Abstract
Samples of whole feces in which Cryptosporidium oocysts were recognized by hospital laboratories were collected from 218 patients with diarrhea. All samples were reexamined by light microscopy, and oocysts were detected in 211 samples. A simple and rapid procedure for the extraction of DNA from whole feces was developed, and this was used to amplify fragments of the Cryptosporidium outer wall protein (COWP), the thrombospondin-related adhesive protein C1 (TRAP-C1), and the 18S rRNA genes by PCR. For seven samples oocysts were not detected by microscopy and DNA failed to be amplified by the three PCR procedures. Among the 211 samples “positive” by microscopy, the sensitivities of PCRs for the 18S rRNA, COWP, and TRAP-C1 gene fragments were 97, 91, and 66%, respectively. The sensitivities of all three PCR procedures increased with increasing numbers of oocysts as observed by microscopy. Two genotypes of the COWP and TRAP-C1 genes can be detected by PCR-restriction fragment length polymorphism analysis. With this series of samples, the same genotypes of the COWP and TRAP-C1 genes always segregated together. A combined genotyping data set was produced for isolates from 194 samples: 74 (38%) were genotype 1 and 120 (62%) were genotype 2. Genotype 2 was detected in a significantly greater proportion of the samples with small numbers of oocysts, and genotype 1 was detected in a significantly greater proportion of the samples with larger numbers of oocysts. There were no significant differences in the distribution of the genotypes by patient sex and age. The distribution of the genotypes was significantly different both in patients with a history of foreign travel and in those from different regions in England.
The coccidian parasite Cryptosporidium parvum is increasingly recognized as a major cause of diarrheal disease worldwide (11): in England and Wales 4,000 to 6,000 cases in humans are reported each year (27a). Infection occurs via the oral route, and large waterborne outbreaks that affect large numbers of people have occurred in the United States and the United Kingdom (16, 36). The exact modes of transmission, however, are often unclear; and the importance of travel, the consumption of foods, beverages, or water, and person-to-person transmission and the role of infected animals in disease transmission remain to be ascertained (9).
Evidence from isoenzyme analysis (2) together with PCR, PCR-restriction fragment length polymorphism (RFLP) analysis, and arbitrary primed PCR analysis of several genes and gene sequences (3, 4, 7, 8, 20–26, 29–34) and sequence analysis of specific genes (7, 26) have identified two major types of C. parvum: one that is isolated exclusively from humans and a second that occurs in both livestock animals and humans. Analysis of the thrombospondin-related adhesive protein C2 (TRAP-C2) similarly indicates that C. parvum comprises two genotypes, and experimental infection of both calves and mice with the TRAP-C2 genotype common to both humans and livestock animals was successful, but infection with the genotype exclusive to humans was not successful (26). It has been suggested that these observations concerning the two genotypes of C. parvum reflect the epidemiology of a parasite with two distinct and exclusive transmission cycles (26) and may even indicate two distinct species of parasite. However, most of the studies mentioned above have been performed with relatively small numbers of samples, standardized reference material has not been available, and although some of the different markers recognize the same two groups (31), the relationships between all the markers described have not been formally established.
A PCR-RFLP technique has been described for the Cryptosporidium outer wall protein (COWP) gene of C. parvum, and this system distinguishes two genotypes (29). The type exclusive to humans was designated genotype 1, and the second type common among livestock and humans was designated genotype 2 (24). We previously reported (24) on a simple and rapid procedure for the extraction of DNA from C. parvum isolates present in stored samples of whole feces and, using this technique, showed that isolates from 91 of 95 (96%) of patients infected during two large waterborne outbreaks were genotype 1. In contrast, COWP genotype 1 comprised 31 of 46 (67%) of the C. parvum isolates from patients with sporadic cases of infection (24). A further polymorphic genetic marker in a structural gene (the thrombospondin-related adhesive protein C1 [TRAP-C1]) also distinguishes two genotypes of C. parvum by a similar PCR-RFLP technique (30).
The current routine laboratory techniques for the diagnosis of cryptosporidiosis relies on the recognition of specific oocyst morphologies by light microscopy in fecal specimens usually stained with modified Ziehl-Neelsen (MZN) stain or phenol-auramine or by immunofluorescence (1, 10). To further improve the means for the identification of this organism, multiple sequence alignment analysis of 18S rRNA gene sequences (25) was used to identify primers which, in combination with universal primers for lower eukaryotic rRNA (28), were specific for C. parvum and Cryptosporidium wrairi and which did not react with other species of Cryptosporidium or other organisms (including five species of two different genera of coccidia).
We report here that the previously described DNA extraction method (24) was less successful when it was applied to recent fecal samples. However, this report further describes the successful modification of the DNA extraction method for amplification of 18S rRNA, COWP, and TRAP-C1 gene fragments from recent samples. We also describe the segregation of the two genotypes of the COWP and TRAP-C1 genes from C. parvum collected from 218 patients with diarrhea in the United Kingdom during the second half of 1998, and we describe a further investigation of the distribution of genotypes from patients in different regions in England.
MATERIALS AND METHODS
Fecal samples.
Whole feces were collected from 218 patients with diarrhea between 1 September and 31 November 1998 whenever Cryptosporidium oocysts were recognized by hospital laboratories by conventional techniques (1, 10). There was no other selection of the samples. The patients comprised 102 females and 107 males; the sexes of 9 patients were not stated. The ages of the patients were as follows: 6 patients were <1 year, 74 patients were between 1 and 5 years, 67 patients were between 6 and 15 years, 38 patients were between 17 and 35 years, and 26 patients were between 36 and 67 years. The ages of the remaining seven patients were not established. All patients were believed to have sporadic cases of infection, and limited additional epidemiological information (recent foreign travel, geographical region, etc.) was collected from the original request form submitted with the sample.
All samples were stored as whole feces at 4°C without preservatives and were examined at between 11 days and 3 months after collection from the patients.
Staining and light microscopy.
All samples were reexamined by light microscopy in the Public Health Laboratory Service (PHLS) Food Hygiene Laboratory both by acid-fast staining with MZN stain and by immunofluorescence as described elsewhere (1, 10, 18). Immunofluorescence staining was performed with an anti-Cryptosporidium-oocyst monoclonal antibody (MAb) (17), which was produced in-house as whole ascitic fluid as described elsewhere (12).
To allow comparisons between the two staining techniques, two smears of approximately 40 μl of the homogenized sample were prepared so that similar and even amounts of fecal material were spread over the surfaces of the glass microscope slides. Oocysts were recognized as described previously (1, 10) with a Standard 14 (Zeiss, Welwyn Garden City, United Kingdom) microscope, and the same ×40 objective and ×10 eye pieces were used for both procedures. The numbers of oocysts on slides stained both with MZN and by immunofluorescence were counted, and the means for 20 fields were calculated. If no oocysts were seen in 20 fields, the entire slide was examined. The numbers of oocysts observed were categorized as follows: −, no oocysts; ±, <1 oocyst per field; +, 1 to 10 oocysts per field; 1+, 11 to 100 oocysts per field; and 3+, >100 oocysts per field. The numbers of oocysts for the ±, +, 2+, and 3+ categories were calculated to be approximately <103, 103 to 104, 104 to 105, and >105 oocysts per g (or per ml) of feces, respectively, by using suspensions of known numbers of oocysts obtained from the PHLS Cryptosporidium Reference Unit, Rhyl, Wales, United Kingdom.
Oocyst disruption and DNA extraction.
Oocyst disruption and DNA purification (method 1) were performed as described previously (24). These procedures comprised shaking with zirconia beads and a DNA extraction procedure modified from that of Boom et al. (5). Method 1 was further modified (method 2) by analysis, whenever possible, of larger (200-μl) volumes of feces, the use of larger-diameter (0.5-mm) zirconia beads, the addition of 60 μl of isoamyl alcohol to inhibit foaming during agitation, the use of a larger volume (100 μl) of activated silica to extract the DNA, and the use of a further DNA purification procedure with polyvinylpyrrolidone (PVP).
The modified extraction procedure (method 2) was as follows. Approximately 200 μl of whole feces was added to 900 μl of 10 M guanidinium thiocyanate in 0.1 M Tris-HCl (pH 6.4)–0.2 M EDTA (pH 8.0)–2% (wt/vol) Triton X-100 together with 0.3 g of 0.5-mm-diameter zirconia beads (Stratech Scientific, Luton, United Kingdom) and 60 μl of isoamyl alcohol. The tube was shaken in a Mini-Beadbeater (Stratech Scientific) for 2 min at maximum speed (5,000 rpm), left at room temperature for 5 min, and centrifuged. Coarse activated silica suspension (100 μl) was added to the supernatant (5), and this mixture was incubated at room temperature for 10 min with gentle agitation. The supernatant was discarded; and the pellet was washed twice with 200 μl of 10 M guanidinium thiocyanate in 0.1 M Tris-HCl (pH 6.4), twice with 200 μl of ice-cold 80% ethanol, and once with 200 μl of acetone. The pellet was then dried at 56°C for 5 min, and the DNA was eluted into 150 μl of water after vortex mixing and incubation at 56°C for 5 min. The supernatant (DNA sample) was recovered by centrifugation and either was used directly for PCR amplification or was stored at −20°C until further use.
For samples for which the PCR amplification (described below) was unsuccessful, a further DNA precipitation with PVP was performed as described by Lawson et al. (14). This comprised the addition of 50 μl of the extracted DNA sample to 150 μl of PVP-TE (10% [wt/vol] PVP in TE buffer) and incubation at room temperature for 10 min. The DNA was then precipitated by the addition of 100 μl of 2 M ammonium acetate and 600 μl of isopropanol at −20°C for 30 min. DNA was recovered by centrifugation (11,000 × g for 10 min), dried, reconstituted in water, and used as described above.
PCR-RFLP analysis.
PCR amplification was performed in 25-μl total volumes and included 2.5 μl of extracted DNA. Positive and negative controls were included in each batch of tests. PCR for the COWP gene fragment with primers CRY-9 and CRY-15 and PCR for the TRAP-C1 gene fragment with primers CP.E and CP.W, followed by restriction digestion with RsaI for both PCRs, were performed as described previously (29, 30).
For the amplification specific for the 18S rRNA fragments of C. parvum and C. wrairi, the primer CP3R (GAG GGA CTT TGT ATG TTT AAT ACA GG) together with universal lower eukaryotic reverse primer B (CCC GGG ATC CAA GCT TGA TCC TTC TGC AGG TTC ACC TAC [28]) gave a 422-bp product (25). Primer CP3R corresponds to bases 1340 to 1365 and 1344 to 1369 of C. parvum sequences with GenBank accession nos. L16996 and L16997, respectively and 1340 to 1365 of the C. wrairi sequence with GenBank accession no. U11440. For this PCR procedure, the following concentrations of the various reagents were used: 1× PCR buffer (Gibco Life Technologies, Paisley, United Kingdom), 1.5 mM MgCl2, 0.1 mM deoxynucleoside triphosphate mixture (Gibco Life Technologies), 15 pmol of primers CP3R and B, and 1.25 U of Taq polymerase (Gibco Life Technologies). The tubes were subjected to 30 cycles of 90°C for 30 s, 65°C for 180 s, and 72°C for 180 s, followed by a final extension at 72°C for 9 min.
A 5-μl aliquot of the PCR products was examined following electrophoresis in 1% agarose gel containing ethidium bromide. Restriction digests of the COWP and TRAP-C1 fragments with RsaI were resolved by electrophoresis in 3.2% typing-grade agarose gels (catalogue no. 14610-018; Gibco Life Technologies) containing ethidium bromide as described previously (29). All gels were recorded by using UV transillumination and Type 52 film (Polaroid Ltd., St. Albans, United Kingdom). The sensitivity of each PCR test was defined as the number of samples positive by PCR/total number of samples in which oocysts were observed by conventional microscopy.
RESULTS
Samples of feces in which cryptosporidial oocysts were recognized by the original laboratory were collected from a total of 218 patients. All samples were subsequently retested in the PHLS Food Hygiene Laboratory. Oocysts were detected in 211 samples by both MZN and immunofluorescence staining but were not detected in the remaining 7 by either microscopy method.
Initial PCR amplification experiments were performed with DNA extracted by method 1, and for samples in which oocysts were detected by microscopy, successful PCR amplification of the COWP gene was achieved for 9 of 38 samples (24%). The DNA extraction method was subsequently improved (method 2), and for PCR for the detection of the COWP gene, DNA was initially amplified from 68% of the samples and was amplified from a further 23% of the samples following PVP purification. DNA was then extracted from all samples by method 2, and all subsequent results were obtained by this procedure.
DNA was not amplified by any of the three primer pairs from the seven samples in which oocysts were not observed by either of the two microscopy methods (Table 1), and these samples are not further considered. Overall, the sensitivities of the PCRs for the detection of the 18S rRNA, COWP, and TRAP-C1 gene fragments were 97, 91, and 66%, respectively (Table 1). For preparations of DNA from the seven samples in which oocysts were seen by microscopy and for which the 18S rRNA gene fragment failed to be amplified, the COWP and TRAP-C1 gene fragments also failed to be amplified. The sensitivities of all three PCR procedures increased as the numbers of oocysts originally observed by immunofluorescence increased (Table 1).
TABLE 1.
Results of PCR amplification of 18S rRNA, COWP, and TRAP-C1 genes of C. parvum from DNA extracted from the feces of 218 patients diagnosed as having cryptosporidiosis
| Gene amplified by PCR and PCR result | No. (%) of patients with the following relative no. of oocysts observed by immunofluorescencea:
|
Total no. of patients positive (sensitivity [%]) by PCRb | ||||
|---|---|---|---|---|---|---|
| None | ± | + | 2+ | 3+ | ||
| 18S rRNA | ||||||
| Positive | 0 | 22 (96) | 98 (95) | 80 (99) | 4 (100) | 204 (97) |
| Negative | 7 | 1 | 5 | 1 | 0 | |
| COWP | ||||||
| Positive | 0 | 17 (74) | 91 (88) | 79 (98) | 4 (100) | 191 (91) |
| Negative | 7 | 6 | 12 | 2 | 0 | |
| TRAP-C1 | ||||||
| Positive | 0 | 14 (61) | 64 (62) | 57 (70) | 4 (100) | 139 (66) |
| Negative | 7 | 9 | 39 | 24 | 0 | |
| Genotypec | ||||||
| 1 | 3 | 32 | 37 | 2 | ||
| 2 | 16 | 60 | 42 | 2 | ||
Percentages were calculated on the basis of the proportion of the samples positive by PCR in each category.
Calculated only for samples in which oocysts were observed by immunofluorescence. Sensitivity is equal to number of samples PCR positive/total number of samples.
Results for 194 samples: 191 samples analyzed for the COWP gene plus an additional 3 samples analyzed for the TRAP-C1 gene (Table 2).
In all 211 samples, oocysts were observed by both MZN and immunofluorescence staining. However, it was noted that there was considerable variation within individual samples in the proportion of oocysts which were acid fast (MZN staining positive) compared to the total number detected by immunofluorescence. Since the objects observed by immunofluorescence may be empty oocysts (for example, following excystation) or oocysts with degraded DNA, the relationship between this and the proportion of samples from which the three cryptosporidial genes could not be amplified by PCR was investigated. Of the 211 microscopy-positive samples, 38, 33, and 29% had >90, 90 to 50, and <50% acid-fast oocysts, respectively; 4% of the samples had <10% acid-fast oocysts. However, there was no association between the proportion of acid-fast oocysts and the ability to amplify the gene fragments by the three PCR procedures. For example, the TRAP-C1 gene fragment was amplified from 72, 59, and 66% of the categories of samples with >90, 90 to 50, and <50% acid-fast oocysts, respectively. In addition, there was no relationship between the proportion of acid-fast oocysts and the genotype of C. parvum detected (data not shown).
All specimens were tested by PCR with the COWP- and TRAP-C1-specific primer pairs. Of the 211 samples in which oocysts were observed by light microscopy, DNA was amplified from 191 (91%) and 139 (66%) of the samples by the PCRs with COWP- and TRAP-C1-specific primers, respectively (Table 1). Amplicons from both these procedures were subjected to RFLP analysis, and the segregation of the two genotypes of the COWP and TRAP-C1 genes is shown in Table 2. COWP genotype 1 always segregated with TRAP-C1 genotype 1, and COWP genotype 2 always segregated with TRAP-C1 genotype 2. No samples infected with isolates of both genotypes were detected. In three samples, a C. parvum genotype was established with the TRAP-C1-specific primer pair but not with the COWP-specific primer pairs (Table 2). Because of the invariable segregation of the two genotypes, a combined set of genotyping results for the 194 samples was compiled from the 191 COWP-specific PCR results and the additional three TRAP-C1-specific PCR results. Overall, among the isolates in the 194 samples, 74 (38%) were genotype 1 and 120 (62%) were genotype 2, and these data are used in the subsequent analysis.
TABLE 2.
Segregation of genotypes identified by PCR-RFLP analysis of COWP and TRAP-C1 genes in DNA extracted from feces of 211 patients with cryptosporidiosis
| TRAP-C1 genotype | No. of patients infected with COWP genotype:
|
||
|---|---|---|---|
| 1 | 2 | Negativea | |
| 1 | 54 | 0 | 1 |
| 2 | 0 | 82 | 2 |
| Negativea | 19 | 36 | 17 |
No amplification by PCR.
Genotype 2 was detected in a greater proportion of the samples with small (± category) numbers of oocysts, and genotype was detected in a greater proportion of samples with larger (2+ category) numbers of oocysts (Table 1). There was a significant difference in the numbers of oocysts observed by immunofluorescence for the two genotypes (χ2 test for trend = 6.84, 1 degree of freedom, P = 0.009). To estimate misclassification bias, 20 samples were blindly retested after 2 to 5 months of storage, and the results of immunofluorescence showed that 19 of the 20 samples would be categorized exactly as they were initially categorized (the remaining sample would have been reclassified from + to 2+). A similar distribution of genotypes was observed when different numbers of oocysts were seen by MZN staining (data not shown).
There were no significant differences between genotype and the sexes and ages of the infected patients. However, a greater proportion of infections were caused by genotype 2 among patients in the 16- to 69-year-old age group, but this was not statistically significant (data not shown).
A recent foreign travel history was recorded for 29 (15%) of the 194 patients in which C. parvum was detected and the genotype was determined and of these, 17 were infected with genotype 1 and 12 were infected with genotype 2. Travel was recorded to 13 different countries: 6 European countries (18 patients), 1 Australasian country (1 patient), 3 countries on the Indian subcontinent (7 patients), 2 African countries (2 patients), and 1 country in the Americas (1 patient). Of the remaining 165 patients, 57 were infected with a genotype 1 strain and 108 were infected with a genotype 2 strain. There was a significant association between infection with a genotype 1 strain and a history of foreign travel (χ2 test = 5.08, 1 degree of freedom, P = 0.024). There were no associations between any of the patients and any single country (data not shown).
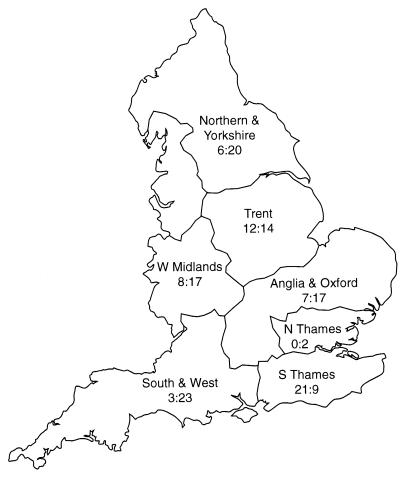
The distributions of both C. parvum genotypes from 159 patients with cryptosporidiosis in England by the regional health authorities of the laboratories performing the original diagnosis are shown in Fig. 1. Since the 29 patients with a recent history of foreign travel were likely to have contracted cryptosporidiosis away from their home regions, these patients have been excluded from the regional distribution shown in Fig. 1. Genotyping results were obtained for a further six C. parvum-infected patients from outside England during the study period: three in Scotland and three in Northern Ireland. All of these infections were due to C. parvum genotype 2. The distribution of genotypes between the regional health authorities of England was significantly different (χ2 test = 26.7, 6 degrees of freedom, P = 0.0002). There was a greater proportion of infections caused by genotype 1 in the South Thames region, almost equal numbers of infections caused by each genotype in Trent, and an excess of infections caused by genotype 2 in northern England and Yorkshire, Anglia and Oxford, West Midlands, and the South and West.
FIG. 1.
Distribution of both C. parvum genotypes from 159 patients with cryptosporidiosis during the second half of 1998 by regional health authorities in England. Patients with a recent history of foreign travel have been excluded. For each region, the name of the regional health authority is given above the number of patients infected with genotype 1: number of patients infected with genotype 2.
DISCUSSION
We report here on an analysis of a large series of sporadic cases of cryptosporidiosis for which genotyping of C. parvum has been performed. These cases, together with other sporadic cases and cases of infection in livestock animals and humans associated with two large waterborne outbreaks that we reported on previously (24), provide data on 335 cases of infection in humans, 60 natural infections in livestock, and a further 2 experimental infections in livestock animals. A summary of the genotyping results from these two studies is shown in Table 3.
TABLE 3.
Distribution of C. parvum genotypes from animals and humans with cryptosporidiosis in the United Kingdom
| Category and outbreak (yr) | No. of isolates
|
Reference or source | |||
|---|---|---|---|---|---|
| Total | Genotype of C. parvum
|
||||
| 1 | 2 | 3 | |||
| Waterborne outbreaks | |||||
| Outbreak 1 (1995) | 49 | 49a | 1a | 0 | 17 |
| Outbreak 2 (1997) | 46 | 43 | 2 | 1 | 17 |
| Sporadic cases | |||||
| Pre-autumn (1998) | 46 | 31a | 16a | 0 | 17 |
| Autumn (1998) | 194 | 74 | 120 | 0 | This study |
| Infections in livestock | |||||
| Cows | 47 | 0 | 47 | 0 | 17 |
| Sheep | 13 | 0 | 13 | 0 | 17 |
| Experimental infections in livestock (Moredun and Iowa strains) | 2 | 0 | 2 | 0 | 17 |
Both genotype 1 and genotype 2 strains were recovered from one patient.
Data suggesting that two of the three PCR-based methods used here are specific for C. parvum have been presented previously (24, 25), although the 18S rRNA gene fragment is also amplified from C. wrairi (25). Further data on the sensitivity and specificity of the PCR for the 18S rRNA sequence used here will be published elsewhere. The methods used here for isolation of DNA from whole feces are relatively simple to perform. PCR-based procedures for the detection of C. parvum in whole feces have been reported elsewhere (22, 37); however, the results presented here demonstrate that the approach used in the present study is applicable to the analysis of relatively large numbers of samples.
Fragments from three cryptosporidial genes were not amplified by PCR from DNA extracted from seven of the samples in which C. parvum oocysts were not observed by microscopy. Of the remaining 211 samples in which oocysts were observed, the sensitivities of the PCRs for the detection of the 18S rRNA, COWP, and TRAP-C1 gene fragments were 97, 91, and 66%, respectively (Table 1). This compares with sensitivities of 100% reported elsewhere from studies in which similar sample preparation procedures were used (22, 37), although those studies used different target genes and tested only 29 and 25 samples containing C. parvum, respectively.
The 18S rRNA, COWP, or TRAP-C1 gene fragments were not amplified from DNA preparations from seven samples in which oocysts were seen by microscopy. Possible reasons for this are degradation of the DNA during storage, copurification of inhibitors of the PCR, the extraction of no or an insufficient amount of cryptosporidial DNA, the presence of no specific target DNA, or the presence of other organisms which produce bodies morphologically similar to cryptosporidial oocysts and which also react with the MAb used here. We believe that degradation of the DNA is unlikely since it has been established in this laboratory that cryptosporidial DNA can be successfully isolated from whole feces by the methods described here after storage at room temperature for 6 weeks or at 4°C for >4 years. We believe that the copurification of inhibitors is also unlikely since, previously, PCR-positive cryptosporidial DNA seeded into extracts from all seven PCR-negative, microscopy-positive samples resulted in successful amplification of the COWP, TRAP-C1, and 18S rRNA sequences by PCR (data not shown). We are investigating further improvements to the DNA extraction and isolation techniques as well as improvements to the means of identification of the organisms by use of additional phenotypic characters, reaction with additional antibodies, and PCR analysis with primers for amplification of other cryptosporidial genes. The results of these studies will be presented elsewhere. DNA sequences for a range of Cryptosporidium species are becoming available (23), and this information will lead to an understanding of the possible role of Cryptosporidium species (other than C. parvum) in human disease.
The 18S rRNA genes occur as five copies in the entire cryptosporidial genome (15), and the TRAP-C1 and COWP genes are present as single copies only (27). The copy number is probably reflected in the increased sensitivity of the 18S rRNA-specific PCR procedure compared to those of the PCRs specific for the TRAP-C1 and COWP genes. Experiments with the seeding of naked DNA into feces has shown that the use of extended periods of agitation in the mini-Beadbeater leads to poor PCR amplification. Therefore, the DNA extraction method described here probably relies on a compromise between the transfer of sufficient mechanical energy to achieve disruption of the oocyst but no excessive fragmentation of the DNA. The sensitivities of all three PCR procedures increased as the numbers of oocysts originally observed by microscopy increased (Table 1), and hence, an increase in sensitivity (at least for the PCR-RFLP genotyping procedure) is desirable. Alternative higher-copy-number gene targets (13) or a nested PCR procedure (37) may increase the sensitivity of the tests, and we are developing a nested PCR for the COWP gene fragment with the aim of attaining at least the sensitivity of the 18S rRNA procedure described here to establish the genotype of C. parvum.
The common human host of the two genotypes of C. parvum, together with their, albeit occasional, occurrence together in the feces of the same individual (24) and the invariable segregation of the COWP and TRAP-C1 genotypes (Table 2), suggests that these two genes either are very closely genetically linked or are not reassorted as a result of genetic recombination. It has recently been shown, however, by two independent studies that the TRAP-C1 gene is located on chromosome I of C. parvum and that the COWP gene is located on chromosome VI (27, 31). This evidence further supports the supposition that the two genotypes of C. parvum represents two reproductively isolated populations which are (or which are behaving as) different species.
The demonstration of significantly different numbers of oocysts in the feces of individuals infected with different genotypes of C. parvum should be interpreted with caution; however, results from repeated blind tests suggest that these results are independent of misclassification bias. The numbers of oocysts clearly varies during the course of infection, and it was not possible to establish the time of onset or course of infection for the samples collected for the present study. However, since C. parvum genotype 1 appears to be host specific for humans, this genotype might be expected to achieve a higher oocyst density in the human gastrointestinal tract than C. parvum genotype 2. Recent studies that used experimental animal infection and growth in in vitro tissue culture (35) demonstrated biological differences between the genotypes, although there was additional intragenotypic variation. Biological differences in the behaviors of the two genotypes of C. parvum may have considerable influences in the epidemiology of this disease, including the potential for transmission.
Since the first recognition of Cryptosporidium as a human pathogen in 1973, great advances have been made in the understanding of the epidemiology of human cryptosporidiosis (9). However, with the present state of knowledge of the epidemiology of this disease, the significance of the different distributions of the two C. parvum genotypes reported here (Table 3 and Fig. 1) and elsewhere (34) is not clear. It is possible that the distribution of the genotypes is a laboratory artifact and, for example, reflects differences in our ability to extract and amplify DNA from the two genotypes. Results of experiments performed in the Food Hygiene Laboratory do not support this proposition. Alternatively, the distributions may reflect differences in the biological properties of the two genotypes. This is certainly possible since material for both in vitro and in vivo tests is almost always derived from livestock with experimental infections, and therefore, almost all laboratory data (i.e., survival in the environment and during water and food processing, sensitivity to disinfectants, infectivity, etc.) reflect the properties of C. parvum genotype 2. The different host ranges between the two genotypes would suggest different transmission cycles which are not yet fully elucidated, but it might also reflect nonwaterborne routes such as transmission through food. The reasons for the differences in the distributions between the genotypes between regions are not clear. Although this may reflect the distribution of rural areas and the distribution of animals in England, it may be of note that the areas, such as the north and southwest of England, where a greater proportion of surface water is used in the public water supply (6) coincide with the regions with the highest proportion of C. parvum genotype 2 (Fig. 1). It may be of note that the large 1995 genotype 1 waterborne outbreak (outbreak 1, Table 3) occurred in the southwest region; in contrast, sporadic cases collected from this region in the second half of 1989 predominantly yielded genotype 2 (Fig. 1).
We previously reported (24) that almost all the patients involved in two large waterborne cryptosporidiosis outbreaks were infected with C. parvum genotype 1 (Table 3). Subsequent characterization of C. parvum from a further four smaller suspected waterborne outbreaks showed that one of these was due to C. parvum genotype 2, but the three remaining outbreaks were due to genotype 1 (27a). Hence, C. parvum from human feces is the likely source of contamination of water in five of these outbreaks. It is not clear why there should be such a large difference in the distributions of the two C. parvum genotypes between sporadic cases and individuals involved in waterborne outbreaks. The application of genetic characterization of C. parvum, together with the use of typing systems with greater discriminatory abilities (19) and field epidemiology, is likely to vastly change our understanding of the transmission and natural history of cryptosporidiosis and lead to more rational approaches to the control of this disease.
ACKNOWLEDGMENTS
We thank colleagues in clinical microbiology laboratories for the donation of specimens, A. J. Lawson (Molecular Biology Unit, Central Public Health Laboratory, PHLS) for initial assistance in the PVP DNA purification method, and A. V. Swan (PHLS Statistics Unit) for statistical advice.
One of us (S.P.-D.) is funded by Biomed Grant PL 962557 from the European Commission and is jointly supervised by the Imperial College of Science and Technology and Medicine, London, United Kingdom.
REFERENCES
- 1.Arrowood M J. Diagnosis. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press, Inc.; 1997. pp. 43–64. [Google Scholar]
- 2.Awad-El-Kariem F M, Robinson H A, Dyson D A, Evans D, Wright S, Fox M T, McDonald V. Differentiation between human and animal strains of Cryptosporidium parvum using isoenzyme typing. Parasitology. 1995;110:129–132. doi: 10.1017/s0031182000063885. [DOI] [PubMed] [Google Scholar]
- 3.Awad-El-Kariem F M, Robinson H A, Petry F, McDonald V, Evans D, Casemore D. Differentiation between human and animal isolates of Cryptosporidium parvum using molecular and biological markers. Parasitol Res. 1998;84:297–301. doi: 10.1007/s004360050399. [DOI] [PubMed] [Google Scholar]
- 4.Bonnin A, Fourmaux M N, Dubremetz J F, et al. Genotyping human and bovine isolates of Cryptosporidium parvum by polymerase chain reaction-restriction fragment length polymorphism analysis of a repetitive DNA sequence. FEMS Microbiol Lett. 1996;137:207–211. doi: 10.1111/j.1574-6968.1996.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 5.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchier I. Cryptosporidium in water supplies: third report of the group of experts. London, United Kingdom: Her Majesty’s Stationery Office; 1998. [Google Scholar]
- 7.Carraway M, Tzipori S, Widmer G. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl Environ Microbiol. 1996;62:712–716. doi: 10.1128/aem.62.2.712-716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carraway M, Tzipori S, Widmer G. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect Immun. 1997;65:3958–3960. doi: 10.1128/iai.65.9.3958-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casemore D P, Wright S E, Coop R L. Cryptosporidiosis: human and animal epidemiology. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press, Inc.; 1997. pp. 65–92. [Google Scholar]
- 10.Casemore D P. Laboratory methods for diagnosing cryptosporidiosis. J Clin Pathol. 1991;44:445–451. doi: 10.1136/jcp.44.6.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fayer R, Speer C A, Dubey J P. The general biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press, Inc.; 1997. pp. 1–41. [Google Scholar]
- 12.Hudson L, Hay F C. Practical immunology. 2nd ed. Oxford, United Kingdom: Blackwell Scientific Publications; 1980. [Google Scholar]
- 13.Khramtsov N V, Woods K M, Nesterenko M V, Dykstra C C, Upton S J. Virus-like, double stranded RNAs in the parasitic protozoan Cryptosporidium parvum. Mol Microbiol. 1997;26:289–300. doi: 10.1046/j.1365-2958.1997.5721933.x. [DOI] [PubMed] [Google Scholar]
- 14.Lawson A J, Linton D, Stanley J, Owen R J. Polymerase chain reaction detection and speciation of Campylobacter uspaliensis and C. helveticus in human faeces and comparison with culture techniques. J Appl Microbiol. 1997;83:375–380. doi: 10.1046/j.1365-2672.1997.00240.x. [DOI] [PubMed] [Google Scholar]
- 15.Le Blancq S M, Khramtsov N V, Zamani F, Upton S J, Wu T W. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 16.MacKenzie W R, Hoxie N J, Proctor M E, et al. A massive outbreak in Milwaukee of cryptosporidium infection transmitted through the public water supply. N Engl J Med. 1994;331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 17.McLauchlin J, Casemore D P, Harrison T G, Gerson P J, Samuel D, Taylor A G. The identification of Cryptosporidium oocysts by monoclonal antibody. Lancet. 1987;i:51. doi: 10.1016/s0140-6736(87)90753-7. [DOI] [PubMed] [Google Scholar]
- 18.McLauchlin J, Black A, Green H T, Nash J Q, Taylor A G. Monoclonal antibodies show Listeria monocytogenes in necropsy tissue samples. J Clin Pathol. 1988;41:983–988. doi: 10.1136/jcp.41.9.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLauchlin J, Casemore D P, Moran S, Patel S. The epidemiology of cryptosporidiosis: application of experimental sub-typing and antibody detection systems to the investigation of water-borne outbreaks. Folia Parasitol. 1988;45:83–92. [PubMed] [Google Scholar]
- 20.Morgan U M, Constantine C C, O’Donoghue P, Meloni B P, O’Brien P A, Thompson R C A. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am J Trop Med Hyg. 1995;52:559–564. doi: 10.4269/ajtmh.1995.52.559. [DOI] [PubMed] [Google Scholar]
- 21.Morgan U M, Constantine C C, Forbes D A, Thompson R C A. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J Parasitol. 1997;83:825–830. [PubMed] [Google Scholar]
- 22.Morgan U M, Pallant L, Dwyer B W, Forbes D A, Rich G, Thompson R C A. Comparison of PCR and microscopy for detection of Cryptosporidium parvum in human fecal specimens: clinical trial. J Clin Microbiol. 1998;36:995–998. doi: 10.1128/jcm.36.4.995-998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan U M, Sargent K D, Deplazes P, Forbes D A, Spano F, Hertzberg H, Elliot A, Thompson R C A. Molecular characterisation of Cryptosporidium from various hosts. Parasitology. 1998;117:31–37. doi: 10.1017/s0031182098002765. [DOI] [PubMed] [Google Scholar]
- 24.Patel S, Pedraza-Díaz S, McLauchlin J, et al. The molecular characterisation of Cryptosporidium parvum from two large suspected waterborne outbreaks. Commun Dis Public Health. 1998;1:231–233. [PubMed] [Google Scholar]
- 25.Patel S M. Ribosomal RNA genes and RAPD for Cryptosporidium species and subspecies discrimination. Ph.D. thesis. Coventry, United Kingdom: Coventry University; 1997. [Google Scholar]
- 26.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S L, MacKenzie W R, Lal A A, Beard C B. Genetic polymorphisms among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piper M, Bankier A T, Dear P H. A HAPPY map of Cryptosporidium parvum. Genome Res. 1998;8:1299–1307. doi: 10.1101/gr.8.12.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Public Health Laboratory Service (London, United Kingdom). Unpublished data.
- 28.Sogin M L. Amplification of ribosomal RNA genes for molecular evolutionary studies. In: Innis M A, Gelfand D A, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 307–314. [Google Scholar]
- 29.Spano F, Putignani L, McLauchlin J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 30.Spano F, Putignani L, Naitza S, Puri C, Wright S, Crisanti A. Molecular cloning and expression analysis of a Cryptosporidium parvum gene encoding a new member of the thrombospondin family. Mol Biochem Parasitol. 1998;92:147–162. doi: 10.1016/s0166-6851(97)00243-0. [DOI] [PubMed] [Google Scholar]
- 31.Spano F, Putignani L, Crisanti A, Sallicandro P, Morgan U M, Le Blancq S, Tchack L, Tzipori S, Widmer G. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sulaiman I M, Xiao L, Yang C, Escalante L, Moore A, Beard C B, Arrowood J, Lal A A. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg Infect Dis. 1998;4:681–685. doi: 10.3201/eid0404.980424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasquez J R, Gooze L, Kim K, Gut J, Peterson C, Nelson R G. Potential antifolate resistance determinants and genetic variation in the bifunctional antifolate reductase-thymidylate synthase gene from human and bovine isolates of Cryptosporidium parvum. Mol Biochem Parasitol. 1996;79:153–156. doi: 10.1016/0166-6851(96)02647-3. [DOI] [PubMed] [Google Scholar]
- 34.Widmer G. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv Parasitol. 1998;40:223–239. doi: 10.1016/s0065-308x(08)60122-0. [DOI] [PubMed] [Google Scholar]
- 35.Widmer G, Tzipori S, Fichtenbaum C J, Griffiths J K. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J Infect Dis. 1998;178:834–840. doi: 10.1086/515373. [DOI] [PubMed] [Google Scholar]
- 36.Willocks L, Crampin A, Milne L, et al. A large outbreak of cryptosporidiosis associated with a public water supply from a deep chalk borehole. Commun Dis Public Health. 1998;1:239–243. [PubMed] [Google Scholar]
- 37.Zhu G, Marchewka M J, Ennis J G, Keithly J S. Direct isolation of DNA from patient stool samples for polymerase chain reaction detection of Cryptosporidium parvum. J Infect Dis. 1998;177:1443–1446. doi: 10.1086/517834. [DOI] [PubMed] [Google Scholar]