Summary
CRISPR-associated proteins (Cas1 and Cas2) integrate foreign DNA at the “leader” end of CRISPR loci. Several CRISPR leader sequences are reported to contain a binding site for a DNA bending protein called Integration Host Factor (IHF). IHF-induced DNA bending kinks the leader of type I-E CRISPRs, recruiting an upstream sequence motif that helps dock Cas1-2 onto the first repeat of the CRISPR locus. To determine the prevalence of IHF-directed CRISPR adaptation, we analyzed 15,274 bacterial and archaeal CRISPR leaders. These experiments reveal multiple IHF binding sites and diverse upstream sequence motifs in a subset of the I-C, I-E, I-F and II-C CRISPR leaders. We identify subtype-specific motifs and show that the phase of these motifs is critical for CRISPR adaptation. Collectively, this work clarifies the prevalence and mechanism(s) of IHF-dependent CRISPR adaptation and suggests that leader sequences and adaptation proteins may coevolve under the selective pressures of foreign genetic elements like plasmids or phages.
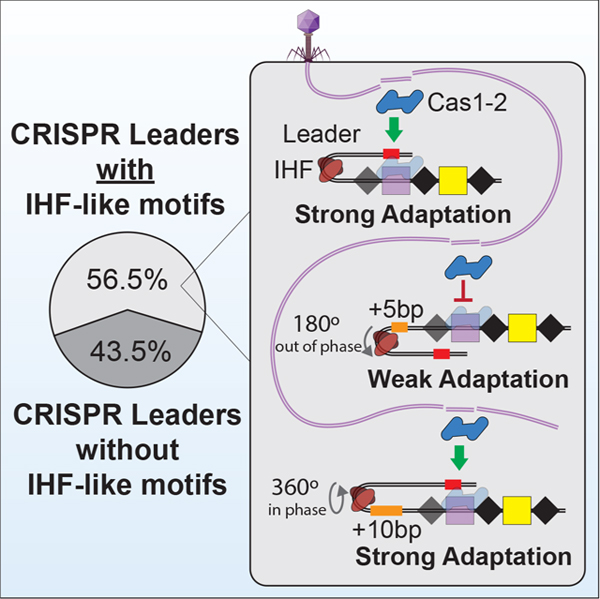
eTOC
Santiago-Frangos et al. determine the prevalence and distribution of DNA sequence motifs that are necessary for the polarized integration of foreign DNA into most CRISPR loci. The spacing between these motifs is critical for maintaining DNA structures that are necessary for efficient CRISPR adaptation.
Graphical Abstract
Introduction
The repeat-spacer-repeat architecture characteristic of all CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) loci was first observed in 19871. However, the biological function of CRISPRs remained obscure until 2005, when three groups independently reported that CRISPR loci frequently contain “spacers” derived from foreign genetic elements2–4. By comparing CRISPR loci from closely related isolatesof Yersinia pestis, Pourcel et al. noted that new phage-derived spacers are preferentially added to one end of the CRISPR4, which is often flanked by an adenine- and thymine-rich (AT-rich) sequence, previously termed the “leader”5. Collectively, these computational observations indicated that CRISPRs are part of an adaptive immune system that maintains a chronological record of previously encountered foreign genetic parasites6,7. A role for CRISPR loci and Cas proteins in adaptive immunity was first established by phage challenge experiments performed by Barrangou et al.8. Polarized integration is crucial in most systems, since spacers at the leader-end of the CRISPR have been shown to provide greater levels of immunity9.
Structural and biochemical experiments aimed at understanding the mechanism(s) of polarized adaptation have resulted in two models for preferential integration of new spacer at the “leader-end”7. In type II systems, the Cas1 proteins have been shown to recognize the leader-repeat junction, while type I-E and I-F systems have been shown to rely on a DNA bending protein called Integration Host Factor (IHF)9–15. In the type I-E CRISPR system of Escherichia coli (K12), IHF binds to a roughly 30-base pair sequence motif that begins 6-base pairs upstream of the first repeat10. IHF-binding kinks DNA in the leader, creating a horseshoe shaped structure that stabilizes the Cas1-2 integrase complex bound to the first repeat of the CRISPR locus11. One of the Cas1 dimers is wedged into the “toe” of the DNA horseshoe, resulting in specific contacts between the Cas1 and IHF proteins, as well as sequence specific interactions between one lobe of Cas1 and an upstream sequence motif11,16.
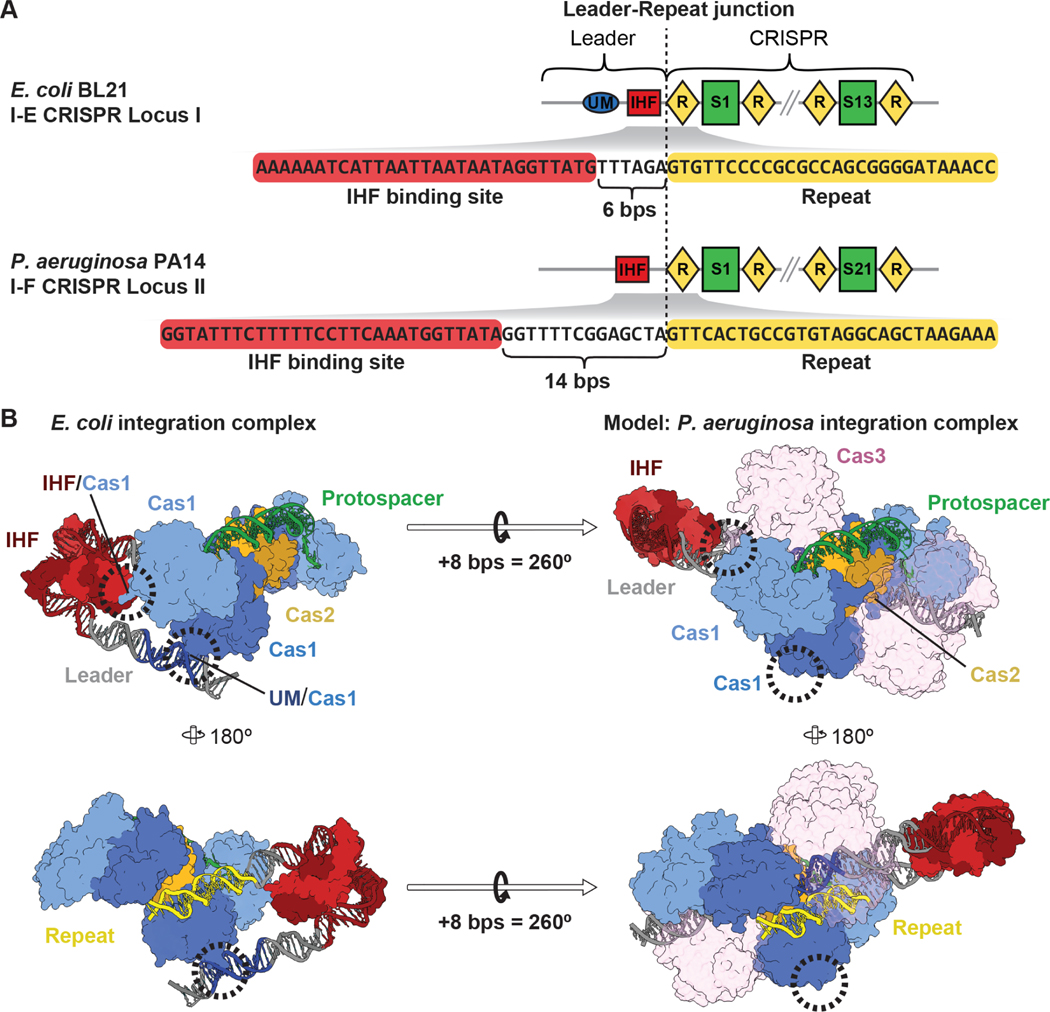
We recently noticed that the IHF binding site in the leader sequence of the type I-F CRISPR locus from Pseudomonas aeruginosa (PA14) is 8-base pairs further from the leader-repeat junction than what was originally observed in the type I-E systems of Escherichia coli (Figure 1). We hypothesized that this seemingly subtle difference has important mechanistic implications for CRISPR adaptation. The helical structure of double stranded DNA (dsDNA), contains ~11-base pairs per 360-degree rotation17,18. Thus, the addition of 8-base pairs not only shifts the IHF binding site ~27 Å away from the leader-repeat junction, which would preclude previously observed Cas1-IHF interactions, but this insertion also introduces a ~260-degree rotation of the upstream DNA. This rotation would position one tip of the “DNA horseshoe” on the opposite side of the Cas1-2 integration complex from what has been previously observed for the E. coli integration complex11.
Figure 1. Spacing of sequence mofits in CRISPR leaders has mechanistic implications for the mechanism of spacer integration.
(A) Schemes of two CRISPR loci from E. coli BL21 (type I-E, top) and P. aeruginosa PA14 (type I-F, bottom), and sequences of the first CRISPR repeat and the upstream IHF binding site in the leaders. Repeating DNA motifs (diamonds) in the CRISPR locus are interspered with unique spacer sequences (rectangles). The IHF binding site in the I-F system from P. aeruginosaPA14 is 8 bps further away from the leader-repeat junction, as compared to the I-E CRISPR locus. (B) Structure of the previously determined type I-E integration intermediate (PDB: 5WFE). The Cas1-2 heterohexamer (blue and yellow) is in the process of integrating a protospacer (green) at the first CRISPR repeat (yellow). IHF kinking of DNA at the IHF binding site recruits an upstream motif (UM, dark blue) to interact with one lobe of a Cas1 subunit, while the second Cas1 subunit in the same dimer makes protein-protein contacts with IHF11. Models of the 8 bp insertion present in P. aeruginosa CRISPR locus II illustrate how this insertion is predicted to disrupt both of these interactions by shifting IHF 27Å away from the integration complex and rotating the upstream motif (UM) 260°. Cas3 domains of the I-F Cas2/3 fusion are shown semi-transparently for clarity. See also Figure S1 and S3.
Here we analyze 14,095 bacterial and 1,179 archaeal CRISPR leaders for conserved sequence motifs. This analysis reveals discretely distributed IHF binding sites and upstream motifs in a subset of I-C, I-E, I-F and II-C CRISPR loci. These leaders frequently contain multiple IHF binding sites and subtype-specific upstream motifs. The sequence, spacing, and orientation between motifs vary within and between subtypes, resulting in the detection of approximately 20 distinct leader architectures (Figure S1). Differences in leaders between closely related strains frequently involve insertions or deletions (indels) of 10–12 bps, which preserves the phase of these motifs, suggesting that phase is more important than distance per se. Additionally, we use in vitro adaptation assays to test the importance of both the sequence and the phase of leader motifs on new spacer integration in the type I-F system from P. aeruginosa. Overall, our data suggest that the mechanisms of polarized CRISPR adaptation are diverse, but all systems that rely on IHF are expected to be phase-dependent.
Results
Identification and distribution of IHF and upstream motifs
IHF binding sites have been identified in the leader sequences of a few type I-E systems and in the type I-F system from Pectobacterium atrosepticum10,12,19. To broadly determine the prevalence and distribution of IHF binding sites in CRISPR leaders, we queried all complete bacterial and archaeal genome sequences available at NCBI for CRISPR loci and associated leader sequences using CRISPRDetect20. In total, we identified 15,274 CRISPR loci, representing most of the major subtypes (Table 1). We queried 200 base pairs upstream of each CRISPR using previously established position weight matrices for IHF binding sites21,22. Sequences identified with this position weight matrix were used to construct more detailed position weight matrices, which were then used to iterate the search (Data S1). In total, we identified 8,631 putative IHF binding sites in 15,274 leader sequences. Next, we culled the list of leaders by eliminating redundant sequences that were greater than 95% identical. This eliminates closely related genomes from a few organisms which were oversampled in our initial dataset (e.g., E. coli), and enables a more accurate representation of the phylogenetic distribution of IHF binding sites in leader sequences. In this subset of 6,533 non-redundant leaders, IHF binding sites were found in 24% of I-C (n=176), 26% of I-E (n=336), 83% of I-F (n=444), and 32% of II-C (n=108) leaders (Table 1, Data S2). Interestingly, most I-F leaders (53%) and several I-E, II-C and I-C leaders (6%, 6%, and 3%, respectively) contain more than one IHF binding site. We differentiate “proximal IHF” from “distal IHF” binding sites according to their position relative to the leader-repeat junction.
Table 1. Subtype distribution of microbial CRISPRs and identified IHF binding sties.
15,274 CRISPR loci were identified in bacterial and archaeal genomes. Closely related microbial genomes, which causes an over-representation of certain CRISPR subtypes (I-B, I-C, I-E, I-F, II-A, II-C, III-A) (“Redundant” column), were systematically removed (“Non-Redundant” columns). The number of non-redundant CRISPR leaders in containing at least one, or at least two unique IHF binding sites is reported. See also Figure S1.
| Number of leaders |
|||||
|---|---|---|---|---|---|
| Redundant | Non-Redundant | ||||
|
|
|||||
| Subtype | All | All | One or more IHF sites | Two or more IHF sites | |
| Type I | I-A | 296 | 187 | 27 | 3 |
| I-B | 1516 | 761 | 238 | 36 | |
| I-C | 1324 | 754 | 178 | 24 | |
| I-D | 152 | 117 | 14 | 2 | |
| I-E | 5068 | 1329 | 335 | 84 | |
| I-F | 1683 | 536 | 444 | 279 | |
| I-U | 157 | 109 | 5 | 0 | |
| I-V | 2 | 2 | 0 | 0 | |
| Type II | II-A | 823 | 178 | 39 | 4 |
| II-B | 34 | 9 | 5 | 0 | |
| II-C | 662 | 337 | 103 | 17 | |
| Type III | III-A | 853 | 242 | 93 | 11 |
| III-B | 409 | 309 | 96 | 12 | |
| III-C | 29 | 23 | 6 | 0 | |
| III-D | 179 | 145 | 31 | 5 | |
| Type V | V-A | 28 | 14 | 6 | 1 |
| V-B | 3 | 3 | 0 | 0 | |
| Type VI | VI-A | 2 | 2 | 1 | 0 |
| VI-B | 56 | 16 | 9 | 2 | |
| VI-C | 2 | 1 | 0 | 0 | |
| NA | 1996 | 1459 | 315 | 32 | |
|
| |||||
| Total | 15274 | 6533 | 1945 | 512 | |
According to the previously established model for type I adaptation, the DNA bending behavior of IHF presents an upstream sequence motif that is recognized by specific amino acids on Cas1 (i.e., R131 and R132)11. In support of this model, the proximal IHF binding sites in I-E leaders are tightly distributed around a midpoint of 20-base pairs from the leader-repeat junction, and I-C and II-C leaders possess similar IHF sites at midpoints of 22 and 21-base pairs from the leader-repeat junction (Figure S1). However, the additional 8-base pairs that separate the IHF binding site from the leader-repeat junction in P. aeruginosa PA14, as compared to E. coli BL21, is preserved in a comparison of all type I-E and I-F leaders that contain IHF binding sites (Figure S1). This ~8-base pair insertion is unique to the type I-F systems and may be an evolutionary adaptation necessary to accommodate the Cas3 domain, which is uniquely fused to the Cas2 protein in I-F CRISPR system (Figure 1)23–25. In addition to making room for Cas3, this insertion also rotates the upstream DNA by ~260-degree. This rotation suggests that either the Cas1-2/3 integration complex relies on a distinct mechanism for recognizing an upstream sequence motif, or that I-F systems do not rely on upstream sequence motifs. To distinguish between these two possibilities and to identify conserved upstream sequence motifs in other IHF containing leaders, we performed a de novo motif search using a combination of MEME and FIMO22,26. MEME identified the upstream motif previously reported for type I-E systems11,19, as well as several new motifs (Figure 2 and Data S1, S2). The sequence and location of these motifs are characteristic of specific subtypes. In addition to the upstream motifs and proximal IHF binding sites, MEME also identified additional upstream IHF binding sites and additional, distally located upstream motifs that are either direct repeats or inverted repeats of the proximal upstream motif (Figure 2). The spacing between these motifs sometimes vary between leaders, but changes in the spacing are almost always restricted to increments of 10–12 base pairs, indicating that phase is conserved (Figure S1). The I-C distal direct repeat and inverted repeat of the usptream motif are exceptions to this rule, which are often shifted by 8-base pairs (Figure S1).
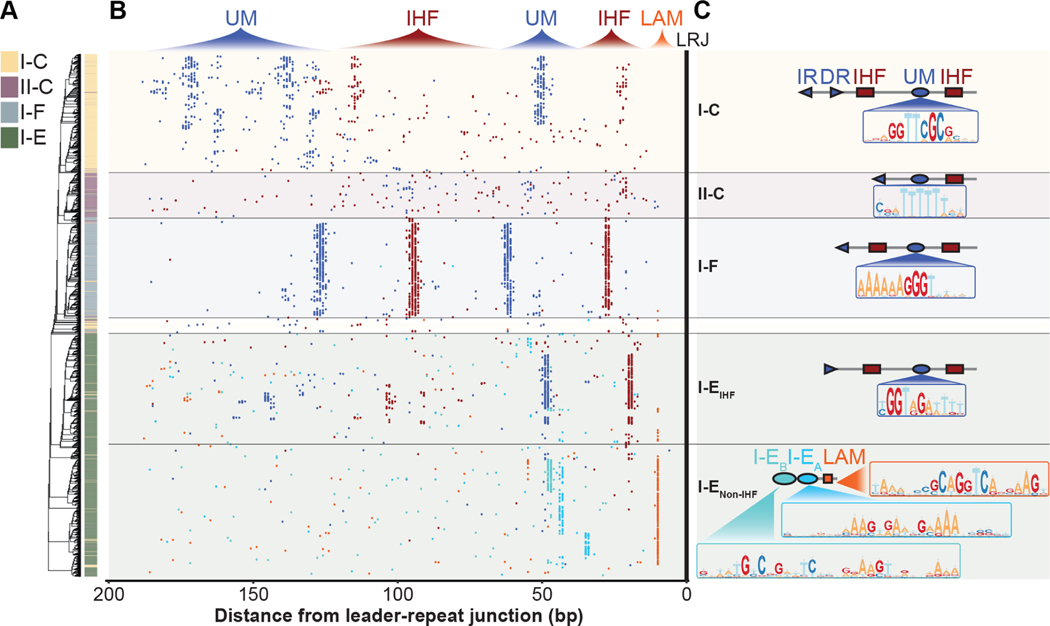
Figure 2. IHF-directed CRISPR adaptation is widespread.
(A) Phylogenetic tree generated from alignment of 200 bp of I-C, II-C, I-F and I-E CRISPR leaders and the first repeat. (B) Distributions of motifs within I-C, II-C, I-F and I-E CRISPR leaders. Each dot represents the midpoint of IHF binding sites (red), subtype specific upstream motifs (UMs) (dark blue), I-EA (cyan), or I-EB (teal), Leader Anchoring Motif (orange). Many of the leaders shown possess proximal IHF and UMs found between 0–70 bp upstream of the leader-repeat junction (LRJ), and distal IHF and UMs found 70–200 bp upstream of the LRJ. (C) Schematic of the prominent architecture for motifs within I-C, II-C, I-F and I-E CRISPR leaders. Position weight matrices are shown for subtype specific upstream motifs, and motifs found in I-E leaders that do not contain IHF binding sites (Data S1). See also Figure S1.
While IHF has been shown to be critical for polarized CRISPR adaptation in the type I-E system from E. coli10,11, we found that roughly 75% of I-E leaders do not contain a canonical IHF binding site. 20% of I-E leaders instead contain a “leader-anchoring motif” that directly abuts the first repeat, in a similar manner to the “leader-anchoring site” reported for several of the type II systems (Figure 2)9,13–15. While proximity of I-E leader-anchoring motifs to the leader-repeat junction is similar to those in type II systems, the sequences themselves are notably different. The type I-E leader-anchoring motifs contain an off-center TCA sequence, and a 5’ to 3’ TTR triplet on the opposite strand, which are hallmarks of IHF and Hbb DNA binding sites (Figure S2)27–31, though these motifs are not detectable using any of the previously established position weight matrices for IHF binding sites (Figure 2). The IHF-like leader-anchoring motif may associate with DNA bending proteins that recruit upstream motifs. In fact, MEME identifies two motifs (I-EA and I-EB) that are unique to I-E leaders that contain “IHF-like leader-anchoring motifs” (Figure 2 and Data S1). If the “IHF-like leader-anchoring motif” is in fact a DNA bending sequence that functions to recruit the unique upstream motifs (i.e., I-EA or I-EB), then the proximity of this motif relative to the leader-repeat junction, would change the phase of the upstream motif relative to what has been observed for the IHF containing leader from E. coli. Thus, we hypothesized that Cas1 proteins associated with these systems may not rely on arginines 131 and 132 (R131 and R132), which are critical for upstream motif recognition by Cas1 in IHF-dependent CRISPRs from E. coli11. To test this hypothesis, we aligned 368 Cas1 sequences from CRISPR systems that contain “IHF-like leader-anchoring motifs”. Unlike I-E Cas1 proteins that are associated with IHF containing leaders, these Cas1 proteins do not maintain R131 and R132, suggesting that the mechanism of adaptation in these CRISPRs is distinct (Table 2). These findings suggest these two types of I-E CRISPR systems may be phylogenetically distinct, indeed IHF-regulated I-E CRISPRs are found in bacteria belonging to the Phylum Proteobacteria, while leader-anchoring motif-regulated I-E CRISPRs are found in Actinobacteria (Data S2). Collectively, these results reveal diverse upstream architectures that include motifs anticipated to interact with DNA bending proteins (e.g., IHF, Hbb, or others) and DNA bending is anticipated to recruit upstream sequences that are critical for polarized adaptation in approximately 56% of all 15,274 CRISPR loci that we analyzed.
Table 2. Cas1 residues that interact with canonical I-E UM are only conserved in Cas1s associated with IHF-regulated I-E CRISPRs.
See also Figure S1.
| R131 | R132 | |||||
|---|---|---|---|---|---|---|
| Subtype | Leader motifs | Number of Cas1s | Identity | Similarity | dentity | Similarity |
| I-E | IHF or UM | 454 | 57.5% | 68% | 71.6% | 72.9% |
| I-E | LAM, I-EA, I-EB | 368 | 3.7% | 5.4% | 11.2% | 13.8% |
| I-F | IHF or UM | 499 | 11% | 11% | 11.9% | 22.4% |
IHF binding sites and upstream motif are critical for efficient integration
To determine if the IHF binding sites and upstream motifs play a direct role in I-F CRISPR adaptation, we performed in vitro integration assays using the leader sequence derived from the CRISPR2 locus of P. aeruginosa PA14. According to our bioinformatic analysis, this leader contains both proximal and distal IHF binding sites, as well as proximal and distal upstream motifs (Figure 2 and 3). To determine which of these motifs participate in new spacer integration, we compared integration efficiencies measured for the wildtype leader sequence, to leaders where we either replaced the proximal IHF binding site with an IHF consensus sequence from E. coli (Opt. IHF prox), mutated key positions in the proximal IHF binding site (Mut. IHF prox), scrambled the proximal upstream motif (Mut. UM prox), deleted the distal IHF site (Del. IHF distal) or scrambled the distal upstream motif (Mut. UM distal) (Figure 3C).
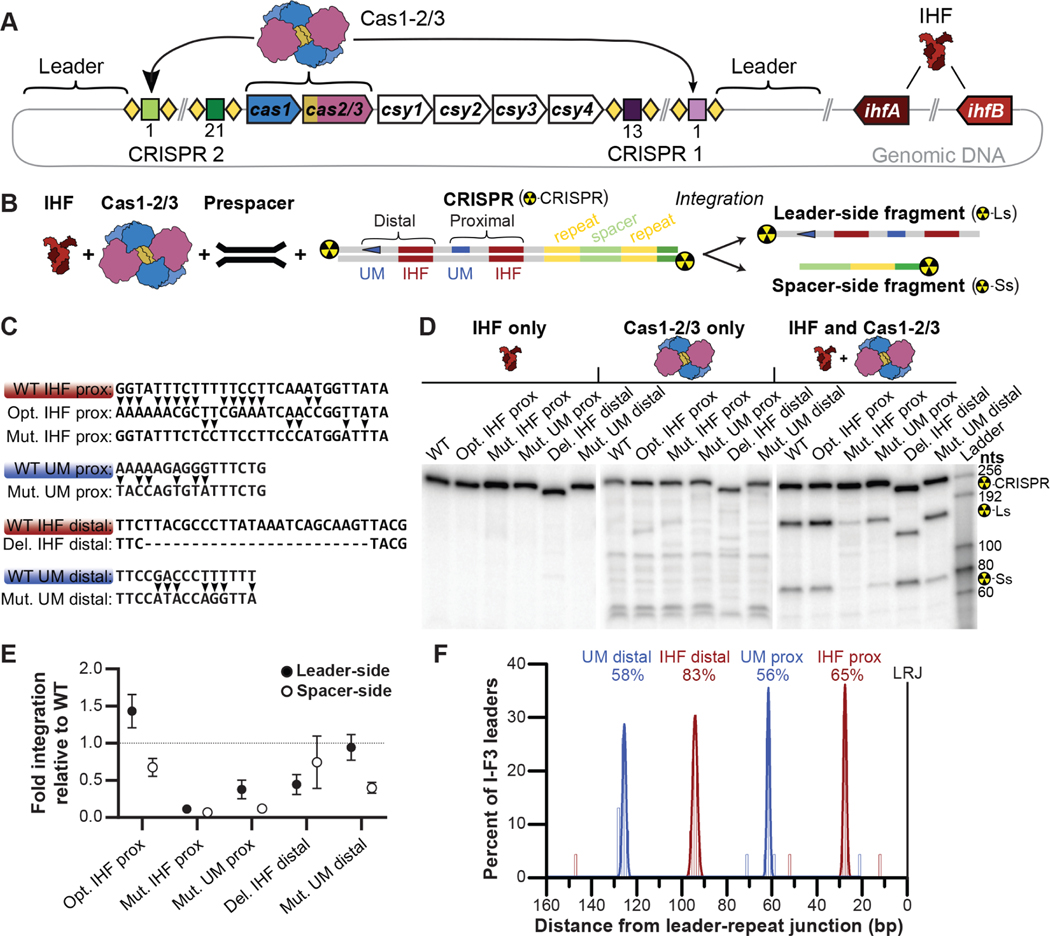
Figure 3. IHF binding sites and upstream motif are critical for efficient integration.
(A) Scheme of I-F CRISPR system from P. aeruginosa PA14. Two CRISPR loci composed of repeats (diamonds) and spacers, flank six cas genes (arrows). Cas1-2/3 assembles into a heterohexameric complex (blue, yellow, and purple, respectively) that catalyzes the integration of new spacers into CRISPR loci. IHF is a heterodimer of two related proteins encoded by ihfA (brown) and ihfB (tan). (B) Scheme for in vitro integration of a prespacer DNA into 32P-labelled DNA derived from the CRISPR2 locus of P. aeruginosa PA14. Leader- and spacer-side transesterification reactions produce a large and a small 32P-labelled DNA fragment respectively, which are separated by denaturing gel electrophoresis. Proximal and distal IHF binding sites and upstream motifs (UMs) in the leader are annotated. (C) IHF binding sites (brown) or UMs (blue) are shown. Two variants (arrows) of the proximal IHF site were tested (i.e. Opt IHF prox and Mut IHF prox.), one variant of the proximal UM (Mut. UM prox.), one variant of the distal IHF (Del. IHF idstal) and one variant of the distal UM (Mut. UM distal) were tested. (D) Representative images of integration assays resolved on denaturing gels. Reactions were performed either in the presence of IHF alone (left), Cas1-2/3 alone (middle), or in the presence of both protein complexes (right). The expected positions of full-length CRISPR substrate (Del. IHF distal variant, 233 nts; WT and other variants, 255 nts), as well as leader side (Ls) (Del. IHF distal variant, 127 nts; WT and other variants, 151 nts) and spacer side (Ss) (all variants, 77 nts) integration products are indicated. (E) Quantification of leader- (dark gray) or spacer-side (light gray) integration. The average (±s.d.) of triplicate reactions is shown. Integration was confirmed via high-throughput sequencing (Figure S3). Uncropped images and replicate gels are provided (Figure S4). (F) Tally of the midpoints of IHF binding site (red) and I-F upstream motifs (blue) identified in 23 non-redundant I-F3 CRISPR leaders36. The area of histograms fit to peaks of motif matches are reported to convey the percentage of leaders with a given motif. Most I-F3 (Tn7-associated) CRISPR leaders possess motifs needed for efficient spacer integration. See also Figure S3-S5.
IHF-binding sites in the P. aeruginosa CRISPR2 leader deviate from the E. coli IHF consensus sequence at the 5’ “A-tract” and central “WATCAR” regions, despite P. aeruginosa and E. coli IHF proteins sharing greater than 70% sequence identity (Figure S2)27–29. To determine whether an “optimized” IHF site would result in more efficient spacer integration, we replaced the IHF binding site in the I-F leader with the consensus IHF binding sequence from E. coli. These two sequences differ at 15 positions over 29 bps. Although optimization of the IHF binding site (Opt. IHF prox) resulted in slightly increased leader-side integration (1.4-fold), there is a concomitant decrease in spacer-side integration (1.5-fold) (Figure 3D, 3E). To verify that the IHF binding sequence is important for integration, we mutated six bases that are critical for IHF recognition (Figure 3C)27,28,32. These mutations (Mut. IHF prox) result in large decreases for both leader- and spacer-side integration (8.7 and 14.7-fold respectively) (Figure 3D, 3E, Figure S3 and S4), which supports previous work from the Fineran lab demonstrating that the IHF protein is necessary for adaptation in the I-F system from P. atrosepticum12. While it is expected that IHF is involved in DNA bending, there have been no prior reports of an upstream motif in I-F CRISPR leaders. To determine if the proximal upstream motif sequence we identified is important for CRISPR adaptation, we scrambled the sequence (Mut. UM prox). This mutation results in a large decrease in both leader- and spacer-side integration efficiency (2.6- and 8.3-fold respectively) (Figure 3C, 3D, 3E, Figure S3 and S4).
Fagerlund et al previously detected several IHF-like sequences upstream of the I-F CRISPR locus in P. atrosepticum12, but the importance of these distal sequences has gone untested. We hypothesized the distal IHF binding site and the distal upstream motif, which both occur at high frequency, would be functionally important for CRISPR adaptation. To test this hypothesis, we deleted the distal IHF site (Del. IHF distal) or scrambled the distal upstream motif (Mut. UM distal). Deletion of the distal IHF site reduces leader-side integration by 2.2-fold while changes to spacer side integration are within experimental error (Figure 2D and 2E). In contrast, scrambling the distal upstream motif sequence does not impact leader-side integration but reduces spacer-side integration by 2.5-fold. Collectively, these results suggest that the distal IHF and upstream motif sequences participate in new spacer integration.
Tn7-associated I-F3 leaders contain motifs needed for efficient integration
Recently, Petassi et al reported that Tn7-associated I-F3 CRISPR systems are accompanied by short CRISPRs33. The spacers in I-F3 CRISPRs have recently been shown to guide transposition in a sequence-specific manner34,35. To determine whether I-F3 CRISPR loci may continue to acquire new spacers that would enable the Tn7-like system to adapt to transpose into novel locations, we analyzed a subset of I-F3 leaders using custom position weight matrices for IHF binding sites and I-F upstream motifs developed above. In 23 non-redundant I-F3 leaders, 65% and 85% possess proximal and distal IHF binding sites (respectively), and 56–58% possess proximal and distal I-F upstream motifs (Figure 3F). Further, all four leader motifs are found in the same positions as in canonical I-F leaders (Figure 2). Since most I-F3 systems are not associated with operons that encode cas1 or cas2/3, it has been hypothesized the integration complex may be provided in trans36. The conservation of IHF binding sites and upstream motifs in a subset of the I-F3 leaders, suggests that trans-acting Cas1-2/3 integration complexes may be compatible with these leaders. One barrier to the spread of mobile genetic elements is compatibility with host factors. Our in vitro integration results show that P. aeruginosa IHF can recognize an E. coli consensus IHF binding site (Figure 3C, 3D, 3E), corroborating previous reports that E. coli IHF may complement the integration of new spacers into I-F CRISPR loci by Cas1-2/337,38. Further, these observations suggest that I-C, II-C, I-E, I-F and Tn7-associated I-F3 CRISPR systems may exchange efficiently between microbes that encode IHF.
Phased leader motifs facilitate CRISPR adaptation
The insertion of 1, 2, or 5-base pairs between the leader-repeat junction and the IHF binding site was previously shown to progressively inhibit new spacer integration into a I-E CRISPR locus11. These results were used to highlight the importance of the IHF-Cas1 interaction, which we have no reason to question. However, we hypothesized that these mutations would also alter the position of the upstream motif and that phase of the upstream motif might be more important than distance per se. To test this hypothesis, we measured the integration efficiency of new spacers into a fragment of a CRISPR from strain 14 ofP. aeruginosa (PA14). Variants of the PA14 leader were designed to either preserve (e.g., +10 bp) or disrupt (e.g., +5 bp) phasing between either the leader-repeat junction and the proximal IHF, or the proximal IHF site and the proximal upstream motif, or both (Figure 4A).
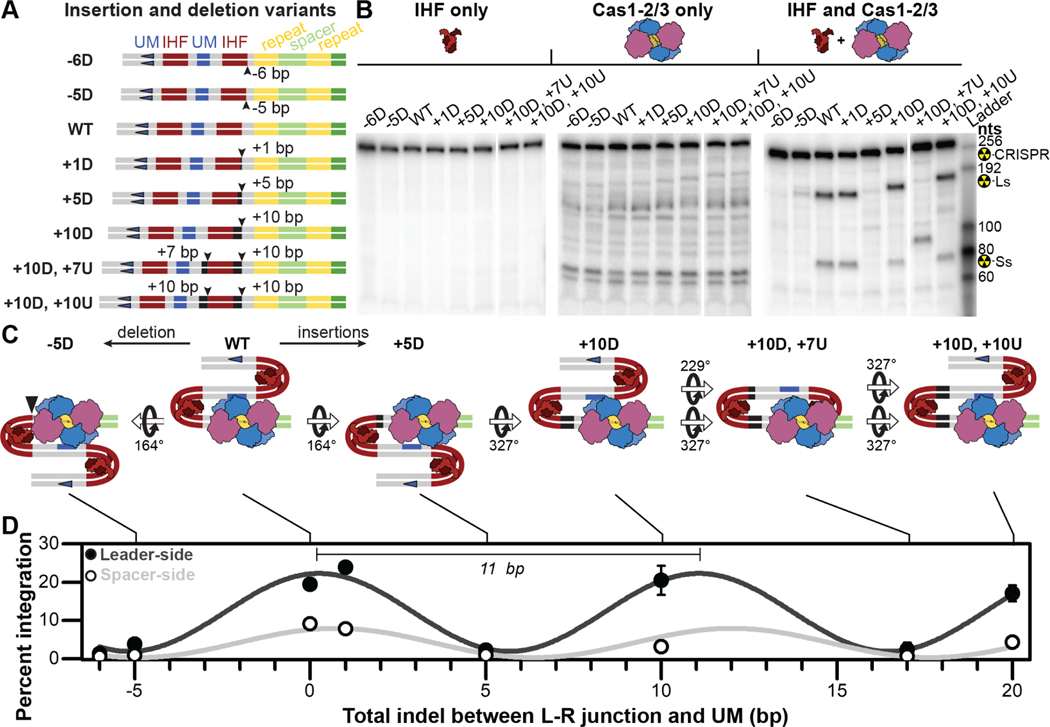
Figure 4. Phase-dependent CRISPR adaptation.
(A) Schematic of insertion and deletion (indel) variants generated to test the impact of distance and phase of leader motifs relative to the CRISPR locus. (B) I-F CRISPR integration is insensitive to indels that maintain phase, (10-base pairs), while indels of less than a full helical turn (five to seven base pairs) result in integration defects. Representative images of integration assays resolved on denaturing gels. Reactions were performed either in the presence of IHF alone (left), Cas1-2/3 alone (middle), or in the presence of both protein complexes (right). The expected positions of full-length substrate (249–275 nts depending on indel), as well as leader-side (145–161 nts depending on indel) and spacer-side (77 nts for all variants) integration products are indicated. (C) Schemes of putative IHF-bound conformations of wildtype and mutant leaders. Estimated rotations of DNA, relative to wildtype, are indicated. (D) Quantification of prespacer integration into all leader indel variants, either at leader- (dark gray) or spacer-side (light gray). The average (±s.d.) of triplicate reactions is shown. Leader- and spacer-side integration data were independently fit to sine waves (R2=0.95 and R2=0.75), of wavelengths 10.8 ± 0.4 and 11.3 ± 0.7 bp, respectively. Integration was confirmed via high-throughput sequencing (Figure S3). Uncropped images and replicate gels are provided (Figure S6). See also Figure S3 and S6.
In the absence of IHF, the Cas1-2/3 proteins from P. aeruginosa inefficiently integrate prespacers into multiple sites along the leader (Figure 3D, 4B). In contrast, the addition of IHF to these reactions facilitates Cas1-2/3–mediated integration of prespacers at the leader- and spacer- side of the first repeat (Figure 3D, 4B). High-throughput sequencing of the integration reactions performed using primers at either end of the CRISPR DNA confirm that IHF decreases off-target spacer integration from 75.2% to 9.0% (Figure S3). Additionally, we observed that leader-side integration is more efficient than spacer-side integration and that 49.8% of unambiguously integrated prespacer substrates are trimmed prior to integration (Figure S3; see methods for explanation of amibugous spacer integration events). These results are similar to those previously reported for E. coli I-E and P. atrosepticum I-F systems12,16,39.
To specifically test the importance of phase, we designed a series of I-F leader mutants that maintain the motif, but perturb the phase of the proximal IHF and proximal upstream motifs to varying degrees (−6D, −5D, +1D, +5D) (Figure 4A). Consistent with our hypothesis, indels between the proximal IHF site and the leader-repeat junction, or the proximal IHF site and proximal upstream motif, both inhibit new spacer integration. The degree to which these changes impact the efficiency of integration correlates with changes to the phase. In other words, the more out of phase, the lower the efficiency of integration. Importantly, insertions that restore the original phase (i.e., +10D, +10U), also restore integration activity (Figure 4). Collectively, the data show that integration efficiencies oscillate with a wavelength of ~11 bp, corresponding to a complete turn of double stranded DNA helix (Figure 4D)17,18. These experiments indicate that the phase of leader motifs, rather than their distance from the leader-repeat junction, is critical for efficient integration of new spacers at the leader end of the CRISPR locus.
Discussion
Since the discovery of regulatory elements in DNA40, there has been an interest in determining the “grammatical rules” of sequence motifs that regulate the storage and retrieval of genetic information. The regulatory influence of a sequence motif is determined by its location and orientation to other cis or trans acting sequence motifs, which often assemble into structures that regulate the activity of neighboring genetic elements41–43.
Specific and efficient integration of new spacers into CRISPR loci is the crucial first step in the adaptive immune response of bacteria and archaea. Here we identify leader sequences and structural elements that regulate CRISPR adaptation. These “grammatical rules” help explain the mechanisms by which CRISPRs evolve and may be important for the design of CRISPR-based data recording applications44,45. Moreover, we anticipate that motifs identified here may be used to improve computational methods designed to identify and assign CRISPR loci to particular subtypes.
While new spacers are generally added to one end of the CRISPR, there are several known examples of non-random integration of spacers outside the CRISPR locus10,12,14,46. To determine whether off-target integration observed inE. coli could be explained by the presence of specific motifs, we queried 200 bps flanking both sides of 697 integration sites to identify I-E repeats, IHF binding sites, and upstream motifs using FIMO22,46. Only 73 of these sites possess a significant match to a I-E repeat, 31 possess an IHF-like binding site, and 44 possessed a canonical I-E upstream motif. Importantly, none of these sites contain all three motifs (Data S3). While the mechanistic basis for integration at these non-CRISPR sites remains unclear, the overexpression of Cas1 and Cas2 may explain some of the promiscuity reported in these experiments46.
Previous studies have demonstrated that some type I CRISPRs rely on leader proximal and distal motifs11,12,16,19,47–49. Conversely, in type II systems, Cas1-2 directly recognizes a leader anchoring motif located directly adjacent to the first repeat9,13–15,50. Our work suggests that the mechanisms for adaptation may be more complex and that some II-C CRISPRs may require IHF, while 20% of type I-E CRISPRs appear to rely on a leader anchoring motif that is unique in sequence but similar in position to what has been observed for the type II systems (Figure 2). Bioinformatic analyses suggest IHF is predominantly restricted to Proteobacteria, although similar type II DNA-binding proteins may be found in other microbes30,51. Perhaps as expected, CRISPR leaders in which we identified phased IHF binding sites and upstream motifs (I-E, I-F, I-C and II-C) are highly represented in Proteobacteria52 (Data S2). Further, the split between I-E leaders that contain IHF sites versus those that contain leader-anchoring motifs roughly coincides with a split between I-E CRISPRs originating from Proteobacteria versus those originating from Actinobacteria (Data S2). These observations point to a continuing co-evolution of CRISPR adaptation complexes with host machinery to support the fidelity of new spacer integration.
Leader sequences are diverse and variations in leader architectures may tune adaptation rates. In fact, many microbes possess multiple CRISPR loci, which evolve at different rates52–54. For example, I-F cas genes in P. aeruginosa are flanked by a short CRISPR locus (CRISPR1) that evolves slowly and a longer CRISPR locus (CRISPR2) that has been shown to evolve more rapidly (Figure S5)54,55. P. atrosepticum SCRI1043 similarly possesses three CRISPR loci that evolve at different rates53. In corroboration with the biochemical data presented here, the fastest evolving I-F CRISPR loci in P. aeruginosa PA14 and P. atrosepticum SCRI1043 contain leaders with proximal and distal IHF binding sites, as well as upstream motifs. Whereas slower evolving CRISPR loci in these bacteria lack the proximal IHF binding site in their respective leaders (Figure S5). Collectively, these data suggest that CRISPR loci with leaders containing the full complement of motifs may adapt rapidly (Data S2). Although rapidly evolving CRISPR loci may enable a quick response to a new pathogen, the dsDNA breaks associated with the integration of a new spacer may cause such loci to be prone to lose previously acquired spacers via homologous recombination between repeats or other DNA repair pathways56,57. Thus, there may be a selective pressure that maintains CRISPR loci that have been tuned for either fast or slow adaptation.
The work presented here started with the observation that there are 8 additional base pairs between the leader-repeat junction and the first repeat of the CRISPR in P. aeruginosa (PA14), as compared to the CRISPRs in E. coli. This 8 bp insertion is not unique to the CRISPR in P. aeruginosa (PA14) but is instead conserved in I-F systems (Figure S1). While we show that IHF binding sites are common in I-E and I-F systems they are certainly not the rule. In fact, 75% of non-redundant I-E leaders do not contain IHF binding sites. Rather, 20% of I-E systems contain a leader-anchoring motif and one or two different unique upstream sequence motifs (Figure 2, Data S2). These observations led us to look more broadly for IHF binding sites and unique upstream motifs, revealing that many leaders contain multiple IHF binding sites and diverse upstream sequence motifs that are characteristic of the I-C, I-E, I-F and II-C leaders. The conservation of relative distances between motifs suggested that phase, rather than distance, might be critical for efficient adaption in IHF-dependent systems. We tested the importance of phase in the I-F system and demonstrate insertions that are in phase with the DNA helix, restore efficient adaption (Figure 4). While leaders are expected to be critical for adaption in most systems, most leaders are also expected to control CRISPR expression58–60. Ultimately, we expect that the motifs identified here are only the tip of the iceberg and that additional bioinformatic studies will provide ongoing insights that will lead to a more comprehensive understanding of how leader sequences integrate signals for expression and adaptation of CRISPRs.
STAR Methods
Resource availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Blake Wiedenheft (bwiedenheft@gmail.com).
Materials Availability
Plasmids generated in this study have been deposited to Addgene and are listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli: Bl21 DE3 competent cells | NEB | Cat# C2527I |
| E. coli: DH5α competent cells | Thermo Fisher Scientific | Cat# 18265017 |
| Chemicals, peptides, and recombinant proteins | ||
| TCEP | Soltec | Cat# M115 |
| Protease inhibitor cocktail | Thermo Fisher Scientific | Cat# 1861278 |
| Q5 DNA Polymerase | NEB | Cat# M0491L |
| T4 Polynucleotide Kinase | NEB | Cat# M0201L |
| γ-32P ATP | PerkinElmer | Cat# NEG502A250UC |
| Desthiobiotin | Sigma-Aldrich | Cat# D1411 |
| Critical commercial assays | ||
| Gel DNA recovery kit | Zymo Research | Cat# D4008 |
| Oligonucleotides | ||
| Sense DNA to make a splayed prespacer substrate: [Alexa546]TACATGCTCTAGCAAAACGACTTGCACAACGAGG | Europhins | N/A |
| Anti-sense DNA to make a splayed prespacer substrate: AAATTAAGTGCAAGTCGTTTTGCTAGAGCTACAT | Europhins | N/A |
| Sense DNA to make a 40 bp prespacer substrate: TCTACATGGTCTAGGAAAAGGACTTGGACAAGGAGGTATA | Europhins | N/A |
| Anti-sense DNA to make a 40 bp prespacer substrate: TATACCTCCTTGTCCAAGTCCTTTTCCTAGACCATGTAGA | Europhins | N/A |
| Forward primer to make 32P-labelled CRISPR integration substrates: CCAATTGCCCGAAGCTTC | Europhins | N/A |
| Reverse primer to make 32P-labelled CRISPR integration substrates: TCCAGAAGTCACCACCCG | Europhins | N/A |
| Primer to amplify integration products, P1: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGACCACCCGGCTTTCTTAG | Europhins | N/A |
| Primer to amplify integration products, P2: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCAATTGCCCGAAGCTTC | Europhins | N/A |
| Primer to amplify integration products, P3: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTGGTCTAGGAAAAGGACTTGGAC | Europhins | N/A |
| Primer to amplify integration products, P4: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCTCCTTGTCCAAGTCCTTTTCC | Europhins | N/A |
| Recombinant DNA | ||
| Plasmid: pCas1-2/3 | 25 | Addgene plasmid # 89240 |
| Plasmid: pHisIHFαIHFβ | This paper | Addgene plasmid # 149384 |
| Plasmid: pStrepIHFβ | This paper | Addgene plasmid # 149385 |
| Plasmid: pCRISPR2_wt | This paper | Addgene plasmid # 149386 |
| Plasmid: pCRISPR2_-6D | This paper | Addgene plasmid # 149387 |
| Plasmid: pCRISPR2_-5D | This paper | Addgene plasmid # 149388 |
| Plasmid: pCRISPR2_+1D | This paper | Addgene plasmid # 149389 |
| Plasmid: pCRISPR2_+5D | This paper | Addgene plasmid # 149390 |
| Plasmid: pCRISPR2_+10D | This paper | Addgene plasmid # 149391 |
| Plasmid: pCRISPR_+10D+7U | This paper | Addgene plasmid # 149393 |
| Plasmid: pCRISPR_+10D+10U | This paper | Addgene plasmid # 149394 |
| Plasmid: pCRISPR2_IHF_Opt | This paper | Addgene plasmid # 149395 |
| Plasmid: pCRISPR2_IHF_Mut | This paper | Addgene plasmid # 149396 |
| Plasmid: pCRISPR2_motif_scram | This paper | Addgene plasmid # 149397 |
| Plasmid: pCRISPR2_IHFdist_rm | This paper | Addgene plasmid # 162318 |
| Plasmid: pCRISPR2_IRdist_scram | This paper | Addgene plasmid # 162319 |
| Software and algorithms | ||
| MEME v5.3.3 | 64 | https://meme-suite.org/meme/tools/meme |
| FIMO v5.3.3 | 22 | https://meme-suite.org/meme/tools/fimo |
| WebLogo v3.7 | 65 | http://weblogo.threeplusone.com/create.cgi |
| 3D-DART | 61 | https://github.com/haddocking/3D-DART |
| Pymol v1.8.2.3 | Schrodinger | https://www.schrodinger.com/downloads/releases |
| ChimeraX v1.1 | UCSF | https://www.rbvi.ucsf.edu/chimerax/download.html |
| R-script for analysis of High-Throughput Sequencing data | This paper | https://github.com/WiedenheftLab/HTS_integration_analysis |
| Python script to extract leader sequences from CRISPRDetect output and perform phylogenetic analysis | This paper | https://github.com/WiedenheftLab/CRISPRleaderget |
| Python script to fetch sequences from a Multi-FASTA file by accession number | This paper | https://github.com/WiedenheftLab/seq_fetch |
| OriginPro | OriginLab | https://www.originlab.com/index.aspx?go=Products/Origin |
| CRISPRDetect v2.4 | 20 | https://github.com/davidchyou/CRISPRDetect_2.4 |
| RStudio | RStudio | https://www.rstudio.com/products/rstudio/ |
| MAFFT v7.429 | 69 | https://mafft.cbrc.jp/alignment/software/ |
| CD-HIT v4.8.1 | 67,68 | https://github.com/weizhongli/cdhit/releases/tag/V4.8.1 |
| MaxAlign v1.1 | 70 | https://services.healthtech.dtu.dk/service.php?MaxAlign-1.2 |
| FastTree v2.1.11 | 71 | http://www.microbesonline.org/fasttree/#Install |
| ggtree | 72–74 | https://github.com/YuLab-SMU/ggtree |
| ggplot2 | 75 | https://github.com/tidyverse/ggplot2 |
| PEAR | 78 | https://github.com/tseemann/PEAR |
| Other | ||
| Spin concentrators | Corning | Cat# 431491 |
| HisTrap HP resin | Cytiva | Cat# 17524701 |
| Microspin G25 columns | Cytiva | Cat# 27-5325-01 |
| HiLoad Superdex 200 26/600 pg | Cytiva | Cat# 28989336 |
| Superdex 75 10/300 GL | Cytiva | Cat# 17-5174-01 |
| HiTrap Heparin HP resin | Cytiva | Cat# 17040701 |
| StrepTrap HP resin | Cytiva | Cat# 28907546 |
Data and Code Availability
The datasets and code generated during this study are available in the published article or at https://github.com/WiedenheftLab.
Experimental Model and Subject Details
Bacterial strains
Escherichia coli DH5α (Thermo Fisher Scientific) cells were used to amplify plasmids used in this paper. E. coli BL21 DE3 (NEB) cells were used to express proteins used in this paper. E. coli were grown in LB (Lennox) media at either 37°C, or 16°C after induction of protein expression with 0.5 mM IPTG (isopropyl-β-D-thiogalactoside), shaking in baffled conical flasks at 200 rpm. The type I-F CRISPR system Cas proteins and CRISPR loci were cloned from P. aeruginosa UCBPP-PA14.
Method Details
Modelling the I-F CRISPR integration intermediate complex
I-F CRISPR leaders contain an 8 bp insertion between the leader-repeat junction and the proximal IHF site relative to IHF site-containing I-E CRISPR leaders. To model the impact of this insertion, 8 bp of idealized B-form dsDNA containing 11 bp/turn was built using the 3D-DART web server61. This 8 bp duplex was inserted immediately downstream of the IHF proximal binding site in the I-E Cas1-2-IHF-CRISPR holo-complex structure (PDB: 5WFE) using PyMOL v1.8.2.3 (Schrödinger, LLC). Next, components of the I-F Cas1-2/3 adapation complex were docked onto the I-E Cas1-2 adaptation complex. Docking was performed by superimposing the I-F Cas1 dimers (PDB: 3GOD) onto each I-E Cas1 dimer (PDB: 5WFE) using the “cealign” command in PyMOL v1.8.2.3 (Schrödinger, LLC). The Cas1 homodimers superimpose with a root mean squared deviation of 1.3 Å over 144 α-carbon atoms. I-F Cas2/3 proteins (PDB: 5B7I) were similarly modeled by superimposing one copy of the I-F Cas2 domain of Cas2/3 onto each of the I-E Cas2 subunits. The Cas2 subunits superimpose with a root mean squared deviation of 1.2 Å over 36 α-carbon atoms. Images of the resulting models were rendered using ChimeraX v1.162,63.
Building a database of CRISPR leaders
A total of 15,567 complete bacterial genomes and 2,658 complete bacterial chromosomes were downloaded from the NCBI RefSeq Assembly database on June 10th of 2019. In addition, 351 complete archaeal genomes and 25 complete archaeal chromosomes were downloaded from the NCBI GenBank Assembly database on the same day. CRISPRDetect v2.420 was used to identify CRISPR loci in all downloaded genomes. Search parameters were set using the following command, “-word_length 11 - minimum_word_repeatation 3 -max_gap_between_crisprs 125 -repeat_length_cutoff 17 - minimum_repeat_length 23 -minimum_no_of_repeats 3 -check_direction 1 -array_quality_score_cutoff 3”. The first repeat and 200 nucleotides upstream of each CRISPR locus (leader) were collected for downstream analyses. Regions corresponding to both sides of the CRISPR were downloaded when CRISPRDetect was not able to reliably determine the directionality (i.e., distinguish the leader from the trailer). Leaders were assigned to a particular CRISPR subtype by CRISPRDetect v2.4, or by proximity to subtype-specific cas genes. The remaining 13% of CRISPR loci (1,996 loci) could not be annotated by CRISPRDetect and lacked nearby cas genes.
Identification of conserved DNA motifs in CRISPR leaders
CRISPR leaders were analyzed using the MEME webserver with default settings64. Custom position weight matrices for I-E leader IHF binding sites, I-F leader IHF binding sites, I-E leader upstream motifs, I-F leader upstream motifs, I-C upstream motifs, II-C upstream motifs, I-EA, I-EB, and the I-E leader-anchoring motif are available in Data S1, and the FIMO-identified sequences found in CRISPR leaders have been submitted to the PRODORIC2 webserver and are available in Data S2. A local copy of FIMO22 was used to find significant matches to each of the position weight matrices. A minimum p-value threshold of 1E-4 was used to define significant position weight matrices matches. The position weight matrix for the I-F leader IHF binding site does not represent the entire protein-occluded region of DNA. Therefore, tallies for the I-F IHF binding site were extended to represent the 29 bps of DNA that are occluded by bound IHF28, in order to calculate IHF binding site midpoints. This analysis allows for comparison of IHF binding site distributions between CRISPR leaders, and for comparison in the position of motifs mapped to the sense and anti-sense strands. Sequence logos for matched DNA sequences were generated with a local copy of WebLogo 3.765.
The microbial genome dataset used here was downloaded from the NCBI RefSeq database on June 10th of 2019. As of April 1st of 2021, an additional 8,703 bacterial genomes, 1,277 bacterial chromosomes, 67 archaeal genomes and 6 archaeal chromosomes have been added to the NCBI RefSeq database. These additions represent about a third of the total number of currently available genomes and chromosomes, with large numbers of genomes orignating from uncultured bacteria and the Phyla Planctomycetes, Ignavibacteria and Candidatus Saccharibacteria. The exclusion of these new genomes is not expected to change the conclusions of this paper, which focuses on CRISPR subtypes (I-E, I-F, I-C and II-C) predominantly found in Actinobacteria, Proteobacteria and Firmicutes66 (Data S2). However, we expect that additional bioinformatic studies will uncover more leader motifs that regulate CRISPR adaptation or transcription.
Phylogenetic analyses
CRISPR leaders containing sequences matching any of the position weight matrices described in this paper (Data S1), were fetched using a Python v2.7 script that uses the Bio SeqIO package. A non-redundant list of CRISPR leaders was generated using CD-HIT v4.8.167,68 with a 95% identity cutoff. Non-redundant CRISPR leaderswere then aligned with a local version of MAFFT v7.42969, using the following command-line options: “--genafpair --maxiterate 1000 --thread 100 --threadit 100 --threadtb 100”. The resulting alignment was then analyzed with MaxAlign v1.170 to find and remove misaligned or non-homologous sequences which introduced a large number of gaps in the alignment. The list of remaining leader sequences were then realigned as above. A maximum-likelihood phylogenetic tree was then generated from realigned leader sequences, using FastTree 2.1.1171. The tree was generated using the following parameters “-quote -gamma -spr 4 -mlacc 2 -slownni -nt”. Trees were visualized in RStudio by overlaying the data of motif distances using ggtree72–74 ggplot275 and tidyverse76.
Plasmid construction
The cas1 and cas2/3 genes from P. aeruginosa UCBPP-PA14 have been previously cloned into a spectinomycin-resistant p2S LIC vector25 (Addgene, #89240). The ihfA and ihfB genes from P. aeruginosa UCBPP-PA14 were PCR-amplified to construct an N-terminal 6x-Histidine tagged variant of IHFα, an untagged IHFβ and an N-terminal Strep-II tagged variant of IHFβ. HRV3C protease cleavage sites were included between the affinity tags and the protein sequence of interest. 6x-His-IHFα and untagged IHFβ were cloned into MCS1 and MCS2 of pACYCDuet-1, respectively. This plasmid (pHisIHFαIHFβ) has been deposited with Addgene (#149384). StrepII-IHFβ was cloned into pET28a (pStrepIHFβ) and has been deposited with Addgene (#149385).
The P. aeruginosa UCBPP-PA14 CRISPR2 locus (position 2935917–2937205, in the genome NC_008463), as well upstream leader DNA (311 bp) and downstream terminus DNA (303 bp) were PCR-amplified and cloned into pHERD30T77, (pCRISPR2_wt, Addgene #149386). Leader sequence variants were generated via site directed mutagenesis using pCRISPR2 as template, followed by ligation of the linearized plasmids. All variants are deposited at Addgene, and are listed in the Key Resources Table (pCRISPR2_−6D, #149387; pCRISPR2_−5D, #149388; pCRISPR2_+1D, #149389, pCRISPR2_+5D, #149390; pCRISPR2_+10D, #149391; pCRISPR_+10D+7U, #149393; pCRISPR_+10D+10U, #149394; pCRISPR2_IHF_Opt, #149395; pCRISPR2_IHF_Mut, #149396; pCRISPR2_motif_scram, #149397; pCRISPR2_IHFdist_rm, #162318; pCRISPR2_IRdist_scram, #162319).
IHF expression and purification
E. coli BL21(DE3)cells were co-transformed with both pHisIHFα-IHFβ and pStrepIHFβ. Cells were grown in LB-Miller media (10 g/L Tryptone, 10 g/L NaCl, 5 g/L yeast extract), supplemented with 34 μg/mL Chloramphenicol and 50 μg/mL Kanamycin, at 37°C and 200 rpm to an OD600 of 0.45. Cultures were then cooled on ice for one hour, without agitation, before the addition of 0.2 mM IPTG. Cells were then grown for an additional 18 hours at 16°C, before centrifugation at 5000 g for 10 minutes. Cell pellets were suspended in Lysis Buffer (25 mM HEPES-NaOH pH 7.5, 500 mM NaCl, 10 mM Imidazole, 1mM TCEP, 5% Glycerol) supplemented with 0.3x Halt™ Protease Inhibitor Cocktail (ThermoFisher), and sonication at 30% amplitude for a total on-time of 6 minutes (1 second on-time with 3 seconds off), at 4°C. Lysate was clarified by two successive centrifugations at 12,000 rpm for 15 minutes each at 4°C. Clarified lysate was then flowed over HisTrap HP resin (Cytiva), to affinity purify His-tagged IHF heterodimers. HisTrap HP resin was washed with 20 column volumes of IHF Lysis Buffer containing 50 mM Imidazole. His-tagged IHF heterodimers were then eluted from the HisTrap HP resin with Lysis Buffer containing 500 mM Imidazole. IHF was then concentrated at 4°C (Corning Spin-X concentrators). 6x-Histidine and StrepII tags were cleaved from IHFα and IHFβ monomers respectively, in the presence of PreScision protease, overnight at 4°C while dialyzing into Lysis Buffer containing no Imidazole. Remaining 6x-His-IHFα and PreScision protease were removed by affinity chromatography using HisTrap HP resin (Cytiva). The IHF heterodimer was concentrated and diluted in buffer to reduce the NaCl concentration to 200 mM. Untagged IHF heterodimer was then purified by affinity chromatography on Heparin Sepharose (GE Healthcare), which was washed with 10 column volumes of Heparin Wash Buffer (25 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 5% Glycerol). IHF was eluted from HiTrap Heparin HP resin (Cytiva) with a linear gradient to Heparin Wash Buffer containing 2 M NaCl. Fractions containing IHF heterodimer were concentrated at 4°C (Corning Spin-X concentrators), before purification on a Superdex 75 size-exclusion column (Cytiva) in Heparin Wash Buffer.
Cas1-2/3 expression and purification
E. coli BL21(DE3) cells were transformed with pCas1Cas23 and grown 2× 1 L LB-Miller media (10 g/L Tryptone, 10 g/L NaCl, 5 g/L yeast extract), supplemented with 50 μg/mL Spectinomycin, at 37°C and 200 rpm to an OD600 of 0.45. Cultures were then cooled on ice for two hours, without agitation, before being induced with 0.2 mM IPTG. Cells were then grown for an additional 18 hours at 16°C, before being centrifuged at 5000 g for 10 minutes. Each cell pellet, originating from 1 L of culture, was resuspended in 20 mLs of Cas1-2/3 Lysis Buffer (50 mM HEPES pH 7.5, 500 mM KCl, 10% Glycerol) supplemented with 0.3x Halt™ Protease Inhibitor Cocktail (ThermoFisher). Cells were lysed via sonication, and lysate was clarified as above. StrepII-tagged Cas1-Cas2/3 was affinity purified on StrepTrap HP resin (GE Healthcare) and eluted with Cas1-2/3 Lysis Buffer containing 3 mM desthiobiotin (Sigma-Aldrich). Eluate was concentrated at 4°C (Corning Spin-X concentrators), before purification over a Superdex 200 size-exclusion column (Cytiva) equilibrated in 10 mM HEPES pH 7.5, 500 mM KCl, and 10% Glycerol.
Nucleic acid preparation
To generate a splayed prespacer with a double-stranded core, sense (5’-[Alexa546]TACATGCTCTAGCAAAACGACTTGCACAACGAGG) and antisense (5’-AAATTAAGTGCAAGTCGTTTTGCTAGAGCTACAT) DNA strands (Europhins) were resuspended in Hybridization Buffer (20 mM Tris-HCl pH 7.5, 100 mM KCl, 5 mM MgCl2) and mixed in equimolar amounts. The bases corresponding to the double-stranded core are underlined. These ssDNAs were annealed to each other by denaturation at 95°C for 5 minutes, followed by slowly cooling to room temperature over 1 hour. Annealed splayed prespacer was purified by electrophoresis through a 8% (w/v) (29:1 mono:bis) polyacrylamide in 1x TBE (100 mM Tris-Borate pH 8.3, 2 mM EDTA) at 4°C. A band corresponding to splayed prespacer was excised from the gel and purified by ethanol precipitation. A 40 bp dsDNA prespacer was made in a similar manner from sense (5’-TCTACATGGTCTAGGAAAAGGACTTGGACAAGGAGGTATA-3’) and antisense (5’-TATACCTCCTTGTCCAAGTCCTTTTCCTAGACCATGTAGA-3’) strands (Europhins).
To make 32P-labelled CRISPR integration substrates, primers (Forward primer: CCAATTGCCCGAAGCTTC-3’; Reverse primer: 5’-TCCAGAAGTCACCACCCG-3’) (Europhins) complementary to far upstream in the leader and to within the 2nd spacer of the CRISPR loci were used to amplify a DNA fragment containing most of the leader and the beginning of the CRISPR locus, from pCRISPR plasmid variants (deposited with Addgene) as templates. These PCR products were purified on a 2% (w/v) agarose native gel and extracted with a gel DNA recovery kit (Zymo Research). 1 pmole of dsDNA, corresponding to 2 pmoles of 5’ ends, was end-labelled on both strands with 4 pmoles of [γ−32P]ATP (PerkinElmer) by polynucleotide kinase (NEB) in 1x PNK buffer at 37°C for 45 minutes. PNK was heat-denatured by incubation at 65°C for 20 minutes. Spin column purification (G-25, GE Healthcare) was used to remove unincorporated radioactive nucleotides and to buffer exchange DNAs into 1x TE (10 mM Tris-HCl pH 8, 1 mM EDTA).
In vitro integration assays
Integration reactions were performed using 300 nM of splayed prespacer (purified as described above), 200 nM Cas1-2/3, ~1 nM of 32P-labelled CRISPR variant fragment and 350 nM IHF, in Integration Buffer (20 mM HEPES pH 7.5, 150 mM Potassium acetate, 5 mM MnCl2, 1 mM DTT, 5% Glycerol), for 1 hour at 37°C. Reactions were stopped by the addition of SDS to 1% and subsequent phenol-chloroform extraction. The nucleic acid containing layer was mixed 1:1 with 2x formamide loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol) and denatured at 95°C for 5 minutes, before resolving full-length 32P-labelled CRISPR strands from those fragmented by an integration event on a 8% (w/v) (29:1 mono:bis) polyacrylamide Urea gel in 1x TBE. Gels were dried and quantified using a Typhoon phosphorimager (GE Healthcare). The intensity of full-length CRISPR variant, leader-side integration fragments, spacer-side integration fragments, non-specific integration events, and background readings were quantified with Multi Gauge v3 (Fujifilm). We then calculated the no integration, leader-side integration, spacer-side integration and non-specific integration events, as percentages of total events.
High-throughput sequencing of in vitro integration products
Integration reactions were performed using 200 nM of a 40 bp dsDNA prespacer (purified as above), 200 nM Cas1-2/3, 1 nM of CRISPR variant fragment, and 350 nM IHF, in Reaction Buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 5 mM MnCl2, 1 mM DTT, 5% Glycerol), for 1 hour at 37°C. Reactions were then phenol-chloroform extracted, and then further purified with a DNA Clean and Concentrator Kit (Zymo Research). 1 μL of eluted DNA was used as a PCR template. Combinations of four primers (P1, 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGACCACCCGGCTTTCTTAG-3’; P2, 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCAATTGCCCGAAGCTTC-3’; P3, 5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTGGTCTAGGAAAAGGACTTGGAC-3’; P4, 5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCTCCTTGTCCAAGTCCTTTTCC-3’) were used to amplify all possible integration products and to simultaneously add Illumina adaptor sequences, using Q5 DNA polymerase (NEB). These PCR products were purified on a 1% (w/v) agarose gel and extracted with a gel DNA recovery kit (Zymo Research). DNA barcoding and paired-end sequencing were performed at the University of Montana Genomics Core, on an Illumina MiSeq 300 V2. Paired end reads were merged in PEAR78 and aligned to integrated and CRISPR substrates in BLAST+79. Downstream sequence analysis was performed in RStudio. The region flanking an integration site often contained 1–3 nucleotides of homology to the end of a prespacer substrate, making it impossible to distinguish if the matching nucleotides came from the prespacer molecule or the CRISPR locus-containing substrate. In the latter event, the prespacer may have been trimmed prior to integration as was seen for about half of the spacers with a clearly discernible integration site. These integration events were therefore marked as ‘ambiguous’, with the range of true integration points spanning up to a three-nucleotide window (Figure S3).
Quantification and Statistical Analysis
A threshold p-value of 1E-4 was used to report significant matches of motif position weight matrices to leader sequences by FIMO22. Autocorrelation analysis of leader motifs was performed in OriginPro (OriginLab). Quantification of gel bands for integration experiments was performed in Multi Gauge (FUJIFILM) image analysis software, from 3 independent reactions and denaturing PAGE gel images. The mean ± 1 standard deviation is reported. In Figure 3, errors were propagated to report the fold integration measured in CRISPR variants relative to wildtype. In Figure 4 the raw mean was plotted ± 1 standard deviation and the data was fit to a sine wave of equation: (in which y0 is the y-axis offset, A is the amplitude, w is the period, and xc is the phase shift). The R2 of the fits were 0.95 for the leader-side integration data and 0.75 for the spacer-side integration data.
Supplementary Material
(A) I-EA motif. (B) I-EB motif. (C) I-E IHF binding site. (D) I-E leader anchoring motif. (E) I-E Upstream motif. (F) I-F IHF binding site. (G) I-F CRISPR repeat. (H) I-F Upstream motif. (I) I-C Upstream motif. (J) II-C Upstream motif.
(A) Significant matches to position weight matrices in all CRISPR leaders analyzed. (B) Tally of the Phyla that I-E, I-F, I-C and II-C CRISPRs originate from and relation to leader motifs. (C) IHF-regulated CRISPR loci predicted to rapidly acquire new spacers. (D) Summary of leader motifs found in Campylobacter and Neisseriaspp. II-C CRISPR loci.
(A) Identification of I-E leader motifs and repeat motifs in sequences flanking both sides of non-canonical integration sites. (B) Summary of top 25 unique integration sites across 4 biological replicates reported in Nivala et al., 2018.
Highlights.
Polarized adaption of many CRISPRs requires IHF binding sites and upstream motifs
IHF bending of CRISPR leaders imposes phase-dependent spacing of upstream motifs
Different CRISPR systems contain upstream motifs specific to a CRISPR subtype
Motif preservation and spacing within leaders correlate with CRISPR adaption rates
Acknowledgements
We thank Metin Balaban and Dr. Niema Moshiri for help using TreeCluster, Dr. Christopher Brown for help with CRISPRDetect. Dr. Ewelina Malecka-Grajek for generous assistance with imaging. Jesse Thomas for maintenance of the phosphor screen imaging system. Dr. Dieter Jahn and Dr. Christian-Alexander Dudek for submission of sequences to PRODORIC. Thanks to Coltran Hophan-Nichols for computational support and members of the B.W. laboratory for critical feedback and thoughtful discussion. A.S-F. is a postdoctoral fellow of the Life Science Research Foundation that is supported by the Simons Foundation. A.S-F. is supported by the Postdoctoral Enrichment Program Award from the Burroughs Wellcome Fund. Research in the Wiedenheft lab is supported by the NIH (R35GM134867), the M.J. Murdock Charitable Trust, a young investigator award from Amgen, and the Montana State University Agricultural Experimental Station (USDA NIFA). P.K. is supported by the National Institute of General Medical Sciences of the NIH (P20GM103474). Funders had no role in designing, performing, interpreting, or submitting the work.
Footnotes
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Declaration of Interests
B.W. is the founder of SurGene, LLC, VIRIS Detection Systems Inc. and is an inventor on patent applications related to CRISPR-Cas systems and applications thereof. A.S-F. is a co-inventor on a patent for a CRISPR-based diagnostic.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol [Internet]. 1987;169(12):5429–33. Available from: https://jb.asm.org/content/169/12/5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology [Internet]. 2005. August 1;151(8):2551–61. Available from: 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- 3.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol [Internet]. 2005. February;60(2):174–82. Available from: 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- 4.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology [Internet]. 2005. March 1;151(3):653–63. Available from: 10.1099/mic.0.27437-0 [DOI] [PubMed] [Google Scholar]
- 5.Jansen R, Embden JDA van, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol [Internet]. 2002. March;43(6):1565–75. Available from: 10.1046/j.1365-2958.2002.02839.x [DOI] [PubMed] [Google Scholar]
- 6.Vale PF, Little TJ. CRISPR-mediated phage resistance and the ghost of coevolution past. Proc R Soc B Biol Sci [Internet]. 2010. July 22;277(1691):2097–103. Available from: 10.1098/rspb.2010.0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson SA, McKenzie RE, Fagerlund RD, Kieper SN, Fineran PC, Brouns SJJ. CRISPR-Cas: Adapting to change. Science (80- ) [Internet]. 2017. April 7;356(6333):eaal5056. Available from: 10.1126/science.aal5056 [DOI] [PubMed] [Google Scholar]
- 8.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science (80- ) [Internet]. 2007. March 23;315(5819):1709–12. Available from: 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 9.McGinn J, Marraffini LA. CRISPR-Cas Systems Optimize Their Immune Response by Specifying the Site of Spacer Integration. Mol Cell [Internet]. 2016. November;64(3):616–23. Available from: 10.1016/j.molcel.2016.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nuñez JK, Bai L, Harrington LB, Hinder TL, Doudna JA. CRISPR Immunological Memory Requires a Host Factor for Specificity. Mol Cell [Internet]. 2016. June;62(6):824–33. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1097276516301034 [DOI] [PubMed] [Google Scholar]
- 11.Wright A V, Liu J-J, Knott GJ, Doxzen KW, Nogales E, Doudna JA. Structures of the CRISPR genome integration complex. Science (80- ) [Internet]. 2017. September 15;357(6356):1113–8. Available from: 10.1126/science.aao0679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagerlund RD, Wilkinson ME, Klykov O, Barendregt A, Pearce FG, Kieper SN, Maxwell HWR, Capolupo A, Heck AJR, Krause KL, et al. Spacer capture and integration by a type I-F Cas1–Cas2–3 CRISPR adaptation complex. Proc Natl Acad Sci [Internet]. 2017. June 13 [cited 2018 Jan 3];114(26):201618421. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28611213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Y, Chesne MT, Terns RM, Terns MP. Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res [Internet]. 2015. February 18;43(3):1749–58. Available from: http://academic.oup.com/nar/article/43/3/1749/2411552/Sequences-spanning-the-leaderrepeat-junction [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright A V, Doudna JA. Protecting genome integrity during CRISPR immune adaptation. Nat Struct Mol Biol [Internet]. 2016. October 5 [cited 2017 Dec 9];23(10):876–83. Available from: 10.1038/nsmb.3289 [DOI] [PubMed] [Google Scholar]
- 15.Xiao Y, Ng S, Nam KH, Ke A. How type II CRISPR–Cas establish immunity through Cas1–Cas2-mediated spacer integration. Nature [Internet]. 2017. October 4 [cited 2017 Nov 7];550(7674):137–41. Available from: 10.1038/nature24020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nuñez JK, Bai L, Harrington LB, Hinder TL, Doudna JA. CRISPR Immunological Memory Requires a Host Factor for Specificity. Mol Cell. 2016;62(6):824–33. [DOI] [PubMed] [Google Scholar]
- 17.Lee DH, Schleif RF. In vivo DNA loops in araCBAD: size limits and helical repeat. Proc Natl Acad Sci [Internet]. 1989. January 1;86(2):476–80. Available from: 10.1073/pnas.86.2.476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang JC. Helical repeat of DNA in solution. Proc Natl Acad Sci [Internet]. 1979. January 1;76(1):200–3. Available from: 10.1073/pnas.76.1.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoganand KNR, Sivathanu R, Nimkar S, Anand B. Asymmetric positioning of Cas1-2 complex and Integration Host Factor induced DNA bending guide the unidirectional homing of protospacer in CRISPR-Cas type I-E system. Nucleic Acids Res [Internet]. 2017. January 9;45(1):367–81. Available from: 10.1093/nar/gkw1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biswas A, Staals RHJ, Morales SE, Fineran PC, Brown CM. CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC Genomics [Internet]. 2016. December 17;17(1):356. Available from: 10.1186/s12864-016-2627-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckweiler D, Dudek C-A, Hartlich J, Brötje D, Jahn D. PRODORIC2: the bacterial gene regulation database in 2018. Nucleic Acids Res [Internet]. 2018. January 4;46(D1):D320–6. Available from: http://academic.oup.com/nar/article/46/D1/D320/4607805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics [Internet]. 2011. April 1;27(7):1017–8. Available from: 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol [Internet]. 2015. November 28;13(11):722–36. Available from: 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter C, Gristwood T, Clulow JS, Fineran PC. In Vivo Protein Interactions and Complex Formation in the Pectobacterium atrosepticum Subtype I-F CRISPR/Cas System. PLoS One. 2012;7(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rollins MF, Chowdhury S, Carter J, Golden SM, Wilkinson RA, Bondy-Denomy J, Lander GC, Wiedenheft B. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc Natl Acad Sci [Internet]. 2017. April 24 [cited 2017 Dec 9];114(26):201616395. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28438998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res [Internet]. 2009. July 1;37(Web Server):W202–8. Available from: 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aeling KA, Opel ML, Steffen NR, Tretyachenko-Ladokhina V, Hatfield GW, Lathrop RH, Senear DF. Indirect Recognition in Sequence-specific DNA Binding by Escherichia coli Integration Host Factor. J Biol Chem [Internet]. 2006. December;281(51):39236–48. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0021925819337901 [DOI] [PubMed] [Google Scholar]
- 28.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal Structure of an IHF-DNA Complex: A Protein-Induced DNA U-Turn. Cell [Internet]. 1996. December;87(7):1295–306. Available from: 10.1016/S0092-8674(00)81824-3 [DOI] [PubMed] [Google Scholar]
- 29.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr Opin Struct Biol [Internet]. 2004. February;14(1):28–35. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0959440X0300188X [DOI] [PubMed] [Google Scholar]
- 30.Mouw KW, Rice PA. Shaping the Borrelia burgdorferi genome: crystal structure and binding properties of the DNA-bending protein Hbb. Mol Microbiol [Internet]. 2007. March;63(5):1319–30. Available from: 10.1111/j.1365-2958.2007.05586.x [DOI] [PubMed] [Google Scholar]
- 31.Kobryn K, Naigamwalla DZ, Chaconas G. Site-specific DNA binding and bending by the Borrelia burgdorferi Hbb protein. Mol Microbiol [Internet]. 2000. July;37(1):145–55. Available from: 10.1046/j.1365-2958.2000.01981.x [DOI] [PubMed] [Google Scholar]
- 32.Connolly M, Arra A, Zvoda V, Steinbach PJ, Rice PA, Ansari A. Static Kinks or Flexible Hinges: Multiple Conformations of Bent DNA Bound to Integration Host Factor Revealed by Fluorescence Lifetime Measurements. J Phys Chem B [Internet]. 2018. December 13;122(49):11519–34. Available from: 10.1021/acs.jpcb.8b07405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petassi MT, Hsieh S-C, Peters JE. Guide RNA Categorization Enables Target Site Choice in Tn7-CRISPR-Cas Transposons. Cell [Internet]. 2020. December;183(7):1757–1771.e18. Available from: 10.1016/j.cell.2020.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klompe SE, Vo PLH, Halpin-Healy TS, Sternberg SH. Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature [Internet]. 2019. July 12 [cited 2019 Aug 6];571(7764):219–25. Available from: http://www.nature.com/articles/s41586-019-1323-z [DOI] [PubMed] [Google Scholar]
- 35.Halpin-Healy TS, Klompe SE, Sternberg SH, Fernández IS. Structural basis of DNA targeting by a transposon-encoded CRISPR–Cas system. Nature [Internet]. 2020. January 18 [cited 2020 Jan 28];577(7789):271–4. Available from: http://www.nature.com/articles/s41586-019-1849-0 [DOI] [PubMed] [Google Scholar]
- 36.Peters JE, Makarova KS, Shmakov S, Koonin E V. Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc Natl Acad Sci [Internet]. 2017. August 29;114(35):E7358–66. Available from: 10.1073/pnas.1709035114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vorontsova D, Datsenko KA, Medvedeva S, Bondy-Denomy J, Savitskaya EE, Pougach K, Logacheva M, Wiedenheft B, Davidson AR, Severinov K, et al. Foreign DNA acquisition by the I-F CRISPR–Cas system requires all components of the interference machinery. Nucleic Acids Res [Internet]. 2015. December 15;43(22):10848–60. Available from: 10.1093/nar/gkv1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiegand T, Semenova E, Shiriaeva A, Fedorov I, Datsenko K, Severinov K, Wiedenheft B. Reproducible Antigen Recognition by the Type I-F CRISPR-Cas System. Cris J [Internet]. 2020. October 1;3(5):378–87. Available from: 10.1089/crispr.2020.0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S, Loeff L, Colombo S, Jergic S, Brouns SJJ, Joo C. Selective loading and processing of prespacers for precise CRISPR adaptation. Nature [Internet]. 2020;579(7797):141–5. Available from: 10.1038/s41586-020-2018-1 [DOI] [PubMed] [Google Scholar]
- 40.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol [Internet]. 1961. June;3(3):318–56. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0022283661800727 [DOI] [PubMed] [Google Scholar]
- 41.Pérez-Martín J, Rojo F, de Lorenzo V. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol Rev [Internet]. 1994;58(2):268–90. Available from: https://mmbr.asm.org/content/58/2/268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schleif R. DNA Looping. Annu Rev Biochem [Internet]. 1992. June 8;61(1):199–223. Available from: 10.1126/science.3353710 [DOI] [PubMed] [Google Scholar]
- 43.Levo M, Segal E. In pursuit of design principles of regulatory sequences. Nat Rev Genet [Internet]. 2014. July 10;15(7):453–68. Available from: http://www.nature.com/articles/nrg3684 [DOI] [PubMed] [Google Scholar]
- 44.Shipman SL, Nivala J, Macklis JD, Church GM. Molecular recordings by directed CRISPR spacer acquisition. Science (80- ) [Internet]. 2016. July 29;353(6298):aaf1175. Available from: 10.1126/science.aaf1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt F, Cherepkova MY, Platt RJ. Transcriptional recording by CRISPR spacer acquisition from RNA. Nature [Internet]. 2018. October 3 [cited 2018 Oct 30];562(7727):380–5. Available from: http://www.nature.com/articles/s41586-018-0569-1 [DOI] [PubMed] [Google Scholar]
- 46.Nivala J, Shipman SL, Church GM. Spontaneous CRISPR loci generation in vivo by non-canonical spacer integration. Nat Microbiol [Internet]. 2018. March 29;3(3):310–8. Available from: 10.1038/s41564-017-0097-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kieper SN, Almendros C, Brouns SJJ. Conserved motifs in the CRISPR leader sequence control spacer acquisition levels in Type I-D CRISPR-Cas systems. FEMS Microbiol Lett [Internet]. 2019. June 1;366(11):2016–20. Available from: 10.1093/femsle/fnz129/5525085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rollie C, Graham S, Rouillon C, White MF. Prespacer processing and specific integration in a Type I-A CRISPR system. Nucleic Acids Res [Internet]. 2018. February 16;46(3):1007–20. Available from: https://watermark.silverchair.com/gkx1232.pdf?token=AQECAHi208BE49Ooan9kkhW_Ercy7Dm3ZL_9Cf3qfKAc485ysgAAAeAwggHcBgkqhkiG9w0BBwagggHNMIIByQIBADCCAcIGCSqGSIb3DQEHATAeBglghkgBZQMEAS4wEQQMpC-UbNUbq09nHfSBAgEQgIIBk1U5V47SDh759Hx7XJ7u3wszWgJQKXxrzaXFzCns8OdJt9i [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40(12):5569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim JG, Garrett S, Wei Y, Graveley BR, Terns MP. CRISPR DNA elements controlling site-specific spacer integration and proper repeat length by a Type II CRISPR–Cas system. Nucleic Acids Res [Internet]. 2019. September 19;47(16):8632–48. Available from: 10.1093/nar/gkz677/5545008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamashev D, Agapova Y, Rastorguev S, Talyzina AA, Boyko KM, Korzhenevskiy DA, Vlaskina A, Vasilov R, Timofeev VI, Rakitina T V. Comparison of histone-like HU protein DNA-binding properties and HU/IHF protein sequence alignment. PLoS One. 2017;12(11):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernheim A, Bikard D, Touchon M, Rocha EPC. Atypical organizations and epistatic interactions of CRISPRs and cas clusters in genomes and their mobile genetic elements. Nucleic Acids Res [Internet]. 2019. November 20;48(2):748–60. Available from: 10.1093/nar/gkz1091/5634034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richter C, Dy RL, McKenzie RE, Watson BNJ, Taylor C, Chang JT, McNeil MB, Staals RHJ, Fineran PC. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42(13):8516–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westra ER, van Houte S, Oyesiku-Blakemore S, Makin B, Broniewski JM, Best A, Bondy-Denomy J, Davidson A, Boots M, Buckling A. Parasite Exposure Drives Selective Evolution of Constitutive versus Inducible Defense. Curr Biol [Internet]. 2015. April;25(8):1043–9. Available from: 10.1016/j.cub.2015.01.065 [DOI] [PubMed] [Google Scholar]
- 55.Heussler GE, Miller JL, Price CE, Collins AJ, O’Toole GA. Requirements for Pseudomonas aeruginosa type I-F CRISPR-Cas adaptation determined using a biofilm enrichment assay. J Bacteriol. 2016;198(22):3080–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kupczok A, Landan G, Dagan T. The Contribution of Genetic Recombination to CRISPR Array Evolution. Genome Biol Evol. 2015;7(7):1925–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gudbergsdottir S, Deng L, Chen Z, Jensen JVK, Jensen LR, She Q, Garrett RA. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol [Internet]. 2011. January;79(1):35–49. Available from: 10.1111/j.1365-2958.2010.07452.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pul Ü, Wurm R, Arslan Z, Geißen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR- cas promoters and their silencing by H-NS. Mol Microbiol [Internet]. 2010. March;75(6):1495–512. Available from: 10.1111/j.1365-2958.2010.07073.x [DOI] [PubMed] [Google Scholar]
- 59.Carte J, Christopher RT, Smith JT, Olson S, Barrangou R, Moineau S, Glover CVC, Graveley BR, Terns RM, Terns MP. The three major types of CRISPR-Cas systems function independently in CRISPR RNA biogenesis in S treptococcus thermophilus. Mol Microbiol [Internet]. 2014. July;93(1):98–112. Available from: 10.1111/mmi.12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Przybilski R, Richter C, Gristwood T, Clulow JS, Vercoe RB, Fineran PC. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol [Internet]. 2011. May 27;8(3):517–28. Available from: 10.4161/rna.8.3.15190 [DOI] [PubMed] [Google Scholar]
- 61.van Dijk M, Bonvin AMJJ. 3D-DART: A DNA structure modelling server. Nucleic Acids Res. 2009;37(SUPPL. 2):235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27(1):14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30(1):70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings Int Conf Intell Syst Mol Biol [Internet]. 1994;2:28–36. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7584402 [PubMed] [Google Scholar]
- 65.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res [Internet]. 2004. June;14(6):1188–90. Available from: ftp://ftp.ncbi.nih.gov/genomes/Bacteria [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pourcel C, Touchon M, Villeriot N, Vernadet JP, Couvin D, Toffano-Nioche C, Vergnaud G. CRISPRCasdb a successor of CRISPRdb containing CRISPR arrays and cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers. Nucleic Acids Res. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics [Internet]. 2006. July 1;22(13):1658–9. Available from: 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 68.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics [Internet]. 2012. December;28(23):3150–2. Available from: 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol [Internet]. 2013. April 1;30(4):772–80. Available from: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gouveia-Oliveira R, Sackett PW, Pedersen AG. MaxAlign: maximizing usable data in an alignment. BMC Bioinformatics [Internet]. 2007. December 28;8(1):312. Available from: 10.1186/1471-2105-8-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Price MN, Dehal PS, Arkin AP. FastTree 2 –Approximately Maximum-Likelihood Trees for Large Alignments. Poon AFY, editor. PLoS One [Internet]. 2010. March 10;5(3):e9490. Available from: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu G, Smith DK, Zhu H, Guan Y, Lam TT. ggtree : an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. McInerny G, editor. Methods Ecol Evol [Internet]. 2017. January 22;8(1):28–36. Available from: 10.1111/2041-210X.12628 [DOI] [Google Scholar]
- 73.Yu G, Lam TT-Y, Zhu H, Guan Y. Two Methods for Mapping and Visualizing Associated Data on Phylogeny Using Ggtree. Battistuzzi FU, editor. Mol Biol Evol [Internet]. 2018. December 1;35(12):3041–3. Available from: https://academic.oup.com/mbe/article/35/12/3041/5142656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu G. Using ggtree to Visualize Data on Tree‐Like Structures. Curr Protoc Bioinforma [Internet]. 2020. March 5;69(1). Available from: 10.1002/cpbi.96 [DOI] [PubMed] [Google Scholar]
- 75.Wickham H. ggplot2 [Internet]. Cham: Springer International Publishing; 2016. (Use R!). Available from: http://www.ncbi.nlm.nih.gov/pubmed/19791908 [Google Scholar]
- 76.Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, et al. Welcome to the Tidyverse. J Open Source Softw [Internet]. 2019. November 21;4(43):1686. Available from: 10.21105/joss.01686 [DOI] [Google Scholar]
- 77.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. PBAD-Based Shuttle Vectors for Functional Analysis of Toxic and Highly Regulated Genes in Pseudomonas and Burkholderia spp. and Other Bacteria. Appl Environ Microbiol [Internet]. 2008. December 1;74(23):7422–6. Available from: https://aem.asm.org/content/74/23/7422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics [Internet]. 2014. March 1;30(5):614–20. Available from: 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics [Internet]. 2009;10(1):421. Available from: http://www.biomedcentral.com/1471-2105/10/421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) I-EA motif. (B) I-EB motif. (C) I-E IHF binding site. (D) I-E leader anchoring motif. (E) I-E Upstream motif. (F) I-F IHF binding site. (G) I-F CRISPR repeat. (H) I-F Upstream motif. (I) I-C Upstream motif. (J) II-C Upstream motif.
(A) Significant matches to position weight matrices in all CRISPR leaders analyzed. (B) Tally of the Phyla that I-E, I-F, I-C and II-C CRISPRs originate from and relation to leader motifs. (C) IHF-regulated CRISPR loci predicted to rapidly acquire new spacers. (D) Summary of leader motifs found in Campylobacter and Neisseriaspp. II-C CRISPR loci.
(A) Identification of I-E leader motifs and repeat motifs in sequences flanking both sides of non-canonical integration sites. (B) Summary of top 25 unique integration sites across 4 biological replicates reported in Nivala et al., 2018.
Data Availability Statement
The datasets and code generated during this study are available in the published article or at https://github.com/WiedenheftLab.