Abstract
The elevated expression of the hyaluronan-mediated motility receptor (HMMR) is known to be highly associated with tumor progression in prostate cancer, but the molecular mechanisms underlying the regulation of HMMR expression remain unclear. Here, we report that mammalian target of rapamycin (mTOR) is a key regulator of HMMR expression, for which its kinase activity is required. Pharmacological inhibitors of mTOR, such as rapamycin and Torin2, markedly suppressed the mRNA level as well as the protein level of HMMR in LNCaP and PC-3 cells. Our data demonstrate that such regulation occurs at the transcription level. HMMR promoter reporter assays revealed that the transcription factor SRF is responsible for the mTOR-mediated transcriptional regulation of HMMR gene. Consistently, the suppression of HMMR expression by Torin2 was noticeably reversed by the overexpression of SRF. Moreover, our findings suggest that the SRF binding sites responsible for the transcriptional regulation of HMMR through the mTOR-SRF axis are located in HMMR promoter sequences carrying the first intron, downstream of the translational start site. Furthermore, the upregulation of HMMR by DHT was abolished by stimulation with rapamycin, prior to DHT treatment, suggesting that mTOR activity is required for the induction of HMMR expression by androgen. Collectively, our study provides new mechanistic insights into the role of mTOR/SRF/AR signaling in HMMR regulation in prostate cancer cells.
Keywords: HMMR, SRF, mTOR, Transcriptional regulation, Androgen receptor, Prostate cancer
INTRODUCTION
Prostate cancer, the second most common cause of death in males, is known to be sensitive to hormone ablation therapy at an early stage, but most prostate cancers eventually acquire castration-resistance, conveying many genetic alterations, copy number changes, dysregulated functions of putative tumor suppressor and tumor promoter genes (Taplin and Balk, 2004), androgen receptor (AR) hypersensitivity, and the activation of alternative pathways for DHT synthesis (Chang et al., 2011). Approximately, 10-20% of all prostate cancers are reported to progress into castration-resistant prostate cancers (CRPC), which are mainly diagnosed as metastatic (mCRPC) (Kirby et al., 2011). Once mCRPC develops, more potent drugs that could further block androgen signaling may help, but the cancer eventually resists those drugs as well. Therefore, CRPC and mCRPC are considered clinically challenging cancers, and no curative clinical options exist yet, especially for mCRPC.
The mammalian target of rapamycin (mTOR), a key downstream molecule of PI3K/Akt, is composed of two distinct complexes: mTORC1 (Tafur et al., 2020) and mTORC2 (Fu and Hall, 2020). These complexes are physically associated with endogenous inhibitors that control their functions and activities (Wang et al., 2018). Because of their important roles in controlling cell survival and growth as well as protein synthesis and metabolism, mTOR complexes have been considered great therapeutic targets, and numerous mTOR inhibitors have been used in clinical trials for treating various human diseases, including prostate cancer (Chen and Zhou, 2020).
Hyaluronan (HA)-mediated motility receptor (HMMR), also known as RHAMM or CD186, has been identified as a breast cancer susceptibility gene (Pujana et al., 2007; Maxwell et al., 2011). HMMR functions as a receptor for HA on the cell surface but also intracellularly binds to the centrosome, actin filaments, microtubules, and the mitotic spindle (Assmann et al., 1999; Turley et al., 2002; Maxwell et al., 2003, 2008), promoting cell proliferation, mitosis, tumorigenesis, and cancer progression. Numerous studies have reported that HMMR expression is tightly controlled in normal tissues but is upregulated in proliferative tissues, contributing to invasiveness and metastasis with a poor prognosis in a variety of human cancers, such as colorectal (Zlobec et al., 2008), breast (Assmann et al., 2001), and prostate cancers (Gust et al., 2009).
The expression or the oncogenic function of HMMR has been challenged by many signaling molecules and pharmacological reagents, being a target of tumor suppressor genes. It has been reported that HMMR expression is differentially regulated during the cell cycle, reaching the maximum of HMMR mRNA level in the G2/M phase, and that tumor suppressor p53 regulates HMMR gene expression through transcriptional suppression (Sohr and Engeland, 2008). Wang et al. (2014) showed that the transcription factor TEAD and the coactivator YAP complex bind to HMMR promoter at specific sites to induce HMMR gene expression. In addition, simvastatin, an inhibitor of the mevalonate pathway for hyperlipidemia, exerts antitumor effects by repressing YAP/TEAD-activated HMMR transcription (Wang et al., 2014). It has also been found that the tumor suppressor complex BRCA1/BARD1 is known to have E3 ubiquitin ligase activity to ubiquitinate HMMR and to attenuate the function of HMMR (Joukov et al., 2006). HMMR is located in the nucleus, as well as in the cytoplasm and on the cell surface. It was demonstrated that E2F1, a regulator of cell cycle progression, acts as a direct inducer of HMMR, and that nuclear HMMR serves as a cofactor of E2F1 to control fibronectin expression, suggesting its role as a transcription coactivator (Meier et al., 2014). Pharmacological interference and genetic modulation studies revealed that HMMR, a chief factor of metastatic potential, is a target gene of Rb/E2F, and the activity of HMMR is sufficient for the metastatic phenotype induced by loss of Rb in prostate cancer. Rb has been reported to act as a transcription repressor of the HMMR gene, thus suppressing the pro-metastatic signaling cascade (Thangavel et al., 2017). Moreover, HMMR expression has been reported to be higher in CRPC, suggesting that an overexpression of HMMR may be associated with the development of hormonal resistance during the progression of prostate cancer (Korkes et al., 2014).
By interacting with HA, HMMR is known to activate or cooperate with many subcellular signaling pathways, thus promoting cell survival and proliferation and cancer cell migration. Some of these signaling pathways activated by HMMR/HA for its oncogenic properties include FAK-ERK1/2 (Kouvidi et al., 2011), Raf-MEK (Hatano et al., 2011), EGFR-AKT-ERK (Song et al., 2019), and ROK/PI3K signaling pathways (Lin et al., 2007). Here, we demonstrate that SRF mediates mTOR-regulated HMMR transcription, and that mTOR activity is required for AR/DHT-mediated HMMR expression. Our findings suggest that HMMR might be a promising therapeutic target for treating hormone-sensitive and -resistant prostate cancers.
MATERIALS AND METHODS
Materials
The sources for the following chemicals, reagents and antibodies were: Rapamycin (WAKO Chemical, Richmond, VA, USA); Torin 2 and Ku0063794 (TOCRIS, Bristol, UK); IGF-1 (BioVision, Milpitas, CA, USA); DHT, LY294002, SP600125, SB202190, PD153035, and PD98059 (Sigma Aldrich, St. Louis, MO, USA); characterized fetal bovine serum (FBS) (ATCC, Manassas, VA, USA); anti-HMMR (Bethyl, Montgomery, TX, USA); anti-p-Rb(S807/811), anti-CDK2, anti-E-cadherin (24E10), anti-vimentin (D21H3), anti-p-Erk1/2 (T202/Y204), and anti-Erk (Cell signaling, Danvers, MA, USA); anti-cyclin D2 (Santa Cruz, Dallas, TX, USA); anti-GAPDH (Bioworld Technology, St. Louis Park, MN, USA); anti-β-actin, anti-FlagM2, and dextran-charcoal stripped fetal bovine serum (DC-FBS) (Sigma Aldrich); characterized fetal bovine serum (FBS) (HyClone, Logan, UT, USA).
Cell lines, culture, and construction of stable PC3+HMMR cell lines
PC-3 and LNCaP, human prostate cancer cell lines, purchased from Korean Cell Line Bank (KCLB, Seoul, Korea) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% FBS at 37°C in a 95% air/5% CO2 environment and passaged every 3-4 days (at subconfluence). All experiments were performed in low serum conditions (DMEM containing 1% FBS or DC-FBS, and 15 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid[HEPES]), unless otherwise specified. In addition, all the experiments in PC3+AR and LNCaP cells were performed in culture medium supplemented with dextran-charcoal stripped fetal bovine serum (DC-FBS). For stable cell line overexpressing HMMR, PC3 cells were transfected with either pCMV6 or pCMV6-flag-HMMR, and stably transfected cells resistant to G418 were then selected. The pool of the resulting colonies (PC3+pCMV6 or PC3+HMMR) was used for experiments.
Western blot analysis
Cells were plated at a density of 2×105 cells/well in 6-well plates containing 2 mL/well of DMEM supplemented with 1% DC-FBS or FBS and 15 mM HEPES and then treated with signaling reagents indicated. Total protein content of cell lysates was quantified by using the BCA protein assay (Thermo Scientific, Rockford, IL, USA), and 20 to 50 µg of protein from whole cell lysates was subjected to western blot analysis as described previously (Aryal et al., 2014).
RNA interference
Cells were transfected with siRNAs (Bioneer, Deajeon, Korea) directed towards specific genes by using Lipofectamine RNAimax (Invitrogen, Carlsbad, CA, USA) in serum-free medium and incubated for 6 h. Cells were then supplemented with serum and treated with the indicated reagents. Total RNAs and lysates were prepared 48 h after siRNA transfection or 24 h after drug treatment, unless otherwise specified. The sequences of siRNAs used in this study were: si-HMMR #1 (sense-CU AGA UAU UGC CCA GUU AG, antisense-CU AAC UGG GCA AUA UCU AG), si-HMMR #2 (sense-CA GGA AAA GUA UGA CAG UA, antisense-UA CUG UCA UAC UUU UCC UC), and si-HMMR #3 (sense-GC UCA CCU UUA UCA CCU CA, antisense-UG AGG UGA UAA AGG UGA GC).
Real-time PCR
Total RNA from PC-3 and LNCaP cells treated with the indicated reagents was extracted using Trizol (Favorgen, Changjhih, Pingtung, Taiwan) according to manufacturer’s protocols and quantitative PCR was performed as described previously (Sun et al., 2020) using the following primers: HMMR (forward5′-GCACACCAAGTCAGCACTAA-3′ and reverse 5′-CTCTTCCTCCTGTTGCTTGA-3′), HAS1 (forward 5′-AACTTGCTGGGTGGT TCTC-3′ and reverse 5′-CCCACACCCTGACCAATAAA-3′), HAS2 (forward 5′-GCTCG ATCTAAGTGCCTTACTG-3′ and reverse 5′-GTACAGCCATTCTCGGAAGTAG-3′), HAS3 (forward 5′-CCTGGTGCTTTCCTTCATCT-3′ and reverse 5′-ACCAGCAGTCTTACCAGA AAC-3′), and GAPDH (forward 5′-GATCATCAGCAATGCCTCCT-3′ and reverse 5′-TGTG GTCATGAGTCCTTCCA-3′). Significant differences in relative luciferase activity were calculated by statistical analysis.
HMMR promoter constructs and Luciferase assay
The human HMMR promoter construct (–520/+3183) cloned into the pGL4.10 firefly-luciferase reporter-gene vector (pRHAMM(–520/+3183)-luc) was generously provided by Dr. Kurt England (Sohr and Engeland, 2008). pRHAMM(–520/+3183)-luc, PSA-luc, or MMTV-luc were co-transfected with cmv-renilla, followed by stimulation of signaling reagents. Separately, cells were transiently transfected with reporter constructs, cmv-renilla (or sv40-renilla), and either control empty vector, pcDNA3.1-myc-mTOR, pCMV-flag-hSRF (Addgene, Watertown, MA, USA), or pCMV6-flag-HMMR (Origene Technologies, Rockville, MD, USA) using Lipofectamine Plus transfection reagent (Invitrogen) as described previously (Aryal et al., 2014; Sun et al., 2020). After 3 h of transfection, the medium was replaced and cells were then cultured in the absence or presence of reagents indicated before lysis or analysis.
Crystal violet staining and cell viability assay
Crystal violet staining assay was performed as described previously (Sun et al., 2020). PC-3 cells were plated at a density of 1×105 cells/well in 12-well plates containing 1 mL of DMEM supplemented with 1% FBS and 15 mM HEPES per well, and transfected with either si-cont or si-HMMR. Separately, PC3+pCMV6 and PC3+HMMR cells plated at density of 5×104 cells/well in 12-well plates were cultured in regular growth medium for additional 1, 3, 5 and 7 days. Then, cells were fixed with 2% formalin/PBS and incubated with 0.2 mg/mL of crystal violet solution to stain the nuclei. After washing twice with PBS the crystal violet dye was eluted by adding 1% Triton X-100/PBS and the absorbance at 550 nm was determined.
Statistical analysis
Data was shown as mean ± SD of triplicate determinations and representative of two or three independent experiments. Statistical significance was calculated with one-way analysis of variance (ANOVA; Tukey’s post hoc analysis), and accepted at the level of p<0.05.
RESULTS
Silencing of HMMR inhibits prostate cancer growth in PC-3 cells
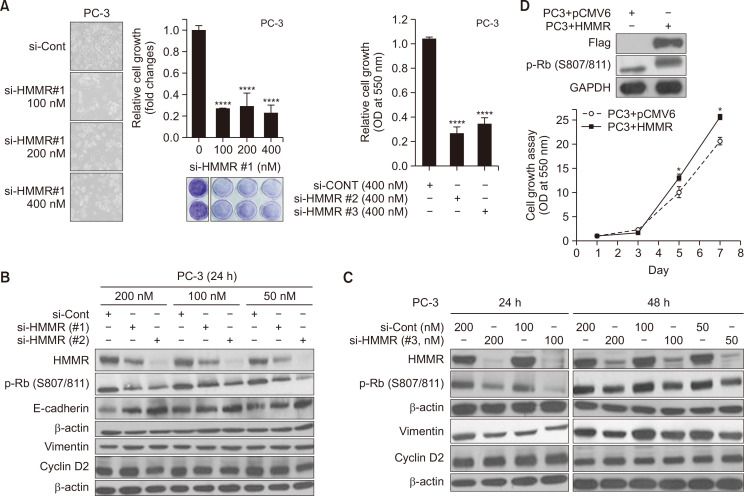
Small interfering RNA interferes with the expression of specific genes with complementary nucleotide sequences by degrading the mRNA after transcription. To test the role of HMMR on cell growth in human prostate cancer cells, we first introduced an siRNA against HMMR to silence its expression. Hormone refractory prostate cancer PC-3 cells were transfected with either control siRNA (si-Cont) or three different doses (100, 200, and 400 nM) of siRNA against HMMR (si-HMMR#1) and further incubated for 4 days before analysis. PC-3 cells were observed using a microscope to determine the morphological changes or the changes in cell death (Fig. 1A, left), and such changes were quantified using spectrophotometry, following crystal violet staining. As shown in Fig. 1A (middle and right), the silencing of HMMR expression using siRNAs significantly blocked cell proliferation by 70%-80%, even at the lowest concentration tested (100 nM), while control siRNA failed to suppress the growth of prostate cancer cells. We performed experiments using si-HMMR#1, #2 and #3 targeting different sites on HMMR to confirm these results at the molecular level. Even though there were variations in the silencing efficiency, all three si-HMMRs significantly downregulated the HMMR protein levels, and si-HMMR #2 and #3 at a dose of 50 nM decreased HMMR protein levels, similar to the results obtained at a dose of 200 nM (Fig. 1B, 1C). We examined the effect of silencing HMMR on the protein expression of cell cycle regulators such as cyclin Ds and phospho-Rb. Our data revealed that silencing HMMR resulted in hypo-phosphorylation of Rb at ser807/811 in PC-3 cells, whereas cyclin D2 levels remained unchanged. In addition, the silencing of HMMR increased the protein level of E-cadherin (Fig. 1B), and vimentin was slightly downregulated by si-HMMR at 24 and 48 h (Fig. 1C). These data suggest that HMMR might play a role in suppressing the cell cycle by dephosphorylating Rb and in blocking migration of cancer cells by controlling E-cadherin and vimentin.
Fig. 1.
Silencing of HMMR suppresses cell growth by dephosphorylating Rb in prostate cancer cells. (A) Human prostate cancer PC-3 cells were transfected with control siRNA (si-Cont) or various doses of si-HMMR(#1) for 6 h, and further cultured for 48 h (A, left panel) or 4 days (A, middle panel). Separately, cells were transfected with si-HMMR(#2) or si-HMMR(#3) (A, right panel). Cells were then either observed under microscope or subjected to crystal violet staining assay at 550 nm (****p<0.0001 vs. control). (B, C) Three siRNAs against HMMR were introduced at various doses and cells were lysed at 24 h or 48 h after transfection, followed by western blot analyses. (D) PC3 cells stably overexpressing pCMV6-Flag-HMMR (PC3+HMMR) and control cells (PC3+pCMV6) were cultured for 7 days, and the rate of cell growth was determined by crystal violet staining assays. Separately, twenty to 40 μg of protein was analyzed by western blotting to measure the protein level of flag-HMMR and pRb. Data shown are the mean ± SD of triplicate determinations and representative of two or three independent experiments (*p<0.05 vs. control).
To further support the role of HMMR in the proliferation of cancer cells, we generated HMMR-overexpressing PC-3 cells and compared the proliferation rate in control (PC3+pCMV6) and HMMR-overexpressing cells (PC3+HMMR). PC3+HMMR cells proliferated faster than the control cells, and the phosphorylation status of the Rb protein at S807/811 also substantially increased in the HMMR-overexpressing PC-3 stable cell line (Fig. 1D). Taken together, the inactivation of Rb might be a key factor mediating the stimulatory effect of HMMR on cancer cell proliferation.
mTOR inhibitors significantly reduced HMMR expression in prostate cancer cells
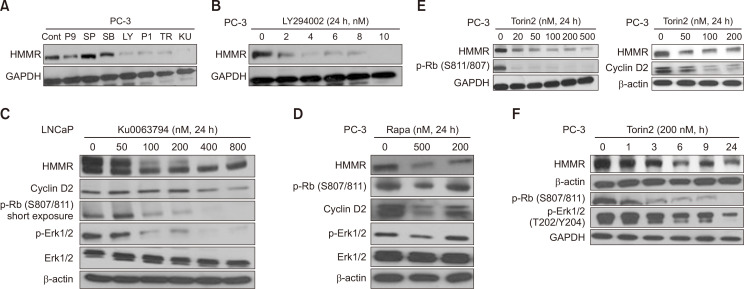
We next tested the signaling pathways that might be involved in regulating HMMR expression. LNCaP and PC-3 cells were exposed to pharmacological inhibitors of various signaling pathways, including MEK (P9), JNK (SP), p38MAPK (SB), PI3K (LY), EGFR (P1), and mTOR (TR, KU, Rapa) inhibitors (Fig. 2). Among these reagents, mTOR inhibitors (Rapa, Rapamycin; TR, Torin2; KU, Ku0063794) and MEK inhibitor (P9, PD98059) were found to significantly downregulate the protein expression of HMMR (Fig. 2A). These data suggest that the PI3K-Akt-mTOR signaling axis could be a key transducer in modulating HMMR expression, stimulating the proliferation and survival of human cancer cells.
Fig. 2.
mTOR activity is critical for regulating the expression of HMMR protein. (A) Cells were exposed to a variety of signaling pathway inhibitors for 24 h and HMMR protein level was measured by immunoblotting analysis. (B-E) PC-3 and LNCaP cells were cultured for 24 h with various doses of LY294002 (0 to 10 nM), KU0063794 (0 to 800 nM), rapamycin (0, 200, and 500 nM), and Torin2 (0 to 500 nM). (F) PC-3 cells were incubated for the indicated time in the absence or presence of 200 nM Torin2. Twenty to 50 μg of protein from cell lysates was subjected to western blot analysis. Data shown are representative of two or three independent experiments (P9; PD98059, MEK inhibitor, SP; SP600125, JNK inhibitor, SB; SB202190, p38 MAPK inhibitor, LY; LY293002, PI3K inhibitor, P1; PD153035, EGFR inhibitor, TR; Torin2, mTOR inhibitor, KU; KU0063794, mTOR inhibitor, Rapa; Rapamycin, mTOR inhibitor).
These data prompted us to further test the role of the PI3K/AKT/mTOR signaling pathway on HMMR expression in prostate cancer cells. Blocking PI3K activity using its pharmacological inhibitor LY294002 significantly reduced HMMR protein expression in a dose-dependent manner (Fig. 2B). Based on the results shown in Fig. 2A, the cells were exposed to various doses of mTOR inhibitors, followed by performing western blot analysis for the protein levels of HMMR. Ku0063794 is known to be a highly specific and potent dual mTOR inhibitor of mTORC1 and mTORC2 (Zhang et al., 2013), resulting in the inhibition of tumor growth, inducing cell cycle arrest and autophagy in cancer cells. As shown in Fig. 2C, an HMMR expression reduction was observed after 24 h of treatment with 50 nM Ku0064793, and HMMR was further decreased in a dose-dependent manner. The maximum effect was reached at 400 nM Ku0064793 in hormone-responsive LNCaP cells. Consistent with other reports, the levels of cell cycle-related proteins, such as cyclin D2 and phosphor-Rb, were suppressed by Ku0064793 in a dose-dependent manner. Moreover, Rb activation by Ku0064793 seemed to be correlated with the suppression of HMMR in LNCaP cells (Fig. 2C), which further supports our finding that HMMR partly induces cell growth through Rb phosphorylation (Fig. 1). Ku0064793 also decreased the phospho-ERK1/2 level in proportion, whereas the total ERK1/2 level remained unchanged (Fig. 2C). An increasing number of studies have shown that HMMR expression activates the ERK1/2 pathway (Kouvidi et al., 2011; Song et al., 2019). Therefore, ERK1/2 activity was suppressed by the mTOR inhibitor, as shown in Fig. 2, which might reflect the reduced levels of HMMR mediated by Ku0064793.
Next, we confirmed these results using other mTOR inhibitors, rapamycin and Torin2, in hormone-insensitive PC-3 cells. Rapamycin is a well-known mTOR inhibitor that has been used as adjuvant therapy in many types of cancers, including prostate cancer. Torin2 is known to be a second-generation ATP-competitive inhibitor and a potent selective inhibitor of mTOR, suppressing T389 phosphorylation of S6K in an mTORC1-dependent manner. In addition to the inhibition of mTOR, Torin2 also has a negative effect on the biochemical and cellular activities of other kinases of PIKK family members, such as ATM, ATR, and DNA-PK, and is known to display noticeable anti-proliferative activity in many cancer cell lines (Liu et al., 2011). Blocking mTOR activity using rapamycin and Torin2 significantly decreased the protein level of HMMR in a dose-dependent manner, markedly suppressing the phosphorylation of Rb, cyclin D2, and the phospho-activation of ERK1/2 (Fig. 2D, 2E). We examined how early mTOR inhibitors could downregulate HMMR expression. For this, the HMMR protein levels were determined at 0, 1, 3, 6, 9, and 24 h after Torin2 stimulation. As shown in Fig. 2F, Torin2 suppressed the HMMR protein levels as early as 3 h after stimulation using 200 nM Torin2, and gradually decreased the HMMR protein levels after 24 h (Fig. 2F).
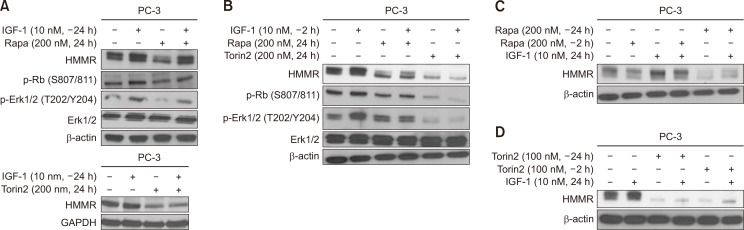
As Fig. 2B shows that PI3K activation increases HMMR expression, we stimulated cells with IGF-1, and found that IGF-1 induces HMMR expression, as expected (data not shown). Therefore, we next studied whether stimulation with IGF-1, a major upstream molecule of mTOR, could reverse the effect of mTOR inhibitors on HMMR expression. Cells were pre-treated with 10 nM IGF-1 for 24 or 2 h, followed by stimulation with rapamycin or Torin2 for an additional 24 h and vice versa. IGF-1 alone (10 nM) increased the protein levels of HMMR in both PC3 (Fig. 3) and LNCaP (data not shown) cells. The protein level of HMMR, which was suppressed by rapamycin, was significantly restored after 24 h of incubation with IGF-1, prior to rapamycin stimulation (Fig. 3A, top). However, the reversal effect on HMMR expression induced by pre-incubation with IGF-1 (2 h) was not as great as that after 24 h of incubation with IGF-1 prior to rapamycin stimulation (Fig. 3B, lane 4). In addition, the pre-incubation with 10 nM IGF-1 did not restore the expression level of HMMR suppressed by Torin2, even when the cells were exposed to IGF-1 for 24 h prior to Torin2 treatment (Fig. 3A, bottom, 3B). When the cells were exposed to rapamycin or Torin2 for 24 h prior to IGF-I treatment, IGF-1 failed to reverse the level of HMMR suppressed by rapamycin or Torin2 (Fig. 3C, 3D). Our data suggest that mTOR could be a key molecule and upstream factor of HMMR expression and that HMMR regulation might occur further downstream of mTOR, possibly at the mRNA or transcription level.
Fig. 3.
HMMR expression induced by IGF-1 is suppressed by pharmacological inhibitors of mTOR. (A, B) PC-3 cells were pre-incubated with or without IGF-1 (10 nM) for 24 h (A) or 2 h (B), followed by treatment with either Rapamycin (200 nM) or Torin-2 (100, 200 nM). (C, D) Separately, cells pre-exposed to Rapamycin (C) or Torin-2 (D) for 24 h or 2 h were then cultured with or without IGF-1 (10 nM) for additional 24 h. Twenty to 50 μg of protein from cell lysates was subjected to western blot analysis, and data shown are representative of two or three independent experiments.
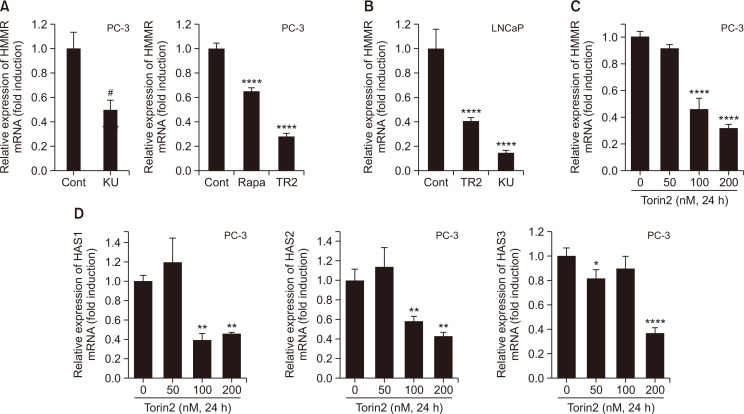
We investigated whether mRNA levels could reflect the HMMR protein levels reduced by mTOR inhibitors. Total RNA was isolated from the cells treated with either Ku0064793, Torin2, or rapamycin for 24 h and was subjected to real-time PCR analysis. Rapamycin, Torin2, and Ku0063794 decreased the mRNA levels of HMMR to 35%, 75%, and 60%, respectively, of control in PC-3 cells (Fig. 4A). Similarly, 60% and 80% of HMMR mRNA was decreased by stimulation with Torin2 and Ku0064793, respectively, in LNCaP cells (Fig. 4B). Torin2-mediated inhibition was dose-dependent in PC3 cells (Fig. 4C). Hyaluronic acid synthases (HAS) are responsible for HA synthesis, and three isozymes (HAS1, 2, and 3) are expressed in mammalian cells. Previously, the simultaneous silencing of HAS2 and HAS3 was reported to suppress HMMR levels and induce the cleavage of PARP, caspase-3 activation, and cell death in lung cancer cells, demonstrating that HA controls the expression of its own receptor (Song et al., 2019). PCR analysis revealed that Torin2 also suppressed all three HAS genes in a dose-dependent manner in PC3 cells (Fig. 4D), suggesting that the suppression of HMMR by Torin2 might be partly due to the reduced levels of HAS1, 2, and 3. Together, our data suggest that mTOR might play a key role in mediating the oncogenic effects of members of the HA signaling pathway (Xu et al., 2006), such as HMMR and HAS. Our results also demonstrate that the downregulation of HMMR proteins by mTOR inhibitors might be due to reduced mRNA expression and could possibly occur through transcriptional mechanisms.
Fig. 4.
Inactivation of mTOR suppresses mRNA levels both HMMR and HAS in dose dependent manner. (A, B) Total RNA was isolated from PC-3 (A) or LNCaP (B) cells stimulated with mTOR inhibitors (KU; KU0063794, Rapa; Rapamycin, TR2; Torin2) for 24 h. (C, D) Cells were exposed to various doses of Torin2 for 24 h. Quantitative real time PCR analysis was performed to determine message levels of HMMR and HAS isoforms. Data shown are the mean ± SD of triplicate determinations and representative of two or three independent experiments (A-D) (#p<0.05 vs. control, *p<0.05, **p<0.01, ****p<0.0001 vs. control).
Androgen upregulates the expression of HMMR mRNA and protein in LNCaP, hormone-responsive prostate cancer cells
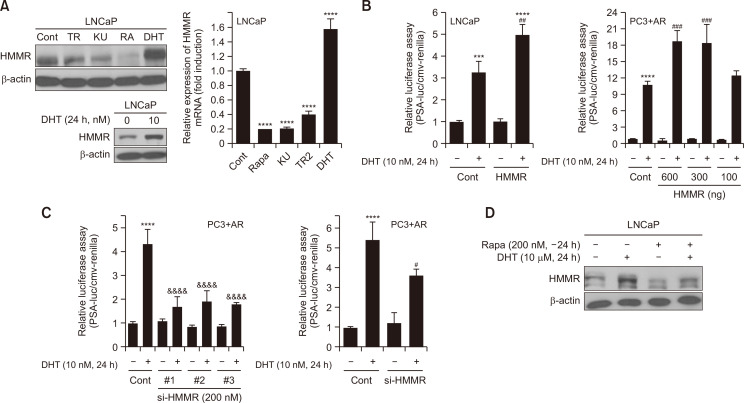
AR plays a critical role in the development and cell growth in prostate cancer, and DHT-AR binding results in the nuclear translocation of AR to control the expression of its target genes (Rahman et al., 2004). It has been reported that the crosstalk between AR and mTOR contributes to the proliferation of prostate cancer cells, showing a DHT-mediated mTORC1 activation (Xu et al., 2006). Because our data showed that mTOR activity is important for HMMR expression, we examined the effect of DHT on HMMR expression. Western blot analysis and a quantitative PCR assay showed that DHT stimulation for 24 h noticeably increased the mRNA levels by about 1.8-fold as well as the protein expression of HMMR in LNCaP cells (Fig. 5A). This result prompted us to test whether HMMR could regulate the androgen responses using PSA-luc and MMTV-luc constructs in LNCaP and PC-3 cells transiently overexpressing AR (PC3+AR). The exposure of LNCaP cells to 10 nM DHT for 24 h increased the PSA-luc activity by 3.2-fold compared with the control, and such DHT induction was further enhanced by introducing ectopic HMMR. We also performed similar experiments in PC-3 cells transiently overexpressing AR (PC3+AR cells). Consistent with LNCaP cells, PSA-luc activity induced by DHT was markedly upregulated by introducing the HMMR-expressing construct, which was dose-dependent (Fig. 5B). To further confirm the role of HMMR in androgen responses, cells were transiently transfected with si-HMMR to silence HMMR gene expression in PC3+AR cells. All three siRNAs against HMMR substantially inhibited PSA-luc and MMTV-luc activities induced by DHT (Fig. 5C).
Fig. 5.
Androgen induces HMMR expression by activating mTOR, and silencing HMMR suppresses androgenic responses in LNCaP cells. (A) LNCaP cells were cultured for 24 h with the indicated pharmacological reagents before lysis (TR; Torin2, KU; KU0063794, RA; Rapamycin, DHT; Dihydrotestosterone). Protein samples and total RNA isolated were subjected to immunoblotting and quantitative real time PCR, respectively to measure HMMR expression. Data shown are the mean ± SD of triplicate determinations and representative of two or three independent experiments (****p<0.0001 vs.Cont). (B) Cells were transiently transfected with 1 μg of total DNA including PSA-luciferase (PSA-luc) construct, cmv-renilla and either pCMV6 or pCMV6-flag-HMMR, followed by DHT (10 nM) treatment for 24 h (***p<0.001, ****p<0.0001 vs. Cont, ##p<0.01, ###p<0.001 vs. DHT alone). (C) Separately, cells were transiently transfected with 1 μg of total DNA including either PSA-luciferase (PSA-luc) or MMTV-luc construct and cmv-renilla, and 200 nM of si-Cont or si-HMMR. Cells were then cultured for 24 h in the absence or presence of DHT (10 nM). Data shown are relative values of firefly luciferase activity normalized to renilla luciferase. Data shown are the mean ± SD of triplicate determinations and representative of two or three independent experiments (****p<0.0001 vs.Cont, #p<0.05, &&&&p<0.0001 vs. DHT alone). (D) LNCaP was stimulated with 200 nM Rapamycin for 24 h, followed by DHT treatment for additional 24 h. The expression level of HMMR protein was measured by western blot analysis. Data shown are the mean ± SD of triplicate determinations and representative of two or three independent experiments (B-D).
We next examined whether mTOR activity is required for the induction of HMMR expression by androgen. When mTOR activity was inhibited by rapamycin, DHT failed to induce the expression level of HMMR in LNCaP hormone-responsive prostate cancer cells (Fig. 5D), and similar results were obtained using KU0063794 and Torin2 (data not shown). These data suggest that HMMR might be required for oncogenic responses by androgen and that mTOR might be a key mediator for regulation of HMMR expression via androgen.
HMMR expression is suppressed by mTOR inhibitors through a transcriptional mechanism in prostate cancer cells
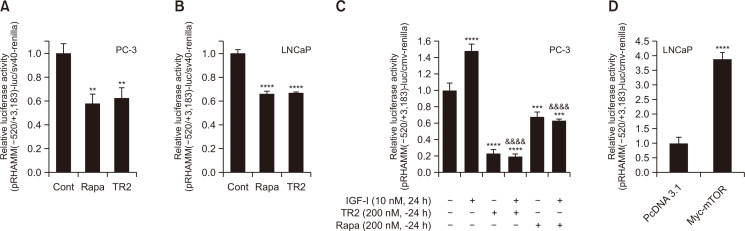
Because the inhibition of mTOR activity suppressed not only the protein levels but also the mRNA levels of HMMR (Fig. 2, 3) and such repression was observed even at 1 or 3 h after stimulation with Torin2 (Fig. 2F), we next examined whether the suppression of HMMR by mTOR inhibitors occurs at the transcriptional level. To do this, we performed luciferase reporter assays using the human HMMR promoter construct (pRHAMM(-520/+3183)-luc), which contains 520 base pairs upstream of the translational start codon, exon 1, and intron 1. The reporter assays showed that HMMR promoter activity was significantly inhibited by Torin2 in both PC-3 and LNCaP cells; similar results were observed using rapamycin (Fig. 6A, 6B). HMMR promoter activity was tested using the pRHAMM(–1830/+1)-luc construct and a similar suppression by mTOR inhibitor was observed (Supplementary Fig. 1A). In addition, the repression by Torin2 or rapamycin of HAMM promoter activity was also observed in HepG2 human hepatoma cells (Supplementary Fig. 1C), suggesting that it might be a general phenomenon in human malignancies. Consistent with the data observed in Fig. 3, HMMR promoter activity was significantly increased by IGF-1, and this induction was markedly blocked by 24 h of pretreatment with both Torin2 and rapamycin (Fig. 6C). As mTOR inhibition significantly downregulated the HMMR promoter activity, we also tested the effect of mTOR on HMMR promoter activity via ectopic expression of the mTOR gene. Consistently, the transient overexpression of mTOR enhanced HMMR promoter activity by 4-fold (Fig. 6D).
Fig. 6.
Downregulation of HMMR gene expression by mTOR inhibitors occur through transcriptional mechanism. (A-C) HMMR promoter reporter construct (pRHAMM(–520/+3183)-luc) construct was introduced to cells along with cmv-renilla. Transient transfection was followed by treatment with mTOR inhibitors indicated for 24 h (A, B). (C) Separately, cells were pre-incubated for 24 h with Torin2 (TR) or Rapamycin (Rapa) after transfection with HMMR promoter constructs and cmv-renilla, and then cultured in the absence or presence of IGF-1 for an additional 24 h. (D) Cells were transfected with 1 μg of total DNA including pRHAMM(-520/+3183)-luc, cmv-renilla and either pcDNA3.1 empty vector or pcDNA3.1-myc-mTOR (myc-MTOR). Data shown are relative values of firefly luciferase activity normalized to renilla luciferase. Data shown are the mean ± SD of triplicate determinations and representative of two or three independent experiments (**p<0.01, ***p<0.001, ****p<0.0001 vs. control, &&&&p<0.0001 vs. IGF-1 alone).
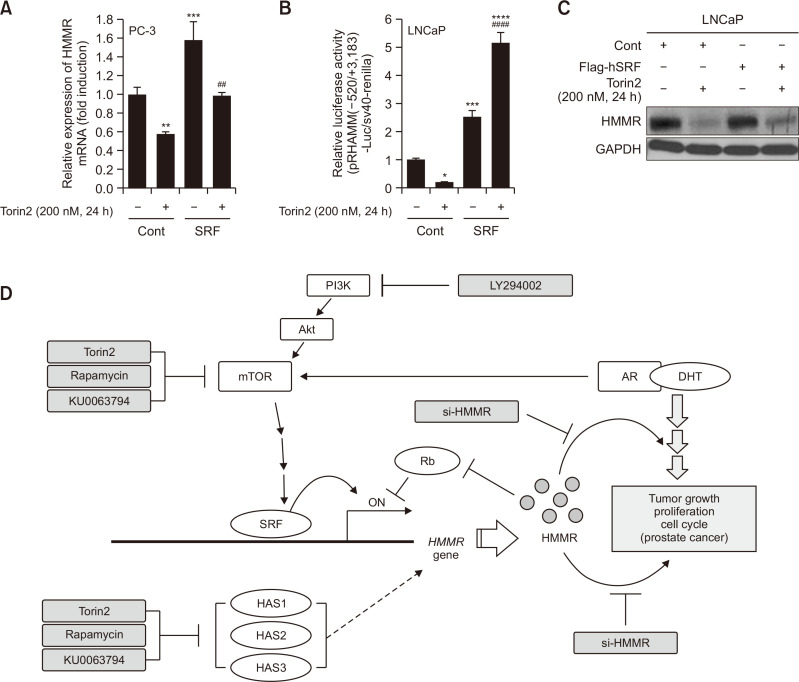
Our reporter activity assay results prompted us to search for possible transcription factors that could be involved in Torin2-mediated suppression of HMMR expression. To this end, the ALIBABA 2.0 software was used to screen the HMMR promoter sequence for transcription factors, and nine SRF-binding sites were found. When cells were transiently transfected with the SRF plasmid, SRF alone significantly increased the basal mRNA levels and the promoter activity of HMMR. In addition, both the mRNA level and the promoter activity of HMMR, which were suppressed by 200 nM Torin2, were completely restored by ectopic SRF in LNCaP cells (Fig. 7A, 7B). Western blot analysis showed that HMMR protein levels were also partly restored after SRF overexpression (Fig. 7C).
Fig. 7.
Transcriptional regulation of HMMR gene expression by mTOR is mediated partly by transcription factor SRF. (A, C) Total RNA (A) and protein (C) samples were prepared from cells transiently transfected with control empty vector or pCMV-flag-human SRF (SRF) followed by Torin2 stimulation. Data shown are the mean ± SD of triplicate determinations and representative of two or three independent experiments (**p<0.01, ***p<0.001 vs. control, ##p<0.01 vs. Torin2). (B) Separately, cells were transfected with 1 μg of total DNA including pRHAMM(-520/+3183)-luc, cmv-renilla and either control empty vector or pCMV-flag-human SRF (SRF), and then incubated for an additional 24 h with or without Torin2. Data shown are relative values of firefly luciferase activity normalized to renilla luciferase. Data shown are the mean ± SD of triplicate determinations and representative of two or three independent experiments (*p<0.05, ***p<0.001, ****p<0.0001 vs. control, ####p<0.0001 vs. Torin2). (D) The schematic diagram of potential mechanism for regulation of HMMR expression by mTOR and androgen, and oncogenic role of HMMR.
In summary, our data suggest that mTOR contributes to the oncogenic properties of HMMR in both hormone-sensitive and hormone-resistant prostate cancers, upregulating HMMR expression through transcriptional machinery, in which SRF might act as a mediator of mTOR signaling. Our findings also demonstrate that HMMR could be a key downstream mediator of the crosstalk between AR and mTOR pathways to promote prostate cancer growth, and that the induction of HMMR expression by androgen might occur through mTOR activation by the AR-DHT complex (Fig. 7D).
DISCUSSION
One of the major clinical challenges regarding prostate cancer therapy is to overcome the resistance to castration therapy. Intensive research has focused on finding the molecular mechanisms involved in the transition from hormone-sensitive to castration-resistant prostate cancer. These studies provide the scientific rationale for developing novel therapeutics or therapeutic candidates that could target both AR- and non-AR signaling pathways in prostate cancer (Watson et al., 2015).
HMMR has been found to be overexpressed in many advanced human cancers and is a valuable survival predictor (Augustin et al., 2015; Koelzer et al., 2015; Wang and Zhang, 2016). However, the mechanism of HMMR regulation in human cancers, including prostate cancer, remains unclear. In the present study, we evaluated the HMMR regulation mechanism in hormone-sensitive and castration-resistant prostate cancer cells, the reciprocal relationship between HMMR and androgen, and the role of mTOR activity and the crosstalk between mTOR/SRF and AR signaling in controlling HMMR expression. The proliferation rate of PC3 cells stably overexpressing HMMR was higher with the hyperphosphorylation of Rb than that of control cells, and the growth of cancer cells was greatly inhibited by silencing HMMR expression (Fig. 1), supporting the role of HMMR as an oncogene.
HA, a tumor promoter and enhancer, has been reported to interact with HMMR, a receptor for HA, resulting in increased invasion and metastasis in prostate cancer (Rizzardi et al., 2014). HA-HMMR signaling has been shown to activate PI3K/Akt signaling, promoting cell proliferation and possibly mediating metastatic castration-resistant prostate cancer transformation (Lin et al., 2007; Lokeshwar et al., 2010; Benitez et al., 2011). Our studies in this manuscript provide a new mechanism by which HMMR is regulated in prostate cancer cells, showing that PI3K/Akt/mTOR signaling contributes to HMMR expression and that mTOR plays a key role in regulating HMMR expression. Experiments using pharmacological inhibitors of mTOR indicate that mTOR activity is necessary for HMMR expression (Fig. 2-4). Studies by other investigators and our own findings suggest that HMMR expression might be regulated within a positive feedback loop between HA-HMMR and PI3K/Akt/mTOR.
Reporter analysis revealed that HMMR regulation by mTOR occurs through SRF-mediated transcription, as demonstrated by the reversal effect of SRF on the suppression of HMMR promoter activity by mTOR inhibition (Fig. 7). It is known that the tumor suppressor p53 represses HMMR expression at the transcriptional level, and the essential HMMR promoter site responsible for maximal repression by p53 is found at positions +2,755 to 2,778 in the first intron, downstream of its translational start site (Sohr and Engeland, 2008). In our studies, promoter analysis was also performed using pRHAMM(–520/+3183mut)-luc carrying a mutation of the p53 binding site (+2755 to +2778). This mutant promoter activity was also markedly inhibited by mTOR inhibitors, and no differences were observed between pRHAMM(–520/+3183)-luc and pRHAMM(–520/+3183mut)-luc (Fig. 7B, Supplementary Fig. 1B). In addition, the similar repressive effect of mTOR inhibitors was also determined using pRHAMM(–1830/+1)-luc, another HMMR promoter, carrying 1,830 bases upstream from the coding sequence and +1 at the translational start site and did not include putative p53 binding sites (Supplementary Fig. 1A). These data suggest that HMMR suppression by mTOR inactivation is not p53-dependent.
Transcription factor SRF-mediated mitogenic cascades have been known to be required for PI3K-dependent cell cycle progression, showing that SRF is a target of PI3K (Poser et al., 2000). We found nine SRF-binding sites in the HMMR promoter, which are mainly located downstream of the translation start site, when screened for transcription factors using ALIBABA 2.0 software. The overexpression of ectopic SRF noticeably inhibited the effect of Torin2 on HMMR gene expression (Fig. 7), demonstrating that SRF is an important mediator of mTOR-mediated HMMR expression. Interestingly, the promoter region carrying 1,830 bases upstream of the coding sequence did not have SRF-binding sites, but by using pRHAMM(–1830/+1)-luc construct, the promoter activity downregulated by Torin2 was completely reversed by SRF overexpression (data not shown). There might be indirect transcription machinery under the influence of SRF in that promoter region, but the role of SRF in the regulation of HMMR expression in the promoter region of 1,830 bases upstream from the translational start site needs to be further investigated.
Rapamycin-sensitive mTORC1 consists of PRAS40, DEPTOR and FKBP38, Tti/Tel2, raptor, and mLST8/GβL. Rapamycin-insensitive mTORC2 shares mTOR, Tti/Tel2, mLST8/GβL, and DEPTOR but contains unique members, such as protor1/2, mSin1, and rictor. These components act as either positive or negative regulators of mTOR activity within the complex (Saxton and Sabatini, 2017a, 2017b). Considering the complexity of mTORC1 and mTORC2, further studies in our laboratory are underway to explore the role of individual components of mTOR complexes in mTOR-mediated HMMR expression.
Recent studies have shown that Rb forms a repressive complex on the HMMR promoter, and that the loss of Rb results in the induction of HMMR function and expression (Thangavel et al., 2017). Here, we provide new insights into the crosstalk between Rb and HMMR signaling by demonstrating that silencing HMMR increases Rb function and that the overexpression of HMMR induces hyperphosphorylated Rb, an inactive form of Rb. This suggests that Rb and HMMR are inversely related to each other’s expression or function, but future studies are required to address the mechanism by which HMMR controls Rb function.
Previously, Lin et al. (2008) demonstrated that DHT, the active androgen, induces HMMR expression and HA-mediated tumorigenicity in cell proliferation, metastasis, and invasion, which are significantly enhanced in prostate cancer cells. Consistent with this, we were also able to detect increased levels of HMMR protein and mRNA by DHT (Fig. 5A). For the first time, we provided evidence that androgen-mediated HMMR expression requires mTOR activity by performing an experiment using a pharmacological inhibitor of mTOR (Fig. 5D). Androgenic responses were significantly suppressed by silencing HMMR gene expression and were further enhanced by HMMR overexpression, as determined according to the PSA-luc and MMTV-luc reporter activities. Recently, it has been reported that rapamycin suppreses AR signaling pathway by interacting with FKBP51 (Zhang et al., 2020). Taken together, the HMMR-mTOR axis might contribute to the oncogenic effect of androgen in hormone-sensitive prostate cancer.
Collectively, our data clearly demonstrate a new mechanism for HMMR regulation at the transcriptional level through the AR-mTOR-SRF axis and provide new insights on the role of HMMR in hormone-sensitive and castration-resistant prostate cancer cells. Given the importance of HMMR expression in prostate cancers, our present findings provide new therapeutic implications for targeting prostate cancer or overcoming castration-resistance in prostate cancer. Drugs that block the expression or activity of HMMR could be promising in treating prostate cancer. Moreover, combination therapy with HMMR inhibitors and either mTOR inhibitors or androgen deprivation might be a more beneficial therapeutic intervention approach.
SUPPLEMENTARY MATERIALS
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (NRF-2018R1D1A1B07050063).
REFERENCES
- Aryal P., Kim K., Park P. H., Ham S., Cho J., Song K. Baicalein induces autophagic cell death through AMPK/ULK1 activation and downregulation of mTORC1 complex components in human cancer cells. FEBS J. 2014;281:4644–4658. doi: 10.1111/febs.12969. [DOI] [PubMed] [Google Scholar]
- Assmann V., Gillett C. E., Poulsom R., Ryder K., Hart I. R., Hanby A. M. The pattern of expression of the microtubule-binding protein RHAMM/IHABP in mammary carcinoma suggests a role in the invasive behaviour of tumour cells. J. Pathol. 2001;195:191–196. doi: 10.1002/path.941. [DOI] [PubMed] [Google Scholar]
- Assmann V., Jenkinson D., Marshall J. F., Hart I. R. The intracellular hyaluronan receptor RHAMM/IHABP interacts with microtubules and actin filaments. J. Cell Sci. 1999;112:3943–3954. doi: 10.1242/jcs.112.22.3943. [DOI] [PubMed] [Google Scholar]
- Augustin F., Fiegl M., Schmid T., Pomme G., Sterlacci W., Tzankov A. Receptor for hyaluronic acid-mediated motility (RHAMM, CD168) expression is prognostically important in both nodal negative and nodal positive large cell lung cancer. J. Clin. Pathol. 2015;68:368–373. doi: 10.1136/jclinpath-2014-202819. [DOI] [PubMed] [Google Scholar]
- Benitez A., Yates T. J., Lopez L. E., Cerwinka W. H., Bakkar A., Lokeshwar V. B. Targeting hyaluronidase for cancer therapy: antitumor activity of sulfated hyaluronic acid in prostate cancer cells. Cancer Res. 2011;71:4085–4095. doi: 10.1158/0008-5472.CAN-10-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K. H., Li R., Papari-Zareei M., Watumull L., Zhao Y. D., Auchus R. J., Sharifi N. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13728–13733. doi: 10.1073/pnas.1107898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhou X. Research progress of mTOR inhibitors. Eur. J. Med. Chem. 2020;208:112820. doi: 10.1016/j.ejmech.2020.112820. [DOI] [PubMed] [Google Scholar]
- Fu W., Hall M. N. Regulation of mTORC2 signaling. Genes. 2020;11:1045. doi: 10.3390/genes11091045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust K. M., Hofer M. D., Perner S. R., Kim R., Chinnaiyan A. M., Varambally S., Moller P., Rinnab L., Rubin M. A., Greiner J., Schmitt M., Kuefer R., Ringhoffer M. RHAMM (CD168) is overexpressed at the protein level and may constitute an immunogenic antigen in advanced prostate cancer disease. Neoplasia. 2009;11:956–963. doi: 10.1593/neo.09694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano H., Shigeishi H., Kudo Y., Higashikawa K., Tobiume K., Takata T., Kamata N. RHAMM/ERK interaction induces proliferative activities of cementifying fibroma cells through a mechanism based on the CD44-EGFR. Lab. Invest. 2011;91:379–391. doi: 10.1038/labinvest.2010.176. [DOI] [PubMed] [Google Scholar]
- Joukov V., Groen A. C., Prokhorova T., Gerson R., White E., Rodriguez A., Walter J. C., Livingston D. M. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Kirby M., Hirst C., Crawford E. D. Characterising the castration-resistant prostate cancer population: a systematic review. Int. J. Clin. Pract. 2011;65:1180–1192. doi: 10.1111/j.1742-1241.2011.02799.x. [DOI] [PubMed] [Google Scholar]
- Koelzer V. H., Huber B., Mele V., Iezzi G., Trippel M., Karamitopoulou E., Zlobec I., Lugli A. Expression of the hyaluronan-mediated motility receptor RHAMM in tumor budding cells identifies aggressive colorectal cancers. Hum. Pathol. 2015;46:1573–1581. doi: 10.1016/j.humpath.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Korkes F., de Castro M. G., de Cassio Zequi S., Nardi L., Del Giglio A., de Lima Pompeo A. C. Hyaluronan-mediated motility receptor (RHAMM) immunohistochemical expression and androgen deprivation in normal peritumoral, hyperplasic and neoplastic prostate tissue. BJU Int. 2014;113:822–829. doi: 10.1111/bju.12339. [DOI] [PubMed] [Google Scholar]
- Kouvidi K., Berdiaki A., Nikitovic D., Katonis P., Afratis N., Hascall V. C., Karamanos N. K., Tzanakakis G. N. Role of receptor for hyaluronic acid-mediated motility (RHAMM) in low molecular weight hyaluronan (LMWHA)-mediated fibrosarcoma cell adhesion. J. Biol. Chem. 2011;286:38509–38520. doi: 10.1074/jbc.M111.275875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. L., Chang D., Chiang A., Ying S. Y. Androgen receptor regulates CD168 expression and signaling in prostate cancer. Carcinogenesis. 2008;29:282–290. doi: 10.1093/carcin/bgm259. [DOI] [PubMed] [Google Scholar]
- Lin S. L., Chang D., Ying S. Y. Hyaluronan stimulates transformation of androgen-independent prostate cancer. Carcinogenesis. 2007;28:310–320. doi: 10.1093/carcin/bgl134. [DOI] [PubMed] [Google Scholar]
- Liu Q., Wang J., Kang S. A., Thoreen C. C., Hur W., Ahmed T., Sabatini D. M., Gray N. S. Discovery of 9-(6-aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyridin-2(1H)-one (Torin2) as a potent, selective, and orally available mammalian target of rapamycin (mTOR) inhibitor for treatment of cancer. J. Med. Chem. 2011;54:1473–1480. doi: 10.1021/jm101520v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokeshwar V. B., Lopez L. E., Munoz D., Chi A., Shirodkar S. P., Lokeshwar S. D., Escudero D. O., Dhir N., Altman N. Antitumor activity of hyaluronic acid synthesis inhibitor 4-methylumbelliferone in prostate cancer cells. Cancer Res. 2010;70:2613–2623. doi: 10.1158/0008-5472.CAN-09-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell C. A., Benitez J., Gomez-Baldo L., Osorio A., Bonifaci N., Fernandez-Ramires R., Costes S. V., Guino E., Chen H., Evans G. J., Mohan P., Català I., Petit A., Aguilar H., Villanueva A., Aytes A., Serra-Musach J., Rennert G., Lejbkowicz F., Peterlongo P., Manoukian S., Peissel B., Ripamonti C. B., Bonanni B., Viel A., Allavena A., Bernard L., Radice P., Friedman E., Kaufman B., Laitman Y., Dubrovsky M., Milgrom R., Jakubowska A., Cybulski C., Gorski B., Jaworska K., Durda K., Sukiennicki G., Lubiński J., Shugart Y. Y., Domchek S. M., Letrero R., Weber B. L., Hogervorst F. B., Rookus M. A., Collee J. M., Devilee P., Ligtenberg M. J., Luijt R. B., Aalfs C. M., Waisfisz Q., Wijnen J., Roozendaal C. E., Easton D. F., Peock S., Cook M., Oliver C., Frost D., Harrington P., Evans D. G., Lalloo F., Eeles R., Izatt L., Chu C., Eccles D., Douglas F., Brewer C., Nevanlinna H., Heikkinen T., Couch F. J., Lindor N. M., Wang X., Godwin A. K., Caligo M. A., Lombardi G., Loman N., Karlsson P., Ehrencrona H., von Wachenfeldt A., Barkardottir R. B., Hamann U., Rashid M. U., Lasa A., Caldés T., Andrés R., Schmitt M., Assmann V., Stevens K., Offit K., Curado J., Tilgner H., Guigó R., Aiza G., Brunet J., Castellsagué J., Martrat G., Urruticoechea A., Blanco I., Tihomirova L., Goldgar D. E., Buys S., John E. M., Miron A., Southey M., Daly M. B., Schmutzler R. K., Wappenschmidt B., Meindl A., Arnold N., Deissler H., Varon-Mateeva R., Sutter C., Niederacher D., Imyamitov E., Sinilnikova O. M., Stoppa-Lyonne D., Mazoyer S., Verny-Pierre C., Castera L., de Pauw A., Bignon Y. J., Uhrhammer N., Peyrat J. P., Vennin P., Fert Ferrer S., Collonge-Rame M. A., Mortemousque I., Spurdle A. B., Beesley J., Chen X., Healey S., Barcellos-Hoff M. H., Vidal M., Gruber S. B., Lázaro C., Capellá G., McGuffog L., Nathanson K. L., Antoniou A. C., Chenevix-Trench G., Fleisch M. C., Moreno V., Pujana M. A. HEBON, author; EMBRACE, author; GEMO Study Collaborators, author; SWE-BRCA, author; BCFR, author; kConFab, author. Interplay between BRCA1 and RHAMM regulates epithelial apicobasal polarization and may influence risk of breast cancer. PLoS Biol. 2011;9:e1001199. doi: 10.1371/journal.pbio.1001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell C. A., Keats J. J., Crainie M., Sun X., Yen T., Shibuya E., Hendzel M., Chan G., Pilarski L. M. RHAMM is a centrosomal protein that interacts with dynein and maintains spindle pole stability. Mol. Biol.Cell. 2003;14:2262–2276. doi: 10.1091/mbc.e02-07-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell C. A., McCarthy J., Turley E. Cell-surface and mitotic-spindle RHAMM: moonlighting or dual oncogenic functions? J. Cell Sci. 2008;121:925–932. doi: 10.1242/jcs.022038. [DOI] [PubMed] [Google Scholar]
- Meier C., Spitschak A., Abshagen K., Gupta S., Mor J. M., Wolkenhauer O., Haier J., Vollmar B., Alla V., Putzer B. M. Association of RHAMM with E2F1 promotes tumour cell extravasation by transcriptional up-regulation of fibronectin. J. Pathol. 2014;234:351–364. doi: 10.1002/path.4400. [DOI] [PubMed] [Google Scholar]
- Poser S., Impey S., Trinh K., Xia Z., Storm D. R. SRF-dependent gene expression is required for PI3-kinase-regulated cell proliferation. EMBO J. 2000;19:4955–4966. doi: 10.1093/emboj/19.18.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana M. A., Han J. D., Starita L. M., Stevens K. N., Tewari M., Ahn J. S., Rennert G., Moreno V., Kirchhoff T., Gold B., Assmann V., Elshamy W. M., Rual J. F., Levine D., Rozek L. S., Gelman R. S., Gunsalus K. C., Greenberg R. A., Sobhian B., Bertin N., Venkatesan K., Ayivi-Guedehoussou N., Solé X., Hernández P., Lázaro C., Nathanson K. L., Weber B. L., Cusick M. E., Hill D. E., Offit K., Livingston D. M., Gruber S. B., Parvin J. D., Vidal M. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- Rahman M., Miyamoto H., Chang C. Androgen receptor coregulators in prostate cancer: mechanisms and clinical implications. Clin. Cancer Res. 2004;10:2208–2219. doi: 10.1158/1078-0432.CCR-0746-3. [DOI] [PubMed] [Google Scholar]
- Rizzardi A. E., Vogel R. I., Koopmeiners J. S., Forster C. L., Marston L. O., Rosener N. K., Akentieva N., Price M. A., Metzger G. J., Warlick C. A., Henriksen J. C., Turley E. A., McCarthy J. B., Schmechel S. C. Elevated hyaluronan and hyaluronan-mediated motility receptor are associated with biochemical failure in patients with intermediate-grade prostate tumors. Cancer. 2014;120:1800–1809. doi: 10.1002/cncr.28646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R. A., Sabatini D. M. mTOR signaling in growth, metabolism, and disease. Cell. 2017a;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R. A., Sabatini D. M. mTOR signaling in growth, metabolism, and disease. Cell. 2017b;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- Sohr S., Engeland K. RHAMM is differentially expressed in the cell cycle and downregulated by the tumor suppressor p53. Cell Cycle. 2008;7:3448–3460. doi: 10.4161/cc.7.21.7014. [DOI] [PubMed] [Google Scholar]
- Song J. M., Im J., Nho R. S., Han Y. H., Upadhyaya P., Kassie F. Hyaluronan-CD44/RHAMM interaction-dependent cell proliferation and survival in lung cancer cells. Mol. Carcinog. 2019;58:321–333. doi: 10.1002/mc.22930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Jiang M., Park P. H., Song K. Transcriptional suppression of androgen receptor by 18beta-glycyrrhetinic acid in LNCaP human prostate cancer cells. Arch. Pharm. Res. 2020;43:433–448. doi: 10.1007/s12272-020-01228-z. [DOI] [PubMed] [Google Scholar]
- Tafur L., Kefauver J., Loewith R. Structural insights into TOR signaling. Genes. 2020;11:885. doi: 10.3390/genes11080885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taplin M. E., Balk S. P. Androgen receptor: a key molecule in the progression of prostate cancer to hormone independence. J. Cell. Biochem. 2004;91:483–490. doi: 10.1002/jcb.10653. [DOI] [PubMed] [Google Scholar]
- Thangavel C., Boopathi E., Liu Y., Haber A., Ertel A., Bhardwaj A., Addya S., Williams N., Ciment S. J., Cotzia P., Dean J. L., Snook A., McNair C., Price M., Hernandez J. R., Zhao S. G., Birbe R., McCarthy J. B., Turley E. A., Pienta K. J., Feng F. Y., Dicker A. P., Knudsen K. E., Den R. B. RB loss promotes prostate cancer metastasis. Cancer Res. 2017;77:982–995. doi: 10.1158/0008-5472.CAN-16-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley E. A., Noble P. W., Bourguignon L. Y. Signaling properties of hyaluronan receptors. J. Biol. Chem. 2002;277:4589–4592. doi: 10.1074/jbc.R100038200. [DOI] [PubMed] [Google Scholar]
- Wang F., Meng M., Mo B., Yang Y., Ji Y., Huang P., Lai W., Pan X., You T., Luo H., Guan X., Deng Y., Yuan S., Chu J., Namaka M., Hughes T., Ye L., Yu J., Li X., Deng Y. Crosstalks between mTORC1 and mTORC2 variagate cytokine signaling to control NK maturation and effector function. Nat. Commun. 2018;9:4874. doi: 10.1038/s41467-018-07277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Zhang T. Prognostic significance of CD168 overexpression in colorectal cancer. Oncol. Lett. 2016;12:2555–2559. doi: 10.3892/ol.2016.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu Y., Wang H., Zhang Y., Mei L., Fang X., Zhang X., Zhang F., Chen H., Liu Y., Jiang Y., Sun S., Zheng Y., Li N., Huang L. Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E89–E98. doi: 10.1073/pnas.1319190110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P. A., Arora V. K., Sawyers C. L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Chen S. Y., Ross K. N., Balk S. P. Androgens induce prostate cancer cell proliferation through mammalian target of rapamycin activation and post-transcriptional increases in cyclin D proteins. Cancer Res. 2006;66:7783–7792. doi: 10.1158/0008-5472.CAN-05-4472. [DOI] [PubMed] [Google Scholar]
- Zhang H., Berel D., Wang Y., Li P., Bhowmick N. A., Figlin R. A., Kim H. L. A comparison of Ku0063794, a dual mTORC1 and mTORC2 inhibitor, and temsirolimus in preclinical renal cell carcinoma models. PLoS ONE. 2013;8:e54918. doi: 10.1371/journal.pone.0054918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wu D., He Y., Li L., Lu J. Z., Gui H., Wang Y., Tao Y., Wang H.Z., Kaushik D., Rodriguez R., Wang Z. Rapamycin inhibits AR signaling pathway in prostate cancer by interacting with the FK1 domain of FKBP51. Biochem. Biophys. Rep. 2020;23:100778. doi: 10.1016/j.bbrep.2020.100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlobec I., Baker K., Terracciano L. M., Lugli A. RHAMM, p21 combined phenotype identifies microsatellite instability-high colorectal cancers with a highly adverse prognosis. Clin. Cancer Res. 2008;14:3798–3806. doi: 10.1158/1078-0432.CCR-07-5103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.