Abstract
Background
Rotavirus A (RVA) causes acute gastroenteritis in children <5 years of age in sub-Saharan Africa. In this study, we described the epidemiology and genetic diversity of RVA infecting Gabonese children and examined the antigenic variability of circulating strains in relation to available vaccine strains to maximize the public health benefits of introducing rotavirus vaccine through the Expanded Programme on Immunization (EPI) in Gabon.
Methods
Stool samples were collected consecutively between April 2018 and November 2019 from all hospitalized children <5 years with gastroenteritis and community controls without gastroenteritis. Children were tested for rotavirus A by quantitative RT-PCR and subsequently sequenced to identify circulating rotavirus A genotypes in the most vulnerable population. The VP7 and VP4 (VP8*) antigenic epitopes were mapped to homologs of vaccine strains to assess structural variability and potential impact on antigenicity.
Findings
Infections were mostly acquired during the dry season. Rotavirus A was detected in 98/177 (55%) hospitalized children with gastroenteritis and 14/67 (21%) of the control children. The most common RVA genotypes were G1 (18%), G3 (12%), G8 (18%), G9 (2%), G12 (25%), with G8 and G9 reported for the first time in Gabon. All were associated either with P[6] (31%) or P[8] (38%) genotypes. Several non-synonymous substitutions were observed in the antigenic epitopes of VP7 (positions 94 and 147) and VP8* (positions 89, 116, 146 and 150), which may modulate the elicited immune responses.
Interpretation
This study contributes to the epidemiological surveillance of rotavirus A required before the introduction of rotavirus vaccination in the EPI for Gabonese children.
Keywords: Rotavirus A, Vaccines, Genotypes, Antigenic epitopes, Acute gastroenteritis, Africa
Research in context.
Evidence before this study
Rotavirus is one of the most important etiologic agents of infantile gastroenteritis, with an estimated 200,000 deaths in children <5 years of age. There are four WHO prequalified Rotavirus vaccines available thus far. We searched PubMed for publications until 2020 using the search terms “rotavirus A” AND “epitopes” AND “children.” We found 10 results describing different studies of rotavirus genetic diversity and characterization of the VP4 and VP7 genes encoding the outer capsid proteins. Eight of the studies compared the antigenic regions of the VP7 and VP4 partial sequences of circulating rotavirus strains with those of Rotarix and RotaTeq vaccines. However, there are no studies from Central Africa, where the Expanded Programme on Immunization (EPI) is scheduled to introduce rotavirus vaccine, particularly in the Republic of Gabon.
Added value of this study
Currently, there is limited data on the genetic similarity of vaccine strains and their relationship with wild-type strains. In this study, we found a high RVA burden in Gabonese children with high antigenic variability in circulating compared to the vaccine strains. This may influence efficacy, as this extensive genetic variability observed in these viruses may evade immune responses induced by prior infection or vaccination through changes in molecular structures by antibodies and/or T cells.
Implications of all the available evidence
The high RVA burden implies an urgent need for the introduction of RVA vaccination for Gabonese children.
Alt-text: Unlabelled box
1. Introduction
Diarrheal diseases remain one of the major cause of illness among children <5 years, causing over 500,000 deaths worldwide each year, mainly in Africa and South Asia [1]. One of the most important etiological agents of gastroenteritis in infants and children is group A rotavirus (RVA). The non-enveloped, triple-layered viral particle has a genome of 11 double-stranded RNA (dsRNA) encoding six structural (VP1-VP4, VP6, VP7) and six non-structural (NSP1-NSP6) proteins [2]. The outer capsid layer is formed by the VP7 glycoprotein (G) and the VP4 spike protease-sensitive (P) protein, both encoded by genomic segments on which a binary taxonomical system of rotaviruses is based at intraspecific level, determining their classification into G and P genotypes [3].
The Rotavirus classification work group of the International Committee on Virus Taxonomy has currently identified 41 G and 57 P genotypes in mammal and bird species worldwide [3,4], although in humans only a few genotypes are responsible for the disease burden (G1-G4, G9, G12, P[4], P[6], P[8]) [5], [6], [7], [8]. Additionally, genotype G5, G6 and G8 are considered relevant to human health in Africa and Asia [7]. In addition to rapid genetic drift caused by the error-prone RNA-dependent RNA polymerase, segment reassortment (genetic shift) is another major driver of rotavirus evolution, leading to new genetic combinations and enabling interspecies transmission [9,10]. Previous studies reported G1P[8] as the most frequent RVA strain globally [5,11], while G2P[4], G3P[8], G4P[8], and G9P[8] were reported to be common strains worldwide in the pre-vaccination era [11].
There are four WHO prequalified Rotavirus vaccines available thus far. Rotarix (GlaxoSmithKline Biologicals, Belgium) is a monovalent vaccine derived from a human G1P[8] strain and RotaTeq (Merck&Co., USA) is a pentavalent (G1-G4 and P[8]) containing five human-bovine RVA reassortant strains [10]. Both vaccines were proven to be safe and efficacious. While Rotarix and Rotateq are used in over 90 countries worldwide [12], WHO recently prequalified Rotavac (Bharat Biotech, India) and ROTASIIL (Serum Institute of India). Both Rotavac and ROTASIIL are in use only in India, whereas Rotavac is used in India and Palestine. Besides pronounced immune responses against the capsid protein, antibody responses against other rotavirus antigens such as VP6, VP2, NSP2 and NSP4 were also detected. However, the mechanisms by which antiviral immunity is acquired are not clearly understood [13]. Because the introduction of RVA vaccination requires a thorough documentation of disease burden, viral diversity, many countries have initiated RVA surveillance. In Africa, the first countries to introduce RVA vaccination were South Africa (2009) and Morocco (2010) [14]. Rotavirus evolution at the genotypic and sub genotypic levels helps to understand transmission dynamics, where new genotypes emerge through recombination of wild-type and vaccine strains, due to selection pressure (post-vaccine strain shift) [15], or due to gene reassortment [16,17]. Therefore, the question remains how antigenic drift can evade adaptive immunity and thus affect vaccine responses [18,19].
In Gabon, a national immunization program for RVA has not yet been implemented. Furthermore, the information on RVA diversity and burden is poorly documented. Some of the pathogenic RVA genotypes (G1, G2, G3, G12, G6, P[4], P[6], P[8]) were previously detected in four different cities of Gabon, as well as one emerging G6P[6] strain [20]. Thus, pursuing national and regional surveillance is important due to rapid evolution including zoonotic transmission with increased odds of emergence of novel strains. To this end, the aim of the present study is to describe epidemiology and genetic diversity of RVA in Gabonese children <5 years old and to investigate antigenic variability of circulating strains in relation to available vaccines.
2. Methods
2.1. Ethics statement
The study protocol was approved by the Institutional Ethical Committee of the Centre de Recherches Médicales de Lambaréné (CERMEL) (CEI-CERMEL: 003/2017). Written informed consent was obtained from parents or legal representatives of the children.
2.2. Study population and sampling
Between April 2018 and November 2019, stool samples were collected from children under 5 years of age who were residents of semi-urban Lambaréné and its surrounding rural area. The population considered here is representative of the general population because children aged 0–5 years who presented to the outpatient clinics of the two major hospitals in Lambaréné were examined and included if they suffered from diarrhoea (defined as three or more liquid stools within 24 h during the previous three days) and lived in the study area (within a radius of approximately 20 km from Lambaréné). The sample size was calculated based on an estimated rotavirus prevalence among Gabonese children with diarrhoea (27.1%) from a previous [20]. The significance level was 0.05 (corresponding to a 95% confidence interval) with a precision of 0.07 for a sample size of 155 children. Rainy seasons (March-May, October-December) of similar length are interspersed between short (January and February) and long (June–September) dry seasons. Children with diarrhea or history of diarrhea within the last 24 h were recruited at outpatient department of two main hospitals (Hôspital Albert Schweitzer and Centre Hospitalier Régional Georges Rawiri de Lambaréné). In addition, stool samples were randomly collected from healthy children of the same age without gastroenteritis who resided in the same compound as index cases. The collected stool samples were immediately transported to the laboratory and were stored in RNAlater at –20°C for further use.
2.3. Rotavirus detection
The suspension of 200 mg stool in 1 ml of RNAlater solution was homogenized by vortexing and centrifuged. Viral RNA was then extracted from 140 µl of the supernatant with the use of QIAamp viral RNA Mini Kit (QIAGEN, Hilden Germany). All steps of the RNA isolation were performed following the manufacturer's instructions. The genomic RNA was eluted in a total volume of 60 µl, concentration was immediately measured on a Qubit dsRNA XR Assay Kit and the Qubit 4.0 fluorometer (Invitrogen, Paisley, UK) and subsequently stored at – 80°C.
The presence of RVA was detected using a one-step reverse transcription real time PCR (RT-qPCR) protocol for the amplification of the NSP4 gene [21]. Briefly, RNA extracts were first diluted and denaturated at 95 C for 1 min, then RT-qPCR was carried out in a total volume of 12 µl using the SuperScript III/Platinum Taq OneStep kit (Invitrogen, Carlsbad, CA). The reaction mix contained PCR buffer, forward primer (RoA 25-s) 5′GCTTTTAAAAGTT-CTGTTCCGAG), and reverse primer (RoA 26a-as) 5′‐ACTCAATGTGTAGTTGAGGTCGG, probe 5′‐VIC‐ATCTTTCCGCACGC‐MGB and Platinum Enzyme mix. Samples with a cycle threshold (CT) ≤ 39 were considered positive.
2.4. Rotavirus genotyping
For the rotavirus G/P typing we used two RT-semi-nested PCR protocols, specific for each type, as described [22,23]. For the first round, pan-specific primers located close to the 3’ and 5’ end of the segments were used. In the second run, one of the pan-specific primer was used as a fluorescence labelled primer (with HEX for G types and FAM for P types). The second primer is specific for each P or G Type. This results in fluorescence labelled amplicons with different fragment length. The amplicons generated in the first and second PCRs were also independently checked using agarose gel electrophoresis. For each sample, the fluorescence labelled amplicons generated in the second PCR (nested-PCR round) were mixed with a fluorescence labelled ladder and then analyzed using capillary sequencer. Thus, specific peaks for G and P type could be determined. Additionally, the amplicons generated in the first round of the G/P typing were sequenced using the PCR primer used in the first PCR round. Those samples with a weak signal in the fragment length analysis and/or negative in the first PCR round, a P or G type specific PCR was additionally performed. Some samples showed a signal in the fragment length analysis, but no amplicons were detectable in the first PCR round. For these samples, a VP4- or VP7-long amplification was attempted to identify their genotypes. All amplicons were Sanger sequenced for further analysis. The RVA nucleotide sequences of this study are available under the following GenBank accession numbers: MZ966335 - MZ966498.
2.5. Sequence data analysis
Generated sequences were analyzed in Geneious Prime v2021.1.1 (Biomatters, Auckland, New Zealand). Genotype was assigned using NCBI BLAST and the VIPR typing tool for rotavirus A genotype determination (https://www.viprbrc.org). Nucleotide sequences of VP7 and VP4 were aligned with MAFFT implemented in Geneious and neighbor-joining phylogenetic trees were reconstructed using MEGA 7.0.26 [24] following substitution model using 1000 bootstrap iterations for evaluation of node support. Amino acid sequence similarity was calculated with the p-distance method. Phylogenetic trees were displayed with iTOL [25] and potential N-linked glycosylation sites were screened with NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/).
2.6. Statistical analysis
Differences of RVA prevalence among children with and without diarrhea were tested using the Chi-square or Fischer exact test, with statistical significance set at two-sided p-value <0.05. Associations of RVA infection with potential risk factors were evaluated by conditional logistic regression and a multiple logistic regression model fitted using a stepwise backward procedure. The variables with a p-value ≥0.30 in the univariate analysis were removed sequentially. We adjusted for potential confounders that may influence the occurrence of RVA (e.g., gender, residence). All statistical tests were performed using R version 4.0.2 [26].
2.7. Role of funding source
The project is part of the GZ EI 1044/1-1 AOBJ 630127 grant funded by DFG. The Funders had any role in study design, data collection, data analyses, interpretation, or writing of report.
3. Results
3.1. Study population
A total of 244 children were recruited in the study. Of these 177 (73%) were symptomatic children aged between 0 and 59 months with a median age of 12 months, whereas the median age of healthy children was 24 months. Most participating children (78%; 190/244) were aged 0-24 months. A total of 152 (62%) participants lived in the semi-urban area. Male children were 57% (101/177) in the symptomatic group and 61% (41/67) in the asymptomatic group.
3.2. RVA Prevalence and seasonal distribution
RVA was detected in 98/177 (55%) of symptomatic children and 14/67 (21%) of controls. High RVA detection rate was associated with age, sex, and residence among symptomatic children (Table 1). Children between 0 and 6 months showed the highest proportion of infection (30/47, 64%), while the lowest detection rate was observed among children between 19 and 24 months (5/16, 31%). Females were more often infected (44/76, 58%) than males (54/101, 54%), with similar observations in children from rural (60%) compared to those from semi-urban settings (53%). In the control group, children between 7 and 12 months showed the highest proportion of infections (4/8, 50%). Males were more likely to be infected (10/41, 24%) than females (4/26, 15%), with similar observations in children from rural (31%) compared to those from semi-urban settings (7%).
Table 1.
Rotavirus burden in the study population.
| RVA in Children with acute gastroenteritis cases n (%) | RVA in Children without acute gastroenteritis cases n (%) | p-value | |
|---|---|---|---|
| Total | 98/177 (55) | 14/67 (21) | <0.0001 |
| Age (months) | |||
| 0-6 | 30/47 (64) | 0/7 (0) | 0.002 |
| 7-12 | 24/47 (51) | 4/8 (50) | 1 |
| 13-18 | 27/45 (60) | 0/4 (0) | 0.0345 |
| 19-24 | 5/16 (31) | 3/16 (19) | 0.6851 |
| 25-59 | 12/22 (55) | 7/32 (22) | 0.0205 |
| Gender | |||
| Female | 44/76 (58) | 4/26 (15) | 0.0002 |
| Male | 54/101 (54) | 10/41 (24) | 0.0016 |
| Residential area | |||
| Rural | 32/53 (60) | 12/39 (31) | 0.0062 |
| Semi-urban | 66/124 (53) | 2/28 (7) | <0.0001 |
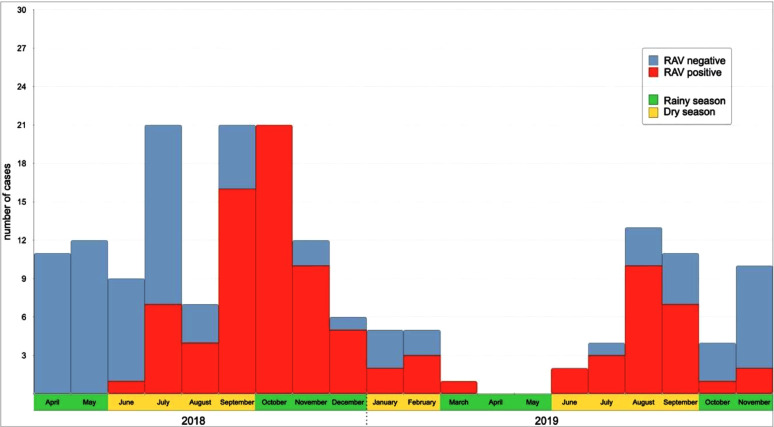
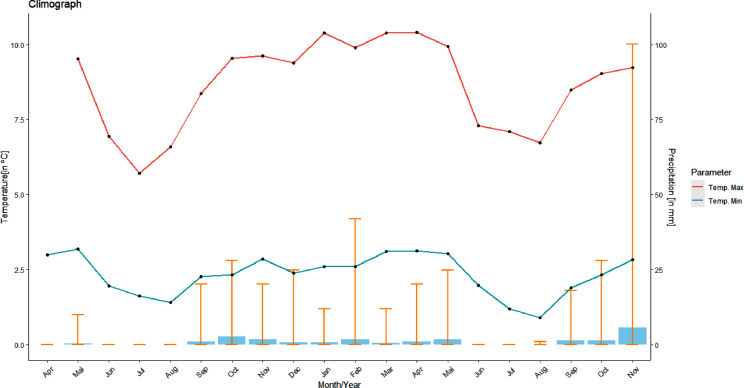
Gabon, a tropical country has both wet and dry seasons. RVA infection among children with diarrhea was not significantly associated with either dry or wet season (χ2 = 0.31, p=0.5). However, a significant difference in RVA positive cases was observed between the cumulated long dry and wet seasons (χ2= 5.06, p=0.02), with peak RVA infection during September and October 2018 (turn of the seasons) and August-September 2019 (dry season) (Fig. 1a, 1b).
Fig. 1.
a. Seasonal burden of rotavirus A among Gabonese children. b. Climate graph with recorded temperature and precipitation data of Lambaréné, during the study period (April 2018 until November 2019)
3.3. Genotype distribution and risk factors
In the 112 RT-qPCR positives, we identified two P types and five G types, either in single or mixed infections (Table 2). The P[8] (38.4%) was predominant, followed by P[6] (31%). The most prevalent G type identified was G12 (25%) followed by G1 (18%), G8 (18%), G3 (12%) and G9 (2%). The G type could not be assigned in 31 cases (26%) cases, while the P type identification failed for 34 positive samples (30%) (Supplementary Table S1). The frequent G/P combinations were G12P[6] (21%), G1P[8] (17%) and G8P[8] (15%) (Table 2).
Table 2.
Rotavirus genotype (G/P type) distribution in Gabonese children.
| G (X) P types n (%) |
||||
|---|---|---|---|---|
| P [6] | P [8] | P[x] | Total | |
| G1 | 0 | 19 (17) | 1 (1) | 20 (18) |
| G3 | 8 (7) | 0 | 1 (1) | 9 (8) |
| G8 | 0 | 17 (15) | 1 (1) | 18 (16) |
| G9 | 0 | 2 (2) | 0 | 2 (2) |
| G12 | 24 (21) | 2 (2) | 0 | 26 (23) |
| Gx | 1 (1) | 1 (1) | 29 (21) | 31 (28) |
| Gmix | 2 (2) | 2 (2) | 2 (2) | 6 (5) |
| Total | 35 (31) | 43 (39) | 34 (30) | 112 (100) |
Mixed types: G1+G3P[8] (1%), G1+G8+G12P[8] (1%), G3+G12P[6] (2%);
Partial G/P mixed types: G3+G8P[x] (1%), G8+G12P[x] (1%).
Crude analysis indicated that nutrition, source of drinking water and diarrhea were associated with RVA. Children nourished with formula milk (OR=1.5; 95% CI: 0.55–4.12, p-value= 0.013) and drinking water from wells (OR=1.29; 95%CI: 0.61–2.80; p-value= 0.009) were at higher risk of acquiring a RVA infection. The RVA detection rate was significantly associated with diarrhea (OR=4.63; 95%CI: 2.45–9.29, p-value = <0.001). After adjusting for potential confounders, RVA infection remained significantly associated with diarrhea (OR=3.5; 95%CI: 1.5–8.58, p-value=0.004), and equally for semi-urban population (OR=0.46; 95%CI: 0.21-0.96, p-value= 0.04) (Supplementary Table S2).
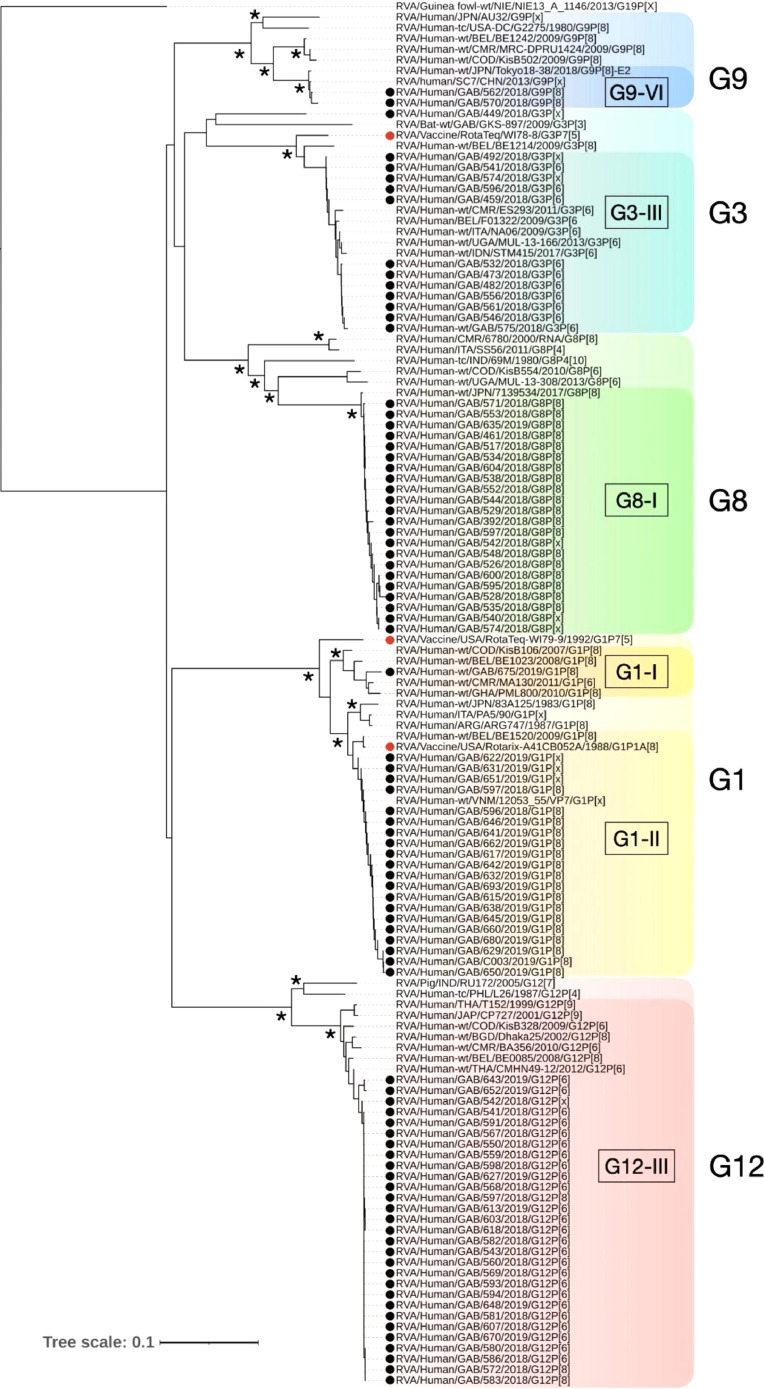
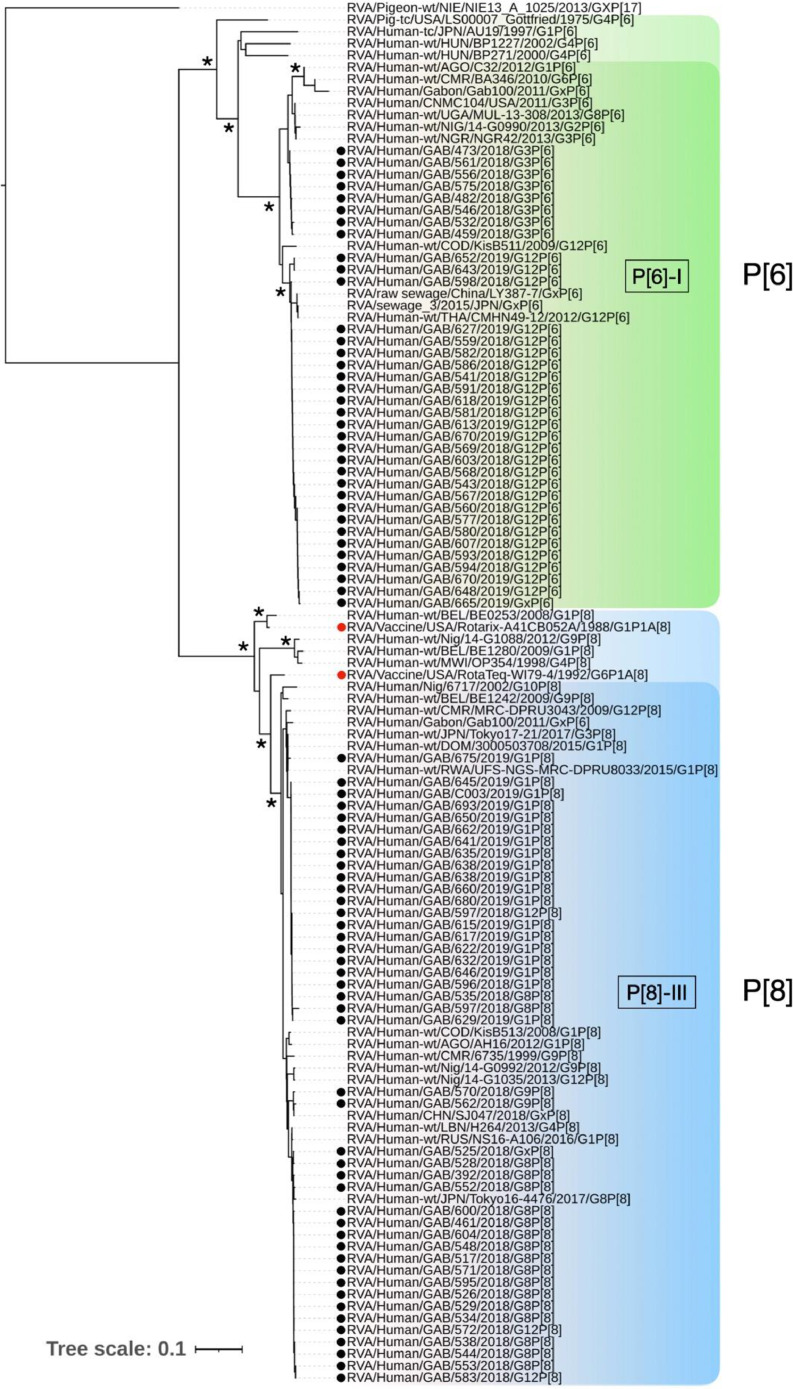
3.4. Phylogenetic analysis with vaccine strains
RVA strains belonging to G8, G9, G12, P[6] and P[8] clustered within a single lineage per genotype (Figs. 2, 3). The Gabonese G1 clustered as lineage I and II, showing genetic homogeneity to African and Asian strains, respectively. The Gabonese G1 (lineage II) revealed a higher amino acid pairwise identity with the Rotarix G1 (98%) than the homologous component of RotaTeq. A low pairwise identity was observed between the protein sequence of Gabonese G3 strains and homologous component of RotaTeq (94-95%), as indicated also by their phylogenetic branching. Gabonese G3 strains grouped in lineage III with European, Indian and African strains. Additionally, we have detected the VP7 nucleotide sequence of a G3 strain (GAB/449) closely related to a bat-borne G3P[3] earlier detected in Gabon. The Gabonese G8 clustered in lineage I, showing a close relationship with Eastern Asian RVA (Japan), similarly to the G9 sequences of lineage VI (China and Japan). The G12 sequences formed a relatively homogenous cluster within lineage III, along African, Asian and European strains.
Fig. 2.
Phylogenetic analysis of representative RVA strains based on a VP7 gene fragment (856 nucleotides). Gabonese strains analyzed are marked by black dots and vaccine strains are marked by red dots. Guineafowl RVA strain NIE13A1146 was used as an outgroup and bootstrap support >80% is indicated by asterisk. Tree scale bar represents number of substitutions per site.
Fig. 3.
Phylogenetic analysis of representative RVA strains based on a fragment of VP4, containing the VP8* sequence (645 nucleotides). Gabonese strains analyzed are marked by black dots and vaccine strains are marked by red dots. Pigeon RVA strain NIE13A1025 was used as an outgroup and bootstrap support >80% is indicated by asterisk. Tree scale bar represents number of substitutions per site.
For both P[6] and P[8] genotypes, their VP4 sequences clustered within lineage I and lineage III, respectively. However, the analysis of all symptomatic children revealed a pattern of P type segregation in Gabonese strains, indicative of their association with different G types (Figs. 2, 3). The Gabonese P[8] strains had 93-96% similarity with RotaTeq strain and 90% with the Rotarix P[8]. Furthermore, the P[8] associated with G1 were more similar with RotaTeq and Rotarix (96% and 90%, respectively) than G8P[8] (93-94% and 89-90%, respectively).
3.5. Analysis of VP7 and VP4 neutralizing epitopes
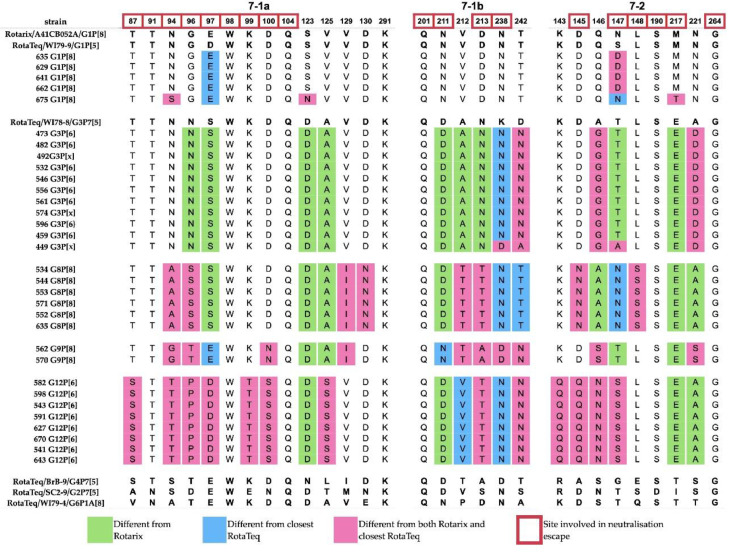
Vaccine efficacy can be undermined if structural differences accumulate in the antigenic epitopes of circulating RVA strains. We used comparative protein analysis to highlight potential antigenic differences between Gabonese RVAs and strains of vaccines that may be introduced. Mutation sites with potential for neutralization escape by monoclonal antibodies reside in the VP7 trimer on two structurally defined antigenic epitopes: 7-1 and 7-2 [27] (Fig. 4). The immunodominant 7-1 epitope is further divided into 7-1a and 7-1b. Overall, the highest number of amino acid differences was observed in the 7-1b subunit of VP7, followed by the 7-2 subunit and the 7-1a subunit. We observed only 5 of 29 amino acid residues completely conserved (W98, Q104, Q201, G264, K291). Most of these sites (W98, Q104, K291) were located on 7-1a. Due to the low viral load and high ct values, we could only obtain only sequence information from only one asymptomatic child.
Fig. 4.
Comparison of VP7 antigenic epitopes of Gabonese and vaccine strains. Amino acids of vaccine strains are indicated in bold.
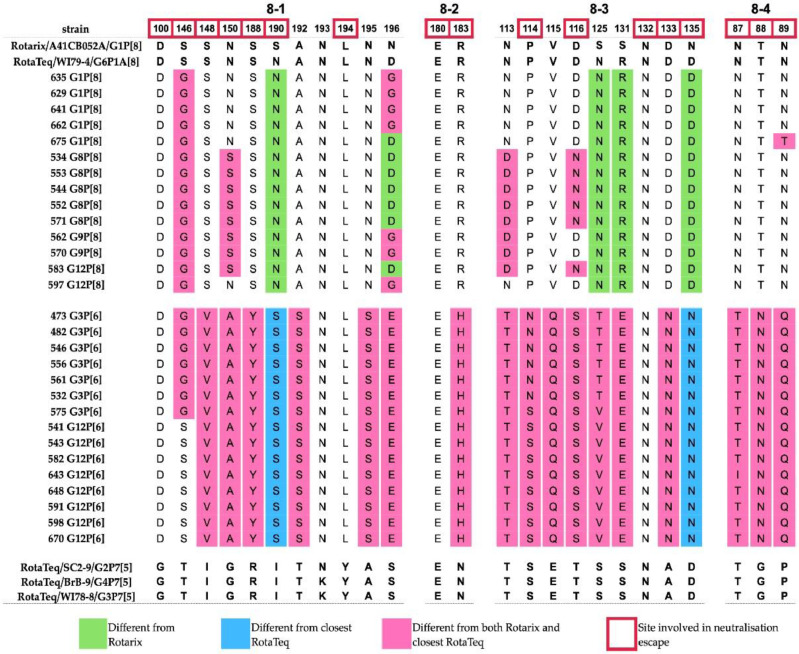
The VP7 antigenic epitopes of Gabonese G1 strains revealed four residue differences relative to Rotarix G1 and five changes compared to the RotaTeq G1. These sites were found on the epitopes 7-1a and 7-2, respectively. The analysis of G3 showed four to five amino acids different from the RotaTeq G3, distributed on 7-1b and 7-2 epitopes and up to 13 differences relative to Rotarix G1. Several exhibited features of escape mutants [28]. The change in the N94S residue was only observed in strain GAB/675, while the entire antigenic region 7-1b of the G3 strains matches the RotaTeq. All G3 strains carried a mutation at position 238 (epitope 7-1), with most having a substitution (K238N) that modulates N-glycosylation, with one exception in the GAB/449 strain (K238D), which was replaced by aspartic acid. In the 7-2 subunit, mutations at positions 147-148 can modulate responsiveness to specific antibodies; however, only position 147 had amino acid substitutions for both G1 and G3 strains. Most G1 strains had dissimilar residues (N/S147D), while GAB/449 (G3), phylogenetically related to the bat RVA, had a T147A substitution previously associated with immune escape [28,29]. In the VP8* region, the P[8] type revealed distinct signatures at positions N135D, S190N that could well differentiate RotaTeq and Rotarix [30], Whereas the amino acid residues associated with immune escape that differ from both RotaTeq and Rotarix P[8] are located on epitope 8-1 (S146G, N150S), epitope 8-3 (D116N) and epitope 8-4 (N89T) [31].
The comparison of G8, G9 and G12 with human G1-G4 and bovine G6 strains of RotaTeq and the G1 strain of Rotarix showed that Gabonese G8 strains contained four amino acids (sites 96, 145, 147, 213) that are not present in any RotaTeq strain, and 11 residue differences compared to the closest (G3) RotaTeq homolog. In comparison to Rotarix, Gabonese G8 strains contained 15 amino acid differences. Gabonese G9 strains had only two residues changes relative to all RotaTeq strains (sites 94 and 242), each located on the two subunits of the 7-1 epitope, but up to 12 amino acid difference in relation to the closest RotaTeq strain (G3). When compared to the Rotarix strain, the Gabonese G9 showed 14 amino acid changes. G12 strains were the most divergent, having a minimum of 9 different amino acids when compared to VP7 epitopes of vaccine strains. Relative to the closest RotaTeq strain (G3), G12 contained 15 residue changes. The most extensive epitope divergence was observed in relation to Rotarix G1, with 17 residues (Fig. 4).
Following the virion's trypsin activation in the intestine, the VP4 spike protein is proteolytically cleaved in two components: the globular head (VP8*) placed on top of the stalk (VP5*). We analyzed the region coding for the VP8* globular head, the main determinant of RVA P type containing four surface-exposed antigenic epitopes: 8-1, 8-2, 8-3 and 8-4 [31]. Among the Gabonese P[8], P[6] and all vaccine strains we found only two completely conserved sites in epitopes 8-2 (E180) and 8-3 (N132), of the 25 present in VP8* (Fig. 5). The Gabonese P[8] contained up to 24 identical amino acids compared to the P[8] of RotaTeq and 19 identical amino acids relative to P[8] of Rotarix. Six amino acids differences located mostly on epitopes 8-1, 8-3 and 8-4 were shared with both P[8] vaccine strains. The highest number of differences (10 residues) was noted between Gabonese G1P[8] strain GAB/675 and Rotarix P[8].
Fig. 5.
Comparison of VP4 (VP8*) antigenic epitopes of Gabonese and vaccine strains. Amino acids of vaccine strains are indicated in bold.
The epitope composition of the Gabonese P[6] strains relative to vaccine VP8* showed extensive dissimilarity, consistent with the levels of sequences divergence. P[6] strains associated with G3 contained 18-20 residue differences, whilst the G12P[6] contained 17-19 different amino acids.
4. Discussion
In preparation for introducing rotavirus vaccine by the EPI for Gabonese children, we investigated the prevalence and genetic characteristics of RVA among children with and without diarrhea over a 20-month period. The RVA detection rate in diarrheal cases is twice as high as previously reported in Gabon [20]. RVA was present to a greater extent in children with diarrhea than in asymptomatic ones, with similar or higher detection rate than in West/East Africa and South Asia [32,33].
As observed in various studies, high infection was observed in most children aged 0–24 months [33,34]. The significant association of RVA infection with the semi-urban environment could be related to population density and contact rates, but also to living conditions. The AOR analysis showed no significant association with pit toilet use and RVA infections, this toilet type was present in about 90% of RVA positive households, in contrast to flush toilets (9%). The use of public taps as a source of drinking water was reported for more than half (55%) of the infected children, as opposed to water from the household tap (13%). Therefore, such factors may discretely contribute to the acquisition of RVA infections, especially in semi-urban areas with high population density. Over the sampling period, the proportion of infections differed statistically only between rainy seasons and cumulative long dry seasons. RVA infections peaked in September-October (2018) or August-September (2019), a pattern consistent with previous observations in Gabon [20], West Africa [35,36], Central Africa [37], Southeast Asia [38] and generally in the tropics [39].
The frequencies of G1 and G3 genotypes in the present study population are higher than previously reported in the country [20] and in neighboring Cameroon [40], while G8 and G9 were detected for the first time in Gabon. In contrast to other Central African countries where G1 is dominant, this genotype was the second most common in our study (18%), while the dominant G12 (25%) was found in similar proportions [34,38]. Although G12 was found at a lower frequency in this study than in Cameroon (67.4%) [40], previous work by Lekana-Douki et al. [20] in Gabon also showed that G12 was less common (11.8%). In contrast to previous work by Lekana-Douki et al. [20], we observed P[8] to be the most frequent P genotype (38%). The other P genotype in our study is P[6] (31%), had a lower detection rate than those previously reported in the country (71.4%). This high incidence of P[8] has been observed not only in Central African countries, but also in Central and South-eastern Europe [41,42]. We observed a high proportion of G1P[8] (17%), although it was significantly lower than in neighboring country (Republic of Congo, 44%) [37]. G12P[8] and G12P[6] were found at lower frequencies compared to Cameroon [40]. In addition, we found G9P[8] and G8P[8], genotypes frequently detected in Southeast Asia [38]. These genotypes considered rare or uncommon, were also reported with low frequencies in other African countries [36]. 28 % of non-typeable types (Gx-Px) were observed and there were many non-typeable types among the P-types. All qPCR-positive samples that were negative in semi-nested RT- PCR were designated as non-typeable. There are two likely reasons that have been raised as limitations of this study. One probable reason is that qPCR has a high sensitivity compared to RT-PCR; this could well be observed in non-typeable samples that gave high ct values in the independently performed tests. The other reason could be due to mutations in the primer binding sites, which impairs annealing. However, the latter explanation is unlikely.
RVA zoonotic genotypes such as G8 (bovine), P[11] (bovine) and P[6] (porcine) are widespread in developing countries and cause infections in humans [43], [44], [45]. High diversity of RVA (including untypeable strains) in human populations, livestock or wildlife [46], [47], [48] are common. In our study, a G3 strain (GAB/449) obtained from a symptomatic child was phylogenetically closer to a Gabonese strain earlier discovered in a giant round bat (H. gigas, GenBank no. MN528121). Poor sanitary conditions and high contact rates with livestock and wildlife may contribute to spillover-spillback transmission patterns. Consequently, the rate of infection is also higher (∼20%) compared to middle- and high-income countries (∼5%) [43].
The phylogenetic analysis of Gabonese G types revealed well supported terminal clades indicating close relatedness with strains of Asian origin. Although P[8] is widespread in Africa, we cannot rule out the possibility of cross-border transmission in Gabon due to the constant influx of migrants from Asia and other countries. The phylogenetic sub-lineage segregation of Gabonese VP4 in discrete clusters following a pattern of G type association (e.g., P[6] with G3 and G12, P[8] with G1 and G8) might be a result of reassortment.
Strains of P[8] contained amino acid disparities in their VP8* epitopes in accordance with their G1, G8 and G9 combinations, although with notable exceptions. G1P[8] was one of the frequent type combinations and most divergent from vaccine cognates, similarly to the less frequent G9P[8]. These genetic and antigenic differences should be considered when planning immunization, since the chosen vaccines will shape the antigenic landscape for circulating RVA. The antigenic G1-VP7 epitopes were mostly conserved and homogeneous with the vaccine strains. Of all three VP7 epitopes, the RVA genome was apparently conserved with vaccine strains, with an exception in G1P[8] (GAB/675), which revealed the N94S substitution associated with immune escape [28,49]. The VP7 epitopes of the Gabonese G3 strains had a relatively low number of different residues compared to RotaTeq G3. Despite this observation, the presence of K238N (epitope 7-1b) may be of concern as it affects glycosylation and has been shown to neutralize RVA in mammals by monoclonal antibodies and hyperimmune sera [46,50]. K238N has also been reported previously in European and North African G3 strains [10,51], with additional sites (70-72) contributing to N-linked glycosylation.
In the case of RVA immunization, a vaccine's failure to eradicate the virus can result in selective pressures that enhance the pathogen's ability to evade host immunity [47]. Viral evolution is often linked to travel and domesticated livestock populations in the community that facilitate the virus shedding and recombination of RVA strains in antigenically naive hosts [48,52]. Although many studies have examined RVA burden in symptomatic patients, this study investigated children without gastroenteritis. The comparison of the RVA genotype distribution between symptomatic and asymptomatic children would have been useful. However, due to the low viral load, we could only obtain a sequence from the control group (asymptomatic children). Another limitation of our study is the small size of the asymptomatic group. This is because sampling began in the second year of the study and therefore precludes observation of the seasonal distribution of RVA cases among asymptomatic children.
The presence of the main pathogenic strains in Gabon, namely G1 and G3 in combination with P[6], the emerging G12 and the less common G9 with antigenic divergence, makes the implementation of vaccination necessary and urgent. This study provides the genetic data needed to assess the epidemiological situation prior to the introduction of vaccination in the Gabonese population, where a wide variety of RVA are present, including strains that are likely to be of zoonotic origin. Additional longitudinal studies at temporal and spatial scales are needed to determine the functional significance of these genetic divergences observed in Gabonese RVA strains and to establish a link with available vaccines, proven to be efficacious.
Contributors
AAA designed, supervised, and coordinated the study procedures. GPM performed all experimental procedures and drafted the manuscript. AT supported experimental procedures, analyzed genetic data, and drafted the manuscript. GPM, MMN, MNM, PANM, GBM, SA and EGR were involved in sampling procedures and collating the baseline data. SN and CTB performed RVA genotyping for independent verification in another laboratory and supported with data analysis. SB, BM, DE, and PGK contributed to the study design and patient recruitment. TPV and AAA contributed to the materials. TPV supervised experimental procedures on RVA screening and genotyping, data analysis, and revised the manuscript from the first draft. All authors read and approved the final version of the manuscript. TPV, AAA, GPM, SN, AT verified the underlying data in this manuscript.
STROBE Statement—Checklist of items that should be included in reports of case-control studies
| Item No | Recommendation | |
|---|---|---|
| Title and abstract | 1 | (a) Indicate the study's design with a commonly used term in the title or the abstract Page 2 – Lines 40–66 |
| (b) Provide in the abstract an informative and balanced summary of what was done and what was found Page 2 – Lines 40–66 | ||
| Introduction | ||
| Background/rationale | 2 | Explain the scientific background and rationale for the investigation being reported Pages 4 and 5 Lines 112–161 |
| Objectives | 3 | State specific objectives, including any prespecified hypotheses Pages 4 and 5 Lines 134–161 |
| Methods | ||
| Study design | 4 | Present key elements of study design early in the paper Page 5, 169–186 |
| Setting | 5 | Describe the setting, locations, and relevant dates, including periods of recruitment, exposure, follow-up, and data collection Page 5, 169–186 |
| Participants | 6 | (a) Give the eligibility criteria, and the sources and methods of case ascertainment and control selection. Give the rationale for the choice of cases and controls Page 5, 169–186 |
| (b) For matched studies, give matching criteria and the number of controls per case Page 5, 169–186 | ||
| Variables | 7 | Clearly define all outcomes, exposures, predictors, potential confounders, and effect modifiers. Give diagnostic criteria, if applicable Supplementary data Table S2 |
| Data sources/ measurement | 8* | For each variable of interest, give sources of data and details of methods of assessment (measurement). Describe comparability of assessment methods if there is more than one group Page 5, 169–186, (cases and controls) Supplementary data Table S2 (cases) |
| Bias | 9 | Describe any efforts to address potential sources of bias Not applicable |
| Study size | 10 | Explain how the study size was arrived at Page 5, 169–186, |
| Quantitative variables | 11 | Explain how quantitative variables were handled in the analyses. If applicable, describe which groupings were chosen and why Page 7, 235–242, |
| Statistical methods | 12 | (a) Describe all statistical methods, including those used to control for confounding Page 7, 235–242, |
| (b) Describe any methods used to examine subgroups and interactions Page 7, 224–233, | ||
| (c) Explain how missing data were addressed Not applicable | ||
| (d) If applicable, explain how matching of cases and controls was addressed | ||
| (e) Describe any sensitivity analyses | ||
| Results | ||
| Participants | 13* | (a) Report numbers of individuals at each stage of study—eg numbers potentially eligible, examined for eligibility, confirmed eligible, included in the study, completing follow-up, and analysed Page 5, 169–186, Page 9, 10, 256–289 |
| (b) Give reasons for non-participation at each stage Not applicable | ||
| (c) Consider use of a flow diagram | ||
| Descriptive data | 14* | (a) Give characteristics of study participants (eg demographic, clinical, social) and information on exposures and potential confounders Page 5, 169–186, (controls and cases) and Supplementary Table S2, (Cases) |
| (b) Indicate number of participants with missing data for each variable of interest Supplementary Table S2 | ||
| Outcome data | 15* | Report numbers in each exposure category, or summary measures of exposure Page 9, 10, 256–289 (controls) and Supplementary Table S2 (cases) |
| Main results | 16 | (a) Give unadjusted estimates and, if applicable, confounder-adjusted estimates and their precision (eg, 95% confidence interval). Make clear which confounders were adjusted for and why they were included Page 9, 10, 256–289 and Supplementary Table S2 |
| (b) Report category boundaries when continuous variables were categorized Supplementary Table S2 | ||
| (c) If relevant, consider translating estimates of relative risk into absolute risk for a meaningful time period Page 9, 10 256–289 | ||
| Other analyses | 17 | Report other analyses done—eg analyses of subgroups and interactions, and sensitivity analyses Page 8–10, 248–371 |
| Discussion | ||
| Key results | 18 | Summarise key results with reference to study objectives Page 10- 371–379 |
| Limitations | 19 | Discuss limitations of the study, taking into account sources of potential bias or imprecision. Discuss both direction and magnitude of any potential bias Page 13, 450–469 |
| Interpretation | 20 | Give a cautious overall interpretation of results considering objectives, limitations, multiplicity of analyses, results from similar studies, and other relevant evidence Pages 11–13, 373–469 |
| Generalisability | 21 | Discuss the generalisability (external validity) of the study results Pages 11–13, 373–469 |
| Other information | ||
| Funding | 22 | Give the source of funding and the role of the funders for the present study and, if applicable, for the original study on which the present article is based Page 7, 244–246 |
*Give information separately for cases and controls.
Note: An Explanation and Elaboration article discusses each checklist item and gives methodological background and published examples of transparent reporting. The STROBE checklist is best used in conjunction with this article (freely available on the Web sites of PLoS Medicine at http://www.plosmedicine.org/, Annals of Internal Medicine at http://www.annals.org/, and Epidemiology at http://www.epidem.com/). Information on the STROBE Initiative is available at http://www.strobe-statement.org.
Data sharing statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
The authors acknowledge Jean Mermoz Ndong Essono Ondong, Elsy Nnoh Dansou, and Judith Makaya Pemba for their support during sampling. We thank Fabrice Lotola Mougeni for his assistance for statistical analysis. TPV, AAA and GPM are members of CANTAM (EDCTP-RegNet2015- 1045) and of PANDORA-ID-Net (EDCTP-RIA2016E-1609) networks both supported by EDCTP and European member states.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103648.
Contributor Information
Thirumalaisamy P. Velavan, Email: velavan@medizin.uni-tuebingen.de.
Ayola Akim Adegnika, Email: aadegnika@cermel.org.
Appendix. Supplementary materials
References
- 1.Troeger C., Forouzanfar M., Rao P.C., Khalil I., Brown A., Reiner R.C. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the global burden of disease study 2015. Lancet Infect Dis. 2017;17(9):909–948. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford S.E., Ramani S., Tate J.E., Parashar U.D., Svensson L., Hagbom M. Rotavirus infection. JAMA Pediatr. 2018;172(SUPPL. 3):50–53. http://repositorio.uchile.cl/bitstream/handle/2250/148901/Rotavirus-infection.pdf?sequence=1&isAllowed=y [Internet]Available from. [Google Scholar]
- 3.Matthijnssens Uniformity of rotavirus strain nomenclature proposed by the rotavirus classification working group (RCWG) jelle. Arch Virol. 2011;156(8):1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matthijnssens J., Ciarlet M., Rahman M., Attoui H., Estes M.K., Gentsch J.R. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol. 2008;153(8):1621–1629. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azaran A., Makvandi M., Teimoori A., Ebrahimi S., Heydari F., Nikfar R. Distribution of rotavirus genotypes circulating in Ahvaz, Iran in 2016. Iran Biomed J. 2018;22(2):107–116. doi: 10.22034/ibj.22.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santos N., Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15(1):29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- 7.Todd S., Page N.A., Steele A.D., Peenze I., Cunliffe N.A. Rotavirus strain types circulating in africa: Review of studies published during 1997–2006. J Infect Dis. 2010;202(SUPPL. 1):34–42. doi: 10.1086/653555. [DOI] [PubMed] [Google Scholar]
- 8.Doro R., Farkas S.L., Martella V., Banyai K. Zoonotic transmission of rotavirus: surveillance and control. Expert Rev Anti Infect Ther. 2015;13(11):1337–1350. doi: 10.1586/14787210.2015.1089171. [DOI] [PubMed] [Google Scholar]
- 9.Gentsch J.R., Laird A.R., Bielfelt B., Griffin D.D., Bányai K., Ramachandran M. Serotype diversity and reassortment between human and animal rotavirus strains: Implications for rotavirus vaccine programs. J Infect Dis. 2005;192(SUPPL. 1):46–59. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- 10.Zeller M., Patton J.T., Heylen E., De Coster S., Ciarlet M., Van Ranst M. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in Belgium and rotaviruses in rotarix and RotaTeq. J Clin Microbiol. 2012;50(3):966–976. doi: 10.1128/JCM.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bányai K., László B., Duque J., Steele A.D., Nelson E.A.S., Gentsch J.R. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: Insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30(SUPPL. 1):122–130. doi: 10.1016/j.vaccine.2011.09.111. [DOI] [PubMed] [Google Scholar]
- 12.Burke R.M., Tate J.E., Kirkwood C.D., Steele A.D., Parashar U.D. Current and new rotavirus vaccines. Curr Opin Infect Dis. 2019;32(5):435–444. doi: 10.1097/QCO.0000000000000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward R.L., Clark H.F., Offit P.A. Influence of potential protective mechanisms on the development of live rotavirus vaccines. J Infect Dis. 2010;202(SUPPL. 1):72–79. doi: 10.1086/653549. [DOI] [PubMed] [Google Scholar]
- 14.Mwenda J.M., Tate J.E., Steele A.D., Parashar U.D. Preparing for the scale-up of rotavirus vaccine introduction in Africa: establishing surveillance platforms to monitor disease burden and vaccine impact. Pediatr Infect Dis J. 2014;33(SUPPL. 1):1–5. doi: 10.1097/INF.0000000000000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roczo-Farkas S., Kirkwood C.D., Cowley D., Barnes G.L., Bishop R.F., Bogdanovic-Sakran N. The impact of rotavirus vaccines on genotype diversity: a comprehensive analysis of 2 decades of australian surveillance data. J Infect Dis. 2018;218(4):546–554. doi: 10.1093/infdis/jiy197. [DOI] [PubMed] [Google Scholar]
- 16.Bucardo F., Rippinger C.M., Svensson L., Patton J.T. Vaccine-derived NSP2 segment in rotaviruses from vaccinated children with gastroenteritis in Nicaragua. Infect Genet Evol. 2012;12(6):1282–1294. doi: 10.1016/j.meegid.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose T.L., da Silva M.F.M., Goméz M.M., Resque H.R., Ichihara M.Y.T., Volotão E de M. Evidence of vaccine-related reassortment of rotavirus, Brazil, 2008–2010. Emerg Infect Dis. 2013;19(11):1843–1846. doi: 10.3201/eid1911.121407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diwakarla C.S., Palombo E.A. Genetic and antigenic variation of capsid protein VP7 of serotype G1 human rotavirus isolates. J Gen Virol. 1999;80(2):341–344. doi: 10.1099/0022-1317-80-2-341. [DOI] [PubMed] [Google Scholar]
- 19.Matthijnssens J., Bilcke J., Ciarlet M., Martella V., Bányai K., Rahman M. Rotavirus disease and vaccination: impact on genotype diversity. Future Microbiol. 2009;4(10):1303–1316. doi: 10.2217/fmb.09.96. [DOI] [PubMed] [Google Scholar]
- 20.Lekana-Douki S.E., Kombila-Koumavor C., Nkoghe D., Drosten C., Drexler J.F., Leroy E.M. Molecular epidemiology of enteric viruses and genotyping of rotavirus A, adenovirus and astrovirus among children under 5 years old in Gabon. Int J Infect Dis. 2015;34:90–95. doi: 10.1016/j.ijid.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 21.Japhet M.O., Famurewa O., Iturriza-Gomara M., Adesina O.A., Opaleye O.O., Niendorf S. Group A rotaviruses circulating prior to a national immunization programme in Nigeria: clinical manifestations, high G12P[8]frequency, intra-genotypic divergence of VP4 and VP7. J Med Virol. 2018;90(2):239–249. doi: 10.1002/jmv.24949. [DOI] [PubMed] [Google Scholar]
- 22.Iturriza-Gómara M., Kang G., Gray J. Rotavirus genotyping: Keeping up with an evolving population of human rotaviruses. J Clin Virol. 2004;31(4):259–265. doi: 10.1016/j.jcv.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Marques A.M., Diedrich S., Huth C., Schreier E. Group A rotavirus genotypes in Germany during 2005/2006. Arch Virol. 2007;152(9):1743–1749. doi: 10.1007/s00705-007-0998-x. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic I., Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R. Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. 2020.
- 27.Aoki S.T., Settembre E., Trask S.D., Greenberg H.B., Stephen C., Dormitzer P.R. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science. 2009;324(5933):1444–1447. doi: 10.1126/science.1170481. 80- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coulson B.S., Kirkwood C. Relation of VP7 amino acid sequence to monoclonal antibody neutralization of rotavirus and rotavirus monotype. J Virol. 1991;65(11):5968–5974. doi: 10.1128/jvi.65.11.5968-5974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyall-Smith M.L., Lazdins I., Tregear G.W., Holmes I.H. Location of the major antigenic sites involved in rotavirus serotype-specific neutralization. Proc Natl Acad Sci USA. 1986;83(10):3465–3468. doi: 10.1073/pnas.83.10.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giammarioli A.M., Mackow E.R., Fiore L., Greenberg H.B., Ruggeri FM. Production and characterization of murine IgA monoclonal antibodies to the surface antigens of rhesus rotavirus. Virology. 1996;225(1):97–110. doi: 10.1006/viro.1996.0578. [DOI] [PubMed] [Google Scholar]
- 31.Dormitzer P.R., Sun Z.Y.J., Wagner G., Harrison SC. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 2002;21(5):885–897. doi: 10.1093/emboj/21.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odimayo M.S., Olanrewaju W.I., Omilabu S.A., Adegboro B. Prevalence of rotavirus-induced diarrhoea among children under 5 years in Ilorin, Nigeria. J Trop Pediatr. 2008;54(5):343–346. doi: 10.1093/tropej/fmn081. [DOI] [PubMed] [Google Scholar]
- 33.Iturriza-Gómara M., Jere K.C., Hungerford D., Bar-Zeev N., Shioda K., Kanjerwa O. Etiology of diarrhea among hospitalized children in blantyre, malawi, following rotavirus vaccine introduction: a case-control study. J Infect Dis. 2019;220(2):213–218. doi: 10.1093/infdis/jiz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shrestha S., Thakali O., Raya S., Shrestha L., Parajuli K., Sherchand J.B. Acute gastroenteritis associated with rotavirus A among children less than 5 years of age in Nepal. BMC Infect Dis. 2019;19(1):1–8. doi: 10.1186/s12879-019-4092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouedraogo N., Ngangas S.M.T., Bonkoungou I.J.O., Tiendrebeogo A.B., Traore K.A., Sanou I. Temporal distribution of gastroenteritis viruses in Ouagadougou, Burkina Faso: seasonality of rotavirus. BMC Public Health. 2017;17(1):1–8. doi: 10.1186/s12889-017-4161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mwenda J.M., Ntoto K.M., Abebe A., Enweronu-Laryea C., Amina I., Mchomvu J. Burden and epidemiology of rotavirus diarrhea in selected african countries: Preliminary results from the african rotavirus surveillance network. J Infect Dis. 2010;202(SUPPL. 1):5–11. doi: 10.1086/653557. [DOI] [PubMed] [Google Scholar]
- 37.Mayindou G., Ngokana B., Sidibé A., Moundélé V., Koukouikila-Koussounda F., Christevy Vouvoungui J. Molecular epidemiology and surveillance of circulating rotavirus and adenovirus in Congolese children with gastroenteritis. J Med Virol. 2016;88(4):596–605. doi: 10.1002/jmv.24382. [DOI] [PubMed] [Google Scholar]
- 38.Lestari F.B., Vongpunsawad S., Wanlapakorn N., Poovorawan Y. Rotavirus infection in children in Southeast Asia 2008-2018: disease burden, genotype distribution, seasonality, and vaccination. J Biomed Sci. 2020;27(1):1–19. doi: 10.1186/s12929-020-00649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy K., Hubbard A.E., Eisenberg J.N.S. Seasonality of rotavirus disease in the tropics: a systematic review and meta-analysis. Int J Epidemiol. 2008;38(6):1487–1496. doi: 10.1093/ije/dyn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ndze V.N., Papp H., Achidi E.A., Gonsu K.H., László B., Farkas S. One year survey of human rotavirus strains suggests the emergence of genotype G12 in Cameroon. J Med Virol. 2013;85(8):1485–1490. doi: 10.1002/jmv.23603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moure U.A.E., Banga-Mingo V., Gody J.C., Mwenda J.M., Fandema J., Waku-Kouomou D. Emergence of G12 and G9 rotavirus genotypes in the Central African Republic, January 2014 to February 2016. BMC Res Notes. 2018;11(1):5. doi: 10.1186/s13104-017-3122-7. [Internet]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tcheremenskaia O., Marucci G., De Petris S., Ruggeri F.M., Dovecar D., Sternak S.L. Molecular epidemiology of rotavirus in Central and Southeastern Europe. J Clin Microbiol. 2007;45(7):2197–2204. doi: 10.1128/JCM.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iturriza Gómara M., Kang G., Mammen A., Jana A.K., Abraham M., Desselberger U. Characterization of G10P[11]rotaviruses causing acute gastroenteritis in neonates and infants in Vellore, India. J Clin Microbiol. 2004;42(6):2541–2547. doi: 10.1128/JCM.42.6.2541-2547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malasao R., Khamrin P., Kumthip K., Ushijima H., Maneekarn N. Complete genome sequence analysis of rare G4P[6]rotavirus strains from human and pig reveals the evidence for interspecies transmission. Infect Genet Evol. 2018;65:357–368. doi: 10.1016/j.meegid.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 45.Matthijnssens J., Ciarlet M., Heiman E., Arijs I., Delbeke T., McDonald S.M. Full genome-based classification of rotaviruses reveals a common origin between human wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol. 2008;82(7):3204–3219. doi: 10.1128/JVI.02257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciarlet M., Hoshino Y., Liprandi F. Single point mutations may affect the serotype reactivity of serotype G11 porcine rotavirus strains: a widening spectrum? J Virol. 1997;71(11):8213–8220. doi: 10.1128/jvi.71.11.8213-8220.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Read A.F., Baigent S.J., Powers C., Kgosana L.B., Blackwell L., Smith L.P. Imperfect vaccination can enhance the transmission of highly virulent pathogens. PLoS Biol. 2015;13(7):1–18. doi: 10.1371/journal.pbio.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pybus O.G., Tatem A.J., Lemey P. Virus evolution and transmission in an ever more connected world. Proc R Soc B Biol Sci. 2015;282(1821):1–10. doi: 10.1098/rspb.2014.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taniguchi K., Hoshino Y., Nishikawa K., Green K.Y., Maloy W.L., Morita Y. Cross-reactive and serotype-specific neutralization epitopes on VP7 of human rotavirus: nucleotide sequence analysis of antigenic mutants selected with monoclonal antibodies. J Virol. 1988;62(6):1870–1874. doi: 10.1128/jvi.62.6.1870-1874.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ciarlet M., Reggeti F., Pina C.I., Liprandi F. Equine rotaviruses with G14 serotype specificity circulate among Venezuelan horses. J Clin Microbiol. 1994;32(10):2609–2612. doi: 10.1128/jcm.32.10.2609-2612.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mouna B.H.F., BenHamida-Rebaï M., Heylen E., Zeller M., Moussa A., Kacem S. Sequence and phylogenetic analyses of human rotavirus strains: comparison of VP7 and VP8* antigenic epitopes between Tunisian and vaccine strains before national rotavirus vaccine introduction. Infect Genet Evol. 2013;18(May):132–144. doi: 10.1016/j.meegid.2013.05.008. [Internet]Available from. [DOI] [PubMed] [Google Scholar]
- 52.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.