Abstract
Small regulatory RNAs (sRNAs) belong to a family of non-coding RNAs, and many of which regulate expression of genes via interaction with mRNA. The recent popularity of high-throughput next generation sequencers have presented abundant sRNA-related data, including sRNAs of several different oral bacterial species. Some sRNA candidates have been validated in terms of their expression and interaction with target mRNAs. Since the oral cavity is an environment constantly exposed to various stimuli, such as fluctuations in temperature and pH, and osmotic pressure, as well as changes in nutrient availability, oral bacteria require rapid control of gene expression for adaptation to such diverse conditions, while regulation via interactions of sRNAs with mRNA provides advantages for rapid adaptation. This review summarizes methods effective for identification and validation of sRNAs, as well as sRNAs identified to be associated with oral bacterial species, including cariogenic and periodontal pathogens, together with their confirmed and putative target genes.
Keywords: Small regulatory RNA, Non-coding RNA, Antisense RNA, Oral bacteria, Streptococcus, Periodontal pathogens
1. Bacterial sRNA introduction
Small regulatory RNA (sRNA) is a member of a family of non-coding RNAs, some of which have regulatory functions [1,2]. As compared with eukaryotic miRNA with sizes ranging from 20 to 30 nucleotides (nt), bacterial sRNAs are structurally heterogenous with a wider range of length from approximately 20–500 nt [[1], [2], [3]], and can be classified into intergenic region sRNAs (IGR sRNA) and antisense sRNAs (asRNAs), depending on their genomic location and orientation [1].
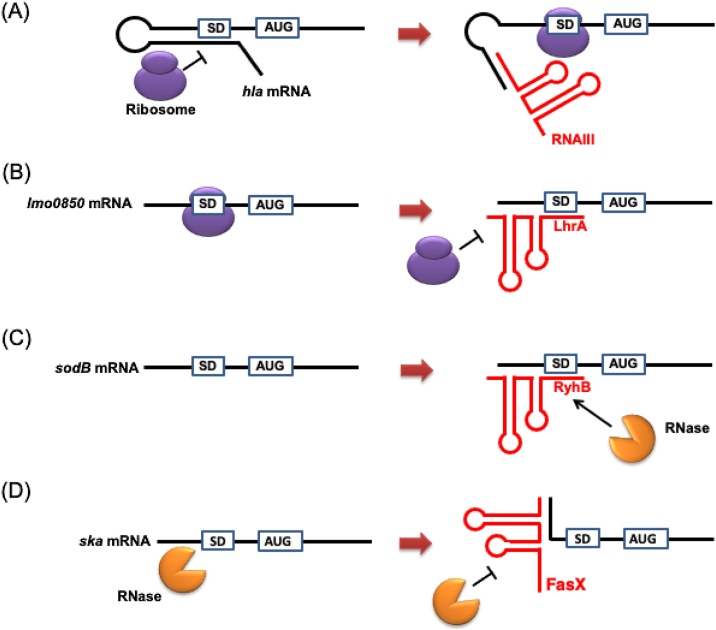
IGR sRNAs are encoded in intergenic regions, which generally contain no protein-coding sequence, and control translation or mRNA stability of target genes situated in various genomic regions (trans-acting regulation). In many instances, IGR sRNAs form a stem-loop structure and regulate expression of target mRNAs in a manner that involves imperfect base-pairing. For example, McaS [4] of Escherichia coli and RNAIII [5] of Staphylococcus aureus mediate translational activation of the target genes flhD (flagellar transcriptional regulator) and hla (α-Hemolysin), respectively. These target mRNAs form a hairpin structure on the 5ʹ untranslated region (UTR) containing the ribosome binding site (RBS), which halts translation due to low accessibility of the ribosome to mRNA. Binding of the sRNAs to target mRNA resolves the hairpin structure and relieves the restricted accessibility (Fig. 1A). The Listeria monocytogenes sRNA LhrA binds to the 5ʹ UTR region containing the RBS of target mRNAs and functions as a translational repressor by preventing ribosome access (Fig. 1B) [6]. Furthermore, mRNA levels of target genes are modulated by binding of IGR sRNA. Also, the sRNA-mRNA complex enhances RNA degradation by endogenous RNases, resulting in decreased expression levels of target genes (Fig. 1C). The Escherichia coli sRNA RyhB regulates the level of sodB encoding superoxide dismutase by RNase E cleavage [7], while the Salmonella typhimurium sRNA MicA binds ompA mRNA encoding an outer membrane protein, and the complex of MicA and target mRNA is cleaved by RNase III [8]. In contrast, some sRNAs are known to enhance target mRNA stability (Fig. 1D), including the Streptococcus pyogenes sRNA FasX that interacts with the 5ʹ UTR of ska mRNA encoding streptokinase [9].
Fig. 1.
Regulatory functions of sRNA.
(A) Translational activation by sRNA. Base-pairing of RNA III, a multifunctional sRNA of Staphylococcus aureus, with the α-hemolysin-encoding hla mRNA modifies the mRNA secondary structure and exposes the RBS to the ribosome. (B) Translational suppression by sRNA. Binding of the lmo0850 mRNA encoding a hypothetical protein to the ribosome is inhibited by base-pairing of the LhrA sRNA in Listeria monocytogenes. (C) mRNA degradation induced by sRNA. In Escherichia coli, base-pairing of the RyhB sRNA and the superoxide dismutase-encoding sodB mRNA recruits endogenous RNase, which cleaves the complex. (D) RNA stabilization by sRNA. Binding of the sRNA FasX to the streptokinase-encoding ska mRNA enhances the mRNA stability in Streptococcus pyogenes.
asRNAs are encoded on the opposite strand of the gene. In many instances, they bind to mRNAs expressed from the antisense strand and mediate regulation (cis-acting regulation) by various mechanisms, such as inhibition/activation of translation as well as alteration of RNA stability [10]. Gene regulation of several different bacterial species, especially Gram-negative bacteria, via sRNAs often requires RNA chaperons, such as Hfq [11], ProQ [12], or CsrA [13]. These chaperons mediate interactions between sRNA and mRNA, which effects the regulation mechanisms of sRNAs [14].
This review summarizes methods used for identification and validation of sRNAs, and also presents findings by which the functions of thus far reported sRNAs and asRNAs of oral bacteria can be confirmed or deduced, including the cariogenic bacterium Streptococcus mutans, and periodontal pathogens Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans. Additionally, various sRNA and asRNA functions are introduced in the context of virulence-related regulation.
2. sRNA identification
The complete genome sequences of many bacterial species have been deposited in databases. However, rather than identification of open reading frames, experimental analysis is basically required for identification of IGR sRNAs and asRNAs. Prior to development of the next-generation sequencing technology, software-based in silico analyses and microarrays covering the entire genome were utilized to predict and identify sRNAs [15,16], while more recently RNA-seq analysis has become the main method for sRNA identification [[17], [18], [19]]. Furthermore, oral bacteria (genus Streptococcus and periodontal pathogens) sRNAs have also been detected by use of this method (Table 1) [[20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30]].
Table 1.
sRNAs of oral bacteria identified by RNA-seq.
| Species | Strain | Source of sequence library | Number of sRNA | References |
|---|---|---|---|---|
| Streptococcus mutans | ATCC 25175 | Small molecules of cDNA were extracted after PAGEa | 922 | 20 |
| UA159 | Small molecules of cDNA were extracted after PAGE | 1879 | 21 | |
| UA159 | Small RNA fraction was extracted using miRNA column | 2125 | 23 | |
| UA159 | Small molecules of cDNA were extracted after PAGE | 736 | 25 | |
| UA159 | Small RNA fraction was extracted using miRNA column | 2749 | 26 | |
| Streptococcus sanguinis | SK36 | Small RNA fraction was extracted using miRNA column | 219 | 24 |
| Aggregatibacter actinomycetemcomitans | 624 | Total RNA-seq without rRNA | 202 | 28 |
| ATCC 33384 | Small RNA fraction was extracted using miRNA column | 59 | 22 | |
| HK1651 | Total RNA was converted to cDNA without fragmentation | 120 | 29 | |
| Porphyromonas gingivalis | ATCC 33277 | Total RNA-seq without rRNA | 2480 | 30 |
| W83 | Small RNA fraction was extracted using miRNA column | 30 | 27 | |
| ATCC 33277 | Small RNA fraction was extracted using miRNA column | 40 | 22 | |
| Treponema denticola | ATCC 35405 | Small RNA fraction was extracted using miRNA column | 11 | 22 |
Polyacrylamide gel electrophoresis.
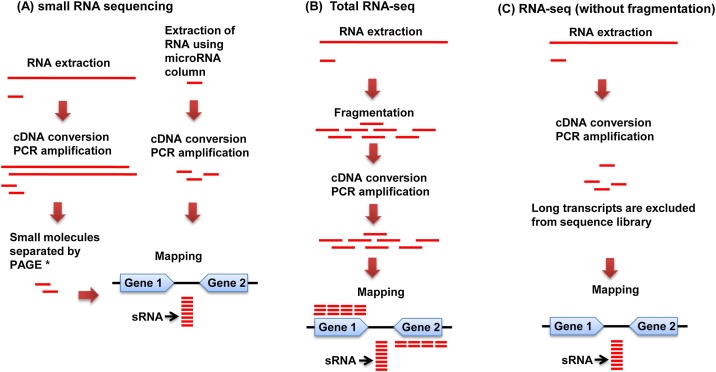
Techniques used for RNA purification and cDNA library construction have effects on the number and class of identifiable sRNAs. Those similar to the method for sequencing eukaryotic miRNA have been used for bacterial sRNA identification, including (1) Small RNA fractions extracted with a column specialized for miRNA and (2) a cDNA library constructed from fractions extracted from a polyacrylamide gel after electrophoresis (Fig. 2A). sRNAs have been identified with these sampling methods by some research groups [[20], [21], [22], [23], [24], [25], [26]] that focused on short sRNAs with a length less than 50 nt, termed miRNA-Size sRNA (msRNAs) (Table 1), as they are not suitable for detection of relatively large sRNA molecules. Representative sRNAs with a large size (>200 nt) that have been well studied include RNAIII of S. aureus [5], VR-RNA of Clostridium perfringens [31], and FasX [32] of Streptococcus pyogenes.
Fig. 2.
Flowchart illustrating bacterial sRNA identification.
(A) Sequence samples of small RNA-seq were prepared from gel extracts after PAGE or from eluates produced by an miRNA specialized column. (B) Total RNA-seq allows for sequences of all transcripts containing sRNA. (C) RNA-seq analysis with non-fragmented RNA allows for effective sequencing of small molecules as well as sRNAs with a relatively large size (>200 nt). *Polyacrylamide gel electrophoresis.
Jorth et al. sequenced all transcripts of strain 624 of the periodontal pathogen A. actinomycetemcomitans, using a total RNA-seq method (Fig. 2B), which provided identification of 202 sRNAs containing long molecules (>200 nt) and defined the transcriptional start sites of all sRNAs [28]. RNA-seq analysis targeting total RNA is appropriate for revealing sRNA repertories, though the data obtained will also contain a large amount of unnecessary sequences, including those related to protein-coding mRNA and ribosomal RNA. In addition, the cost for such a sequencing procedure is higher than that for sequencing only small molecules. A library of fragmented RNA is generally used for RNA-seq analysis (Fig. 2B), because the sequence platform limits the size (for example, <600 nt with an Illumina sequencer).
Previously, one of the present authors reported sRNAs of A. actinomycetemcomitans strain HK1651 [29]. In that study, total RNA was intentionally not fragmented and long transcripts were excluded in the following step of cDNA library construction by PCR (Fig. 2C). Sequencing was performed using an illumina MiSeq platform and a total of 120 sRNAs with lengths of 35–386 nt were identified. Most of the obtained reads were 5s rRNA, thus elimination of 5s rRNA from a sequence library may be useful for efficient sRNA identification. Several rRNA depletion kits are commercially available, including MICROBExpress Bacterial mRNA Enrichment Kit (Thermo Fisher Scientific, Waltham, MA, USA), RiboMinus Transcriptome Isolation Kit, bacteria (Thermo Fisher Scientific), and Ribo-Zero rRNA Depletion Kit (Illumina, San Diego, CA, USA), though only the Ribo-Zero kit contains 5S rRNA-specific probes that can be used to eliminate 5S rRNA by RNA hybridization. Alternatively, terminator 5ʹ monophosphate-dependent exonuclease (TEX) can be utilized to remove rRNA from total RNA. TEX is a processive 5ʹ → 3ʹ exonuclease and specifically digests RNA with 5ʹ monophosphate, while it does not degrade RNA with the 5′-triphosphate, 5ʹ -cap, or 5ʹ-hydroxyl group. The primary bacterial transcripts contain a triphosphate at the 5ʹ terminus. On the other hand, rRNA and RNA degradant have a mono-phosphate at the 5ʹ terminus, which can be digested by TEX. Unlike eukaryotic miRNA, several bacterial sRNAs contain a triphosphate at the 5ʹ terminus, and TEX treatment can be used to eliminate rRNA and RNA degradants.
Co-immunoprecipitation with RNA chaperones has been applied for identification of RNA molecules bound to those chaperones [33,34]. Since several functional sRNAs form the mRNA-sRNA-protein complex, sequencing of RNAs co-precipitated with chaperones can be used to identify such sRNAs [[11], [12], [13]]. In particular, Hfq, a bacterial member of the Sm family of RNA-binding proteins, has been used for identification of RNA, including sRNAs and their target mRNA [35,36].
3. sRNA validation
The detected expression levels of sRNAs vary, depending on the strain and culture conditions. Furthermore, previously identified sRNAs were listed by use of criteria determined by individual researchers, which were mainly based on expression levels and genomic locations. Therefore, the numbers of sRNAs identified differ considerably among studies, even of the same species (Table 1). In addition, there is a possibility of false-positive results that might have been caused by sequencing RNA degradation products even though expression level was high. Thus, sRNA validation in additional experiments is important to reconfirm expression and orientation, as well as the encoded chromosomal locations. Northern blot analysis has been frequently used for validation of sRNA candidates [37,38] as well as reverse transcription PCR (RT-PCR) [37], though use of Taq-man probes is necessary to distinguish transcription orientations. Additionally, it is also difficult to design primers for short IGR sRNA, thus rapid amplification of cDNA ends (RACE) analysis is suitable for validation. In a previous study, 70 of 120 sRNA candidates were validated by use of 3ʹ RACE analysis [29], as that is useful for determination of the transcriptional start site and 3ʹ terminal end of sRNA.
4. Oral Streptococcus sRNAs
The genus Streptococcus is comprised of Gram-positive cocci and dominant in the oral cavity. RNA-dependent regulation of virulence, competence, and bacterial immunity has been examined in studies of pathogenic streptococci [[39], [40], [41]]. Organisms belonging to a subset of oral Streptococcus species, such as mitis group Streptococcus, are known to be primary colonizers on tooth surfaces and promote dental biofilm formation. Mutans group Streptococcus is involved in dental caries development by their activities to metabolize carbohydrates and produce acids. These oral commensals also occasionally cause systemic infections such as endocarditis by gaining access to the bloodstream. Streptococcus mutans and Streptococcus sanguinis sRNAs have been investigated, with a brief discussion of those findings following. Table 2 summarizes information about their notable sRNAs.
Table 2.
Notable sRNAs found in oral streptococi.
| sRNA name | Species | Strain | Expression | Targets | Putative phenotype | References |
|---|---|---|---|---|---|---|
| srn884837 | Streptococcus mutans | UA159 | Decreased in low pH | glnQ, glnM, brpA, relA | Acid tolerance | 20 |
| srn133480 | Streptococcus mutans | UA159 | Decreased in low pH | ffh, brpA, relA | Acid tolerance | 20 |
| sRNA133474 | Streptococcus mutans | UA159 and clinical strains | Decreased in low pH | liaR, ciaR, covR | Acid tolerance | 49 |
| msRNA 1701 | Streptococcus mutans | UA159 | Decreased in rnc mutant | vicK | Biofilm formation | 58 |
| msRNA 3405 | Streptococcus mutans | UA159 | Decreased in rnc mutant | vicR | Biofilm formation | 58 |
| msRNA 1657 | Streptococcus mutans | UA159 | Decreased in rnc mutant | vicK | Biofilm formation | 58 |
| ASvicR | Streptococcus mutans | UA159 | Decreased expression in biofilm | vicR | Biofilm formation | 59 |
| sRNA0187 | Streptococcus mutans | UA159 and clinical strains | Increased in high sucrose | Biofilm formation | 25 | |
| sRNA0593 | Streptococcus mutans | UA159 and clinical strains | Decreased in high sucrose | Biofilm formation | 25 | |
| irvA 5ʹ UTR | Streptococcus mutans | UA159 | gpbC | Dextran-dependent aggregation | 66 | |
| srSm | Streptococcus mutans | UA159 | fst-Sm | Toxin-antitoxin system | 68 | |
| S.S-1964 | Streptococcus sanguinis | SK36 | Decreased in skim milk | SSA0513 | Conversion of cobalamin | 24 |
| csRNA1-1 | Streptococcus sanguinis | SK36 | Decreased in ciaRH mutant | pilT | Biofilm formation | 40 |
| csRNA1-2 | Streptococcus sanguinis | SK36 | Decreased in ciaRH mutant | pilT | Biofilm formation | 40 |
4.1. Streptococcus mutans
S. mutans is the major pathogen responsible for dental caries, which arises from decalcification of the tooth tissues under an acidic condition (less than pH 5.5) [42]. This bacterium can metabolize a variety of carbohydrates by fermentation and produces acid as a metabolic byproduct, leading to decalcification. The acid tolerance of this bacterium has been noted as a crucial factor related with its cariogenic activity and ability to survive within acidic carious lesions [43].
Liu et al. identified small-sized sRNAs (18−50 nt) in S. mutans strain UA159 cultured under an acidic condition and further examined two sRNAs abundantly expressed (referred to as srn884837 and srn133480, with length of 29 and 27 nt, respectively) [21]. Using RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) [44], target mRNAs were predicted to encode factors involved in acid tolerance, including the glutamate transporter components GlnQ and GlnM [45], Ffh, homologous to the signal recognition particle of E. coli [46], the surface-associated protein BrpA, homologous to LytR-CpsA-Psr family proteins [47], and RelA responsible for synthesis and hydrolysis of guanosine tetraphosphate/pentaphosphate [48] (glnQ, glnM, brpA, and relA for srn884837; ffh, brpA, and relA for srn133480). The expression levels of the two sRNAs were decreased with lowered pH (pH 6.5, 5.5, 4.5), while those of the corresponding putative target genes were increased, implying that these sRNAs have negative effects on expression of acid tolerance related genes.
Expression of sRNA molecules containing both the srn133480 sequence and 6 additional bases at the 5ʹ end (referred to as sRNA133474) was also shown to be decreased along with a decrease in pH in ten tested clinical isolates [49]. Among the examined strains, the expression levels of sRNA133474 varied, while highly acid-tolerant strains showed lower expression levels of srn133480 as compared with low acid-tolerant strains. Furthermore, target mRNAs predicted with use of the RNApredator system (http://rna.tbi.univie.ac.at/RNApredator) [50] included mRNAs of two-component system (TCS) genes, including liaS, liaR, comE, covR, and ciaR [[51], [52], [53], [54]], with the expression levels of liaR, covR, and ciaR shown to be negatively correlated with that of sRNA133474. The authors suggested that sRNA133474 could be utilized for adaptation to acidic conditions via TCS modulation.
Other TCS genes are also under the control of an asRNA and sRNAs. The VicRK TCS is highly conserved in Gram-positive bacteria and is related to virulence-associated traits, such as biofilm formation, genetic competence, bacteriocin production, and exopolysaccharide synthesis [[55], [56], [57]]. Mao et al. reported that three putative sRNAs (referred to as msRNA 1701, msRNA 3405, and msRNA 1657) potentially target vicRK mRNA, and their expression levels were negatively correlated with that of vicRK and positively correlated with that of rnc encoding RNase III [58]. Consequently, the authors inferred that RNase III post-transcriptionally reduced vicRK mRNA levels through sRNAs. Furthermore, VicR protein level was shown to be negatively regulated by asRNA of vicR (ASvicR), which was confirmed by Northern blot analysis findings [59]. Overexpression of ASvicR was reported to delay bacterial growth, and compromised the ability of the organisms to synthesize exopolysaccharide and form carious lesions on teeth in a rat infection model [59]. Furthermore, transcriptome analysis indicated that ASvicR inhibits galactose and glucose metabolism. Thus, the elaborate interplay between VicRK TCS genes, ASvicR, putative sRNAs, and RNase III could be related to the cariogenic properties of S. mutans.
Adhesion to tooth surfaces and synthesis of water-insoluble glucan are also determinants for S. mutans cariogenicity [60]. Liu et al. reported that sRNAs participate in regulation of bacterial adherence and glucan synthesis in response to extracellular sucrose [23]. Of 2125 sRNA candidates expressed under a 1% or 5% sucrose condition, 22 were differentially expressed depending on the sucrose concentration, six of which were validated by quantitative RT-PCR analysis. Target mRNAs predicted with an RNAhybrid algorithm were found to include spaP, gtfB, and gtfC, and shown to encode antigen I/II family surface proteins, protein antigen C (PAc) [61], and two types of glucosyltransferases (GTFs) responsible for synthesis of extracellular water-insoluble glucan [62,63]. However, details regarding direct interaction of sRNAs with mRNAs of these putative targets and the potential consequences remain unknown. A subset of sRNAs were also reported to be induced in response to extracellular glucose [26]. In addition, expression of two sRNAs (referred to as sRNA0187 and sRNA0593) were correlated with bacterial adherence to a Petri dish surface in the presence of 1% sucrose [25].
The 5ʹ UTR of the irvA gene encoding a putative repressor [64] was shown to interact with glucan-binding protein C (GbpC)-encoding mRNA [65], also to modulate mRNA stability as well as GbpC production [66]. Furthermore, the protein encoding irvA mRNA has an ability to function as a trans-RNA, while sRNAs have also been reported to be involved in bacterial toxin-antitoxin (TA) systems [67]. This system consists of two components, a peptide toxin and cognate antitoxin. Of six classes of bacterial TA systems, S. mutans strain UA159 possesses the type I TA system, where the untranslated antisense small RNA (∼70 nt) serves as the antitoxin [68]. Base-pairing of the toxin mRNA and cis-encoded small antitoxin sRNA may suppress the toxin translation.
Although several S. mutans sRNAs have been reported, the exact mechanisms related to their functions remain largely elusive. Elucidation of the detailed functions and critical roles of those sRNAs in regulatory networks may provide a molecular basis for therapeutic approaches for treatment of dental caries.
4.2. Streptococcus sanguinis
S. sanguinis, a commensal oral bacterium, is a member of mitis group Streptococcus and known as an early colonizer on tooth surfaces, thus is involved in development of dental plaque [69]. In addition, this bacterium has frequently been isolated from endocarditis lesions [70]. Using RNA-seq analysis, Choi et al. identified 219 msRNAs and then focused on an asRNA (referred to as S.S-1964) expressed from the antisense strand corresponding to the 3ʹ UTR of the gene encoding ATP:cob(I)alamin adenosyltransferase (SSA0513), an enzyme involved in conversion of cobalamin (vitamin B12) to its coenzyme form. Expression of SSA0513 was increased following addition of skim milk to the culture, while the expression of asRNA was decreased. The authors also showed that sRNAs including S.S-1964 were secreted by extracellular membrane vesicles. However, the interaction of asRNA and ssa0513 mRNA was not elucidated in that study [24].
Streptococcus pneumoniae, a member of the mitis group, was shown to produce five cia-dependent small RNAs (csRNA), of which expression is regulated by CiaRH TCS [71]. Thereafter, homology searches revealed six csRNA genes present in S. sanguinis, including csRNA1-1, csRNA1-2, csRNA1-3, csRNA2, csRNA7, and csRNA8 [72]. Ota et al. performed target prediction of those csRNAs using TargetRNA2, which showed the pilT gene encoding a component of the type IV pilus to be a target of csRNA1-1 [40,73]. While S. sanguinis strains, such as strain SK36, produce cell wall anchored pili that bind to salivary amylase [74,75], type IV pili are produced in only a few of those strains [76]. Type IV pili mediate a variety of cellular processes, such as twitching motility, genetic competence, and biofilm formation [77]. Furthermore, Ota et al. demonstrated that csRNA1-1 directly binds to pilT mRNA using RNA-RNA electrophoretic mobility shift assay, while mutation of both csRNA1-1 and csRNA1-2 compromised the ability to form biofilm [40].
5. Periodontal pathogen sRNAs
Periodontal disease is a polymicrobial disease caused by a variety of bacterial virulence factors in concert with the immune response [78,79]. Currently, periodontal pathogens are recognized as pathobionts related with the onset of systemic diseases [80]. In the following section, reported sRNAs of P. gingivalis, A. actinomycetemcomitans, and Treponema denticola are introduced. Table 3 summarizes information about the notable sRNAs.
Table 3.
Notable sRNAs found in periodontal bacteria.
| sRNA name | Species | Strain | Expression | Targets | Putative phenotype | References |
|---|---|---|---|---|---|---|
| sRNA PG_RS02100 | Porphyromonas gingivalis | W83 | Decreased in stationary phase cell | Oxidative stress resistance and heme accumulation | 81 | |
| sRNA JA03 | Aggregatibacter actinomycetemcomitans | HK1651 | Increased in iron-chelated medium | hitC | Iron acquisition | 83 |
| AaHKsRNA042 | Aggregatibacter actinomycetemcomitans | HK1651 | ltxD | Cytolysin production | 29 | |
| AaHKsRNA093 | Aggregatibacter actinomycetemcomitans | HK1651 | cdtA | Cytolethal distending toxin production | 29 | |
| AaHKsRNA051 | Aggregatibacter actinomycetemcomitans | HK1651 | flp-1 | Adhesin to host cell | 29 |
5.1. P. gingivalis
P. gingivalis is a Gram-negative anaerobe that belongs to the phylum Bacteroidetes. Hirano et al. performed RNA-seq analysis of P. gingivalis strain ATCC 33277 and confirmed 11 sRNAs based on 5ʹ RACE analysis results [30], while Philips et al. identified 30 sRNAs in P. gingivalis strain W83 using RNA-seq and microarray analyses [27]. Thereafter, the latter group constructed an inactivated mutant strain of sRNA PG_RS02100 and demonstrated that the mutation rendered bacteria more resistant to oxidative stress. The color of the mutant strain colonies grown on blood agar plates was darker than that of those of the wild type. The authors suggested that sRNA regulates genes involved in oxidative stress resistance and heme accumulation [81].
5.2. A. actinomycetemcomitans
The major periodontal pathogen A. actinomycetemcomitans is a Gram-negative facultative anaerobe that belongs to the phylum Proteobacteria. Jorth et al. validated nine sRNAs in strain VT1169 from sRNA candidates predicted by bioinformatics [82]. Additionally, the authors also performed a more systematic screening of sRNA in strain 624 using RNA-seq and identified 202 sRNAs [28]. Utilizing bioinformatic analysis and strain HK1651, Amarasinghe et al. identified four sRNAs expressed under iron-limiting conditions and found that overexpression of the sRNA JA03 significantly repressed expression of hitC encoding an ATP-binding protein, a component of the Fe(III) ABC transporter [83]. Oogai et al. examined strain HK1561 and reported a total of 120 sRNAs (90 IGR-sRNAs, 30 asRNAs) [29]. Distribution of the identified sRNAs among five A. actinomycetemcomitans strains was analyzed by RT-PCR and intraspecies sRNA variations were noted. Target mRNA prediction of identified sRNAs using intaRNA [84] showed interactions of sRNAs and mRNAs encoding major virulence factors (cdtA, lktD, and flp-1 encoding cytolethal distending toxin subunit A, leukotoxin export protein LtxD, and fimbrial protein Flp 1, respectively), suggesting that A. actinomycetemcmoitans virulence is controlled by sRNAs.
5.3. T. denticola and outer membrane vesicles of periodontal pathogens
T. denticola is an oral spirochete associated with chronic periodontitis. Choi et al. identified 11 msRNAs from the T. denticola strain ATCC 35405, three of which (T.D2161, T.D15612, and T.D16563) were detected in secreted outer membrane vesicles (OMVs) by Northern blotting [22]. Additionally, they also found 40 msRNAs from P. gingivalis strain ATCC 33277 and 59 msRNAs from A. actinomycetemcomitans strain ATCC 33384. Furthermore, msRNAs were also detected in OMVs of these species, including an msRNA (A.A_20050) from A. actinomycetemcomitans and three msRNAs (P.G_45033, P.G_4378, P.G_122) from P. gingivalis. When OMVs from the three species were purified and separately added to NIH3T3 fibroblastic cells, penetration of all tested msRNAs into cytoplasm of those cells was detected. Also, transfection of a synthetic msRNA oligonucleotide, such as A.A_20050, P.G_45033, or T.D_2161, into Jurkat T cells decreased expression levels of interleukin (IL) -5, IL-13, and IL-15. Together, these findings indicated that msRNAs delivered to mammalian cells by OMVs potentially modulate host immune responses through RNA interference.
6. Conclusion
Over the recent decade, numerous sRNAs of oral bacteria have been identified. As shown in the above sections, the sRNA repertoire obtained from RNA-seq analyses of the same bacterial species (even the same strain) among various conducted studies have revealed great diversity. The differences can be mainly attributed to strain-specific expression due to genomic diversity as well as the criteria used for analysis. To better understand the relationship of sRNAs with virulence-related phenotypes of oral bacteria, detailed experimental analyses together with the computational validations [85,86] are anticipated. It will be necessary to confirm the expression mode, precise chromosomal locations encoded, target mRNA, and mechanisms of action. sRNAs demonstrate posttranscriptional regulation of genes important for cellular events, including adaptation to environmental cues, biofilm formation, and virulence [87]. Although several studies thus far presented have predicted that sRNAs of oral pathogens interact with virulence gene mRNAs, functional information regarding sRNAs is very limited. Additional studies of oral bacteria sRNAs together with metatranscriptome data of the oral microbiome [88,89] should provide additional insight into the regulatory networks and exact roles of sRNAs related to the pathogenesis of oral diseases. Since the sRNA-mediated artificial regulation has been utilized for modulating gene expression and engineering metabolic status in bacteria [90,91], application of sRNAs, such as delivering effective synthetic sRNAs into distinct oral bacteria, may provide a new strategy to combat oral diseases.
Conflicts of interest
The authors have no conflicts of interest to declare in regard to this study.
References
- 1.Waters L.S., Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fröhlich K.S., Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomason M.K., Fontaine F., De Lay N., Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morfeldt E., Taylor D., von Gabain A., Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen J.S., Lei L.K., Ebersbach T., Olsen A.S., Klitgaard J.K., Valentin-Hansen P. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 2010;38:907–919. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Afonyushkin T., Vecerek B., Moll I., Bläsi U., Kaberdin V.R. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viegas S.C., Silva I.J., Saramago M., Domingues S., Arraiano C.M. Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res. 2010;39:2918–2930. doi: 10.1093/nar/gkq1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramirez-Peña E., Treviño J., Liu Z., Perez N., Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol. 2010;78:1332–1347. doi: 10.1111/j.1365-2958.2010.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saberi F., Kamali M., Najafi A., Yazdanparast A., Moghaddam M.M. Natural antisense RNAs as mRNA regulatory elements in bacteria: a review on function and applications. Cell Mol Biol Lett. 2016;21:6. doi: 10.1186/s11658-016-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geissmann T.A., Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Attaiech L., Boughammoura A., Brochier-Armanet C., Allatif O., Peillard-Fiorente F., Edwards R.A. Silencing of natural transformation by an RNA chaperone and a multitarget small RNA. Proc Natl Acad Sci USA. 2016;113:8813. doi: 10.1073/pnas.1601626113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vakulskas C.A., Potts A.H., Babitzke P., Ahmer B.M.M., Romeo T. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev. 2015;79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quendera A.P., Seixas A.F., dos Santos R.F., Santos I., Silva J.P.N., Arraiano C.M. RNA-binding proteins driving the regulatory activity of small non-coding RNAs in bacteria. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pichon C., Felden B. Small RNA gene identification and mRNA target predictions in bacteria. Bioinformatics. 2008;24:2807–2813. doi: 10.1093/bioinformatics/btn560. [DOI] [PubMed] [Google Scholar]
- 16.Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 17.Shinhara A., Matsui M., Hiraoka K., Nomura W., Hirano R., Nakahigashi K. Deep sequencing reveals as-yet-undiscovered small RNAs in Escherichia coli. BMC Genomics. 2011;12:428. doi: 10.1186/1471-2164-12-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gómez-Lozano M., Marvig R.L., Molin S., Long K.S. Genome-wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ Microbiol. 2012;14:2006–2016. doi: 10.1111/j.1462-2920.2012.02759.x. [DOI] [PubMed] [Google Scholar]
- 19.Carroll R.K., Weiss A., Broach W.H., Wiemels R.E., Mogen A.B., Rice K.C. Genome-wide annotation, identification, and global transcriptomic analysis of regulatory or small RNA gene expression in Staphylococcus aureus. mBio. 2016;7 doi: 10.1128/mBio.01990-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H.-J., Hong S.-H. Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. FEMS Microbiol Lett. 2012;326:131–136. doi: 10.1111/j.1574-6968.2011.02441.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu S., Tao Y., Yu L., Zhuang P., Zhi Q., Zhou Y. Analysis of small RNAs in Streptococcus mutans under acid stress—a new insight for caries research. Int J Mol Sci. 2016;17:1529. doi: 10.3390/ijms17091529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi J.W., Kim S.C., Hong S.H., Lee H.J. Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J Dent Res. 2017;96:458–466. doi: 10.1177/0022034516685071. [DOI] [PubMed] [Google Scholar]
- 23.Liu S.S., Zhu W.H., Zhi Q.H., Liu J., Wang Y., Lin H.C. Analysis of sucrose-induced small RNAs in Streptococcus mutans in the presence of different sucrose concentrations. Appl Microbiol Biotech. 2017;101:5739–5748. doi: 10.1007/s00253-017-8346-x. [DOI] [PubMed] [Google Scholar]
- 24.Choi J.-W., Kwon T.-Y., Hong S.-H., Lee H.-J. Isolation and characterization of a microRNA-size secretable small RNA in Streptococcus sanguinis. Cell Biochem Biophy. 2018;76:293–301. doi: 10.1007/s12013-016-0770-5. [DOI] [PubMed] [Google Scholar]
- 25.Zhu W., Liu S., Liu J., Zhou Y., Lin H. High-throughput sequencing identification and characterization of potentially adhesion-related small RNAs in Streptococcus mutans. J Med Microbiol. 2018;67:641–651. doi: 10.1099/jmm.0.000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S., Zhou Y., Tao Y., Zhuang P., Pang L., Zhi Q. Effect of different glucose concentrations on small RNA levels and adherence of Streptococcus mutans. Curr Microbiol. 2019;76:1238–1246. doi: 10.1007/s00284-019-01745-1. [DOI] [PubMed] [Google Scholar]
- 27.Phillips P., Progulske-Fox A., Grieshaber S., Grieshaber N. Expression of Porphyromonas gingivalis small RNA in response to hemin availability identified using microarray and RNA-seq analysis. FEMS Microbiol Lett. 2014;351:202–208. doi: 10.1111/1574-6968.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorth P., Trivedi U., Rumbaugh K., Whiteley M. Probing bacterial metabolism during Infection using high-resolution transcriptomics. J Bacteriol. 2013;195:4991–4998. doi: 10.1128/JB.00875-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oogai Y., Gotoh Y., Ogura Y., Kawada-Matsuo M., Hayashi T., Komatsuzawa H. Small RNA repertoires and their intraspecies variation in Aggregatibacter actinomycetemcomitans. DNA Res. 2017;25:207–215. doi: 10.1093/dnares/dsx050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirano T., Beck D., Demuth D., Hackett M., Lamont R. Deep sequencing of Porphyromonas gingivalis and comparative transcriptome analysis of a LuxS mutant. Front Cell Infect Microbiol. 2012;2 doi: 10.3389/fcimb.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimizu T., Yaguchi H., Ohtani K., Banu S., Hayashi H. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol Microbiol. 2002;43:257–265. doi: 10.1046/j.1365-2958.2002.02743.x. [DOI] [PubMed] [Google Scholar]
- 32.Kreikemeyer B., Boyle M.D., Buttaro B.A., Heinemann M., Podbielski A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol Microbiol. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 33.Chao Y., Papenfort K., Reinhardt R., Sharma C.M., Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dugar G., Svensson S.L., Bischler T., Wäldchen S., Reinhardt R., Sauer M. The CsrA-FliW network controls polar localization of the dual-function flagellin mRNA in Campylobacter jejuni. Nat Commun. 2016;7:11667. doi: 10.1038/ncomms11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saadeh B., Caswell C.C., Chao Y., Berta P., Wattam A.R., Roop R.M., 2nd Transcriptome-wide identification of Hfq-associated RNAs in Brucella suis by Deep Sequencing. J Bacteriol. 2016;198:427–435. doi: 10.1128/JB.00711-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sittka A., Sharma C.M., Rolle K., Vogel J. Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol. 2009;6:266–275. doi: 10.4161/rna.6.3.8332. [DOI] [PubMed] [Google Scholar]
- 37.Grüll M.P., Peña-Castillo L., Mulligan M.E., Lang A.S. Genome-wide identification and characterization of small RNAs in Rhodobacter capsulatus and identification of small RNAs affected by loss of the response regulator CtrA. RNA Biol. 2017;14:914–925. doi: 10.1080/15476286.2017.1306175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan D., Jenniches L., Reichardt S., Barquist L., Westermann A.J. A high-resolution transcriptome map identifies small RNA regulation of metabolism in the gut microbe Bacteroides thetaiotaomicron. Nat Commun. 2020;11:3557. doi: 10.1038/s41467-020-17348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakata M., Sumitomo T., Patenge N., Kreikemeyer B., Kawabata S. Thermosensitive pilus production by FCT type 3 Streptococcus pyogenes controlled by Nra regulator translational efficiency. Mol Microbiol. 2020;113:173–189. doi: 10.1111/mmi.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ota C., Morisaki H., Nakata M., Arimoto T., Fukamachi H., Kataoka H. Streptococcus sanguinis noncoding cia-dependent small RNAs negatively regulate expression of type IV pilus retraction ATPase PilT and biofilm formation. Infect Immun. 2018;86 doi: 10.1128/IAI.00894-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamada S., Slade H.D. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welin-Neilands J., Svensäter G. Acid tolerance of biofilm cells of Streptococcus mutans. Appl Environ Microbiol. 2007;73:5633–5638. doi: 10.1128/AEM.01049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krastel K., Senadheera D.B., Mair R., Downey J.S., Goodman S.D., Cvitkovitch D.G. Characterization of a glutamate transporter operon, glnQHMP, in Streptococcus mutans and its role in acid tolerance. J Bacteriol. 2010;192:984–993. doi: 10.1128/JB.01169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kremer B.H., van der Kraan M., Crowley P.J., Hamilton I.R., Brady L.J., Bleiweis A.S. Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J Bacteriol. 2001;183:2543–2552. doi: 10.1128/JB.183.8.2543-2552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen Z.T., Baker H.V., Burne R.A. Influence of BrpA on critical virulence attributes of Streptococcus mutans. J Bacteriol. 2006;188:2983–2992. doi: 10.1128/JB.188.8.2983-2992.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemos J.A., Brown T.A., Jr., Burne R.A. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect Immun. 2004;72:1431–1440. doi: 10.1128/IAI.72.3.1431-1440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu W., Liu S., Zhuang P., Liu J., Wang Y., Lin H. Characterization of acid-tolerance-associated small RNAs in clinical isolates of Streptococcus mutans: potential biomarkers for caries prevention. Mol Med Rep. 2017;16:9242–9250. doi: 10.3892/mmr.2017.7751. [DOI] [PubMed] [Google Scholar]
- 50.Eggenhofer F., Tafer H., Stadler P.F., Hofacker I.L. RNApredator: fast accessibility-based prediction of sRNA targets. Nucleic Acids Res. 2011;39:W149–W154. doi: 10.1093/nar/gkr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suntharalingam P., Senadheera M.D., Mair R.W., Lévesque C.M., Cvitkovitch D.G. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J Bacteriol. 2009;191:2973–2984. doi: 10.1128/JB.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y.H., Lau P.C., Lee J.H., Ellen R.P., Cvitkovitch D.G. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alves L.A., Nomura R., Mariano F.S., Harth-Chu E.N., Stipp R.N., Nakano K. CovR regulates Streptococcus mutans susceptibility to complement immunity and survival in blood. Infect Immun. 2016;84:3206–3219. doi: 10.1128/IAI.00406-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahn S.J., Wen Z.T., Burne R.A. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun. 2006;74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fabret C., Feher V.A., Hoch J.A. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Senadheera M.D., Guggenheim B., Spatafora G.A., Huang Y.-C.C., Choi J., Hung D.C.I. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol. 2005;187:4064–4076. doi: 10.1128/JB.187.12.4064-4076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senadheera D.B., Cordova M., Ayala E.A., Chávez de Paz L.E., Singh K., Downey J.S. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol. 2012;194:1307–1316. doi: 10.1128/JB.06071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mao M.-Y., Yang Y.-M., Li K.-Z., Lei L., Li M., Yang Y. The rnc gene promotes exopolysaccharide synthesis and represses the vicRKX gene expressions via microRNA-size small RNAs in Streptococcus mutans. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lei L., Stipp R.N., Chen T., Wu S.Z., Hu T., Duncan M.J. Activity of Streptococcus mutans VicR is modulated by antisense RNA. J Dent Res. 2018;97:1477–1484. doi: 10.1177/0022034518781765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamashita Y., Bowen W.H., Burne R.A., Kuramitsu H.K. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect Immun. 1993;61:3811–3817. doi: 10.1128/iai.61.9.3811-3817.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okahashi N., Sasakawa C., Yoshikawa M., Hamada S., Koga T. Molecular characterization of a surface protein antigen gene from serotype c Streptococcus mutans, implicated in dental caries. Mol Microbiol. 1989;3:673–678. doi: 10.1111/j.1365-2958.1989.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 62.Aoki H., Shiroza T., Hayakawa M., Sato S., Kuramitsu H.K. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect Immun. 1986;53:587–594. doi: 10.1128/iai.53.3.587-594.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanada N., Kuramitsu H.K. Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans. Infect Immun. 1988;56:1999–2005. doi: 10.1128/iai.56.8.1999-2005.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merritt J., Kreth J., Shi W., Qi F. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Mol Microbiol. 2005;57:960–969. doi: 10.1111/j.1365-2958.2005.04733.x. [DOI] [PubMed] [Google Scholar]
- 65.Sato Y., Yamamoto Y., Kizaki H. Cloning and sequence analysis of the gbpC gene encoding a novel glucan-binding protein of Streptococcus mutans. Infect Immun. 1997;65:668–675. doi: 10.1128/iai.65.2.668-675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu N., Niu G., Xie Z., Chen Z., Itzek A., Kreth J. The Streptococcus mutans irvA gene encodes a trans-acting riboregulatory mRNA. Mol Cell. 2015;57:179–190. doi: 10.1016/j.molcel.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarpong D.D., Murphy E.R. RNA regulated toxin-antitoxin systems in pathogenic bacteria. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.661026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koyanagi S., Lévesque C.M. Characterization of a Streptococcus mutans intergenic region containing a small toxic peptide and its cis-encoded antisense small RNA antitoxin. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawamura Y., Hou X.G., Sultana F., Miura H., Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- 70.Dyson C., Barnes R.A., Harrison G.A. Infective endocarditis: an epidemiological review of 128 episodes. J Infect. 1999;38:87–93. doi: 10.1016/s0163-4453(99)90074-9. [DOI] [PubMed] [Google Scholar]
- 71.Halfmann A., Kovács M., Hakenbeck R., Brückner R. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol Microbiol. 2007;66:110–126. doi: 10.1111/j.1365-2958.2007.05900.x. [DOI] [PubMed] [Google Scholar]
- 72.Marx P., Nuhn M., Kovács M., Hakenbeck R., Brückner R. Identification of genes for small non-coding RNAs that belong to the regulon of the two-component regulatory system CiaRH in Streptococcus. BMC Genomics. 2010;11:661. doi: 10.1186/1471-2164-11-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kery M.B., Feldman M., Livny J., Tjaden B. TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. 2014;42:W124–W129. doi: 10.1093/nar/gku317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okahashi N., Nakata M., Sakurai A., Terao Y., Hoshino T., Yamaguchi M. Pili of oral Streptococcus sanguinis bind to fibronectin and contribute to cell adhesion. Biochem Biophys Res Commun. 2010;391:1192–1196. doi: 10.1016/j.bbrc.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 75.Okahashi N., Nakata M., Terao Y., Isoda R., Sakurai A., Sumitomo T. Pili of oral Streptococcus sanguinis bind to salivary amylase and promote the biofilm formation. Microb Pathog. 2011;50:148–154. doi: 10.1016/j.micpath.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 76.Gurung I., Spielman I., Davies M.R., Lala R., Gaustad P., Biais N. Functional analysis of an unusual type IV pilus in the Gram-positive Streptococcus sanguinis. Mol Microbiol. 2016;99:380–392. doi: 10.1111/mmi.13237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Melville S., Craig L. Type IV pili in Gram-positive bacteria. Microbiol Mol Biol Rev. 2013;77:323–341. doi: 10.1128/MMBR.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holt S.C., Ebersole J.L. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 79.Gholizadeh P., Pormohammad A., Eslami H., Shokouhi B., Fakhrzadeh V., Kafil H.S. Oral pathogenesis of Aggregatibacter actinomycetemcomitans. Microb Pathog. 2017;113:303–311. doi: 10.1016/j.micpath.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 80.Carrizales-Sepúlveda E.F., Ordaz-Farías A., Vera-Pineda R., Flores-Ramírez R. Periodontal disease, systemic inflammation and the risk of cardiovascular disease. Heart Lung Circ. 2018;27:1327–1334. doi: 10.1016/j.hlc.2018.05.102. [DOI] [PubMed] [Google Scholar]
- 81.Phillips P.L., Reyes L., Sampson E.M., Murrell E.A., Whitlock J.A., Progulske-Fox A. Deletion of a conserved transcript PG_RS02100 expressed during logarithmic growth in Porphyromonas gingivalis results in hyperpigmentation and increased tolerance to oxidative stress. PLoS One. 2018;13 doi: 10.1371/journal.pone.0207295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jorth P., Whiteley M. Characterization of a novel riboswitch-regulated lysine transporter in Aggregatibacter actinomycetemcomitans. J Bacteriol. 2010;192:6240–6250. doi: 10.1128/JB.00935-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amarasinghe J.J., Connell T.D., Scannapieco F.A., Haase E.M. Novel iron-regulated and Fur-regulated small regulatory RNAs in Aggregatibacter actinomycetemcomitans. Mol Oral Microbiol. 2012;27:327–349. doi: 10.1111/j.2041-1014.2012.00645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Busch A., Richter A.S., Backofen R. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008;24:2849–2856. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu S.H., Vogel J., Förstner K.U. ANNOgesic: a Swiss army knife for the RNA-seq based annotation of bacterial/archaeal genomes. Gigascience. 2018;7 doi: 10.1093/gigascience/giy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leonard S., Meyer S., Lacour S., Nasser W., Hommais F., Reverchon S. APERO: a genome-wide approach for identifying bacterial small RNAs from RNA-Seq data. Nucleic Acids Res. 2019;47:e88. doi: 10.1093/nar/gkz485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chakravarty S., Massé E. RNA-dependent regulation of virulence in pathogenic bacteria. Front Cell Infect Microbiol. 2019;9:337. doi: 10.3389/fcimb.2019.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duran-Pinedo A.E., Yost S., Frias-Lopez J. Small RNA transcriptome of the oral microbiome during periodontitis progression. Appl Environ Microbiol. 2015;81:6688–6699. doi: 10.1128/AEM.01782-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ram-Mohan N., Meyer M.M. Comparative metatranscriptomics of periodontitis supports a common polymicrobial shift in metabolic function and identifies novel putative disease-associated ncRNAs. Front Microbiol. 2020;11:482. doi: 10.3389/fmicb.2020.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Na D., Yoo S.M., Chung H., Park H., Park J.H., Lee S.Y. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol. 2013;31:170–174. doi: 10.1038/nbt.2461. [DOI] [PubMed] [Google Scholar]
- 91.Villa J.K., Su Y., Contreras L.M., Hammond M.C. Synthetic biology of small RNAs and riboswitches. Microbiol Spectr. 2018;6 doi: 10.1128/microbiolspec.rwr-0007-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]