Graphical abstract
Abbreviations: BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, Kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma
Keywords: Protein profiling, Cross-talk, microRNA (miRNA), isomiR, Long non-coding RNA (lncRNA), Cholangiocarcinoma (CCA)
Highlights
-
•
Protein profiling identified crucial genes that were used to screen related ncRNAs.
-
•
Both miRNAs and isomiRs were involved in coding-non-coding RNA regulatory network.
-
•
IsomiR-associated ceRNA networks implied the complex interactions among RNAs.
Abstract
Cholangiocarcinomas (CCAs) are tumors that arise from the cholangiocytes. Although some genes have been shown with important roles in pathological process, interactions or cross-talks among different RNAs are important to understand the detailed molecular mechanisms in cancer development, especially discussing cross-talks among isomiRs and other RNAs. Herein, to characterize crucial genes in CCA, the protein expression profile was performed to survey potential crucial mRNAs and related non-coding RNAs (ncRNAs) in mRNA-ncRNA network, mainly including miRNAs/isomiRs and lncRNAs. Deregulated mRNAs were firstly obtained if consistent expression patterns were found at protein and mRNA levels, and related miRNAs/isomiRs were screened according to regulatory relationships. Diverse isomiRs from a given miRNA locus also contributed to interactions between the small RNAs and target mRNAs, and miRNAs were further used to survey related lncRNAs to expand the interactions. Thus, several groups of RNAs were constructed as candidate competitive endogenous RNA (ceRNA) networks. Finally, we found that RAB11FIP1:miR-101-3p:MIR3142HG may be a potential ceRNA network, and the interactions among them may be more complex due to variety of isomiRs. Simultaneously, RAB11FIP1 and miR-194-5p were also detected other related lncRNAs (FBXL19-AS1, SNHG1 and PVT1) that may be crucial in coding-non-coding RNA regulatory network. Our results show that diverse isomiRs with sequence and expression heterogeneities contribute to ceRNA regulatory network that may have crucial roles in CCA, which will expand our understanding of interactions among diverse RNAs and their contributions in cancer development.
1. Introduction
Cholangiocarcinoma (CCA) is a devastating malignancy that is difficult to diagnose, and its incidence and mortality drastically increase annually, despite CCA is a kind of rare disease [1]. CCA may be mainly caused by multiple risk factors as well as contributions of genetic factors. Currently, surgery and chemotherapy are main treatment approaches, but they are still not effective treatments for CCA [2], [3]. It has been estimated that patients suffer from less than 10% of 5-year survival due to rapid progression or tumor metastasis and poor prognosis [4]. It is urgent to explore the potential novel diagnostic or therapeutic targets in clinical.
Some genes may have crucial roles during the development and progression of CCA. For example, SIRT3 may act via an anti-Warburg effect on the downstream pathway HIF1α/PDK1/PDHA1 in the inhibition of CCA progression [5], TRIM59 may inhibit proliferation via the PI3K/AKT/mTOR signalling pathway [6], the long non-coding RNA (lncRNA), CASC15, may promote intrahepatic cholangiocarcinoma through inducing PRDX2/PI3K/AKT axis [7], and miR-26a can promote cholangiocarcinoma growth by activating β-catenin [8]. These studies have shown that some genes, especially for those non-coding RNAs (ncRNAs), mainly including small microRNAs (miRNAs) and lncRNAs, have potential crucial roles in pathologic processes of CAA. Indeed, cross-talk among RNAs may show more implication in relevant biological pathways, which will provide references for study of cancer progression, prognosis and future cancer treatment. Some studies have shown the potential interactions among differentially expressed RNAs [9], [10], implying their contributions to cancer via complex interactions, especially between mRNAs and ncRNAs. Further, the regulatory pattern of miRNA:mRNA may be disturbed if lncRNA can also bind to the same miRNA, which may protect target mRNA from degradation by acting as a competitive endogenous RNA (ceRNA) [11], [12]. CeRNA network has been widely studied in diverse cancer types [13], [14], [15], and identified potential regulatory axes may be potential promising targets for cancer treatment. However, it is unclear that whether and how different molecules co-contribute to the complex process, particularly complex interaction among multiple miRNA variants (also termed isomiRs) and other RNAs. In recent years, multiple isomiRs have been found in small RNAs via in-depth analysis of high-throughput sequencing data [16], [17], [18], [19], [20], and these varied isomiRs may contribute to disturbing coding-non-coding-RNA regulatory network via obtaining or losing target mRNAs than their canonical miRNAs [21]. From a given miRNA locus, multiple isomiRs may be generated via alternative cleavage or 3′ addition events [18], and the classical single miRNA sequence is only a specific member in these isomiRs with heterogeneities of length and sequence. Compared with the canonical miRNAs, some of their isomiRs also have distinct targets and functions [22], and relevant studies have been concerned in recent years [23], [24], [25]. It is necessary to perform analysis from the isomiR levels to understand the interactions among different RNAs, which will enrich our understanding of regulatory networks and their potential contributions during the development and progression of cancers.
Herein, to understand the potential interactions among RNAs, mainly including mRNAs, miRNAs especially for their multiple isomiRs, and lncRNAs, we firstly screened abnormally expressed genes from protein profiling and mRNA profiling, respectively, and then to search potential interactions among mRNAs and ncRNAs. We aim to discuss the complex interactions of mRNAs and their small flexible regulators, including levels of miRNAs and isomiRs, and further screen potential mRNA:isomiR interactions. Then, miRNAs were used to screen related lncRNAs to explore the potential cross-talks among RNAs as ceRNA networks, which will provide implications to understand their potential roles in the occurrence and development of CCA. This study may contribute to understanding of the potential regulatory roles of multiple isomiRs with their targeted mRNAs and lncRNAs as a ceRNA network, especially in the pathological and physiological processes of cancer, which will provide candidate targets for precision medicine.
2. Materials and methods
2.1. Protein profiling
A total of 16 pairs of tumor and normal (adjacent-to-tumor samples) tissues from 16 male patients (all of these patients had similar ages and disease processes) diagnosed with CCA were collected from the First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu, China. Of these, 11 pairs were performed protein profiling, and another 5 pairs were used to experimental validation. This study was conducted in accordance with the declaration of Helsinki and approval from the Ethics Committee of Nanjing Medical University, and written informed consent was obtained from each participant.
Expression profiles of protein were measured with Triple TOF5600 System (AB SCIEX, Concord, ON) fitted with a Nanospray III source (AB SCIEX, Concord, ON) at LC-BIO Technologies (Hangzhou) CO., LTD. Proteins were removed if they were not detected in more than 12 samples (n = 22). Differentially expressed proteins were firstly analyzed for the paired data, and only those proteins with both p < 0.05 (corrected p value) and FC (fold change) value > 1.20 or < 0.83 were primarily obtained as candidate abnormal proteins. These proteins were further performed in-depth analysis to discuss their potential biological function in cancer.
2.2. Data resource for further analysis
Based on the screened differentially expressed proteins, further knowledge for these proteins at RNA levels would contribute to understanding their potential correlations with pathophysiological process of cancer. Moreover, the detailed knowledge in diverse cancer types may provide references for revealing the potential associations with cancer. Therefore, we obtained sequencing data in The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/tcga/) using the “TCGAbiolinks” package [26] (https://doi.org/10.1093/nar/gkv1507) to perform a comprehensive analysis from multiple molecular levels, mainly including RNA sequencing data (mRNA and miRNA levels) and clinical data. Differentially expressed genes from sequencing data, mainly including mRNAs, miRNAs/isomiRs and lncRNAs, were obtained using DESeq2 [27]. Genes were identified as abnormally expressed if |log2FC| > 1.5 and padj < 0.05. Candidate mRNAs were collected if they had consistent expression patterns at protein and mRNA levels.
2.3. Function enrichment analysis
In order to understand the potential biological roles of the screened deregulated mRNAs via protein profiling, functional enrichment analysis was performed using The Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.8 [28]. Simultaneously, they were also investigated distributions of genes associated with hallmarks of cancer [29] (http://software.broadinstitute.org/gsea/msigdb/) to estimate the potential gene features. The similar method was used to evaluate the potential roles in multiple biological processes according to screened gene sets.
2.4. miRNA:mRNA interactions at miRNA and isomiR levels
Screened differentially expressed genes were used to search potential interacted small ncRNAs that are important regulatory molecules. Firstly, the experimentally validated miRNA-mRNA interactions were collected from the miRNet [30], and involved miRNAs were further queried for their expression patterns based on the most dominant isomiR in each miRNA locus. The multiple isomiRs from gene locus were mainly identified according to the location annotations of mature miRNA in the miRBase database [31]. If interacted miRNA and mRNA had opposite dyregulated expressions, the detailed isomiR expression levels were analyzed. Co-expression analysis was also used to validate the expression correlation, which would contribute to validation of interaction between isomiR and target mRNA.
Moreover, to understand the difference of target mRNAs between the canonical seed and novel shifted seed from the specific miRNA locus, their target mRNAs were also predicted using TargetScan 7.0 [32].
2.5. Sequence analysis
For relevant homologous miRNAs that were interacted with candidate mRNAs, we also performed sequence analysis to understand their sequence relationships among classical miRNAs and multiple isomiRs, especially for the potential correlations with their biological function. First, sequences of miRNAs, mainly including pre-miRNAs (precursor miRNAs) and mature miRNAs, were collected from the miRBase database [31]. Second, Homologous sequences and multiple isomiRs were firstly aligned with ClustalX 2.1 [33]. A phylogenetic tree of miRNA genes were reconstructed using Neighbor-Net method in SplitsTree 4.14 [34], and phylogenetic networks for the mature miRNAs and isomiRs were obtained using Network 10.2.00 (Copyright Fluxus Technology Ltd., 2004–2020, https://www.fluxus-engineering.com) based on the median-joining (MJ) method [35]. Finally, sequence logos were estimated using WebLogo [36] to assess the potential sequence features among multiple isomiRs from homologous miRNA loci.
2.6. Interactions with lncRNAs based on screened miRNAs
As a class of ncRNA, lncRNA have been widely concerned because of its potential regulatory role in coding-non-coding RNA network. To further understand the potential cross-talks among different RNAs, associated lncRNAs were collected according to the screened related miRNAs using starbase database [37], [38]. Further, we also queried for the related lncRNAs from the protein level. Then, according to the correlations among different RNAs, ceRNA network was constructed using “networkD3” package (https://CRAN.R-project.org/package = networkD3).
2.7. Quantitative real-time PCR (qRT-PCR) validates mRNA expression patterns
In order to validate expression patterns of screened genes, qPCR was performed in another 5 paired normal and tumor tissues. We selected down-regualted ADH1B and APOC2, and up-regulated VCAN to validate whether there were consistent expression trends at mRNA and protein levels. Total RNA was extracted from tumor and normal tissues using TRIzol reagent (Invitrogen, Carlsbad, CA), and cDNA was then generated from total RNA using a reverse transcription kit (Takara, Dalian, China). Gene expression was measured by qPCR (Lightcycler96, Roche, Basel, Switzerland) using a SYBR green kit (Yeasen, Shanghai, China). Used primers were shown in supplemental Table S1. GAPDH was used to as a control to normalize mRNA levels, and each gene was detected six times in each tissue.
2.8. Statistical analysis and network visualization
Paired t-test and the Wilcoxon rank sum test were used to estimate differentially expressed proteins and genes based on paired samples, and unpaired t test and the Wilcoxon rank-sum test were used for the unpaired samples. A trend test was used to perform hypothesis testing to understand expression patterns among diverse isomiRs from the specific miRNA locus. For interactions between related genes, especially for among different RNAs, further network visualization was presented using Cytoscape 3.6.0 [39]. A Pearson or spearman correlation coefficient was estimated to assess expression relationship. All of these statistical analyses were analyzed using R programming language (version 3.4.3), and venn distributions were performed with a publicly available tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
3. Results
3.1. Overview of protein profiling
A total of 70 candidate proteins with abnormal expression levels were firstly collected from protein profiling, including 40 up-regulated and 30 down-regulated proteins (Table S2). These proteins were matched 68 genes (both PKM and VCAN were matched with 2 proteins), and these genes were mainly significantly enriched some gene ontology (GO) terms (Fig. S1A), including haptoglobin binding, cell adhesion, hemoglobin complex, and etc. The results showed that these screened proteins were involved in multiple biological processes, implying their potential roles in CCA.
3.2. Integrative analysis of protein and mRNA expression
In order to understand expression trends at mRNA levels, candidate 70 abnormal proteins were further queried for their expression patterns at mRNA levels using sequencing data in cholangiocarcinoma obtained from The Cancer Genome Atlas (TCGA) database (cholangiocarcinoma was named CHOL). A total of 25 genes (matched to 27 proteins), including 6 significant down-regulated and 19 up-regulated genes, were collected from both protein and mRNA expression levels (Table S3 and Fig. S1B), and all of these were dominantly and abnormally expressed. Compared with expression distributions at mRNA levels, expression patterns of proteins were more concentrate (Fig. S1C). These involved genes were dominantly expressed in bile duct tissue, implying their potential roles in pathophysiological process of cholangiocarcinoma. To validate whether there were consistent expression patterns at mRNA and protein levels, several genes, including down-regulated ADH1B and APOC2, and up-regulated VCAN, were performed qRT-PCR experiment (Fig. 1A). Compared with the detailed protein expression patterns via protein profiling, mRNA expression showed consistent patterns. These results indicated that protein profiling were consistent with the mRNA expression levels in CCA patients.
Fig. 1.
Expression and functional analysis of screened genes. Consistent results of protein profiling and qPCR validation (n = 5). Each gene is detected 6 times in each sample. Paired analysis in CHOL (n = 9). The log2FC and p values are also presented for each gene based on paired t-test. T: tumor samples; N: paired normal samples. C. Expression patterns of 21 genes across diverse cancer types. Freq indicates frequency. D. Distribution of involved hallmark of cancer. The above picture shows involved hallmark of cancer based on 21 screened genes, and the below picture shows distributions for each gene. Tissue Invasion: Tissue invasion and metastasis; Self-sufficiency: Self-sufficiency in growth signals; Insensitivity: Insensitivity to antigrowth signals; Evading: Evading apoptosis; Reprogramming: Reprogramming energy metabolism.
For these 25 genes, paired analysis (n = 9) in cholangiocarcinoma was performed to understand the detailed expression patterns. We found that 21 of them were also identified as significantly up-regulated or down-regulated genes (Fig. 1B), which were consistent with above analysis. In order to understand the potential roles of them in different cancers, they were further queried for the detailed expression distributions across diverse cancer types. Many genes were dominantly expressed and showed consistent expression patterns in diverse tissues (Fig. 1C and Fig. S1D), especially for ADH1B (significantly down-regulated in most cancer types) and COL10A1 (significantly up-regulated in many cancers). These abnormal genes implicated their potential roles in pathophysiological process. For example, ADH1B may be involved in multiple biological pathways and contribute to alcohol abuse and carcinogenesis [40], and COL10A1 might play a crucial role in gastric cancer progression [41]. Some genes were specifically abnormally expressed in cholangiocarcinoma, such as RAB11FIP1 (up-regulated) and APOC2 (down-regulated), indicating their potential specific biological roles in CCA.
3.3. Functional enrichment analysis shows potential roles of screened genes
To understand the potential biological roles of the screened 21 deregulated genes, they were further queried for their potential roles in the hallmarks of cancer and pathways. We found that 11 of them were involved in hallmarks of cancer, and 5 of them were involved in self-sufficiency in growth signals and tissue invasion and metastasis (Fig. 1D), indicating the role in hallmarks of cancer. Similarly, 9 of them were involved in 21 KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, especially for ECM receptor interaction, focal adhesion and PPAR signaling pathway, and some genes (such as ADH1B contributed to 6 pathways, and CD36 and LAMA3 contributed to 4 pathways) were involved in multiple pathways (Fig. S2A). These results indicated that screened abnormal genes may contribute to important biological processes. Moreover, ASNS and PKM were identified as core essential genes according to the common data of Hart et al. [42], Blomen et al. [43] and Wang et al. [44], implying their roles in basic biological processes. Indeed, dual covalent inhibition of PKM and IMPDH targets metabolism in cutaneous metastatic melanoma [45], modulation of PKM alternative splicing by PTBP1 can promote gemcitabine resistance in pancreatic cancer cells [46], and anti-cancer fatty-acid derivative can induce autophagic cell death via modulation of PKM isoform expression profile mediated by bcr-abl in chronic myeloid leukemia [47]. These studies showed an important role of PKM in cancer.
3.4. miRNA:mRNA interactions from both miRNA and isomiR levels
Based on experimentally validated miRNA:mRNA interactions, the most dominant isomiR from each miRNA locus was considered as canonical miRNA to query for its expression patterns (Fig. S2B). These interactions showed the potential regulatory relationship between the small RNAs and mRNAs, and some pairs were screened based on the opposite expression patterns (Fig. S2B). The screened interactions included 4 mRNAs and 8 miRNAs from 9 interactions, and all of these involved miRNAs or mRNAs were dominantly expressed in cholangiocarcinoma. These 8 miRNAs were dynamically expressed across diverse cancer types (Fig. 2A), and most of them were found with inconsistent expression patterns across different tissues. For example, miR-101-3p was up-regulated in COAD, but down-regulated in CHOL, LUSC, UCEC and HNSC, while miR-122-5p was not detected in some cancer types (Fig. 2A). These various expression patterns showed that the small RNAs were spatiotemporal expressed, and the flexible expression patterns may contribute to complex regulatory patterns. Except for miR-122-5p, other miRNAs were abundantly enriched, especially in cholangiocarcinoma (Fig. 2B and 2C), implicating that these abnormal miRNAs may directly or indirectly contribute to pathophysiological process. SP1-induced upregulation of lncRNA SPRY4-IT1 could exert oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma [48], SNHG6 promoted proliferation and angiogenesis of cholangiocarcinoma cells through sponging miR-101-3p and activation of E2F8 [49], and these studies indicated that the potential role of miRNA in cancer, especially cross-talks with lncRNAs and mRNAs.
Fig. 2.
Screened interacted miRNAs. A. Expression patterns of screened interacted validated miRNAs across diverse cancer types based on the most dominant isomiR. The detailed baseMean values are also presented in CHOL to show the relative expression distributions. In miR-10b-5p, 29–48 indicates hg38:chr2:176150329–176150348:+; in miR-101-3p, 45–69 indicates hg38:chr9:4850345–4850369:+; in miR-122-5p, 89–109 indicates hg38:chr18:58451089–58451109:+; in miR-181a-5p, 480–498 indicates hg38:chr9:124692480–124692498:+; in miR-181b-5p, 26–47 indicates hg38:chr9:124693726–124693747:+; in miR-181c-5p, 25–46 indicates hg38:chr19:13874725–13874746:+; in miR-194-5p, 404–427 indicates hg38:chr11:64891404–64891427:-; in miR-625-5p, 53–74 indicates hg38:chr14:65471153–65471174:+. B. Expression levels for each involved miRNA across different cancers. C. Expression levels for each cancer type based on all involved miRNAs.
Then, the detailed isomiR repertoires for the screened 8 miRNAs were queried to understand the expression patterns of miRNA isoforms. Several miRNAs, including miR-101-3p, miR-194-5p, miR-181a-5p and miR-181b-5p, were found that they could be yielded from 2 multicopy pre-miRNAs (precursor miRNAs). In order to understand the detailed expression patterns, we firstly screened abundant miRNA variants (reads per million mapped reads (RPM) > 50) to perform expression analysis. Homologous pre-miRNAs usually had similar expression patterns (Fig. S3A and S3B), indicating that these multiple miRNA variants could be further analyzed based on each pre-miRNA. Indeed, it is difficult to determine the detailed productions from these multicopy pre-miRNAs, and we selected the miRNA loci that could generate more kinds of isomiR types with higher enrichment levels than other homologous miRNA loci (isomiRs in miR-101-3p was estimated from mir-101–2 locus, isomiRs in miR-181a-5p was estimated from mir-181a-2 locus, isomiRs in miR-181b-5p was estimated from mir-181b-2, and isomiRs in miR-194-5p was estimated from mir-194–2 locus). If the cutoff values of expression levels were reduced, more variants were involved (Fig. S3C and S3D).
3.5. IsomiR expression profiles and sequence relationships
For every miRNA locus, we found that multiple isomiRs could be detected, but only several of them were highly enriched and many did not possess higher expression percentages (Fig. 3A). For example, isomiRs in miR-10b-5p locus, one isomiR had absolute expression advantage (p < 0.05 based on trend test), and similar result could be found in other gene loci, while two dominant isomiRs with higher expression patterns were detected in miR-181a-5p locus (p < 0.05) (Fig. 3A). Generally, among the 8 miRNA loci, 1–2 isomiRs could possess>30% total expression (based on variants with RPM > 20), and the most dominant isomiR showed diverse expression percentage across different miRNA loci (Fig. 3B). Except for miR-181b-5p locus, the most dominant isomiRs in other loci possessed more than 50%, and even more than 80% in miR-625-3p locus. Further analysis based on tumor and normal samples showed that the most dominant isomiRs from many gene loci had consistent sequences (Fig. 3C). These results indicated that isomiR expression profiles were always stable, and the most dominant isomiR may have absolute expression advantage. However, other isomiRs, should be not ignored because they may also have unexpectedly expression levels, especially those isomiRs with abnormal expression in tumor samples. In order to understand the detailed expression patterns, a pan-cancer expression analysis was performed. IsomiRs showed dynamic expression across diverse cancers as well as their canonical miRNAs, and different isomiRs also had inconsistent expression patterns (Fig. 4A).
Fig. 3.
IsomiR expression patterns across different miRNA loci in CHOL. A. The detailed expression distribution for miRNA variants based on involved miRNA loci (all of these variants with RPM > 20), and number of variants are also presented. For the expression distributions in the specific miRNA locus, p value based on trend test is presented. The canonical miRNA sequence is highlighted with red. Note: the canonical miR-194-5p is not detected. B. Distribution of dominantly expressed isomiRs on the left (if isomiR possesses>30% total expression for the specific miRNA locus), and distribution of the most dominant isomiR is presented on the right based on individual sample. The mean expression percentage and standard deviation are also presented. C. Distribution of the most dominant isomiR in tumor and normal samples, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Expression patterns across diverse cancer types and sequence analysis. A. Expression patterns of involved isomiRs across diverse cancer types, and distributions of seeds are also presented below the image. Although multiple isomiRs are detected from miRNA loci, only 1–2 seeds are found. The distribution for the total seeds is also presented on the right. B. A phylogenetic tree of homologous miRNA genes in mir-181 gene family. mir-181d is also simultaneously analyzed despite it is not involved in validated interactions with candidate mRNAs. C. Phylogenetic networks for miR-181-5p and miR-181-3p, respectively. Red circle indicates median vector. D. Sequence features for involved isomiRs in homologous miR-181 loci and phylogenetic network of these isomiRs. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Although multiple isomiRs could be detected, most of them were not involved in varied seeds (nucleotides 2–8) (Fig. 4A). Generally, only 1–2 seeds were detected in these miRNA loci based on dominantly expressed isomiRs, indicating that the stable seed sequences could ensure the miRNA:mRNA interactions. Seed from miR-181-5p (including homologous miRNAs in miR-181 gene family) was even derived from 16 different isomiRs. Among dominantly expressed seeds, only 11 novel seeds were involved in “seed shifting” events, and others were annotated canonical seeds. To understand the expression patterns of isomiRs with novel seeds, we further screened several novel seeds and found their dominant expression patterns (Fig. S4B). Compared with canonical seeds, isomiRs with novel seeds generated from seed shifting (shifting to 5’ or 3’ terminus) might gain the new mRNA targets or lose some mRNA targets via the new 5’ terminus. Thus, consistent seeds from multiple isomiRs with sequence and expression heterogeneities can contribute to interactions between small RNAs and target mRNAs, which maybe not perturb coding-non-coding RNA regulatory network although it is unclear whether length variations affect regulation efficiency, while novel seeds may perturb regulatory network by gaining or losing target mRNAs.
Interestingly, we found that 3 miRNAs were derived from homologous miRNA loci, including miR-181a-5p, miR-181b-5p and miR-181c-5p. Indeed, there are 4 members in miR-181 gene family (miR-181a, miR-181b, miR-181c and miR-181d), while miR-181d was not identified as candidate interacted miRNAs with screened mRNAs. Among the 4 homologous miRNA genes, two clades were clustered (Fig. 4B), indicating the close phylogenetic relationships among mir-181a (including 2 multicopy genes) and mir-181c loci, and mir-181b (including 2 multicopy genes) and mir-181d loci, respectively. The mature miRNAs, mainly including miR-181-5p and miR-181-3p, showed different phylogenetic relationships (Fig. 4C). Network of miR-181-3p showed the larger genetic distances, and more median vectors were involved in constructing the phylogenetic network. miR-181-3p was ever considered as star miRNA, and it was not well-conserved among different homologous miRNAs. For the well-conserved miR-181-5p, relevant isomiRs showed complex network than their canonical miRNA sequences (Fig. 4C and D). Herein, no varied nucleotides were considered and only variations in 5′ or 3′ ends were involved to search and identify isomiRs, and thus the varied nucleotides were mainly derived from variation among homologous miRNAs.
3.6. Validation of target mRNAs for isomiRs with novel seeds
To understand the potential change of regulatory pattern when more isomiRs were involved in analysis, according to dominant expression patterns, we selected down-regulated isomiRs from miR-101-3p and miR-194-5p loci to predict their target mRNAs. Except for the 2 canonical miRNA seeds, there were 2 novel seeds in miR-194-5p and 1 novel seed in miR-101-3p, and they showed significant difference in gaining or losing of target mRNAs (Fig. 5A and B). Although isomiRs with shifted seeds were only involved in one-nucleotide difference, they might remain the same targets with their canonical miRNAs, lose targets, or even gain novel target mRNAs (Fig. 5B). The dynamic fluctuation of isomiR:mRNA interactions perturbed and complicated the original miRNA:mRNA interactions [21], especially for those new interactions, showing the potential roles in relevant biological processes.
Fig. 5.
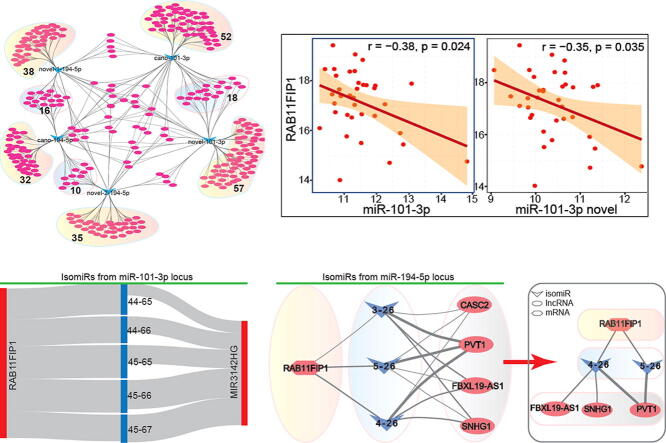
IsomiR:mRNA interaction and expression patterns. A. Expression distributions of screened predicted target mRNAs and prediction scores using TargetScan algorithm for canonical seeds and novel seeds from miR-101-3p and miR-194-5p loci. A scatter plot shows screened significantly up-regulated target mRNAs, and distribution of baseMean values and predicted miRNA:mRNA scores are also presented. B. The interaction network among target mRNAs of canonical seeds and novel seeds showing gain or loss of target mRNAs. The specific target mRNAs are circled and their numbers are also presented. C. The binding sites of isomiRs in miR-101-3p and target RAB11FIP1 according to TargetScan (http://www.targetscan.org/cgi-bin/targetscan/vert_72/view_gene.cgi?rs=ENST00000287263.4&taxid=9606&members=miR-194-5p&showcnc=0&shownc=0&shownc_nc=&showncf1=&showncf2=&subset=1#miR-101-3p.1). D. The consistent expression pattern in CHOL based on sequencing data for the isomiR and mRNA levels. For isomiRs, the Y-axis is log2RPM; for mRNA, the Y-axis is log2FPKM. E. The significant negative correlations between isomiRs in miR-101-3p and RAB11FIP1.
Here, based on the dynamic target mRNAs between the canonical seed and novel seed, we mainly focused on whether they could simultaneously bind to the same target RAB11FIP1 that was screened via protein profiling. According to interaction between small RNA and RAB11FIP1, both the shifted isomiR and its canonical miRNA could bind to the same target RAB11FIP1 (Fig. 5C). They showed opposite expression patterns: RAB11FIP1 was significantly up-regulated, while both canonical miR-101-3p and its isomiR with the novel seed were significantly down-regulated (Fig. 5D). The opposite expression implied interactions between the small RNAs and RAB11FIP1. IsomiR with novel seed showed binding of the whole seed and RAB11FIP1, while canonical miRNA showed only interaction between part of seed (nucleotides 2–7) and RAB11FIP1. Shifted seed led to the stable binding of isomiR and target mRNA, strongly indicating the potential regulatory roles of multiple isomiRs. Simultaneously, the significant negative correlations could be found between isomiRs and RAB11FIP1 (Fig. 5E), and the similar correlations were also detected via all dominant isomiRs from miR-101-3p gene locus (Fig. S4C). Further experimental validation showed that isomiRs with novel seeds in miR-101-3p locus could inhibit RAB11FIP1 than its canonical miRNA [50]. These results indicated that isomiR with shifted seed also had biological role as well as the canonical miRNA, but the varied expression pattern and sequence, especially for the varied seed, would complicate the interactions between the small RNA and their target mRNA.
3.7. Associated lncRNAs based on screened miRNAs
According to the screened 8 miRNAs, a total of 207 related lncRNAs were firstly collected. miR-194-5p loci was detected 88 related lncRNAs (Fig. S4D), and some lncRNAs were prone to locate on several chromosomes, such as chr17 (8.43%) and chr11 (8.43%). Of these, the same numbers of related lncRNAs (57) were detected in homologous miR-181-5p loci, indicating these homologous miRNAs could interact with the same lncRNAs, which mainly derived from similar miRNA sequences.
Based on relative enrichment levels and interactions among different RNAs (if miRNA was significantly up-regulated, its related lncRNA was significantly down-regulated), 9 significantly deregulated lncRNAs were finally obtained (CASC2, FBXL19-AS1, FENDRR, LINC00265, LINC02015, LINC02027, MIR3142HG, PVT1, and SNHG1) (Fig. 6A). Three of them were significantly down-regulated, while other 6 genes were significantly up-regulated. Many of these lncRNAs showed association with other RNAs. CASC2 acts as a tumor suppressor and potentially as a competing endogenous RNA for miR-21, and plays important role in IDH1 wild type glioma pathogenesis and patients' outcomes [51], LINC00265 can promote colorectal tumorigenesis via ZMIZ2 and USP7-mediated stabilization of beta-catenin [52], and PVT1 exon 9 regulates claudin 4 expression and migration in CL TNBC cells, and may have clinical implications in CL TNBC [53]. A pan-cancer analysis showed that some were also deregulated in other cancer types (Fig. 6B), indicating that they might be correlated with the pathological processes. Of these, LINC02015 was significantly down-regulated in CHOL and KICH, but it was up-regulated in other cancer types. Fewer studies focused on this gene, but the diverged expression pattern may implicate its role in diverse tissues. Furthermore, we attempted to obtain related lncRNAs with screened proteins, but no related lncRNAs were found.
Fig. 6.
Screening of related lncRNAs and potential interactions among different RNAs. A. The expression distributions of related lncRNAs in tumor samples, and fold change and adjust p-values are also presented. B. Expression patterns of lncRNAs in diverse cancer types. * sig indicates abnormal expression, log2FC > 1.5 and padj < 0.05, or log2FC < -1.5 and padj < 0.05. C. Paired analysis of screened lncRNAs based on paired samples. D. Interactions among different RNAs based on multiple isomiRs from miR-101-3p locus. Red column shows up-regulated expression, blue column shows down-regulated expression, and link indicates the absolute value of correlation coefficient. E. Interactions among different RNAs based on multiple isomiRs from miR-194-5p locus. Red column shows up-regulated expression, blue column shows down-regulated expression, and link indicates the absolute value of correlation coefficient. The right interaction shows negative correlation (only negative correlations between isomiRs and PVT1 are significant). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.8. The cross-talks among diverse RNAs
To further understand the detailed expression distributions of screened lncRNAs, a paired analysis was then performed. We found that FENDRR and LINC02015 were not significantly deregulated (Fig. 6C), and thus 7 lncRNAs were finally obtained that were related with miR-101-3p, miR-194-5p, or miR-625-3p. There were potential interactions between deregulated mRNAs, miRNAs and lncRNAs (Fig. S4F), and miRNAs were crucial RNAs that interacted mRNAs and lncRNAs as regulatory molecules.
Based on the potential roles of multiple isomiRs in miRNA locus, dominantly expressed isomiRs were queried for correlations with other RNAs. Due to the lower enrichment level of LINC02027, further analysis would focus on other gene loci. In miR-101-3p locus, 5 isomiRs containing 2 seeds were involved, and the interactions indicated a potential ceRNA network between diverse RNAs (Fig. 6D). Similar to classical miRNA, isomiR with the shifted seed also could bind to mRNAs and lncRNAs (Fig. S5), indicating the potential biological roles of isomiRs with sequence and expression diversity. Similarly, multiple isomiRs in the miR-194-5p locus also indicated their biological interactions with other mRNAs and lncRNAs (Fig. 6E). Of these, due to the small sample size, the correlation coefficient may be not significant, but the interactions still provided the potential cross-talk among different RNAs, especially between mRNAs and ncRNAs. These interactions may contribute to the pathological process, and further complicate the coding-non-coding RNA interaction network.
4. Discussion
miRNAs have been widely studied as a class of important inhibitory regulators at the post-transcriptional level via interactions with specific target mRNAs. Increasing evidences have shown that miRNAs contribute to diagnosis, prognosis and stratification of cancer treatment in CCA [54], [55], [56], [57], and complex interactions with other molecules are also widely studied [58], [59], especially interactions contribute to cancer pathology via ceRNA regulatory network [9], [60], [61], [62]. However, many studies only focus on the classical annotated miRNA despite miRNA is not a single sequence, but it is a series of multiple isomiRs with expression and sequence diversity [16], [17], [18], [19], [20]. Interactions among different RNAs are more complex than we thought, because these multiple isomiRs generated from a given miRNA locus extremely complicate the coding-non-coding RNA regulatory network. These isomiRs with heterogeneities of sequence and expression are not random events in miRNA maturation process, which may be a strategy to select dominant sequence in specific species [16]. Varied isomiRs involved in “seed shifting” events may lead to gain or loss of target mRNAs [50], which will perturb the coding-non-coding RNA interactions that might be crucial in the occurrence and development of cancers. Current related studies always ignore these varied miRNA isoforms, although these multiple isomiRs are not random and they also have potential contributions to cancer. Therefore, it is necessary to explore the interactions among different RNAs from the isomiR level, which will help understanding of the detailed regulatory pattern in vivo.
Herein, based on protein profiling and biological interactions among RNAs, especially the intermediate link of small ncRNAs (at miRNA and isomiR levels), we obtained two groups of potential ceRNA networks (Fig. 6D and E). IsomiRs from the miR-101-3p locus are involved in the shifted seeds, but these deregulated isomiRs may also contribute to abnormal expression patterns of their target mRNAs. Experimental validation indicates the regulating role of isomiR with shifted seed [50], and varied seeds expand and complicate the miRNA repression process [63]. The isomiR-associated ceRNA network could be used to predict interactions among different RNAs, and lncRNAs may perturb isomiR:mRNA network that further control mRNA expression. If the expression balance is disturbed, further abnormal expression patterns may directly or indirectly contribute to further perturbing of biological pathways. Previous studies have shown that lncRNA SPRY4-IT1 can positively regulate the expression of EZH2 through sponging miR-101-3p in cholangiocarcinoma [48], and lncRNA SOX2OT contributes to gastric cancer progression by sponging miR-194-5p from AKT2 [64]. RAB11FIP1, also known as RCP, is a RAB-coupling protein. It may be a human breast cancer-promoting gene with Ras-activating function [65], and can mediate epithelial-mesenchymal transition and invasion in esophageal cancer [66]. Some lncRNAs are also concerned due to their crucial roles in cancer. For example, PVT1 can promote cell proliferation and migration by silencing ANGPTL4 expression [67], and SNHG1 may promote malignancy of CCA by regulating the transcription of CDKN1A [68]. Most screened genes have been verified with crucial roles in multiple biological processes, and further studies should be focus on their correlations using experimental validation.
Taken together, based on protein profiling, we firstly screened potential crucial proteins to search consistent mRNAs, then to survey related miRNAs and lncRNAs. Interactions between deregulated RNAs are further queried at the isomiR levels, indicating that isomiRs with shifted seeds also contribute to the coding-non-coding RNA regulatory network. We finally obtained several potential ceRNA networks associated with RAB11FIP1, and these interactions imply complex cross-talks among different RNAs, especially interactions at the isomiR level. Further experimental validation should be focus on their biological function and potential roles in the occurrence and development of cancer, particularly the potential values in cancer diagnosis and prognosis, and precision medicine.
Author contributions
Li Guo: project design, data analyses, manuscript writing. Yuyang Dou: data analyses. Yifei Yang: data analyses. Shiqi Zhang: data analyses. Yihao Kang: data analyses. Lulu Shen: experimental validation. Lihua Tang: data analyses. Yaodong Zhang: data analyses. Changxian Li: data analyses. Jun Wang: data analyses. Tingming Liang: project design, data analyses, manuscript writing. Xiangcheng Li: project design.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Nos. 62171236, 61771251 and 81670570), Sponsored by NUPTSF (No. NY220041), the Qinglan Project in Jiangsu Province, and the Priority Academic Program Development of Jiangsu Higher Education Institution (PAPD).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.10.014.
Contributor Information
Tingming Liang, Email: tmliang@njnu.edu.cn.
Xiangcheng Li, Email: drxcli@njmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Petrowsky H., Hong J.C. Current surgical management of hilar and intrahepatic cholangiocarcinoma: the role of resection and orthotopic liver transplantation. Transplant Proc. 2009;41(10):4023–4035. doi: 10.1016/j.transproceed.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Squadroni M., Tondulli L., Gatta G., Mosconi S., Beretta G., Labianca R. Cholangiocarcinoma. Crit Rev Oncol Hematol. 2017;116:11–31. doi: 10.1016/j.critrevonc.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Torgeson A., Lloyd S., Boothe D., Cannon G., Garrido-Laguna I., Whisenant J. Chemoradiation therapy for unresected extrahepatic cholangiocarcinoma: a propensity score-matched analysis. Ann Surg Oncol. 2017;24(13):4001–4008. doi: 10.1245/s10434-017-6131-9. [DOI] [PubMed] [Google Scholar]
- 4.Skipworth J.R., Olde Damink S.W., Imber C., Bridgewater J., Pereira S.P. Review article: surgical, neo-adjuvant and adjuvant management strategies in biliary tract cancer. Aliment Pharmacol Ther. 2011;34:1063–1078. doi: 10.1111/j.1365-2036.2011.04851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu L., Li Y., Zhou L., Dorfman R.G., Liu L. SIRT3 elicited an anti-Warburg effect through HIF1alpha/PDK1/PDHA1 to inhibit cholangiocarcinoma tumorigenesis. Cancer Med. 2019;8:2380–2391. doi: 10.1002/cam4.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen H., Zhang J., Zhang Y., Feng Q., Wang H., Li G. Knockdown of tripartite motif 59 (TRIM59) inhibits proliferation in cholangiocarcinoma via the PI3K/AKT/mTOR signalling pathway. Gene. 2019;698:50–60. doi: 10.1016/j.gene.2019.02.044. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., Zhang L., Lu S., Xiang Y., Zeng C., He T. Long Non-coding RNA CASC15 Promotes Intrahepatic Cholangiocarcinoma Possibly through Inducing PRDX2/PI3K/AKT Axis. Cancer Res Treat. 2021;53(1):184–198. doi: 10.4143/crt.2020.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J., Han C., Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating beta-catenin. Gastroenterology. 2012;143(246–256) doi: 10.1053/j.gastro.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Hu K.B., Zhang Y.Q., Yang C.J., Yao H.H. Comprehensive analysis of aberrantly expressed profiles of lncRNAs, miRNAs and mRNAs with associated ceRNA network in cholangiocarcinoma. Cancer Biomark. 2018;23(4):549–559. doi: 10.3233/CBM-181684. [DOI] [PubMed] [Google Scholar]
- 10.Zhu H., Zhai B.o., He C., Li Z., Gao H., Niu Z. LncRNA TTN-AS1 promotes the progression of cholangiocarcinoma via the miR-320a/neuropilin-1 axis. Cell Death Dis. 2020;11(8):637. doi: 10.1038/s41419-020-02896-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X., Lin B., Luo M., Chu W., Li P., Liu H. Identifying circRNA- and lncRNA-associated-ceRNA networks in the hippocampi of rats exposed to PM2.5 using RNA-seq analysis. Genomics. 2021;113(1):193–204. doi: 10.1016/j.ygeno.2020.12.025. [DOI] [PubMed] [Google Scholar]
- 14.Chen W., Yu J., Xie R., Zhou T., Xiong C. Roles of the SNHG7/microRNA95p/DPP4 ceRNA network in the growth and (131)I resistance of thyroid carcinoma cells through PI3K/Akt activation. Oncol Rep. 2021:45. doi: 10.3892/or.2021.7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding Y., Li T., Yan X., Cui M., Wang C., Wang S. Identification of hub lncRNA ceRNAs in multiple sclerosis based on ceRNA mechanisms. Mol Genet Genomics. 2021;296(2):423–435. doi: 10.1007/s00438-020-01750-1. [DOI] [PubMed] [Google Scholar]
- 16.Guo L., Liang T. MicroRNAs and their variants in an RNA world: implications for complex interactions and diverse roles in an RNA regulatory network. Brief Bioinform. 2018;19:245–253. doi: 10.1093/bib/bbw124. [DOI] [PubMed] [Google Scholar]
- 17.Cloonan N., Wani S., Xu Q., Gu J., Lea K., Heater S. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011;12(12):R126. doi: 10.1186/gb-2011-12-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neilsen C.T., Goodall G.J., Bracken C.P. IsomiRs–the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012;28(11):544–549. doi: 10.1016/j.tig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Tan G.C., Chan E., Molnar A., Sarkar R., Alexieva D., Isa I.M. 5' isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014;42(14):9424–9435. doi: 10.1093/nar/gku656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telonis A.G., Magee R., Loher P., Chervoneva I., Londin E. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017;45:2973–2985. doi: 10.1093/nar/gkx082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang T., Han L., Guo L.i. Rewired functional regulatory networks among miRNA isoforms (isomiRs) from let-7 and miR-10 gene families in cancer. Comput Struct Biotechnol J. 2020;18:1238–1248. doi: 10.1016/j.csbj.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Kwast R.V.C.T., Woudenberg T., Quax P.H.A., Nossent A.Y. MicroRNA-411 and Its 5'-IsomiR have distinct targets and functions and are differentially regulated in the vasculature under ischemia. Mol Ther. 2020;28(1):157–170. doi: 10.1016/j.ymthe.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dika E., Broseghini E., Porcellini E., Lambertini M., Riefolo M., Durante G. Unraveling the role of microRNA/isomiR network in multiple primary melanoma pathogenesis. Cell Death Dis. 2021;12(5):473. doi: 10.1038/s41419-021-03764-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhardwaj A., Singh H., Trinidad C.M., Albarracin C.T., Hunt K.K., Bedrosian I. The isomiR-140-3p-regulated mevalonic acid pathway as a potential target for prevention of triple negative breast cancer. Breast Cancer Res. 2018;20(1):150. doi: 10.1186/s13058-018-1074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karali M., Persico M., Mutarelli M., Carissimo A., Pizzo M., Singh Marwah V. High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res. 2016;44(4):1525–1540. doi: 10.1093/nar/gkw039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D., Weinberg R. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Chang L., Zhou G., Soufan O., Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48:W244–W251. doi: 10.1093/nar/gkaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 34.Huson D.H. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14(1):68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 35.Bandelt H.J., Forster P., Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 36.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J.H., Li J.H., Shao P., Zhou H., Chen Y.Q. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polimanti R., Gelernter J. ADH1B: From alcoholism, natural selection, and cancer to the human phenome. Am J Med Genet B Neuropsychiatr Genet. 2018;177:113–125. doi: 10.1002/ajmg.b.32523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li T., Huang H., Shi G., Zhao L., Zhang Z. TGF-beta1-SOX9 axis-inducible COL10A1 promotes invasion and metastasis in gastric cancer via epithelial-to-mesenchymal transition. Cell Death Dis. 2018;9:849. doi: 10.1038/s41419-018-0877-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K., MacLeod G. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163(6):1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 43.Blomen V.A., Majek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350(6264):1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 44.Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J. Identification and characterization of essential genes in the human genome. Science. 2015;350(6264):1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zerhouni M., Martin A.R., Furstoss N., Gutierrez V.S., Jaune E., Tekaya N. Dual covalent inhibition of PKM and IMPDH targets metabolism in cutaneous metastatic melanoma. Cancer Res. 2021;81(14):3806–3821. doi: 10.1158/0008-5472.CAN-20-2114. [DOI] [PubMed] [Google Scholar]
- 46.Calabretta S., Bielli P., Passacantilli I., Pilozzi E., Fendrich V., Capurso G. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. 2016;35(16):2031–2039. doi: 10.1038/onc.2015.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shinohara H., Taniguchi K., Kumazaki M., Yamada N., Ito Y., Otsuki Y. Anti-cancer fatty-acid derivative induces autophagic cell death through modulation of PKM isoform expression profile mediated by bcr-abl in chronic myeloid leukemia. Cancer Lett. 2015;360(1):28–38. doi: 10.1016/j.canlet.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 48.Xu Y.i., Yao Y., Jiang X., Zhong X., Wang Z., Li C. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J Exp Clin Cancer Res. 2018;37(1) doi: 10.1186/s13046-018-0747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H., Wang L.i., Tang L., Luo J., ji H., Zhang W. Long noncoding RNA SNHG6 promotes proliferation and angiogenesis of cholangiocarcinoma cells through sponging miR-101-3p and activation of E2F8. J Cancer. 2020;11(10):3002–3012. doi: 10.7150/jca.40592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo L., Li Y., Cirillo K.M., Marick R.A., Su Z. mi-IsoNet: systems-scale microRNA landscape reveals rampant isoform-mediated gain of target interaction diversity and signaling specificity. Brief Bioinform. 2021 doi: 10.1093/bib/bbab091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skiriute D., Stakaitis R., Steponaitis G., Tamasauskas A., Vaitkiene P. The Role of CASC2 and miR-21 interplay in glioma malignancy and patient outcome. Int J Mol Sci. 2020;21(21):7962. doi: 10.3390/ijms21217962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Y., Gu L., Lin X., Cui K., Liu C. LINC00265 promotes colorectal tumorigenesis via ZMIZ2 and USP7-mediated stabilization of beta-catenin. Cell Death Differ. 2020;27:1316–1327. doi: 10.1038/s41418-019-0417-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levine F., Ogunwobi O.O. Targeting PVT1 Exon 9 Re-Expresses Claudin 4 protein and inhibits migration by claudin-low triple negative breast cancer cells. Cancers (Basel) 2021;13(5):1046. doi: 10.3390/cancers13051046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puik J.R., Meijer L.L., Le Large T.YS., Prado M.M., Frampton A.E., Kazemier G. miRNA profiling for diagnosis, prognosis and stratification of cancer treatment in cholangiocarcinoma. Pharmacogenomics. 2017;18(14):1343–1358. doi: 10.2217/pgs-2017-0010. [DOI] [PubMed] [Google Scholar]
- 55.Chen Q., Wang C., Zhang H., Li Y., Cao Y., Zhang Y. Expression levels of serum miRNA-195 in different types of patients with cholangiocarcinoma and its value to determine the prognosis thereof. Oncol Lett. 2018:5947–5951. doi: 10.3892/ol.2018.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loeffler M.A., Hu J., Kirchner M., Wei X., Xiao Y. miRNA profiling of biliary intraepithelial neoplasia reveals stepwise tumorigenesis in distal cholangiocarcinoma via the miR-451a/ATF2 axis. J Pathol. 2020;252:239–251. doi: 10.1002/path.5514. [DOI] [PubMed] [Google Scholar]
- 57.Ursu S., Majid S., Garger C., de Semir D., Bezrookove V., Desprez P.-Y. Novel tumor suppressor role of miRNA-876 in cholangiocarcinoma. Oncogenesis. 2019;8(8):42. doi: 10.1038/s41389-019-0153-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He Y., Liu C., Song P., Pang Z., Mo Z., Huang C. Investigation of miRNA- and lncRNA-mediated competing endogenous RNA network in cholangiocarcinoma. Oncol Lett. 2019:5283–5293. doi: 10.3892/ol.2019.10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwon H., Song K., Han C., Zhang J., Lu L.u., Chen W. Epigenetic Silencing of miRNA-34a in human cholangiocarcinoma via EZH2 and DNA methylation: impact on regulation of notch pathway. Am J Pathol. 2017;187(10):2288–2299. doi: 10.1016/j.ajpath.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang Z., Guo L., Zhu Z., Qu R. Identification of prognostic factors for intrahepatic cholangiocarcinoma using long non-coding RNAs-associated ceRNA network. Cancer Cell Int. 2020;20:315. doi: 10.1186/s12935-020-01388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang B., Zhou M., Zou L., Miao J., Wang Y. Long non-coding RNA LOXL1-AS1 acts as a ceRNA for miR-324-3p to contribute to cholangiocarcinoma progression via modulation of ATP-binding cassette transporter A1. Biochem Biophys Res Commun. 2019;513:827–833. doi: 10.1016/j.bbrc.2019.04.089. [DOI] [PubMed] [Google Scholar]
- 62.Long J., Xiong J., Bai Y.i., Mao J., Lin J., Xu W. Construction and investigation of a lncRNA-Associated ceRNA regulatory network in cholangiocarcinoma. Front Oncol. 2019;9:649. doi: 10.3389/fonc.2019.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bofill-De Ros X., Yang A., Gu S. IsomiRs: expanding the miRNA repression toolbox beyond the seed. Biochim Biophys Acta Gene Regul Mech. 2020;1863(4):194373. doi: 10.1016/j.bbagrm.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qu F., Cao P. Long noncoding RNA SOX2OT contributes to gastric cancer progression by sponging miR-194-5p from AKT2. Exp Cell Res. 2018;369(2):187–196. doi: 10.1016/j.yexcr.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 65.Zhang J., Liu X., Datta A., Govindarajan K., Tam W.L. RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest. 2009;119:2171–2183. doi: 10.1172/JCI37622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang Q., Lento A., Suzuki K., Efe G., Karakasheva T. Rab11-FIP1 mediates epithelial-mesenchymal transition and invasion in esophageal cancer. EMBO Rep. 2021;22 doi: 10.15252/embr.201948351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu Y., Zhang M., Liu J., Xu B., Yang J., Wang N.i. Long Non-coding RNA PVT1 promotes cell proliferation and migration by silencing ANGPTL4 expression in cholangiocarcinoma. Mol Ther Nucleic Acids. 2018;13:503–513. doi: 10.1016/j.omtn.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu Y., Zhang M., Wang N.i., Li Q., Yang J., Yan S. Epigenetic silencing of tumor suppressor gene CDKN1A by oncogenic long non-coding RNA SNHG1 in cholangiocarcinoma. Cell Death Dis. 2018;9(7):746. doi: 10.1038/s41419-018-0768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.