., Wang V.Y., Chang A.C.Y., Guo H. A human iPSC double-reporter system enables purification of cardiac lineage subpopulations with distinct function and drug response profiles. Cell Stem Cell. 2019;24:802–811.e5. doi: 10.1016/j.stem.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article includes all datasets generated or analyzed during this study. No new data have been generated for this protocol.