Abstract
The rigid core of intracellular tau filaments from Alzheimer's disease (AD), Pick's disease (PiD), and Corticobasal disease (CBD) brains has been shown to differ in their cryo-EM atomic structure. Despite providing critical information on the intimate arrangement of a fraction of htau molecule within the fibrillar scaffold, the cryo-EM studies neither yield a complete picture of tau fibrillar assemblies structure nor contribute insights into the surfaces that define their interactions with numerous cellular components. Here, using proteomic approaches such as proteolysis and molecular covalent painting, we mapped the exposed amino acid stretches at the surface and those constituting the fibrillar core of in vitro-assembled fibrils of human htau containing one N-terminal domain and three (1N3R) or four (1N4R) C-terminal microtubule-binding repeat domains as a result of alternative splicing. Using limited proteolysis, we identified the proteolytic fragments composing the molecular “bar-code” for each type of fibril. Our results are in agreement with structural data reported for filamentous tau from AD, PiD, and CBD cases predigested with the protease pronase. Finally, we report two amino acid stretches, exposed to the solvent in 1N4R not in 1N3R htau, which distinguish the surfaces of these two kinds of fibrils. Our findings open new perspectives for the design of highly specific ligands with diagnostic and therapeutic potential.
Keywords: tau fibrils, tauopathies, protein conformation, structural proteomics, mass spectrometry, limited proteolysis, chemical surface modification
Abbreviations: AD, Alzheimer's disease; AEBSF, 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride; AGD, argyrophilic grain disease; CBD, corticobasal degeneration; CID, collision-Induced dissociation; cryo-EM, cryo-electron microscopy; DTT, 1,4-dithiotheritol; FDR, false discovery rate; htau, human tau; MALDI-TOF-TOF, matrix assisted laser desorption/ionization-time-of-flight-time-of-flight; MS, mass spectrometry; MS2, fragmentation mass spectrum; nanoLC-MS/MS, nanoflow liquid chromatography combined to tandem mass spectrometry bromide; NHS-biotin, N-hydroxysulfosuccinimide biotin; N-TOP, N-terminal-oriented proteomics; PBS, phosphate buffered saline; PiD, Pick's disease; PSP, progressive supranuclear palsy; PVDF, polyvinylidene difluoride; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; TD, tangle disease; TEM, transmission electron microscopy; TFA, trifluoroacetic acid; TMPP(, N-succinimidyloxycarbonylmethyl)-tris(2,4,6-trimethoxyphenyl)-phosphonium
Tau has been discovered as a microtubule-associated protein that is enriched in the axonal compartment (1). It binds neuronal microtubule and regulates microtubule dynamics and stability (1). Recent studies have shown intracellular sorting of tau in different subcellular compartments such as the nucleus, plasma membrane, and dendrites, suggesting distinct subcellular functions of tau and its role in neuronal physiology such as cytoskeletal dynamics, axonal transport, synaptic transmission and plasticity, nuclear transport (2, 3). In the normal adult human brain, six tau isoforms are produced by alternative mRNA splicing of the single MAPT tau gene (4, 5). MAPT gene exons 2 and 3 define whether tau lacks (0N) or bears one (1N) or two (2N) N-terminal domains, involved in modulating tau interaction with the cell membranes and tyrosine kinases (6, 7). The presence or absence of MAPT exon 10 defines whether tau contains the second (R2) of four C-terminal microtubule-binding repeats (4R) or not (3R). Similar levels of 3R and 4R tau isoforms are expressed in the normal adult human brain, with 1N3R and 1N4R most represented (8).
Intraneuronal tau inclusions are the hallmark of tauopathies, a range of neurodegenerative diseases including Alzheimer's disease (AD) (9, 10, 11). The neuronal populations where tau aggregates form and the tau isoform content in intracellular inclusions define the type of tauopathy (12, 13, 14, 15). Unlike AD and Tangle Disease (TD) where both 3R-/4R-tau are found within tau intracellular inclusions, aggregated 4R-tau isoform is predominant in progressive supranuclear palsy (PSP), corticobasal degeneration (CBD) and argyrophilic grain disease (AGD) while 3R-tau is predominant in Pick's disease (PiD) (9, 16). Evidences for the existence of distinct tau strains in different tauopathies have been brought, and it has been shown that distinct tau strains trigger well defined tauopathies by imprinting their intrinsic structure in vitro and in vivo to monomeric tau (17, 18, 19, 20, 21, 22). Altogether, these findings have led to the view that the aggregation, spread, and amplification of fibrillar tau assemblies from cell to cell within the central nervous system contribute to the progression of tauopathies.
Recent reports have brought insight into the rigid structure of the protease resistant core of distinct tau filaments purified from the brain of individuals with either AD (23, 24, 25), CBD (25, 26), or PiD (27). Tau protease resistant core was shown to span amino acid residues 304–380 (2N4R tau numbering) in AD, 274–380 in 4R htau fibrils from CBD cases, and 254–378 in 3R htau fibrils from PiD (28). The structures, determined by cryo-electron microscopy (cryo-EM), define the architecture of fibrillar tau isoforms and the amino acid residues that are excluded from the surface of the pathogenic assemblies. Despite being essential, the cryo-EM structures neither provide a complete picture of the structure nor information on fibrillar tau amino acid residues stretches that are exposed to the solvent (29). Such information is critical as fibrillar tau surfaces play a crucial role in tau fibrils binding to neurons, a key step in their prion-like propagation process.
Information pertaining to the amino acid residues constituting the fuzzy coat of pathogenic tau filaments comes from immuno-gold staining. Antibodies directed against tau R1 and R2 domains (first and second microtubule-binding repeats, respectively) have been reported to bind tau fibrils prior to pronase or trypsin treatment, suggesting they are accessible (23, 30, 31, 32).
Here we map the amino acid residue stretches that constitute human tau (htau) 1N3R and 1N4R fibrillar core and those that are exposed at the surface of the fibrillar form of the two tau isoforms using two complementary mass spectrometry (MS)-based proteomic approaches we implemented. The first, relying on limited proteolysis by the protease GluC, allowed us to identify the amino acid residue stretches constituting the core of htau 1N3R and 1N4R fibrils and establish a specific molecular “bar-code” for the fibrillar forms of the two tau isoforms. The second relying on molecular painting through covalent modification with NHS-biotin allowed us to identify amino acid residue stretches that are exposed at the surface of htau 1N3R and 1N4R fibrils. We demonstrate that the structure of the fibrillar form of full-length 1N3R and 1N4R htau is different from that previously reported for fibrillar 2N3R and 2N4R htau isoforms assembled in the presence of heparin (33). We also show that the structure of the fibrils we made of 1N3R and 1N4R recombinant tau resembles that of pathogenic fibrillar assemblies isolated from the brains of patients who developed AD, PiD, and CBD, in contrast with recent reports on 2N3R and 2N4R tau fibrils generated under different experimental conditions (33). The resolution we reached is unprecedented to our best knowledge. It allows identifying similarities and differences between 1N3R and 1N4R htau fibrils. Tau N-terminal and C-terminal ends are exposed to the solvent both in 1N3R and 1N4R fibrils, whereas the C-terminal region spanning residues 223–380 is protected (2N4R numbering). The conformation of htau 1N3R and 1N4R within the fibrils differs significantly. The differences we report are located within tau N-terminal and C-terminal domains, spanning amino acid residues 118–133 and 381–402, respectively. The demonstration that fibrillar htau 1N3R and 1N4R are structurally unlike opens the way for the design of highly specific ligands with diagnostic and therapeutic potential.
Results
Mapping of solvent-exposed surfaces in htau 1N3R and 1N4R fibrils by limited proteolysis
Fibrillar tau surfaces define their interactions with numerous cellular components ranging from proteins, to phospholipids, and the extracellular matrix (34, 35). Tau 1N3R and 1N4R are the most abundant tau isoforms expressed in adult brain, representing 26% and 23% of tau proteins in the adult human cortex, respectively (8). We therefore implemented a structural MS-based proteomics approach to map full-length human recombinant htau 1N3R and 1N4R fibrils solvent-accessible amino acid stretches. Proteolysis has been used to identify or generate highly structured, solvent-protected domains in tau assemblies for biochemical and structural studies (24, 32, 33, 36). Limited proteolysis provides information on tau surfaces that are exposed to the solvent and accessible to proteases (37). 1NR3 and 1N4R htau (40 μM) in monomeric and fibrillar forms (Fig. 1, A and B) were subjected to proteolysis. Seven specific proteolytic enzymes (Trypsin, Chymotrypsin, GluC, AspN, LysC, LysN, ArgC) whose theoretical cleavage sites are distributed along tau amino acid sequence were used.
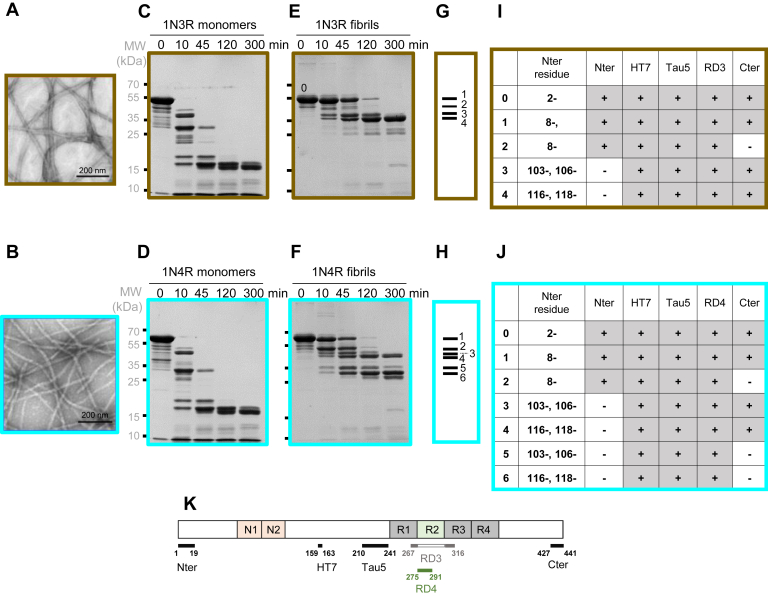
Figure 1.
Limited proteolysis of 1N3R and 1N4R htau monomers and fibrils using the endoproteinase GluC. Negatively stained transmission electron micrographs (TEMs) of 1N3R (A) and 1N4R (B) htau fibrils. GluC proteolytic degradation patterns of 1N3R and 1N4R htau (40 μM monomer concentration) monomers (C and D, respectively) and fibrils (E and F, respectively) were monitored over time on Coomassie stained SDS-PAGE (12%). Time (min) and molecular weight markers (MW, kDa) are indicated on the top and left side of the gels. The most prominent proteolytic fragments generated upon degradation of 1N3R and 1N4R htau fibrils at time 45 min are represented as bar codes made of four or six bars, the thickness of which reflects the relative intensity of each proteolytic fragment for 1N3R and 1N4R htau, respectively (G and H). Characterization of fibrillar 1N3R and 1N4R htau polypeptides prior to (band 0) or after GluC treatment for 45 min (bands 1–4 [I] and 1 to 6 [J] from fibrillar 1N3R and 1N4R htau, respectively). All polypeptides N-termini were identified by both Edman sequencing and MS after in-gel digestion (with and without N-terminal TMPP-labeling). The proteolytic fragments were also identified by Western blot analysis using the antibodies Nter, HT7, Tau5, RD3, RD4, and Cter, the epitopes of which span the amino acid stretches 1–19, 159–163, 210–241, 267–316, 275–291, and 427–441 as depicted (K).
The proteolytic patterns analyzed by SDS-PAGE we obtained led us to select the endoprotease GluC for detailed characterization of the cleavage sites using an enzyme-to-monomeric and fibrillar htau 1N3R and 1N4R ratio of 1:150 (w/w) after 10, 45, 120, and 300 min digestion. No major difference was observed upon comparing the proteolytic patterns of monomeric 1N3R and 1N4R htau (Fig. 1, C and D). We conclude from this observation that monomeric htau 1N3R and 1N4R solvent accessibility is similar with complete degradation of full-length tau as early as 10 min after addition of GluC. In contrast, the proteolytic patterns of fibrillar 1N3R and 1N4R htau differed significantly (Fig. 1, E and F). The very distinctive proteolytic patterns we report at time 45 min for fibrillar 1N3R and 1N4R htau reflect conformational differences that can be schematized as molecular bar codes (Fig. 1, G and H). GluC treatment of fibrillar 1N3R htau generated four proteolytic fragments with apparent molecular weights 55–35 kDa (labeled 1–4 in Fig. 1G), whereas six polypeptides with apparent molecular weights 60–25 kDa were observed for 1N4R htau fibrils (labeled 1–6 in Fig. 1H).
We identified the N-termini of the proteolytic polypeptides resulting from GluC cleavage after E residues exposed to the solvent in fibrillar 1N3R and 1N4R htau by classical in-gel digestion combined to MS identification, Edman sequencing, and the N-TOP (N-Terminal-Oriented Proteomics) strategy. The latter is based on chemical derivatization of the N-terminus of the polypeptides fragments isolated on SDS-PAGE with TMPP ((N-succinimidyloxycarbonylmethyl)- tris(2,4,6-trimethoxyphenyl)- phosphonium bromide) (38). The results are presented in Figure 1, I and J. For the sake of clarity and to facilitate comparison between tau isoforms, all the data presented in this work follow the 2N4R amino acid numbering. Thus, as the second N-terminal repeat (amino acid D74-T102) lacks in 1N3R and 4R tau, the amino acid residue preceding A103 is E73 in Figure 2, Figure 3, Figure 4, Figure 5, Figure 6. Proteolytic polypeptides with the N-terminal amino acid residues F8, A103, A106, D116, and A118 were identified upon treatment of both fibrillar 1N3R and 1N4R htau by GluC (Fig. 1, I and J). They result from cleavages at the solvent-accessible residues E7 (polypeptide bands 1 and 2 for both 1N3R and 1N4R), E73 and E105 (polypeptide bands 3 for 1N3R, and 3 and 5 for 1N4R), and E115 and E117 (polypeptide bands 4 for 1N3R, and 4 and 6 for 1N4R). MS analysis of the fragments generated from 10 min GluC digestion clearly confirms that E7, E73, E105, E115, and E117 are accessible to solvent both in 1N3R and 1N4R (Fig. S1). Less abundant cleavages at residues E9, E12, and E45 were detected by MS, not by Edman sequencing. Interestingly, E36, E53, E57, E58, E62 appear not as exposed to the solvent at the surface of 1N3R and 1N4R htau fibrils as an amino acid residues stretch of over 16 residues spanning E73, E105, E115, and E117. These results suggest that the N-terminal domains of 1N3R and 1N4R htau are folded similarly within the fibrillar form of both proteins. We further characterized the GluC cleavage products using antibodies directed against epitopes spanning over 1N3R and 1N4R htau primary structure (depicted in Fig. 1K): N-ter [residues 1–19], HT7 [residues 159–163], Tau5 [residues 210–241], RD3 [residues 267–316, specific to 3R htau], RD4 [residues 275–291, specific to 4R htau] and C-ter [residues 427–441]. Combining the sequencing and immuno-characterization data allows defining the N- and C-terminal boundaries of fibrillar 1N3R and 1N4R htau GluC cleavage products (Figs. 1, I and J and S2). All fibrillar 1N3R htau-derived polypeptide bands, with the exception of the minor band 2, retain their C-terminal end (Fig. 1I), whereas three out of six fibrillar 1N4R htau-derived polypeptide bands lack their C-terminal end (Fig. 1J, bands 2, 5, and 6 and Fig. S2). We further assessed the identity of each polypeptide by nanoLC-MS/MS after in-gel tryptic digestion. Fibrillar 1N3R htau proteolytic product bands 1, 3, and 4 exhibit two C-terminal ends corresponding to residues E431 and L441. Fibrillar 1N4R htau GluC proteolytic product bands 1, 3, and 4 also present two C-terminal ends (residues E431and L441), while bands 2, 5, and 6 end at residue E372. Solvent accessibility of the C-terminal region of 1N4R was confirmed by MS analysis of the proteolytic products from 10 min GluC digestion indicating both efficient cleavage at E372 and additional cleavage sites in the C-terminal region spanning from T373 to L441 at residues E380, E391, and E431 (Fig. S1). Interestingly MS analysis showed no cleavage within tau 1N3R C-terminal region after 10 min GluC digestion.
Figure 2.
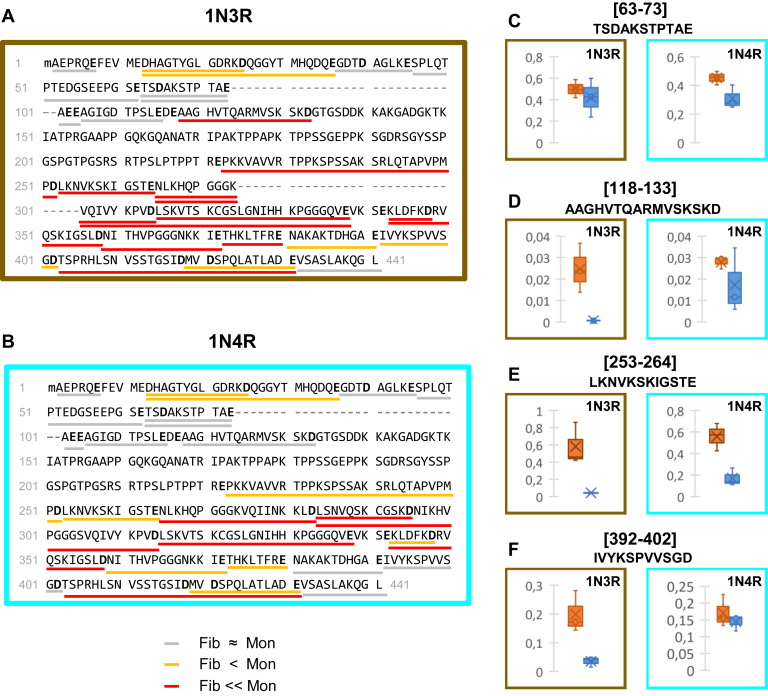
Changes in the exposure to the solvent assessed through GluC accessibility upon assembly of monomeric 1N3R and 1N4R htau into fibrils.The proteolytic peptides resulting from in-solution digestion of monomeric and fibrillar htau 1N3R (A) and 1N4R (B) by GluC were identified and quantified by nanoLC-MS/MS. Changes in enzyme accessibility were measured by calculating the fold changes (Fc) between fibrillar (F) and monomeric (M) htau, for each GluC proteolytic peptide (FcF/Mpeptide 1 = Intensity Fpeptide 1/Intensity Mpeptide 1 using ion intensities normalized to the total amount of htau), for both 1N3R (A) and 1N4R (B) isoforms. Changes in the exposure to the solvent assessed through GluC accessibility upon htau assembly into fibrils are represented: peptides with 0.1 < Fc F/M ≤ 0.55 (underlined in orange) are at least 1.8 times less abundant in fibrillar samples; peptides with FcF/M ≤ 0.1 (underlined in red) are at least ten times less abundant or not even detected after proteolysis of fibrils. Peptides at least as abundant in GluC treated htau monomers and fibrils are considered as accessible and underlined in gray (FcF/M > 0.55). The cleavage sites delineating each quantified peptides are in bold. The complete sequence coverage and the changes measured for each individual polypeptide are presented in supplementary information (Figs. S3 and S4, Tables S1 and S2). Changes in the accessibility of GluC upon htau 1N3R (brown panel) and 1N4R (cyan panel) assembly of monomers (orange) into fibrils (blue) (C–F). The normalized intensities of representative GluC peptides from n = 3 experiments are represented as boxplots: (C), 63–73 (no significant change), (D), 118–133 (no change for 1N4R, significant change for 1N3R), (E), 253–264 (significant change for both 1N3R and 1N4R, but to different extents), and (F), 392–402 (no change for 1N4R, significant change for 1N3R). The data for all the other peptides are presented in Figure S4, a and b.
Figure 3.
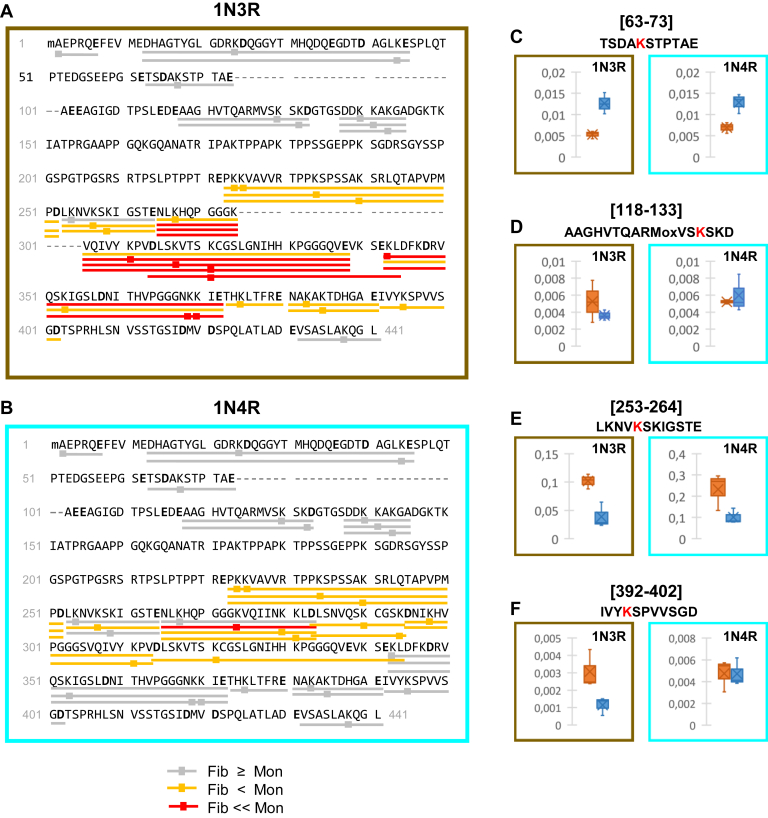
Changes in lysine residues exposure to the solvent upon assembly of monomeric 1N3R and 1N4R htau into fibrils. To measure changes in the accessibility of lysine residues and the N-terminal residue, 1N3R (A) and 1N4R (B) htau monomers and fibrils were exposed in solution to NHS-Biotin. The samples were next denatured, ran on SDS-PAGE, and the protein bands were subjected to a complete in-gel digestion with GluC and AspN. The resulting biotinylated peptides and the biotinylation sites were identified and quantified by nanoLC-MS/MS. Changes in lysine accessibility were measured by calculating the fold changes of biotinylated peptide intensity between fibrillar (F) and monomeric (M) 1N3R (A) and 1N4R (B) Tau: (FcF/M Biotinylated Peptide 1= IntensityF Biotinylated Peptide 1/IntensityM Biotinylated Peptide 1 using ion intensities normalized to the total amount of htau). The changes in the exposure to the solvent assessed through Lysine residues biotinylation upon htau assembly into fibrils are represented. Peptides where lysine residues are biotinylated to at least a similar extent in monomeric and fibrillar htau (Fc > 0.55) considered as accessible are underlined in gray; those where lysine residues are biotinylated to a significant lesser extent (0.1 < Fc ≤ 0.55) or not accessible (Fc ≤ 0.1) in fibrillar htau peptides are underlined orange and red, respectively. The biotinylated lysine residues are marked by squares. The complete sequence coverage and the changes measured for each individual polypeptide are presented in supporting information (Figs. S8–S10 and Tables S3 and S4). Changes in Lys residues biotinylation upon htau 1N3R (brown panel) and 1N4R (cyan panel) assembly of monomers (orange) into fibrils (blue) (C–F). The normalized intensities of representative GluC peptides from n = 3 experiments are represented as boxplots: (C), 63–73 (no significant change), (D), 118–133 (no change for 1N4R, significant change for 1N3R), (E), 253–264 (significant change for both 1N3R and 1N4R, but to different extents), and (F), 392–402 (no change for 1N4R, significant change for 1N3R). The data for all the other peptides are presented in Figure S10, a and b.
Figure 4.
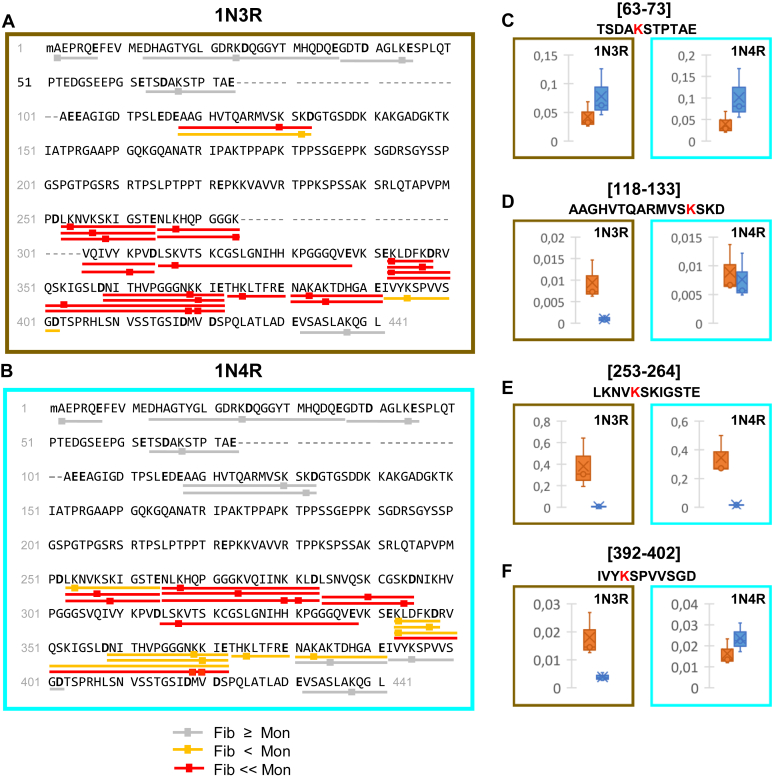
Changes in the exposure to the solvent assessed through GluC accessibility and lysine residues biotinylation upon assembly of monomeric 1N3R and 1N4R htau into fibrils. Changes in the accessibility of GluC and in the biotinylation of lysine residues and the N-terminal amino acid residue upon assembly of 1N3R (A) and 1N4R (B) hau into fibrils were obtained after in-solution digestion of biotinylated htau, and comparison of the intensities of biotinylated GluC peptides generated from htau fibrils or monomers, as described in Figure 3. The resulting biotinylated peptides, together with their site(s) of biotinylation(s) (squares), were identified and quantified by nanoLC-MS/MS. The changes in the exposure to the solvent assessed using GluC cleavage and lysine residues biotinylation upon htau assembly into fibrils are represented following the same color code used in Figure 3. The complete sequence coverage and the changes measured for each individual polypeptide are presented in supplementary information (Figs. S11 and S12 and Tables S5 and S6). Changes in the accessibility of GluC and surface labeling upon htau 1N3R (brown panel) and 1N4R (cyan panel) assembly of monomers (orange) into fibrils (blue) (C–F). The normalized intensities of representative GluC peptides from n = 3 experiments are represented as boxplots: (C), 63–73 (no significant change), (D), 118–133 (no change for 1N4R, significant change for 1N3R), (E), 253–264 (significant change for both 1N3R and 1N4R, but to different extents), and (F), 392–402 (no change for 1N4R, significant change for 1N3R). The data for all the other peptides are presented in Figure S12, a and b.
Figure 5.
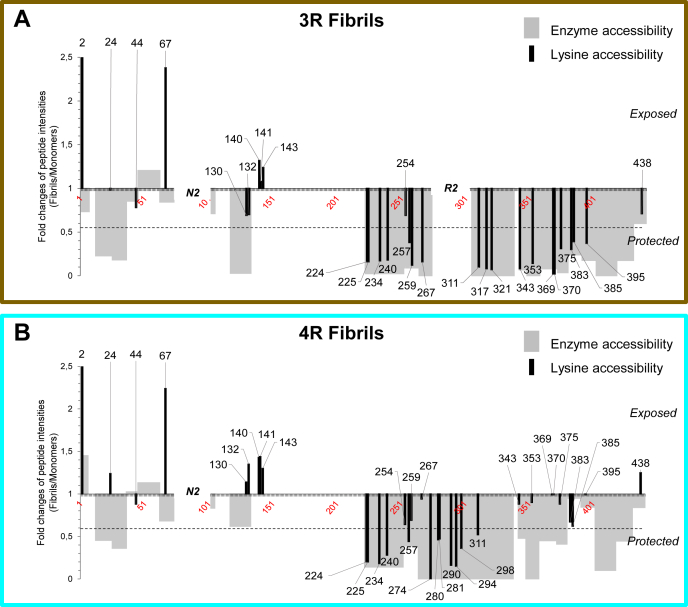
Changes in the exposure of 1N3R and 1N4R htau to the solvent upon assembly into fibrils. Graphic representation of the changes in solvent accessibility upon assembly of 1N3R (A) and 1N4R (B) htau into fibrils. The primary structure and amino acid residues numbering used is that of 2N4R htau. The gray bars represent the changes in GluC accessibility only (derived from Fig. 2). The black lines represent the changes in lysine and the N-terminal amino acid residue biotinylation only (derived from Fig. 3). The fold changes (Fc F/M) correspond to the ratio of the intensity (normalized to the total amount of htau protein) of peptides from fibrillar (F) htau relative to the intensity of the same peptides from monomeric (M) htau. The additional N-terminal domain N2 (spanning from D74 to T102) and microtubule-binding repeat R2 (spanning from residues V275 to S305) specific to 1N4R htau locations within the primary structure are indicated. As a consequence of the primary sequence difference between 1N3R and 1N4R htau, K317 and K321 were detected only for 1N3R htau, and K274, K280, K281, K290, K294, K298 only for 1N4R htau. The dotted lines indicate the threshold of significance for fold changes 0.55 and 1.8, corresponding to peptides or residues, exposure or protection of which is increased by a factor 1.8 or more in htau fibrils compared to monomers. For a good visibility of all peptides, the scale has been cropped over 2.5 a.u, for fold changes in accessibility to biotinylation of peptides [2–7], with fold changes of 3.44 and 2.63 for 1N3R and 1N4R htau, respectively.
Figure 6.
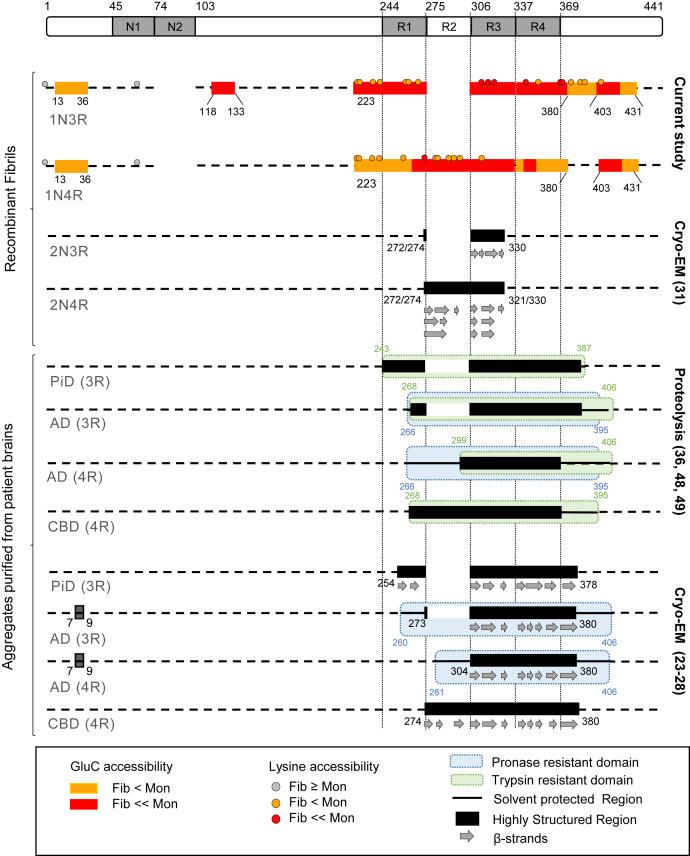
Comparison of all structural data obtained to date for distinct htau fibrillar assemblies. The primary structure of the longest htau isoform (2N4R) is represented at the very top. Structural data obtained for htau fibrils assembled in the presence of heparin are shown in the top part of the figure. First are the solvent protected regions identified in the current study for 1N3R and 1N4R htau fibrils, using two complementary approaches for assessing surface accessibility (orange and red boxes/circles correspond to respectively significantly and highly protected regions/Lysines identified by enzyme accessibility/NHS-biotin labeling). Data obtained by cryo-EM for htau fibrils assembled in the presence of heparin are summarized next (33). Data reported for different htau filaments purified from AD, PiD, and CBD patient's brain are summarized in the bottom part of the figure. Structural information obtained by proteolysis and identification of proteolysis products using N-terminal sequencing, MS analysis and immunodetection are presented above (36, 48, 49) data obtained by cryo-EM using the pronase resistant core of htau filaments (23, 24, 25, 26, 27, 28).
Identification of protease-resistant polypeptides within htau 1N3R and 1N4R fibrils by proteolysis
To further define the architecture of htau fibrils, we exposed monomeric and fibrillar 1N3R and 1N4R htau, in solution, to GluC for 15 h at an enzyme-to-protein ratio 1:50 (w:w). No proteolytic fragment with apparent molecular weights over 15 kDa was visible upon SDS-PAGE analysis after proteolysis (not shown). All proteolytic polypeptides below 6 kDa were next identified and quantified by nanoLC-MS/MS analysis. First, proteolytic polypeptides were identified by tandem mass spectrometry and a database search using the 1N3R and 1N4R htau primary structures. The primary structure coverages were 75% and 74% for monomeric 1N3R and 1N4R htau respectively, 48% and 62% for fibrillar 1N3R and 1N4R htau, respectively (Fig. S3). We next quantified each identified GluC proteolytic polypeptide extracted ion chromatogram intensity of identical GluC polypeptides ion peaks in the, matched across samples and replicates, using their fragmentation mass spectrum (MS2) spectrum, m/z, and retention time. For each sample, the MS intensity of each GluC peptide was normalized to the amount of total tau protein using the intensity of the highly accessible, both in 1N3R and 1N4R htau fibrils, GluC peptide A106-E117 as reference as previously reported (39). Enzymatic cleavage efficiency was determined, in experiments performed in triplicate, by expressing the average MS intensity of each normalized GluC peptide generated from tau fibrils to the one generated from tau monomers.
The fold changes (Fc) in enzyme accessibility upon assembly of htau monomers into fibrils were calculated by dividing the normalized MS intensity of fibrillar htau GluC-derived polypeptides by the normalized MS intensity of monomeric htau GluC-derived polypeptides (Fc F/M peptide = Intensity Fib peptide/Intensity Mono peptide). We sorted the polypeptides in two categories: the first category gathers polypeptides 1.8-fold less abundant upon GluC proteolysis of 1N3R and 1N4R htau fibrils than monomers (0.1 < Fc F/M ≤ 0.55, in orange); the second category represents polypeptides 10-fold less abundant upon GluC proteolysis of 1N3R and 1N4R htau fibrils than monomers or even not detected (Fc F/M ≤ 0.1, in red). Polypeptide intensities and the corresponding fold changes Fc F/M are given in Tables S1 and S2. The accessibility to the solvent of the identified polypeptides is represented in Figure 2. Polypeptides as accessible to GluC in fibrillar and monomeric 1N3R and 1N4R htau are underlined in gray. Those slightly less or much less accessible in htau fibrils are underlined in orange and red, respectively. Changes measured for each individual polypeptide are presented in boxplots in Figure S4, a and b. No significant difference in GluC accessibility between fibrillar and monomeric 1N3R and 1N4R htau for amino acid residues stretches A2-E7 and G37-E115 was observed. The amino acid residues stretch D13-E36 appears significantly protected from GluC cleavage both in 1N3R and 1N4R htau fibrils, as the Fc F/M of the proteolytic peptides D13-D25 and D13-E36 values are Fc F/M 1N3R [13-25] = 0.23, Fc F/M 1N3R [13-36] = 0.18; Fc F/M 1N4R [13-25]= 0.45, Fc F/M 1N4R [13-36] = 0.36 (Figs. 2 and S4a, Tables S1 and S2). These results agree with those we obtained from the limited proteolysis approach (Figs. 1 and S1). The amino acid stretch A118-D133 was remarkably protected in 1N3R, not in 1N4R htau fibrils, Fc F/M 1N3R [118-133] =0.03; Fc F/M 1N4R [118-133] = 0.62.
The 1N3R htau amino acid stretch P223-E380, comprising the second part of the htau Proline rich region, the R1, R3, and R4 repeats from the Microtubule-Binding Region (MTBR) and 12 additional amino acids after the R4 repeat, and peptide T403-E431 exhibited extreme protection (Fig. 2A). Part of the corresponding region in 1N4R htau, amino acid stretch N265-D358, spanning the second half of the R1 repeat, the R2 and R3 repeats, the first half of the R4 repeat, and the amino acid stretch T403-E431 exhibited extreme protection (Fig. 2B). Interestingly, the amino acid stretches N381-D402, N359-E380, and P223-E264 in 1N4R htau fibrils were significantly more exposed to the solvent than their counterparts in 1N3R fibrils (Fig. 2). N359-E380 and P223-E264 appeared protected to a higher extent in 1N3R fibrils as compared with 1N4R fibrils, whereas the protected N381-D402 amino acid residues stretch in 1N3R fibrils was accessible in 1N4R fibrils. Finally, the C-terminal amino acid stretch V432-L441 was exposed similarly in 1N3R and 1N4R htau fibrils. Altogether, these results bring additional insight into the differential exposure of fibrillar 1N3R and 1N4R htau stretches to the solvent.
Mapping of the surfaces of 1N3R and 1N4R htau fibrils by molecular painting
To challenge our findings and further strengthen our conclusions, we implemented a molecular painting strategy based on covalent surface modification of htau 1N3R and 1N4R fibrils with NHS-biotin and mass spectrometry analysis. Indeed, the 27 kDa protease GluC we used to assess solvent accessibility has a binding/cleavage site that spans at least ten amino acid residues (40). NHS-biotin (341 Da) that is used to footprint solvent accessibility of Lys residues is a small probe and expected to provide a higher resolution in mapping solvent-exposed surfaces of tau fibrils. Molecular painting, a footprinting method applied to analyze higher-order structures of proteins (41), has been used previously to assess surface accessibility of amino acid residues of proteins, alone or in complex, when conventional structural methods were unsuccessful (42, 43, 44).
We first optimized the labeling conditions so that a similar level of biotinylation is achieved for monomeric and fibrillar htau 1N3R and 1N4R. Biotinylation yields with increasing reaction times (1–30 min) and lysine to NHS-biotin molar ratio (40:1–1:1) were measured. For a labeling time of 10 min at a K:NHS-Biotin molar ratio of 2.5:1, the average total number of bound biotin molecules ranged from one to seven per tau molecule (Fig. S5). We next subjected labeled monomeric and fibrillar htau 1N3R and 1N4R htau to proteolysis to identify and quantify the biotinylated peptides by MS. Biotinylated monomeric and fibrillar htau 1N3R and 1N4R htau were subjected, in-gel after SDS-PAGE and in-solution under nondenaturing conditions, to digestion by GluC or AspN to distinguish surface from enzyme accessibility.
As expected, we obtained a high primary structure coverage, ranging from 79% to 92%, after GluC or AspN in-gel digestion for biotinylated and nonbiotinylated monomeric and fibrillar 1N3R and 1N4R htau (Figs. S6–S9). NHS-Biotin reacts strongly with primary amines (K residues and the N-terminus of proteins) and mildly with hydroxyl or sulfhydryl groups of T, Y, S, and C (45). In agreement with this, minimal amounts of T, Y, S, and C residues were observed, leading us to only consider htau N-terminus and K residues biotinylation. We identified and quantified 29 biotinylated amines (28 K and the N-terminus) out of 39 amines in 1N3R tau polypeptides and 34 biotinylated amines (33 K and the N-terminus) out of 44 amines in 1N4R tau polypeptides (Tables S3 and S4). Biotinylated polypeptides obtained after GluC treatment were quantified as described for Figure 2, using as reference for normalization the polypeptide A106-E117 given it contains no biotinylation site. Similarly, the polypeptide D421-A429 was used as reference for normalization of biotinylated polypeptides obtained after AspN treatment. The fold changes for each biotinylated polypeptide upon assembly of monomeric 1N3R and 1N4R htau into fibrils were obtained by dividing its normalized MS intensity in the fibrillar form of htau by that in monomeric htau (Fc F/M Biotinylated Peptide at K position= Intensity Fib Biotinylated Peptide/Intensity Mono Biotinylated Peptide). The fold changes for each biotinylated polypeptide are given in Tables S3 and S4, and the boxplots for each individual biotinylated polypeptide are presented in Figure S10, a and b.
We sorted the biotinylated polypeptides in three categories (Fig. 3). Polypeptides biotinylated at least to a similar extent in monomeric and fibrillar htau (Fc F/M > 0.55) are underlined in gray, those significantly (0.1 < Fc F/M ≤ 0.55) or extremely (Fc F/M ≤ 0.1) less biotinylated in fibrillar htau are underlined in orange and red, respectively.
Amino acid residues A2 (N-terminus) and K67 were found better exposed to the solvent and therefore reactive in 1N3R and 1N4R htau fibrils than in the monomeric form of the protein, (FC F/M 1N3R (A2) = 3.44 and FC F/M 1N4R (A2) =2.63; FC F/M 1N3R (K67) = 2.38 and FC F/M 1N4R (K67) = 2.24). This higher accessibility of some Lys residues might result from a structural rearrangement upon assembly of htau into fibrils with, as a consequence, increased flexibility and/or exposure to the solvent. However, as these peptides are accessible both in monomeric and fibrillar htau, we indexed them in accessible polypeptides category. Amino acid residues K24, K44, K130, K132, K140, K141, K143 reacted to the same extent with NHS-biotin in 1N3R and 1N4R htau fibrils and monomers suggesting that the amino acid stretches D13-E45 and A118-A145 are exposed to the solvent (Figs. 3 and S10, Tables S3 and S4). This contrasts with the finding that GluC cleavage sites E36 and D133 were significantly or extremely protected both in 1N3R and 1N4R htau fibrils (Figs. 2 and S1) and may be due to differential accessibility of a small NHS-biotin (341 Da) compared with the enzyme GluC (27 kDa and a binding site exceeding ten amino acid residues). The exposure of K residues within the amino acid residues stretch D146-E222 could not be assessed as this region contains only one GluC and one AspN cleavage site yielding long peptides. The amino acid stretch spanning residues P223-E252 was found significantly protected within htau 1N3R and 1N4R fibrils (FC F/M comprised between 0.18 and 0.28) (Fig. 3, Tables S3 and S4) in agreement with the significant or extreme protection reported for this region in Figure 2.
Very significant differences in the exposure of K residues within the region bearing the microtubule binding repeats (polypeptide stretch Q244-N368, where R1, R2, R3, and R4 span residues Q244-K274, V275-S305, V306-Q336, and V337-N368, respectively) were observed either following assembly of htau into fibrils or upon comparison of fibrillar 1N3R and 1N4R htau or both.
In the first microtubule-binding repeat (R1, spanning Q244-K274), K254 in 1N3R and 1N4R monomers and fibrils (FC F/M 1N3R (K254) = 0.69; FC F/M 1N4R (K254) = 0.64) were similarly exposed to the solvent, while K257 exhibited moderate protection, comparable in 1N3R and 1N4R htau fibrils (FC F/M 1N3R (K257) = 0.38; FC F/M 1N4R (K257) = 0.44), and K259 and K267 were significantly protected in 1N3R, not in 1N4R fibrils (FC F/M 1N3R (K259) = 0.24 and FC F/M 1N4R (K259) = 0.69; FC F/M 1N3R (K267) = 0.16 and FC F/M 1N4R (K267) = 0.94). All K residues within or surrounding the 1N4R htau-specific R2 repeat (spanning V275-S305) exhibited extreme (FC F/M (K274) = 0) or significant protection upon fibrils formation (FC F/M (K280, 281, 290, 294, 298) from 0.15 to 0.47). Regarding the R3 repeat (spanning V306-Q336), the comparison between 1N3R and 1N4R htau was made difficult as we were unable to assess the reactivity of all K residues because of differences in primary structure and distribution of D and E residues in this repeat resulting in different length of GluC proteolytic peptides for 1N3R and 1N4R. Indeed, K311, K317, and K321 appear highly protected (FC F/M 1N3R from 0.07 to 0.1) in 1N3R htau fibrils while only K311 and K321 in 1N4R fibrillar tau was detected and exhibited moderate protection upon assembly (FC F/M 1N4R (K311) = 0.52, FC F/M 1N4R (K311) = 0.31) (Fig. 3). Within or at the limit of the R4 domain (spanning V337-N368), K343, K353, K369, K370 exhibited extreme or significant protection upon assembly of 1N3R htau into fibrils (FC F/M 1N3R (K343) = 0.08; FC F/M 1N3R (K353) = 0.14; FC F/M 1N3R (K369/370) = 0.02) while the equivalent K from 1N4R did not (FC F/M 1N4R (K343) = 0.88; FC F/M 1N4R (K353) = 0.90; FC F/M 1N4R (K369/370) = 0.99). K375, K383, K385, and K395 from the C-terminal end of 1N3R htau exhibited moderate protection upon assembly of 1N3R htau into fibrils unlike the equivalent K from 1N4R htau (FC F/M 1N3R (K375, 383, 385, 395) = 0.30 to 0.39; FC F/M 1N4R (K375, 383, 385, 395) = 0.62–0.99) as shown in Fig. 3, Tables S3 and S4 and Fig. S10, a and b. Finally, K438 from the C-terminal end was as exposed in monomeric and fibrillar 1N3R and 1N4R htau, respectively, (FC F/M 1N3R (K438) = 0.71; FC F/M 1N4R (K438) = 1.25). Overall, these results agree with the protease accessibility data showing accessibility from N381 to D402 in 1N4R not 1N3R htau fibrils presented in Figures 1 and 2.
We conclude from these measurements that the N-terminal domain, the first microtubule-binding repeat, and the C-terminus are exposed similarly to the solvent in 1N3R and 1N4R htau fibrils. Highly significant differences in exposure to solvent are observed for the polypeptide stretch spanning K274-K395 in 1N3R and 1N4R htau fibrils.
Mapping of the surfaces of fibrillar 1N3R and 1N4R htau using protease accessibility after molecular painting
We next exposed biotinylated monomeric and fibrillar 1N3R and 1N4R htau in solution, under native condition to GluC for 15 h in-solution digestion, in order to assess enzyme accessibility of surface-labeled protein samples. The primary structure coverage we obtained after in-solution GluC treatment for biotinylated monomeric htau 1N3R and 1N4R was 73% while that of the biotinylated fibrillar forms of the proteins was 45% and 62%, respectively (Fig. S11). The changes in accessibility of GluC to surface-labeled proteins we measured (Tables S5 and S6 and Fig. S12, a and b) are summarized in Figure 4. The data are in excellent agreement with the results presented in Figures 2 and 3. As biotinylation of Lys residues is per se identical to the experiments presented in Figure 3 and digestion is identical to the experiments presented in Figure 2, the differences we report through this experimental strategy result predominantly from Glu residues accessibility in the native form of tau fibrils as compared with the denatured form in Figure 3 to the protease GluC and represent only differences observed for biotinylated peptides. With the exception of the polypeptide stretch A118-D133 in 1N3R fibrils, a good accessibility to solvent, as assessed by NHS-biotin accessibility, was observed for the N-terminal domains and for the C-terminus of both in 1N3R and 1N4R fibrils. The microtubule-binding repeat regions (spanning Q244-N368) exposure to the solvent decreased significantly in fibrillar 1N3R and 1N4R htau but to different extents, 1N3R fibrils exhibiting an extreme protection. In addition, the polypeptide stretch spanning K375-K395 was much less exposed to the solvent in 1N3R than 1N4R htau fibrils.
Discussion
Spreading of tau aggregates and tau pathology in the brain through a prion-like mechanism involves several crucial steps including abnormal tau aggregation, multiplication of the aggregates by recruitment of endogenous tau, and the spread of tau seeds from a donor to a recipient cell, which requires binding of tau seeds to a naïve cell and their uptake (34, 46). Identifying fibrillar tau surfaces involved in fibrils interaction with partner molecules, at the surface or within the cell, is key for a better understanding of tau aggregates prion-like propagation and the changes in cell proteostasis they trigger upon multiplication. We implemented a combination of MS-based proteomic methods to identify the surfaces of full-length 1N3R and 1N4R htau fibrils assembled in vitro in the presence of heparin.
Limited proteolysis of 1N3R and 1N4R htau fibrils with GluC yields different proteolytic profiles comparable to molecular “bar-codes” characteristic of each type of fibrils. The identification of the proteolytic polypeptides revealed two regions accessible to the proteases to the same or different extents within fibrillar htau 1N3R and 1N4R. The N-terminal amino-acid stretch and the end of tau N1 projection domain spanning residues 2–10 and 103–118, respectively, together with the C-terminus corresponding to the amino-acid stretch 432–441 were found highly accessible to GluC. The exposure to the solvent of these two parts within fibrillar 2N4R tau has been reported based on hydrogen-deuterium exchange and mass spectrometry measurements (47). In contrast, the C-terminal moiety spanning the microtubule binding repeats and the C-terminal regions was found poorly accessible. In addition, significant differences in the accessibility of GluC were observed within htau 3R and 4R C-terminal moieties.
To further delineate protected regions at the best possible resolution, we further quantified proteolytic peptides generated by advanced proteolysis using mass spectrometry and implemented a surface modification approach using biotinylation of accessible amines (lysine and N-terminus) with the small modifier NHS-biotin. Changes in the accessibility and exposure of amino acid residues upon assembly of monomeric htau 1N3R and 1N4R into fibrils are represented in Figure 5. Overall, the exposure to solvent of the polypeptide stretch located at the end of the N1 projection domain and in the C-terminal moiety, spanning residues N381 and D402, was found remarkably lower in 1N3R as compared with 1N4R fibrils. The core of tau filaments has been delineated by limited proteolysis (30, 32, 36, 48, 49), and its structure within deposits characteristic of AD, PiD, CBD, and 2N tau fibrils assembled de novo has been obtained by cryo-EM after pronase degradation (23, 28, 33, 50). We compared our results to those published previously. The comparison is displayed in Figure 6 where amino acid numbering is that of 2N4R htau. The N-terminal moiety of htau that has been proposed to constitute the so-called “fuzzy coat” (51) appears overall exposed to the solvent and available to establish interactions with partner molecules (6, 7, 14, 52). We identify two amino acid stretches which exposure to the solvent decreases significantly or extremely upon assembly of monomeric tau into fibrils. Peptide D13-E36 is common to 1N3R and 1N4R htau while A118-D133 is specific to 1N3R htau. The conformation of those peptides, and as a consequence their exposure to solvent, has not been previously reported to change upon assembly of tau into fibrils. These results bring molecular information on the accessibility to the solvent of the so-called fuzzy coat of tau fibrils (24, 51) where very little molecular information is available.
The cryo-EM density maps of the core of 2N tau fibrils made de novo (33) provide no information on tau N-terminal polypeptides we demonstrate to adopt a compact conformation upon assembly of monomeric tau into fibrils. Interestingly however, the cryo-EM structures of AD patients-derived tau fibrils exhibited densities surrounding the rigid core of the protein (24). This density was first attributed to the polypeptide 7-EFE-9 that was proposed to interact with the microtubule-binding domain (K317 and K321) then to ubiquitin molecules linked to lysine residues. This density may originate from the stacking of tau N-terminal polypeptides D13-E36 and/or A118-D133 to the core of the fibrils.
Changes in the conformation of monomeric tau C-terminal domain are widely believed to modulate tau interaction with microtubules and subsequent tau aggregation. We demonstrate here moderate to very significant changes in the exposure to the solvent of polypeptides within the amino-acid stretch encompassing P223-E380 upon monomeric tau assembly into fibrils. Recent cryo-EM structural studies performed on tau fibrils purified from AD, PiD, or CBD brain cases or made de novo suggest significant differences in the fold fibrils core adopts (23, 28, 33). Indeed, the core of fibrils purified by centrifugation in the presence of detergents from patient brains comprises amino acid residues G273-E380 for 3R and G304-E380 for 4R tau for AD cases, K254-F378 for 3R tau for PiD cases, and K274-E380 for CBD cases (Fig. 6). In contrast, the cryo-EM-derived structural models for 2N3R and 2N4R fibrils assembled de novo show that the core of 2N3R and 2N4R tau fibrils is formed by residues G272/274-H330 and G272-H330 or G274-K321, respectively (33) (Fig. 6). Despite the structural differences that may originate from the lack of posttranslational modifications (phosphorylation, acetylation, and proteolytic cleavage) on recombinant fibrils and the need to use cofactors such as heparin (53), arachidonic acid (54), or RNA (55) to trigger tau aggregation, our results are compatible with the cryo-EM data obtained from tau fibrils extracted from AD, PiD, or CBD patient brains. We further bring additional insight into the structure of htau fibrils. Indeed, our results demonstrate that the amino acid stretch P223-E380 is highly protected in 1N3R htau assembled in vitro. Our results also show that the amino acid stretches D13-E36 and A118-D133 as well as P223-E431 from the N-and C-terminal moieties of the protein are also significantly or highly protected within 1N3R htau fibrils. In addition, we show that the amino acid stretches P223-E380 and T403-E431 besides D13-E36 are significantly or highly protected within 1N4R htau fibrils (Fig. 6). Overall, our results are significantly compatible with previous assessments of the amyloid core of tau assemblies purified from patient brains subjected to pronase treatment using proteolytic and immunodetection as well as cryo-EM strategies. Indeed, those strategies have demonstrated that the core of 3R htau filaments that characterize PiD spans residues 243–387 while that of tau filaments in AD spans residues 260–406 and 266–406 for 3R and 4R tau, respectively (Fig. 6). 1N tau isoforms are predominantly expressed in adult brain (8) representing about 50% of tau isoforms, compared with 40% of 0N tau and 10% of 2N tau. The results we present are of added value besides the cryoEM structures described for 2N tau fibrils from patient brains.
Limited pronase digestion of tau filaments from PiD cases revealed a protease resistant C-terminal polypeptide that is recognized by the Tau46 antibody directed against the peptide 404–441 (25). It is worth noting that densities observed by cryo-EM surrounding the highly structured core were not sufficiently resolved to be attributed unambiguously to the amino acid stretch 7-EFE-9, to posttranslational modifications or to another polypeptide from tau. Our results suggest that this density may originate from the stacking of amino acid stretches D13-E36 and/or A118-D133.
Our findings uncover major differences between 1N3R and 1N4R htau fibrils. The amino acid stretch N381-D402, located in the C-terminal region just after the R4 (spanning V337-N368), appears highly protected in 1N3R but not in 1N4R htau fibrils. This is in agreement with the observation that proteolytic cleavage at E391 is drastically reduced at late PiD stages compared with early stages of the pathology when fewer aggregates are observed and to AD (56). These differences may also account for the significant mechanical differences between 4R and 3R tau fibrils we reported recently upon assessment of their persistence length (57).
In conclusion and despite the claim that heparin-induced tau fibrils differ structurally from brain-derived tau aggregates (33), we show that the MS-based proteomic methods we implemented to map the surfaces of htau fibrils assembled in vitro yield results compatible with data generated after proteolysis and immunodetection and cryo-EM imaging of tau filaments purified from AD, PiD, and CBD cases. Thus, in vitro assembled tau fibrils appear as a relevant tool to document aggregated tau prion-like propagation. Our results bring insight, most likely lost following pronase treatment, into the polypeptides exposed to the solvent, e.g., the surfaces of tau fibrils, and those that are protected, which are likely to be involved in fibrils core. We identify two amino acid stretches (118-AAGHVTQARMVSKSKD-133 and 381-NAKAKTDHGAEIVYKSPVVSGD-402) that are exposed to the solvent in 1N4R not in 1N3R htau that may allow discriminating the two kinds of fibrils. We finally report differences in the exposure to the solvent within the C-terminal domain indicating that the structures of 1N3R and 1N4R htau fibrils are different, in agreement with the models derived from the assessment of filaments purified from AD, PiD, and CBD cases. The differences in the surface accessibility we report are important as the two peptide stretches differentially exposed in 3R and 4R fibrils we identify were described as belonging to the fuzzy coat of tau fibrils (24, 51). Our findings suggest for the first time, to our best knowledge, a contribution of the so-called fuzzy coat to distinct fibrillar polymorphs intrinsic organization. They also open the way for designing specific molecular tools with diagnostic and therapeutic potential and new perspectives for understanding the physiopathology of distinct tauopathies. These results also suggest that identification of common and specific protein interactors will contribute to a better understanding of tauopathies and the design of molecular tools able to interfere with their pathogenic functions and propagation. Future characterization of accessible/protected surfaces of pathogenic tau fibrils derived and amplified (58, 59) from patient's brain with the strategy we implemented may provide insights into different tauopathies and contribute to optimize the design of molecular tools targeting specifically or in a generic manner tau fibrils involved in disease.
Experimental procedures
Preparation of human 1N3R and 1N4R htau
Full-length human 1N3R and 1N4R htau was expressed and purified as previously described (60). Final monomeric hTau fraction was dialyzed against PBS buffer containing 1 mM DTT before storage of aliquots at –80 °C. The concentration of monomeric htau was determined spectrophotometrically using an extinction coefficient at 280 nm of 7450 M−1 cm−1 and 7575 M−1 cm−1 for htau 1N3R and 1N4R, respectively.
Htau 1N3R and 1N4R were assembled into fibrils at 40 μM in the presence of 10 μM heparin, in PBS buffer containing 1 mM DTT under continuous shaking (600 rpm) for 5 days at 37 °C in an Eppendorf Thermomixer. After assembly, fibrils were spun at 20 °C and 50,000 rpm for 30 min. The pelleted fibrils were resuspended in PBS buffer with 1 mM DTT at 40 μM equivalent monomeric tau concentration as reported previously (60).
The quality of htau 1N3R and 1N4R fibrils produced after 5 days was assessed by ThioflavinT binding, sedimentation, and transmission electron microscopy (TEM) performed as reported previously (60).
Limited proteolysis
Monomeric and fibrillar htau 1N3R and 1N4R were digested with endoprotease GluC (Roche) at a 1/150 E/S (w/w) ratio, at 37 °C under agitation (300 rpm). The proteolysis reaction was stopped at 0, 2, 10, 45, 120, and 300 min by addition of AEBSF (10 mM final concentration) and denaturation at 95 °C in Laemmli buffer 3× (vol:vol). Five microliters of sample was separated on 12% Tris-Glycine SDS-PAGE. Coomassie Blue stained gels were imaged using ChemiDoc MP (BioRad) and analyzed with ImageLab version 5.2.1.
Identification of the proteolytic fragments generated by limited proteolysis
Western blots
After SDS-PAGE separation, proteolytic polypeptides were transferred to a nitrocellulose membrane (Cat #10600002 Protran, Amersham) and stained with Ponceau Red. Immunodetection was performed with Nter (epitope [1–15]) and Cter (epitope [427–441]) rabbit polyclonal antibodies diluted at 1/10,000 (Gift from Luc Buée, INSERM), HT7 (epitope [159–163]) mouse monoclonal antibody diluted at 1/4000 (Cat #MN1000, Thermo Fisher), Tau5 (epitope [210–241] mouse monoclonal antibody diluted at 1/20,000 (Cat #AHB0042 Thermo Fisher), anti-3R tau (3-repeat isoform RD3, epitope [267–316]) mouse monoclonal antibody clone 8E6/C11 diluted at 1/10,000 (Cat #05-803 Merck Millipore), anti-4R tau (4-repeat isoform RD4, epitope [275–291]) mouse monoclonal antibody, clone 1E1/A6 diluted at 1/1000 (Cat #05-804 Merck Millipore). Immunodetection was developed with Pierce ECL Western Blotting Substrate (Cat #32106) and imaged on a Chemidoc MP Imaging System (Biorad).
N-terminal identification of proteolytic fragments using N-terminal labeling and mass spectrometry
After SDS-PAGE separation and Coomassie blue staining of an equivalent of 5 μg of digested htau, gel slices corresponding to the different proteolytic polypeptides were excised and in-gel N-terminal protein derivatization was performed as described (38) with the following modifications: Reduction and alkylation of cysteines were performed respectively with 10 mM DTT (56 °C, 30 min) and 55 mM Iodoacetamide (30 min room temperature, in the dark). N-terminal derivatization was performed by addition of 1.5 μl of 100 mM TMPP solution ((N-succinimidyloxycarbonylmethyl)- tris(2,4,6-trimethoxyphenyl)- phosphonium bromide) to each reaction buffer (50 μl Tris HCl 100 mM pH 8.2) for 1 h under agitation at room temperature. The reactions were stopped by addition of 5 μl of 16 M hydroxylamine. After three washes with 25 mM NH4HCO3 and acetronitrile, trypsin (Promega Gold; Promega) digestion was performed overnight in 50 mM NH4HCO3 at 37 °C under agitation. Peptides were extracted by addition of first 1.2 volumes of 60% acetonitrile, 0.1% formic acid, and second two volumes of acetonitrile. Extracted peptides were vacuum dried and stored at −20 °C.
Tryptic peptides were resuspended in 10 μl 0.1% TFA and 2 μl was analyzed by nanoLC-MS/MS using a LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific) in the data-dependent acquisition mode as previously described (61) with the following change: the reversed-phase gradient was 5–60% solvent B for 70 min.
NanoLC-MS/MS data were processed automatically using the Thermo Proteome Discoverer software (version 1.4) to generate the peak lists identified with the Sequest search engine (version 1.2) and a database composed of the two 1N3R and 1N4R htau amino acid sequences. Peptides were identified using semispecific trypsin cleavage with up to four missed cleavages and the following chemical modifications: cysteine carbamidomethylation (+57.021 Da) as fixed modification, methionine oxidation (+15.995 Da) and TMPP labeling of any N-terminus (+572.181 Da) as variable modifications. Mass tolerance was set to 10 ppm for precursor ions and 0.6 Da fragment ions respectively. Individual spectra were accepted using unambiguous Peptide Spectrum Matches (PSMs) identified with a target decoy false discovery rate (FDR) set to 0.05. Fragmentation mass spectra of TMPP labeled N-terminal peptides were further checked manually.
N-terminal identification of proteolytic fragments peptides by Edman sequencing
Proteolytic fragments were transferred onto 0.2 μm PVDF membranes (Applied Biosystems), using a Trans-Blot Turbo Transfer System (Biorad) in 10 min (25 V 1.3 A) and the transfer buffer: 50 mM Tris, 50 mM Boric Acid, 0.1% SDS, 10% Ethanol, pH 8.3. Bands corresponding to proteolytic polypeptides were excised after Coomassie blue staining.
N-terminal sequences for each proteolytic peptide were obtained by automated Edman degradation using a sequencer (PPSQ-1 Shimadzu). The PVDF membranes were washed three times in Ethanol/water (90/10), dried, and loaded in the cartridge of the Edman sequencer Shimadzu PPSQ 31B. Ten Edman degradation cycles were performed.
C-terminal identification of proteolytic fragments by mass spectrometry
The proteolytic polypeptides were separated and digested using trypsin as described previously, with and without the N-terminal derivatization step. To determine the sequence coverage of each GluC polypeptide and identify its C-terminal end, the resulting tryptic peptides were analyzed and identified by nanoLC-MS/MS using either a LTQ-Orbitrap velos as described previously or a Triple TOF 4600 as described below.
The data were confirmed by MALDI-MS analysis of the entire masses of small and large GluC proteolytic products generated after 10 min GluC digestion, as described in Figure S1.
Chemical surface labeling of monomeric and fibrillar tau
Monomeric and fibrillar htau 1N3R and 1N4R were labeled with N-hydroxysulfosuccinimide biotin (EZ-Link Sulfo-NHS-Biotin [Thermo Scientific]) at [Lysines/NHS-Biotine] ratio of 2.5/1 during 10 min at room temperature. The reaction was stopped by addition of Tris pH7.5 at final molar concentration of 50 mM. Fibrillar tau was sedimented by ultracentrifugation (30 min, 50.000 rpm, 20 °C), and the supernatant was replaced by a volume of 50 mM Tris pH7.5 equivalent to that withdrawn.
The average level of biotinylation of monomeric and fibrillar htau was determined by MALDI-MS. Samples diluted to 4–8 μM in 1% TFA milliQ water were mixed (vol:vol) with α-Cyano-4-hydroxycinnamic acid (20 mg/ml in acetonitrile 70%, formic acid 5%) as matrix. MALDI-MS measurements were performed with a MALDI-TOF-TOF 4800 (ABSciex) mass spectrometer in the linear mode, after external calibration using recombinant alpha-synuclein. T2D spectra were Gaussian smoothed on PeakView Software 1.2. AB SCIEX.
In-solution digestion under native conditions
Monomeric and fibrillar htau 1N3R and 1N4R (10 μM), biotinylated or not, were digested for 15 h at 37 °C, under agitation with endoprotease GluC (Roche) at a E/S (w/w) ratio of 1/50. Digestion was stopped by flash freezing in liquid nitrogen and storage at –80 °C for further analysis. Samples were diluted to a final concentration of 4 μM in 0.4% TFA before nanoLC-MS/MS analysis using a Triple TOF 4600 mass spectrometer. Three replicate experiments were performed on fibrils assembled independently.
In-gel digestion under denaturating conditions
Control and biotinylated monomeric and fibrillar htau 1N3R and 1N4R (4 μg, 100 pmol) were denatured in Laemmli buffer and loaded on a SDS-PAGE as described above. The corresponding protein bands were excised for in-gel digestion, as described above, but without the N-terminal labeling step. GluC and AspN digestions were performed at 1/20 Enzyme-to-Substrate ratios during 15 h at 37 °C under agitation. Proteolytic peptides were extracted and prepared as described above for nanoLC-MS/MS analysis using a Triple TOF 4600 mass spectrometer. Three replicate experiments were performed on fibrils assembled independently.
nanoLC-MS/MS analysis of proteolytic peptides
Proteolytic peptides resulting from in-solution and in-gel digestion of control and/or biotinylated monomeric and fibrillar htau 1N3R and 1N4R were analyzed by nanoLC-MS/MS using a Triple-TOF 4600 mass spectrometer (ABSciex) as previously described (35) with the following changes: the gradient of 5–35% solvent B was developed in 40 min, and the data-dependent acquisition method selected for CID fragmentation the 20 most intense precursor ions, with charge states from 1 to 5 and above an intensity threshold of 1000. The MS Data Converter software (version 1.3) included in the PeakView software (version 1.2, AB Sciex) was used to process raw data and convert them into mgf data files.
Peptide identifications were performed using the Mascot search engine (Matrix Science; version 2.4.1) against the two 1N3R and 1N4R htau amino acid sequences, including a decoy database search. Peptides were identified using specific digestion with GluC (C-terminus of E or D) and AspN (N-terminus of D) with up to five missed cleavages and following modifications: oxidation of methionine (+15.99 Da); biotinylation of N-ter, Lys and Cys, Ser, Thr, Tyr (+226.08 Da) were specified as variable modifications; carbamidomethylation of Cys (+57.02 Da) as constant modification for in-gel digestion. Precursor and fragment mass tolerances were set to 40 ppm and 0.05 Da, respectively. FDR was set to 1%. Peptide mascot score threshold for accepting peptide identification was set to 20.
nanoLC-MS/MS data analysis
A list of peptides, identified in the MASCOT search with a peptide score above 20 and at least two identifications, was used to extract each peptide ion intensity from the more intense mass-to-charge ratio for each replicate data set using the extracted ion chromatogram (XIC) manager of the PeakView Software (version 1.2, AB SCIEX), within a mass and retention time window of 0.1 Da and 0.3 to 0.6 min, respectively. The accordance between the MASCOT peptide identification and the MS2 spectrum of the extracted intensity was checked. The peptide ion intensity values were normalized to the total amount of htau in each sample, using as reference peptides, A106-E117 or D421-D429 for GluC or AspN digestion, respectively, as performed previously (39). The reference peptides contain no lysine and cover an accessible region as confirmed by limited proteolysis. The average normalized intensity of the three replicates is represented in boxplots (Fig. S4, a and b, S10, a and b, S12, a and b). Fold change (Fc F/M) between fibrillar (F) and monomeric (M) htau, for a given peptide, was calculated as the ratio of the averaged normalized peptide intensity for the fibrils to the averaged normalized peptide intensity for the monomers. Standard deviation was calculated using the normalized intensities. Statistical significance of the Fc was assessed by a nonparametric Kruskal–Wallis Test (p-value < 0.050).
Data availability
All data are included in the article and supporting information. Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (62) partner repository with the dataset identifiers PXD027403.
Dedications
The authors dedicate this work to late Mrs Viviane Morin.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge the Proteomic-Gif (SICaPS) platform (supported by IBiSA, Ile de France Region, Plan Cancer, CNRS and Paris-Sud University) and the Microscopy platform from the I2BC (CNRS UMR9198, Gif-sur-Yvette, France). We thank David Cornu and Laila Sago for assistance in mass spectrometric analysis, Pascal Mansuelle for the Edman sequencing service (IMM, Marseille, France), Tracy Bellande for technical assistance, Luc Bousset for assistance in electron microscopy imaging, and Luc Buée for the gift of anti-Tau antibodies (Inserm, Lille, France). This work was supported by grants from the Fondation pour la Recherche Medicale (contract DEQ. 20160334896 and ALZ201912009776), the EU Joint Programme on Neurodegenerative Disease Research and Agence National de la Recherche (contracts PROTEST-70, ANR-17-JPCD-0005-01 and Trans-PathND, ANR-17-JPCD-0002-02), and Vaincre Alzheimer Foundation (FR-15047, EC, PhD grant).
Author contributions
E. C., V. R., and R. M. conceptualization; E. C., V. R., and R. M. data curation; E. C. and V. R. formal analysis; V. R., and R. M. funding acquisition; E. C., V. R., and K. M. investigation; E. C., V. R., and R. M. methodology; R. M. project administration; V. R., and R. M. supervision; E. C. and V. R. validation; E. C., V. R., and R. M. writing–original draft.
Edited by Paul Fraser
Supporting information
References
- 1.Weingarten M.D. A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. U. S. A. 1975;72:1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Mandelkow E. Tau in physiology and pathology. Nat. Rev. Neurosci. 2016;17:5–21. doi: 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- 3.Sotiropoulos I., Galas M.C., Silva J.M., Skoulakis E., Wegmann S., Maina M.B., Blum D., Sayas C.L., Mandelkow E.-M., Mandelkow E., Spillantini M.G., Sousa N., Avila J., Medina M., Mudher A. Atypical, non-standard functions of the microtubule associated Tau protein. Acta Neuropathol. Commun. 2017;5:91–101. doi: 10.1186/s40478-017-0489-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goedert M., Spillantini M.G., Potier M.C., Ulrich J., Crowther R.A. Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: Differential expression of tau protein mRNAs in human brain. EMBO J. 1989;8:393–399. doi: 10.1002/j.1460-2075.1989.tb03390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu F., Gong C.X. Tau exon 10 alternative splicing and tauopathies. Mol. Neurodegener. 2008;3:3–13. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandt R., Leger J., Lee G. Interaction of tau with the neural plasma membrane mediated by tau's amino-terminal projection domain. J. Cell Biol. 1995;131:1327–1340. doi: 10.1083/jcb.131.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee G., Thangavel R., Sharma V., Litersky J., Bhaskar K., Fang S.M., Do L.H., Andreadis A., Van Hoesen G., Ksiezak-Reding H. Phosphorylation of tau by fyn: Implications for Alzheimer's disease. J. Neurosci. 2004;24:2304–2312. doi: 10.1523/JNEUROSCI.4162-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trabzuni D., Wray S., Vandrovcova J., Ramasamy A., Walker R., Smith C., Luk C., Gibbs J.R., Dillman A., Hernandez D.G., Arepalli S., Singleton A.B., Cookson M.R., Pittman A.M., de Silva R. MAPT expression and splicing is differentially regulated by brain region: Relation to genotype and implication for tauopathies. Hum. Mol. Genet. 2012;21:4094–4103. doi: 10.1093/hmg/dds238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee V.M., Goedert M., Trojanowski J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 10.De Silva R., Lashley T., Gibb G., Hanger D., Hope A., Reid A., Bandopadhyay R., Utton M., Strand C., Jowett T., Khan N., Anderton B., Wood N., Holton J., Revesz T. Pathological inclusion bodies in tauopathies contain distinct complements of tau with three or four microtubule-binding repeat domains as demonstrated by new specific monoclonal antibodies. Neuropath. Appl. Neurobiol. 2003;29:288–302. doi: 10.1046/j.1365-2990.2003.00463.x. [DOI] [PubMed] [Google Scholar]
- 11.Kovacs G.G. Invited review: Neuropathology of tauopathies: Principles and practice. Neuropathol. Appl. Neurobiol. 2015;41:3–23. doi: 10.1111/nan.12208. [DOI] [PubMed] [Google Scholar]
- 12.Buée L., Delacourte A. Comparative biochemistry of tau in progressive supranuclear palsy, corticobasal degeneration, FTDP-17 and Pick's disease. Brain Pathol. 1999;9:681–693. doi: 10.1111/j.1750-3639.1999.tb00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee G., Leugers C.J. Tau and tauopathies. Prog. Mol. Biol. Transl. Sci. 2012;107:263–293. doi: 10.1016/B978-0-12-385883-2.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sergeant N., Delacourte A., Buée L. Tau protein as a differential biomarker of tauopathies. Biochim. Biophys. Acta. 2005;1739:179–197. doi: 10.1016/j.bbadis.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Goedert M., Spillantini M.G. Propagation of tau aggregates. Mol. Brain. 2017;10:18–27. doi: 10.1186/s13041-017-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams D.R. Tauopathies: Classification and clinical update on neurodegenerative diseases associated with microtubule-associated protein tau. Intern. Med. J. 2006;36:652–660. doi: 10.1111/j.1445-5994.2006.01153.x. [DOI] [PubMed] [Google Scholar]
- 17.Sanders D.W., Kaufman S.K., DeVos S.L., Sharma A.M., Mirbaha H., Li A., J Barker S.J., Foley A.C., Thorpe J.R., Serpell L.C., M Miller T.M., Grinberg L.T., Seeley W.W., Diamond M.I. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron. 2014;82:1271–1288. doi: 10.1016/j.neuron.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaufman S.K., Sanders D.W., Thomas T.L., Ruchinskas A.J., Vaquer-Alicea J., Sharma A.M., Miller T.M., Diamond M.I. Tau prion strains dictate patterns of cell pathology, progression rate, and regional vulnerability in vivo. Neuron. 2016;92:796–812. doi: 10.1016/j.neuron.2016.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J.L., Narasimhan S., Changolkar L., He Z., Stieber A., Zhang B., Gathagan R.J., Iba M., McBride J.D., Trojanowski J.Q., Lee V.M.Y. Unique pathological tau conformers from Alzheimer's brains transmit tau pathology in nontransgenic mice. J. Exp. Med. 2016;213:2635–2654. doi: 10.1084/jem.20160833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narasimhan S., Guo J.L., Changolkar L., Stieber A., McBride J.D., Silva L.V., He Z., Zhang B., Gathagan R.J., Trojanowski J.Q., Lee V.M.Y. Pathological tau strains from human brains recapitulate the diversity of tauopathies in nontransgenic mouse brain. J. Neurosci. 2017;37:11406–11423. doi: 10.1523/JNEUROSCI.1230-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clavaguera F., Akatsu H., Fraser G., Crowther R.A., Frank S., Hench J., Probst A., Winkler D.T., Reichwald J., Staufenbiel M., Ghetti B., Goedert M., Tolnay M. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9535–9540. doi: 10.1073/pnas.1301175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clavaguera F., Hench J., Goedert M., Tolnay M. Invited review: Prion-like transmission and spreading of tau pathology. Neuropath. Appl. Neurobiol. 2015;41:47–58. doi: 10.1111/nan.12197. [DOI] [PubMed] [Google Scholar]
- 23.Falcon B., Zhang W., Schweighauser M., Murzin A.G., Vidal R., Garringer H.J., Ghetti B., Scheres S.H.W., Goedert M. Tau filaments from multiple cases of sporadic and inherited Alzheimer's disease adopt a common fold. Acta Neuropathol. 2018;136:699–708. doi: 10.1007/s00401-018-1914-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzpatrick A.W.P., Falcon B., He S., Murzin A.G., Murshudov G., Garringer H.J., Crowther R.A., Ghetti B., Goedert M., Scheres S.H.W. Cryo-EM structures of tau filaments from Alzheimer's disease. Nature. 2017;547:185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arakhamia T., Lee C.E., Carlomagno Y., Duong D.M., Kundinger S.R., Wang K., Williams D., DeTure M., Dickson D.W., Cook C.N., Seyfried N.T., Petrucelli L., Fitzpatrick A.W.P. Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell. 2020;180:633–644. doi: 10.1016/j.cell.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W., Tarutani A., Newell K.L., Murzin A.G., Matsubara T., Falcon B., Vidal R., Garringer H.J., Shi Y., Ikeuchi T., Murayama S., Ghetti B., Hasegawa M., Goedert M., Scheres S.H.W. Novel tau filament fold in corticobasal degeneration. Nature. 2020;580:283–287. doi: 10.1038/s41586-020-2043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falcon B., Zhang W., Murzin A.G., Murshudov G., Garringer H.J., Vidal R., Crowther R.A., Ghetti B., Scheres S.H.W., Goedert M. Structures of filaments from Pick's disease reveal a novel tau protein fold. Nature. 2018;561:137–140. doi: 10.1038/s41586-018-0454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheres S., Zhang W., Falcon B., Goedert M. Cryo-EM structures of tau filaments. Curr. Opin. Struct. Biol. 2020;64:17–25. doi: 10.1016/j.sbi.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Lyumkis D. Challenges and opportunities in cryo-EM single-particle analysis. J. Biol. Chem. 2019;294:5181–5197. doi: 10.1074/jbc.REV118.005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wischik C.M., Novak M., Thogersen H.C., Edwards P.C., Runswick M.J., Jakes R., Walker J.E., Milstein C., Roth M., Klug A. Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc. Natl.Acad. Sci. U.S.A. 1988;85:4506–4510. doi: 10.1073/pnas.85.12.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novak M., Kabat J., Wischik C.M. Molecular characterization of the minimal protease resistant tau unit of the Alzheimer's disease paired helical filament. EMBO J. 1993;12:365–370. doi: 10.1002/j.1460-2075.1993.tb05665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Von Bergen M., Barghorn S., Müller S.A., Pickhardt M., Biernat J., Mandelkow E.-M., Davies P., Aebi U., Mandelkow E. The core of tau-paired helical filaments studied by scanning transmission electron microscopy and limited proteolysis. Biochemistry. 2006;45:6446–6457. doi: 10.1021/bi052530j. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W., Falcon B., Murzin A.G., Fan J., Crowther R.A., Goedert M., Scheres S.H. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases. Elife. 2019;8 doi: 10.7554/eLife.43584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shrivastava A.N., Aperia A., Melki R., Triller A. Physico-pathologic mechanisms involved in neurodegeneration: Misfolded protein-plasma membrane interactions. Neuron. 2017;95:33–50. doi: 10.1016/j.neuron.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 35.Shrivastava A.N., Redeker V., Pieri L., Bousset L., Renner M., Madiona K., Mailhes-Hamon C., Coens A., Buée L., Hantraye P., Triller A., Melki R. Clustering of Tau fibrils impairs the synaptic composition of α3-Na+/K+-ATPase and AMPA receptors. EMBO J. 2019;38 doi: 10.15252/embj.201899871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasegawa M., Watanabe S., Kondo H., Akiyama H., Mann D.M.A., Saito Y., Murayama S. 3R and 4R tau isoforms in paired helical filaments in Alzheimer's disease. Acta Neuropathol. 2014;127:303–305. doi: 10.1007/s00401-013-1191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontana A., de Laureto P.P., Spolaore B., Frare E., Picotti P., Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim. Pol. 2004;51:299–321. [PubMed] [Google Scholar]
- 38.Ayoub D., Bertaccini D., Diemer H., Wagner-Rousset E., Colas O., Cianférani S., Van Dorsselaer A., Beck A., Schaeffer-Reiss C. Characterization of the N-terminal heterogeneities of monoclonal antibodies using in-gel charge derivatization of α-amines and LC-MS/MS. Anal. Chem. 2015;87:3784–3790. doi: 10.1021/ac504427k. [DOI] [PubMed] [Google Scholar]
- 39.Barthélemy N.R., Gabelle A., Hirtz C., Fenaille F., Sergeant N., Schraen-Maschke S., Vialaret J., Buée L., Junot C., Becher F., Lehmann S. Differential mass spectrometry profiles of tau protein in the cerebrospinal fluid of patients with Alzheimer's disease, progressive supranuclear palsy, and dementia with lewy bodies. J. Alzheimers Dis. 2016;51:1033–1043. doi: 10.3233/JAD-150962. [DOI] [PubMed] [Google Scholar]
- 40.Hubbard S.J. The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta. 1998;1382:191–216. doi: 10.1016/s0167-4838(97)00175-1. [DOI] [PubMed] [Google Scholar]
- 41.Liu X.R., Zhang M.M., Gross M.L. Mass spectrometry-based protein footprinting for higher-order structure analysis: Fundamentals and applications. Chem. Rev. 2020;120:4355–4454. doi: 10.1021/acs.chemrev.9b00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carven G.J., Stern L.J. Probing the ligand-induced conformational change in HLA-DR1 by selective chemical modification and mass spectrometric mapping. Biochemistry. 2005;44:13625–13637. doi: 10.1021/bi050972p. [DOI] [PubMed] [Google Scholar]
- 43.Gabant G., Augier J., Armengaud J. Assessment of solvent residues accessibility using three sulfo-NHS-biotin reagents in parallel: Application to footprint changes of a methyltransferase upon binding its substrate. J. Mass Spectrom. 2008;43:360–370. doi: 10.1002/jms.1328. [DOI] [PubMed] [Google Scholar]
- 44.Dreger M., Bo W.L., Brownlee G.G., Deng T. A quantitative strategy to detect changes in accessibility of protein regions to chemical modification on heterodimerization. Protein Sci. 2009;18:1448–1458. doi: 10.1002/pro.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mädler S., Bich C., Touboul D., Zenobi R. Chemical cross-linking with NHS esters: A systematic study on amino acid reactivities. J. Mass Spectrom. 2009;44:694–706. doi: 10.1002/jms.1544. [DOI] [PubMed] [Google Scholar]
- 46.Mudher A., Colin M., Dujardin S., Medina M., Dewachter I., Alavi Naini S.M., Mandelkow E.M., Mandelkow E., Buée L., Goedert M., Brion J.P. What is the evidence that tau pathology spreads through prion-like propagation? Acta Neuropathol. Commun. 2017;5:99–119. doi: 10.1186/s40478-017-0488-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang R.Y.-C., Iacob R.E., Sankaranarayanan S., Yang L., Ahlijanian M., Tymiak A.A., Chen G. Probing conformational dynamics of tau protein by hydrogen/deuterium exchange mass spectrometry. J. Am. Soc. Mass Spectrom. 2018;29:174–182. doi: 10.1007/s13361-017-1815-8. [DOI] [PubMed] [Google Scholar]
- 48.Jakes R., Novak M., Davison M., Wischik C.M. Identification of 3- and 4-repeat tau isoforms within the PHF in Alzheimer's disease. EMBO J. 1991;10:2725–2729. doi: 10.1002/j.1460-2075.1991.tb07820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taniguchi-Watanabe S., Arai T., Kametani F., Nonaka T., Masuda-Suzukake M., Tarutani A., Murayama S., Saito Y., Arima K., Yoshida M., Akiyama H., Robinson A., Mann D.M.A., Iwatsubo T., Hasegawa M. Biochemical classification of tauopathies by immunoblot, protein sequence and mass spectrometric analyses of sarkosyl-insoluble and trypsin-resistant tau. Acta Neuropathol. 2016;131:267–280. doi: 10.1007/s00401-015-1503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goedert M., Falcon B., Zhang W., Ghetti B., Scheres S.H.W. Distinct conformers of assembled tau in Alzheimer's and Pick's diseases. Cold Spring Harb. Symp. Quant. Biol. 2018;83:163–171. doi: 10.1101/sqb.2018.83.037580. [DOI] [PubMed] [Google Scholar]
- 51.Ksiezak-Reding H., Yen S.H. Structural stability of paired helical filaments requires microtubule-binding domains of tau: A model for self-association. Neuron. 1991;6:717–728. doi: 10.1016/0896-6273(91)90169-z. [DOI] [PubMed] [Google Scholar]
- 52.Brandt R., Trushina N.I., Bakota L. Much more than a cytoskeletal protein: Physiological and pathological functions of the non-microtubule binding region of tau. Front. Neurol. 2020;11:590059. doi: 10.3389/fneur.2020.590059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goedert M., Jakes R., Spillantini M.G., Hasegawa M., Smith M.J., Crowther R.A. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature. 1996;383:550–553. doi: 10.1038/383550a0. [DOI] [PubMed] [Google Scholar]
- 54.Wilson D.M., Binder L.I. Free fatty acids stimulate the polymerization of tau and amyloid beta peptides. In vitro evidence for a common effector of pathogenesis in Alzheimer's disease. Am. J. Pathol. 1997;150:2181–2195. [PMC free article] [PubMed] [Google Scholar]
- 55.Kampers T., Friedhoff P., Biernat J., Mandelkow E.M., Mandelkow E. RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett. 1996;399:344–349. doi: 10.1016/s0014-5793(96)01386-5. [DOI] [PubMed] [Google Scholar]
- 56.Mondragón-Rodríguez S., Mena R., Binder L.I., Smith M.A., Perry G., Garcia-Sierra F. Conformational changes and cleavage of tau in Pick bodies parallel the early processing of tau found in Alzheimer pathology. Neuropathol. Appl. Neurobiol. 2008;34:62–75. doi: 10.1111/j.1365-2990.2007.00853.x. [DOI] [PubMed] [Google Scholar]
- 57.Makky A., Bousset L., Madiona K., Melki R. Atomic force microscopy imaging and nanomechanical properties of six tau isoform assemblies. Biophys. J. 2020;119:2497–2507. doi: 10.1016/j.bpj.2020.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saijo E., Groveman B.R., Kraus A., Metrick M., Orrù C.D., Hughson A.G., Caughey B. Ultrasensitive RT-QuIC seed amplification assays for disease-associated tau, alpha-synuclein, and prion aggregates. Methods Mol. Biol. 2019;1873:19–37. doi: 10.1007/978-1-4939-8820-4_2. [DOI] [PubMed] [Google Scholar]
- 59.Xu H., O'Reilly M., Gibbons G.S., Changolkar L., McBride J.D., Riddle D.M., Zhang B., Stieber A., Nirschl J., Kim S.J., Hoxha K.H., Brunden K.R., Schellenberg G.D., Trojanowski J.Q., Lee V.M. In vitro amplification of pathogenic tau conserves disease-specific bioactive characteristics. Acta Neuropathol. 2021;141:193–215. doi: 10.1007/s00401-020-02253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tardivel M., Bégard S., Bousset L., Dujardin S., Coens A., Melki R., Buée L., Colin M. Tunneling nanotube (TNT)-mediated neuron-to neuron transfer of pathological Tau protein assemblies. Acta Neuropathol. Commun. 2016;4:117–130. doi: 10.1186/s40478-016-0386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Redeker V., Pemberton S., Bienvenut W., Bousset L., Melki R. Identification of protein interfaces between alpha-synuclein, the principal component of Lewy bodies in Parkinson disease, and the molecular chaperones human Hsc70 and the yeast Ssa1p. J. Biol. Chem. 2012;287:32630–32639. doi: 10.1074/jbc.M112.387530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the article and supporting information. Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (62) partner repository with the dataset identifiers PXD027403.