Highlights
-
•
Glioblastoma organoid cultures preserve diversity of proliferative cell phenotypes.
-
•
Heterogeneous 3D cultures recapitulate resistance to clinical GBM therapeutics.
-
•
Patient specimens show different behavior depending on 2D vs 3D growth.
Keywords: Glioblastoma, Organoids, Tumor microenvironments, Therapeutic resistance, Chemotherapy
Abstract
Background
Glioblastoma (GBM) is the most common primary brain tumor with a dismal prognosis. The inherent cellular diversity and interactions within tumor microenvironments represent significant challenges to effective treatment. Traditional culture methods such as adherent or sphere cultures may mask such complexities whereas three-dimensional (3D) organoid culture systems derived from patient cancer stem cells (CSCs) can preserve cellular complexity and microenvironments. The objective of this study was to determine if GBM organoids may offer a platform, complimentary to traditional sphere culture methods, to recapitulate patterns of clinical drug resistance arising from 3D growth.
Methods
Adult and pediatric surgical specimens were collected and established as organoids. We created organoid microarrays and visualized bulk and spatial differences in cell proliferation using immunohistochemistry (IHC) staining, and cell cycle analysis by flow cytometry paired with 3D regional labeling. We tested the response of CSCs grown in each culture method to temozolomide, ibrutinib, lomustine, ruxolitinib, and radiotherapy.
Results
GBM organoids showed diverse and spatially distinct proliferative cell niches and include heterogeneous populations of CSCs/non-CSCs (marked by SOX2) and cycling/senescent cells. Organoid cultures display a comparatively blunted response to current standard-of-care therapy (combination temozolomide and radiotherapy) that reflects what is seen in practice. Treatment of organoids with clinically relevant drugs showed general therapeutic resistance with drug- and patient-specific antiproliferative, apoptotic, and senescent effects, differing from those of matched sphere cultures.
Conclusions
Therapeutic resistance in organoids appears to be driven by altered biological mechanisms rather than physical limitations of therapeutic access. GBM organoids may therefore offer a key technological approach to discover and understand resistance mechanisms of human cancer cells.
Keypoints
GBM organoid cultures preserve diversity of proliferative cell phenotypes.
Heterogeneous 3D cultures recapitulate resistance to clinical GBM therapeutics.
Importance of the study
Glioblastoma (GBM) is the most common primary brain tumor with a dismal prognosis. Although various therapies have shown efficacy in preclinical studies, treatments are often poorly effective in patients, creating a disconnect between ease of treatment in lab and the clinical reality of drug resistance. GBM's inherent cellular diversity and complex network of interactions within tumor microenvironments pose a significant challenge to finding effective treatment. Patient tumor specimens grown as 3D organoid cultures retain high cellular diversity which may drive GBM therapeutic response and adaptation. Patient-derived GBM organoids may play a role in uncovering in vivo therapeutic resistance mechanisms and have potential applications in predicting personalized drug sensitivities.
Introduction
Glioblastoma (GBM) is the most common primary brain tumor with a dismal prognosis: median survival is approximately 15 months and the 5-year survival rate is below 5% [32]. Difficulty in effectively treating GBM is attributed in part to its cellular diversity and heterogeneous microenvironments [20,29,30,33]. Within glioblastoma, this may include a subset of self-renewing cancer stem cells (CSCs) that give rise to this heterogeneity and contribute to the complexity of intratumoral interactions [2,10,13,43].
Recreating these diverse tumor microenvironments is challenging in the in vitro setting. Researchers must make choices about culture conditions that differ from the original tumor and that can impact the complex network of cellular responses [18,22,23]. Traditional culture models can fail to properly model tumor stemness or the genetic and cellular diversity seen in GBM [5,23,35,49]. Many therapeutics currently in use are highly effective in cultured samples, yet have an incomplete response in human patients and inevitably lead to resistance (reviewed in [12,52]. Indeed, many cultured cancer lines have poor transcriptional fidelity to their clinical tumor counterparts, and this is exemplified by cultured brain tumor models where no low grade glioma and only ∼5% of GBM models can be correctly classified as brain tumors [34]. The use of single-cell sequencing analysis, genetically engineered mouse models, or tumor organoid models dramatically improved fidelity to clinical disease, with all tested GBM organoid models being transcriptionally classified as GBM with high confidence [34].
The use of three-dimensional (3D) organoids may help overcome limitations of traditional sphere culture, by recapitulating the cellular diversity seen in patient tumors and modeling of tumor microenvironments and microenvironmental gradients [18,26,41]. There has been work done in the past decade with both stem-cell-derived cerebral organoids and patient-CSC-derived GBM organoids. Cerebral organoids that mimic healthy brain tissue but with oncogenic mutations introduced have been shown to invade brain tissue in mice [3,31]. Human GBM CSCs have been co-cultured with pluripotent stem-cell-derived brain organoids, which allows for interactions between cancer and non-cancer cells [1,6]. Linkous et al. also demonstrated that such co-cultured organoids formed a network of microtubule transport that mimics how GBM invades healthy brain tissue in patients [26]. GBM organoids established directly from patient tumor specimens and subsequently grown in rodent brains recreate indicative features of that patient's GBM [19]. Single cell RNA sequencing of GBM organoids has shown that diversity of cell types is recreated compared to sets of patient GBMs [28,36]. Lenin et al. also grew GBM organoids directly from specimens without the use of a 3D matrix for rapid throughput analysis and saw differences between these organoids and 2D sphere culture in response to various drugs [25].
Organoid cultures grown from patient-derived GBM CSCs recreate hypoxic and nonhypoxic niches, allowing for the coexistence of diverse tumor microenvironments as seen in clinical tumors [18,19]. Here, we used three-dimensional (3D) organoid culture methods to ask whether 3D growth recapitulates aspects of biological drug resistance observed in clinical practice.
Materials and methods
Cell and organoid culture
GBM samples were obtained directly from adult and pediatric surgical specimens in accordance with protocol #2559 approved by the Cleveland Clinic Institutional Review Board or provided by collaborators. Details of patient specimens are provided in Supplementary Table 1. Sphere cultures or patient-derived organoids were maintained in Neurobasal media supplemented with EGF (R&D systems, Minneapolis, MN), bFGF (R&D systems), B27 (Invitrogen, Carlsbad, CA), glutamax (Invitrogen), sodium pyruvate (Invitrogen), and antibiotics (Antibiotic-Antimicotic, Invitrogen), termed “NBM complete”. Spheres were maintained in 10 cm plates in a 37 °C incubator with NBM complete media. Sphere cultures were dissociated with Accutase into single cell suspension and replated approximately once weekly (depending on growth rate to prevent large unhealthy sphere growth) as previously described [8,18,40,41]. Adherent cultures were grown by plating dissociated GBM sphere cells on glass chamber slides using a laminin coating and the same media conditions as above. Unless otherwise noted, GBM organoids were formed by suspending tumor cells in Matrigel (BD Biosciences, San Jose, CA) and forming 20 µL pearls, prior to culture in 6-well or 10 cm plates shaking in NBM complete media, as previously described [18]. Organoids created from several patient specimens were subjected to 0.1% DMSO with or without 3 Gy radiation alone, 250 uM temozolomide, and combination temozolomide plus radiotherapy. Matched sphere cultures from the same patient specimen were treated identically to the corresponding organoids. Doxorubicin, a chemotherapy agent that is inherently fluorescent, was used to visualize permeation through organoids for 24 or 48 h total.
Creation of organoid microarrays
Organoids were initially fixed in formalin, followed by 70% ethanol. Organoids formed from various cell specimens were embedded in paraffin blocks to create microarrays for medium-throughput analysis, which were sectioned onto slides for histology and staining. Slides were stained for hematoxylin and eosin (H&E) or immunohistochemistry (IHC) was performed with phospho-histone H3 (pHH3, Ser10 rabbit, Cell Signaling Technology 9701S), a marker specific for proliferating cells, or Cleaved Caspase-3 (CC-3, Asp175, Cell Signaling Technology, 9664S). We imaged whole slides using Leica Aperio AT2 digital slide scanner (Leica Biosystems, Vista, CA) and visualized images with Aperio Imagescope software (Leica Biosystems, Vista, CA). For quantitation of images, 3–4 non-overlapping uniformly sized high power fields were captured and total positive cell numbers were counted and averaged. Student's t-test was used to calculate p-values.
DNA content analysis
Organoids were regionally labeled and subsequent cell isolation from organoid layers was achieved with a 20 µM final concentration of CellTracker Blue CMAC Dye (Invitrogen, #C2110) in NBM complete media as previously described [41]. Briefly, the mature organoids were incubated with dye for 2 h at 37 °C with shaking to allow outer layer labeling. Elevated levels of CMAC dye indicate cells in the organoid rim, while low levels of CMAC dye indicate organoid core cells. Then, organoids were finely minced and dissociated using Accutase (FisherSci, #ICN1000449) at 4 °C for 15 min and then heated to 37 °C for an additional 10 min. GBM spheres were pelleted by centrifugation and dissociated using Accutase. The cells were then labelled with 1:2000 TO-PRO-3 iodide (Invitrogen, #T3605) per the manufacturer's protocols, single-cell filtered, and analyzed using a BD LSR Fortessa flow cytometer. DNA content analysis was performed using ModFit LT (Verity Software House, Topsham, ME).
Viability assays with CTG for spheres and organoids
GBM spheres were dissociated into single-cell suspension and grown adherently in laminin coated 96-well plates. In addition to a 0.1% DMSO control, all cells received a range of drug concentrations for 6 days total. The drugs used included temozolomide, ibrutinib, lomustine, and ruxolitinib. The range of drug concentration was based on the existing literature. CellTiter-Glo (CTG; Promega), a luminescent cell viability assay, was mixed 1:1 with phosphate buffered saline (PBS) and added to all wells (emptied of culture media) at the end of 6 days. The plates were shaken for 2 min and incubated for 10 min, then read using a luminometer (Cytation5, BioTek, Winooski, VT). The data was normalized to each specimen's DMSO control average on the same plate. From these cultures, an optimal drug concentration was chosen to treat organoids. Organoids received 0.1% DMSO, 250 uM temozolomide, 6.25 uM ibrutinib, 25 uM lomustine, or 50 uM ruxolitinib for a total of 6 days. CellTiter-Glo 3D (CTG-3D) was mixed 1:1 with PBS and added to individual organoids (without culture media), which then were manually triturated (twice; 5 min incubation in between) to help with cell lysis. Lysates were then diluted 1:4, in CTG-3D mixed 1:1 with PBS, in 96-well plates. The plates were shaken for 2 min and incubated for an additional 10 min prior to reading with our luminometer.
Immunofluorescence and imaging
Entire organoids were fixed with 10% neutral buffered formalin, followed by 70% ethanol, embedded in paraffin, sectioned at 5um, and then probed with antibodies against SOX2 (R&D systems, AF2018) and Cleaved Caspase-3 (CC-3, Asp175, Cell Signaling Technology, 9664S) for immunofluorescence. DNA was detected with DAPI (1:10,000) and all images were acquired with the Leica DM5500B upright microscope and Leica DFC 7000 GT monochrome camera (Leica Biosystems).
Senescence-associated beta-galactosidase activity assay
GBM organoids were treated with the indicated drugs for 6 days as described above, and were finely chopped and dissociated using Accutase at 4 °C for 15 min and then warmed to 37 °C for 10 min. Senescence-associated beta-galactosidase activity was assayed as previously described [7]. Briefly, dissociated organoid cells were single-cell filtered and resuspended in 1:1000 Bafilomycin A1 (#B1793) and incubated for 1 hour at 37 °C. Then C12FDG was added to each sample and incubated for an additional 90 min, followed by addition of 1:2000 TO-PRO-3 (Invitrogen, #T3605). Stained cells were counted using a Cytek™ Aurora spectral cell analyzer (Cytek Biosciences, Fremont, CA). Population analysis was performed using FlowJo Software (BD Biosciences).
Results
Organoid microarrays increase throughput while preserving diverse niches and pathology
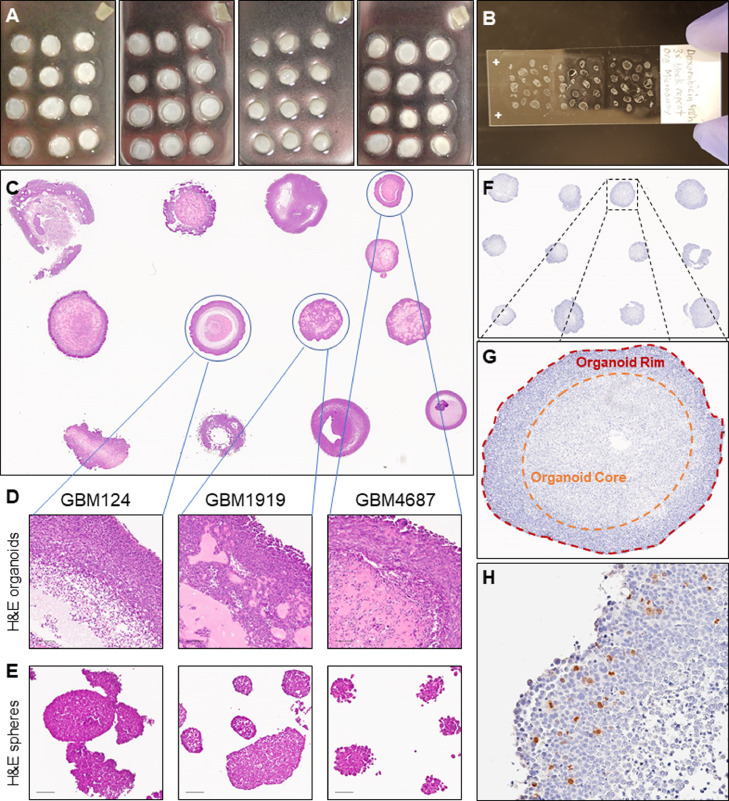
Although organoids can offer significant insight into the heterogeneous tumor environments of glioblastoma, a significant shortcoming of this culture system is low throughput over long timelines. We therefore established organoid microarrays to allow for medium throughout analysis of multiple specimens at once, and direct comparison and visualization of different specimens and treatments on the same slide (Fig. 1A). Depending upon the organoid size and number per block, a single slide can accommodate 12 – 60 single specimens (Fig. 1B). After testing organoid growth in multiwell plates using shaking culture, we selected the smallest well size (6-well plates) that had a relatively low impact on organoid growth compared to 10 cm plates (Supplementary Figure 1). Interspecimen and intraspecimen morphologic diversity is preserved using these methods (Fig. 1C,D), unlike the relative morphologic homogeneity observed in GBM sphere cultures grown in identical media (Fig. 1E). As with tissue microarrays, IHC staining can be employed to investigate regional differences in protein expression while having experiments internally controlled on the same slide (Fig. 1F–H). This allows investigators to see diverse niches and pathology in a broader array of specimens than was previously feasible.
Fig. 1.
Creation of organoid microarrays for medium-throughput analysis. A Microarrays of multiple organoids embedded in paraffin. B Slide with multiple preserved organoid specimen section. C H&E stain of GBM organoids of various patient specimens. D-E Zoomed H&E views of three GBM organoids and matched sphere cultures, scale bar = 100 um. F-H IHC staining of organoid microarray for pHH3 marking dividing cells in separate organoid niches.
GBM organoids display interspecimen and intraspecimen diverse proliferative phenotypes
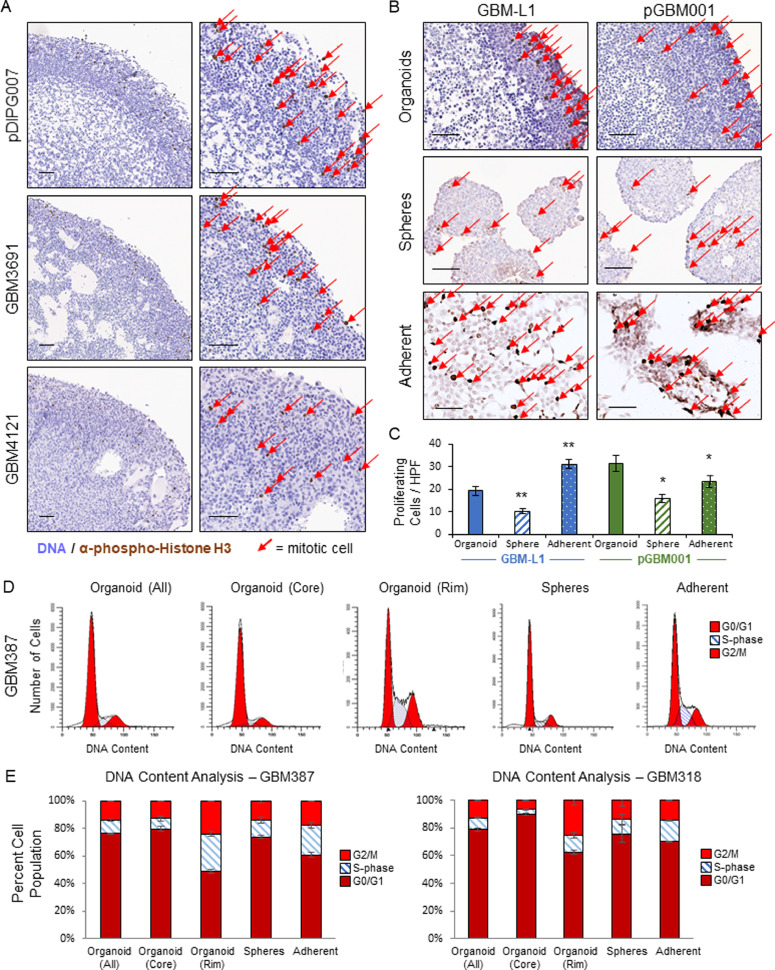
Studying the proliferative profile of tumor specimens is important for understanding both tumor propagation and for predicting effectiveness of therapeutic agents. Using the microarrays developed above, we stained multiple specimens for pHH3 to visualize diversity of regional cell proliferation in organoid culture (Figs. 2A, Supplementary 2). We see broad and consistent proliferation in the organoid rim, compared with the core that only shows rare divisions by pHH3 staining, consistent with prior studies [18]. We also compared pHH3 staining for matched specimens growing in organoid, sphere, or adherent culture and were able to see interspecimen and intraspecimen variations in cell proliferation. Sphere and adherent cultures did not demonstrate the stark regional variation of dividing cells found in organoids (Fig. 2B). We quantified the number of dividing cells per high powered field (HPF) and found less division in sphere cultures compared to adherent culture or to organoids. (Fig. 2C). We also noticed that the majority of this proliferation was localized to the organoid outer rim. To further quantify this phenomenon, we performed DNA content analysis of matched organoid, sphere, and adherent cultured specimens, including a spatial dye to separate organoid rim and core cells [41]. This confirmed greater proliferation in the organoid rim and less in the core, and sphere and adherent culture both showed less proliferation compared to the organoid rim yet more than the organoid core cells (Fig. 2D,E). This underscores the wider degree of phenotypic heterogeneity in organoid culture compared to alternate methods.
Fig. 2.
GBM organoids show spatial and quantitative proliferative phenotypes not demonstrated in matched spheres. A Multiple GBM organoid specimens IHC stained for pHH3, scale bar = 100 um. B IHC staining of two GBM organoids, matched sphere samples, and adherent cultures for pHH3. Red arrows indicate cells staining for pHH3, scale bar = 100 um. C Graph of number of proliferating cells per high powered field (HPF) for the two GBM organoids, matched sphere specimens, and adherent cultures in B. D DNA content analysis of GBM organoid, matched spheres, and adherent cultures showing G0/G1, S, and G2/M phases.
E Graph for DNA content analysis of two GBM organoids, matched sphere specimens, and adherent cultures.
Compared to patient-matched sphere cultures, organoids are resistant to GBM standard of care therapy
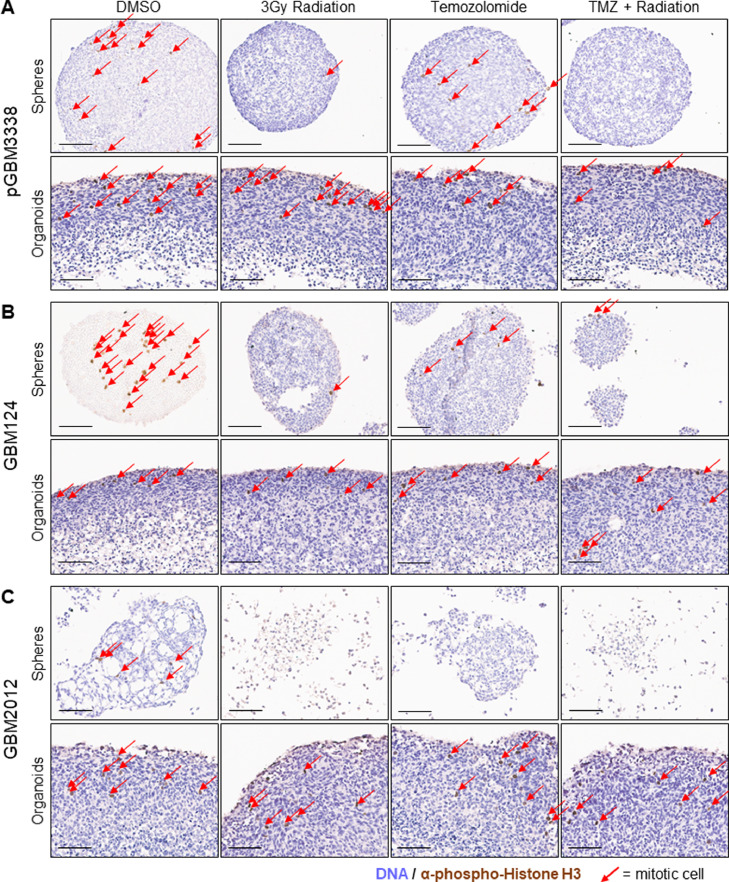
To provide clinical context to the significant resistance of organoids to therapeutic options, we exposed matched sphere and organoid cultures to both temozolomide and/or radiation, mimicking the currently accepted standard of care combination therapy used for patients with GBM [44]. Both sphere and organoid cultures received radiation, temozolomide, or a combination of both prior to fixation and IHC analysis (Fig. 3A–C). The sphere cultures showed near complete reduction in proliferative staining using pHH3. Meanwhile, the corresponding organoid culture showed a more blunted effect, demonstrated by moderately frequent pHH3 staining and multiple visibly mitotic cells in the organoid rim, indicating cellular proliferation. Organoid specimens were also able to maintain their structure and overall viability compared to the matched spheres. Since DNA alkylating agents (including temozolomide) and radiation damage both asymmetrically target rapidly dividing cells, we initially expected the organoid rim to be more highly impacted by therapy. However, we see that the organoids continued to have proliferation in the rim, evidenced by pHH3 staining (Fig. 3A–C). To rule out diffusion as a reason for the lack of drug response, we evaluated the penetration of doxorubicin (a naturally fluorescent small molecule chemotherapeutic, comparable in size to temozolomide) into organoids and saw distribution throughout each organoid in less than 48 h (Supplementary Fig. 3). Furthermore, if limited drug diffusion were driving the lackluster response, we would expect to see the strongest effect near the organoid rim where concentrations of drug would be highest, yet we continue to see proliferation in this region. It was striking, especially for a proliferation-targeted therapeutic combination, that even though the organoid rim region is more highly proliferative than matched bulk sphere cultures (Fig. 2D,E), this region was less affected by therapy compared to sphere cultures. This suggests an altered biologic response underlying this drug resistance in 3D cultures. Maintaining patient tumor specimens as organoids may therefore offer information about drug sensitivity that is otherwise unassessed in traditional adherent or sphere culture growth. To expand on this hypothesis, we explored the impact of additional chemotherapeutic drugs on organoids versus sphere culture.
Fig. 3.
Organoids demonstrate resistance to standard of care therapies for GBM. A H&E and IHC staining for pHH3 of matched sphere and organoid specimens of sample GBM3338 receiving DMSO, radiation alone, temozolomide alone, and combination radiation and temozolomide therapy. Red arrows indicate cells staining for pHH3, scale bar = 100 um. B H&E and IHC staining for pHH3 of matched sphere and organoid specimens of sample GBM124 receiving DMSO, radiation alone, temozolomide alone, and combination radiation and temozolomide therapy. Red arrows indicate cells staining for pHH3, scale bar = 100 um. C H&E and IHC staining for pHH3 of matched sphere and organoid specimens of sample GBM2012 receiving DMSO, radiation alone, temozolomide alone, and combination radiation and temozolomide therapy. Red arrows indicate cells staining for pHH3, scale bar = 100 um.
Organoids demonstrate widespread therapeutic resistance when compared to matched 2D sphere culture
We tested the effect of various chemotherapeutic drugs on matched organoid and sphere culture across multiple patient specimens. Four drugs were chosen to span different mechanisms of action, including both standard of care treatments and novel chemotherapies undergoing clinical trials: temozolomide, ibrutinib, lomustine, ruxolitinib. Temozolomide is part of current standard of care therapy for patients with GBM and is a DNA alkylating agent that inhibits cellular replication. Lomustine is a DNA and RNA alkylating agent, commonly used to treat recurrent glioblastoma at present. Ibrutinib is a small molecule drug that is a BTK/BMX tyrosine kinase inhibitor, traditionally used to treat hematological malignancies, but currently undergoing clinical trials for use in GBM [11]. Ruxolitinib is a small molecule inhibitor of JAK1 and JAK2, typically used to treat myelofibrosis, that is currently in clinical trials for patients with GBM [16,38]. JAK2 is responsible for activating the STAT3 pathway, which is an important part of promoting CSC maintenance in GBM. Normally, JAK2 is inhibited by SOCS3. BMX is able to bypass this JAK2 activation of STAT3 and instead, directly activate STAT3 [42]. Ibrutinib and ruxolitinib therefore both target different components of the JAK/STAT3 pathway which ultimately promotes CSC maintenance. Both organoids and spheres were treated with each drug for 6 days total, followed by quantification of cell viability. All specimens were normalized to their own DMSO control. DMSO levels showed no detectable toxicity and, due to our controlled formation and growth processes, cell viability measurement was highly repeatable between both technical and biological replicate organoids from multiple different GBM specimens (Supplementary Fig. 4). For the majority of treatments and specimens, GBM organoids demonstrated significantly greater overall resistance after treatment with each test drug, compared to identically treated matched sphere cultures (Fig. 4A).
Fig. 4.
GBM organoids demonstrate widespread, significant resistance to clinical therapeutics compared to matched 2D sphere culture. A Graphs of six different GBM patient specimens comparing normalized cell viability of both organoid culture and sphere culture after undergoing CTG dissociation for temozolomide, ibrutinib, lomustine, and ruxolitinib. * indicates p < 0.05, ** indicates p < 0.001, *** indicates p < 10^−10. Colored data points represent organoid culture, gray data points represent sphere culture. p in patient specimen indicates pediatric specimen. B Graph of organoid to sphere response ratio for each specimen and each drug on logarithmic scale. Values greater than 1 indicate ‘resistance’ and less than 1 indicates ‘sensitive’. C Graph of difference between sphere and organoid normalized cell viability.
We found both intraspecimen and interspecimen variability in the extent of organoid resistance to chemotherapeutic drug. The normalized ratios of organoid to sphere viability demonstrated that nearly every organoid specimen showed some amount of drug resistance, but further evaluating by both drug and specimen, we see both general trends and exceptions to these trends (Fig. 4B). No specimens showed organoids being significantly more sensitive to drug compared to the matched sphere culture. The delineation between statistical significance and meaningful difference is important to consider as well. Specimen GBM3832 shows a statistically significant resistance of organoids compared to spheres for temozolomide, lomustine, and ruxolitinib, however the trends of effect are the same in each case and we do not feel these organoid cultures provide meaningfully different information compared to the spheres. This demonstrates one of several cases where 3D culture did not provide a notably different result compared to traditional methods. In contrast, the majority of specimen-drug combinations provided meaningfully differing results in organoid versus sphere viability. This both demonstrates the importance of 3D culture methods as a tool to accurately test therapeutics and also cautions that 3D culture is not always required and, in some cases, will produce the same results as other more efficient growth methods.
Looking at the absolute difference in cell viability, we see temozolomide demonstrates a dramatic difference in efficacy between organoid and sphere culture in two of the six patient specimens tested (Fig. 4C). By contrast, tumor cytotoxicity after exposure to lomustine and ruxolitinib demonstrated significant difference in resistance between GBM spheres and organoid conditions in four specimens out of six cases. Ibrutinib has a consistent resistance (statistical and large absolute differences) across specimens, except for GBM3832. pDIPG007 demonstrated a nearly identical response to ruxolitinib and ibrutinib in sphere culture, but demonstrated significant resistance to ibrutinib and sensitivity to ruxolitinib in organoid culture. Since ibrutinib and ruxolitinib both target components of the STAT3 signaling pathway, this reveals a previously unreported 3D specific response to two therapies targeting the same molecular pathway.
Organoids demonstrate blunted proliferative effect and varied induction of apoptotic activity in response to clinical therapies
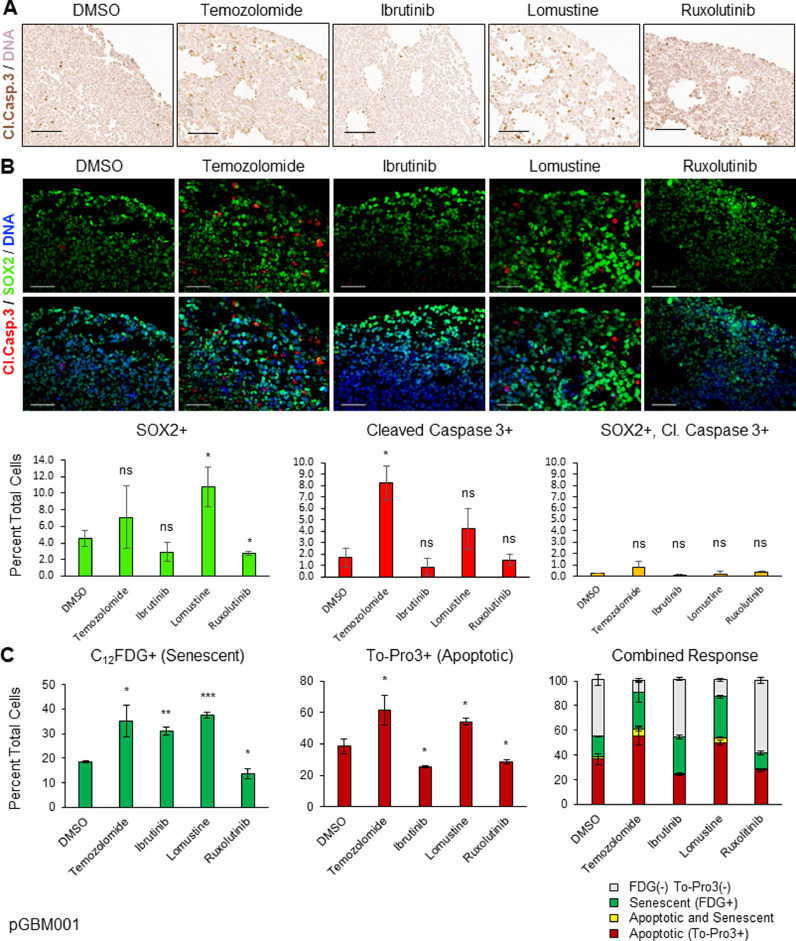
To better understand how organoids maintain viability compared to spheres, we evaluated both proliferation and apoptosis in drug-treated organoids. We used IHC staining of pHH3 to measure active proliferation of multiple organoid specimens after treatments with each of the above therapies (Fig. 5). Most organoids demonstrated at least a blunted antiproliferative response to each therapy, which is consistent with our above findings (Fig. 3). However, despite organoids showing generally decreased proliferation after exposure to chemotherapeutic drugs, this did not translate to decreased organoid viability measurements as shown in Fig. 4. For example, the organoid specimens with perhaps the most dramatic reduction in pHH3 staining were pGBM001/ibrutinib and GBM3832/lomustine (Fig. 5). However, according to ATP-based cell viability data (Fig. 4), pGBM001 shows resistance to ibrutinib whereas GBM3832 is incongruently sensitive to lomustine. Quantification of pHH3 staining compared to the DMSO control shows a generally antiproliferative response, but there are some instances where this is not the case. In specimen GBM3832, we see a notable difference in pHH3 staining between temozolomide and lomustine, both DNA alkylating agents. We theorized that although these chemotherapies blunt proliferation, this may not be resulting in apoptosis and might explain the measured therapeutic resistance based upon total cell viability. We therefore performed IHC staining of each treated organoid specimen for Cleaved Caspase-3 (CC-3) to assess apoptotic activity. As expected, there was variable response across therapies and specimens. However, organoid specimens with greater drug sensitivity such as GBM3832/Ibrutinib, pDIPG007/Ruxolitinib, and GBM3832/lomustine (Fig. 4) were found to have increased apoptotic activity after treatment with that drug (Figs. 6A, Supplemental 5). Taken together, our data demonstrates that while chemotherapy can inhibit active cellular proliferation at least modestly for specimens in 3D organoid culture, it is more difficult to induce cell death consistently or strongly enough to render the organoid sensitive to the drug.
Fig. 5.
Proliferative staining of multiple organoids demonstrates blunted response to cell division. Immunohistochemistry staining of phospho-Histone H3 and hematoxylin. * indicates p < 0.05, ** p < 0.01, ns non-significant, scale bar = 100um.
Fig. 6.
Organoids demonstrate varied induction of apoptotic and senescence activity in response to clinical therapies. A Apoptotic staining of pGBM001 organoids treated with chemotherapeutics for Cleaved caspase 3, scale bar = 100um. B Double immunolabeling of pGBM001 organoids for markers of stemness (SOX2) and apoptosis (Cleaved caspase 3), and graph of percent of SOX2+, Cleaved Caspase 3+ and SOX2+, Cl.Casp.3+ double positive cells, scale bar = 50um. C Graph for flow cytometric analysis of pGBM001 organoids for senescence marker C12FDG and DNA (apoptosis) marker To-Pro3. * indicates p < 0.05, ** p < 0.001, *** p < 0.0001, ns non-significant.
More “stem-like” GBM cells are known to have greater resistance to therapy [2,9,15,21,47]. To see whether the apoptotic cells were enriched or depleted for CSCs, we treated pGBM001 organoids with the above drugs for 6 days and double immunostained for SOX2 and CC-3. Although we could visualize SOX2+ cells in all organoids and induction of CC-3 cleavage, we observed that very few of the CC-3+ cells were also SOX2+ (Fig. 6B). This appeared true for all treatments, albeit to varying degrees, and reflects the previously published response of GBM organoid cellular populations to radiation therapy [18]. Quantifying our results also demonstrated that drugs with similar mechanisms of action tended to have similar trends (Supplemental Figs. 5, 6B). The alkylating agents temozolomide and lomustine both show increased SOX2 staining as well as increased CC3 staining, whereas the JAK/STAT inhibitors have similar or slightly decreased SOX2 and CC3 staining compared to DMSO control.
In addition to proliferative and apoptotic cell populations, we have previously shown that GBM organoids also harbor senescent cell populations [18], which may have a very different response to therapy than cycling cells, particularly to antiproliferative chemotherapies such as our alkylating agents. Senescent cells remaining in organoids would contribute to total signal after treatment as quantified by our ATP-based viability measurement experiments in Fig. 4. We therefore explored whether treatment of pGBM001 organoids with our drug panel resulted in induction of senescence. We used senescence-associated beta-galactosidase (SA-βgal), measured by 5-dodecanoylaminofluorescein di-β-d-galactopyranoside (C12FDG) fluorescence [7], as a marker of senescent cells. Because this readout is FACS-based, we also included marking of late-apoptoptic cells with a propidium iodide based apoptotic dye (TO-PRO-3) (Fig. 6C). Indeed, we observed the same trends of strong apoptotic induction from alkylating agents as we described when quantifying CC-3 cleavage by immunohistochemistry (Supplemental Fig. 5) and by immunofluorescence (Fig. 6B). In addition, we also found strong induction of SA-βgal activity by these drugs, and strong senescence induction by ibrutinib which did not create a parallel apoptotic response. Interestingly, we again found a dichotomous response between the two JAK/STAT inhibitors where BMX inhibition through ibrutinib resulted in increased senescent cells in organoid culture but parallel JAK2 inhibition with ruxolitinib did not. Further research would need to be conducted to dissect the contribution of these different facets of JAK/STAT signaling to senescence in GBM. Interestingly, we observe that remarkably few GBM organoid cells are both senescent and apoptotic. This raises the intriguing possibility that assumption of a senescent or senescent-like state may be a strategy for GBM cells to avoid apoptosis induced by clinical therapeutics. Future research will be required to further investigate such a hypothesis. Taken together, our data suggests that 3D GBM culture methods may be a useful platform through which to investigate diverse tumor-like responses to potential therapies and may be leveraged for additional molecular insight in to GBM.
Discussion
GBM organoids represent, in some respects, a remarkably inefficient system in which to conduct research. The high volumes of expensive media required combined with the comparatively very long timelines to conduct a single experiment make this model system a cumbersome tool compared to sphere or adherent culture of the same specimens. The primary strength of organoid culture is the ability to recapitulate cellular diversity and maintain various tumor microenvironments by allowing cellular self-organization within its 3D structures. We have detailed many relative benefits and drawbacks of such culture systems elsewhere [18,37] and strongly feel that the culture system used must be properly matched to the scientific question to be answered. In other cancers, such as biliary tract, endometrial, and pancreatic carcinomas, patient-derived organoids are being used to test various treatments and organoid-specific phenotypes are observed [4,17,39]. Patient-derived organoids of metastatic gastrointestinal tumors receiving treatment have even been shown to mimic patient response to therapies [45]. Specifically in GBM, organoids are able to mimic phenomena seen in tumors that are not captured in the more homogeneous sphere culture [18,35,49]. This ability to model CSC properties that are thought to contribute to therapeutic resistance is valuable for the future of personalized medicine. Loong et al. demonstrated that not only did GBM organoids grown from initial surgery mimic the temozolomide resistance the patient later developed, but that genetic evaluation of those organoids led to targeted drug therapy with everolimus that led to clinical reduction of tumor size [27]. Indeed, GBM, organoids treated with combination temozolomide and radiation therapy have shown no significant change in size [19,25], consistent with our detailed findings of blunted proliferation and growth (Fig. 3, Fig. 4, Fig. 5) and induction of senescence (Fig. 6C) response in organoids treated with standard of care therapies.
We saw that organoids demonstrate resistance to standard of care therapy and sought to explore this further by testing additional drugs and studying potential mechanisms of resistance, including proliferation, senescence, or apoptosis. The use of organoids has a complementary role to traditional culture in revealing drug response in a different way than may be seen in 2D. Beyond the organoid cultures simply being harder to kill, comparison between 2D/sphere and 3D behaviors could be used to investigate deeper biologic mechanisms of signaling and drug resistance in GBM patients. For instance, specimen pDIPG007 demonstrated sensitivity as sphere culture to both ibrutinib and ruxolitinib, which each affect different aspects of the JAK/STAT3 pathway. However, organoid culture unveiled 3D resistance to ibrutinib, without loss of sensitivity to the same dose of ruxolitinib. This finding could be expanded further to mechanistically pursue what parts of the JAK/STAT3 pathway are most likely to be effective targets for new therapeutics, and therefore most likely to be effective upon translation to clinical trials.
Clinically, one often cited reason for difficulty effectively treating GBM is drug delivery and diffusion of seemingly successful therapeutic agents into a patient's tumor mass. One obvious way GBM organoids may be resistant to a drug would be if there is lack of delivery to the targeted cells. Because organoids are vastly larger than corresponding sphere culture, it is tempting to blame limited drug diffusion to tumor cells within the mass. There are several reasons we do not believe this is the driving force behind resistance shown here. If drug delivery to the center of organoids were a limiting factor in killing proliferative cells that maintain the tumor, we might expect to see the fewest proliferating cells in the outer rim with relatively more remaining proliferative cells in the core. However, when we evaluate IHC staining for pHH3, we see that the outside rim closest to the drug containing media still has the greatest number of proliferating cells remaining after treatment, including cells proliferating right near the media boundary (Fig. 5), suggesting that this effect is biological and not diffusion-based. Diffusion is also not a variable for radiation therapy which impacts all cells equally at this scale, yet we see clear resistance of organoids compared to matched spheres when receiving only radiation therapy (Fig. 3). To prove whether small molecules such as our drugs can successfully penetrate the core of the organoids, we evaluated cancer therapeutic doxorubicin which is naturally fluorescent at 590 nm (Supplementary Fig. 3). We see that at 24–48 h, doxorubicin has diffused within the organoid mass. Taken together, we believe these data demonstrate that 3D grown GBM cells demonstrate a biologically different response to the tested therapeutics compared to sphere cultures. Organoid culture allows for interactions between tumor cells and extracellular matrix, and between cells of different microenvironments; both types of interactions may contribute to this differing response to chemotherapy.
We demonstrate that chemotherapy can blunt cell proliferation in organoid specimens, however this does not translate to cell death. By comparison, decreased viability is seen in matched sphere specimens. We therefore infer from our findings that organoids display an unknown protective mechanism against cell death. In the literature, growth of cells in 3D architecture such as Matrigel or hydrogel is well known to be protective against cell death, and this may be a likely contributing factor to the drug resistance demonstrated here [14,24,46,48,50,51]. For example, Wang et al. showed that cancer cells developed resistance to alkylating agents when grown in a 3D porous scaffold. Tumor cells maintained in a hydrogel demonstrate increased survival against a variety of drugs [24]. This was thought to be due to microglial cells that release cytokines which leads to activation of signaling pathways that maintain GSC stemness and contribute to resistance against cytotoxic agents. Overall, these diverse studies using various methods of forming 3D structures converge on a similar conclusion that live 3D modeling is likely to be the key technological approach underpinning the discovery of resistance mechanisms of human cancer cells.
In our above data, we see general trends and ‘rules’, but also many exceptions. This variable response may not be explained simply with one molecular behavior and underscores the dramatic complexity of cancer growth and evolution. Further research in this arena is necessary to accurately predict which specimens will show specific drug resistance. Importantly, patient tumor specimens grown in sphere culture versus organoid culture can paint a different picture of that tumor and how to attack it. As we have demonstrated above, a specimen may seem effectively treated with a therapeutic agent in sphere culture, yet that same specimen as an organoid can show significant resistance to the same dose of the same therapeutic. Three-dimensional organoid culture can provide unique insight into the complex network of interactions and serve as a complementary method in addition to sphere culture to help better understand what makes GBM such a clinically evasive disease.
CRediT authorship contribution statement
Swetha J. Sundar: Visualization, Data curation, Writing – review & editing. Sajina Shakya: Visualization, Data curation, Writing – review & editing. Austin Barnett: Data curation, Investigation. Lisa C. Wallace: Data curation. Hyemin Jeon: Data curation. Andrew Sloan: Investigation, Writing – review & editing. Violette Recinos: Investigation, Writing – review & editing. Christopher G. Hubert: Visualization, Data curation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank our patients for being willing to provide tissue samples for research. We thank Mary McGraw, Pengjing Huang, and Sadie Johnson of the Rose Ella Burkhardt Brain Tumor Bank, as well as Dr. Steven Dombrowski and Dr. Daniel Silver (Cleveland Clinic) for coordinating and providing brain tumor specimens and cultures. We thank Jeongwu Lee, Katrina Fife, and Maya Camhi of the Cleveland Clinic Biomedical Engineering histology core for excellent technical assistance in the development of organoid microarrays.
Funding
This work was supported in part by National Institutes of Health grants NIH/NCATS CTSA KL2 TR0002547 (C.G.H.). Work was also supported by an American Brain Tumor Association Discovery Grant #DG1800016 in memory of Dr. Joseph Weiss (C.G.H.), grant #IRG-16–186–21 to the Case Comprehensive Cancer Center from the American Cancer Society (C.G.H.), and grant funding from the Center for Transformative Nanomedicine (C.G.H). A.E.S. is supported by R21 CA256573, the Peter D. Cristal Chair in Neurosurgery, the DC Austin Fund, the Arsouze Fund, and the Kimble Family, Ferry Family, and Gerald T. Kaufman Jr. Foundations.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101251.
Appendix. Supplementary materials
References
- 1.Azzarelli R., Ori M., Philpott A., Simons B.D. Three-dimensional model of glioblastoma by co-culturing tumor stem cells with human brain organoids. Biol. Open. 2021:10. doi: 10.1242/bio.056416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response'. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 3.Bian S., Repic M., Guo Z., Kavirayani A., Burkard T., Bagley J.A., Krauditsch C., Knoblich J.A. Author correction: genetically engineered cerebral organoids model brain tumor formation. Nat. Methods. 2018;15:748. doi: 10.1038/s41592-018-0118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boretto M., Maenhoudt N., Luo X., Hennes A., Boeckx B., Bui B., Heremans R., Perneel L., Kobayashi H., Van Zundert I., Brems H., Cox B., Ferrante M., Uji I.H., Koh K.P., D'Hooghe T., Vanhie A., Vergote I., Meuleman C., Tomassetti C., Lambrechts D., Vriens J., Timmerman D., Vankelecom H. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 2019;21:1041–1051. doi: 10.1038/s41556-019-0360-z. [DOI] [PubMed] [Google Scholar]
- 5.Cheng L., Huang Z., Zhou W., Wu Q., Donnola S., Liu J.K., Fang X., Sloan A.E., Mao Y., Lathia J.D., Min W., McLendon R.E., Rich J.N., Bao S. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–152. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Silva B., Mathew R.K., Polson E.S., Williams J., Wurdak H. Spontaneous glioblastoma spheroid infiltration of early-stage cerebral organoids models brain tumor invasion. SLAS Discov. 2018;23:862–868. doi: 10.1177/2472555218764623. [DOI] [PubMed] [Google Scholar]
- 7.Debacq-Chainiaux F., Erusalimsky JD., Campisi J., Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 8.Flavahan W.A., Wu Q., Hitomi M., Rahim N., Kim Y., Sloan A.E., Weil R.J., Nakano I., Sarkaria J.N., Stringer B.W., Day B.W., Li M., Lathia J.D., Rich J.N., Hjelmeland A.B. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu J., Liu Z.G., Liu X.M., Chen F.R., Shi H.L., Pangjesse C.S., Ng H.K., Chen Z.P. Glioblastoma stem cells resistant to temozolomide-induced autophagy'. Chin. Med. J. (Engl.) 2009;122:1255–1259. [PubMed] [Google Scholar]
- 10.Galli R., Binda E., Orfanelli U., Cipelletti B., Gritti A., De Vitis S., Fiocco R., Foroni C., Dimeco F., Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 11.Heddleston J.M., Li Z., McLendon R.E., Hjelmeland A.B., Rich J.N. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8:3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heffernan J.M., Sirianni R.W. Modeling microenvironmental regulation of glioblastoma stem cells: a biomaterials perspective. Front. Mater. 2018;5:7. [Google Scholar]
- 13.Hemmati H.D., Nakano I., Lazareff J.A., Masterman-Smith M., Geschwind D.H., Bronner-Fraser M., Kornblum H.I. Cancerous stem cells can arise from pediatric brain tumors. Proc. Natl. Acad. Sci. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrera-Perez M., Voytik-Harbin S.L., Rickus J.L. Extracellular matrix properties regulate the migratory response of glioblastoma stem cells in three-dimensional culture. Tissue Eng. Part A. 2015;21:2572–2582. doi: 10.1089/ten.tea.2014.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hitomi M., Chumakova A.P., Silver D.J., Knudsen A.M., Pontius W.D., Murphy S., Anand N., Kristensen B.W., Lathia J.D. Asymmetric cell division promotes therapeutic resistance in glioblastoma stem cells. JCI Insight. 2021:6. doi: 10.1172/jci.insight.130510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hjelmeland A.B., Lathia J.D., Sathornsumetee S., Rich J.N. Twisted tango: brain tumor neurovascular interactions. Nat. Neurosci. 2011;14:1375–1381. doi: 10.1038/nn.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L., Holtzinger A., Jagan I., BeGora M., Lohse I., Ngai N., Nostro C., Wang R., Muthuswamy L.B., Crawford H.C. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell–and patient-derived tumor organoids'. Nat. Med. 2015;21:1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubert C.G., Rivera M., Spangler L.C., Wu Q., Mack S.C., Prager B.C., Couce M., McLendon R.E., Sloan A.E., Rich J.N. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016;76:2465–2477. doi: 10.1158/0008-5472.CAN-15-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob F., Salinas R.D., Zhang D.Y., Nguyen P.T.T., Schnoll J.G., Wong S.Z.H., Thokala R., Sheikh S., Saxena D., Prokop S., Liu D.A., Qian X., Petrov D., Lucas T., Chen H.I., Dorsey J.F., Christian K.M., Binder Z.A., Nasrallah M., Brem S., O'Rourke D.M., Ming G.L., Song H. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204. doi: 10.1016/j.cell.2019.11.036. .e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lathia J.D., Heddleston J.M., Venere M., Rich J.N. Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell Stem Cell. 2011;8:482–485. doi: 10.1016/j.stem.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lathia J.D., Mack S.C., Mulkearns-Hubert E.E., Valentim C.L., Rich J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ledur P.F., Onzi G.R., Zong H., Lenz G. Culture conditions defining glioblastoma cells behavior: what is the impact for novel discoveries? Oncotarget. 2017;8:69185–69197. doi: 10.18632/oncotarget.20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W., Park J.K., Fine H.A. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 24.Leite D.M., Zvar Baskovic B., Civita P., Neto C., Gumbleton M., Pilkington G.J. A human co-culture cell model incorporating microglia supports glioblastoma growth and migration, and confers resistance to cytotoxics. FASEB J. 2020;34:1710–1727. doi: 10.1096/fj.201901858RR. [DOI] [PubMed] [Google Scholar]
- 25.Lenin S., Ponthier E., Scheer K.G., Yeo E.C.F., Tea M.N., Ebert L.M., Oksdath Mansilla M., Poonnoose S., Baumgartner U., Day B.W., Ormsby R.J., Pitson S.M., Gomez G.A. A drug screening pipeline using 2D and 3D patient-derived in vitro models for pre-clinical analysis of therapy response in Glioblastoma. Int. J. Mol. Sci. 2021:22. doi: 10.3390/ijms22094322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linkous A., Balamatsias D., Snuderl M., Edwards L., Miyaguchi K., Milner T., Reich B., Cohen-Gould L., Storaska A., Nakayama Y., Schenkein E., Singhania R., Cirigliano S., Magdeldin T., Lin Y., Nanjangud G., Chadalavada K., Pisapia D., Liston C., Fine H.A. Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 2019;26:3203–3211. doi: 10.1016/j.celrep.2019.02.063. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loong H.H., Wong A.M., Chan D.T., Cheung M.S., Chow C., Ding X., Chan A.K., Johnston P.A., Lau J.Y., Poon W.S., Wong N. Patient-derived tumor organoid predicts drugs response in glioblastoma: a step forward in personalized cancer therapy? J. Clin. Neurosci. 2020;78:400–402. doi: 10.1016/j.jocn.2020.04.107. [DOI] [PubMed] [Google Scholar]
- 28.Luo X., Weiss W.A. Utility of human-derived models for glioblastoma. Cancer Discov. 2020;10:907–909. doi: 10.1158/2159-8290.CD-20-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer M., Reimand J., Lan X., Head R., Zhu X., Kushida M., Bayani J., Pressey J.C., Lionel A.C., Clarke I.D., Cusimano M., Squire J.A., Scherer S.W., Bernstein M., Woodin M.A., Bader G.D., Dirks P.B. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc. Natl. Acad. Sci. 2015;112:851–856. doi: 10.1073/pnas.1320611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neftel C., Laffy J., Filbin M.G., Hara T., Shore M.E., Rahme G.J., Richman A.R., Silverbush D., Shaw M.L., Hebert C.M., Dewitt J., Gritsch S., Perez E.M., Gonzalez Castro L.N., Lan X., Druck N., Rodman C., Dionne D., Kaplan A., Bertalan M.S., Small J., Pelton K., Becker S., Bonal D., Nguyen Q.D., Servis R.L., Fung J.M., Mylvaganam R., Mayr L., Gojo J., Haberler C., Geyeregger R., Czech T., Slavc I., Nahed B.V., Curry W.T., Carter B.S., Wakimoto H., Brastianos P.K., Batchelor T.T., Stemmer-Rachamimov A., Martinez-Lage M., Frosch M.P., Stamenkovic I., Riggi N., Rheinbay E., Monje M., Rozenblatt-Rosen O., Cahill D.P., Patel A.P., Hunter T., Verma I.M., Ligon K.L., Louis D.N., Regev A., Bernstein B.E., Tirosh I., Suva M.L. An integrative model of cellular states, plasticity, and genetics for Glioblastoma. Cell. 2019;178:835–849. doi: 10.1016/j.cell.2019.06.024. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogawa J., Pao G.M., Shokhirev M.N., Verma I.M. Glioblastoma model using human cerebral organoids. Cell Rep. 2018;23:1220–1229. doi: 10.1016/j.celrep.2018.03.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostrom Q.T., Patil N., Cioffi G., Waite K., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013-2017. Neuro Oncol. 2020;22:iv1–iv96. doi: 10.1093/neuonc/noaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel AP., Tirosh I., Trombetta JJ., Shalek AK., Gillespie SM., Wakimoto H., Cahill DP., Nahed BV., Curry WT., Martuza RL., Louis DN., Rozenblatt-Rosen O., Suvà ML., Regev A., Bernstein BE. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng D., Gleyzer R., Tai W.H., Kumar P., Bian Q., Isaacs B., da Rocha E.L., Cai S., DiNapoli K., Huang F.W., Cahan P. Evaluating the transcriptional fidelity of cancer models. Genome Med. 2021;13:73. doi: 10.1186/s13073-021-00888-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perrin S.L., Samuel M.S., Koszyca B., Brown M.P., Ebert L.M., Oksdath M., Gomez G.A. Glioblastoma heterogeneity and the tumour microenvironment: implications for preclinical research and development of new treatments. Biochem. Soc. Trans. 2019;47:625–638. doi: 10.1042/BST20180444. [DOI] [PubMed] [Google Scholar]
- 36.Pine A.R., Cirigliano S.M., Nicholson J.G., Hu Y., Linkous A., Miyaguchi K., Edwards L., Singhania R., Schwartz T.H., Ramakrishna R., Pisapia D.J., Snuderl M., Elemento O., Fine H.A. Tumor microenvironment is critical for the maintenance of cellular states found in primary Glioblastomas. Cancer Discov. 2020;10:964. doi: 10.1158/2159-8290.CD-20-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pontius W.D., Wallace L.C., Fife K., Hubert C.G. In: Brain Tumors. Seano G., editor. Springer US; New York, NY: 2021. Human glioblastoma organoids to model brain tumor heterogeneity ex vivo. [Google Scholar]
- 38.Rauf Y., Hufsey R., Robinson K., Suh J.H., Chao S.T., Murphy E.S., Yu J.S., Peereboom D.M., Singh Ahluwalia M., Wei W. Phase I study of ruxolitinib with radiation and temozolomide in patients with newly diagnosed grade III gliomas and glioblastoma. J. Clin. Oncol. 2021;39:2060. 60. [Google Scholar]
- 39.Saito Y., Muramatsu T., Kanai Y., Ojima H., Sukeda A., Hiraoka N., Arai E., Sugiyama Y., Matsuzaki J., Uchida R., Yoshikawa N., Furukawa R., Saito H. Establishment of patient-derived organoids and drug screening for biliary tract carcinoma. Cell Rep. 2019;27:1265–1276. doi: 10.1016/j.celrep.2019.03.088. e4. [DOI] [PubMed] [Google Scholar]
- 40.Schonberg D.L., Miller T.E., Wu Q., Flavahan W.A., Das N.K., Hale J.S., Hubert C.G., Mack S.C., Jarrar A.M., Karl R.T., Rosager A.M., Nixon A.M., Tesar P.J., Hamerlik P., Kristensen B.W., Horbinski C., Connor J.R., Fox P.L., Lathia J.D., Rich J.N. Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell. 2015;28:441–455. doi: 10.1016/j.ccell.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shakya S., Gromovsky A.D., Hale J.S., Knudsen A.M., Prager B., Wallace L.C., Penalva L.O.F., Alex Brown H., Kristensen B.W., Rich J.N., Lathia J.D., Mark Brown J., Hubert C.G. Altered lipid metabolism marks glioblastoma stem and non-stem cells in separate tumor niches. Acta Neuropathol. Commun. 2021;9:101. doi: 10.1186/s40478-021-01205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y., Guryanova O.A., Zhou W., Liu C., Huang Z., Fang X., Wang X., Chen C., Wu Q., He Z., Wang W., Zhang W., Jiang T., Liu Q., Chen Y., Wang W., Wu J., Kim L., Gimple R.C., Feng H., Kung H.F., Yu J.S., Rich J.N., Ping Y.F., Bian X.W., Bao S. Ibrutinib inactivates BMX-STAT3 in glioma stem cells to impair malignant growth and radioresistance. Sci. Transl. Med. 2018:10. doi: 10.1126/scitranslmed.aah6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B. Identification of human brain tumour initiating cells'. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 44.Stupp R., Mason WP., Bent MJ., Weller M., Fisher B., Taphoorn M.J.B., Belanger K., Brandes A.A., Marosi C., Bogdahn U., Curschmann J., Janzer R.C., Ludwin S.K., Gorlia T., Allgeier A., Lacombe D., Gregory Cairncross J., Eisenhauer E., Mirimanoff R.O. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 45.Vlachogiannis G., Hedayat S., Vatsiou A., Jamin Y., Fernández-Mateos J., Khan K., Lampis A., Eason K., Huntingford I., Burke R., Rata M., Koh D.M., Tunariu N., Collins D., Hulkki-Wilson S., Ragulan C., Spiteri I., Moorcraft S.Y., Chau I., Rao S., Watkins D., Fotiadis N., Bali M., Darvish-Damavandi M., Lote H., Eltahir Z., Smyth E.C., Begum R., Clarke P.A., Hahne J.C., Dowsett M., de Bono J., Workman P., Sadanandam A., Fassan M., Sansom O.J., Eccles S., Starling N., Braconi C., Sottoriva A., Robinson S.P., Cunningham D., Valeri N. 'Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C., Tong X., Yang F. Bioengineered 3D brain tumor model to elucidate the effects of matrix stiffness on glioblastoma cell behavior using PEG-based hydrogels. Mol. Pharm. 2014;11:2115–2125. doi: 10.1021/mp5000828. [DOI] [PubMed] [Google Scholar]
- 47.Wang J., Wakeman T.P., Lathia J.D., Hjelmeland A.B., Wang X.F., White R.R., Rich J.N., Sullenger B.A. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28:17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang K., Kievit F.M., Erickson A.E., Silber J.R., Ellenbogen R.G., Zhang M. Culture on 3D chitosan-hyaluronic acid scaffolds enhances stem cell marker expression and drug resistance in human glioblastoma cancer stem cells. Adv. Healthc. Mater. 2016;5:3173–3181. doi: 10.1002/adhm.201600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Prager B.C., Wu Q., Kim L.J.Y., Gimple R.C., Shi Y., Yang K., Morton A.R., Zhou W., Zhu Z., Obara E.A.A., Miller T.E., Song A., Lai S., Hubert C.G., Jin X., Huang Z., Fang X., Dixit D., Tao W., Zhai K., Chen C., Dong Z., Zhang G., Dombrowski S.M., Hamerlik P., Mack S.C., Bao S., Rich J.N. Reciprocal signaling between glioblastoma stem cells and differentiated tumor cells promotes malignant progression. Cell Stem Cell. 2018;22:514–528. doi: 10.1016/j.stem.2018.03.011. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao W., Wang S., Zhang R., Sohrabi A., Yu Q., Liu S., Ehsanipour A., Liang J., Bierman R.D., Nathanson D.A., Seidlits S.K. Bioengineered scaffolds for 3D culture demonstrate extracellular matrix-mediated mechanisms of chemotherapy resistance in Glioblastoma. Matrix Biol. 2020;85-86:128–146. doi: 10.1016/j.matbio.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao W., Zhang R., Sohrabi A., Ehsanipour A., Sun S., Liang J., Walthers C.M., Ta L., Nathanson D.A., Seidlits S.K. Brain-mimetic 3D culture platforms allow investigation of cooperative effects of extracellular matrix features on therapeutic resistance in Glioblastoma. Cancer Res. 2018;78:1358–1370. doi: 10.1158/0008-5472.CAN-17-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zanders ED., Svensson F., Bailey DS. Therapy for glioblastoma: is it working? Drug Discov. Today. 2019;24:1193–1201. doi: 10.1016/j.drudis.2019.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.