Abstract
Mesothelin is a glycosylphosphatidylinositol-linked glycoprotein highly expressed in mesothelial cells, mesotheliomas, and ovarian cancer, but the biological function(s) of the protein is not known. We have analyzed the expression of the mouse mesothelin gene in different developmental stages and in various adult tissues by Northern hybridization. The 2.5-kb mesothelin transcript was detected in the mRNA of E 7.0, E 15.0, and E 17.0 stages of mouse development. In adult tissues the mesothelin gene was expressed in lung, heart, spleen, liver, kidney, and testis. To directly assess the function of the mesothelin in vivo, we generated mutant mice in which the mesothelin gene was inactivated by replacing it with the neomycin resistance gene. In homozygous mutant mice neither mesothelin mRNA nor the protein product was detected. Null mutant mice were obtained in accordance with Mendelian laws, and both males and females produced offspring normally. No anatomical or histological abnormalities were detected in any tissues where mesothelin was reportedly expressed in wild-type mice. Our results demonstrate that mesothelin function is not essential for growth or reproduction in mice.
Mesothelin, a differentiation antigen of mesothelial cells, is a 40-kDa glycosylphosphatidylinositol (GPI)-linked glycoprotein. It is synthesized as a precursor of molecular mass 69 kDa which is then proteolytically processed into an N-terminal secreted form of 32 kDa, and a membrane-bound form of 40 kDa.
The cDNA for mesothelin was cloned by two independent groups using two different approaches. In our laboratory mesothelin cDNA was cloned as an antigen for the monoclonal antibody K1 raised against ovarian cancer cells (3). K1 antibody reacts with the 40-kDa membrane-bound fragment of mesothelin. On the other hand (5), the gene was cloned as the cDNA for a 32-kDa megakaryocyte-potentiating factor. The 40-kDa GPI-linked form of human mesothelin is predominantly present on the surfaces of mesothelial cells, mesotheliomas, epithelial ovarian cancers, and some squamous cell carcinomas (2, 3). Although the 32-kDa megakaryocyte-potentiating factor can stimulate the megakaryocyte colony-forming activity of murine interleukin-3 in mouse bone marrow cell culture (5, 9), the biological functions of mesothelin are not known. Mesothelin is very abundant in normal mesothelial cells. These cells are extremely flat in shape and regulate the traffic of molecules and cells in and out of the peritoneal cavity. Mesothelin might have a role in these processes. Mesothelial cells can transform into mesotheliomas and cystadenocarcinomas, and mesothelin function might be involved in these activities. One of the most promising approaches to the identification of gene function in vivo involves the generation of mice carrying a null mutation within a specific gene.
In this study we have disrupted the mouse mesothelin gene by homologous recombination and present the characterization of the resulting mutant animals. They were born and grew as normally as wild-type mice. There were no apparent abnormalities in mutant mice in terms of growth and reproduction compared to their wild-type littermates.
MATERIALS AND METHODS
Preparation of recombinant m-mesothelin protein.
The mouse mesothelin (m-mesothelin) gene cDNA fragment (from bp 84 to 1450) was cloned by PCR amplification from PCR-ready mouse lung cDNA (Clontech, Palo Alto, Calif.) using a primer pair derived from the published m-mesothelin sequences. The DNA encoding amino acids 115 to 483 of m-mesothelin, which covers both the membrane-bound and the secretory forms, was amplified from the m-mesothelin cDNA using primer pair T145 (TTT CAT ATG GAA CAA GCC AAG GGG CTG GCT) and T147 (TTT AAG CTT GCT GAA GTC ACA TAG ATA GCT TAA CGG). The PCR product was gel purified, digested with NdeI and HindIII, and ligated into an NdeI-HindIII-digested pET23b vector (Novagen, Inc., Madison, Wis.). The resulting plasmid, pTKB6.9, encodes amino acids 115 to 483 of mesothelin with six histidine residues at the carboxy terminus encoded by the vector to facilitate purification of the protein. The recombinant protein was then expressed in Escherichia coli and purified using a Ni-nitrilotriacetic acid matrix following the supplier's instructions (Qiagen Inc., Chatsworth, Calif.).
Production of polyclonal anti-m-mesothelin antibodies in rabbit and purification of IgG from antisera.
A purified m-mesothelin protein fragment was diluted to 100 μg/200 μl and injected into rabbits with complete Freund's adjuvant for the first immunization and with incomplete Freund's adjuvant for subsequent immunizations. Sera were collected after the fourth, fifth, and sixth immunizations and titrated against the purified recombinant m-mesothelin protein. The immunoglobulin G antibodies (IgGs) from the rabbit antisera were then purified on an immobilized protein A matrix (Pierce Chemical, Rockford, Ill.) following the supplier's instructions.
Generation of ES cells heterozygous for mesothelin.
A 129 SVJ mouse genomic lambda FIX II phage library (Stratagene, La Jolla, Calif.) was screened with a probe derived from the 5′ end of the m-mesothelin cDNA. A clone containing an insert of approximately 12 kb was subcloned in pBluescript II S/K plasmid (Stratagene), and the restriction map of the insert was determined. The locations of exons were mapped by Southern blotting. The precise exon-intron boundaries were determined by DNA sequencing.
The targeting plasmid was constructed in several steps. First, a 7.2-kb EcoRI-SalI fragment was inserted between the neomycin resistance gene and the thymidine kinase gene within the pJMM4 vector (a gift from Lino Tessarollo, National Cancer Institute). Next, the 2.0-kb EcoRV-HindIII region was amplified by PCR with NotI and XbaI sites at the 5′ and the 3′ ends, respectively. The resulting fragment was digested with NotI and XbaI enzymes and inserted into the NotI-XbaI site, which is upstream of the neo marker in the targeting vector. As a result, a 1.0-kb HindIII-EcoRI fragment consisting of part of exon 1 to part of intron 3 (amino acids 20 to 100 of mesothelin) was deleted in the final vector and was replaced by the neo marker. The final targeting plasmid was designated pJMM10A (Fig. 2A).
FIG. 2.
Targeted disruption of the mesothelin gene. (A) Strategy used for mesothelin gene targeting. Construction of the targeting vector, organization of the m-mesothelin gene, and the structure of the targeted genome are shown. Restriction sites are indicated as follows: Bm, BamHI; E, EcoRI; Ev, EcoRV; H, HindIII, N, NotI; S, SalI; Xb, XbaI; Xh, XhoI. The DNA fragments used as 5′ and 3′ probes are indicated. WT, wild type; TK, thymidine kinase. (B) Southern blot analysis of BamHI-digested genomic DNA from representative pups. +/+, wild type; +/−, heterozygote; −/−, homozygote.
Embronic stem (ES) cells from 129 SVJ mice were electroporated and transfected with the targeting plasmid pJMM10A linearized with NotI. Ninety-five individual neomycin-resistant clones were picked, grown, and analyzed. Genomic DNA was extracted from each clone (6), digested with BamHI, run on a 0.9% agarose gel, and blotted onto BioDyne membranes (Life Technologies, Gaithersburg, Md.) for Southern analysis. The membranes were hybridized with a radiolabeled 5′ probe which anneals upstream of the targeting region (5′ BamHI-EcoRV fragment). The wild-type allele gives a band of 10.2 kb, whereas the correctly targeted mutant allele is represented by a band of 3.0 kb. Twelve clones were identified as correctly targeted and were reanalyzed by using a 3′ internal probe. DNAs from two of these clones were prepared and injected into blastocysts from C57BL/6 mice for generation of chimeric mice. Chimeric males were crossed with C57BL/6 females, and the agouti-colored offspring were analyzed for transmission of the mesothelin mutation. Heterozygous animals were intercrossed to generate homozygous mutated animals. Wild-type siblings obtained from the offspring of these crosses were used as control animals in the experiments. All animal work was performed in accordance with guidelines established by the National Institutes of Health.
Northern hybridization.
Northern blots containing 2 μg of poly(A) mRNA from mouse tissues (Clontech) were hybridized with randomly primed 32P-labeled DNA fragments under high-stringency conditions (1). Membranes were blocked for >4 h in hybridization solution (Oncor, Gaithersburg, Md.) and then hybridized for 15 h with a probe at 55°C. The probed blots were rinsed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS), and washed twice with 2× SSC–0.1% SDS at room temperature, with a final wash at 65°C in 0.2× SSC–0.1% SDS.
RT-PCR analysis.
To look at expression from the mutant allele, RNA was isolated from adult lung tissues with Trizol (Life Technologies) reagent for reverse transcription-PCR (RT-PCR). First-strand cDNA was synthesized by using a first-strand cDNA synthesis kit (Amersham Pharmacia Biotech, Piscataway, N.J.), and PCR was performed following the manufacturer's instructions. The primers used for PCR amplification were T75 (TCA GAG TCA TTG TTA TCC ACA GAC), which is located upstream of the HindIII site, and T76 (AGT GTG GCC TCC TGG CTT GTC TTT), located within the deleted portion of exon 1.
Histological analysis and immunohistochemistry.
Wild-type and mutant mice were maintained in the same colony in accordance with the appropriate animal care and handling guidelines. Fifteen mice from each group (3 months old) were euthanized by CO2 inhalation, and complete necropsies were performed. Different tissues from each animal were dissected out and fixed in 10% buffered formalin. A portion of lung tissue from each animal was snap-frozen in dry ice for Northern and RT-PCR analysis. Sections (5 μm thick) were cut from the formalin-fixed tissues and stained with hematoxylin and eosin. To visualize mesothelin protein by immunohistology, protein A-purified rabbit anti-mesothelin polyclonal antibody was used as the primary antibody and goat anti-rabbit IgG-biotin and avidin peroxidase (ABC kit; Vector Labs, Burlingame, Calif.) were used as the secondary antibody. Mouse slides were prepared and stained with the polyclonal antibody at 1:5,000 dilution by Molecular Histology, Inc. (Gaithersburg, Md.).
RESULTS
Expression of mouse mesothelin in different tissues.
Analysis of the expression pattern of a gene during development and adulthood can provide valuable insights regarding its function. As a prelude to disrupting the gene for mesothelin, we have analyzed the expression of the mesothelin gene in developmental stages and in different adult tissues by Northern hybridization and immunohistochemistry. The 2.5-kb mesothelin transcript can be detected in the mRNA of E7.0 embryos. The message disappears by day 11 of development, reappears by day 15, and is elevated at day 17 of mouse development (Fig. 1A). In adult tissues the expression of the mesothelin gene was predominant in lung and heart tissues, and lower expression was observed in spleen, liver, kidney, and testis (Fig. 1B). To demonstrate the cell types expressing the mesothelin protein, we performed immunohistochemical staining of mouse lung tissue using rabbit antimesothelin polyclonal antibodies. Very strong mesothelin expression was observed in the mesothelial cell lining of the lung and in the peritoneal wall, a pattern which resembles the expression pattern of mesothelin protein in human tissue (Fig. 4).
FIG. 1.
Tissue- and development-specific expression of mesothelin transcripts in mice. Shown are the results of Northern blot analysis of mesothelin expression in developmental stages (A) and eight adult tissues (B). The filters were obtained from Clontech and contained 2 μg of poly(A)+ RNA in each lane. Beta actin, blots hybridized with the β-actin probe; Sk muscle, skeletal muscle.
FIG. 4.
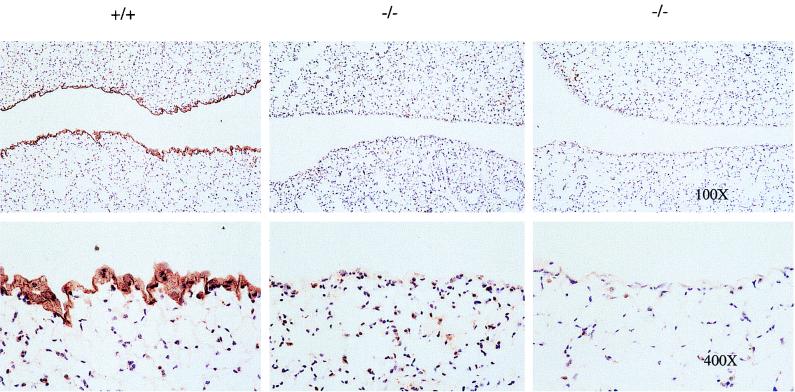
Immunohistochemical detection of mesothelin in lung tissue sections from wild-type (+/+) and mutant (−/−) adult mice with antimesothelin antibodies. Tissues were fixed and embedded in paraffin, and serial sections 5 to 6 μm in width were cut. The sections were stained with rabbit polyclonal antibodies to m-mesothelin. Unlike what was observed in mesothelial cells in wild-type lung tissue, no staining was observed in mutant tissue. Magnifications are indicated.
Targeted disruption of m-mesothelin gene.
To study the functional role of mesothelin in mouse development and growth, we inactivated the mesothelin gene. A 12-kb isogenic genomic fragment extending from the 5′-flanking DNA of the mouse mesothelin gene was isolated from a mouse genomic library (129 SVJ mouse genomic library; Stratagene). This genomic fragment was used to create the construct for homologous recombination in ES cells. The strategy used to construct the targeting vector is illustrated in Fig. 2A. As shown in Fig. 2A, a 1.0-kb HindIII-EcoRI fragment containing part of exon 1 to part of intron 3 was replaced with a neomycin resistance gene cassette. As a result amino acids 20 to 100 of the mesothelin cDNA were deleted in the targeted allele. Of 95 ES cell clones analyzed by Southern analysis (Fig. 2B), 12 clones indicated homologous recombination. DNA from two different ES cell clones heterozygous for the mesothelin gene mutation were independently injected into blastocysts and gave rise to germ line-transmitting chimeric mice that were used to breed homozygous mutant progeny.
Mesothelin-negative mutant mice have normal growth and reproductive function.
To determine the phenotype of homozygous mutant mice, heterozygous mice were intercrossed. The expected Mendelian distribution of wild-type mice and heterozygous and homozygous mutants for mesothelin was obtained. The homozygous mutant mice did not exhibit a particular phenotype and appeared similar to their heterozygous or homozygous wild-type littermates. Moreover, both homozygous mutant males and females gave rise to litters of normal size that were composed of apparently healthy mice.
Mesothelin (−/−) is a null mutation.
Since the mice homozygous for the mesothelin gene mutation [mesothelin (−/−) mice] developed and bred normally, it was essential to show unambiguously that the disrupted mesothelin allele introduced in the mice resulted in a null mutation. To test for the generation of true null mutants, we analyzed the mesothelin mRNA expression by Northern and RT-PCR analyses in wild-type and mutant mice. Total RNA was extracted from adult lung that previously had been shown to produce large amounts of mesothelin, and the amount of mesothelin mRNA was then determined by Northern blot analysis using a radiolabeled mouse mesothelin cDNA as the probe. As expected, the 2.5-kb mesothelin mRNA was detected in the RNA from wild-type mice but not in RNA from the mutant mice (Fig. 3A). To further confirm the Northern data, we performed RT-PCR analysis of the RNA samples from wild-type and mutant mice. As shown in Fig. 3B, a 125-bp fragment can be amplified from reverse-transcribed RNA samples from wild-type mice but not from mutant mice. Although Northern blot and RT-PCR analyses indicated the absence of mesothelin expression at the mRNA level, it was important to look in situ at both the actual cells or tissues that normally express mesothelin in these organs and at the possible effect of the absence of mesothelin at the tissue or cell level. Examination of histological sections of numerous organs that reportedly contain mesothelin-expressing cells did not detect differences in histological appearance between wild-type and mutant mice (Fig. 5). Furthermore, when antibodies to mesothelin were applied to these sections, it was clear that in the mesothelin (−/−) mice, cells normally expressing mesothelin were lacking it (Fig. 4) but nevertheless retained their normal histological appearance (Fig. 5).
FIG. 3.
Expression of mesothelin transcript in mutant null mice. (A) Northern blot analysis of the mesothelin transcript. Total RNA was prepared from lung tissues of wild-type (+/+) and mutant (−/−) mice. RNA (20 μg from each) was separated on a 1.2% agarose formaldehyde gel and hybridized with a mouse cDNA mesothelin probe covering most of the mesothelin coding sequence (top). Bottom, methylene blue staining of total RNA transferred onto the nylon membrane shown at the top prior to hybridization. (B) RT-PCR analysis of the mesothelin transcript. RNAs from wild type and mutant mice were reverse transcribed, amplified by PCR using the primer pair T75 and T76 (see Materials and Methods), and analyzed in 2% agarose gel.
FIG. 5.
Histological analysis of selected tissues from wild-type and mutant mice. Tissues were fixed in 4% paraformaldehyde, sectioned, and stained with hematoxylin and eosin. Magnification, ×100.
Platelet counts in mesothelin (−/−) mice.
The secretory portion of the mesothelin protein has been shown to exhibit megakaryocyte-potentiating activity in cell culture experiments (9). Megakaryocytes are the progenitors for platelet cells in the blood, which are important in blood clotting. To determine the effect of mesothelin on megakaryocyte growth in vivo, we analyzed platelet numbers in blood from wild-type and mesothelin (−/−) mice. There was no statistical difference in platelet counts between wild-type and mesothelin (−/−) mice (data not shown).
DISCUSSION
This study reports that mice harboring a null mutation in the mesothelin gene did not exhibit a particular phenotype and appeared similar to their heterozygous or homozygous wild-type littermates.
Mesothelin is one of many proteins and glycoproteins that are attached to the cell surface by GPI. GPI-linked proteins have a wide variety of functions in different cells. Some are receptors involved in cell signaling; others are involved in cellular recognition and adhesion (4, 7, 8). Mesothelin is very abundant in normal mesothelial cells, which are extremely flat and regulate the traffic of molecules and cells into and out of the peritoneal cavity. Mesothelial cells are major components of the mesothelium, which lines the serous membranes of the pleural, pericardial, and peritoneal spaces. Histological analysis of these tissues showed no difference between wild-type and mutant mice. One possible explanation may be that there are other proteins which are functionally similar to the mesothelin in cells and thus take over the functions normally provided by mesothelin. However, there are no reports of such proteins in the literature. It has also been reported that the 32-kDa secretory portion of mesothelin can stimulate the megakaryocyte colony-forming activity of murine interleukin-3 in mouse bone marrow cell culture (5, 9). Megakaryocytes are the progenitors for the platelet cells in the blood. When we analyzed the platelet numbers in both wild-type and mesothelin (−/−) mice, we found no difference, suggesting that mesothelin is not required for megakaryocyte growth and differentiation in vivo.
ACKNOWLEDGMENTS
We thank Lino Tessarollo for helping us generate the knockout mice; Wilfred Vieira for technical assistance; Jayati Bera for genotype analysis; Maria Gallo, Glenn Merlino, Partha Chowdhury, and Magnus Essand for critical reading of the manuscript; and Jennie Evans for editorial assistance.
REFERENCES
- 1.Brinkmann U, Brinkmann E, Bera T K, Wellmann A, Pastan I. Tissue specific alternatively spliced variants of hCSE1/CAS may be regulators of nuclear transporter of tissue specific proteins. Genomics. 1999;58:41–49. doi: 10.1006/geno.1998.5700. [DOI] [PubMed] [Google Scholar]
- 2.Chang K, Pai L H, Pass H, Pogrebniak H W, Tsao M S, Pastan I, Willingham M C. Monoclonal antibody K1 reacts with epithelial mesothelioma but not with lung adenocarcinoma. Am J Surg Pathol. 1992;16:259–268. doi: 10.1097/00000478-199203000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc Natl Acad Sci USA. 1996;93:136–140. doi: 10.1073/pnas.93.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dustin M L, Selvara P, Mattaliano R J, Springer T A. Anchoring mechanisms for lfa-3 cell-adhesion glycoprotein at membrane-surface. Nature. 1987;329:846–848. doi: 10.1038/329846a0. [DOI] [PubMed] [Google Scholar]
- 5.Kojima T, Oheda M, Hattori K, Taniguchi Y, Tamura M, Ochi N, Yamaguchi N. Molecular-cloning and expression of megakaryocyte potentiating factor cDNA. J Biol Chem. 1995;270:21984–21990. doi: 10.1074/jbc.270.37.21984. [DOI] [PubMed] [Google Scholar]
- 6.Laird P W, Zijderveld D A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey A, Shao H N, Marks R M, Polverini P J, Dixit V M. Role of B61, the ligand for the eck receptor tyrosine kinase, in tnf-alpha-induced angiogenesis. Science. 1995;268:567–569. doi: 10.1126/science.7536959. [DOI] [PubMed] [Google Scholar]
- 8.Stefanova I, Horejsi V, Ansotegui I J, Knapp W, Stockinger H. GPI-anchored cell-surface molecules complexed to protein tyrosine kinases. Science. 1991;254:1016–1019. doi: 10.1126/science.1719635. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi N, Hattori K, Oheda M, Kojima T, Imai N, Ochi N. A novel cytokine exhibiting megakaryocyte potentiating activity from a human pancreatic tumor-cell line HPC-Y5. J Biol Chem. 1994;269:805–808. [PubMed] [Google Scholar]