Abstract
Madurella mycetomatis is the commonest cause of eumycetoma in Sudan and other countries in tropical Africa. Currently, the early diagnosis of mycetoma is difficult. In attempting to improve the identification of M. mycetomatis and, consequently, the diagnosis of mycetoma, we have developed specific oligonucleotide primers based on the sequence of the internal transcribed spacer (ITS) regions spacing the genes encoding the fungal ribosomal RNAs. The ITS regions were amplified with universal primers and sequenced, and then two sets of species-specific primers were designed which specifically amplify parts of the ITS and the 5.8S ribosomal DNA gene. The new primers were tested for specificity with DNA isolated from human mycetoma lesions and DNA extracted from cultures of M. mycetomatis reference strains and related fungi as well as human DNA. To study the genetic variability of the ITS regions of M. mycetomatis, ITS amplicons were obtained from 25 different clinical isolates and subjected to restriction fragment length polymorphism (RFLP) analysis with CfoI, HaeIII, MspI, Sau3AI, RsaI, and SpeI restriction enzymes. RFLP analysis of the ITS region did not reveal even a single difference, indicating the homogeneity of the isolates analyzed during the current study.
Mycetoma is a chronic granulomatous subcutaneous infection caused by true fungi (eumycetoma) or higher bacteria of the genus Actinomadura (actinomycetoma) (6, 15). The disease is endemic in tropical and subtropical areas and is the major mycological health problem in Sudan (6, 15). The majority of cases of mycetoma in Sudan are caused by Madurella mycetomatis. The pathogens involved are found in the environment in certain types of soil and are directly inoculated into the subcutaneous tissues, commonly in the foot, through minor trauma or a thorn prick (9, 14). Mycetoma has a prolonged, progressive, and indolent course and, if untreated, ultimately leads to destruction of deeper tissues and bone, resulting in deformity and disability which may necessitate amputation. The triad of a subcutaneous painless mass, sinuses, and grains discharged through the sinuses is the hallmark of mycetoma (6, 15). Diagnosis may be more difficult in early stages, especially prior to the appearance of the sinuses and grains. At this stage the disease may be difficult to distinguish from a variety of soft tissue tumors and granulomata (11, 23).
Currently, the available diagnostic tools for mycetoma are few and have many limitations. Diagnosis is based on the identification of the grains in the discharge of the sinuses or biopsies from the lesions. Staining grains allows the identification of the causative organism, but this is of limited value in the differentiation of true fungi (19). Culture is always necessary for definitive diagnosis. However, culturing clinical specimens is cumbersome, time-consuming, prone to secondary bacterial contamination (1), always needs deep surgical biopsy under general anesthesia, and is not always practical or cost-effective in areas of endemicity (19). Serodiagnosis is hampered by cross-reactivity among the multiple species of actinomycetes as well as by the lack of standardized antigen preparations (8, 20).
Comparative studies of the nucleotide sequences of the rRNA genes provide a means for analyzing phylogenetic relationships over a wide range of taxonomic levels and to assist in the development of identification assays for fungal species (24). The small-subunit ribosomal rDNA sequences evolve relatively slowly and are useful for studying distantly related organisms (24). The internal transcribed spacer (ITS) regions 1 and 2 can be amplified with the primers ITS5 and ITS4, which are located in the conserved regions of the 18S and 28S genes, respectively. Together with the intergenic spacer of the nuclear rRNA repeat units, this region evolves faster and may vary among the different species within a genus or even among cells found in a single population (24). rDNA sequences have been utilized by many investigators for the determination of species identity for a multitude of yeasts and fungi (2, 7, 10, 12, 25). It has also been reported that ITS ribotyping is a simple method that can distinguish among most of the Saccharomyces species (17), and ITS sequences can differentiate between the closely related strains in the Trichophyton mentagrophytes complex (16).
The aim of the present study was to supplement current diagnostic tools for mycetoma with the development of a species-specific PCR assay for identification of M. mycetomatis, based on the nucleotide sequence of the ITS regions in the rDNA operon. The study also addressed the taxonomic position of the causative organisms and tested for the genetic variability of different M. mycetomatis isolates.
MATERIALS AND METHODS
Clinical specimens.
Clinical specimens were collected from 48 consecutive patients presenting with black grain mycetoma at the Mycetoma Research Center, University of Khartoum, Sudan, during the period between November 1997 and August 1998. The patients were from different regions of the country. Following written consent from the patients, deep-excision biopsy specimens with visible grains were collected.
Fungal isolates.
For isolation of the fungus, some grains were collected from the biopsy specimens, washed twice in physiological saline containing 1% chloramphenicol, inoculated into Sabouraud’s agar (Difco, Amsterdam, The Netherlands), and incubated at 37°C for 3 to 4 weeks. Potential M. mycetomatis cultures were identified morphologically, and the fungal mycelia were scraped and stored in 9% glycerol broth at −80°C and shipped to the Department of Medical Microbiology and Infectious Diseases, Erasmus University Medical Center Rotterdam, Rotterdam, The Netherlands. Control strains from related fungal species were obtained from the Centraalbureau voor Schimmelcultures, The Netherlands. The following eight reference strains were included: Madurella grisea CBS 331-50 and CBS 332-50, M. mycetomatis CBS 247.48 and CBS 868.95, Pyrenochaeta mackinnonii CBS 674.75, Pyrenochaeta romeri CBS 252.60, Pyrenochaeta unguis-homini CBS 378.92, and Chaetosphaeronema larense CBS 640.73.
DNA extraction and purification.
Prior to DNA extraction the fungi were subcultured on Sabouraud’s agar and incubated at 37°C for 3 weeks. The mycelia were scraped from the culture medium and homogenized with sterile pestles and a mortar. The homogenized mycelia were then snap frozen in liquid nitrogen, thawed and refrozen twice, and rehomogenized in 2 ml of lysis buffer containing 4 M guanidinium isothiocyanate, 0.1 M Tris-HCl (pH 6.4), 0.2 M EDTA, and 0.1% Triton X-100. The DNA was purified by Celite affinity chromatography (Janssen Pharmaceuticals, Beerse, Belgium) as described before (3). Two other DNA purification protocols were tested for the destruction of the cell wall of the fungus, using either lysis buffer with lyticase enzymes followed by proteinase K treatment (21) or cetyltrimethylammonium bromide (Janssen Pharmaceuticals) buffer at 56°C (22).
PCR amplification.
DNA extracts of 25 different M. mycetomatis isolates were amplified with primers ITS4 and ITS5 (24). The sequences of the two primers were 5′-TCCTCCGCTTATTGATATGC-3′ and 5′-GGAAGTAAAAGTCGTAACAAGG-3′, respectively. The PCRs were performed in 50-μl reaction volumes containing 0.2 U of Taq polymerase (Super Taq; HT Biotechnology, Cambridge, United Kingdom) and 5 ng of template DNA. Cycling was performed in a model 60 thermocycler (Biomed, Theres, Germany) with the following temperature trajectory: 40 cycles of alternating denaturation (94°C for 1 min), annealing of primers (58°C for 1 min), and enzymatic extension by the thermostable polymerase (72°C for 2 min). The PCR products were examined by electrophoresis in 1% agarose gels stained with ethidium bromide.
Cloning, sequencing, and primer design.
Amplimers obtained with primers ITS4 and ITS5 were cloned into the plasmid pCRII with the Topo-TA cloning kit (Invitrogen, Leek, The Netherlands) and sequenced commercially (Eurogentec, Seraing, Belgium). The sequences obtained were aligned, adjusted, and compared for homology with the sequences derived from other species and deposited in the various databases. Two potentially M. mycetomatis-specific sets of primers were designed (primers 26.1A [5′-AATGAGTTGGGCTTTAACGG-3′] and 28.3A [5′-TCCCGGTAGTGTAGTGTCCCT-3′] and primers 26.1B [5′-GCAACACGCCCTGGGCGA-3′] and 28.3B [5′-TCCGCGGGGCGTCCGCCGGA-3′]). The newly designed primers were tested for sensitivity and specificity with a PCR protocol that was identical to the one described above except that the extension step was shortened to 1 min.
Detection of ITS polymorphisms.
Restriction fragment length polymorphism (RFLP) in the M. mycetomatis ITS regions was assessed by analysis of the PCR products generated by primers ITS4 and ITS5 with CfoI, MspI, HaeIII, RsaI, Sau3A, and SpeI restriction enzymes (Boehringer-Mannheim, Mannheim, Germany). The enzymes were used as recommended by the manufacturer. RFLP was determined by electrophoresis in 3% Nusieve GTG agarose gels (Biozym, Landgraaf, The Netherlands).
Nucleotide sequence accession number.
The sequence of the M. mycetomatis ITS region has been deposited in GenBank under accession no. AF162133.
RESULTS
From the 48 patients, 45 isolates were successfully cultured. Of these 45 isolates, only 25 survived storage at −80°C and transportation from the Sudan to The Netherlands. DNA isolation from fungal mycelia was initially performed in a comparative fashion. The three different protocols used for DNA purification gave virtually the same yield of DNA.
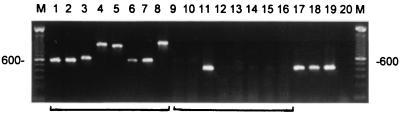
The amplified DNA fragments of the ITS regions (with ITS4 and ITS5 primers) of M. mycetomatis and the control strains were found to be between 600 and 1,200 bp in length (Fig. 1). Note that there appears to be a small difference in length when the amplicons obtained for the two M. mycetomatis reference strains are compared. PCR products of identical size (approximately 630 bp) were obtained when DNA extracted from the clinical isolates of M. mycetomatis was amplified (result not shown). The size of this fragment equaled that obtained for M. mycetomatis CBS 247.48. Cloning and sequencing of this type of ITS fragment for M. mycetomatis revealed that the fragment was 624 bp in length, which includes the ITS4 and ITS5 primers. The potentially species-specific primer sets 26.1A and 28.3A and 26.1B and 28.3B were found to be specific for M. mycetomatis DNA (Fig. 1). When the 26.1A-28.3A combination was used, a fragment of about 420 bp was synthesized, which is in agreement with expectations on the basis of the ITS nucleotide sequence. Similar-sized fragments were obtained as well when DNA isolated from the clinical strains was used as a template (n = 25). The primers did not amplify human DNA.
FIG. 1.
PCR amplification of the ITS for M. mycetomatis and related species. Lanes 1 to 8 show the amplicons obtained by using the universal primers ITS4 and ITS5. DNAs from the following species were amplified: M. grisea CBS331-50 (lane 1) and CBS 332-50 (lane 2), M. mycetomatis CBS 247.48 (lane 3) and CBS 868.95 (lane 4), P. mackinnonii CBS 674.75 (lane 5), P. romeri CBS 252.60 (lane 6), P. unguis-homini CBS 378.92 (lane 7), and C. larense CBS 640.73 (lane 8). In lanes 9 to 16 PCR results obtained with the primer combination 26.1A and 28.3A are displayed; the strains are ordered similarly from left to right. Note that only M. mycetomatis CBS 247.48 yielded a positive signal. Lanes 17 to 19 show the representative PCR products obtained for all of the clinical M. mycetomatis isolates. Lane 20 contains the negative PCR control. On the left and right the molecular size of the intensely fluorescing 600-bp-long fragment in the 100-bp ladder is indicated.
When the sequences determined for the ITS regions of two different M. mycetomatis isolates were compared, they were found to be identical with only minor ambiguities in two nucleotide positions. Running the M. mycetomatis spacer sequence through the GenBank data depository did not highlight any closely related sequence homologues from other fungal species. Ranking of homologous sequences indicated that the ITS sequence as determined for a nonspeciated isolate from the genus Phialophora came closest to the Madurella sequence. However, the homology appeared to be mainly restricted to the ribosomal gene sequences. In ITS1 several mutations were documented, whereas in ITS2 a 110-nucleotide stretch showing no significant homology was encountered. Consequently, the generation of informative parsimony trees will have to await the determination of ITS sequences for more closely related fungal species, such as the ones used in the control panel described above.
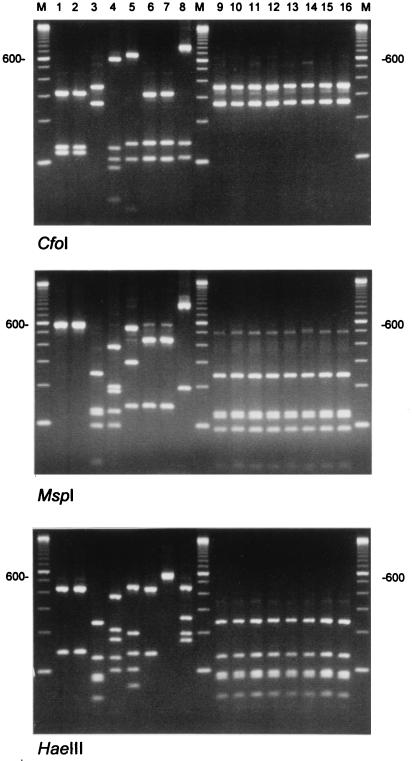
The RFLP pattern generated by six different restriction enzymes produced banding profiles that were in full agreement with the nucleotide sequence. The RFLP patterns generated for the control species were clearly different (Fig. 2). The two M. grisea isolates could not be discriminated but clearly differed from the M. mycetomatis isolates. Furthermore, the three species of Pyrenochaeta could be distinguished, since P. romeri and P. unguis-homini differed only in a single HaeIII site (Fig. 2, bottom panel). The RFLP pattern obtained for M. mycetomatis (CBS 247.48) matched the RFLP pattern of the clinical isolates. In all assays, M. mycetomatis (CBS 868.95) showed profiles completely different from those of all other M. mycetomatis strains, raising doubts about the species status of this isolate (see Discussion).
FIG. 2.
RFLP analysis of the ITS amplicons obtained for M. mycetomatis and related species. Lanes 1 to 8 show the results obtained for M. grisea CBS331-50 (lane 1) and CBS 332-50 (lane 2), M. mycetomatis CBS 247.48 (lane 3) and CBS 868.95 (lane 4), P. mackinnonii CBS 674.75 (lane 5), P. romeri CBS 252.60 (lane 6), P. unguis-homini CBS 378.92 (lane 7), and C. larense CBS 640.73 (lane 8). The three panels were generated with the restriction enzymes indicated. Lanes 9 to 16 show the results obtained for eight of the clinical M. mycetomatis isolates, indicating the high degree of ITS homogeneity. Lanes M contain the 100-bp size marker ladder; the position of the 600-bp-long fragment is indicated.
DISCUSSION
Successful medical treatment or surgical excision of mycetoma lesions depends on the accurate diagnosis of the type of mycetoma and determination of the extent of the lesion. The latter is often difficult preoperatively, since the available diagnostic tools are not sensitive or specific (6, 15). In addition to the laboratory procedures for staining, cultivation, and serology already in use for detection of M. mycetomatis, aspiration cytology of mycetoma was recently described, but in the absence of grains the test is of little value (5). Histopathological examination is useful, but it carries a substantial risk of spreading mycetoma, since a deep surgical biopsy is always required (2). Furthermore, it is less reliable than a positive culture (19). Also, with histopathology it is not possible to differentiate among the different clinically important species, such as M. mycetomatis and M. grisea (13). In conclusion, no simple test is currently available for the diagnosis of mycetoma, other than clinical assessment and the invasive procedure of surgical biopsy. Furthermore, assessment of response to treatment is difficult, as is the prediction of cure or relapse in the absence of a reliable diagnostic test.
Our results showed that M. mycetomatis can be easily isolated from clinical specimens, but its survival in the glycerol stock cultures is quite poor. This explains the lack of available archival stocks of M. mycetomatis, which has been described previously (18). It appeared that the protocol employing freeze-thaw cycles in combination with the guanidinium lysis procedure was most convenient for DNA isolation in our laboratory setting. It should be mentioned that the yield equalled that of the other procedures, implying that the alternative procedures can be used as effectively.
Here we present the first nucleotide sequence information for the fungal species M. mycetomatis. RFLP analyses of the ITS region for which the primary structure was elucidated demonstrate a high degree of homogeneity among clinical isolates of M. mycetomatis. The sizes of the different restriction fragments generated precisely match expectations based on simple analysis of the primary structure of the ITS region (analysis not shown). These results seem to be in conflict with an earlier supposition by De Hoog et al. (4) that agents of mycetoma would be highly diverse. The present data indicate that mycetomata are caused by endemic species, possibly with a limited geographical distribution or—given the identity of the Caribbean strain with the Sudanese isolates—at least a local preponderance. Imported cases of mycetoma, such as those in The Netherlands (4), comprise cases from diverse localities and hence are likely to show a higher species diversity. Indeed, the M. mycetomatis-like strains from The Netherlands proved to represent a very different taxon. The geographical origin of the patients is thus an important factor in the anamnesis of cases of mycetoma. Several potential explanations for the genetic homogeneity among the clinical isolates of M. mycetomatis can be considered: the entire species may be clonal, there may be a type of M. mycetomatis that has spread through Sudan, or there may be an intimate link between infection and fungal type. The fact that preliminary randomly amplified polymorphic DNA studies have demonstrated at least a certain degree of genetic heterogeneity in regions other than the ribosomal operons indicates that at present this question cannot be answered and that future studies, involving additional, geographically diverse strains of M. mycetomatis, are mandatory.
On the basis of the noncoding ITS regions, we designed PCR primers for the identification of M. mycetomatis, the major cause of eumycetoma in the Sudan. The differentiation of the two clinically relevant Madurella species is feasible with our newly described assay in a manner that could not be achieved by currently used immunological techniques, such as Western blotting (26). By using the newly designed primers, future diagnosis of eumycetoma caused by M. mycetomatis may be quicker and simpler, even in the early stages of the infection. We have already been able to detect fungal DNA in biopsy specimens of mycetoma lesions, but serum samples or regional lymph node biopsy specimens still require additional testing (preliminary observations). To conclude, all M. mycetomatis organisms isolated during the course of this study belong to a single species. In addition, the clinical value of the newly designed PCR RFLP test for the identification of M. mycetomatis can now be assessed for early case detection, assessment of subclinical infections, and follow-up of patients and for determination of cure or relapse.
REFERENCES
- 1.Ahmed A O A, Abugroun E A M, Fahal A H, Zijlstra E E, van Belkum A, Verbrugh H A. Unexpected high prevalence of secondary bacterial infection in patients with mycetoma. J Clin Microbiol. 1998;36:850–851. doi: 10.1128/jcm.36.3.850-851.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baleiras Couto M M, Vogels J T, Hofstra H, Huis in ’t Veld J H, Vossen J M. Random amplified polymorphic DNA and restriction enzyme analysis of PCR amplified rDNA in taxonomy: two identification techniques for food borne yeasts. J Appl Bacteriol. 1995;79:525–535. doi: 10.1111/j.1365-2672.1995.tb03173.x. [DOI] [PubMed] [Google Scholar]
- 3.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Hoog G S, Buiting A, Tan C S, Stroebel A B, Ketterings C, de Boer E J, Naafs B, Brimicombe R, Nohlmans-Paulssen M K E, Fabius G T J, Klokke A H, Visser L G. Diagnostic problems with imported cases of mycetoma in The Netherlands. Mycoses. 1993;36:81–87. doi: 10.1111/j.1439-0507.1993.tb00693.x. [DOI] [PubMed] [Google Scholar]
- 5.El Hag I A, Fahal A H, Khalil E A G. Fine needle aspiration cytology of mycetoma. Acta Cytol. 1996;40:461–464. doi: 10.1159/000333899. [DOI] [PubMed] [Google Scholar]
- 6.Fahal A H, Hassan M A. Mycetoma. Br J Surg. 1992;79:1139–1141. doi: 10.1002/bjs.1800791107. [DOI] [PubMed] [Google Scholar]
- 7.Fujita S, Lasker B A, Lott T J, Reiss E, Morrison C J. Microtitration plate enzyme immunoassay to detect PCR amplified DNA from Candida species in blood. J Clin Microbiol. 1995;33:962–967. doi: 10.1128/jcm.33.4.962-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gumaa S A, Mahgoub E S. Counter immuno-electrophoresis in the diagnosis of mycetoma and its sensitivity as compared to immuno-diffusion. Sabouraudia. 1975;13:309–315. doi: 10.1080/00362177585190541. [DOI] [PubMed] [Google Scholar]
- 9.Hay R J, Mahgoub E S, Leon G, Al Sogair S, Welsh O. Mycetoma. J Med Vet Mycol. 1992;30:41–49. doi: 10.1080/02681219280000751. [DOI] [PubMed] [Google Scholar]
- 10.Johnston C G, Aust S D. Detection of Phanerochaete chrysosporium in soil by PCR and restriction enzyme analysis. Appl Environ Microbiol. 1994;60:2350–2354. doi: 10.1128/aem.60.7.2350-2354.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi G S. Actinomycetoma: the fungus-like bacteria. In: Davis B D, editor. Microbiology. Philadelphia, Pa: Harper and Row; 1980. pp. 298–321. [Google Scholar]
- 12.Kumeda Y, Asao T. Single-strand confirmation polymorphism analysis of PCR-amplified ribosomal DNA internal transcribed spacer to differentiate species of Apergillus Section Flavi. Appl Environ Microbiol. 1996;62:2947–2952. doi: 10.1128/aem.62.8.2947-2952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machado L A, Rivitti M C, Cuce L C, Saelebian A, da Lacaz C, Heins-Vaccari E M, Belda W, Jr, de Melo N T. Black grain eumycetoma due to Madurella grisea: a report of two cases. Rev Inst Med Trop Sao Paulo. 1992;34:569–580. [PubMed] [Google Scholar]
- 14.Magana M. Mycetoma. Int J Dermatol. 1984;23:221–236. doi: 10.1111/j.1365-4362.1984.tb01238.x. [DOI] [PubMed] [Google Scholar]
- 15.Mahgoub E S. Mycetoma. Int J Dermatol. 1985;24:230–239. [Google Scholar]
- 16.Makimura K, Mochizuki T, Hasegawa A, Uchida K, Saito H, Yamaguchi H. Phylogenetic classification of Trichophyton mentagrophytes complex strains based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Clin Microbiol. 1998;36:2629–2633. doi: 10.1128/jcm.36.9.2629-2633.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCullough M J, Clemons K V, McCusker J H, Stevens D A. Intergenic transcribed spacer PCR ribotyping for differentiation of Saccharomyces species and interspecific hybrids. J Clin Microbiol. 1998;36:1035–1038. doi: 10.1128/jcm.36.4.1035-1038.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasarell L, McGinnis M R. Viability of fungal cultures maintained at −70°C. J Clin Microbiol. 1992;30:1000–1004. doi: 10.1128/jcm.30.4.1000-1004.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rippon J W. Medical mycology. The pathogenic fungi and the pathogenic Actinomycetes. 3rd ed. Philadelphia, Pa: W. B. Saunders; 1988. pp. 80–118. [Google Scholar]
- 20.Taha A. A serological survey of antibodies of Streptomyces somaliensis and Actinomadura madurae in Sudan using enzyme-linked immunosorbent assay (ELISA) Trans R Soc Trop Med Hyg. 1983;77:49–50. doi: 10.1016/0035-9203(83)90011-1. [DOI] [PubMed] [Google Scholar]
- 21.Van Deventer A J M, Goessens W H F, van Belkum A, van Vliet H J A, van Etten E W M, Verbrugh H A. Improved detection of Candida albicans by PCR in blood of neutropenic mice with systemic candidiasis. J Clin Microbiol. 1995;33:625–628. doi: 10.1128/jcm.33.3.625-628.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells O. Mycetoma. Semin Dermatol. 1993;1:290–295. [PubMed] [Google Scholar]
- 24.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to method and applications. San Diego, Calif: Academic Press; 1996. pp. 315–322. [Google Scholar]
- 25.Williams D W, Wilson M J, Lewis M A, Potts A J. Identification of Candida species by PCR and restriction fragment length polymorphism analysis of intergenic spacer regions of ribosomal DNA. J Clin Microbiol. 1995;33:2476–2479. doi: 10.1128/jcm.33.9.2476-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaini F, Moore M K, Hathi D, Hay R J, Noble W C. The antigenic composition and protein profiles of eumycetoma agents. Mycoses. 1991;34:19–28. doi: 10.1111/j.1439-0507.1991.tb00614.x. [DOI] [PubMed] [Google Scholar]