Abstract
Introduction
To investigate the genomic profiles of patients with lung cancer with idiopathic pulmonary fibrosis (IPF-LC), mechanism of carcinogenesis, and potential therapeutic targets.
Methods
We analyzed 29 matched, surgically resected, cancerous and noncancerous lung tissues (19 IPF-LC and 10 non–IPF-LC) by whole-exome sequencing and bioinformatics analysis and established a medical-engineering collaboration with the Department of Engineering of the Tokyo University of Science.
Results
In IPF-LC, CADM1 and SPC25 were mutated at a frequency of 47% (9 of 19) and 53% (10 of 19), respectively. Approximately one-third of the IPF-LC cases (7 of 19; 36%) had both mutations. Pathway analysis revealed that these two genes are involved in transforming growth factor-β1 signaling. CADM1 and SPC25 gene mutations decreased the expression of CADM1 and increased that of SPC25 revealing transforming growth factor-β1–induced epithelial-to-mesenchymal transition and cell proliferation in lung cancer cells. Furthermore, treatment with paclitaxel and DNMT1 inhibitor suppressed SPC25 expression.
Conclusions
CADM1 and SPC25 gene mutations may be novel diagnostic markers and therapeutic targets for IPF-LC.
Keywords: CADM1, SPC25, Whole-exome sequencing, Lung cancer, Idiopathic pulmonary fibrosis
Introduction
Idiopathic pulmonary fibrosis (IPF) is associated with an increased risk of lung cancer (LC), with cumulative incidence rates of 3.3% and 15.4% at 1 and 5 years of follow-up, respectively.1 The prognosis for patients with LC with IPF (IPF-LC) is worse than that for patients with IPF alone, mainly because of the progression of LC and complications after treatment.2, 3, 4, 5, 6
Specific germline mutations predispose toward both IPF and LC, leading to imbalance between oncogenes and tumor suppressor genes, and ultimately carcinogenesis within the fibrotic lungs. Aberrant epigenetic regulation (e.g., methylation, histone modifications, and deregulation of noncoding RNAs) represents a possible pathogenic link between the two disease paradigms.7 Previously, we reported on the allelic loss of FHIT in peripheral precancerous lesions and IPF-LC.8 We also revealed frequent hypermethylation of the SMAD4 promoter and reduced SMAD4 expression in IPF-LC compared with the IPF lung tissue.9 Nevertheless, the specific molecular mechanism involved in carcinogenesis with this lung disease remains unknown. In addition, promising therapeutic targets for IPF-LC have not been identified.
Whole-exome sequencing (WES) by the next-generation sequencing method can identify all the protein-coding genes in the human genome. Nevertheless, WES data are complex and massive for analysis using conventional techniques. Therefore, it is difficult to perform an effective analysis, unless a certain level of knowledge in both medicine and engineering is achieved. To overcome this issue, we collaborated with the Department of Engineering of Tokyo University of Science (Tokyo, Japan) for the bioinformatics analysis.
In this study, we aimed to characterize the gene mutations underlying IPF-LC by WES and bioinformatics analysis.
Materials and Methods
Clinical Samples
This study analyzed 29 frozen primary NSCLC tissues with or without IPF and corresponding noncancerous lung tissue samples extracted from the same patients (19 IPF-LC and 10 non–IPF-LC). All participants provided informed written consent before their enrollment in the study. Subsequently, samples were obtained from patients who underwent lung surgery from 2013 to 2017 at the Nippon Medical School Hospital (Tokyo, Japan). Corresponding noncancerous lung tissues were obtained from the surgical specimen at the maximum possible distance from the tumor by a certified pathologist, avoiding contamination of the tumor and background lung. The pathologic diagnosis of IPF was confirmed for all IPF-LC samples, but not for the non–IPF-LC samples. In addition, of the 19 IPF-LC cases, seven (nonsquamous carcinoma) tested for EGFR mutations and only one patient had EGFR mutation. The study was approved by the institutional review board at the Nippon Medical School (Number 160804).
DNA Extraction and WES
DNA was extracted from frozen samples and cell lines using the QIAmp DNA Mini Kit (Qiagen, Hilden, Germany) in accordance with the instructions provided by the manufacturer. WES was performed for cancerous and noncancerous genomic DNA samples using the SureSelect Human All Exon exome capture kit version 5 (Agilent Technologies Japan, Tokyo, Japan) on the HiSeq 2500 platform (Illumina, San Diego, CA). WES data have been deposited in the Sequence Read Archive under the accession number PRJNA708255.
Machine Learning Method
The selection of candidate gene alterations discriminating IPF-LC and non–IPF-LC was conducted using machine learning methods, such as random forests and cross-validation, in collaboration with the Department of Industrial Administration, Tokyo University of Science. We trained random forests and deleted the lower 1% of the feature repeatedly until the top 1% of all the features were extracted. We finally adopted cross-validation to rank the top gene mutations and validate the random forest classifier.
Data Mining Using the Kaplan–Meier Plotter
The association between the expression of the mutated gene and overall survival (OS) in patients with LC was evaluated by data mining using the Kaplan–Meier plotter (www.kmplot.com).
RNA Extraction and Real-Time Quantitative Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted using TRIzol Reagent (Invitrogen, Tokyo, Japan); quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) was performed using the THUNDERBIRD Probe qPCR Mix (Toyobo Co., Osaka, Japan) and 7500 FAST system (Applied Biosystems, Foster City, CA), according to the instructions provided by the manufacturers. Gene expression was quantified by normalizing the levels to those of glyceraldehyde-3-phosphate dehydrogenase using the 2−ΔΔCt method.
Cell Lines and Small Interfering RNA
A549 and PC10 cells were purchased from the Health Science Research Resource Bank (Osaka, Japan) and Immuno-Biological Laboratories (Fujioka, Japan). Transfections were performed with Lipofectamine iMAX (Invitrogen) and Opti-MEM (Gibco, Grand Island, NY). Cells at 50% to 60% confluency in complete medium were subjected to serum starvation for 24 hours and treated with transforming growth factor (TGF)-β1 (5 ng/mL) for different time periods in the presence or absence of cell transfection. The small interfering-CADM1, small interfering-SPC25, and small interfering-negative control were synthesized by Ambion (Austin, TX).
Western Blotting
Protein extraction, two-dimensional polyacrylamide gel electrophoresis, and transfer to nitrocellulose membranes were performed as previously described.10,11 The antibody against CADM1 was purchased from Medical & Biological Laboratories (Woburn, MA). The antibody against SPC25 was purchased from Proteintech (Rosemont, IL). The antibody against CD133 was purchased from Abcam (Cambridge, United Kingdom). The antibodies against E-cadherin, VIM, AKT, phosphorylated-AKT, ERK, phosphorylated-ERK, phosphorylated-histone-H3, and cleaved PARP were purchased from Cell Signaling Technology (Danvers, MA). The antibody against glyceraldehyde-3-phosphate dehydrogenase was obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Immunohistochemical Staining
The sections were incubated with the following primary antibodies at the appropriate dilution: anti-CADM1 (1:50 dilution; Medical & Biological Laboratories) and rabbit polyclonal anti-SPC25 antibody (1:400 dilution; Proteintech). Subsequently, the sections were incubated with the Histofine Simple Stain MAX-PO Kit (Nichirei Biosciences, Tokyo, Japan) as the secondary antibody with peroxidase for 30 minutes. Peroxidase activity was detected using a solution of 3,3′-diaminobenzidine and hydrogen peroxide, with Mayer’s hematoxylin used as the counterstain. Immunohistochemistry (IHC) staining was evaluated by two independent board-certified pulmonologists using the H-score, as previously described.12
Cell Proliferation Assay and Treatment With Anticancer Drugs
Proliferation was evaluated at 72 hours. Cell proliferation was quantified using the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan), according to the instructions provided by the manufacturer. Cells (5.0 × 103 cells) were subjected to serum starvation for 24 hours before treatment with 10 μM carboplatin (CBDCA), paclitaxel (PTX), nintedanib, pemetrexed (PEM), 5-fluorouracil (5-FU), and 5-azacytidine (5-AZA) (Sigma-Aldrich Japan, Tokyo, Japan).
Statistical Analysis
All statistical analyses were performed using Statistical Package for the Social Sciences version 17.0 (SPSS Inc., Chicago, IL). The data are expressed as the mean plus or minus SD. Student’s t test was used to compare the values of the test and control samples. A p value less than 0.05 denoted a statistically significant difference. Kaplan–Meier curves of disease-free survival after initial surgery were generated using R version 4.0.0.
Results
WES and Data Mining
The characteristics of the 29 patients analyzed (comprising 19 IPF-LC and 10 non–IPF-LC) are illustrated in Table 1. Sex, smoking status, histology, pathologic stage, and respiratory function were not significantly different between the IPF-LC and non–IPF-LC groups. Disease-free survival in the IPF-LC group was significantly worse than that noted in the non–IPF-LC group (Fig. 1A). We performed WES using genomic DNA extracted from the 29 cancerous and noncancerous samples. We initially analyzed 8868 gene mutations using recursive feature elimination by random forests and deleted the lower 1% at every iteration (Fig. 1B and C). By training random forests and deleting unimportant mutations, we eventually identified 85 gene mutations associated with IPF-LC (Supplementary Fig. 1A and B). Of those, 25 mutations were specifically expressed in IPF-LC (Fig. 1B and D). The prognostic potential of these 25 genes in NSCLC was further evaluated using the Kaplan–Meier plotter. For 15 of those genes, differences in expression (i.e., high and low) resulted in significant differences in OS (Supplementary Fig. 2). We ultimately identified CADM1 and SPC25 as candidate gene mutations associated with IPF-LC. Gene mutations previously associated with IPF-LC (e.g., FHIT, TP53, and TP63, which are related to smoking or squamous cell carcinoma) were not significantly different between IPF-LC and non–IPF-LC (Supplementary Fig. 3). In tumor tissues extracted from patients with IPF-LC, CADM1 and SPC25 were mutated at a frequency of 47% (9 of 19) and 53% (10 of 19), respectively (Fig. 1E). Approximately one-third of the patients (7 of 19; 36%) with IPF-LC had both mutations (Fig. 1E). Interestingly, in fibrotic tissue extracted from patients with IPF-LC, CADM1 and SPC25 were mutated at a frequency of 26% (5 of 19) and 32% (6 of 19), respectively (Fig. 1E). In contrast, there was no mutation of CADM1 or SPC25 found in cancerous and noncancerous tissues obtained from patients without IPF-LC. Details of the CADM1 (c.1026-1027insACC) and SPC25 (c.551-4_551-2delCTA) mutations are found in Figure 1F and G. In one case, we were able to microscopically extract normal lung background, IPF, and tumor tissues from a single surgical sample. Regarding SPC25, both the IPF and tumor specimens had the mutation. Nevertheless, the normal lung background tissue was wild-type, which eliminates the possibility that SPC25 is a germline mutation.
Table 1.
Clinicopathologic Characteristics of Patients With and Without IPF-LC
| Variables | IPF-LC (n = 19) | Non–IPF-LC (n = 10) | p Value |
|---|---|---|---|
| Age at surgery, y | 71 (range: 56–84) | 68 (range: 43–84) | 0.10 |
| Sex | |||
| Male | 14 | 7 | 0.12 |
| Female | 5 | 3 | |
| Smoking status | |||
| Nonsmoker | 1 | 0 | 0.10 |
| Smoker | 17 | 9 | |
| Pack per ya | 515 ± 82.3 | 558 ± 168.0 | 0.79 |
| Histology | |||
| Nonsquamous | 8 | 2 | 0.10 |
| Squamous | 11 | 8 | |
| Pathologic stage | |||
| I | 10 | 7 | 0.17 |
| II + III | 9 | 3 | |
| %FEV1.0 | 72.3 (range: 54.9–84.5) | 72.5 (range: 54.8–82.3) | 0.51 |
| %DLCo | 79.5 (range: 66.1–107.3) | 89.5 (range: 46.7–143.9) | 0.28 |
Note: Tumor stages were evaluated according to the seventh American Joint Commission on Cancer guidelines. Histologic subtypes of lung cancer were defined according to the 1999 WHO Classification. The diagnosis of IPF was based on the American Thoracic Society/European Respiratory Society/Japanese Respiratory Society/Latin American Thoracic Association Statement.
DLCo, diffusing capacity for carbon monoxide; FEV1.0, forced expiratory volume in 1 second; IPF-LC, patients with lung cancer with idiopathic pulmonary fibrosis.
Data are presented as the mean ± SD. The p values were derived from the comparison between IPF-LC and non–IPF-LC using Student’s t test. Data are presented as numbers unless otherwise noted.
Figure 1.
Background and outline of the study. (A) DFS in the IPF-LC and non–IPF-LC groups. Kaplan–Meier curve revealing DFS in IPF-LC and non–IPF-LC. The p values were determined using the log-rank test. (B) Outline of the study design for the identification of candidate genes in IPF-LC. (C) Algorithm of the random forest method. (D) Of the 85 gene mutations identified, 40 were specifically observed in IPF-LC. The figure reveals the distribution of the 40 gene mutations in all 29 cases. The distribution of all 85 gene mutations is found in Supplementary Figure 1B. Red: both N/T mutated; yellow: T mutated. (E) Mutation status of CADM1 and SPC25 in all 29 cases. (F) Schema of CADM1 gene mutation (c.1026_1027insACC). (G) Schema of SPC25 gene mutation (c.551-4_551-2delCTA). Random forests are an ensemble machine learning method consisting of a multitude of decision trees. Random forests classify positive and negative samples and estimate the importance of each feature in classification problems. Using random forests, recursive feature elimination was applied to the 8868 mutations. Because we focused on the presence and absence of gene alternations, we substituted negative 1 for reference and positive 1 for Het or Homo and converted our data to a binary feature. DFS, disease-free survival; HGVS, Human Genome Variation Society; IPF-LC, patients with lung cancer with idiopathic pulmonary fibrosis; mt, mutant; N, normal; No., number; qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction; T, tumor; UTR, untranslated region; wt, wild-type.
Low CADM1 and High SPC25 Expression in the Tumor Site of IPF-LC
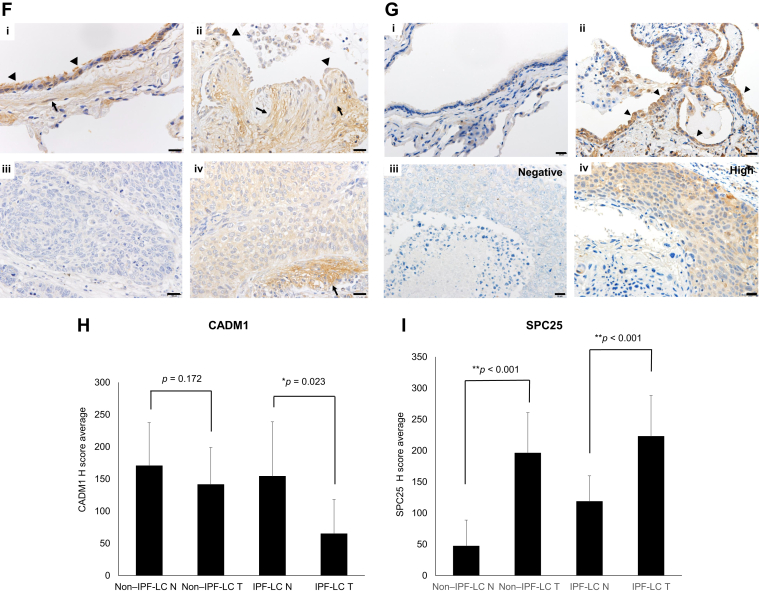
We next evaluated the correlation between the mutation status and expression of CADM1 and SPC25 genes. CADM1 is recognized as a tumor suppressor gene and frequently inactivated by promoter methylation. We performed a quantification analysis of CADM1 methylation using methylation-specific PCR coupled with bisulfite sequencing. Aberrant promoter methylation of the CADM1 gene was observed in cancerous and noncancerous tissues of both patients with and without IPF-LC, except for one IPF-LC case (Supplementary Fig. 4). We evaluated the expression of CADM1 in clinical samples through qRT-PCR. Its expression was lower in tumor tissues, particularly in the IPF-LC group (Fig. 2A). In addition, CADM1 mutants expressed lower levels of CADM1; however, the difference was not statistically significant (Fig. 2B). IPF-LC may occur through a two-hit inactivation of promoter methylation and CADM1 mutations. In contrast, SPC25 is recognized as an oncogene. The expression of SPC25 was higher in tumor tissues than the corresponding normal tissues. Notably, this increase was significantly greater in the IPF-LC group, as revealed by qRT-PCR (Fig. 2C). SPC25 mutants had higher SPC25 expression, whereas SPC25 wild-type cases had lower SPC25 expression; however, the difference was not statistically significant (Fig. 2D). Low CADM1 and high SPC25 expression levels were frequently found in IPF-LC. We next evaluated the prognostic and functional significance of low CADM1 and high SPC25 expression in LC. The Kaplan–Meier plotter was used to evaluate the prognostic potential of CADM1 and SPC25. The analysis indicated that, of a total of 1925 patients with LC (865 adenocarcinoma, 675 squamous cell carcinoma), patients with LC with lower CADM1 and higher SPC25 expression had shorter OS (Fig. 2E). Finally, we investigated the expression status of CADM1 and SPC25 by IHC analysis in 24 testable cases. Typical staining (positive and negative) is illustrated in Figure 2F and G. There was no correlation between the expression detected by qRT-PCR and IHC in individual cases. Nevertheless, when we evaluated the protein status (Histoscore, H-score) between the groups, the results were similar to those yielded by qRT-PCR. Of note, CADM1 expression was significantly lower in IPF-LC tumor tissues compared with IPF background and other non–IPF-LC tumor tissues and background (Fig. 2H). Regarding SPC25, the H-score was significantly higher in IPF tumor tissues compared with IPF and non-IPF tumor tissues and background (Fig. 2I). We next compared the H-score in tumor tissues of the IPF and non–IPF-LC groups. CADM1 expression was significantly lower in the IPF-LC group (Supplementary Fig. 5A). Regarding SPC25, IPF-LC cases had higher SPC25 expression; however, the difference was not statistically significant (Supplementary Fig. 5B).
Figure 2.
CADM1 and SPC25 expression in IPF-LC and non–IPF-LC. The mRNA expression of CADM1 and SPC25 was detected using RT-PCR. The changes in expression levels are represented as fold increase compared with the control. The p values were determined using Student’s t test. (A) CADM1 expression between non–IPF-LC and IPF-LC. (B) CADM1 expression between CADM1 mutants and wild-type cases. (C) SPC25 expression between non–IPF-LC and IPF-LC. (D) SPC25 expression between SPC25 mutants and wild-type cases. (E) Prognostic value of CADM1 and SPC25 derived from the Kaplan–Meier plotter. Data were derived from a total of 1925 patients with lung cancer (865 adenocarcinoma, 675 squamous cell carcinoma). The OS data were based on caArray, GSE 14814, GSE 19188, GSE 29013, GSE 30219, GSE 31210, GSE 3141, GSE 31908, GSE 37745, GSE 43580, GSE 4573, GSE 50081, GSE 8894, and TCGA. (F) Representative CADM1 protein expression in normal lung (i) and lung tissues from patients with associated UIP (ii) and NSCLC (iii, iv). Ciliated bronchiolar epithelium and smooth muscle cells had moderate intensity in normal lung (i). Regenerated epithelial cells (arrows) and fibroblasts (arrow heads) in fibroblastic foci from patients with UIP had weak-to-moderate staining (ii). Large cell neuroendocrine carcinoma had negative immunostaining (iii). Squamous cell carcinoma had moderate immunostaining, whereas interstitial cells (arrows) of the desmoplasia had intense immunostaining (iv). Scale bars represent 20 μm. (G) Representative SPC25 protein expression in normal lung (i) and lung tissues from patients with associated UIP (ii) and NSCLC (iii, iv). Metaplastic bronchiolar epithelial cells lining honeycomb lungs had increased signals (arrow heads) (ii) compared with the controls (i) and NSCLC (iv). The staining status is indicated (iii: negative; iv: high). Scale bars represent 20 μm. (H) The average H-score of CADM1 between non–IPF-LC and IPF-LC. (I) The average H-score of SPC25 between non–IPF-LC and IPF-LC. CADM1 and SPC25 expression levels were scored using the following scale: no expression, 0; low expression, 1+; and high expression, 2+ and 3+. The score was based on the fraction of positive cells (0%–100%). The total score was calculated by multiplying the intensity score and the fraction score, producing a total range of 0 to 300. GSE, Gene Expression Omnibus Series; H-score, histoscore; HR, hazard ratio; IPF-LC, patients with lung cancer with idiopathic pulmonary fibrosis; mt, mutant; N, normal; OS, overall survival; RT-PCR, reverse-transcriptase polymerase chain reaction; T, tumor; TCGA, The Cancer Genome Atlas; UIP, usual interstitial pneumonia; wt, wild-type.
TGF-β1 Signal Involved in CADM1 and SPC25 Expression
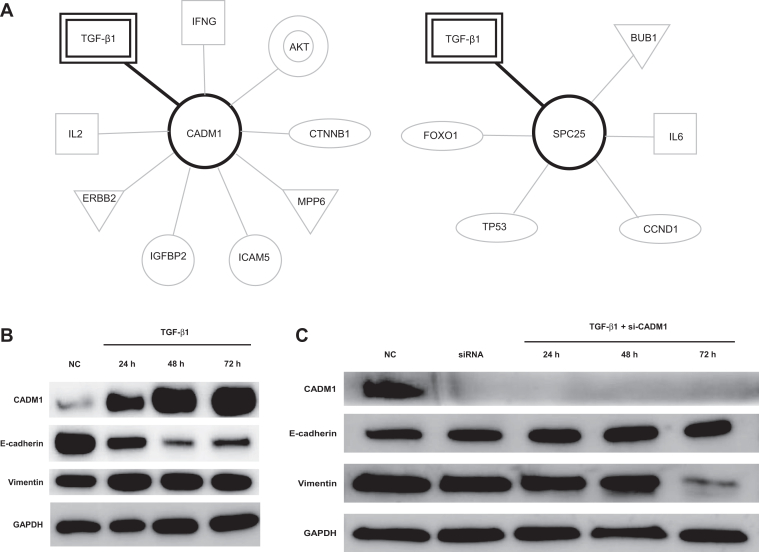
We performed ingenuity pathway analysis to clarify the signaling pathway associated with CADM1 and SPC25. It was found that TGF-β1 was a common signaling pathway involved in CADM1 and SPC25 expression (Fig. 3A). TGF-β1 signaling, a major profibrotic pathway and mediator of epithelial-to-mesenchymal transition (EMT), also provides mitogenic stimuli to LC cells.
Figure 3.
CADM1 and SPC25 involved in TGF-β1 signaling. (A) CADM1 and SPC25 IPA pathway. Pathway analysis revealed that both CADM1 and SPC25 are involved in TGF-β1 signaling. (B) Exposure to TGF-β1 induced CADM1 expression and EMT. A549 cells were incubated with TGF-β1 (5 ng/mL) for different time periods. Exposure to TGF-β1 induced CADM1 expression. The expression of epithelial marker E-cadherin was down-regulated by TGF-β1 stimulation in a time-dependent manner. The expression of mesenchymal marker VIM was up-regulated. (C) CADM1 inhibition suppressed the TGF-β1–induced EMT. Pooled synthetic siRNA duplexes targeting CADM1 were transfected into A549 cells (25 nM). At 24 h after transfection, the cells were stimulated with TGF-β1 (5 ng/mL) in serum-free Opti-MEM for a further 72 h before harvest. CADM1 inhibition suppressed TGF-β1–induced EMT. (D) CADM1 inhibition activated cell proliferation. A549 cells were transfected with siRNA duplexes targeting CADM1 (25 nM). CADM1 inhibition activated cell proliferation. (E) Exposure to TGF-β1 induced SPC25 expression. A549 cells were incubated with TGF-β1 (5 ng/mL) for different time periods. Exposure to TGF-β induced SPC25 expression. (F) SPC25 inhibition suppressed the TGF-β1–induced EMT and proliferation. Pooled synthetic siRNA duplexes targeting SPC25 were transfected into A549 cells (25 nM). At 24 h after transfection, the cells were stimulated with TGF-β1 (5 ng/mL) in serum-free Opti-MEM for a further 72 h before harvest. (G) At 72 h after transfection with si-SPC25, CCK-8 solution (10 μL) was added to each well to evaluate cell proliferation using the MTT assay. Inhibition of SPC25 suppressed cell proliferation. CCK-8, Cell Counting Kit—8; EMT, epithelial-to-mesenchymal transition; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; h, hour; IFNG, interferon gamma; IL2, interleukin 2; IL6, interleukin 6; IPA, ingenuity pathway analysis; IPF-LC, patients with lung cancer patients with idiopathic pulmonary fibrosis; MEM, minimal essential medium; NC, negative control; p-AKT, phosphorylated-AKT; p-ERK, phosphorylated-ERK; si-CADM1, small interfering-CADM1; siRNA, small interfering RNA; si-SPC25, small interfering-SPC25.
We evaluated the correlation between TGF-β1 signaling and CADM1 expression. CADM1 expression was up-regulated after stimulation with TGF-β1 and accompanied by decreased E-cadherin and increased VIM expression (Fig. 3B). Inhibition of CADM1 by small interfering-CADM1 suppressed TGF-β1–induced EMT in A549 cells (Fig. 3C). Furthermore, CADM1 knockdown markedly accelerated the proliferation of A549 cells (Fig. 3D). SPC25 was also overexpressed in response to the EMT phenomenon after exposure to TGF-β1 (Fig. 3E). Knockdown of SPC25 using small interfering RNA increased the expression of E-cadherin and decreased that of VIM, revealing TGF-β1–induced EMT (Fig. 3F). In addition, inhibition of SPC25 reduced the TGF-β1–induced expression of CD133, which is associated with cancer stem cell (CSC) properties (Fig. 3F). Moreover, knockdown of SPC25 by small interfering RNA strongly inhibited the proliferation of A549 cells, suggesting SPC25 is a therapeutic target for IPF-LC (Fig. 3G).
Therapeutic Agent for SPC25 in IPF-LC
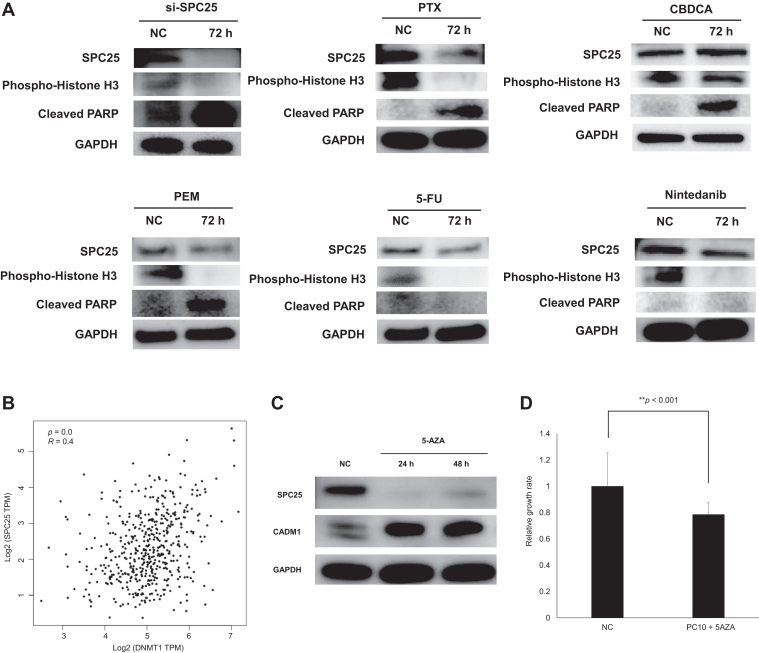
Finally, we attempted to identify therapeutic agents for IPF-LC. SPC25 is involved in kinetochore-microtubule interactions and spindle checkpoint activity associated with G2/M arrest. In fact, inhibition of SPC25 by small interfering-SPC25 suppressed phospho-histone-H3 (indicating G2/M arrest) and up-regulated cleaved PARP (indicating apoptotic activity) (Fig. 4A). We evaluated whether SPC25 expression is reduced by treatment with standard agents used in patients with NSCLC and IPF, including the microtubule inhibitor PTX, CBDCA, PEM, 5-FU, and nintedanib. Treatment with PEM, 5-FU, and nintedanib slightly decreased the expression of SPC25 (Fig. 4A). Increased levels of cleaved PARP were observed after treatment with CBDCA and PEM (Fig. 4A). In contrast, treatment with PTX significantly suppressed the expression of SPC25 and phospho-histone H3, whereas it increased that of cleaved PARP (Fig. 4A). These results suggest that the combination of PTX and CBDCA may be a reasonable treatment option for IPF-LC.
Figure 4.
Effects of therapeutic agents on SPC25 in IPF-LC. (A) Effects of therapeutic agents on SPC25 in IPF-LC. A549 cells were transfected with pooled synthetic siRNA duplexes targeting SPC25 (25 nM). A549 cells were treated with 10 μM PTX, CBDCA, PEM, 5-FU, or nintedanib. Treatment with PTX, PEM, 5-FU, and nintedanib significantly down-regulated the expression of SPC25; however, this effect was not observed after treatment with CBDCA. (B) Correlation between SPC25 and DNMT1. GEPIA2 database analysis revealed that the expression levels of SPC25 and DNMT1 were positively correlated in NSCLC. (C) 5-AZA suppressed the expression of SPC25 and increased that of CADM1. PC-10 cells were treated with 5-AZA (10 μM). Treatment of PC-10 cells with 5-AZA down-regulated the expression of SPC25 and reactivated CADM1. (D) Treatment with 5-AZA suppressed the proliferation of PC-10 cells. 5-AZA, 5-azacytidine; 5-FU, 5-fluorouracil; CBDCA, carboplatin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GEPIA2, Gene Expression Profiling Interactive Analysis 2; h, hour; IPF-LC, patients with lung cancer with idiopathic pulmonary fibrosis; NC, negative control; PEM, pemetrexed; PTX, paclitaxel; siRNA, small-interfering RNA; si-SPC25, small interfering-SPC25; TPM, transcript count per million.
Treatment of CSCs with DNMT1 inhibitor and HDAC inhibitor revealed that inhibition of chromatin modifiers repressed growth-promoting signaling molecules, such as SPC25.13 Using evidence from the Gene Expression Profiling Interactive Analysis 2 database, a positive correlation between SPC25 and DNMT1 expression was found in NSCLC (Fig. 4B). Treatment of PC-10 LC cells with DNMT1 inhibitor 5-AZA successfully reduced the expression of SPC25 and reactivated CADM1 (Fig. 4C). Treatment with 5-AZA significantly inhibited the proliferation of PC-10 cells (Fig. 4D). Hence, 5-AZA may be a promising therapeutic agent for IPF-LC.
Discussion
Using WES and medical-engineering cooperation machine learning methods, we identified that CADM1 and SPC25 gene mutations were associated with IPF-LC.
Of the IPF-LC cases, 36% had both CADM1 and SPC25 mutations. Interestingly, one-third of the patients with IPF-LC had CADM1 and SPC25 mutations in noncancerous fibrotic tissues. Pathway analysis revealed that CADM1 and SPC25 mutations are involved in TGF-β1 signaling. TGF-β1 is a multifunctional cytokine that inhibits epithelial cell proliferation, induces EMT, and activates the development of both LC and pulmonary fibrosis.14 There is a correlation between decreased susceptibility to the antiproliferative effects of TGF-β1 and malignant progression. TGF-β1 is highly expressed in the lung tissues of patients with IPF.15,16 Tumor cells often escape the antiproliferative effects of TGF-β1 by mutational inactivation or dysregulated expression of components of the TGF-β1 signaling pathway.17 Therefore, loss of the growth inhibitory response to TGF-β1 may be crucial in promoting tumor development in the lungs of patients with IPF, where TGF-β1 is highly expressed. In the TGF-β1 signaling pathway in IPF-LC, CADM1 and SPC25 may be key molecules for the development of lung neoplasms.
CADM1 is located on a region of chromosome 11q and encodes a transmembrane protein of the immunoglobulin superfamily involved in cell-cell interactions between epithelial cells.18,19 As a tumor suppressor gene, reduced expression of CADM1 or promoter methylation has been found in lung, cervical, nasopharyngeal, esophageal, and breast cancers.20,21 CADM1 methylation was also associated with pack-years in smokers.22 Therefore, we can hypothesize that tobacco smoking induces aberrant CADM1 gene methylation in the precursors or tumor cells of NSCLC. In addition to DNA methylation in patients with IPF, CADM1 mutations may be involved in carcinogenesis. The function of the CADM1 gene mutation (c.1026_1027insACC) identified in this study remains unclear. We hypothesized that the insertion of ACC may have caused a frameshift, resulting in the shedding of the extracellular domain of the transmembrane protein CADM1 and a reduction in CADM1 expression.
SPC25 is a protein involved in kinetochore-microtubule interactions and spindle checkpoint activity.23 Carcinogenesis can result from genetic instability in the cell cycle owing to incorrect chromosome segregation, and kinetochores play an important role in this process.23 Recent studies have reported that SPC25 is overexpressed in LC and associated with carcinogenesis and metastasis.24 SPC25 gene mutation (c.551-4_551-2delCTA) with a deletion of CTA at the splice site of SPC25 has been detected. We hypothesized that this splice site mutation may have left one or more introns in the mature RNA, leading to aberrant production of SPC25. Overexpression of SPC25 has been implicated in carcinogenesis in IPF-LC and may be an appropriate therapeutic target. IPF-LC is considered a high-risk condition, is linked to poor prognosis, and has been excluded from clinical trials.3,6,7 Combination of PTX (a microtubule inhibitor) with CBDCA is one of the treatment options for IPF-LC.3,5 In this study, we revealed that SPC25 is regulated by the administration of PTX. PTX reduced the expression of SPC25, resulting in the induction of G2/M arrest and apoptotic activity. CBDCA also induced apoptotic activity. These results confirm that the combination of PTX with CBDCA is a key regimen for the treatment of IPF-LC.
We have revealed the promising potential of the DNMT inhibitor 5-AZA as an agent in the treatment of IPF-LC. 5-AZA is an effective demethylating agent that can inhibit cell cycle progression, induce apoptosis, promote differentiation, and reduce tumor cell invasion and metastasis.25 Previous studies have revealed that transformation of immortalized bronchial epithelial cells owing to exposure to carcinogens is associated with increased DNMT1 protein expression and de novo methylation of tumor suppressor genes.26 In addition, DNMT1 plays an important role in the maintenance of mammary stem and progenitor cells and CSCs. In combination with HDAC inhibitors, the DNMT inhibitor 5-AZA significantly reduced the presence of CSCs and prolonged OS in a mouse model of mammary tumors.27 It has been found that treatment of CSCs with 5-AZA and HDAC inhibitors inhibits chromatin-modifying factors, such as SPC25. We confirmed that SPC25 expression was reduced after the administration of 5-AZA in LC cells. We also successfully reactivated the expression of CADM1. Hence, 5-AZA may be an effective treatment option for IPF-LC.
This study had several limitations. First, the sample size of IPF-LC was small. Only patients with good lung function were enrolled. Second, the biological significance of CADM1 (c.1026_1027insACC) and SPC25 (c.551-4_551-2delCTA) remains unclear. Nevertheless, we are currently working on gene editing and plan to perform further functional analysis in the future.
In summary, we analyzed the genomic profile of IPF-LC and identified relevant mutations in the CADM1 and SPC25 genes. Functional analyses revealed that CADM1 and SPC25 are potential diagnostic and therapeutic markers for IPF-LC. It is essential to validate the clinical significance of CADM1 and SPC25 in an independent cohort of patients.
CRediT Authorship Contribution Statement
Aya Fukuizumi: Conceptualization, Investigation, Writing - original draft.
Rintaro Noro, Masahiro Seike: Conceptualization, Writing - review & editing, Supervision.
Akihiko Miyanaga, Yuji Minegishi: Conceptualization.
Miwako Omori, Mamiko Hirao, Kuniko Matsuda: Validation.
Shinobu Kunugi: Formal analysis, Resources.
Kazutaka Nishiwaki, Masahiro Morimoto, Haruka Motohashi, Hayato Ohwada: Software, Formal analysis.
Jitsuo Usuda: Resources.
Akihiko Gemma: Supervision.
Acknowledgments
This study was supported in part by the Clinical Rebiopsy Bank Project for Comprehensive Cancer Therapy Development from the Ministry of Education, Culture, Sports, Science and Technology Supported Program for the Strategic Research Foundation at Private Universities (grant S1311022 to Gemma and Seike). The authors thank Mrs. C. Hoshino of Nippon Medical School (Tokyo, Japan) for excellent assistance in sample collection.
Footnotes
Disclosure: Seike reports receiving honoraria and research funding from Boehringer Ingelheim Pharmaceuticals, Taiho Pharmaceutical, and Eli Lilly Japan K.K. Usuda reports receiving research funding from Taiho Pharmaceutical and Eli Lilly Japan K.K. Gemma reports receiving honoraria from Boehringer Ingelheim Pharmaceuticals. The remaining authors declare no conflict of interest.
Cite this article as: Fukuizumi A, Noro R, Seike M, et al. CADM1 and SPC25 gene mutations in lung cancer patients with idiopathic pulmonary fibrosis. JTO Clin Res Rep. 2021;2:100232.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org/ and at https://doi.org/10.1016/j.jtocrr.2021.100232.
Supplementary Data
References
- 1.Ozawa Y., Suda T., Naito T. Cumulative incidence of and predictive factors for lung cancer in IPF. Respirology. 2009;14:723–728. doi: 10.1111/j.1440-1843.2009.01547.x. [DOI] [PubMed] [Google Scholar]
- 2.Tomassetti S., Gurioli C., Ryu J.H. The impact of lung cancer on survival of idiopathic pulmonary fibrosis. Chest. 2015;147:157–164. doi: 10.1378/chest.14-0359. [DOI] [PubMed] [Google Scholar]
- 3.Fukuizumi A., Minegishi Y., Omori M. Weekly paclitaxel in combination with carboplatin for advanced non-small-cell lung cancer complicated by idiopathic interstitial pneumonias: a single-arm phase II study. Int J Clin Oncol. 2019;24:1543–1548. doi: 10.1007/s10147-019-01516-9. [DOI] [PubMed] [Google Scholar]
- 4.Minegishi Y., Kuribayashi H., Kitamura K. The feasibility study of carboplatin plus etoposide for advanced small cell lung cancer with idiopathic interstitial pneumonias. J Thorac Oncol. 2011;6:801–807. doi: 10.1097/JTO.0b013e3182103d3c. [DOI] [PubMed] [Google Scholar]
- 5.Minegishi Y., Gemma A., Homma S. Acute exacerbation of idiopathic interstitial pneumonias related to chemotherapy for lung cancer: nationwide surveillance in Japan. ERJ Open Res. 2020;6:00184–2019. doi: 10.1183/23120541.00184-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimoto T., Okazaki T., Matsukura T. Operation for lung cancer in patients with idiopathic pulmonary fibrosis: surgical contraindication? Ann Thorac Surg. 2003;76:1674–1678. doi: 10.1016/s0003-4975(03)00966-4. [DOI] [PubMed] [Google Scholar]
- 7.Tzouvelekis A., Gomatou G., Bouros E., Trigidou R., Tzilas V., Bouros D. Common pathogenic mechanisms between idiopathic pulmonary fibrosis and lung cancer. Chest. 2019;156:383–391. doi: 10.1016/j.chest.2019.04.114. [DOI] [PubMed] [Google Scholar]
- 8.Uematsu K., Yoshimura A., Gemma A. Aberrations in the fragile histidine triad (FHIT) gene in idiopathic pulmonary fibrosis. Cancer Res. 2001;61:8527–8533. [PubMed] [Google Scholar]
- 9.Takenaka K., Gemma A., Yoshimura A. Reduced transcription of the Smad4 gene during pulmonary carcinogenesis in idiopathic pulmonary fibrosis. Mol Med Rep. 2009;2:73–80. doi: 10.3892/mmr_00000064. [DOI] [PubMed] [Google Scholar]
- 10.Seike M., Goto A., Okano T. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugano T., Seike M., Noro R. Inhibition of ABCB1 overcomes cancer stem cell-like properties and acquired resistance to MET inhibitors in non-small cell lung cancer. Mol Cancer Ther. 2015;14:2433–2440. doi: 10.1158/1535-7163.MCT-15-0050. [DOI] [PubMed] [Google Scholar]
- 12.Zou F., Seike M., Noro R., Kunugi S., Kubota K., Gemma A. Prognostic significance of ABCB1 in stage I lung adenocarcinoma. Oncol Lett. 2017;14:313–321. doi: 10.3892/ol.2017.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathania R., Ramachandran S., Elangovan S. DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nat Commun. 2015;6:6910. doi: 10.1038/ncomms7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eser P.Ö., Jänne P.A. TGFβ pathway inhibition in the treatment of non-small cell lung cancer. Pharmacol Ther. 2018;184:112–130. doi: 10.1016/j.pharmthera.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Khalil N., O’Connor R.N., Unruh H.W. Increased production and immunohistochemical localization of transforming growth factor-beta in idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol. 1991;5:155–162. doi: 10.1165/ajrcmb/5.2.155. [DOI] [PubMed] [Google Scholar]
- 16.Khalil N., O’Connor R.N., Flanders K.C., Unruh H. TGF- beta 1, but not TGF- beta 2 or TGF- beta 3, is differentially present in epithelial cells of advanced pulmonary fibrosis: an immunohistochemical study. Am J Respir Cell Mol Biol. 1996;14:131–138. doi: 10.1165/ajrcmb.14.2.8630262. [DOI] [PubMed] [Google Scholar]
- 17.de Caestecker M., Piek E., Roberts A.B. Role of transforming growth factor- beta signaling in cancer. J Natl Cancer Inst. 2000;92:1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- 18.Gomyo H., Arai Y., Tanigami A. A 2-Mb sequence-ready contig map and a novel immunoglobulin superfamily gene IGSF4 in the LOH region of chromosome 11q23.2. Genomics. 1999;62:139–146. doi: 10.1006/geno.1999.6001. [DOI] [PubMed] [Google Scholar]
- 19.Masuda M., Yageta M., Fukuhara H. The tumor suppressor protein TSLC1 is involved in cell-cell adhesion. J Biol Chem. 2002;277:31014–31019. doi: 10.1074/jbc.M203620200. [DOI] [PubMed] [Google Scholar]
- 20.Murakami Y., Nobukuni T., Tamura K. Localization of tumor suppressor activity important in nonsmall cell lung carcinoma on chromosome 11q. Proc Natl Acad Sci U S A. 1998;95:8153–8158. doi: 10.1073/pnas.95.14.8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukami T., Fukuhara H., Kuramochi M. Promoter methylation of the TSLC1 gene in advanced lung tumors and various cancer cell lines. Int J Cancer. 2003;107:53–59. doi: 10.1002/ijc.11348. [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi S., Yamada D., Fukami T. Hypermethylation of the TSLC1/IGSF4 promoter is associated with tobacco smoking and a poor prognosis in primary nonsmall cell lung carcinoma. Cancer. 2006;106:1751–1758. doi: 10.1002/cncr.21800. [DOI] [PubMed] [Google Scholar]
- 23.Tooley J., Stukenberg P.T. The Ndc80 complex: integrating the kinetochore’s many movements. Chromosome Res. 2011;19:377–391. doi: 10.1007/s10577-010-9180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong J., Keum S., Kim D. Spindle pole body component 25 homolog expressed by ECM stiffening is required for lung cancer cell proliferation. Biochem Biophys Res Commun. 2018;500:937–943. doi: 10.1016/j.bbrc.2018.04.205. [DOI] [PubMed] [Google Scholar]
- 25.Chai G., Li L., Zhou W. HDAC inhibitors act with 5-aza-2'-deoxycytidine to inhibit cell proliferation by suppressing removal of incorporated abases in lung cancer cells. PLoS One. 2008;3:e2445. doi: 10.1371/journal.pone.0002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damiani L.A., Yingling C.M., Leng S., Romo P.E., Nakamura J., Belinsky S.A. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res. 2008;68:9005–9014. doi: 10.1158/0008-5472.CAN-08-1276. [DOI] [PubMed] [Google Scholar]
- 27.Pathania R., Ramachandran S., Mariappan G. Combined inhibition of DNMT and HDAC blocks the tumorigenicity of cancer stem-like cells and attenuates mammary tumor growth. Cancer Res. 2016;76:3224–3235. doi: 10.1158/0008-5472.CAN-15-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.