Abstract
Human phospholipase A2s (PLA2) constitute a superfamily of enzymes that hydrolyze the sn-2 acyl-chain of glycerophospholipids, producing lysophospholipids and free fatty acids. Each PLA2 enzyme type contributes to specific biological functions based on its expression, subcellular localization, and substrate specificity. Among the PLA2 superfamily, the cytosolic cPLA2 enzymes, calcium-independent iPLA2 enzymes, and secreted sPLA2 enzymes are implicated in many diseases, but a central issue is the preference for double-bond positions in polyunsaturated fatty acids (PUFAs) occupying the sn-2 position of membrane phospholipids. We demonstrate that each PLA2 has a unique preference between the specific omega-3 fatty acids eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) and the omega-6 arachidonic acid (AA), which are the precursors of most proinflammatory and anti-inflammatory or resolving eicosanoids and related oxylipins. Surprisingly, we discovered that human cPLA2 selectively prefers AA, whereas iPLA2 prefers EPA, and sPLA2 prefers DHA as substrate. We determined the optimal binding of each phospholipid substrate in the active site of each PLA2 to explain these specificities. To investigate this, we utilized recently developed lipidomics-based LC-MS/MS and GC/MS assays to determine the sn-2 acyl chain specificity in mixtures of phospholipids. We performed μs timescale molecular dynamics (MD) simulations to reveal unique active site properties, especially how the precise hydrophobic cavity accommodation of the sn-2 acyl chain contributes to the stability of substrate binding and the specificity of each PLA2 for AA, EPA, or DHA. This study provides the first comprehensive picture of the unique substrate selectivity of each PLA2 for omega-3 and omega-6 fatty acids.
Supplementary key words: phospholipase A2, arachidonic acid, glycerophospholipid, fatty acid, omega-3, omega-6, substrate specificity, EPA, DHA, AA
Abbreviations: AA, arachidonic acid; DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; LPC, lysophosphatidylcholine; PLA2, phospholipase A2; PUFA, polyunsaturated fatty acid
Glycerophospholipids are a major component of cell membranes and are the defining component of the phospholipid bilayer. They contain a glycerol backbone with esterified fatty acids on the sn-1 and sn-2 positions and a phosphodiester headgroup on the sn-3 position. Phospholipase A2 (PLA2) is an enzyme that hydrolyzes the sn-2 acyl chain of glycerophospholipids and produces lysophospholipids and free fatty acids as products. PLA2 constitutes a superfamily of enzymes composed of six different types of enzymes, and some types consist of various groups and subgroups based on structural features (1). PLA2s contribute numerous biological functions by hydrolyzing membrane glycerophospholipids producing lysophospholipids and free fatty acids, which are converted into bioactive mediators that have important downstream effects especially in inflammatory processes (2). The function of each PLA2 type or group predominantly depends on its expression, subcellular localization, and substrate specificity. Therefore, to understand the complex functions of PLA2s in lipid metabolism, it is essential to elucidate the complete specificity of each enzyme at the molecular level.
Furthermore, elucidating the mechanism of each PLA2’s substrate specificity provides the basis for developing specific inhibitors and enables one to predict unknown substrate specificity, functions, and role in disease. Among the human PLA2 superfamily, the cytosolic cPLA2, calcium-independent iPLA2, and secreted sPLA2 are the three essential types of PLA2. In this study, we sought to elucidate the substrate specificity and detailed binding mode of the Group IVA (GIVA) cPLA2, Group VIA (GVIA) iPLA2, and Group V (GV) sPLA2 as each is responsible for the production of bioactive mediators derived from fatty acids (1, 3, 4, 5).
A very large number of specific glycerophospholipid molecular species exist because of the combinatorial possibilities of two acyl chains and a headgroup, and each molecular species exhibits distinct chemical and structural features. Fatty acyl chains are quite variable based on the length of the carbon chain and the position and number of double bonds. Furthermore, we have found that each PLA2 has a unique acyl chain specificity for the cleaved fatty acid at the sn-2 position, and the specificity contributes significantly to its function. In contrast to the variety of acyl chains in mammalian phospholipids, the headgroup species are more limited, but PLA2s also have a unique preference for the specific headgroup (6). For example, GV sPLA2 shows more than twice the activity toward phosphatidylglycerol comparing to the other headgroups in mixtures of various headgroups (7). While it has been shown that each PLA2 has a different acyl chain specificity and different affinity to various headgroups (8, 9, 10), it has not been clear if there is an interaction between the head group specificity and the acyl chain specificity of each PLA2.
Among the products of PLA2s, lysophospholipids act as lipid second messengers involved in signal transduction, and especially it is well known that lysophosphatidylcholine and lysophosphatidic acid directly bind to G-protein-coupled receptors and activate downstream signaling pathways (11, 12). Also, the free fatty acid products are involved in various cellular responses. Of special note is the fact that PLA2s release free arachidonic acid (20:4, AA), which is the key omega-6 fatty acid, which is enzymatically converted into numerous eicosanoids regulating homeostatic and inflammatory processes (13, 14). Similarly, omega-3 fatty acids, especially eicosapentaenoic acid (20:5 EPA) and docosahexaenoic acid (22:6, DHA), are enzymatically converted into related oxygenated polyunsaturated fatty acids (PUFAs), which are bioactive oxylipins responsible for resolution and anti-inflammatory effects (15). Therefore, PLA2 is a crucial player in inflammation and its resolution as well as other diseases including cardiovascular consequences depending whether AA, EPA, or DHA is released (16). Moreover, the human brain enriches DHA in membrane phospholipids, and DHA is suggested to be involved in brain development and various neurological disorders (17, 18). Therefore, understanding the specificity of each PLA2 type toward AA, EPA, and DHA is important for understanding disease implications as well as PLA2 function.
In our previous in vitro study with pure human PLA2s, we employed lipidomics-based LC-MS/MS to quantify the lysophospholipids produced from conventional glycerophospholipids, and we observed the activity of PLA2s toward AA and DHA on the sn-2 position (7), but comparative phospholipids containing EPA were not available. In the present study, we took advantage of the lipidomics-based LC-MS/MS assay and a newly developed GC-MS assay, which allowed us to investigate the sn-2 acyl chain specificity in mixtures of phospholipids. We found each enzyme showed a unique preference for AA, EPA, or DHA on the sn-2 position and molecular dynamics (MD) simulations enabled elucidation of underlying binding conformations and resulting substrate selectivity.
Materials and methods
Expression and purification of GIVA cPLA2 and GVIA iPLA2
The human GIVA cPLA2 and GVIA iPLA2 were expressed and purified by the following previously established method using Sf9 insect cells and baculovirus (19). The serum-free cultured Sf9 cells were infected with baculovirus encoding a C-terminal 6×His tag conjugated human GIVA cPLA2 or N-terminal 6×His tag conjugated human GVIA iPLA2 with MOI of 0.1 and 2.0 respectively. The expression was induced for 72 h at 28°C with shaking. The cells were harvested and lysed in the lysis buffer (25 mM Tris–HCl pH 8.0, 100 mM NaCl, 10 mM β-mercaptoethanol, and protease inhibitors; in the case of iPLA2, 2 mM ATP was added). The supernatant was incubated with Ni-NTA agarose for 1 h at 4°C. The slurry was loaded on an open column and washed with the wash buffer (25 mM Tris–HCl pH 8.0, 250 mM NaCl, and 10 mM imidazole). Finally, the enzyme was eluted with elution buffer (25 mM Tris-HCl pH 7.5, 50 mM NaCl, 250 mM imidazole, 30% glycerol; in the case of iPLA2, 2 mM ATP was included in the buffer).
Expression, purification, and refolding of GV sPLA2
The human GV sPLA2 was expressed using the E. coli expression system and refolded by following and modifying the dilution method reported previously (20). The plasmid encoding C-terminal 6×His tag conjugated human GV sPLA2 was transfected into BL21 (DE-3) Gold E. coli and precultured in the LB medium for 16 h at 37°C. Twenty milliliter of the precultured cells was inoculated into 1 L of LB medium and cultured until the OD600 reached 0.7–1.0 at 37°C. Then, 0.1 mM IPTG was added and expression was induced for 4 h at 27°C. The cell pellet was harvested and frozen at –80°C. The pellet was melted and resuspended with lysis buffer (25 mM Tris-HCl pH 8.0, 100 mM NaCl, 10 mM β-mercaptoethanol, 5 mM EDTA, 0.5 mg/ml lysozyme) and incubated on ice for 1 h. The solution was sonicated six times with a 30s on 30s off-cycle and centrifuged (11,000 g for 20 min). The pellet was washed with 40 ml of buffer A (25 mM Tris-HCl pH 8.0, 10 mM β-mercaptoethanol, 5 mM EDTA) and centrifuged (11,000 g for 20 min). The supernatant was discarded, and the pellet was washed with 40 ml of 2 M urea in buffer A. The solution was centrifuged (11,000 g for 20 min), and the pellet was incubated with 40 ml of 6 M guanidine hydrochloride (GuHCl) contained buffer A for 16 h at 4°C. The solution was centrifuged (14,000 g for 30 min) and 10 mM MgCl2 was added to chelate EDTA in the supernatant. Then, the solution was incubated with Ni-NTA agarose for 1 h at 4°C. The slurry was loaded onto an open column and washed with 50 ml of wash buffer (25 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM β-mercaptoethanol, 5 mM imidazole, 1 M GuHCl). The denatured GV sPLA2 protein was eluted with elution buffer (25 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM β-mercaptoethanol, 250 mM imidazole, 6 M GuHCl). The eluted solution was dialyzed against dialyzing buffer (50 mM Tris-HCl pH 8.5, 100 mM NaCl, 6 M GuHCl), and the 0.2 mg/ml protein solution was diluted dropwise with refolding buffer (50 mM Tris-HCl pH 8.5, 8.2 mM oxidized glutathione, 9.3 mM reduced glutathione, 12 mM CaCl2 12% glycerol) at a 1: 7 dilution ratio. The diluted solution was incubated for 7 days at 4°C. Finally, the solution was dialyzed against enzyme buffer (50 mM Tris-HCl pH 8.0, 50 mM NaCl, 10 mM CaCl2, 20% glycerol) to remove GuHCl completely resulting in the GV sPLA2.
Lipidomics-based LC-MS/MS assay
The lipidomics-based LC-MS/MS assay was performed as previously described (7). In brief, we prepared a mixed micelle substrate containing 100 μM phospholipid, 400 μM C12E8 surfactant, and 2.5 μM 17:0 lysophosphatidylcholine (LPC) as an internal standard. For cPLA2, 3 μM of porcine brain PIP2 was mixed with 97 μM phospholipid to enhance the activity. The reaction was started by adding 5 μl of enzyme solution to 95 μl of substrate solution in a 96-well plate, and the plate was incubated for 30 min at 40°C with shaking. The reaction was quenched by adding 120 μl of methanol/acetonitrile (80/20, v/v), and the sample was directly injected into the HPLC-MS/MS system. Mass spectrometry of lysophospholipids was performed on a 4000 QTRAP® (AB Sciex, Framingham, MA). The product produced was quantified using a standard curve for each product, and the activity was calculated based on protein concentration.
Lipidomics-based GC-MS assay
Phospholipids with various sn-2 acyl-chains were equally mixed to 100 μM, and the substrate was prepared by the same method as employed with the LC-MS/MS assay. For the GC-MS assay, 250 ng of deuterated fatty acids were used as internal standards (16:0-d3 for 16:0, 18:1-d2 for 18:1 and 18:2, 20:4-d8 for 20:4, 20:5, and 22:6). Twenty-five microliter of the purified enzyme was incubated with 475 μl of the substrate for 30 min at 40°C. The reaction was quenched by adding 2.5 ml of Dole’s regent (isopropyl alcohol/heptane/H2SO4/H2O: 300/75/0.42/14.6). Silica gel, 1.5 ml of H2O, and 1.5 ml of heptane were added to the solution, and the solution was mixed well. The upper layer of the solution was passed through the silica gel column and dried down. The dried sample was derivatized with 50 μl of 0.5% pentafluorobenzyl bromide and 50 μl of 0.5% diisopropylethylamine in acetonitrile for 20 min. After derivatization, the sample was dried down and dissolved with 50 μl of iso-octane. The free fatty acid in the sample was analyzed using GC-MS (Agilent 6890N Gas Chromatograph and Agilent 5975 Mass Selective Detector). The amount of the products was calculated with a standard curve for each product, and the activity was calculated based on the protein concentration.
Molecular dynamics (MD) simulations
Enzyme-substrate complexes
Initial complexes of each enzyme with their studied phospholipids were generated using our previously published model based on the crystal structure of GIVA cPLA2 (21), our previously published homology model of iPLA2 based on patatin (7, 22), and a homology model of GV sPLA2 based on the crystal structure of GIIA sPLA2 (7). Phospholipids were docked in the active site of each enzyme using the Glide software implemented in the Schrödinger suite using a previously published docking protocol (7, 19, 23, 24).
Enzyme-membrane systems
The Membrane Builder implemented in CHARMM-GUI was employed to generate enzyme-membrane models for the MD simulations (25, 26). The membrane patch consisted of POPC, SAPC, POPE, POPA, POPG, POPS, SAPI(4,5)P2, and cholesterol. The average ratios of the phospholipids were chosen to be 0.48 for PC, 0.27 for PE, 0.10 for PI(4,5)P2, 0.06 for PS, and 0.09 for PA and PG. The average cholesterol/lipid ratio was chosen to be 0.40. These ratios are the average ratio of the nuclear, mitochondrial, and plasma membranes, which are the major cellular membranes and where cPLA2, iPLA2, and sPLA2 are localized and acting (6). The membrane composition was selected based on the localization of each enzyme on cellular membranes according to previously published studies (27, 28, 29). Each system was solvated with TIP3P water molecules and neutralized with 150 mM sodium chloride (NaCl) using the Visual Molecular Dynamics (VMD) package (30).
Equilibration and production runs
MD simulations were carried out using NAMD 2.12 (31). The following minimization and equilibration protocol was performed: a minimization of 80,000 steps was initially performed by applying harmonic constraints on the enzyme-ligand-membrane that were gradually turned off using a constraint scaling factor, followed by a second 120,000 steps minimization without constraints. An initial equilibration of 10,000 steps was performed by also applying harmonic constraints on the enzyme-ligand-membrane that were gradually turned off using the same constraint scaling factor, followed by a second 10,000 steps equilibration without constraints.
During the equilibration, each system was slowly heated and held to 310 K using temperature reassignment with a reassignment frequency of 500 timesteps (1,000 fs) and a reassignment increment of 1 K. The above minimization and equilibration protocol were sufficient to induce the appropriate disorder of a fluid-like bilayer, avoid unnatural atomistic positions, and failure of the simulations by atoms moving at exceedingly high velocities. Each system was finally subjected to a 1 μs production run. For each production run, the temperature was maintained at 310 K using the Langevin thermostat with Langevin coupling coefficient of 1/ps (32).
The NPT ensemble was employed and the pressure was kept constant at 1.01325 kPa using the Langevin piston method with the “useGroupPressure,” “useFlexibleCell,” and “useConstantArea” parameters turned on (33). A time step of 2 fs was used in combination with the SHAKE algorithm to hold the bonds of hydrogen atoms similarly constrained (34). Nonbonded interactions and full electrostatics were calculated every 1 and 2 time steps, respectively. Switching functions were used to smoothly take electrostatic and van der Waals interactions to zero with a switching distance of 10 Å and a cutoff of 12 Å. Long-range electrostatic forces in the periodic system were evaluated using the Particle Mesh Ewald (PME) Sum method with grid spacing 1/Å (35). The CHARMM General Force Field (CGenFF) and the CHARMM36 all-atom additive force field and parameters were used for the simulations (36, 37).
Statistical analysis
All error bars show the standard deviation (SD). A one-way ANOVA followed by the Tukey–Kramer multiple comparison test was carried out when more than three groups were compared using BellCurve for Excel Version 3.20. A P-value of less than 0.05 was considered to be significant.
Results
Effect of the headgroup on the sn-2 acyl-chain specificity
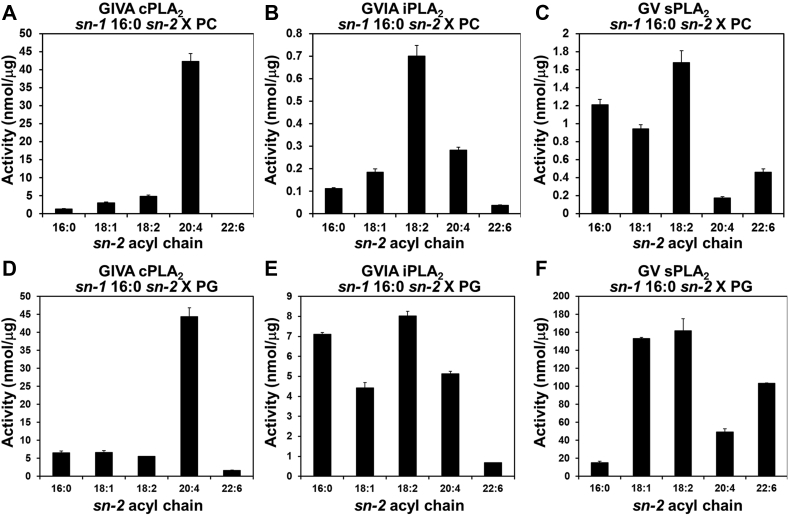
First, we employed the LC-MS/MS assay to confirm the PLA2 sn-2 acyl-chain specificity toward sn-1 16:0 (palmitic acid, PA), sn-2 X PC (PC 16:0/X) since PC is the most abundant headgroup in mammalian membranes. X was PA, 18:1 (oleic acid, OA), 18:2 (linoleic acid, LA), 20:4 (arachidonic acid, AA), and 22:6 (docosahexaenoic acid, DHA). The substrate specificity of all three enzymes was consistent with our earlier study (7). In short, cPLA2 showed high specificity toward sn-2 20:4 and no activity toward sn-2 22:6 (Fig. 1A). iPLA2 showed the highest activity toward sn-2 18:2, less activity toward 16:0, 18:1, and 20:4, and the lowest activity toward 22:6 (Fig. 1B). sPLA2 showed the highest activity toward 18:2 and did not show good activity toward 20:4 (Fig. 1C). Notably, only sPLA2 showed relatively high activity toward PC 16:0/22:6 among the three PLA2s.
Fig. 1.
The activity of human PLA2s toward PC and PG. The activity of (A, D) GIVA cPLA2, (B, E) GVIA iPLA2, and (C, F) GV sPLA2 is shown toward sn-1 16:0 sn-2 X PC and PG, where X is 16:0 (PA), 18:1 (OA), 18:2 (LA), 20:4 (AA), or 22:6 (DHA). The activity was determined and quantified using authentic lyso-PC and lyso-PG primary and internal standards. The values are mean ± SD. N = 3.
To determine the effect of the head group on the sn-2 acyl-chain specificity of the three PLA2 enzymes, we tested the activity of them toward sn-1 16:0 sn-2: X phosphatidylglycerol (PG 16:0/X), which is anionic and the best headgroup for both iPLA2 and sPLA2 (7). The sn-2 acyl chain specificity of cPLA2 was not affected by the substitution of PG for PC (Fig. 1D). In contrast, although iPLA2 showed the highest activity toward sn-2 18:2, 16:0 was also a good substrate in the case of PG (Fig. 1E). sPLA2 showed high activity toward both sn-2: 18:1 and 18:2 in the case of PG, and 16:0 was the worst substrate in the PG in contrast to PC (Fig. 1F). Notably, the overall activity of iPLA2 and sPLA2 was significantly higher in the case of PG. These results indicated that PG somewhat affects the sn-2 acyl-chain specificity of iPLA2 and sPLA2.
Acyl-chain specificity toward PE
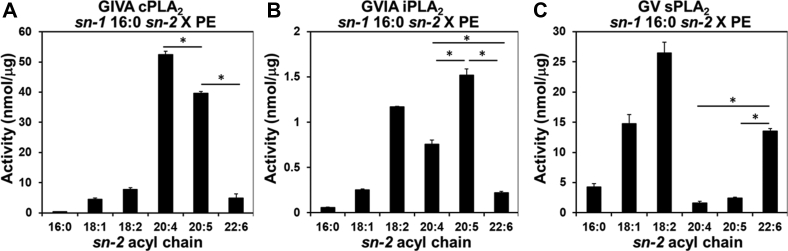
In order to examine the specificity toward EPA in the sn-2 acyl-chain of all three PLA2s, we tested PE 16:0/20:5 as well as other sn-1 16:0 sn-2: X phosphatidylethanolamine (PE 16:0/X), which is the second most abundant species in humans. Overall, the sn-2 acyl-chain specificity of all three enzymes was similar to the specificity toward PCs, that is, cPLA2 showed the highest activity toward sn-2 AA and iPLA2 and sPLA2 showed significant high activity toward sn-2 18:2 (Fig. 2A–C). Interestingly, especially focusing on the AA, EPA, and DHA, cPLA2 showed high activity toward sn-2 EPA, but the activity was not as high as the activity toward sn-2 AA (Fig. 2A). iPLA2 showed the highest activity toward sn-2 EPA, and the activity was much higher than that toward sn-2 AA (Fig. 2B). With both cPLA2 and iPLA2, the activity toward DHA was the lowest of the three sn-2 acyl chains (Fig. 2A, B). In contrast, sPLA2 showed a clear preference for sn-2 DHA in the three acyl chains and did not show activity toward EPA similar to AA (Fig. 2C).
Fig. 2.
The activity of human PLA2s toward PE. The activity of (A) GIVA cPLA2, (B) GVIA iPLA2, and (C) GV sPLA2 is shown toward sn-1 16:0 sn-2 X PE, where X is 16:0 (PA), 18:1 (OA), 18:2 (LA), 20:4 (AA), 20:5 (EPA), or 22:6 (DHA). The activity was determined and quantified using authentic lyso-PE primary and internal standards. The values are mean ± SD. N = 3. ∗P < 0.05 (one-way ANOVA followed by Tukey-Kramer post hoc test).
Acyl-chain specificity toward mixtures of phospholipids
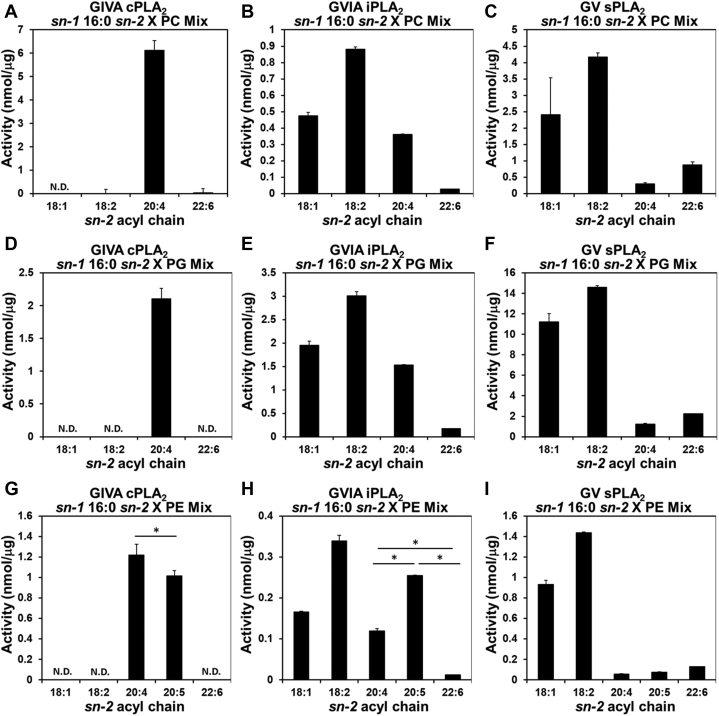
Phospholipid membranes allosterically activate PLA2s and the enzyme extracts the specific substrate phospholipid from the lipid membrane (22). Therefore, we investigated whether these enzymes show a different sn-2 acyl-chain specificity in mixtures of phospholipids containing different sn-2 acyl-chains. However, since the LC-MS/MS assay follows the lysophospholipid products, we utilized a GC-MS to directly measure the free fatty acid products. We used deuterated fatty acids as internal standards generating standard curves (Fig. S1). All three enzymes showed linearity with protein and time with their best substrate, PC 16:0/20:4 for cPLA2, PG 16:0/18:2 for iPLA2 and sPLA2 (Fig. S2). We also determined the activity of the three enzymes toward PC 16:0/20:4 by both the LC-MS/MS and GC-MS assay, and the two assays gave comparable activity (Fig. S3). Additionally, we determined the sn-2 acyl-chain specificity of iPLA2 using single PG phospholipid substrates with the GC-MS assay to confirm that the two assay methods gave similar results with the same substrate. (Fig. S4).
With the GC-MS assay established, we determined the sn-2 acyl-chain specificity of the three enzymes using a mixture of PCs, PGs, and PEs and found it overall consistent with the specificity from the single phospholipid substrates, but some minor differences were observed. Notably, in the mixture, cPLA2 showed little or no detectable activity toward other than AA and EPA (Fig. 3A, D, G). The activity of iPLA2 toward 18:1 and 18:2 became slightly better than that toward 20:4 and 20:5, respectively, in contrast to the result with the LC-MS/MS assay (Fig. 3B, E, H). In the case of sPLA2, the activity toward DHA was relatively lower than that of the LC-MS/MS assay with all headgroups (Fig. 3C, F, I). Due to the remarkably higher activity of sPLA2 toward 18:1 and 18:2 in the sn-2 position, the difference among AA, EPA, DHA in that position was not statistically significant when compared with the other sn-2 acyl chains (Fig. 3I). However, the difference of these three sn-2 acyl chains was statistically significant when we compared only the three of them with one another by one-way ANOVA. Importantly, all three enzymes showed their unique preference for AA, EPA, or DHA even in mixtures of phospholipids (Fig. 3G, H, I).
Fig. 3.
The specificity of human PLA2s toward mixtures of phospholipids. The activity of (A, D, G) GIVA cPLA2, (B, E, H) GVIA iPLA2, and (C, F, I) GV sPLA2 toward sn-1 16:0 sn-2 X PC, PG, and PE in equal proportions with 100 μM total phospholipid. X is 18:1 (OA), 18:2 (LA), 20:4 (AA), and 22:6 (DHA) for PC and PG, 18:1 (OA), 18:2 (LA), 20:4 (AA), 20:5 (EPA), and 22:6 (DHA) for PE. The activity was calculated using a standard curve for each fatty acid. N.D.: not detected. The values are mean ± SD. N = 3. ∗P < 0.05 (one-way ANOVA followed by Tukey–Kramer post hoc test).
Binding mode of phospholipids in cPLA2
To explain the lipidomics preferences toward AA, EPA, and DHA, we analyzed the binding between each PLA2 and PE 16:0/20:5 or 16:0/22:6 using μs scale MD simulations as previously reported for PE 16:0/20:4 (7).
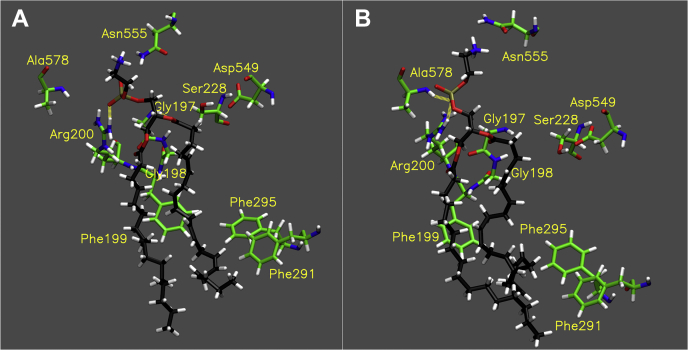
The active site of cPLA2 is a deep and narrow hydrophobic funnel enriched with aromatic amino acids (21). The binding mode of PE 16:0/20:4 revealed that the entire phospholipid was accommodated in the active site of cPLA2, and the AA on the sn-2 position was held by aromatic amino acids (Movie S1). We could see Ser228 and Asp549 composing the catalytic dyad, Asp200 contributing the binding of the phosphate group, and Gly197 and Gly198 forming the oxyanion hole, which stabilizes the negatively charged transition state of the reaction, are essential (38, 39). The simulation showed that the phosphate and headgroup were accommodated in the polar pocket, especially, Ala578, Asn555, and Arg200 formed hydrogen bonds with high occupancy (Movie S2A and Table S1). Further, the carbonyl group of the sn-2 acyl chain frequently interacted with Gly197 and 198, which forms an oxyanion hole (Movie S2A and Table S1). Notably, the fact that Arg200 formed a highly stable interaction with the phosphate group of every substrate throughout the simulation was consistent with the previous report (40).
The binding mode of PE 16:0/20:5 was similar to the PE 16:0/20:4 (Movie S3 and Fig. 4A). The headgroup and phosphate were also accommodated in the polar pocket in a similar manner as with PE 16:0/20:4, but Ala578 did not participate in the binding (Movie S2B and Table S1). Also, Gly197 and 198 interacted with the carbonyl group of the sn-2 EPA with relatively high occupancy (Movie S2B and Table S1). However, the overall occupancy of hydrogen bonds was lower than that of PE 16:0/20:4. In contrast to AA and EPA, PE 16:0/22:6 in the active site was not settled, and DHA on the sn-2 position moved significantly during the simulation (Movie S4). Further, the sn-2 DHA was a distance from the catalytic dyad in the optimal binding mode (Fig. 4B). Indeed, the mean of the distance from the catalytic serine to the carbonyl group of the sn-2 acyl chain of AA, EPA, and DHA during the simulation was 3.03 ± 0.1 Å, 3.09 ± 0.1 Å, and 6.13 ± 1.2 Å, respectively. The hydrogen bond analysis revealed that although Arg200 frequently interacted with the phosphate group, the occupancy of the binding of Gly197 and 198 to the sn-2 carbonyl group was significantly low due to its distance from the active site (Movie S2C and Table S1).
Fig. 4.
Binding of cPLA2. Image of the optimal binding mode of (A) PE 16:0/20:5 and (B) PE 16:0/22:6 in the cPLA2 active site. The images are created by the final frame of the 1 μs simulations. Movies S3 and S4 show the result of 1 μs simulation for PE 16:0/20:5 and PE 16:0/22:6, respectively. The yellow dashed lines show hydrogen binds (distance cutoff: 3.2 Å, angle cutoff: 30°).
We repeated the 1 μs simulations for EPA and DHA, and these simulations were quite consistent with the first simulations and in vitro activity (Movies S5 and S6). In brief, even in the second simulation, the sn-2 EPA chain settled in the active site during the simulation, but not sn-2 DHA. Importantly, the mean of distances between the catalytic serine and the sn-2 carbonyl during the simulation was consistent with the first simulation (EPA: 2.91 ± 0.1 Å and DHA: 6.06 ± 0.8 Å).
We analyzed the root-mean-square deviation (RMSD) of ligands, which represents the conformational fluctuations of the ligands to monitor the movement of substrates in the active site during the simulations. Thus, a low RMSD indicates less movement of the ligand. The RMSD clearly indicated that PE 16:0/22:6 was more flexible than PE 16:0/20:4 and PE 16:0/20:5 in the active site of cPLA2 during the MD simulations (Fig. S5), indicating that this flexibility of the sn-2 acyl chain results in the lower selectivity toward DHA. From the above, the pocket accommodating the tail of the sn-2 acyl chain seems to be contributing to the stability of substrates. The optimal binding mode revealed that the tails of AA and EPA were accommodated in the narrow aromatic and aliphatic cavity (Fig. S6A, B). However, the tail of DHA did not fit and settle in the cavity, and during the simulation, the tail drifted around the hydrophobic regions (Movies S4 and S6 and Fig. S6C).
Finally, we calculated binding free energy by the MM-GBSA method using the final frame of the 1 μs simulation as an optimal binding mode of each ligand. The calculation revealed that the binding stability of the ligands was AA > EPA > DHA based on the binding free energy (Table S2), which were consistent with the in vitro selectivity. These results clearly showed that the fitness of the sn-2 acyl chain in the hydrophobic binding site affects the binding of the headgroup to the hydrophilic pocket and the binding stability creating the strict substrate specificity of GIVA cPLA2.
Binding mode of phospholipids in iPLA2
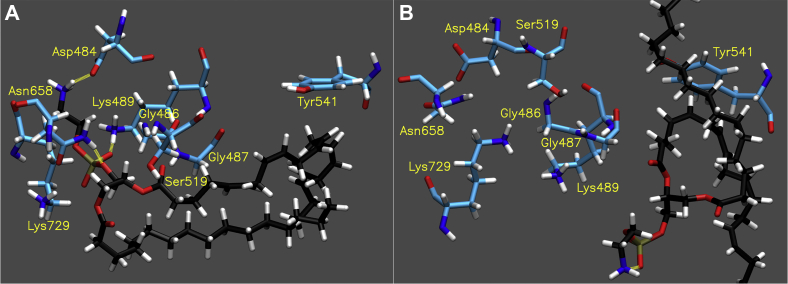
The active site of iPLA2 has a wide and flexible cavity providing its permissive specificity in contrast to cPLA2 (7). Also, it is well known that iPLA2 contains the Ser519/Asp652 catalytic dyad, and Gly487 and Gly486 form the oxyanion hole. In the iPLA2 active site, the AA tail of PE 16:0/20:4 moved around, but the AA chain had settled at the end of the simulation (Movie S7). Moreover, the headgroup seemed to be stable in the hydrophilic pocket by forming hydrogen bonds, despite the movement of the acyl chains (Movie S7). The simulation of PE 16:0/20:4 showed that Asp484, Lys729, Lys489, and Asn658 mainly formed hydrogen bonds with the phosphate and head group ethanolamine and seemed to be critical (Movie S8A and Table S3). Also, Gly487 and Gly486, which form the oxyanion hole, interacted with a carbonyl group of AA, although Gly486 only forms weak hydrogen bonds (Movie S8A and Table S3).
The binding mode of PE 16:0/20:5 was similar to the one of PE 16:0/20:4 (Movie S9 and Fig. 5A). However, the sn-2 EPA showed less movement and seemed to be more stable than AA. The simulation revealed that although there are some minor differences, similar residues interacted with the phosphate and headgroup, and most of the residues formed hydrogen bonds with higher occupancy compared with the case of AA (Movie S8B and Table S3), indicating that PE16:0/20:5 fits the active site better than PE 16:0/20:4. The binding of PE 16:0/22:6 was different from the other two. At first, PE 16:0/22:6 made a similar pose to that of the other two substrates; however, finally, the substrate dissociated from the catalytic residues and flipped in the active site (Movie S10 and Fig. 5B). The mean distance from the catalytic serine to the carbonyl group of the sn-2 DHA (5.42 ± 2.8 Å) was too great for it to hydrolyze the acyl chain. The distance of AA was slightly longer than that of EPA (3.23 ± 0.4 Å and 3.12 ± 0.1 Å, respectively). The RMSD of each ligand indicated that the dramatic conformational change of DHA after 600 ns was observed in contrast to the movement of AA and EPA in the active site (Fig. S7). Importantly, it is consistent with the in vitro specificity of iPLA2 that EPA showed the lowest binding free energy among the three substrates (Table S2).
Fig. 5.
Binding of iPLA2. Image of the optimal binding mode of (A) PE 16:0/20:5 and (B) PE 16:0/22:6 in the iPLA2 active. The images are created by the final frame of the 1 μs simulations. Movies S9 and S10 show the result of 1 μs simulation for PE 16:0/20:5 and PE 16:0/22:6, respectively. The yellow dashed lines show hydrogen binds (distance cutoff: 3.2 Å, angle cutoff: 30°).
In the second simulation, PE 16:0/20:5 showed a similar binding mode and hydrogen bond pairs with the first simulation (Movie S11), and PE 16:0/22:6 showed fewer hydrogen bonds and dissociated from the active site similar to the first simulation (Movie S12). The mean distances from the catalytic serine to the sn-2 carbonyl of EPA and DHA in the second simulation were 3.06 ± 0.1 Å and 7.08 ± 2.5 Å, respectively. These results clearly indicated that the binding of EPA in the iPLA2 active site is more stable than AA and DHA and reasonable for hydrolysis, which is consistent with the in vitro specificity of GVIA iPLA2.
Our previous study analyzed the optimal binding mode of PC 16:0/14:0 and PC 16:0/18:1, which are good substrates for iPLA2 in the iPLA2 active site. The binding mode revealed that iPLA2 utilizes two different hydrophobic pockets surrounded by either aliphatic or aromatic amino acids to accommodate the sn-2 acyl chain (Fig. S8A, B) (7). Next, we focused on the hydrophobic pockets and explored how EPA adjusts to the hydrophobic pocket. In PE 16:0/20:4, the tail of AA deeply penetrated and interacted with similar residues in the pocket that accommodates 18:1 (Fig. S8C). In contrast, the tail of EPA was not accommodated in the pocket accommodating 14:0, 18:1, and 20:4, but was surrounded by the shallow hydrophobic cavity (Fig. S8D). The additional double bond at the omega end of EPA bent the chain to fit the cavity. Indeed, the RMSD of AA and EPA on the sn-2 acyl chain indicated that EPA was more rigid than AA (Fig. S9). Similarly, the tail of DHA was accommodated in the shallow hydrophobic cavity during the first 500 ns of simulation, but the cavity did not retain the tail because of its longer chain length, resulting in dissociation from the active site (Fig. S8C). From these results, it is clear that the formation of these hydrophobic pockets stabilizing the sn-2 acyl chain is critical for the sn-2 acyl chain specificity of iPLA2.
Binding mode of phospholipids in sPLA2
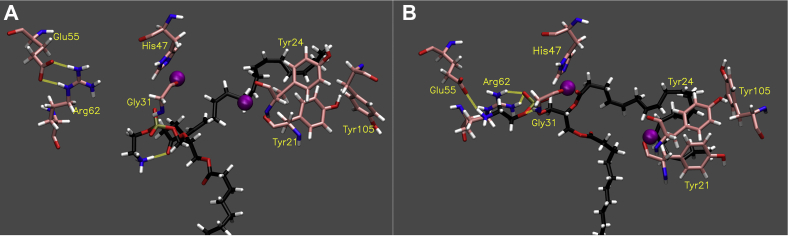
As MD simulations of sPLA2 with PE 16:0/20:4 show, in contrast to the other PLA2s, the active site of sPLA2 is a shallow cavity that accommodates only the sn-2 acyl chain, phosphate, and head group (Movie S13). For the head group and phosphate binding, Gly31, Arg62, and Glu55 contribute to it by forming hydrogen bonds (Movie S14A and Table S4). sPLA2 has a His47/Asp48 catalytic dyad and requires Ca2+ for the catalytic activity. It is believed that the activity of sPLA2 is strongly affected by the chain length of the sn-2 acyl chain because of the capacity of the cavity. However, in our in vitro assay, sPLA2 showed relatively high activity toward sn-2 DHA among AA, EPA, and DHA, although DHA has the longest chain.
In the case of PE 16:0/20:5, the substrate gradually moved away from the active site, and in the optimal binding mode, the sn-2 carbonyl group was a distance from the catalytic histidine, although the sn-2 acyl chain stayed in the active site (Movie S15 and Fig. 6A). Although Gly31 and Arg62 formed hydrogen bonds with the phosphate, the occupancy was low due to the dissociation of the head group from the active site (Movie S14B and Table S4). Notably, in the second simulation of PE 16:0/20:5, the substrate was completely dissociated from the active site by 500 ns (Movie S16). These results confirm that the active site of sPLA2 does not retain PE 16:0/20:5 in a stable manner.
Fig. 6.
Binding of sPLA2. Image of the optimal binding mode of (A) PE 16:0/20:5 and (B) PE 16:0/22:6 in the sPLA2 active site. The images are created by the final frame of the 1 μs simulations. Movies S15 and S17 show the result of 1 μs simulation for PE 16:0/20:5 and PE 16:0/22:6, respectively. The purple particles show Ca2+, and the yellow dashed lines show hydrogen binds (distance cutoff: 3.2 Å, angle cutoff: 30°).
However, surprisingly, PE 16:0/22:6 stays in the active site, and the sn-2 acyl chain stays close to the catalytic histidine during the simulation (Movie S17 and Fig. 6B). The phosphate and headgroup of PE 16:0/22:6 frequently formed hydrogen bonds with Gly31, Arg62, and Glu55 despite the longer sn-2 acyl chain (Movie S14C and Table S4). The mean and standard deviation of the distance from the catalytic histidine to the carbonyl group of DHA (3.62 ± 0.3 Å) were significantly shorter than that of AA (6.97 ± 1.4 Å) and EPA (6.81 ± 2.4 Å). Overall, PE 16:0/22:6 sustained a lower RMSD value than the other two (Fig. S10). Indeed, in contrast to the other sn-2 chains, DHA was fixed in the active site by folding the acyl chain back in to fit in the cavity (Movie S17 and Fig. 6B). Importantly, in the second simulation for PE 16:0/22:6, the head group interacted with the same residues as during the first run and showed less movement (Movie S18). Also, the distance between the sn-2 carbonyl group and the catalytic histidine was still significantly shorter (4.01 ± 0.3 Å) than with the other two fatty acids.
The hydrophobic cavity, composed of Tyr21, Tyr24, and Tyr105, accommodates the sn-2 acyl chain by interacting with double bonds in DHA, and this region appeared to allow DHA to fold back and fit in the cavity (Fig. S11C). In the binding of AA and EPA, the orientation of these residues appeared to not be optimal for accommodating the acyl chains (Fig. S11A, B).
Despite the nature of its binding mode, PE 16:0/22:6 is suitable for enzymatic hydrolysis and the binding free energy was slightly higher than for the other two substrates (Table S2). In other words, PE 16:0/20:4 and PE 16:0/20:5 have an advantage in the binding stability of their optimal binding mode, but the binding poses are not optimal for enzymatic hydrolysis. These results indicate that sPLA2 accommodates DHA properly by folding its acyl chain back in the active site to hydrolyze it, but the binding stability is lower due to its longer acyl chain. This observation may explain why the activity toward DHA was not as high as that toward 18:2, which is the best substrate. Indeed, the binding free energy of the optimal binding mode of PC 16:0/18:2 in the sPLA2 active site from the 1 μsec simulation we previously observed (7) was much lower than that of omega-3 and omega-6 fatty acids in the sn-2 position (-80.2 kcal/mol), although the headgroup is different.
Discussion
In this study, we found a detailed and unique substrate specificity for each of the three important human PLA2s employing lipidomics-based LC-MS/MS and GC-MS assays. Furthermore, we found that the binding mode of each PLA2 during MD simulations revealed unique active site properties, especially the cavity accommodating the sn-2 acyl chain tails; this results in unique sn-2 acyl chain specificity for each PLA2 toward AA, EPA, and DHA.
For this current study, we used glycerophospholipids which contain a 16:0 in the sn-1 position since this is one of the major species in human membranes. We previously compared the substrate specificity of c, i, and sPLA2 toward PC 16:0/X and 18:0/X, and found that there was little difference in the sn-2 acyl chain specificity (7). Therefore, the effect of the major saturated sn-1 acyl chains on the sn-2 acyl chain specificity should be minimal, although the effect of the less abundant unsaturated sn-1 acyl-chains and the linkage of the sn-1 acyl chain (alkyl ether and vinyl ether) on the sn-2 acyl chain specificity may be more significant and is the subject of current studies.
The main limitation of MD simulations is that the starting position of the enzymes and substrates could affect the results. Docking of the membrane and the single substrate phospholipid with each PLA2 is based on hydrogen-deuterium exchange mass spectrometry (HDX/MS) (41). Based on the resulting models, we have shown the extraction of a single phospholipid substrate into the active site of GIVA cPLA2, and GVIA iPLA2 (22). The starting position was based on these results. Furthermore, we repeated all MD simulations at least twice to confirm that the results were reproducible, and we found that the results were always consistent with the first simulations and in vitro specificity of all PLA2s. Also, a rigorous minimization and equilibration protocol was performed to ensure that the entire system (enzyme, membrane, and solvent) was well equilibrated, as shown by the RMSD versus time plots (Fig. S12) confirming that the models and MD simulation results are reliable.
We analyzed the optimal binding mode of PE 16:0/20:4, 16:0/20:5, and 16:0/22:6 in the cPLA2, iPLA2, and sPLA2 active site. However, the difference in the preference for AA, EPA, and DHA in the sn-2 position seemed to not be simply derived from its chain length and number of double bonds, but the positions of the double bonds are also important. It has been reported that the double bond at the Δ5 position on the sn-2 acyl chain is critical for the activity of cPLA2 (42). Indeed, both AA and EPA have that double bond at the Δ5 position, but DHA does not contain one and has the double bond at the Δ4 position instead. The RMSD analysis of aromatic amino acids in the cPLA2 active site indicated that Phe199, Phe291, Phe295, and Phe681 show significant movement in the case of DHA (Fig. S13). Especially among them, Phe199 is the closest to the Δ5 double bond (within 5 Å), which suggests that the Phe199 plays an important role for selectivity toward the Δ5 double bond. However, recently, it has been reported that cPLA2 contributes to mobilization of adrenic acid (22:4 ω-6), which possesses a Δ7 double bond instead of a Δ5 double bond (43). This might suggest the hypothesis that double bonds on the odd position are critical for the activity of cPLA2 though this conclusion would require further in vitro study. In addition, it was reported that GIVC cPLA2 (also known as cPLA2γ) has less specificity toward the sn-2 AA; however, Phe291 and Phe681 are substituted (21). These facts are consistent with our results from MD simulations suggesting that these aromatic regions are critical for the specificity of cPLA2 based on the double bond number and position.
Interestingly, iPLA2 showed high activity toward EPA, and the activity was even higher than that toward AA. Also, in the LC-MS/MS assay, EPA was the best sn-2 acyl chain over 18:2, which was believed to be the best sn-2 acyl chain. MD simulations revealed that the shallow hydrophobic cavity for accommodating the sn-2 EPA tail contributes to the stability of binding, and an additional double bond of EPA contributed to the fitness of EPA to the cavity. Indeed, root mean square fluctuation (RMSF) analysis of the iPLA2 active site revealed that residues such as 541–549, 555–560, 634–638, and 771–775, which form a shallow hydrophobic cavity, are more stable than with AA (Fig. S14). However, similarly, overall RMSF values of DHA were small despite that the optimal binding mode was different from that of AA and EPA. Indeed, the binding mode of PE 16:0/22:6 in the first 500 ns of the simulation seemed to be stable, which means the binding mode was at a local minimum (Movie S10 and Fig. S7). However, the optimal binding mode as a global minimum at the end of the simulation was not suitable for hydrolysis (Fig. 5B). This global minimum binding mode can be a reason for the gap between activity and stability of binding after the first 500 ns.
Several in vivo and ex vivo studies have reported the contribution of PLA2s to specific fatty acid release and metabolism. It is known that DHA is highly abundant in the brain (44, 45, 46). Several studies using iPLA2 knock-out mice suggested that iPLA2 is involved in DHA metabolism in the brain (47, 48, 49). However, among AA, EPA, and DHA, although iPLA2 showed some activity toward DHA in mixtures of phospholipids, the activity was significantly lower compared with other acyl chains. Similarly, it was also reported that cPLA2 knock-out mice showed impaired brain DHA as well as AA metabolism (50), although cPLA2 did not show significant activity toward DHA in our studies on mixtures of phospholipids. Substrate availability might contribute to this gap between in vitro and in vivo; however, further study is needed to explain this difference. In previous studies, it was observed that purified rabbit cPLA2 releases EPA (51). However, the precise in vitro activity of human cPLA2, iPLA2 and sPLA2 toward EPA and the selectivity of these PLA2s toward various PUFAs especially omega-3 fatty acids were not known (10). In the present study, we demonstrated that iPLA2 and cPLA2 have high activity toward EPA and their molecular basis of selectivity toward EPA can be explained. Therefore, we raise the possibility that the low activity toward DHA does not prevent DHA metabolism in the brain since these enzymes release EPA, and EPA can be subsequently actively metabolized into DHA, although several important questions require further clarification. For example, it is well known that EPA is only a very minor component of human brain. Therefore, exactly how the activity of the various PLA2s toward EPA outside of the brain affects the metabolism of DHA in the brain needs further clarification.
Furthermore, it has been recognized that GV sPLA2 engages in AA mobilization and eicosanoid production (52, 53). However, the in vitro activity of GV sPLA2 toward AA was significantly lower than toward other substrates. Though, it is also suggested that lipopolysaccharide (LPS)-induced AA release predominantly depends on GIVA cPLA2 it is noted that LPS stimulation increases GV sPLA2 expression in macrophages (54). Since GV sPLA2 is a secreted enzyme and acts both intracellularly and extracellularly, the pool of phospholipids available depends on the stimulation and localization. This variety might obscure in vivo functions of GV sPLA2. In this study, we demonstrated that GV sPLA2 prefers DHA though this has not been previously addressed in the context of GV sPLA2 function (55). Therefore, our results suggest that further investigation of the in vivo functions of GV sPLA2 is warranted.
The sn-2 acyl chain specificity of each of the three PLA2s toward mixtures of phospholipids was slightly different from the specificity toward the single phospholipid substrates. For example, cPLA2 showed more strict specificity toward AA and EPA alone (Fig. 3). Since phospholipids exist in membranes, PLA2s are required to extract the most desirable phospholipid substrate from the membrane (22). We found that the composition of phospholipids in mixtures affects the specificity observed in the GC-MS assay, perhaps reflecting the in vivo activity more accurately.
In contrast to cPLA2 and iPLA2, sPLA2 is a very small enzyme, and six disulfide bonds make the structure rigid. Therefore, it was generally thought that sPLA2 does not act on substrates with a longer acyl chain, such as ones containing more than 18 carbons due to the small size of the active site. Indeed, our simulations demonstrated that AA and EPA do not fit well in the active site. Interestingly, sPLA2 showed greater activity toward DHA than AA and EPA, although DHA has an even longer chain. This led to the surprising observation that DHA fits the shallow cavity by folding back the acyl chain on itself. This observation suggests that sPLA2s can hydrolyze substrates that have more than 18 carbons on the sn-2 chain if they contain appropriate double bonds.
We have previously described numerous active site inhibitors for each PLA2 type based on conventional radiolabeled-based assays and hydrogen/deuterium exchange mass spectrometry coupled with MD simulations (41, 56). In the present study, we demonstrated that unique active site geometries, especially the binding pocket accommodating the sn-2 acyl chain, conferred unique substrate specificity toward AA, EPA or DHA where a double bond could be critical. Previously, we defined the specific binding sites for optimal inhibitors of i and cPLA2s, and the current studies on substrate specificity confirm that the most potent and specific inhibitors bound primarily in what we now have defined as the sn-2 acyl chain binding sites. This suggests that future inhibitor design should focus on these sites to create more specific inhibitors. Furthermore, the lipidomics-based assays described herein can now be used for higher-throughput inhibitor screening analysis (57).
Conclusion
We previously demonstrated that the unique active site conformation of c, i, and sPLA2 generally controls the unique substrate specificity of each enzyme (7). However, we have now shown how the unique and specific hydrophobic sn-2 fatty acid active site geometries of each of these important human PLA2s confer a unique substrate selectivity toward precise double bond positions and chain length. This illustrates the enormous power of a hydrophobic site cavity to show the unexpected exquisite specificity and sensitivity generally attributed to charged and polar sites in proteins. This is a novel finding that constitutes a new paradigm that should be of great interest to those studying protein interactions with membranes including peripheral protein and especially with the hydrophobic chains of phospholipids in the membrane (58).
In the present study, we focused on phospholipid-containing PUFAs including omega-6 AA and omega-3 EPA and DHA, and this revealed that each of the three major PLA2s shows a unique substrate specificity toward these critically important fatty acids. This new information should allow investigators to now determine the precise role of each PLA2 in specific diseases where production of AA has very different consequences than production of EPA or DHA. This information should be useful in the design of more potent and specific inhibitors to explore important neurological and other diseases.
Data availability
The crystal structure used in this manuscript is available in the Protein Data Bank (PDB, http://www.wwpdb.org) with the following codes: 1cjy. All remaining data are contained within the main text or supplemental data.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgment
We sincerely thank Prof. Ostwald Quehenberger and Aaron Armando for their support and help with the LC-MS/MS and GC-MS systems in our laboratory. We are grateful to Prof. Andrew J. McCammon for his support and counsel and use of his computational facilities, which contributed greatly to our molecular dynamics studies.
Author contributions
D. H., formal analysis; D. H. and V. D. M., investigation; E. A. D., supervision; D. H., V. D. M., and E. A. D writing–review and editing.
Funding and additional information
This work was supported by NIH grants RO1 GM20501-45 and R35 GM139641-01 (E. A. D.) and by a postdoctoral fellowship from The Uehara Memorial Foundation, Japan (D. H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplemental data
References
- 1.Dennis E.A., Cao J., Hsu Y.H., Magrioti V., Kokotos G. Phospholipase A2 enzymes: Physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 2011;111:6130–6185. doi: 10.1021/cr200085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dennis E.A., Norris P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015;15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leslie C.C. Cytosolic phospholipase A2: Physiological function and role in disease. J. Lipid Res. 2015;56:1386–1402. doi: 10.1194/jlr.R057588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramanadham S., Tomader A., Ashley J.W., Bone R.N., Hancock W.D., Lei X. Calcium-independent phospholipases A2 and their roles in biological processes and diseases. J. Lipid Res. 2015;56:1643–1668. doi: 10.1194/jlr.R058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakami M., Sato H., Miki Y., Yamamoto K., Taketomi Y. A new era of secreted phospholipase A2. J. Lipid Res. 2015;56:1248–1261. doi: 10.1194/jlr.R058123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mouchlis V.D., Chen Y., Andrew McCammon J., Dennis E.A. Membrane allostery and unique hydrophobic sites promote enzyme substrate specificity. J. Am. Chem. Soc. 2018;140:3285–3291. doi: 10.1021/jacs.7b12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batchu K.C., Hänninen S., Jha S.K., Jeltsch M., Somerharju P. Factors regulating the substrate specificity of cytosolic phospholipase A2-alpha in vitro. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2016;1861:1597–1604. doi: 10.1016/j.bbalip.2016.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Vasquez A.M., Mouchlis V.D., Dennis E.A. Review of four major distinct types of human phospholipase A2. Adv. Biol. Regul. 2018;67:212–218. doi: 10.1016/j.jbior.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Astudillo A.M., Balboa M.A., Balsinde J. Selectivity of phospholipid hydrolysis by phospholipase A2 enzymes in activated cells leading to polyunsaturated fatty acid mobilization. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019;1864:772–783. doi: 10.1016/j.bbalip.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Post X.S.R., Diego S., Jolla L. Ventricular Zone Gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J. Cell Biol. 1996;135:1071–1083. doi: 10.1083/jcb.135.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun J., Hla T., Lynch K.R., Spiegel S., Moolenaar W.H. International Union of Basic and Clinical Pharmacology. LXXVIII. Lysophospholipid Receptor Nomenclature. J. Clin. Invest. 2010;62:579–587. doi: 10.1124/pr.110.003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funk C.D. Prostaglandins and leukotrienes: Advances in eicosanoid biology. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- 14.Buczynski M.W., Dumlao D.S., Dennis E.A. An integrated omics analysis of eicosanoid biology. J. Lipid Res. 2009;50:1015–1038. doi: 10.1194/jlr.R900004-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serhan C.N. Resolution phase of inflammation: Novel endogenous anti-inflammatory and proresolving lipid mediators and pathways. Annu. Rev. Immunol. 2007;25:101–137. doi: 10.1146/annurev.immunol.25.022106.141647. [DOI] [PubMed] [Google Scholar]
- 16.Connor W.E. Importance of n-3 fatty acids in health and disease. Am. J. Clin. Nutr. 2000;71:171–175. doi: 10.1093/ajcn/71.1.171S. [DOI] [PubMed] [Google Scholar]
- 17.Salem N., Litman B., Kim H.Y., Gawrisch K. Mechanisms of action of docosahexaenoic acid in the nervous system. Lipids. 2001;36:945–959. doi: 10.1007/s11745-001-0805-6. [DOI] [PubMed] [Google Scholar]
- 18.Lauritzen L., Brambilla P., Mazzocchi A., Harsløf L.B.S., Ciappolino V., Agostoni C. DHA effects in brain development and function. Nutrients. 2016;8:6. doi: 10.3390/nu8010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mouchlis V.D., Morisseau C., Hammock B.D., Li S., McCammon J.A., Dennis E.A. Computer-aided drug design guided by hydrogen/deuterium exchange mass spectrometry: A powerful combination for the development of potent and selective inhibitors of Group VIA calcium-independent phospholipase A2. Bioorg. Med. Chem. 2016;24:4801–4811. doi: 10.1016/j.bmc.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giordanetto F., Pettersen D., Starke I., Nordberg P., Dahlström M., Knerr L., Selmi N., Rosengren B., Larsson L.O., Sandmark J., Castaldo M., Dekker N., Karlsson U., Hurt-Camejo E. Discovery of AZD2716: a novel secreted phospholipase A2 (sPLA2) inhibitor for the treatment of coronary artery disease. ACS Med. Chem. Lett. 2016;7:884–889. doi: 10.1021/acsmedchemlett.6b00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dessen A., Tang J., Schmidt H., Stahl M., Clark J.D., Seehra J., Somers W.S. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell. 1999;97:349–360. doi: 10.1016/s0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- 22.Mouchlis V.D., Bucher D., McCammon J.A., Dennis E.A. Membranes serve as allosteric activators of phospholipase a2, enabling it to extract, bind, and hydrolyze phospholipid substrates. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E516–E525. doi: 10.1073/pnas.1424651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mouchlis V.D., Limnios D., Kokotou M.G., Barbayianni E., Kokotos G., McCammon J.A., Dennis E.A. Development of potent and selective inhibitors for group VIA calcium-independent phospholipase A2 guided by molecular dynamics and structure-activity relationships. J. Med. Chem. 2016;59:4403–4414. doi: 10.1021/acs.jmedchem.6b00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 25.Wu E.L., Cheng X., Jo S., Rui H., Song K.C., Dávila-Contreras E.M., Qi Y., Lee J., Monje-Galvan V., Venable R.M., Klauda J.B., Im W. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y., Jo S., Pande V.S., Case D.A., Brooks C.L., MacKerell A.D. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirai Y., Balsinde J., Dennis E.A. Localization and functional interrelationships among cytosolic Group IV, secreted Group V, and Ca2+-independent Group VI phospholipase A 2s in P388D1 macrophages using GFP/RFP constructs. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2005;1735:119–129. doi: 10.1016/j.bbalip.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Song H., Bao S., Lei X., Jin C., Zhang S., Turk J., Ramanadham S. Evidence for proteolytic processing and stimulated organelle redistribution of iPLA2β. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2010;1081:547–558. doi: 10.1016/j.bbalip.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams S.D., Gottlieb R.A. Inhibition of mitochondrial calcium-independent phospholipase A2 (iPLA2) attenuates mitochondrial phospholipid loss and is cardioprotective. Biochem. J. 2002;362:23–32. doi: 10.1042/0264-6021:3620023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 31.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adelman S.A., Doll J.D. Generalized Langevin equation approach for atom/solid-surface scattering: General formulation for classical scattering off harmonic solids. J. Chem. Phys. 1976;64:2375–2388. [Google Scholar]
- 33.Feller S.E., Zhang Y., Pastor R.W., Brooks B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 34.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 35.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 36.Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., Mackerell A.D. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klauda J.B., Venable R.M., Freites J.A., O’Connor J.W., Tobias D.J., Mondragon-Ramirez C., Vorobyov I., MacKerell A.D., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dessen A. Structure and mechanism of human cytosolic phospholipase A2. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2000;1488:40–47. doi: 10.1016/s1388-1981(00)00108-6. [DOI] [PubMed] [Google Scholar]
- 39.Grandits M., Oostenbrink C. Selectivity of cytosolic phospholipase A2 type IV toward arachidonyl phospholipids. J. Mol. Recognit. 2015;28:447–457. doi: 10.1002/jmr.2462. [DOI] [PubMed] [Google Scholar]
- 40.Pickard R.T., Grace Chiou X., Strifler B.A., DeFelippis M.R., Hyslop P.A., Tebbe A.L., Yee Y.K., Reynolds L.J., Dennis E.A., Kramer R.M., Sharp J.D. Identification of essential residues for the catalytic function of 85- kDa cytosolic phospholipase A2. Probing the role of histidine, aspartic acid, cysteine, and arginine. J. Biol. Chem. 1996;271:19225–19231. doi: 10.1074/jbc.271.32.19225. [DOI] [PubMed] [Google Scholar]
- 41.Hsu Y.H., Bucher D., Cao J., Li S., Yang S.W., Kokotos G., Woods V.L., McCammon J.A., Dennis E.A. Fluoroketone inhibition of Ca2+-independent phospholipase A 2 through binding pocket association defined by hydrogen/deuterium exchange and molecular dynamics. J. Am. Chem. Soc. 2013;135:1330–1337. doi: 10.1021/ja306490g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanel A.M., Schüttel S., Gelb M.H. Processive interfacial catalysis by mammalian 85-kilodalton phospholipase A2 enzymes on product-containing vesicles: Application to the determination of substrate preferences. Biochemistry. 1993;32:5949–5958. doi: 10.1021/bi00074a005. [DOI] [PubMed] [Google Scholar]
- 43.Monge P., Garrido A., Rubio J.M., Magrioti V., Kokotos G., Balboa M.A., Balsinde J. The contribution of cytosolic group IVA and calcium-independent group via phospholipase a2s to adrenic acid mobilization in murine macrophages. Biomolecules. 2020;10:542. doi: 10.3390/biom10040542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C.T., Liu Z., Ouellet M., Calon F., Bazinet R.P. Rapid β-oxidation of eicosapentaenoic acid in mouse brain: An in situ study. Prostaglandins Leukot. Essent. Fat. Acids. 2009;80:157–163. doi: 10.1016/j.plefa.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 45.Sinclair A.J. Long-chain polyunsaturated fatty acids in the mammalian brain. Proc. Nutr. Soc. 1975;34:287–291. doi: 10.1079/pns19750051. [DOI] [PubMed] [Google Scholar]
- 46.Brenna J.T., Diau G.Y. The influence of dietary docosahexaenoic acid and arachidonic acid on central nervous system polyunsaturated fatty acid composition. Prostaglandins Leukot. Essent. Fat. Acids. 2007;77:247–250. doi: 10.1016/j.plefa.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheon Y., Kim H.W., Igarashi M., Modi H.R., Chang L., Ma K., Greenstein D., Wohltmann M., Turk J., Rapoport S.I., Taha A.Y. Disturbed brain phospholipid and docosahexaenoic acid metabolism in calcium-independent phospholipase A2-VIA (iPLA2β)- knockout mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2012;1821:1278–1286. doi: 10.1016/j.bbalip.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beck G., Sugiura Y., Shinzawa K., Kato S., Setou M., Tsujimoto Y., Sakoda S., Sumi-Akamaru H. Neuroaxonal dystrophy in calcium-independent phospholipase A2β deficiency results from insufficient remodeling and degeneration of mitochondrial and presynaptic membranes. J. Neurosci. 2011;31:11411–11420. doi: 10.1523/JNEUROSCI.0345-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basselin M., Rosa A.O., Ramadan E., Cheon Y., Chang L., Chen M., Greenstein D., Wohltmann M., Turk J., Rapoport S.I. Imaging decreased brain docosahexaenoic acid metabolism and signaling in iPLA2β (VIA)-deficient mice. J. Lipid Res. 2010;51:3166–3173. doi: 10.1194/jlr.M008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenberger T.A., Villacreses N.E., Contreras M.A., Bonventre J.V., Rapoport S.I. Brain lipid metabolism in the cPLA2 knockout mouse. J. Lipid Res. 2003;44:109–117. doi: 10.1194/jlr.m200298-jlr200. [DOI] [PubMed] [Google Scholar]
- 51.Shikano M., Masuzawa Y., Yazawa K., Takayama K., Kudo I., Inoue K. Complete discrimination of docosahexaenoate from arachidonate by 85 kDa cytosolic phospholipase A2 during the hydrolysis of diacyl- and alkenylacylglycerophosphoethanolamine. Biochim. Biophys. Acta. 1994;1212:211–216. doi: 10.1016/0005-2760(94)90255-0. [DOI] [PubMed] [Google Scholar]
- 52.Balboa M.A., Balsinde J., Winstead M.V., Tischfield J.A., Dennis E.A. Novel group V phospholipase A2 involved in arachidonic acid mobilization in murine P388D1 macrophages. J. Biol. Chem. 1996;271:32381–32384. doi: 10.1074/jbc.271.50.32381. [DOI] [PubMed] [Google Scholar]
- 53.Muñoz N.M., Kim Y.J., Meliton A.Y., Kim K.P., Han S.K., Boetticher E., O’Leary E., Myou S., Zhu X., Bonventre J.V., Leff A.R., Cho W. Human group V phospholipase A2 induces group IVA phospholipase A2-independent cysteinyl leukotriene synthesis in human eosinophils. J. Biol. Chem. 2003;278:38813–38820. doi: 10.1074/jbc.M302476200. [DOI] [PubMed] [Google Scholar]
- 54.Kessen U.A., Schaloske R.H., Stephens D.L., Lucas K.K., Dennis E.A. PGE2 release is independent of upregulation of Group V phospholipase A2 during long-term stimulation of P388D1 cells with LPS. J. Lipid Res. 2005;46:2488–2496. doi: 10.1194/jlr.M500325-JLR200. [DOI] [PubMed] [Google Scholar]
- 55.Balestrieri B., Arm J.P. Group V sPLA2: Classical and novel functions. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2006;1761:1280–1288. doi: 10.1016/j.bbalip.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 56.Burke J.E., Babakhani A., Gorfe A.A., Kokotos G., Li S., Woods V.L., McCammon J.A., Dennis E.A. Location of inhibitors bound to group IVA phospholipase A2 determined by molecular dynamics and deuterium exchange mass spectrometry. J. Am. Chem. Soc. 2009;131:8083–8091. doi: 10.1021/ja900098y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mouchlis V.D., Armando A., Dennis E.A. Substrate-specific inhibition constants for phospholipase A 2 acting on unique phospholipid substrates in mixed micelles and membranes using lipidomics. J. Med. Chem. 2019;62:1999–2007. doi: 10.1021/acs.jmedchem.8b01568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller M.P., Jiang T., Sun C., Lihan M., Pant S., Mahinthichaichan P., Trifan A., Tajkhorshid E. Characterization of lipid-protein interactions and lipid-mediated modulation of membrane protein function through molecular simulation. Chem. Rev. 2019;119:6086–6161. doi: 10.1021/acs.chemrev.8b00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structure used in this manuscript is available in the Protein Data Bank (PDB, http://www.wwpdb.org) with the following codes: 1cjy. All remaining data are contained within the main text or supplemental data.