Abstract
PCR assays targeting rRNA genes were developed to identify species (genomovars) within the Burkholderia cepacia complex. Each assay was tested with 177 bacterial isolates that also underwent taxonomic analysis by whole-cell protein profile. These isolates were from clinical and environmental sources and included 107 B. cepacia complex strains, 23 Burkholderia gladioli strains, 20 Ralstonia pickettii strains, 10 Pseudomonas aeruginosa strains, 8 Stenotrophomonas maltophilia strains, and 9 isolates belonging to nine other species. The sensitivity and specificity of the 16S rRNA-based assay for Burkholderia multivorans (genomovar II) were 100 and 99%, respectively; for Burkholderia vietnamiensis (genomovar V), sensitivity and specificity were 87 and 92%, respectively. An assay based on 16S and 23S rRNA gene analysis of B. cepacia ATCC 25416 (genomovar I) was useful in identifying genomovars I, III, and IV as a group (sensitivity, 100%, and specificity, 99%). Another assay, designed to be specific at the genus level, identified all but one of the Burkholderia and Ralstonia isolates tested (sensitivity, 99%, and specificity, 96%). The combined use of these assays offers a significant improvement over previously published PCR assays for B. cepacia.
Burkholderia cepacia is a plant pathogen that is generally nonpathogenic for healthy humans. However, chronic colonization of the respiratory tract of persons with cystic fibrosis (CF) occurs and is associated with increased rates of morbidity and mortality (17). Because most strains exhibit broad-range antimicrobial resistance, therapeutic options are limited; therefore, prevention of acquisition is a major goal of patient management (10). The resultant stringent infection control measures place an enormous psychosocial and economic burden on the CF community. Accurate laboratory identification of B. cepacia underlies such infection control programs; however, misidentification of this and related nonfermenting gram-negative species is relatively common (1, 5).
Recent taxonomic analyses have demonstrated that bacteria identified as B. cepacia actually comprise at least five distinct genomic species, or genomovars, referred to collectively as the B. cepacia complex (19). The name Burkholderia multivorans has been proposed for genomovar II, while genomovar V has been identified as the previously named species Burkholderia vietnamiensis (4). The remaining three species are referred to as genomovars I, III, and IV pending further taxonomic study. Although all five species have been recovered from CF sputum culture, B. multivorans and genomovar III account for the majority of CF isolates (9, 19). Preliminary studies also indicate that most epidemic B. cepacia isolates are genomovar III and that this species is associated with greater morbidity and mortality than other members of the B. cepacia complex (14). To expand these observations and to improve the ability to accurately identify B. cepacia, simple and reliable assays for all B. cepacia complex species are needed. We report the development of rRNA gene-targeted PCR assays to identify bacteria within the B. cepacia complex.
MATERIALS AND METHODS
Bacteria.
For 16S rRNA gene sequence determination, B. cepacia complex type strains were obtained from the American Type Culture Collection or the Belgian Coordinated Collections of Microorganisms-Laboratorium voor Microbiologie at the University of Ghent Culture Collection (Ghent, Belgium). Strains ATCC 25416, LMG 14293, LMG 12614, LMG 14294, and LMG 10929T represented B. cepacia genomovars I through V, respectively.
For examination of PCR assays, a total of 177 isolates were tested. These included 107 B. cepacia complex, 23 Burkholderia gladioli, 20 Ralstonia pickettii, 10 Pseudomonas aeruginosa, and 8 Stenotrophomonas maltophilia strains. Among these, 132 were recovered from CF sputum culture and referred from clinical laboratories in the United States, 24 were recovered from patients without CF, and 12 were obtained from environmental cultures. In addition, nine strains representing species that may be encountered in CF sputum (3) were obtained from the American Type Culture Collection, including Pseudomonas fluorescens, Pseudomonas stutzeri, Brevundimonas vesicularis, Comamonas testosteroni, Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, Proteus mirabilis, and Klebsiella pneumoniae.
Taxonomic analyses.
After routine isolation and identification by referring laboratories, species (genomovar) identification of all clinical and environmental isolates was confirmed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis of whole-cell proteins as described elsewhere (19). In brief, after an incubation period of 48 h, whole-cell protein extracts were prepared and SDS-polyacrylamide gel electrophoresis was performed. The densitometric analysis, normalization and interpolation of the protein profiles, and numerical analysis were performed by using the GelCompar software package version 4.2 (Applied Maths, Kortrijk, Belgium). In previous studies, whole-cell protein analysis has provided excellent taxonomic resolution comparable to that obtained by using DNA-DNA hybridization (19, 20).
Cloning and sequence determination of 16S rRNA genes.
The 16S rRNA gene of each B. cepacia complex type strain was amplified by using PCR with a primer pair (UFPL and URPL) based on published B. cepacia rRNA gene sequences (Table 1). The resultant 1.5-kbp fragment was cloned into pGEM-T (Promega, Madison, Wis.), and the nucleotide sequence was determined with multiple internal primers by dideoxy chain termination in an ABI PRISM 373A automated sequencer (PE Applied Biosystems, Foster City, Calif.). The 16S rRNA gene of each type strain was cloned, and the sequence was determined in duplicate; the sequences of both ribosomal DNA (rDNA) strands were determined for one clone of each type strain. DNA sequences were assembled manually and analyzed by using the Genetics Computer Group Wisconsin package (Madison, Wis.).
TABLE 1.
PCR primers
| Primer | Sequence (5′-3′) | Target | Nucleotide positiona |
|---|---|---|---|
| UFPL | AGTTTGATCCTGGCTCAG | 16S rDNA—kingdom Bacteria | 9–26 |
| URPL | GGTTACCTTGTTACGACTT | 16S rDNA—kingdom Bacteria | 1482–1500 |
| BC-GII | AGGCGGTCTGTTAAGACA | 16S rDNA—B. multivorans | 578–595 |
| BC-GV | TAATACCGCATACGATCTAT | 16S rDNA—B. vietnamiensis | 165–184 |
| BC-R | AGCACTCCCGAATCTCTT | 16S rDNA—B. multivorans and B. vietnamiensis | 1005–1022 |
| PC-SSF | TCGGAATCCTGCTGAGAGGC | 16S rDNA—B. cepacia genomovar I | 994–1013 |
| PC-SSR | GCCATGGATACTCCAAAAGGA | 23S rDNA—B. cepacia genomovar I | NA |
| RHG-F | GGGATTCATTTCCTTAGTAAC | 16S rDNA—genus Burkholderia | 835–851 |
| RHG-R | GCGATTACTAGCGATTCCAGC | 16S rDNA—genus Burkholderia | 1324–1345 |
Numbering corresponds to 16S rDNA sequences in GenBank whose accession numbers are provided in the text. NA, not applicable.
PCR primer design.
An alignment of the five type strain 16S rRNA gene sequences was used to design primers (BC-GII, BC-GV, and BC-R) specific for species within the B. cepacia complex (Table 1). A primer pair (PC-SSF and PC-SSR) that had been previously designed (12) based on 16S and 23S rRNA gene sequences of ATCC 25416 (genomovar I) and that was putatively specific for B. cepacia was also examined. Analysis of Burkholderia 16S rRNA gene sequences available in National Center for Biotechnology Information databases was used to develop genus-specific primers (RHG-F and RHG-R).
Genomic DNA purification.
Genomic DNA was prepared from test bacteria by using the Easy-DNA Kit (Invitrogen, Carlsbad, Calif.) with the following modifications. The initial cell pellet was suspended in 200 μl of 50 mM Tris HCl–20 mM EDTA (pH 8.0), centrifuged, and resuspended in 200 μl of 50 mM Tris HCl–2 mM EDTA (pH 8.0). Lysozyme was added to 10 mg/ml, and the suspension was placed on ice for 15 min before addition of solution A. After incubation at 65°C for 15 min, 5 μl of a 10-mg/ml proteinase K solution and 25 μl of 10% SDS were added, and the suspension was mixed and incubated in a 37°C water bath for 1 h. After completion of DNA isolation and precipitation according to the manufacturer’s instructions, the DNA was suspended in 50 μl of UV-irradiated sterile water containing 50 μg of RNase per ml.
PCR.
PCR assays were performed in 50-μl reaction mixtures containing 50 ng of DNA template, 1 U of Taq DNA polymerase (Promega), 20 pmol of each primer, 200 μM (each) deoxyribonucleoside triphosphate (Promega), and 2 mM MgCl2 in a buffer with 10 mM Tris-HCl (pH 9.0) and 50 mM KCl (Promega). After the initial denaturation at 95°C for 3 min, 30 amplification cycles were completed in a PTC-100 programmable thermal cycler (MJ Research, Watertown, Mass.). Each cycle consisted of 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 1 min of extension at 72°C. For the last cycle, the extension step was 4 min.
Analysis of test bacteria.
All 177 test bacteria were examined by PCR employing primer pairs BC-GII and BC-R, BC-GV and BC-R, PC-SSF and PC-SSR, and RHG-F and RHG-R. In addition, all test bacteria were examined with two previously published primer pairs in PCR assays as described elsewhere (2). The first pair, PSR and PSL, amplifies a conserved 313-bp 16S rRNA gene segment from all bacteria and was used as a positive control. The second primer pair, PSR1 and PSL1, was designed to be specific for B. cepacia and was tested for comparison with the new primers. Negative control PCRs with all reaction mixture components except template DNA were employed for every experiment.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences for ATCC 25416, LMG 14293, LMG 12614, LMG 14294, and LMG 10929 are deposited in the GenBank database at the National Center for Biotechnology Information under accession no. AF097530, AF097531, AF097532, AF097533, and AF097534, respectively.
RESULTS
Taxonomic analyses.
Among the 107 isolates confirmed as B. cepacia complex, 10 were genomovar I, 39 were B. multivorans, 31 were genomovar III, 4 were genomovar IV, and 23 were B. vietnamiensis (data not shown).
Sequence analyses and primer selection.
Multiple sequence alignment of the 16S rRNA genes cloned from the five B. cepacia complex type strains revealed a high degree of identity; overall, the five sequences had 98.2% identity. Despite this high degree of identity, species-level sequence signatures were detected at positions 595 for B. multivorans and 184 for B. vietnamiensis, and these were incorporated into the 3′ end of forward primers BC-GII and BC-GV, respectively. These two species also shared sequences that differed from genomovars I, III, and IV between positions 1005 and 1013. A reverse primer, BC-R, was designed based on these differences to be specific for both B. multivorans and B. vietnamiensis.
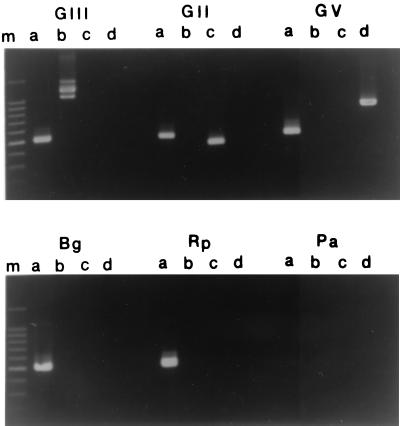
Sequence differences among genomovars I, III, and IV were insufficient to allow design of primers specific for these species. However, in preliminary experiments a primer pair, PC-SSF and PC-SSR, previously designed based on ATCC 25416 (genomovar I) sequence data, amplified genomovar I, III, and IV strains. These primers target 16S and 23S sequences, respectively, and their use in PCR results in amplification of polymorphic fragments of 16S-23S intergenic spacer region DNA. The putative genus-specific primer pair, RHG-F and RHG-R, amplified both Burkholderia and Ralstonia species in preliminary tests. Figure 1 illustrates the results of PCR with these various primer pairs; products are of the predicted sizes.
FIG. 1.
PCR analysis of B. cepacia complex type strains and related species. Lanes m, DNA markers; lanes a, primer pair RHG-F and RHG-R; lanes b, primer pair PC-SSF and PC-SSR; lanes c, primer pair BC-GII and BC-R; lanes d, primer pair BC-GV and BC-R. GIII, LMG 12614 (B. cepacia genomovar III); GII, LMG 14293 (B. multivorans); GV, LMG 10929T (B. vietnamiensis); Bg, B. gladioli; Rp, R. pickettii; Pa, P. aeruginosa.
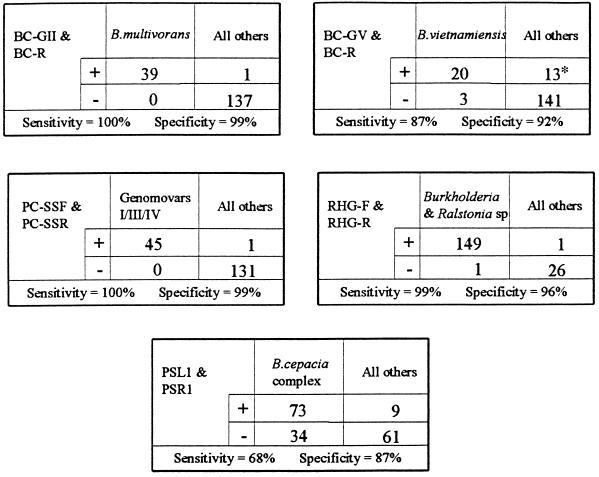
Sensitivity and specificity of PCR assays.
Each of the 177 test bacteria was examined by PCR with the following primer pairs: (a) BC-GII and BC-R, (b) BC-GV and BC-R, (c) PC-SSF and PC-SSR, (d) RHG-F and RHG-R, and (e) PSR1 and PSL1. The results are detailed in Fig. 2.
FIG. 2.
Sensitivity and specificity of PCR assays. *, all 13 are B. multivorans.
When test isolates were stratified by species, then sensitivity and specificity (respectively) of the primer pair PSR1 and PSL1 for each group were as follows: for B. multivorans, 56 and 56%; for B. vietnamiensis, 26 and 50%; and for genomovars I, III, and IV (as a group), 100 and 71%.
DISCUSSION
Several important questions regarding the epidemiology, natural history, and virulence of B. cepacia infection in CF remain unanswered. Accurate identification of B. cepacia and related species underlies studies to address these issues and is critical to clinical management and implementation of rational infection control measures. Because B. cepacia can be transmitted person to person among CF patients (11) and because there are currently no effective therapies to eradicate pulmonary colonization, the medical and psychosocial consequences of identifying B. cepacia in sputum culture are enormous.
However, accurate identification of B. cepacia has been problematic. Previous studies have demonstrated misidentification rates as high as 20% (1, 5, 13). In an analysis of nearly 1,100 recent CF sputum isolates received from over 100 laboratories, we have found that approximately 10% of bacteria identified as B. cepacia are, in fact, not members of the B. cepacia complex based on combined phenotypic and genotypic analyses (9). There are likely several reasons for misidentification. Specific protocols, including use of selective media, for processing of CF respiratory secretions do not always yield unequivocal results (7), and the degree to which they are used varies among clinical microbiology laboratories (16). Our incomplete understanding of the taxonomy of the B. cepacia complex also contributes to the difficulty of accurate species identification.
Previous work by others to develop genotypic methods of identifying B. cepacia has yielded a number of candidate PCR assays (2, 6, 15, 18). However, these studies were conducted before the recognition that several species comprise the B. cepacia complex, and most relied on published DNA sequence data derived from analyses of culture collection strains that, in retrospect, are poorly representative of CF sputum isolates. Most culture collection B. cepacia strains of environmental origin (e.g., the ATCC species type strain 25416) are genomovar I. Preliminary data from our laboratory indicate that the great majority of CF isolates are either B. multivorans or genomovar III (9).
To design PCR assays to identify all members of the B. cepacia complex, we sought species-level signature sequences in the 16S rRNA genes of the five complex type strains. Anticipating a high degree of identity among these genes, we employed a redundant sequencing strategy; 16S rRNA genes from each type strain were cloned in duplicate in independent experiments, multiple internal primers that provided overlapping sequence data were used, and both DNA strands from one of the two clones from each type strain were completely sequenced.
As expected, our analyses revealed a high degree of identity that offered few opportunities to design species-specific primers. However, by capitalizing on sequences shared by both B. multivorans and B. vietnamiensis, a reverse anchor primer, BC-R, that allowed identification of these species when paired with species-specific forward primers was designed. Although the assay for B. vietnamiensis amplified a subset of B. multivorans isolates, the assay for the latter species demonstrates excellent sensitivity and specificity. Thus, the combined use of these assays provides accurate identification of both species.
The sequence identity among the remaining three species (genomovars I, III, and IV) was too great to allow design of species-specific primers. However, this sequence conservation allowed another primer pair, PC-SSF and PC-SSR, to demonstrate excellent sensitivity and specificity in identifying these three species as a group. This primer pair had been previously developed to amplify B. cepacia 16S-23S rRNA intergenic regions (12). Because there are multiple rRNA operons, this primer pair has the potential to yield multiple polymorphic fragments, allowing for single-step identification and PCR ribotyping (8). This primer pair (also referred to as G1-G2) recently proved useful in detecting B. cepacia DNA directly in sputum specimens from CF patients (21).
In the current study, the previously described primer pair PSR1 and PSL1 demonstrated relatively poor sensitivity and specificity for B. cepacia complex isolates. These primers were designed based on the published 16S rRNA gene sequence from strain ATCC 25416, a genomovar I strain (2). When test isolates were stratified by species, this primer pair showed excellent sensitivity for genomovars I, III, and IV (as a group) but still suffered from poor specificity.
Primer pair RHG-F and RHG-R was developed before several members of the genus Burkholderia (including Burkholderia pickettii) were reclassified as Ralstonia species. In the present study, this primer pair was excellent in identifying the Burkholderia and Ralstonia species investigated. In additional preliminary studies, we have found this primer pair to be useful in a screening PCR assay; nonfermenting, nonenteric CF sputum isolates that are PCR negative do not require further testing by the species-specific PCR assays described above.
In summary, the combined use of primer pairs BC-GII and BC-R, BC-GV and BC-R, and PC-SSF and PC-SSR allows for identification of all five species currently assigned to the B. cepacia complex and offers a significant improvement over previously published assays. Ongoing studies will identify targets for the design of PCR assays to distinguish B. cepacia genomovars I, III, and IV. The development of PCR assays for the identification of the bacterial species most commonly confused with B. cepacia complex is also under way and will significantly enhance our ability to contribute to the care of persons with CF.
ACKNOWLEDGMENTS
This work was supported by grants from the Cystic Fibrosis Foundation (United States) (to J.J.L. and T.L.S.), the Cystic Fibrosis Trust (United Kingdom) (grant RS15), and the Belgische Vereniging voor Strijd tegen Mucoviscidose. P.V. is indebted to the Fund for Scientific Research-Flanders (Belgium) for a position as postdoctoral research fellow. T.C. acknowledges the support received from the Vlaams Instituut voor Bevordering van Wetenschappelijk-technologisch onderzoek in de Industrie (Belgium) in the form of a bursary for advanced study. T.L.S. and P.W.W. acknowledge the support of the Children’s Medical Research Institute.
J.J.L. acknowledges Sherif S. Abdelhak, David W. McConnell, and Nancy A. Wynstra, whose leadership and academic vision had a significant impact on the progress of this work.
REFERENCES
- 1.Burdge D R, Noble M A, Campbell M E, Krell V L, Speert D P. Xanthomonas maltophilia misidentified as Pseudomonas cepacia in cultures of sputum from patients with cystic fibrosis: a diagnostic pitfall with major clinical implications. Clin Infect Dis. 1995;20:445–448. doi: 10.1093/clinids/20.2.445. [DOI] [PubMed] [Google Scholar]
- 2.Campbell P W, Phillips III J A, Heidecker G J, Krishnamani M R S, Zahorchak R, Stull T L. Detection of Pseudomonas (Burkholderia) cepacia using PCR. Pediatr Pulmonol. 1995;20:44–49. doi: 10.1002/ppul.1950200109. [DOI] [PubMed] [Google Scholar]
- 3.Gilligan P H. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev. 1991;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillis M, Van T V, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an amended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 5.Henry D A, Campbell M E, LiPuma J J, Speert D P. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J Clin Microbiol. 1997;35:614–619. doi: 10.1128/jcm.35.3.614-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karpati F, Jonasson J. Polymerase chain reaction for the detection of Pseudomonas aeruginosa, Stenotrophomonas maltophilia and Burkholderia cepacia in sputum of patients with cystic fibrosis. Mol Cell Probes. 1996;10:397–403. doi: 10.1006/mcpr.1996.0055. [DOI] [PubMed] [Google Scholar]
- 7.Kiska D L, Kerr A, Jones M C, Caracciolo J A, Eskridge B, Jordan M, Miller S, Hughes D, King N, Gilligan P H. Accuracy of four commercial systems for identification of Burkholderia cepacia and other gram-negative nonfermenting bacilli recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:886–891. doi: 10.1128/jcm.34.4.886-891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostman J R, Edlind T D, LiPuma J J, Stull T L. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol. 1992;30:2084–2087. doi: 10.1128/jcm.30.8.2084-2087.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LiPuma, J. J. Unpublished data.
- 10.LiPuma J J. Burkholderia cepacia epidemiology and pathogenesis: implications for infection control. Curr Opin Pulm Med. 1998;4:337–341. doi: 10.1097/00063198-199811000-00005. [DOI] [PubMed] [Google Scholar]
- 11.LiPuma J J, Dasen S E, Nielson D W, Stern R C, Stull T L. Person-to-person transmission of Pseudomonas cepacia between patients with cystic fibrosis. Lancet. 1990;336:1094–1096. doi: 10.1016/0140-6736(90)92571-x. [DOI] [PubMed] [Google Scholar]
- 12.LiPuma J J, Heidekcer G J, Stull T L. PCR-based detection and typing of Pseudomonas cepacia, abstr. 2352. Pediatr Res. 1994;35:394A. [Google Scholar]
- 13.LiPuma J J, Henry D, Mehar F, Speert D, Saiman L. Misidentification of Burkholderia cepacia, abstr. 1810. Pediatr Res. 1997;41:304A. [Google Scholar]
- 14.Mahenthiralingam, E. Personal communication.
- 15.O’Callaghan E M, Tanner M S, Boulnois G J. Development of a PCR probe test for identifying Pseudomonas aeruginosa and Pseudomonas (Burkholderia) cepacia. J Clin Pathol. 1994;47:222–226. doi: 10.1136/jcp.47.3.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shreve M R, Butler S, Kaplowitz H J, Rabin H R, Stokes D, Light M, Regelmann W E. Impact of microbiology practice on cumulative prevalence of respiratory tract bacteria in patients with cystic fibrosis. J Clin Microbiol. 1999;37:753–757. doi: 10.1128/jcm.37.3.753-757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tablan O C, Martone W J, Doershuk C F, Stern R C, Thomassen M J, Klinger J D, White J W, Carson L A, Jarvis W R. Colonization of the respiratory tract with Pseudomonas cepacia in cystic fibrosis. Risk factors and outcomes. Chest. 1987;91:527–532. doi: 10.1378/chest.91.4.527. [DOI] [PubMed] [Google Scholar]
- 18.Tyler S D, Strathdee C A, Rozee K R, Johnson W M. Oligonucleotide primers designed to differentiate pathogenic pseudomonads on the basis of the sequencing of genes coding for the 16S-23S rRNA internal transcribed spacers. Clin Diagn Lab Immunol. 1995;2:448–453. doi: 10.1128/cdli.2.4.448-453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients: proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 20.Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitby P W, Dick H L N, Campbell III P W, Tullis D E, Matlow A, Stull T L. Comparison of culture and PCR for detection of Burkholderia cepacia in sputum samples of patients with cystic fibrosis. J Clin Microbiol. 1998;36:1642–1645. doi: 10.1128/jcm.36.6.1642-1645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]