Abstract
Malaria is a global health threat that requires immediate attention. Malaria is caused by the protozoan parasite Plasmodium, the most severe form of which is Plasmodium falciparum. The methylerythritol phosphate (MEP) pathway of isoprenoid biosynthesis is essential to the survival of many human pathogens, including P. falciparum, but is absent in humans, and thus shows promise as a new antimalarial drug target. The enzyme 1-deoxy-d-xylulose 5-phosphate reductoisomerase (IspC) catalyzes the first committed step in the MEP pathway. In addition to a divalent cation (Mg2+), the enzyme requires the substrates 1-deoxy-D-xylulose 5-phosphate (DXP) and NADPH to catalyze its reaction. We designed N-alkoxy and N-acyl fosmidomycin analogs to inhibit the activity of P. falciparum IspC in a bisubstrate manner. Enzyme assays reveal that the N-alkoxy fosmidomycin analogs have a competitive mode of inhibition relative to both the DXP- and NADPH-binding sites, confirming a bisubstrate mode of inhibition. In contrast, the N-acyl fosmidomycin analogs demonstrate competitive inhibition with respect to DXP but uncompetitive inhibition with respect to NADPH, indicating monosubstrate inhibitory activity. Our results will have a positive impact on the discovery of novel antimalarial drugs.
Introduction
In 2017, an estimated 219 million cases of malaria occurred worldwide, with an estimated 435 000 malaria-associated deaths.1 Of the various species of Plasmodium that cause malaria, Plasmodium falciparum is the most virulent.2 This unicellular protozoan parasite is transmitted to humans via an infected mosquito, allowing the parasite to pass from the mosquito’s saliva to the human bloodstream.3 Once in the bloodstream, the parasites are carried to the liver, where they infect the liver cells. After multiplying in liver cells for 5–30 days, the parasite re-enters the bloodstream to continue its lifecycle.4 Symptoms of malaria include fever, headache, and chills and if left untreated can result in death.5 Currently, artemisinin-based combination therapies (ACTs) are first-line antimalarial treatments worldwide.6 However, P. falciparum resistance to ACTs is spreading rapidly.6 Therefore, there is an urgent need to develop new antibiotics to treat malaria.
The methylerythritol phosphate (MEP) pathway of isoprenoid biosynthesis has been identified as an important drug target, as it is essential for the survival of many human pathogens, including P. falciparum. Mammalian cells use a distinct isoprenoid pathway, known as the mevalonate (MVA) pathway, for the biosynthesis of isopentenyl pyrophosphate (IPP) and its isomer dimethylallyl pyrophosphate (DMAPP). IPP and DMAPP are building blocks of isoprenoids, molecules involved in several essential cellular functions.7−9
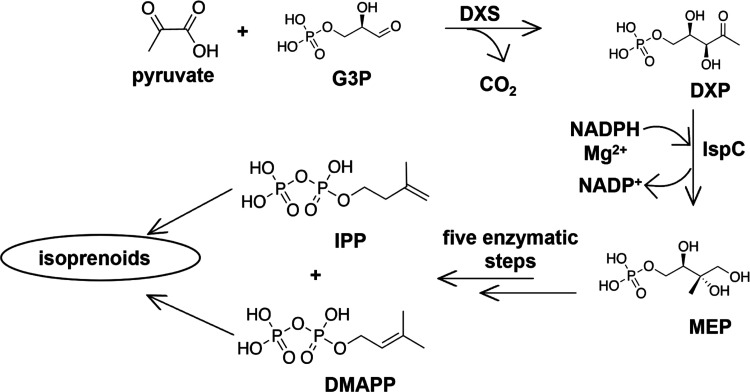
The MEP pathway consists of seven enzymatically catalyzed steps (Figure 1) and is initiated by DXP synthase (DXS), catalyzing the formation of 1-deoxy-d-xylulose 5-phosphate (DXP) from pyruvate and glyceraldehyde 3-phosphate. 1-Deoxy-d-xylulose 5-phosphate reductoisomerase (IspC or DXR) is the second enzyme in the MEP pathway, catalyzing the first committed step of isoprenoid biosynthesis. Using NADPH and a divalent cation (Mg2+), IspC converts DXP into 2-C-methylerythritol 4-phosphate (MEP). Sequentially, five additional enzymes convert MEP to the final MEP pathway products, IPP and DMAPP.9,10
Figure 1.
MEP pathway of isoprenoid biosynthesis. G3P: glyceraldehyde 3-phosphate; DXS: DXP synthase; IspC: 1-deoxy-d-xylulose 5-phosphate reductoisomerase; DXP: 1-deoxy-d-xylose 5-phosphate; MEP: 2-C-methyl-d-erythritol 4-phosphate; IPP: isopentenyl pyrophosphate; and DMAPP: dimethylallyl pyrophosphate.
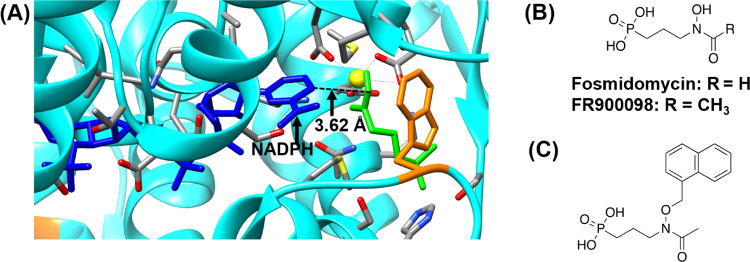
In IspC, the NADPH-binding site is located adjacent to the DXP-binding site (Figure 2A). For the enzyme to carry out its activity, NADPH must bind first and induce a conformational change that facilitates the binding of DXP.11 Fosmidomycin (FOS) and FR900098 (Figure 2B), natural products from Streptomyces lavendulae and Streptomyces rubellomurinus, respectively, are potent inhibitors of P. falciparum IspC (PfIspC) and effectively inhibit the growth of P. falciparum.12 These inhibitors are competitive with respect to DXP (Figure 2A), targeting the DXP-binding site in IspC (i.e., they are monosubstrate inhibitors). Accordingly, FOS and FR900098 are uncompetitive inhibitors with respect to NADPH; NADPH must bind to the NADPH site before FOS/FR900098 can bind to the DXP site.10,13,14 Although FOS and FR900098 are potent inhibitors of PfIspC, they suffer from low bioavailability, short serum half-life, and malaria recrudescence.10,12 Therefore, there is a need for novel analogs of FOS/FR900098 to overcome the shortcomings of these inhibitors.
Figure 2.
(A) Active site of P. falciparum IspC (PDB ID: 3AU9). NADPH is shown in blue, fosmidomycin in green, Mg2+ in yellow, and Trp29615 in orange. FR900098 makes a bindentate interaction with Mg2+. Amino acid labels are removed for clarity. The image was prepared using UCSF Chimera.16 (B) Monosubstrate PfIspC inhibitors fosmidomycin and FR900098 (later labeled as 1a and 2a in Figure 3, respectively). (C) Previously reported bisubstrate inhibitor of IspC.10 This N-alkoxy FR900098 analog is a more potent inhibitor of Mycobacterium tuberculosis, compared to the monosubstrate inhibitor FR900098.10 Bisubstrate inhibitors are designed to bind to both the DXP site and the NADPH site.
We have reported previously the effectiveness of a bisubstrate inhibitor, designed to target the adjacent NADPH- and DXP-binding sites in IspC (Figure 2C).10,13 The bisubstrate inhibitor demonstrated improved antibacterial activity relative to the monosubstrate inhibitors FOS/FR900098 (MIC = 25–200 μg/mL compared to MIC > 500 μg/mL, respectively).10 Mechanistic investigations have revealed a “flip-and-lock” mechanism associated with the bisubstrate inhibitor, wherein inhibitor binding to the NADPH site causes a conformation change (“flip”) in the active site of IspC, thereby forming the DXP site, which is then bound by the phosphonate side of the inhibitor (“lock”).10,11 Thus, a bisubstrate inhibitor is competitive with respect to both NADPH and DXP.10
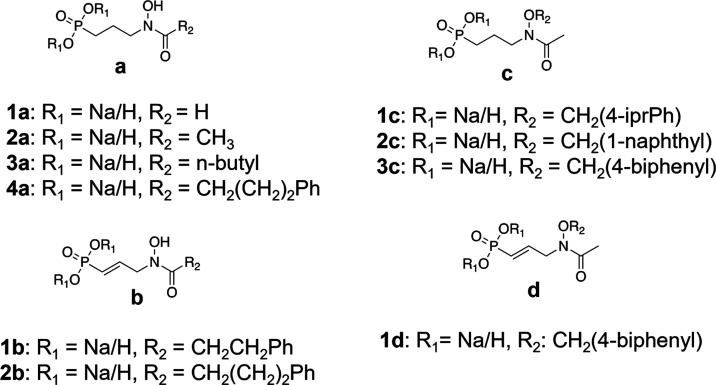
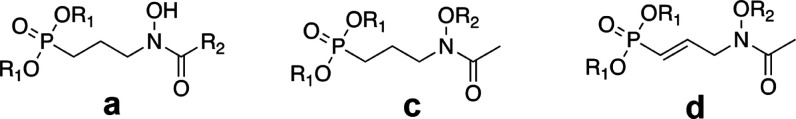
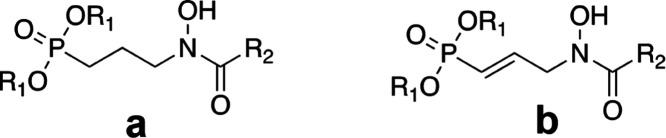
The aim of the current study was to expand upon this initial work by rationally designing and synthesizing bisubstrate inhibitors of PfIspC. Two classes of inhibitors were prepared: N-acyl and N-alkoxy FOS analogs (Figure 3). Each class was designed to act as a bisubstrate inhibitor, i.e., to be competitive with respect to both NADPH and DXP binding. As detailed herein, through the mechanism of inhibition assays, we ascertained that the N-alkoxy FOS analogs undergo a bisubstrate mode of inhibition. However, in contrast, the rationally designed N-acyl FOS analogs demonstrated a monosubstrate mode of inhibition, competitive with respect to DXP but uncompetitive with respect to NADPH. While the aromatic substituents of the N-acyl FOS analogs were expected to orient their N-acyl group toward the NADPH-binding site in IspC, our mechanism of inhibition data clearly indicates that the expected binding mode does not occur. Further, molecular modeling studies also support our findings, that the N-acyl FOS analogs preferentially position the N-acyl groups away from the NADPH site and thus are uncompetitive with respect to NADPH.
Figure 3.
P. falciparum IspC inhibitors: N-acyl (a, b) and N-alkoxy (c, d) fosmidomycin analogs. Natural products 1a = fosmidomycin (FOS) and 2a = FR900098. The N-acyl FOS analog 4a is a saturated version of 2b. Similarly, the N-alkoxy FOS analog 3c is a saturated version of 1d. All synthesized compounds were designed to act as bisubstrate inhibitors of IspC and tested as the monosodium salts.
Results and Discussion
Chemical Synthesis
The synthesis of compounds 4a, 1c, 2c, 3c, and 1d was performed, as described previously.12,13,17,18 The synthesis of compound 3a is shown in Scheme 1. Briefly, acylation of compound 5(10) was achieved using valeryl chloride and TEA to afford compound 6. Debenzylation of 6 with boron trichloride gave hydroxyl amine 7. Deprotection of phosphonate ester 7 using trimethylsilylbromide in the presence of BSTFA yielded desired target compound 3a. Characterization of compounds 1b and 2b is given. The synthetic details will be reported elsewhere.
Scheme 1. (a) Valeryl Chloride, TEA, and CH2Cl2; (b) 1 M BCl3, CH2Cl2; and (c) TMSBr, N,O-Bis(trimethylsilyl)trifluoroacetamide, and CH2Cl2.

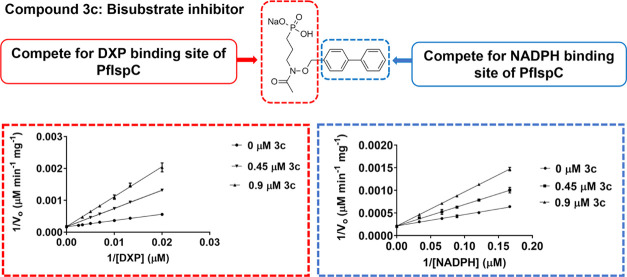
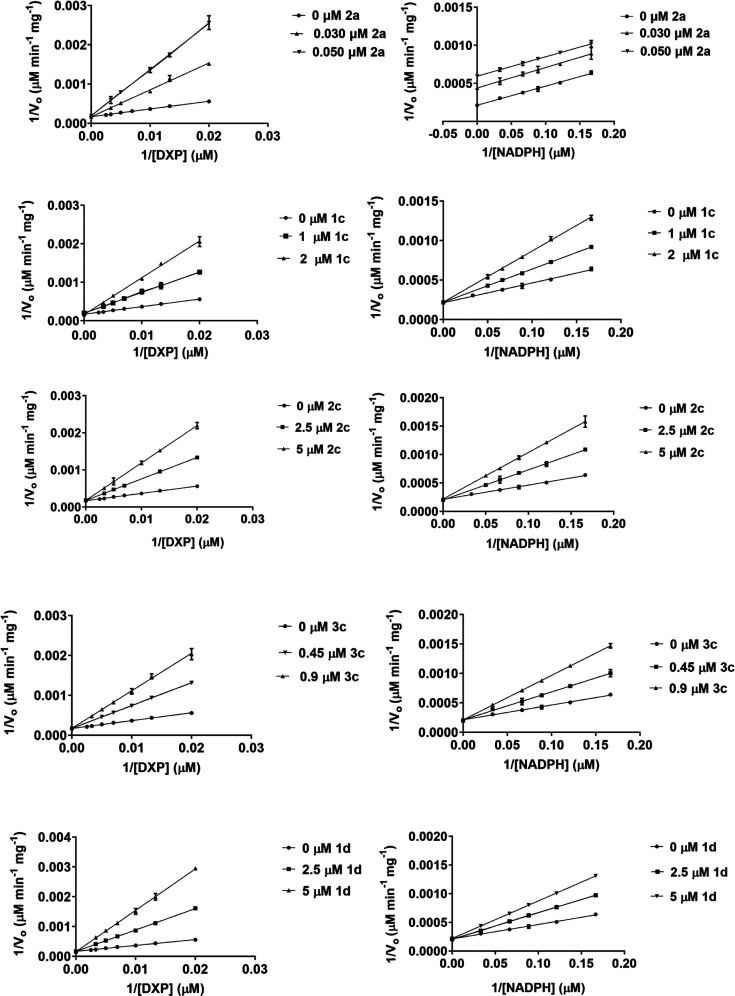
Mechanism of Inhibition of N-Alkoxy Fosmidomycin Analogs
To determine the mechanism of inhibition for these analogs, enzyme assays were performed using purified recombinant P. falciparum IspC and each inhibitor is listed in Figure 3. The results of the assays were plotted, as shown in Figure 4. As seen, the double reciprocal plots reveal that all of the N-alkoxy analogs (1c, 2c, 3c, and 1d) behave as competitive inhibitors with respect to both DXP and NADPH, indicating that they function as bisubstrate inhibitors. Among these analogs, the biphenyl compound (3c) is the most potent, with an inhibition constant (Ki) of 0.240 μM relative to DXP (Table 1). On the other hand, compound 1d, the α,β-unsaturated version of compound 3c, has a Ki value of 0.719 μM with respect to DXP (3-fold higher than that of 3c). Similarly, saturated compound 1c has a more potent Ki value (0.480 μM relative to DXP) than the structurally similar α,β-unsaturated compound, 1d (0.719 μM). Therefore, it appears likely that the poorer Ki value of compound 1d is, at least in part, due to the restriction imposed by the α,β-double bond, interfering with the binding of the phosphonate moiety of the inhibitor to the enzyme. Relative to NADPH, compound 3c yields the lowest Ki value (Ki = 0.440 μM), while compound 1d has the highest (Ki = 3.145 μM), again revealing the preference for the more flexible saturated backbone.
Figure 4.
Lineweaver–Burk plots of the N-alkoxy fosmidomycin analogs. All four analogs (1c, 2c, 3c, and 1d) demonstrate a competitive mode of inhibition with respect to both DXP and NADPH, indicating a bisubstrate mode of inhibition. FR900098 (2a), an N-acyl fosmidomycin analog, is presented for comparison; FR900098 is competitive with respect to DXP and uncompetitive with respect to NADPH. All assays were performed at least in triplicates with purified recombinant P. falciparum IspC.
Table 1. Half-Maximal Inhibitory Concentration (IC50) and Inhibition Constant (Ki) Values of N-Alkoxy Inhibitors Assayed Against P. falciparum IspC.
| compound | R1 | R2 | IC50 (μM) | Kia (μM) | Kib (μM) |
|---|---|---|---|---|---|
| 1a = fosmidomycin | Na/H | H | 0.03412 | ND | NA |
| 2a = FR900098 | Na/H | CH3 | 0.02412 | 0.009 | NA |
| 1c | Na/H | CH2(4-iprPh) | 1.411 | 0.480 | 1.217 |
| 2c | Na/H | CH2(1-naphthyl) | 3.361 | 1.166 | 2.195 |
| 3c | Na/H | CH2(4-biphenyl) | 0.804 | 0.240 | 0.440 |
| 1d | Na/H | CH2(4-biphenyl) | 5.670 | 0.719 | 3.145 |
Relative to DXP.
Relative to NADPH; ND: not determined; NA: not applicable. The inhibition constant (Ki) values are the results of the mechanism of inhibition (MoI) enzymatic assays performed at least in triplicate.
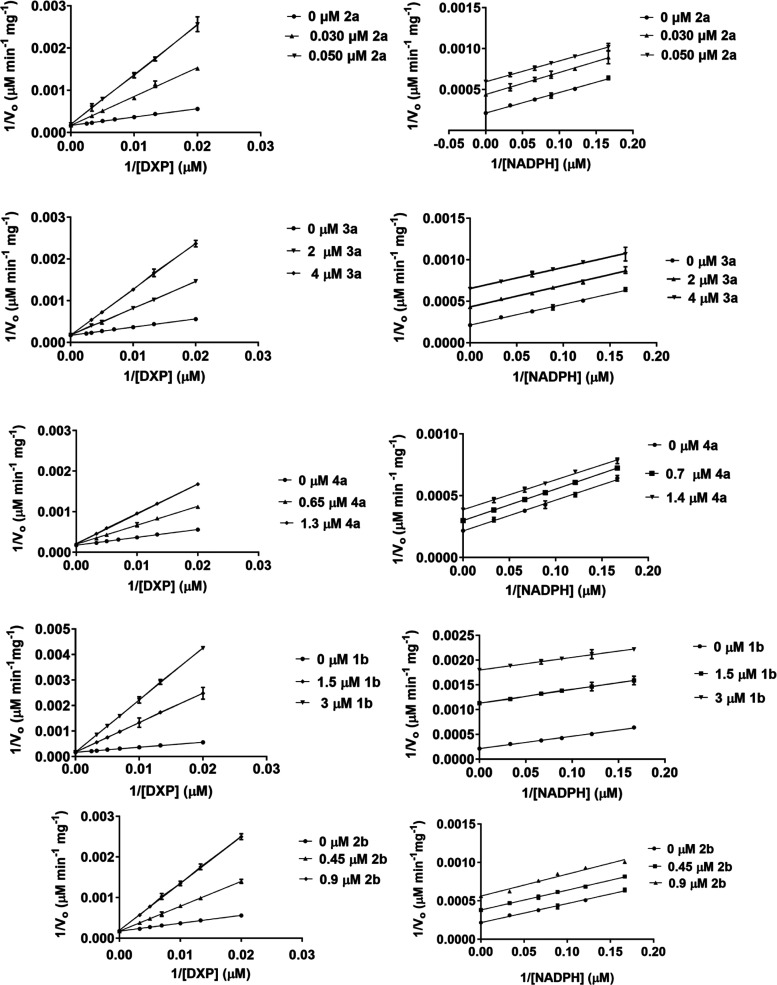
Mechanism of Inhibition of N-Acyl Fosmidomycin Analogs
The mechanism of inhibition experiments was carried out for the N-acyl fosmidomycin analogs in the same manner as for the N-alkoxys. As expected, all N-acyl fosmidomycin analogs (FR900098, 3a, 4a, 1b, and 2b) showed a competitive mode of inhibition relative to DXP (Figure 5). However, unlike the N-alkoxy analogs, all of the N-acyl analogs demonstrated an uncompetitive mode of inhibition relative to NADPH. Hence, NADPH binds to the enzyme before these inhibitors bind to the DXP site. While we hypothesized that the N-acyl moieties on compounds 3a, 4a, 1b, and 2b would extend into the NADPH site of P. falciparum IspC, it appears that they have adopted an alternative conformation (as discussed below).
Figure 5.
Lineweaver–Burk plots of the N-acyl fosmidomycin analogs. All five inhibitors (2a = FR900098, 3a, 4a, 1b, and 2b) show a competitive mode of inhibition against the DXP site of P. falciparum IspC but an uncompetitive mode of inhibition relative to the NADPH site. Bisubstrate inhibition was not observed for any of these N-acyl analogs. All assays were performed in at least in triplicate with purified recombinant P. falciparum IspC.
FR900098 (2a) shows potent inhibition of IspC; the Ki value relative to the DXP site is 0.009 μM. Looking at the structure–activity relationships among the N-acyl analogs, substitution of the FR900098 methyl group with an n-butyl group (3a) significantly diminished the Ki relative to the DXP site (Ki = 0.009 μM for FR900098 vs Ki = 0.860 μM for 3a). Interestingly, an even larger substituted group (4a) had less deleterious effect (Ki = 0.470 μM for 4a vs 0.860 μM for 3a). And, in contrast to the N-alkoxy analogs, which are less potent with α,β-unsaturation, the unsaturated N-acyl analogs performed better than their saturated counterparts (1b and 2b vs 3a and 4a) (Table 2).
Table 2. Half-Maximal Inhibitory Concentration (IC50) and Inhibition Constant (Ki) Values of N-Acyl Inhibitors Assayed Against P. falciparum IspC.
| compound | R1 | R2 | IC50 (μM) | Kia (μM) | Kib (μM) |
|---|---|---|---|---|---|
| 1a = fosmidomycin | Na/H | H | 0.03412 | ND | NA |
| 2a = FR900098 | Na/H | CH3 | 0.02412 | 0.009 | NA |
| 3a | Na/H | n-butyl | 2.448 | 0.860 | NA |
| 4a | Na/H | CH2(CH2)2Ph | 1.344 | 0.470 | NA |
| 1b | Na/H | CH2CH2Ph | 1.309 | 0.338 | NA |
| 2b | Na/H | CH2(CH2)2Ph | 0.900 | 0.162 | NA |
Relative to DXP.
Relative to NADPH; ND = not determined; NA = not applicable. The inhibition constant (Ki) values are the results of the mechanism of inhibition (MoI) enzymatic assays performed at least in triplicate.
Docking Studies
Previously determined X-ray crystal structures of P. falciparum IspC have illustrated the structural flexibility of the active site loop in response to alternative binding modes of fosmidomycin analogs with bulky substituents.19−21 In particular, positions of Trp296 and His293 change in response to binding analogs with bulky groups in the α- and β-positions (see Figure S6 and Table S1). Given this flexibility, we utilized four P. falciparum IspC crystal structures (PDB codes: 3AUA, 3WQR, 4Y67, and 4Y6R) with variable positioning of the loop and Trp296/His293 residues to conduct molecular docking studies to rationalize the difference in competitive vs uncompetitive binding with respect to NADPH for N-alkoxy and N-acyl inhibitors. The former two structures contain NADPH in the active site, while the latter two do not. Thus, docking models were created with and without NADPH for the former two structures giving six docking models.
To represent the N-alkoxy and N-acyl analogs, we selected compounds 3c and 4a, respectively, for docking. Briefly, the Glide docking routine in Schrodinger Maestro22 was used to conduct flexible ligand docking in a rigid protein in an unbiased manner (i.e., no required contacts or binding with Mg2+ or other residues). Although docking of N-alkoxy 3c produced reasonable poses in 5 of 6 docking models (Figure S7), it failed to produce any docked poses leading to bidentate binding of the inhibitor with Mg2+ except when docked in the 3AUA structure without the presence of NADPH (Figure 6). In this model, the N-alkoxy side chain binds in a mode that would be competitive with NADPH. Docking with unsaturated analog N-alkoxy 1d produced virtually identical results and docked poses, as observed with 3c (not shown). That supports the biochemical data showing 1d binds in a mode competitive with NADPH. The α,β-unsaturated bond of 1d located on the main chain of the inhibitor is one bond away from the phosphorus atom forming the phosphonate moiety, also located on the main chain of the inhibitor. However, this double bond is far from the N-alkoxy side chain (Figure 3). In addition, the N-alkoxy side chain of 1d is not binding to the main chain of the inhibitor but to an oxygen atom with a single bond that allowed flexibility of the side chain. As a result, we anticipated that the α,β-unsaturated bond of 1d would affect the binding of the phosphonate moiety of the inhibitor to the DXP-binding site but not significantly influence the orientiation of the N-alkoxy side chain relative to the NADPH-binding site. The Ki of 3c and 1d relative to the DXP are 0.240 and 0.719 μM, respectively. The observed difference in Ki relative to the DXP site between 3c, the saturated N-alkoxy FOS analog, and 1d is likely attributed to the presence of the α,β-unsaturated bond of 1d. In agreement with our anticipation, the biochemical assays and modeling studies revealed that both N-alkoxy compounds 3c and 1d, saturated and unsaturated, respectively, acted competitively with NADPH.
Figure 6.
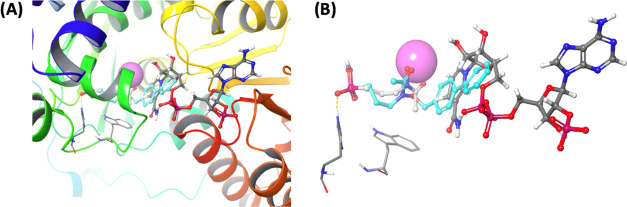
Docking model of N-alkoxy fosmidomycin analog 3c. Analog 3c was docked in P. falciparum IspC FR900098 structure 3AUA with NADPH pre-removed. FR900098, Trp296 and His293 are colored light gray. NADPH is colored dark gray. Mg2+ is colored magenta. Docked N-alkoxy analog 3c is colored cyan. (A) Top scoring pose of 3c overlaid on 3AUA structure. (B) Protein ribbons removed for clarity.
Docking of N-acyl 4a produced satisfactory binding poses in all six docking models (Figure S8). Five of the docking models maintained the bidentate Mg2+ binding, including all four models where the NADPH site was unavailable for docking. Docking with the unsaturated analog N-acyl 2b produced virtually identical results and docked poses, as observed with 4a (example shown in Figure S9). This supports the biochemical data showing 4a and 2b to be uncompetitive with NADPH (i.e., they can bind by occupying the DXP site in the presence of NADPH (Figures 7 and S9)). The α,β-unsaturated bond of 2b does not seem to affect the orientation of the N-acyl side chain relative to NADPH, since both the saturated 4a and the unsaturated 2b show uncompetitive mode relative to NADPH. It is likely that the orientation of the N-acyl side chains relative to the NADPH-binding site is mainly affected by the amide bond the side chains form.
Figure 7.
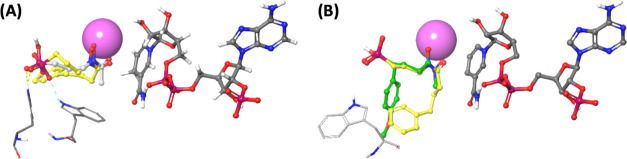
Docking models of N-acyl fosmidomycin analog 4a. N-acyl analog 4a was docked in P. falciparum IspC/FR900098 structure 3AUA with NADPH present (A) and 3WQR/α-Ph analog with NADPH present (B). FR900098, Trp296, and His293 are colored light gray. α-Ph analog from 3WQR is colored green. NADPH is colored dark gray. Mg2+ is colored magenta. Docked N-acyl analog 4a is colored yellow.
It should be noted that we cannot make any definitive statements regarding precise positioning of N-alkoxy and N-acyl side chains other than to say it is likely that N-alkoxy compounds such as 3c likely cannot form bidentate interaction with the Mg2+ except through displacement of NADPH (i.e., when the lipophilic group is buried deep in the binding site), while N-acyl compounds such as 4a can adopt an alternative binding mode that may mimic other bulky substituents off the central propyl chain extending toward solvent.
Conclusions
Artemisinin-based combination therapies (ACTs) are already facing drug resistance by P. falciparum, therefore, there is an urgent need for novel antibiotics. Fosmidomycin is a potent inhibitor of P. falciparum IspC and P. falciparum growth, and clinical trials have shown that fosmidomycin is a remarkably safe drug in combination therapies. Nonetheless, fosmidomycin suffers from low bioavailability, short serum-life, and malaria recrudescence. We have designed, synthesized, and determined the mechanisms of inhibition of a series of N-acyl and N-alkoxy fosmidomycin analogs. We have shown that N-acyl fosmidomycin analogs do not inhibit P. falciparum IspC in a bisubstrate manner, possibly due to restrictions imposed by the amide bond. Molecular modeling revealed the N-acyl fosmidomycin analogs position their N-acyl groups away from the NADPH site and thus are uncompetitive relative to NADPH. We have shown that the N-alkoxy fosmidomycin analogs act in a bisubstrate mode of inhibition. Further study is warranted to develop additional compounds against P. falciparum IspC and to establish structure–activity relationships with various N-alkoxy and N-acyl moieties.
Materials and Methods
Reagents and Chemical Synthesis
General
1H and 13C NMR spectra were recorded in CDCl3, CD3OD, or D2O on an Agilent spectrometer at 400 and 100 MHz, respectively. 1H and 13C NMR spectra for one compound (3a) were recorded on an Agilent spectrometer at 200 and 50 MHz, respectively. TMS, H2O, or solvent signal was used as internal standard. Chemical shifts are given in parts per million (ppm). Spin multiplicities are given with the following abbreviations: s (singulet), br s (broad singulet), d (doublet), dd (doublet of doublets), ddd (doublet of doublets of doublets), t (triplet), dt (doublet of triplets), ddt (doublet of doublet of triplets), q (quadruplet), qt (quintuplet), and m (multiplet). Mass spectra were measured in the ESI mode on an HPLC-MS (Agilent 1100) or in the EI mode on a GC-MS (Shimadzu GC-MS-QP2010S). Thin-layer chromatography (TLC) was performed on Baker-flex Silica Gel IB2-F silica plates and flash column chromatography was carried out using SiliCycle SiliaFlash P60 silica gel (40–63 μm). All reagents were purchased from commercial suppliers and used without further purification. Anhydrous solvents were filtered by the MBRAUN MB-SPS solvent purification system before use. All air-sensitive reactions were carried out under a nitrogen atmosphere. The purity of synthesized compounds (>95%) was determined by 1H/13C NMR in combination with HPLC-MS (Agilent 1100). Column: Thermo Fisher Scientific Hypersil GOLD aQ C-18 3 μm particle (250 mm × 4.6 mm). Mobile phase (containing 0.1% formic acid as the additive): linear gradient of acetonitrile (50–100%) in water at a flow rate of 0.8 mL/min over 12.5 min, followed by 100% acetonitrile that was maintained for another 12.5 min. The UV detection wavelengths were 210 nm and 254 nm. High-resolution mass spectroscopy spectra (HRMS) were recorded in positive or negative ESI mode on a Waters Q-TOF Ultima mass spectrometer (UIUC Mass Spectrometry Laboratory) or in positive FAB mode on a VG Analytical VG70SE magnetic sector mass spectrometer (JHU Mass Spectrometry Facility). Compound 5 was prepared, as previously described.10
1b: Sodium Hydrogen [(1E)-3-(N-hydroxy-3-phenylpropanamido)prop-1-en-1-yl]phosphonate
Light yellow solids (51 mg, 92%). 1H NMR (400 MHz, D2O) δ 7.26–7.09 (m, 5H), 6.29–6.13 (m, 1H), 5.77–5.59 (m, 1H), 4.22–4.11 (m, 2H), 2.85–2.66 (m, 4H). 13C NMR (101 MHz, D2O) δ 175.3, 140.7, 139.3 (d, J = 5.3 Hz), 128.7, 128.3, 126.4, 124.0 (d, J = 177.7 Hz), 50.8 (d, J = 23.6 Hz), 33.1, 30.2. LC-MS (ESI+): 571.2 m/z [2M – 2Na + 3H]+. HRMS (FAB+) calculated for C12H15NNaO5P, 307.0586; found, 308.0655 [M + H]+.
2b: Sodium Hydrogen [(1E)-3-(N-hydroxy-4-phenylbutanamido)prop-1-en-1-yl]phosphonate
Light yellow solids (28 mg, 77%). 1H NMR (400 MHz, CDCl3) δ 7.12–7.25 (m, 5H), 6.30–6.41 (m, 1H), 5.75–5.85 (m, 1H), 4.22 (bs, 2H), 2.52–2.57 (m, 2H), 2.25 (t, J = 7.3 Hz, 2H), 1.76–1.83 (m, 2H). 13C NMR (101 MHz, D2O) δ 176.5, 142.1, 133.0 (d, J = 5.5 Hz), 130.4, 128.8, 126.3, 121.9 (d, J = 180.7 Hz), 50.9 (d, J = 23.5 Hz), 34.6, 31.2, 26.1. LC-MS (ESI+): 300.0 m/z [M – Na + 2H]+, 599.2 m/z [2M – 2Na + 3H]+, 898.2 m/z [3M – 3Na + 4H]+. HRMS (FAB+) calculated for C13H17NNaO5P, 321.0742; found, 322.0862 [M + H]+.
3a: Sodium Hydrogen-3-(N-hydroxypentanamido)propylphosphonate
N,O-Bis(trimethylsilyl)trifluoroacetamide (0.15 mL, 0.145 g, 0.56 mmol) was added under a nitrogen atmosphere to diethyl 3-(N-hydroxypentanamido)propylphosphonate (7) (0.050 g, 0.17 mmol) in CH2Cl2 (0.75 mL) and stirred at room temperature for 20 min. The reaction mixture was cooled to 0°C and bromotrimethylsilane (0.22 mL, 0.255 g, 1.67 mmol) was added dropwise to the reaction. The reaction was warmed to room temperature and stirred overnight under a nitrogen atmosphere. Ethyl bromide and excess silylating agent were removed under reduced pressure and the residue was dissolved in aqueous NaOH (0.86 mL, 7.8 mg/mL) and stirred for a 2nd night. The reaction mixture was partitioned between H2O and CH2Cl2 to remove any residual impurities/organics. The aqueous fractions were combined, and the solvent was removed by lyophilization to give 3a (0.049 g, 0.18 mmol, Quantitative) as a pale-yellow solid. 1H NMR (200 MHz, acetone-d6/D2O) δ (ppm): 3.72–3.62 (m, 2H), 2.55 (t, J = 7.7 Hz, 2H),2.07–1.77 (m, 2H), 1.61 (p, J = 8.3, 7.8 Hz, 4H), 1.39 (h, J = 13.8, 7.1 Hz, 2H), 0.95 (t, J = 7.4 Hz, 3H). 13C NMR (50 MHz, acetone-d6/D2O) δ (ppm): 176.33, 49.00 (d, J = 18.8 Hz), 32.01, 27.02, 24.11, 22.30, 21.09, 13.52. LC-MS (ESI) m/z 240.1 (M + H). HRMS (ESI) m/z calcd for C8H17NO5P (M – Na): 238.0838, found: 238.0833.
7: Diethyl 3-(N-hydroxypentanamido)propylphosphonate
A solution of 6 (0.208 g, 0.54 mmol) in dry CH2Cl2 (10.0 mL) was cooled to −50 °C and a 1 M solution of BCl3 in CH2Cl2 (3.5 mL, 3.5 mmol) was added dropwise. After stirring for 4 h at −50 °C, the reaction was quenched with saturated NaHCO3 (aq, 16 mL) and warmed to room temperature. The aqueous reaction mixture was extracted with CH2Cl2 (×3). The organic fractions were combined, dried over MgSO4, and filtered and the solvent was removed under reduced pressure. The resulting crude mixture was purified using an Isolera Flash Chromatography System (EtOAc/MeOH; 5–10% MeOH) to yield 7 (0.125 g, 0.42 mmol, 78.5%) as a yellow oil. 1H NMR (200 MHz, CDCl3) δ (ppm): 9.53 (s, 1H), 4.06 (p, J = 6.9 Hz, 4H), 3.74 (t, J = 5.6 Hz, 2H), 2.51 (t, J = 7.7 Hz, 2H), 2.10–1.90 (m, 2H), 1.88–1.71 (m, 2H), 1.61 (p, J = 7.4 Hz, 2H), 1.47–1.36 (m, 2H), 1.32 (t, J = 7.1 Hz, 7H), 0.91 (t, J = 7.3 Hz, 3H). 13C NMR (50 MHz, CDCl3) δ (ppm): 175.31, 62.16 (d, J = 7.2 Hz), 47.81, 32.30, 26.89, 23.72, 22.66, 19.34, 16.42 (dd, J = 3.2 Hz), 13.92. LC-MS (ESI) m/z 296.2 (M + H).
6: Diethyl 3-[N-(benzyloxy)pentanamido]propylphosphonate
To a stirred solution of 5 (0.203 g, 0.66 mmol) and triethylamine (0.2 mL, 0.14 g, 1.3 mmol) in CH2Cl2 (3 mL) at room temperature and under an argon atmosphere was added valeryl chloride (0.1 mL, 0.097 g, 0.8 mmol) dropwise. The reaction mixture was stirred at room temperature overnight. The reaction mixture was extracted with water and brine. The aqueous layer was back-extracted with CH2Cl2 (×3) and dried over anhydrous magnesium sulfate. The magnesium sulfate was filtered from the organic layer and the solvent was removed under reduced pressure. The crude residue was purified using an Isolera Flash Chromatography (100% EtOAc) to give 6 (0.207 g, 0.54 mmol, 81.4%) as a yellow oil. 1H NMR (CDCl3, 200 MHz), δ (ppm): 7.38 (s, 5Harom), 4.81 (s, 2H), 4.08 (p, J = 7.3 Hz, 4H), 3.71 (t, J = 6.8 Hz, 2H), 2.39 (t, J = 7.6, 2H), 2.03–1.48 (m, 6H), 1.75–1.60 (m, 2H), 1.30 (t, J = 6.2 Hz, 6H), 1.37–1.24 (m, 2H). LC-MS (ESI) m/z 486.2 (M + H).
Expression and Purification of P. falciparum IspC
The cloning, expression, and purification of P. falciparum IspC were performed, as described previously.10,12,17 Briefly, the P. falciparum IspC gene was cloned into a pET100/D-TOPO vector to facilitate the expression of an N-terminal His6-tagged protein. The recombinant protein was expressed in Escherichia coli Rosetta2(DE3) cells obtained from Novagen (San Diego, CA). E. coli was cultured at 37 °C in Luria-Bertani media supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol with constant shaking at 250 rpm. Agar (1.5% w/v) was added to prepare solid media. Protein was isolated and purified from the cells via chemical lysis and TALON affinity chromatography (Clontech Laboratories, Mountain View, CA).
P. falciparum IspC Inhibition Assays
The IspC activity was assayed at 37 °C by spectrophotometrically monitoring the enzyme-catalyzed oxidation of NADPH, as described previously.12,13,17 Briefly, the assay mixture contained 100 mM Tris pH 7.8, 25 mM MgCl2, and 0.86 μM P. falciparum IspC. To determine the half-maximal inhibition (IC50) concentrations, the enzyme was preincubated with the inhibitor for 10 min at 37 °C prior to the addition of 150 μM NADPH. The reaction mixture was further incubated for 5 min at 37 °C. The reaction was initiated with the addition of 144 μM DXP (Echelon Biosciences, Salt Lake City, UT) to the assay mixture. The oxidation of NADPH was monitored at 340 nm using an Agilent 8453 UV–visible spectrophotometer equipped with a temperature-regulated cuvette holder. One unit of P. falciparum IspC activity is defined as the amount of enzyme that catalyzes the oxidation of 1 μM NADPH per min per mg of the enzyme. Half-maximal inhibition (IC50) of enzyme activity was determined using a plot of fractional enzyme activity as a function of inhibitor concentration. Nonlinear regression of the sigmoidal dose–response curve was generated using GraphPad Prism version 7.00 for Windows (GraphPad Software Inc., San Diego, CA). All enzymatic assays performed to determine half-maximal inhibition concentration (IC50) of an inhibitor were performed in duplicate. All IC50 values were validated by a second enzymologist performing an inhibition assay (data not shown) with the inhibitor at the IC50 concentration, confirming 50% residual enzyme activity (±5%).
To determine the mechanism of inhibition (MoI), the enzyme assay was performed as described above with varying concentrations of DXP and NADPH. To determine the MoI with respect to the DXP-binding site, the DXP concentration was varied between 50 and 400 μM, while the NADPH concentration was kept constant at 150 μM. To determine the MoI with respect to the NADPH-binding site, the NADPH concentration was varied between 6 and 30 μM, while the DXP concentration was kept constant at 144 μM. The MoI was determined using Lineweaver–Burk plots; linear regression of the data was generated using GraphPad Prism. The inhibitor constants (Ki values) were determined by generating secondary plots from the Lineweaver–Burk plots; linear regression of the data was generated using GraphPad Prism. All of the MoI assays were performed at least in triplicate.
Molecular Modeling Studies
Modeling studies were conducted with Schrodinger Maestro Release 2019-4.22 Crystal structures 3AUA, 3WQR, 4Y6R, and 4Y67 were imported from the Protein Data Bank (PDB)23 and prepared using Protein Preparation Wizard. Glide docking grids were generated from these structures removing the bound ligand while placing no restrictions or constraints on the ligand binding protocol. For structures 3AUA and 3WQR, a second Glide docking grid was generated wherein the NAPDH and associated water molecules were removed before grid generation. Glide docking was performed using these six docking grids under the XP precision mode and all other defaults with the exception of increasing the number of output structures to three per docked compound.
Acknowledgments
This work was generously supported by the George Mason University Department of Chemistry and Biochemistry, the U.S. Army MRDC (W81XWH-17-C-0066), the Military Infectious Disease Research Program (W0161_15_WR), the NIH (5R01AI123433-04), and by an appointment to the Student Research Participation Program at the Walter Reed Army Institute of Research, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and USAMRDC.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c01711.
The authors declare no competing financial interest.
Supplementary Material
References
- WHO. World Malaria Report 2018. http://www.who.int/malaria/publications/world-malaria-report-2018/report/en/ (accessed October 31, 2019).
- Hayward R. E.; Tiwari B.; Piper K. P.; Baruch D. I.; Day K. P. Virulence and Transmission Success of the Malarial Parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 4563–4568. 10.1073/pnas.96.8.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. About Malaria – Biology. https://www.cdc.gov/malaria/about/biology/index.html (accessed November 2, 2019).
- Soulard V.; Bosson-Vanga H.; Lorthiois A.; Roucher C.; Franetich J.-F.; Zanghi G.; Bordessoulles M.; Tefit M.; Thellier M.; Morosan S.; Le Naour G.; Capron F.; Suemizu H.; Snounou G.; Moreno-Sabater A.; Mazier D. Plasmodium falciparum Full Life Cycle and Plasmodium ovale Liver Stages in Humanized Mice. Nat. Commun. 2015, 6, 7690 10.1038/ncomms8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Malaria Report 2019. https://www.who.int/publications-detail-redirect/world-malaria-report-2019 (accessed June 30, 2020).
- Ouji M.; Augereau J.-M.; Paloque L.; Benoit-Vical F. Plasmodium falciparum Resistance to Artemisinin-Based Combination Therapies: A Sword of Damocles in the Path toward Malaria Elimination. Parasite 2018, 25, 24 10.1051/parasite/2018021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuston S.; Begley M.; Gahan C. G. M.; Hill C. Isoprenoid Biosynthesis in Bacterial Pathogens. Microbiology 2012, 158, 1389–1401. 10.1099/mic.0.051599-0. [DOI] [PubMed] [Google Scholar]
- Holstein S. A.; Hohl R. J. Isoprenoids: Remarkable Diversity of Form and Function. Lipids 2004, 39, 293–309. 10.1007/s11745-004-1233-3. [DOI] [PubMed] [Google Scholar]
- Hoshino Y.; Gaucher E. A. On the Origin of Isoprenoid Biosynthesis. Mol. Biol. Evol. 2018, 35, 2185–2197. 10.1093/molbev/msy120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Jose G.; Jackson E. R.; Haymond A.; Johny C.; Edwards R. L.; Wang X.; Brothers R. C.; Edelstein E. K.; Odom A. R.; Boshoff H. I.; Couch R. D.; Dowd C. S. Structure–Activity Relationships of the MEPicides: N-Acyl and O-Linked Analogs of FR900098 as Inhibitors of Dxr from Mycobacterium tuberculosis and Yersinia pestis. ACS Infect. Dis. 2016, 2, 923–935. 10.1021/acsinfecdis.6b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppisch A. T.; Fox D. T.; Blagg B. S. J.; Poulter C. D. E. coli MEP Synthase: Steady-State Kinetic Analysis and Substrate Binding. Biochemistry 2002, 41, 236–243. 10.1021/bi0118207. [DOI] [PubMed] [Google Scholar]
- Wang X.; Edwards R. L.; Ball H.; Johnson C.; Haymond A.; Girma M.; Manikkam M.; Brothers R. C.; McKay K. T.; Arnett S. D.; Osbourn D. M.; Alvarez S.; Boshoff H. I.; Meyers M. J.; Couch R. D.; Odom John A. R.; Dowd C. S. MEPicides: α,β-Unsaturated Fosmidomycin Analogues as DXR Inhibitors against Malaria. J. Med. Chem. 2018, 61, 8847–8858. 10.1021/acs.jmedchem.8b01026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Jose G.; Jackson E. R.; Uh E.; Johny C.; Haymond A.; Lundberg L.; Pinkham C.; Kehn-Hall K.; Boshoff H. I.; Couch R. D.; Dowd C. S. Design of Potential Bisubstrate Inhibitors against Mycobacterium tuberculosis (Mtb) 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr)-Evidence of a Novel Binding Mode. MedChemComm 2013, 4, 1099–1104. 10.1039/C3MD00085K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E. R.; Dowd C. S. Inhibition of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase (Dxr): A Review of the Synthesis and Biological Evaluation of Recent Inhibitors. Curr. Top. Med. Chem. 2012, 12, 706–728. 10.2174/156802612799984599. [DOI] [PubMed] [Google Scholar]
- RCSB PDB 3AU9: Crystal Structure of the Quaternary Complex-1 of an Isomerase. https://www.rcsb.org/structure/3AU9 (accessed August 11, 2020).
- UCSF Chimera Home Page. https://www.cgl.ucsf.edu/chimera/ (accessed July 10, 2020).
- Haymond A.; Johny C.; Dowdy T.; Schweibenz B.; Villarroel K.; Young R.; Mantooth C. J.; Patel T.; Bases J.; Jose G. S.; Jackson E. R.; Dowd C. S.; Couch R. D. Kinetic Characterization and Allosteric Inhibition of the Yersinia Pestis 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (MEP Synthase). PLoS One 2014, 9, e106243 10.1371/journal.pone.0106243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Dowd C. S. The Methylerythritol Phosphate Pathway: Promising Drug Targets in the Fight against Tuberculosis. ACS Infect. Dis. 2018, 4, 278–290. 10.1021/acsinfecdis.7b00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chofor R.; Sooriyaarachchi S.; Risseeuw M. D. P.; Bergfors T.; Pouyez J.; Johny C.; Haymond A.; Everaert A.; Dowd C. S.; Maes L.; Coenye T.; Alex A.; Couch R. D.; Jones T. A.; Wouters J.; Mowbray S. L.; Van Calenbergh S. Synthesis and Bioactivity of β-Substituted Fosmidomycin Analogues Targeting 1-Deoxy-d-Xylulose-5-Phosphate Reductoisomerase. J. Med. Chem. 2015, 58, 2988–3001. 10.1021/jm5014264. [DOI] [PubMed] [Google Scholar]
- Umeda T.; Tanaka N.; Kusakabe Y.; Nakanishi M.; Kitade Y.; Nakamura K. T. Molecular Basis of Fosmidomycin’s Action on the Human Malaria Parasite Plasmodium falciparum. Sci. Rep. 2011, 1, 9 10.1038/srep00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzuch S.; Umeda T.; Held J.; Hähn S.; Brücher K.; Lienau C.; Behrendt C. T.; Gräwert T.; Bacher A.; Illarionov B.; Fischer M.; Mordmüller B.; Tanaka N.; Kurz T. Binding Modes of Reverse Fosmidomycin Analogs toward the Antimalarial Target IspC. J. Med. Chem. 2014, 57, 8827–8838. 10.1021/jm500850y. [DOI] [PubMed] [Google Scholar]
- Schrödinger. Announcing Schrödinger Software Release 2019-4. https://www.schrodinger.com/user-announcement/announcing-schrodinger-software-release-2019-4 (accessed June 1, 2020).
- RCSB PDB: Homepage. https://www.rcsb.org/ (accessed November 29, 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.