Abstract
The central melanocortin-3 and melanocortin-4 receptors (MC3R, MC4R) are key regulators of body weight and energy homeostasis. Herein, the discovery and characterization of first-in-class small molecule melanocortin agonists with selectivity for the melanocortin-3 receptor over the melanocortin-4 receptor are reported. Identified via “unbiased” mixture-based high-throughput screening approaches, pharmacological evaluation of these pyrrolidine bis-cyclic guanidines resulted in nanomolar agonist activity at the melanocortin-3 receptor. The pharmacological profiles at the remaining melanocortin receptor subtypes tested indicated similar agonist potencies at both the melanocortin-1 and melanocortin-5 receptors, and antagonist or micromolar agonist activities at the melanocortin-4 receptor. This group of small molecules represents a new area of chemical space for the melanocortin receptors with mixed receptor pharmacology profiles that may serve as novel lead compounds to modulate states of dysregulated energy balance.
Graphical Abstract
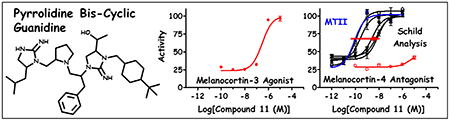
Introduction
Appetite and body weight are highly regulated processes in the human body. The discovery of new pharmacological therapies targeting key regulatory points in this system, such as the melanocortin-3 and -4 receptors, have implications in the treatment of numerous conditions ranging from cachexia and anorexia to obesity. Identifying new melanocortin ligands with novel receptor subtype pharmacological profiles may prove essential in the development of innovative therapies to treat energy disorder states.
To date, five melanocortin receptor subtypes, members of the seven transmembrane G protein-coupled receptor (GPCR) superfamily, have been cloned and characterized. The melanocortin-1 receptor (MC1R) is found primarily in the skin and is important for skin and hair pigmentation.1, 2 The melanocortin-2 receptor (MC2R) is found primarily in the adrenal glands and is essential in the hypothalamic-pituitary-adrenal pathway for steroidogenesis.2 The melanocortin-3 and melanocortin-4 receptors (MC3R and MC4R) are centrally expressed in the brain hypothalamus and are implicated in energy regulation.3–8 While the melanocortin-5 receptor (MC5R) has been implicated in exocrine function in a mouse knockout model, the function of this receptor has yet to be fully elucidated in humans.9 The melanocortin receptors are controlled by several endogenous agonists (derived from the pro-opiomelanocortin gene transcript) and the antagonists agouti (ASP) and agouti-related protein (AGRP). While these melanocortin receptors primarily signal through the Gαs signaling pathway, there are reports suggesting the melanocortin receptors can signal in tissue/cell specific mechanisms through additional pathways including Gi/o, MAPK, and the Kir7.1 ion channel.10–12
Activation of the MC3R and MC4R decreases food intake, while antagonism at these receptors increases food intake.13, 14 Mouse knockout (KO) models suggest a synergistic mechanism between the two-receptor subtypes. The MC4R KO mice are obese and have an increased body length compared to wildtype littermates.7 While the MC3R KO mice maintain a similar body weight to control wild type littermates, they possess increased fat mass and decreased lean mass.6, 8 Double MC3R/MC4R KO mice are heavier than MC4R KO mice alone and possess increased fat mass, suggesting the MC3R and MC4R have distinct, but synergistic, roles in regulating body weight.8, 15–17 A similar phenotype as the MC4RKO mouse (regarding weight gain, hyperphagia, and disrupted endocrine hormone pathways) is evident in humans deficient in MC4R signaling, as previously reviewed,18 making single nucleotide polymorphisms (SNPs) within the MC4R one of the largest monogenetic origins of obesity.19–21
While targeting the MC4R is a rational approach for modulating body weight based upon MC4R KO mice and human genetic studies, administration of purported MC4R-selective compounds identified several side effects including hypertension, sexual arousal, and skin darkening.22–25 The hypertensive effects of MC4R compounds may be transient, molecule-specific,26–30 and/or related to human genetic composition. In 2013, the structurally similar cyclic octapeptides Setmelanotide (Ac-Arg-c[Cys-DAla-His-DPhe-Arg-Trp-Cys]-NH2) and LY2112688 (Ac-DArg-c[Cys-Glu-His-DPhe-Arg-Trp-Cys-NH2) were evaluated in rhesus macaques, and only LY2112688 significantly increased heart rate and blood pressure.26 To avoid undesired side effects, one postulated alternate approach would be the targeting of the centrally located MC3R, which also plays a role in food intake and fat disposition.6, 8, 13 Selective agonist compounds for the MC3R may bypass the noted side effects of MC4R selective candidates in the search for weight modulating therapies, but remains to be proven experimentally.
In the search for selective and potent melanocortin ligands, many traditional peptide technologies have been applied. These include structural determination of the endogenous peptides,31, 32 identification of essential amino acids via alanine scans,33–35 truncation studies,36–38 and synthesis of focused libraries using peptide modifications that evoke specific turns and motifs.39–44 While peptides have the potential for exquisite selectivity and potency, they can (but not always) possess weaknesses including chemical instability, relatively short half-lives, and typically limited bio-availability compared to small molecules, as reviewed.45 Several classes of small molecule melanocortin ligands have been disclosed in the literature and reviewed.46–48 The earliest work featured β-turn-inducing thioether cyclized peptidomimetics.49, 50 Merck introduced the THIQ-substituted piperidine scaffold with several reported derivatizations.51–53 Additional scaffolds that have been explored include formaldehyde cyclized cyclophanes,54 tripeptide-based ureas,55, 56 and diazepines.57, 58 As most of these scaffolds were optimized for MC4R agonist activity, few possessed potencies at the MC3R of less than 1 μM and none were selective (greater than 10-fold more potent) for the MC3R over the MC4R.49–58
In attempts to identify novel small molecule scaffolds with activity at the MC3R, a mixture-based high throughput screen examining numerous scaffolds is reported herein. Further examination of two scaffolds, penta-amines and pyrrolidine bis-cyclic guanidines, demonstrated that the pyrrolidine bis-cyclic guanidines possessed nanomolar potencies at the MC3R and were greater than 10-fold selective for the MC3R over the MC4R. This novel MC3R small molecule scaffold contains four points of substitution and is amenable to further optimization aimed at improving DMPK properties.
Results and Discussion
The mixture-based combinatorial library technology has been extensively reviewed.59–61 This approach has previously been applied to the melanocortin system to identify tetrapeptide ligands which rescued the function of known “dysfunctional” MC4R single nucleotide polymorphic receptors as well as ligands which possessed a novel mouse (m)MC3R agonist and mouse (m)MC4R antagonist mixed receptor poly-pharmacology.42–44 For this study, it was hypothesized that mixture-based scanning techniques would identify novel small molecule ligands targeting the MC3R, similar to a previous screening that identified MC3R-selective agonist ligands.42, 43 The “Scaffold Ranking Library” (available for collaborations at the time) contained 69 distinct scaffold mixtures and was selected for this screening campaign. There were three discrete stages to the reported screen and mixtures/compounds were triaged based upon activity, selectivity, and cytotoxicity concerns. Figure 1 illustrates the overall workflow including the general yes/no decision criteria. Briefly, two structurally related scaffolds were selected for initial follow-up from among 69 scaffold mixtures after assaying them for agonist activity and selectivity for the mMC3R over the mMC4R. A mixture-based positional scan for both scaffolds was performed at the mMC3R and mMC4R to rank functionalities at each position. In the last stage, two sets of individual compounds were synthesized based on the possible permutations from the top ranked functionalities at each position for both scaffolds and were evaluated at the mMC1R, mMC3R, mMC4R, and mMC5R.
Figure 1:
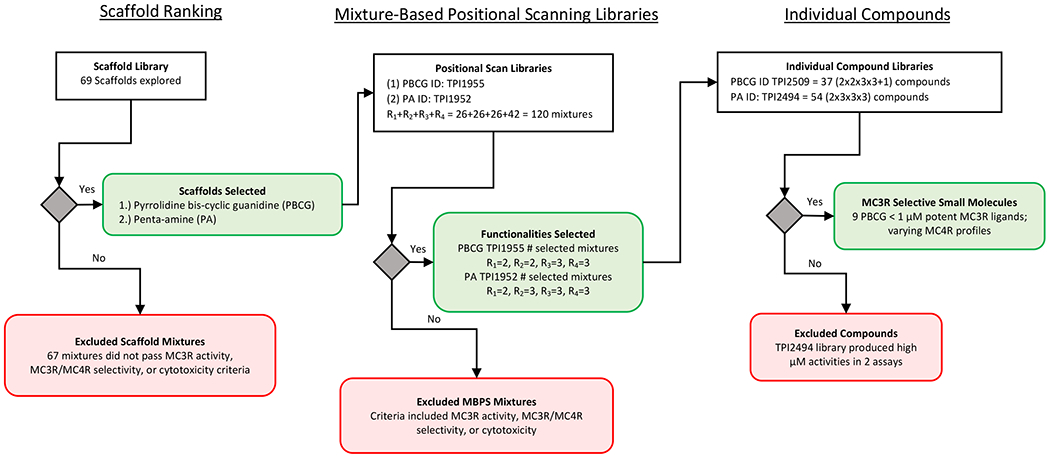
Screening Workflow and Checkpoint Criteria
Scaffold Ranking
A total of 69 scaffold mixtures, representing all the small molecule and peptide scaffolds available in the collection at the time, were evaluated for activity using a gain-of-signal β-galactosidase assay which indirectly measures cellular cAMP levels in HEK293 cells stably expressing the mMC3R or mMC4R.62 The mixtures were assayed in a dose-response manner at concentrations of 50, 25, and 12.5 μg/mL with a single replicate in at least two independent experiments. On each plate, negative controls (assay media without compound) and positive controls (7-point dose response curve of the synthetic melanocortin agonist NDP-MSH and 10−5 M forskolin) were included. Each well of the 96-well plate was visually inspected before and after compound stimulation to account for cell toxicity and assigned a score on a 4-point scale. The activity of each mixture was normalized to the maximal response of NDP-MSH (10−7 to 10−6 M) in addition to the protein content in each well, and then selectivity for the mMC3R over the mMC4R was assessed. While a few scaffold samples were shown to be active in the mMC3R screen, sample TPI1955, a pyrrolidine bis-cyclic guanidine, was the most active (mMC3R) and selective (mMC3R vs mMC4R) sample and was therefore selected for positional scanning deconvolution. TPI1952, a pyrrolidine penta-amine, was also selected for positional scanning deconvolution. TPI1952 showed some activity in mMC3R but was not selective for mMC3R over mMC4R. Additionally, TPI1955 and TPI1952 were developed from a common polyamine scaffold that utilized the same set of amino acid and carboxylic acid reagents (Scheme 1 and SI Table 1) for the R group functionalities, thus potentially allowing for structural activity relationship (SAR) studies across the scaffolds.
Scheme 1: Synthesis of the Pyrrolidine Bis-Cyclic Guanidine and Penta-Amine Libraries.
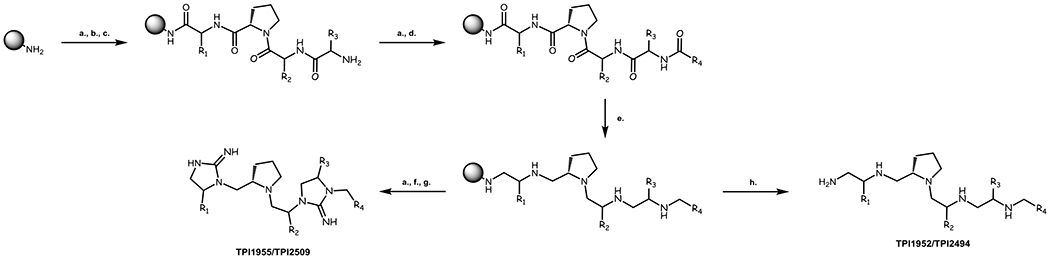
Start with MBHA resin. a. 3 x DCM (1 min); 3 x 5% DIEA/DCM (2 min); 3 x DCM (1 min); b. Boc-AA-OH (6 eq), DIC (6 eq), HOBt (6 eq), 0.1 M in DMF (1 h); c. 3 x DMF (1 min); 3 x DCM (1 min); 1 x 55% TFA/DCM (30 min); 2 x DCM (1 min); 2 x IPA (1 min); d. R-COOH (10 eq), DIC (10 eq), HOBt (10 eq), 0.1 M in DMF (1 h); e. Borane/THF (40 eq), anhydrous conditions, 65°C (72 h); piperidine, 65°C (24 h); f. under anhydrous conditions, 2 x anhydrous DCM (1 min); 1 x Cyanogen Bromide (3 eq per site), 0.1 M in anhydrous DCM (3 h); 2 x anhydrous DCM (1 min); g. HF, 0°C, (1.5 h); h. HF, 0°C, (7 h). The crude bis-cyclic guanidines were obtained at >90% theoretical yields and individual crude compounds at >70% purity by LCMS; the individual compounds were further purified to >95% by LCMS as described in the SI. The crude polyamines were obtained at >80% theoretical yields and individual crude compounds at >70% purity by LCMS; the individual compounds were further purified to >95% by LCMS as described in the SI.
It has been reported that the pyrrolidine bis-cyclic guanidine template was used to identify novel compounds with antinociceptive activity (lasting up to 5 h) and possesses anti-microbial activity to drug-resistant Gram-positive pathogens (Figure 2).63, 64 In addition, both tri- and tetra-amine templates have been used to identify potent, MOR-selective compounds.65 The exact structures and side-chain substitutions were different within the scaffold mixtures and devoid of reactive functionalities. This indicates these templates/scaffolds are not simply pan-assay interference compounds (PAINS) that can lead to false positives.66 Due to the activity in multiple biological systems, these templates may be considered “privileged” structures and prove useful with other receptor systems.
Figure 2: Prior Examples of the Pyrrolidine Bis-Cyclic Guanidine Scaffold.
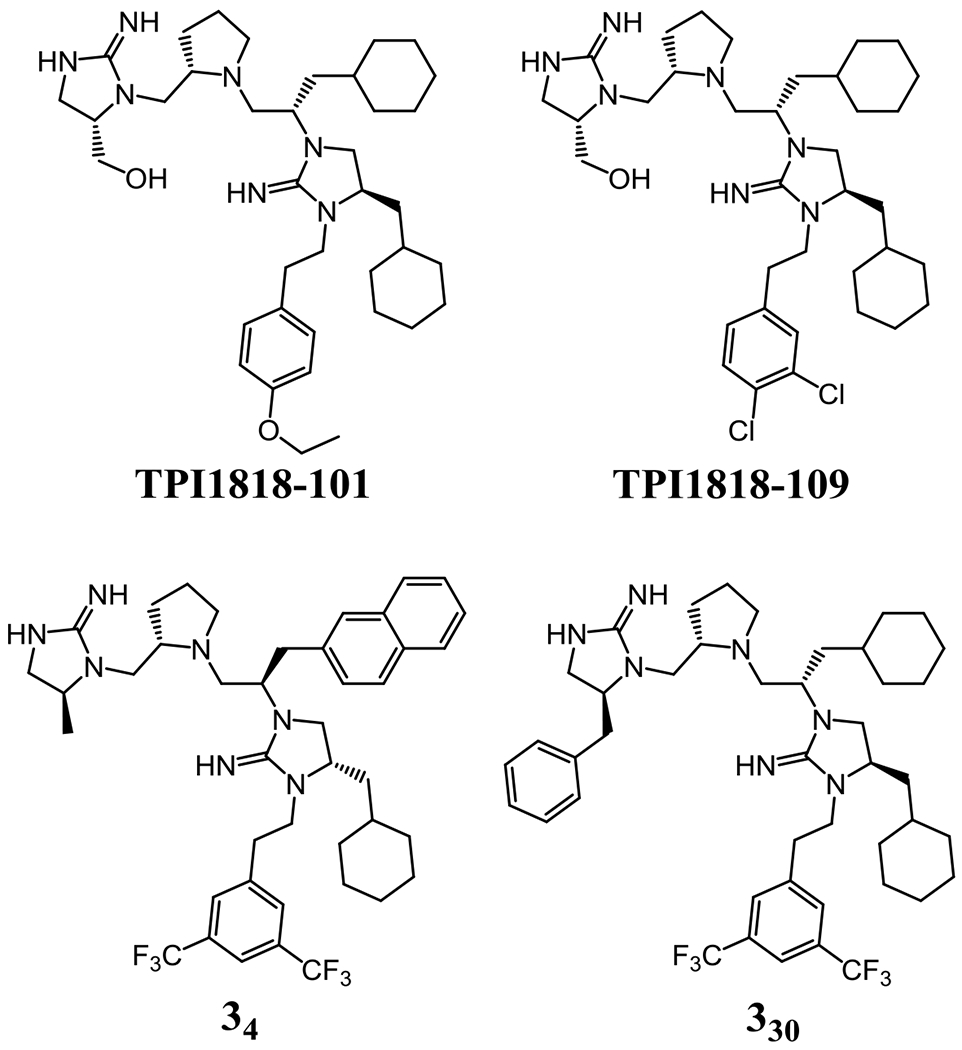
Compounds TPI1818-101 and TPI1818-109 were identified screening pyrrolidine bis-cyclic guanidine scaffold mixtures in an in vivo antinociception assay paradigm.63 Compounds 34 and 330 were identified from screening pyrrolidine bis-cyclic guanidine scaffold mixtures against methicillin-resistant Staphylococcus aureus, and also showed bactericidal activity against vancomycin-resistant Enterococcus faecalis.64
Mixture-Based Positional Scan
A mixture-based positional scan was conducted around the pyrrolidine bis-cyclic guanidine (TPI1955) and the penta-amine (TPI1952) scaffolds. The constituents in each mixture were in approximate equimolar amounts. The mixtures for each scaffold were synthesized on solid support using N-α-Boc compatible chemistry, shared a common peptidic intermediate, and utilized previously reported methodology (Scheme 1).67 Both templates were derived through the backbone modification of a capped tetrapeptide. The backbone amides were transformed to the corresponding amines through the use of an established BH3 reduction without epimerizing the α-carbons sidechain functionalities.68, 69 Control sequences were cleaved after reduction and assessed by LCMS and NMR to confirm completion of reduction. Following the reduction, an on-resin cyclization of the polyamine with cyanogen bromide yielded the desired bis-cyclic guanidine. The final product was globally deprotected and cleaved off the polystyrene support with anhydrous HF. In contrast, the penta-amine mixtures were deprotected and cleaved off the polystyrene support via anhydrous HF following the reduction. After removal of excess HF, the mixtures were assayed without further purification.
The positional scan was comprised of 120 mixtures for each template with the same chiral pool of 26 amino acid building blocks for the R1, R2, and R3 positions, and 42 carboxylic acid building blocks at the R4 position. The mixture numbers with the position and building block scanned along with the corresponding functionality are tabulated (SI Table 1). The common feature for the constituents in each mixture was a single functionality at one position and an equimolar mixture of the other functionalities X at each of the remaining positions. The mixtures were labeled by the position and functionality. The first mixture (TPI1955.001, Ala-XXX) contained 28,392 (1x26x26x42) compounds all possessing an R1 S-methyl functionality. The second mixture (TPI1955.002, Phe-XXX) also contained 28,392 (1x26x26x42) compounds all with an R1 S-benzyl functionality. The twenty-seventh mixture (TPI1955.027, X-Ala-XX) contained 28,392 (26x1x26x42) compounds all possessing an R2 S-methyl functionality. The 120 mixtures were the combination of all the building blocks (26+26+26+42) tested at each of the four positions (SI Table 1).
The amino acids and capping carboxylic acids sampled a variety of electronic and steric functionalities. Additionally, both D and L amino acids were used in positions R1, R2, and R3. The mixtures were tested for activity at the mMC3R and mMC4R in a dose-response manner at concentrations of 50, 25, and 12.5 μg/mL (in at least two independent experiments with two or more replicates per experiment), using the same β-galactosidase assay as in the scaffold-ranking assay. The results were normalized to maximal efficacy of NDP-MSH in addition to protein levels. Like the scaffold ranking screen, the cells were visually assessed for signs of stress before and after the compound stimulation in the bioassay to aid in the elimination of cytotoxic compounds. Activity at the mMC3R, selectivity for the mMC3R over the mMC4R, and cell health after compound stimulation were considered when choosing the compounds for deconvolution.
Pyrrolidine Bis-Cyclic Guanidine Positional Scan Results
The mean normalized activities were plotted and the relative potency and selectivity profiles for each of the MC3R and MC4R subtypes were compared (Figure 3). Positions R1, R3, and R4 showed mixtures with relative potencies greater than 60% at the mMC3R while position R2 did not show any mixture with activities greater than 60%. The most potent mixtures at the R1, R2, and R3 positions gravitated towards functionalities which possessed stereochemistry derived from D amino acid building blocks. Additionally, these D amino acid-derived functionalities also tended to possess the greatest selectivity for the mMC3R over the mMC4R. For this reason, D amino acid building blocks were selected for positions R1, R2, and R3 for the follow-up compounds synthesized individually.
Figure 3: Illustration of the Results Obtained from the Pyrrolidine Bis-Cyclic Guanidine Mixture-Based Positional Scan (TPI1955, 50 μg/mL).
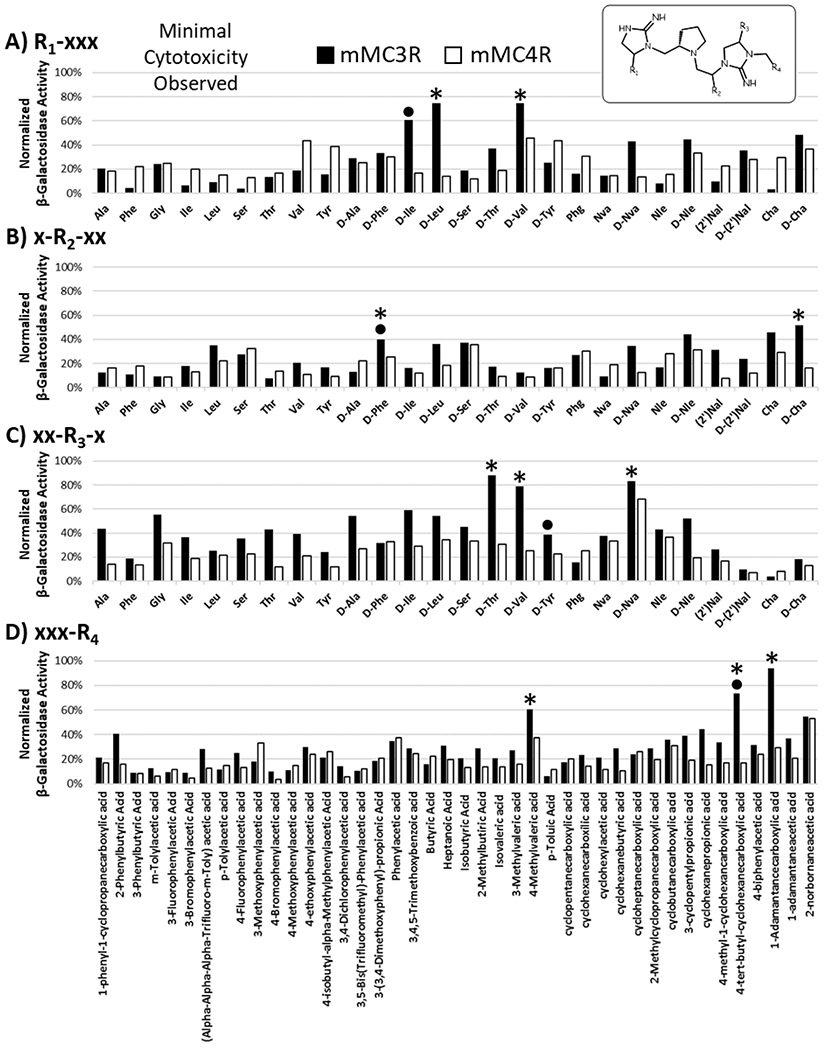
A total of 120 mixtures were assayed in the mixture-based positional scan around the pyrrolidine bis-cyclic guanidine template (inset). Mixtures were synthesized through the backbone modification and subsequent cyclization of a the tetrapeptide scaffold R4-R3-R2-Pro-R1-NH2, and a total of 26 amino acid functionalities were scanned at positions R1, R2, and R3 while 42 carboxylic acid derivatives were scanned at position R4. HEK293 cells stably transfected with the desired melanocortin receptor subtype were stimulated with 50 μg/mL a mixture in a CRE reporter β-galactosidase assay in a 96-well plate format. The results were normalized to well protein content and the maximal activity achieved by NDP-MSH. After mixture stimulation, the health of the cells was visually assessed under a microscope, and it was noted that little to no visual cell cytotoxicity was observed. Mixtures with optimal potency and selectivity for the mMC3R over the mMC4R were selected (*) for the positional scan deconvolution which resulted in the synthesis of 36 (2x2x3x3) individual compounds. One additional compound was synthesized based on the results (•) obtained from a closely related mixture-based positional scan on the penta-amine template (TPI1952 scaffold, TPI2494 deconvolution library) for comparative purposes.
The chemical properties of the selected mixtures tended to be consistent within each R group. Two small aliphatic side chains, R-isobutyl (D-Leu) and R-isopropyl (D-Val), were selected for the R1 position. The R-cyclohexyl-methyl (D-cyclohexylalanine) and the aromatic R-benzyl (D-Phe) side chains were chosen for the R2 position. The small aliphatic side chains R-propyl (D-norvaline) and R-isopropyl (D-Val), as well as the small nucleophilic side chain (S,R)-1-hydroxyethyl (D-Thr) were picked for the R3 position. For MC3R potency and selectivity, the R4 position favored the bulky 4-tert-butyl-cyclohexyl-methyl (4-tert-butyl-cyclohexanecarboxylic acid) and adamantan-1-yl-methyl (1-adamantanecarboxylic acid) functionalities and a 4-methyl-pentane side chain derived from 4-methylvaleric acid.
At the R4 position, activity was modulated with minor structure alterations. For example, shifting one methyl group from 4-methyl-pentane (4-methylvaleric acid, 60% relative potency, Figure 3) to 3-methyl-pentane (3-methylvaleric acid, <30% relative potency, Figure 3) decreased receptor activity. A larger decrease was observed for a methylene group extension when comparing adamantan-1-yl-methyl (1-adamantanecarboxylic acid, >90% relative potency, Figure 3) compared to adamantan-1-yl-ethyl (1-adamantaneacetic acid, <40% relative potency, Figure 3).
Figure 4 illustrates the building blocks which were selected based on the results from the mixture-based positional scan for the synthesis of the 36 individual compounds (2x2x3x3), with one additional compound added as described below. The selected building blocks induced little to no visual cell cytotoxicity at the highest concentration assayed (50 μg/mL).
Figure 4: Building Blocks for the Pyrrolidine Bis-Cyclic Guanidine Scaffold.
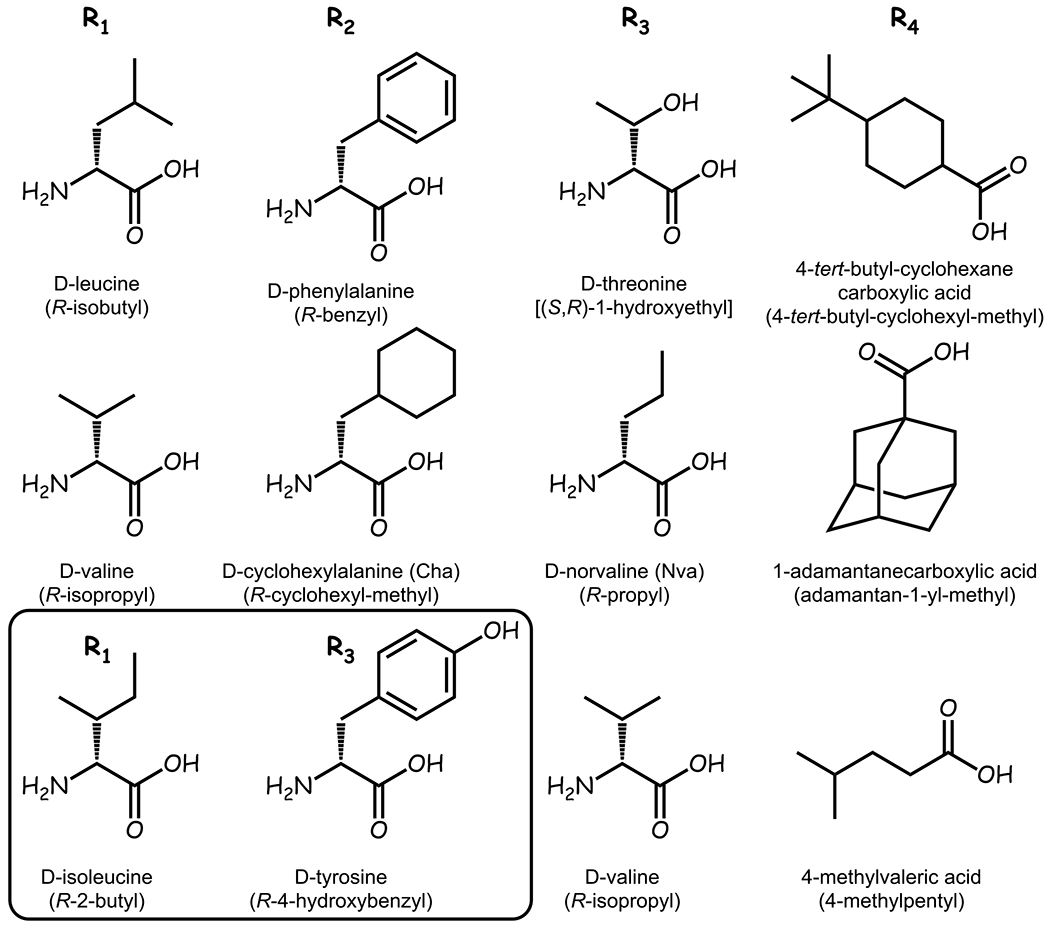
A combinatorial library of individual compounds using the pyrrolidine bis-cyclic guanidine template were synthesized using the building blocks selected from the mixture-based positional results, and the corresponding functionality is indicated under each building block. Positions R1, R2, and R3 contained exclusively D amino acid building blocks. Inset contains two additional building blocks which were selected for mixture deconvolution based upon hits in the penta-amine mixture-based positional scan.
Penta-Amine Positional Scan Results
The mixture-based positional scan around the penta-amine template utilized the same 26 amino acids (R1, R2, and R3), 42 carboxylic acid derivatives (R4), and the same dose-response concentrations compared to the pyrrolidine bis-cyclic guanidine positional scan. The normalized results obtained for the 50 μg/mL concentration for both the mMC3R and mMC4R are presented in Figure 5. During the assay, each well was visually assessed under a microscope as an indicator of cytotoxicity. Unlike the pyrrolidine bis-cyclic guanidine compounds, exposure to the penta-amine small molecules resulted in substantial apparent cellular stress, observed as cells balling up and/or lifting from the plate surface. The relative amounts of apparent cell stress are represented in Figure 5 with increasing stress resulting in increased red content of the cell health circle. These observations were considered during the deconvolution analysis in the design of the individual compound library.
Figure 5: Illustration of the Results Obtained from the Penta-Amine Mixture-Based Positional Scan (TPI1952, 50 μg/mL).
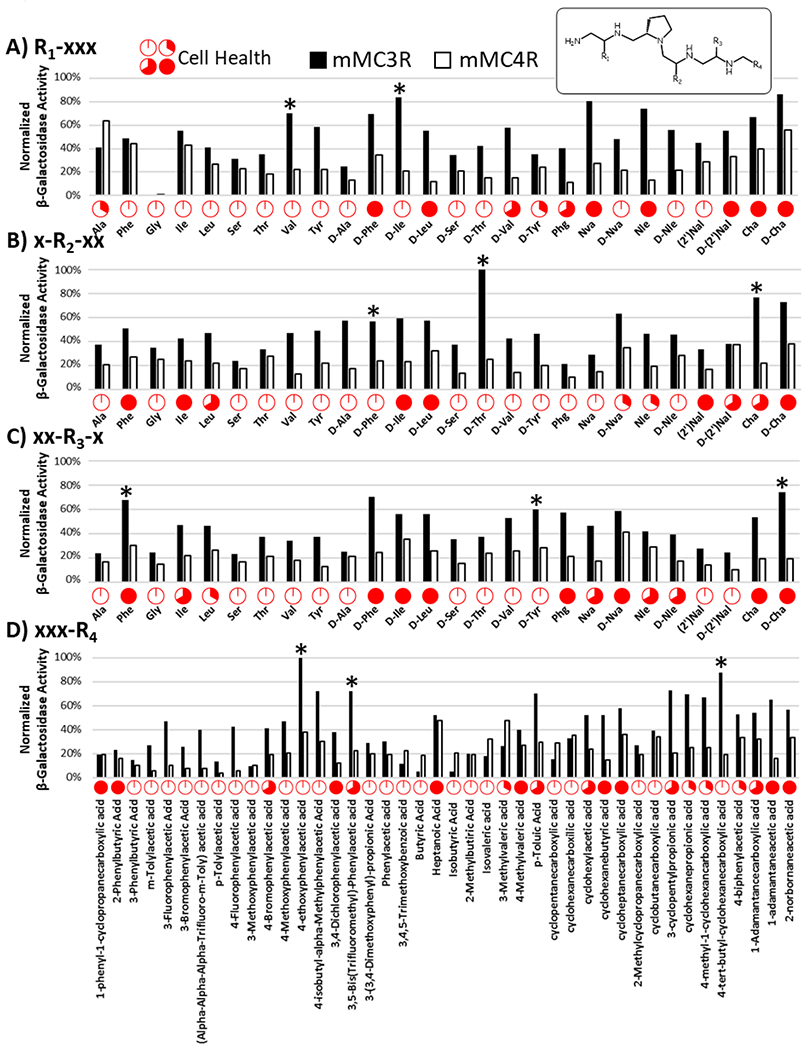
A mixture-based positional scan was conducted around a poly-amine template (inset) based on the reduced tetrapeptide scaffold R4-R3-R2-Pro-R1-NH2 where R1, R2, and R3 were based on 26 amino acid building blocks and R4 was based on 42 carboxylic acid derivatives. HEK293 cells stably transfected with the desired melanocortin receptor subtype were stimulated with 50 μg/mL a mixture in a CRE reporter β-galactosidase assay in a 96-well plate format. The results were normalized to well protein content and the maximal activity achieved by NDP-MSH. After mixture stimulation, the health of the cells was visually assessed under a microscope, and the fraction of wells which appeared stressed was approximated and reported above via a pie chart for the mMC3R expressing cells. Unlike the pyrrolidine bis-cyclic guanidine mixture-based positional scan, a substantial number of wells appeared stressed which is a possible indicator of cytotoxicity. The building blocks picked for follow up mixture deconvolution were indicated (*) above each of the selected building blocks, and from these results a combinatorial library of 54 (2x3x3x3) individual compounds was constructed.
Overall, the mixtures trended towards increased potency at the mMC3R compared to the mMC4R, yet cell stress was noted for many of the most potent and/or selective MC3R mixtures (Figure 5). The two functionalities selected for the R1 position were the smaller hydrophobic functionalities S-isopropyl (L-Val) and R-2-butyl (D-Ile). The three functionalities selected for the R2 position included the R-benzyl (D-Phe), S-cyclohexyl-methyl (L-Cha), and (S,R)-1-hydroxyethyl (D-Thr) functionalities. For the R3 position, the two aromatic functionalities S-benzyl (L-Phe) and R-4-hydroxybenzyl (D-Tyr) were selected. Additionally, the R-cyclohexyl-methyl (D-Cha) was also chosen. The functionalities selected for the R4 position consisted of substituted phenyl and cyclohexyl rings with 2-(4-ethoxy-phenyl)-ethyl (4-ethoxyphenylacetic acid), 2-(3,5-bis-trifluoromethyl-phenyl)-ethyl [3,5-Bis(trifluoromethyl)-phenylacetic acid], and 4-tert-butyl-cyclohexyl-methyl (4-tert-butyl-cyclohexanecarboxylic acid). The position substitutions (Figure 6) selected led to the synthesis of 54 (2x3x3x3) individual compounds.
Figure 6: Building Blocks for the Penta-Amine Scaffold.
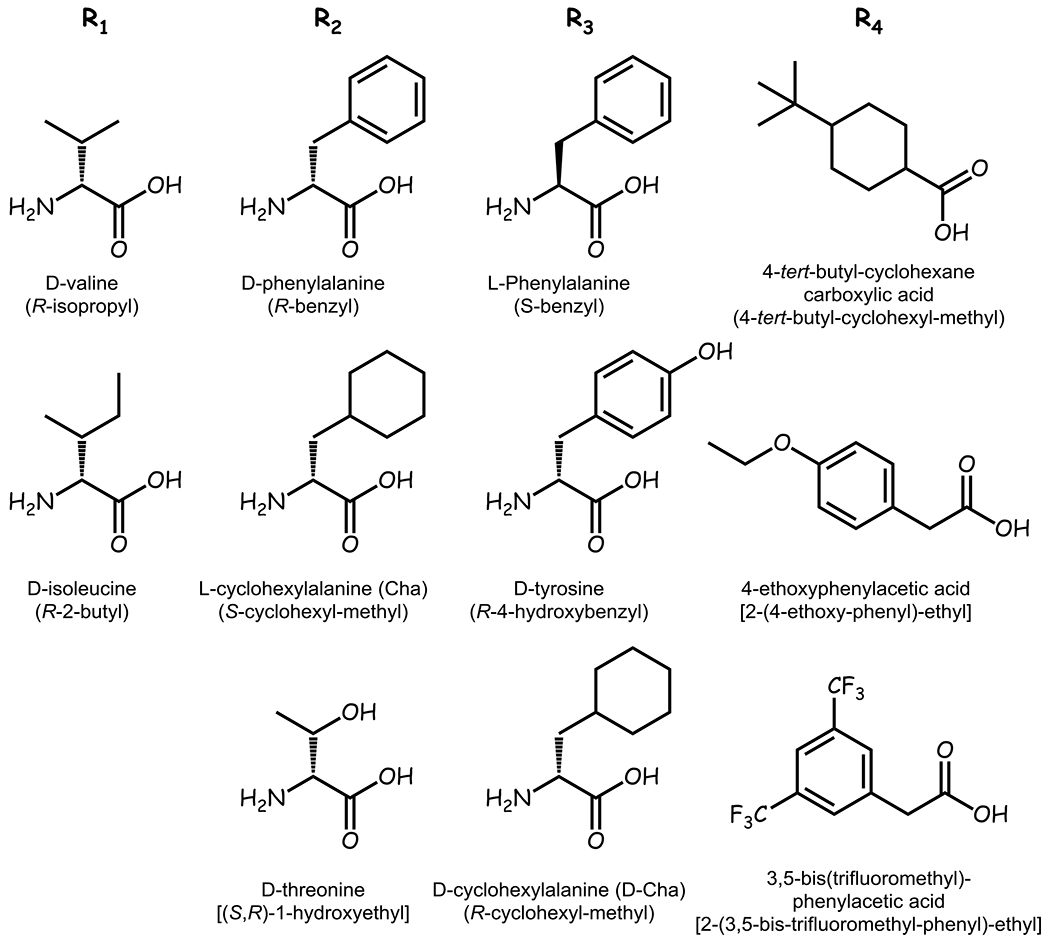
A combinatorial library of individual compounds using the penta-amine template were synthesized using the building blocks selected from the mixture-based positional results, and the corresponding functionality is indicated under each building block.
Common to both positional scan active mixtures were the R-benzyl (D-Phe) at the R2 position and 4-tert-butyl-cyclohexyl-methyl (4-tert-butyl-cyclohexanecarboxylic acid) at R4. To probe potential differences between the two scaffolds, an additional compound was synthesized containing R-2-butyl (D-Ile) functionality at the R1 position, R-benzyl (D-Phe) at R2, R-4-hydroxybenzyl (D-Tyr) at R3, and 4-tert-butyl-cyclohexyl-methyl (4-tert-butyl-cyclohexanecarboxylic acid) at R4 in the bis-cyclic guanidine template. These positional scan mixtures were selected with the penta-amine template and were observed to have a moderate activity profile with the pyrrolidine bis-cyclic guanidine template (Figure 3).
Individual Compound Library
Compound Synthesis:
Two series of individual compounds were synthesized and assayed based upon the MC3R activity and selective mixture data from the pyrrolidine bis-cyclic guanidines and penta-amines positional scanning libraries. Each compound was synthesized on solid support using similar methodology as the mixture-based positional scan (Scheme 1). Individual compounds were synthesized with single chemical building blocks (Figures 3 and 5) as opposed to equireactive mixtures of building blocks used in the generation of the positional scanning libraries. Each individual compound was purified to ≥95% pure using preparative HPLC following HF cleavage from the solid support. The compounds were characterized by 1H NMR and LC/MS methods. Characterization information is provided for the pyrrolidine bis-cyclic guanidine analogues (TPI2509) and penta-amine analogues (TPI2494) in the supplemental information. The penta-amine analogues (TPI2494) did not result in compounds with sub-micromolar potencies in the initial in vitro screening and were triaged as illustrated in the screening workflow and checkpoint criteria (Figure 1).
In Vitro Evaluation
Penta-amine Analogue Series Primary Screen
The results from the penta-amine library screen led to the synthesis of 54 compounds which yielded micromolar agonists at all four of the selected melanocortin receptor subtypes assayed (data not shown). The compounds were assayed in the β-galactosidase assay utilized in the scaffold ranking and positional scans, as well as in an orthogonal “AlphaScreen” assay that directly measures cAMP levels.70–72 The penta-amine compounds tended to have higher amounts of apparent cellular cytotoxicity (Figure 5) whereas the related pyrrolidine bis-cyclic guanidines tended to have little toxicity present, observable sometimes only at the highest concentrations tested. Since the β-galactosidase values were normalized to protein levels, and toxic compounds produced lower protein levels (decrease cell count will result in less protein), a low signal may be artificially enhanced as a false positive for toxic compounds. This may explain why the individual penta-amine compounds yielded minimal to no activity upon mixture deconvolution. In a prior screen using a penta-amine scaffold, individual compounds were tested for cytotoxicity using a MTT assay in VERO cells.67 Compounds were reported to decrease viability to less than 50% at concentrations of 1.03 to 4.23 μg/mL (1.7 to 6.5 μM),67 supporting the observed toxicity for the penta-amine scaffold in the present study. Although this library of compounds could be construed as negative data, it was included to provide a comparative example of a mixture-based positional scanning library. The MC3R selective results obtained from the pyrrolidine bis-cyclic guanidine compounds indicate that the MC3R activity is specific to this class of ligands and that similar libraries do not result in positive hits, supporting the activity of the pyrrolidine bis-cyclic guanidine compounds.
Pyrrolidine Bis-Cyclic Guanidine Analogue Overview and SAR Summary
Each of the 37 compounds based on the pyrrolidine bis-cyclic guanidine template were evaluated for functional activity at the mMC1R, mMC3R, mMC4R, and mMC5R using the β-galactosidase bioassay. The compounds were initially screened for agonist activity from 10−4 to 10−10 M, and the range was adjusted accordingly if the compound EC50 values were more potent and not within the middle of this concentration range. The reported results are the means of duplicate replicates of at least three independent experiments (Table 1). For select compounds, the 100 μM concentration resulted in toxicity. For these compounds, the 100 μM concentration was not used in the sigmoidal dose-response fitting. If efficacy of at least 90% compared to NDP-MSH controls was observed for at least two concentrations, the EC50 (nM) values are reported. For compounds where EC50 values could not be calculated due to the absence of a sigmoidal dose-response curve, the percent activity at 10 μM, as compared to the maximal NDP-MSH response, is reported and the ligands binned into two groups (A = 10-50% NDP-MSH, B = 51-90% NDP-MSH). Compounds that did not possess any agonist potency or antagonist activity were reported as >100,000. Three compounds (1, 2, and 28) possessed partial agonist efficacy with a sigmoidal dose-response curve at the MC4R (Figure 7). The apparent EC50 values and percent of maximal NDP-MSH signal are indicated for these three compounds. Additional dose-response antagonist experiments and pA2 values73 were determined at the MC4R for compounds that were not full agonists at the MC4R, were full agonists at the MC3R, and possessed EC50 values < 1 μM at the MC3R.
Table 1:
Summary of the Pyrrolidine Bis-Cyclic Guanidine Pharmacology at the Selected Mouse Melanocortin Receptors
| ID | R1 | R2 | R3 | R4 | EC50(nM) |
|||
|---|---|---|---|---|---|---|---|---|
| mMC1R | mMC3R | mMC4R | mMC5R | |||||
| NDP-MSH [(Nle4, DPhe7)α-MSH] | 0.09 ± 0.03 | 0.36 ± 0.04 | 0.12 ± 0.01 | 2.6 ± 0.6 | ||||
| α-MSH | 0.64 ± 0.16 | 0.56 ± 0.06 | 1.6 ± 0.2 | 0.7 ± 0.2 | ||||
| Ac-His-DPhe-Arg-Trp-NH2 | 13.4 ± 3.9 | 31 ± 9 | 5.5 ± 0.8 | 4.2 ± 1 | ||||
| THIQ | 100 ± 20 | 260 ± 20a | 0.9 ± 0.2 | 0.49 ± 0.08 | ||||
| 1 | R-isobutyl | R-cyclohexyl-methyl | (S,R)-1-hydroxyethyl | adamantan-1-yl-methyl | 330 ± 110 | 240 ± 40 | 650 ± 120 (55%) pA2= 5.5 ± 1.1 |
340 ± 140 |
| 2 | R-isobutyl | R-cyclohexyl-methyl | (S,R)-1-hydroxyethyl | 4-tbutyl-cyclohexyl-methyl | 300 ± 90 | 210 ± 50 | 270 ± 70 (45%) pA2 = 5.8 ± 0.1 |
140 ± 70 |
| 3 | R-isobutyl | R-cyclohexyl-methyl | (S,R)-1-hydroxyethyl | 4-methylpentyl | 7300 ± 4400 | 58% @ 10 μM (B) | (A) | 630 ± 50 |
| 4 | R-isobutyl | R-cyclohexyl-methyl | R-propyl | adamantan-1-yl-methyl | 70% @ 10 μM (B) | 67% @ 10 μM (B) | 51% @ 10 μM (B) | 470 ± 90 |
| 5 | R-isobutyl | R-cyclohexyl-methyl | R-propyl | 4-tbutyl-cyclohexyl-methyl | 500 ± 50 | 75% @ 10 μM (B) | 65% @ 10 μM (B) | 530± 50 |
| 6 | R-isobutyl | R-cyclohexyl-methyl | R-propyl | 4-methylpentyl | 2300 ± 90 | 58% @ 10 μM (B) | 6700 ± 2400 | 1800 ± 130 |
| 7 | R-isobutyl | R-cyclohexyl-methyl | R-isopropyl | adamantan-1-yl-methyl | 570 ± 150 | 70% @ 10 μM (B) | 50% @ 10 μM (A) | 500 ± 40 |
| 8 | R-isobutyl | R-cyclohexyl-methyl | R-isopropyl | 4-tbutyl-cyclohexyl-methyl | 54% @ 10 μM (B) | 69% @ 10 μM (B) | >100,000 | 560 ± 50 |
| 9 | R-isobutyl | R-cyclohexyl-methyl | R-isopropyl | 4-methylpentyl | 50% @ 10 μM (B) | 33% @ 10 μM (A) | (B) | 1850 ± 800 |
| 10 | R-isobutyl | R-benzyl | (S,R)-1-hydroxyethyl | adamantan-1-yl-methyl | 41% @ 10 μM (A) | 360 ± 40 | 58% @ 10 μM (B) pA2 = 5.6 ± 0.2 |
810 ± 280 |
| 11 | R-isobutyl | R-benzyl | (S,R)-1-hydroxyethyl | 4-tbutyl-cyclohexyl-methyl | 58% @ 10 μM (B) | 310 ± 60 | (A) pA2 = 7.0 ± 0.7 |
530 ± 50 |
| 12 | R-isobutyl | R-benzyl | (S,R)-1-hydroxyethyl | 4-methylpentyl | 62% @ 10 μM (B) | >100,000 | >100,000 | 77% @ 10 μM (B) |
| 13 | R-isobutyl | R-benzyl | R-propyl | adamantan-1-yl-methyl | 7000 ± 6100 | 74% @ 10 μM (B) | 1330 ± 510 | 610 ± 160 |
| 14 | R-isobutyl | R-benzyl | R-propyl | 4-tbutyl-cyclohexyl-methyl | 38% @ 10 μM (A) | 83% @ 10 μM (B) | 28% @ 10 μM (A) | 480 ± 170 |
| 15 | R-isobutyl | R-benzyl | R-propyl | 4-methylpentyl | >100,000 | >100,000 | (B) | 2180 ± 625 |
| 16 | R-isobutyl | R-benzyl | R-isopropyl | adamantan-1-yl-methyl | 15500 ± 14000 | 67% @ 10 μM (B) | 8200 ± 3550 | 440 ± 110 |
| 17 | R-isobutyl | R-benzyl | R-isopropyl | 4-tbutyl-cyclohexyl-methyl | 6800 ± 600 | 62% @ 10 μM (B) | >100,000 | 560 ± 170 |
| 18 | R-isobutyl | R-benzyl | R-isopropyl | 4-methylpentyl | >100,000 | (A) | (A) | 1260 ± 130 |
| 19 | R-isopropyl | R-cyclohexyl-methyl | (S,R)-1-hydroxyethyl | adamantan-1-yl-methyl | 200 ± 80 | 370 ± 90 | 49% @ 10 μM (A) pA2 = 5.8 ± 0.1 |
600 ± 210 |
| 20 | R-isopropyl | R-cyclohexyl-methyl | (S,R)-1-hydroxyethyl | 4-tbutyl-cyclohexyl-methyl | 300 ± 120 | 220 ± 30 | 41% @ 10 μM (A) pA2 = 5.9 ± 0.1 |
150 ± 20 |
| 21 | R-isopropyl | R-cyclohexyl-methyl | (S,R)-1-hydroxyethyl | 4-methylpentyl | 700 ± 80 | 5200 ± 1540 | >100,000 | 740 ± 270 |
| 22 | R-isopropyl | R-cyclohexyl-methyl | R-propyl | adamantan-1-yl-methyl | 550 ± 170 | 2800 ± 1800 | 45% @ 10 μM (A) | 590 ± 180 |
| 23 | R-isopropyl | R-cyclohexyl-methyl | R-propyl | 4-tbutyl-cyclohexyl-methyl | (B) | 350 ± 20 | 50% @ 10 μM (A) pA2 = 5.8 ± 0.2 |
540 ± 50 |
| 24 | R-isopropyl | R-cyclohexyl-methyl | R-propyl | 4-methylpentyl | (B) | (B) | 40% @ 10 μM (A) | 1040 ± 150 |
| 25 | R-isopropyl | R-cyclohexyl-methyl | R-isopropyl | adamantan-1-yl-methyl | 300 ± 30 | 3700 ± 880 | 40% @ 10 μM (A) | 580 ± 160 |
| 26 | R-isopropyl | R-cyclohexyl-methyl | R-isopropyl | 4-tbutyl-cyclohexyl-methyl | 290 ± 20 | 4300 ± 3300 | 28% @ 10 μM (A) | 510 ± 50 |
| 27 | R-isopropyl | R-cyclohexyl-methyl | R-isopropyl | 4-methylpentyl | 6100 ± 4700 | (B) | (B) | 1700 ± 520 |
| 28 | R-isopropyl | R-benzyl | (S,R)-1-hydroxyethyl | adamantan-1-yl-methyl | 440 ± 140 | 360 ± 10 | 580 ± 130 (75%) pA2 = 5.8 ± 0.1 |
180 ± 90 |
| 29 | R-isopropyl | R-benzyl | (S,R)-1-hydroxyethyl | 4-tbutyl-cyclohexyl-methyl | 600 ± 340 | 420 ± 120 | (A) pA2 = 6.5 ± 0.5 |
490 ± 155 |
| 30 | R-isopropyl | R-benzyl | (S,R)-1-hydroxyethyl | 4-methylpentyl | (B) | >100,000 | >100,000 | 40% @ 10 μM (A) |
| 31 | R-isopropyl | R-benzyl | R-propyl | adamantan-1-yl-methyl | 6700 ± 4700 | 2000 ± 850 | 1400 ± 760 | 6600 ± 5740 |
| 32 | R-isopropyl | R-benzyl | R-propyl | 4-tbutyl-cyclohexyl-methyl | 2300 ± 1000 | 6200 ± 5400 | 27% @ 10 μM (A) | 940 ± 340 |
| 33 | R-isopropyl | R-benzyl | R-propyl | 4-methylpentyl | 22900 ± 10000 | >100,000 | 41% @ 10 μM (A) | 2030 ± 980 |
| 34 | R-isopropyl | R-benzyl | R-isopropyl | adamantan-1-yl-methyl | 47800 ± 23100 | 4000 ± 1900 | 54% @ 10 μM (B) | 660 ± 130 |
| 35 | R-isopropyl | R-benzyl | R-isopropyl | 4-tbutyl-cyclohexyl-methyl | 29100 ± 20000 | 68% @ 10 μM (B) | >100,000 | 600 ± 170 |
| 36 | R-isopropyl | R-benzyl | R-isopropyl | 4-methylpentyl | 10220 ± 6570 | >100,000 | 21% @ 10 μM (A) | 1600 ± 460 |
| 37 | R-2-butyl | R-benzyl | R-4-hydroxybenzyl | 4-tbutyl-cyclohexyl-methyl | 290 ± 20 | 65% @ 10 μM (B) | 23% @ 10 μM (A) | 380 ± 50 |
The compounds were screened at the selected receptor subtypes for agonist activity from 10−4 to 10−10 M and the results are tabulated as the mean EC50 values of duplicate values for at least three independent experiments. For select compounds, the 100 μM concentration resulted in cytotoxicity and was excluded from the sigmoidal dose-response fittings. For these compounds, the activity observed at 10 μM relative to maximal expression was tabulated to aide in the differentiation between partial agonists which have both a reported EC50 and percent activation and compounds that exhibited some receptor activation at 10 μM. These compounds were binned as A (10-50% receptor activation) or B (51-90% receptor activation). Follow up antagonist experiments and corresponding pA2 values, via a Schild analysis,73 were determined for the mMC4R if the compound possessed a nanomolar EC50 value (< 1 μM) at the mMC3R and produced little to no activity at the mMC4R (< 20% of maximal activity). Included as positive controls were α-MSH, the α-MSH analogue NDP-MSH, the tetrapeptide Ac-His-DPhe-Arg-Trp-NH2, and the THIQ small molecule.
The efficacy of THIQ was observed to be 90-95% at the mMC3R compared to the maximal response of α-MSH.
Figure 7: Illustration of Partial Agonist Pharmacology.
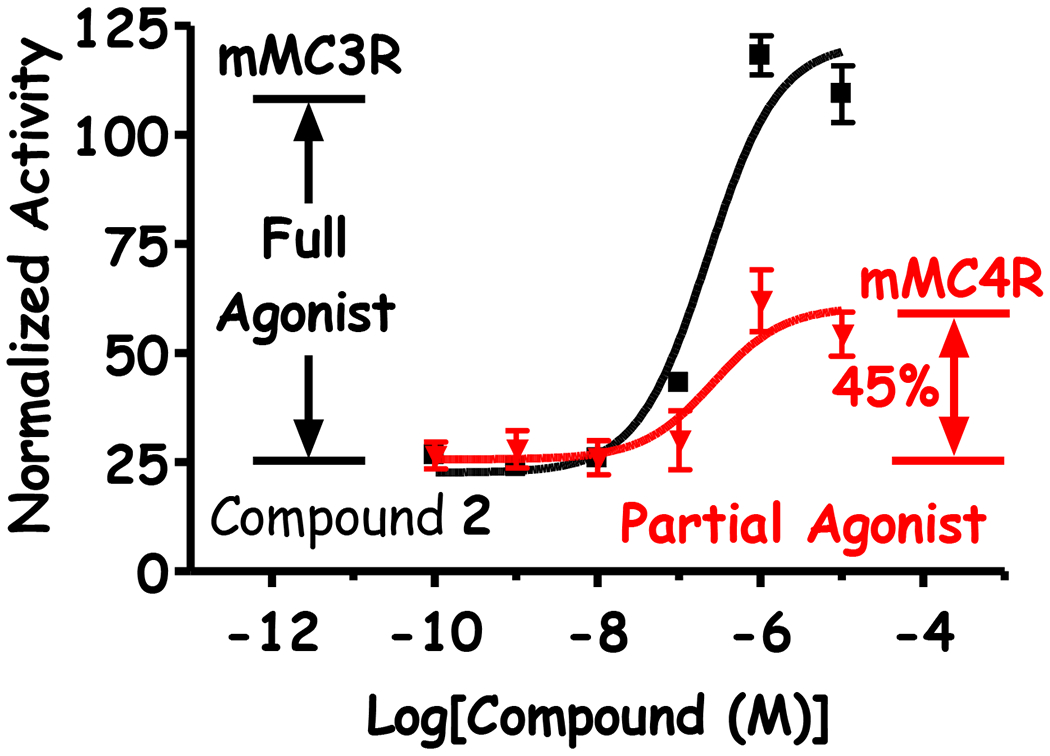
Three compounds demonstrated partial agonist activity at the MC4R. These compounds were described with both an EC50 and a percent activity relative to NDP-MSH (positive control and full agonist) where compound activity leveled off. For compound 2, full agonist activity was observed for the mMC3R (EC50 = 210 nM) whereas partial agonist activity was observed at the mMC4R (EC50 = 270 nM, 45% activity).
The functionalities for the synthesis of the individual compounds were selected for MC3R agonist potency and selectivity for the MC3R over the MC4R. The results from the individual compound library resulted in pharmacological and SAR patterns that were consistent with these selections. Out of the 37 compounds, 24% (9/37) possessed full agonist efficacies and potencies < 1 μM at the MC3R, while 8% (3/37) were partial agonists at the MC4R (efficacies 45-75% NDP-MSH) with potencies < 1 μM. An additional 18% (7/37) and 11% (4/37) possessed full agonist efficacy and potencies >1 μM at the MC3R and MC4R, respectively (Table 1).
Specific substitutions at R3 and R4 resulted in the observed sub-μM potencies at the MC3R. Of the 9 compounds that possessing EC50 < 1 μM at the MC3R, 8 contained the (S,R)-1-hydroxyethyl (D-Thr) group at the R3 position (1, 2, 10, 11, 19, 20, 28, and 29), while one possessed the R-propyl (D-norvaline) substitution at R3 (23). The (S,R)-1-hydroxyethyl (D-Thr) group is capable of hydrogen bonding, unlike the related aliphatic R-propyl (D-norvaline) and R-isopropyl (D-Val) substitutions which are capable of weaker Van der Waals interactions with the receptor. All 9 of the compounds with potencies < 1 μM also possessed either an adamatan-1-yl-methyl (1-adamantanecarboxylic acid) or 4-tbutyl-cyclohexyl-methyl (4tBu-cyclohexanecarboxylic acid) at the R4 position and none possessed the small 4-methyl-pentane (4-methylvaleric acid) capping group. This may indicate the more sterically bulky groups are preferred at the R4 position to increase MC3R potency.
Comparison of Results Obtained for mMC3R and mMC4R
Since 9 compounds possessed full agonist efficacy at the MC3R with potencies < 1 μM, and none of these compounds was a full agonist at the MC4R, the antagonist activity of these 9 compounds was evaluated at the MC4R using a Schild paradigm73 and the synthetic MTII as the agonist. Three of these compounds that possessed partial agonist activity at the MC4R (1, 2, and 28) and four that resulted in partial activation of the MC4R (10, 19, 20, and 23) all possessed > 1 μM (pA2 < 6) antagonist potencies at the MC4R. The two compounds that resulted in <20% activation of the MC4R at 10 μM concentrations (11 and 29, Figure 8) possessed sub-μM antagonist potencies at the MC4R (7.0 and 6.5 for 11 and 29, respectively, Figure 8). Thus, compounds that possessed sub-μM MC3R agonism were found that could also partially stimulate the MC4R at sub-μM concentrations, stimulate the MC4R at 10 μM concentrations, or have <20% stimulatory activity but sub-μM antagonist potencies at the MC4R. Further refinement of these small molecule leads may lead to the development of more MC3R selective or MC3R agonist/MC4R antagonist polypharmacology compounds that could be administered in vivo to help elucidate the roles of the centrally expressed MC3R and MC4R.
Figure 8: Comparison of Compound 11 and 29 at the Central Melanocortin-3 and -4 Receptors.
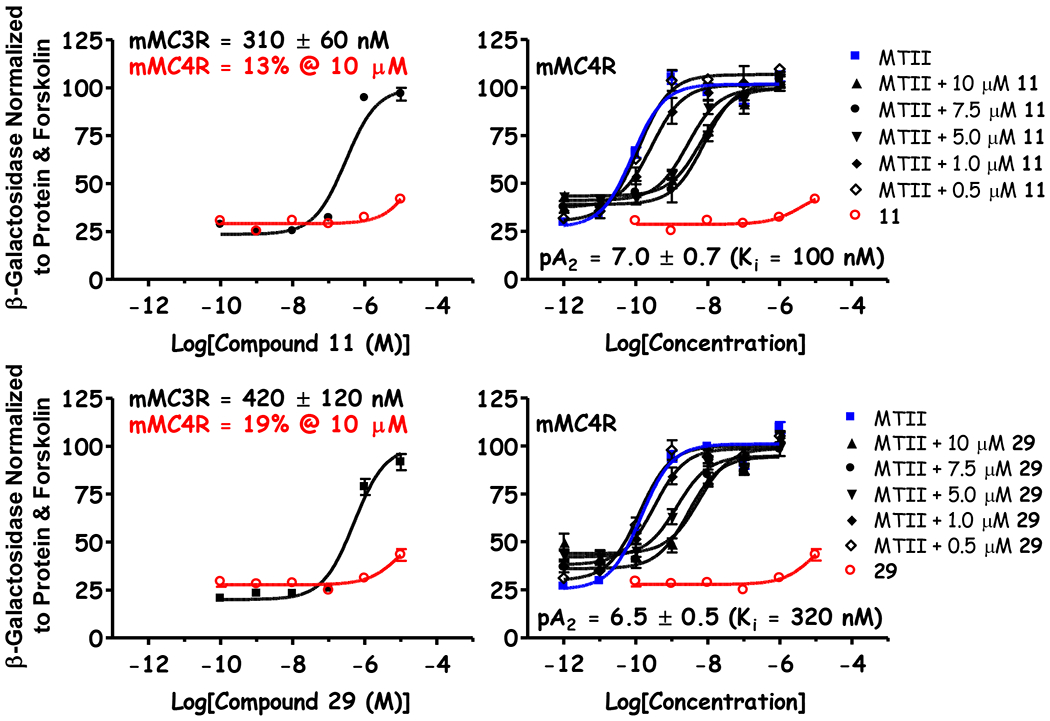
Results derived from the mixture-based positional scan and subsequent mixture deconvolution produced selective mMC3R agonists with nanomolar EC50 values (EC50 < 1 μM) where little to no agonist activity was observed for the mMC4R at the highest concentrations tested (left panel). Additional follow-up experiments indicated these inactive mMC4R agonists produced some agonist activity (pA2 < 7.0) as measure by a competitive agonist displacement assay and Schild analysis (right panel).73
Pharmacological Evaluation at the mMC1R and mMC5R
While certain functionalities resulted in MC3R agonism, the individual pyrrolidine bis-cyclic guanidine compounds yielded minimal structural trends at the mMC1R and mMC5R (Table 1). Results for the agonist screen at the mMC1R gave rise to 13 full agonists which possessed potencies less than 1 μM, while 26 were observed for the MC5R. Of the 25 compounds possessing adamantan-1-yl-methyl or 4-tbutyl-cyclohexyl-methyl at the R4 position, 24 were sub-μM agonists at the MC5R, suggesting bulky groups at this position may increase MC5R potency in this scaffold. All 9 compounds possessing < 1 μM potency at the MC3R also possessed < 1 μM potency at the MC5R, indicating these compounds did not achieve selectivity over the MC5R. Three compounds from this set did not fully activate the MC1R at 10 μM concentrations (10, 11, and 23). These compounds may be useful as probes and therapeutic lead compounds that do not result in pigmentary effects that have been indicated for other melanocortin ligands that have been in clinical trials.28, 29
The most potent compounds identified in this research possessed sub-200 nM potencies at the MC5R (2, 20, and 28). These compounds were less than 3-fold selective between the MC5R and the MC1R and MC3R, but indicate that structural modification to this series may also identify MC5R-selective compounds. While the MC5R has been shown to be involved in exocrine gland function in mice,9 its exact role in humans has not been identified, although it has been hypothesized to be involved in diverse physiological functions including glucose uptake in muscles74 and sebogenesis (as reviewed).75 Developing lead ligands selective for the MC5R, such as 14 (480 nM potency at the MC5R, partial receptor activation of the MC1R, MC3R, and MC4R at 10 μM) that can be further optimized may be important in clarifying the biological roles of the MC5R.
Comparison to Other Related Compounds/Targets
Apparent cell cytotoxicity was observed for the pyrrolidine bis-cyclic guanidine compounds only when assayed at the highest concentrations of 100 μM (more than 100-fold higher than the observed EC50 values for the nanomolar potent mMC3R compounds). There are reports that the pyrrolidine bis-cyclic guanidine scaffold possesses antimicrobial properties in both Gram-positive and -negative bacteria, in addition to Mycobacterium tuberculosis.64, 67 A different study identified pyrrolidine bis-cyclic guanidine compounds that inhibit Holliday junctions in the bacterial DNA repair pathway and possessed antibacterial activity.76 Additional experiments evaluated eukaryotic cell toxicity, via an MTT assay, and confirmed that pyrrolidine bis-cyclic guanidine based compounds can decrease cell viability to less than 65% at concentrations of 64 μg/mL (approximately 75 μM).76 An associated report demonstrated that closely-related compounds derived from a common butyl bis-cyclic guanidine core possessed a broader spectrum of activity against the ESKAPE pathogens than compounds with the pyrrolidine-based core.77 Additional MTT assay results indicated these compounds had similar toxicity at 100 μM concentrations.77 These results suggest the observed toxicity at 100 μM concentrations are common to this scaffold.
Conclusions
The design of the present study focused on the identification of potent nM MC3R agonist compounds with a selectivity profile over the MC4R. Using a series of selection criteria allowed for the rapid evaluation of millions of compounds to identify a handful of lead compounds. Following a scaffold scan of potential MC3R agonist ligands, two mixture-based positional scans were performed. The pyrrolidine bis-cyclic guanidine scaffold yielded the first small molecule chemotype with sub-μM agonist potencies at the MC3R and >10-fold selectivity for the MC3R over the MC4R. The resulting sub-μM potent MC3R compounds displayed relatively minimal agonist activity at the MC4R and, in some instances, antagonist activity at the MC4R. This group of small molecules represent a new area of chemical space for the melanocortin receptors with unique profiles that may have implications for pharmaceutical intervention to treat obesity and other states of dysregulated energy homeostasis.
Experimental Section
General
The THIQ small molecule was obtained through a material transfer agreement with Dr. Lex Van Der Ploeg at Merck Research Laboratories.51
Beta-Galactosidase Bioassay
This living whole cell assay indirectly measures intracellular cAMP using a beta-galactosidase reporter gene,62 and in our hands is compatible with the reported mixture-based screening approach.44 All mixtures and individual compounds were dissolved in 10% DMSO:90% H2O and stored at −20°C until assayed. The mixtures were stored at an approximate concentration of 1,000 μg/mL whereas individual compounds were stored at a concentration of 10−2 M. The compounds and mixtures were assayed in HEK293 cells stably expressing the mMC1R, mMC3R, mMC4R, and mMC5R as previously reported.78 Cells were plated onto 10 cm plates with media (Dulbecco’s Modified Eagle Medium [DMEM] supplemented with 10% Bovine Serum and 1% Penicillin Streptomycin) and incubated for 24 hours at 37 °C and 5% CO2. Post incubation, the plates were approximately 40% confluent. Cells were transiently transfected with 4 μg CRE-PBKS per 10 cm dish using the calcium phosphate method79 and incubated for 24 hours at 35°C and 3% CO2. The cells were plated onto collagen treated Nunclon Delta Surface 96-well plates (Thermo Fisher Scientific) with fresh media and incubated for 24 hours at 37°C and 5% CO2. Forty-eight to seventy-two hours post transient transfection, the media was aspirated. For individual small molecules, wells were treated with 50 μL of compound at a concentration of 10−4 to 10−12 M, depending on compound potency, in assay media (1.0 mL 1% bovine serum albumin [BSA] in phosphate buffered saline [PBS] and 1.0 mL 100x isobutylmethylxanthine [IBMX] in 98.0 mL DMEM). In the case of the scaffold ranking and mixture-based positional scan, wells were treated with 40 μL of each mixture at the approximate concentrations of 50, 25, and 12.5 μg/mL. Controls included NDP-MSH (10−6 to 10−12 M), forskolin (10 μM) and plain assay media for all bioassays. The plates were incubated at 37 °C with 5% CO2 for six hours. Post stimulation, the media was aspirated and 50 μL of lysis buffer (250 mM Tris-HCl pH = 8.0, 740 mL DD H2O, 10 mL 10% Triton X-100 in water) was added. In addition, the health of the cells was visually inspected under a microscope for the mixture bioassays before cell lysis. The plates were stored at −80 °C for up to two weeks. After the plates were thawed, they were assessed for protein content and a 2-nitrophenyl β-D-galactopyranoside substrate was added to measure beta-galactosidase activity. Protein content was assessed by adding 10 μL of cell lysate to 200 μL of BioRad dye solution (1:4 dilution with water) in a 96 well plate, and the absorbance was read using a plate reader (Molecular Devices) at λ = 595 nm. To the remaining cell lysate, 40 μL of 37 °C, 0.5% BSA in PBS was added in addition to 150 μL of the β-galactosidase substrate (120 mM Na2HPO4, 2 mM MgCl2, 20 mM KCl, 100 mM 2-mercaptoethanol, and 13 mM 2-nitrophenyl β-D-galactopyranoside). The plates were incubated at 37 °C and periodically read on the 96 well plate reader until the absorbance at λ = 405 nm reached approximately 1.0 relative absorbance units for the positive controls. The β-galactosidase activity was normalized to both protein content and maximal response of the positive controls. Screening experiments are indicated throughout the manuscript in terms of replicates and independent experiments performed at each stage. Individual ≥95% pure compound-melanocortin receptor pharmacological characterization experiments were performed with at least two replicates and in at least three independent experiments. Agonist EC50 and antagonist pA2 values, and their associated standard errors or the mean (SEM), were determined by fitting the data to a nonlinear least-squares analysis using the PRISM program (GraphPad Inc.).
Compound Synthesis
The compounds were synthesized utilizing the “tea-bag” method80 wherein p-methylbenzhydrylamine (MBHA) resin was sealed in a mesh “tea-bag,” washed with dichloromethane (DCM) (3 x 1 min), neutralized with 5% diisopropylethylamine (DIEA) in DCM (3 x 2 min), and then swelled with additional DCM washes (3 x 1 min). Boc-amino acids R1 (6 eq) were coupled using a standard coupling protocol with N,N’-diisopropylcarbodiimide (DIC) (6 eq) and hydroxybenzotriazole (HOBt) (6 eq) in dimethylformamide (0.1 M) for 60 min. Following DMF (3 x 1 min) and DCM (3 x 1 min) washes, the Boc protecting group was removed with 55% trifluoroacetic acid (TFA) in DCM (1 x 30 min) and the resin was washed with DCM (2 x 1 min) and isopropyl alcohol (IPA) (2 x 1 min). The resin was washed and neutralized with the same protocol (DCM 3 x 1 min, 5% DIEA/DCM 3 x 2 min, DCM 3 x 1min). The amide bond coupling, Boc protecting group removal, and neutralization steps with identical equivalents were repeated for the remaining Boc-L-Proline-OH, Boc-amino acid R2, and Boc-amino acid R3. Following the Boc removal and neutralization from the R3 amino acid, the carboxylic acid R4 was coupled with the same protocol but with an increase in equivalents of the carboxylic acid (10 eq), DIC (10 eq), and HOBt (10 eq). The amide bonds were reduced to the corresponding polyamines without the racemization of the side chains in the presence of 1.0 M borane in tetrahydrofuran (THF) (40 eq per amide bond) using anhydrous conditions and heated to 65 °C for 72 hours. The solution was removed and the bags washed with THF and methanol, and subsequently treated with piperidine at 65 °C for 24 hours. The cyclization to the corresponding bis-cyclic guanidine moieties was performed on solid support with a 0.1 M cyanogen bromide (5 eq per cyclization) in anhydrous DCM at room temperature overnight. The bags were rinsed with DMF and DCM, and then the desired compounds were removed from the solid support with HF in the presence of anisole in an ice bath at 0 °C for 1.5 hours. Excess HF was removed with N2 gas and the product was removed from the reaction vessel with 95% acetic acid in water, frozen, and lyophilized. Individual compounds were purified using the protocol below and lyophilized an additional three times.
Compound Purification
Individual compounds were purified using preparative HPLC with a dual pump Shimadzu LC-20AB system equipped with a Luna C18 preparative column (21.5 x 150 mm, 5 micron) at λ = 214 nm, with a mobile phase of (A) H2O (+0.1% formic acid)/(B) acetonitrile (ACN) (+0.1% formic acid) at a flow rate of 15 mL/min; gradients varied by compound based on hydrophobicity. The purities of synthesized compounds were confirmed to be greater than 95% by LC/MS analysis on a Shimadzu LCMS-2010 instrument with ESI Mass Spec and SPD-20A Liquid Chromatograph equipped with a Luna C18 column (50 x 4.6 mm, 5micron) with a mobile phase of (A) H2O (+0.1% formic acid)/(B) ACN (+0.1% formic acid) (5-95% over 6 min with a 4 min rinse). 1H NMR spectra were recorded in DMSO-d6 or Chloroform-d on a Bruker Ascend 400 MHz spectrometer at 400.14 MHz.
Supplementary Material
Funding Sources
This work was supported in part by NIH R01 grants R01DK091906, RO1DK124504 (C.H.L.) and R01DA031370 (R.A.H.), the State of Florida, Executive Office of the Governor’s Office of Tourism, Trade, and Economic Development (R.A.H.). We would also like to acknowledge the receipt of a 2017 Wallin Neuroscience Discovery Fund Award and Engebretson Drug Design and Development Grant (C.H.L.).
Abbreviations Used
- AGRP
agouti-related protein
- MCR
melanocortin receptor
- KO
knockout
- PAINS
pan-assay interference compounds
- Cha
cyclohexylalanine
- DMEM
Dulbecco’s modified eagle medium
- IBMX
isobutylmethylxanthine
- DIEA
diisopropylamine
- MBHA
methylbenzhydrylamine
- DIC
N,N’-diisopropylcarbodiimide
- HOBt
hydroxybenzotriazole
- IPA
isopropyl alcohol
- ACN
acetonitrile
Footnotes
The authors declare no competing financial interest.
Supporting Information
Compound synthesis and characterization, HPLC of key compounds, scaffold functional building block functionality, and SMILES structures are supplied in the supporting information. Additionally, a CSV file for the pyrrolidine bis-cyclic guanidine compounds containing the SMILES structures and functional data at the mMC1R, mMC3R, mMC4R, and mMC5R is supplied in the supporting information.
References
- 1.Chhajlani V; Wikberg JE, Molecular cloning and expression of the human melanocyte stimulating hormone receptor cDNA. FEBS Lett. 1992, 309, 417–420. [DOI] [PubMed] [Google Scholar]
- 2.Mountjoy KG; Robbins LS; Mortrud MT; Cone RD, The cloning of a family of genes that encode the melanocortin receptors. Science 1992, 257, 1248–1251. [DOI] [PubMed] [Google Scholar]
- 3.Roselli-Rehfuss L; Mountjoy KG; Robbins LS; Mortrud MT; Low MJ; Tatro JB; Entwistle ML; Simerly RB; Cone RD, Identification of a receptor for γ melanotropin and other proopiomelanocortin peptides in the hypothalamus and limbic system. Proc. Natl. Acad. Sci. U. S. A 1993, 90, 8856–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mountjoy KG; Mortrud MT; Low MJ; Simerly RB; Cone RD, Localization of the melanocortin-4 receptor (MC4-R) in neuroendocrine and autonomic control circuits in the brain. Mol. Endocrinol 1994, 8, 1298–1308. [DOI] [PubMed] [Google Scholar]
- 5.Gantz I; Miwa H; Konda Y; Shimoto Y; Tashiro T; Watson SJ; DelValle J; Yamada T, Molecular cloning, expression, and gene localization of a fourth melanocortin receptor. J. Biol. Chem 1993, 268, 15174–15179. [PubMed] [Google Scholar]
- 6.Butler AA; Kesterson RA; Khong K; Cullen MJ; Pelleymounter MA; Dekoning J; Baetscher M; Cone RD, A unique metabolic syndrome causes obesity in the melanocortin-3 receptor-deficient mouse. Endocrinology 2000, 141, 3518–3521. [DOI] [PubMed] [Google Scholar]
- 7.Huszar D; Lynch CA; Fairchild-Huntress V; Dunmore JH; Fang Q; Berkemeier LR; Gu W; Kesterson RA; Boston BA; Cone RD; Smith FJ; Campfield LA; Burn P; Lee F, Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 1997, 88, 131–141. [DOI] [PubMed] [Google Scholar]
- 8.Chen AS; Marsh DJ; Trumbauer ME; Frazier EG; Guan XM; Yu H; Rosenblum CI; Vongs A; Feng Y; Cao LH; Metzger JM; Strack AM; Camacho RE; Mellin TN; Nunes CN; Min W; Fisher J; Gopal-Truter S; MacIntyre DE; Chen HY; Van der Ploeg LHT, Inactivation of the mouse melanocortin-3 receptor results in increased fat mass and reduced lean body mass. Nat. Genet 2000, 26, 97–102. [DOI] [PubMed] [Google Scholar]
- 9.Chen W; Kelly MA; Opitz-Araya X; Thomas RE; Low MJ; Cone RD, Exocrine gland dysfunction in MC5-R-deficient mice: Evidence for coordinated regulation of exocrine gland function by melanocortin peptides. Cell 1997, 91, 789–798. [DOI] [PubMed] [Google Scholar]
- 10.Büch TRH; Heling D; Damm E; Gudermann T; Breit A, Pertussis toxin-sensitive signaling of melanocortin-4 receptors in hypothalamic GT1-7 cells defines agouti-related protein as a biased agonist. J. Biol. Chem 2009, 284, 26411–26420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mo XL; Tao YX, Activation of MAPK by inverse agonists in six naturally occurring constitutively active mutant human melanocortin-4 receptors. Biochim. Biophys. Acta, Mol. Basis Dis 2013, 1832, 1939–1948. [DOI] [PubMed] [Google Scholar]
- 12.Ghamari-Langroudi M; Digby GJ; Sebag JA; Millhauser GL; Palomino R; Matthews R; Gillyard T; Panaro BL; Tough IR; Cox HM; Denton JS; Cone RD, G-protein-independent coupling of MC4R to Kir7.1 in hypothalamic neurons. Nature 2015, 520, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irani BG; Xiang ZM; Yarandi HN; Holder JR; Moore MC; Bauzo RM; Proneth B; Shaw AM; Millard WJ; Chambers JB; Benoit SC; Clegg DJ; Haskell-Luevano C, Implication of the melanocortin-3 receptor in the regulation of food intake. Eur. J. Pharmacol 2011, 660, 80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan W; Boston BA; Kesterson RA; Hruby VJ; Cone RD, Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature 1997, 385, 165–168. [DOI] [PubMed] [Google Scholar]
- 15.Atalayer D; Robertson KL; Haskell-Luevano C; Andreasen A; Rowland NE, Food demand and meal size in mice with single or combined disruption of melanocortin type 3 and 4 receptors. Am. J. Physiol.: Regul., Integr. Comp. Physiol 2010, 298, R1667–R1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowland NE; Fakhar KJ; Robertson KL; Haskell-Luevano C, Effect of serotonergic anorectics on food intake and induction of Fos in brain of mice with disruption of melanocortin 3 and/or 4 receptors. Pharmacol., Biochem. Behav 2010, 97, 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowland NE; Schaub JW; Robertson KL; Andreasen A; Haskell-Luevano C, Effect of MTII on food intake and brain c-Fos in melanocortin-3, melanocortin-4, and double MC3 and MC4 receptor knockout mice. Peptides 2010, 31, 2314–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinney A; Volckmar AL; Knoll N, Melanocortin-4 receptor in energy homeostasis and obesity pathogenesis. Prog. Mol. Biol. Transl. Sci 2013, 114, 147–191. [DOI] [PubMed] [Google Scholar]
- 19.Farooqi IS; Keogh JM; Yeo GS; Lank EJ; Cheetham T; O’Rahilly S, Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N. Engl. J. Med 2003, 348, 1085–1095. [DOI] [PubMed] [Google Scholar]
- 20.Vaisse C; Clement K; Guy-Grand B; Froguel P, A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat. Genet 1998, 20, 113–114. [DOI] [PubMed] [Google Scholar]
- 21.Yeo GS; Farooqi IS; Aminian S; Halsall DJ; Stanhope RG; O’Rahilly S, A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat. Genet 1998, 20, 111–112. [DOI] [PubMed] [Google Scholar]
- 22.Martin WJ; McGowan E; Cashen DE; Gantert LT; Drisko JE; Hom GJ; Nargund R; Sebhat I; Howard AD; Van der Ploeg LHT; MacIntyre DE, Activation of melanocortin MC4 receptors increases erectile activity in rats ex copula. Eur. J. Pharmacol 2002, 454, 71–79. [DOI] [PubMed] [Google Scholar]
- 23.Hadley ME, Discovery that a melanocortin regulates sexual functions in male and female humans. Peptides 2005, 26, 1687–1689. [DOI] [PubMed] [Google Scholar]
- 24.Greenfield JR; Miller JW; Keogh JM; Henning E; Satterwhite JH; Cameron GS; Astruc B; Mayer JP; Brage S; See TC; Lomas DJ; O’Rahilly S; Farooqi IS, Modulation of blood pressure by central melanocortinergic pathways. N. Engl. J. Med 2009, 360, 44–52. [DOI] [PubMed] [Google Scholar]
- 25.Van der Ploeg LHT; Martin WJ; Howard AD; Nargund RP; Austin CP; Guan XM; Drisko J; Cashen D; Sebhat I; Patchett AA; Figueroa DJ; DiLella AG; Connolly BM; Weinberg DH; Tan CP; Palyha OC; Pong SS; MacNeil T; Rosenblum C; Vongs A; Tang R; Yu H; Sailer AW; Fong TM; Huang C; Tota MR; Chang RS; Stearns R; Tamvakopoulos C; Christ G; Drazen DL; Spar BD; Nelson RJ; MacIntyre DE, A role for the melanocortin 4 receptor in sexual function. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 11381–11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kievit P; Halem H; Marks DL; Dong JZ; Glavas MM; Sinnayah P; Pranger L; Cowley MA; Grove KL; Culler MD, Chronic treatment with a melanocortin-4 receptor agonist causes weight loss, reduces insulin resistance, and improves cardiovascular function in diet-induced obese rhesus macaques. Diabetes 2013, 62, 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen KY; Muniyappa R; Abel BS; Mullins KP; Staker P; Brychta RJ; Zhao X; Ring M; Psota TL; Cone RD; Panaro BL; Gottesdiener KM; Van der Ploeg LHT; Reitman ML; Skarulis MC, RM-493, a melanocortin-4 receptor (MC4R) agonist, increases resting energy expenditure in obese individuals. J. Clin. Endocrinol. Metab 2015, 100, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhnen P; Clement K; Wiegand S; Blankenstein O; Gottesdiener K; Martini LL; Mai K; Blume-Peytavi U; Gruters A; Krude H, Proopiomelanocortin deficiency treated with a melanocortin-4 receptor agonist. N. Engl. J. Med 2016, 375, 240–246. [DOI] [PubMed] [Google Scholar]
- 29.Clement K; Biebermann H; Farooqi IS; Van der Ploeg L; Wolters B; Poitou C; Puder L; Fiedorek F; Gottesdiener K; Kleinau G; Heyder N; Scheerer P; Blume-Peytavi U; Jahnke I; Sharma S; Mokrosinski J; Wiegand S; Muller A; Weiss K; Mai K; Spranger J; Gruters A; Blankenstein O; Krude H; Kuhnen P, MC4R agonism promotes durable weight loss in patients with leptin receptor deficiency. Nat. Med 2018, 24, 551–555. [DOI] [PubMed] [Google Scholar]
- 30.Collet TH; Dubern B; Mokrosinski J; Connors H; Keogh JM; Mendes de Oliveira E; Henning E; Poitou-Bernert C; Oppert JM; Tounian P; Marchelli F; Alili R; Le Beyec J; Pepin D; Lacorte JM; Gottesdiener A; Bounds R; Sharma S; Folster C; Henderson B; O’Rahilly S; Stoner E; Gottesdiener K; Panaro BL; Cone RD; Clement K; Farooqi IS; Van der Ploeg LHT, Evaluation of a melanocortin-4 receptor (MC4R) agonist (Setmelanotide) in MC4R deficiency. Mol. Metab 2017, 6, 1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Obeidi F; Castrucci AMD; Hadley ME; Hruby VJ, Potent and prolonged acting cyclic lactam analogs of α-melanotropin: Design based on molecular-dynamics. J. Med. Chem 1989, 32, 2555–2561. [DOI] [PubMed] [Google Scholar]
- 32.Al-Obeidi F; Hadley ME; Pettitt BM; Hruby VJ, Design of a new class of superpotent cyclic α-melanotropins based on quenched dynamic simulations. J. Am. Chem. Soc 1989, 111, 3413–3416. [Google Scholar]
- 33.Todorovic A; Ericson MD; Palusak RD; Sorensen NB; Wood MS; Xiang Z; Haskell-Luevano C, Comparative functional alanine positional scanning of the α-melanocyte stimulating hormone and NDP-melanocyte stimulating hormone demonstrates differential structure-activity relationships at the mouse melanocortin receptors. ACS Chem. Neurosci 2016, 7, 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahm UG; Olivier GW; Branch SK; Moss SH; Pouton CW, Synthesis and biological evaluation of α-MSH analogues substituted with alanine. Peptides 1994, 15, 1297–1302. [DOI] [PubMed] [Google Scholar]
- 35.Sahm UG; Qarawi MA; Olivier GW; Ahmed AR; Branch SK; Moss SH; Pouton CW, The melanocortin (MC3) receptor from rat hypothalamus: photoaffinity labelling and binding of alanine-substituted alpha-MSH analogues. FEBS Lett. 1994, 350, 29–32. [DOI] [PubMed] [Google Scholar]
- 36.Haskell-Luevano C; Sawyer TK; Hendrata S; North C; Panahinia L; Stum M; Staples DJ; Castrucci AM; Hadley MF; Hruby VJ, Truncation studies of α-melanotropin peptides identify tripeptide analogues exhibiting prolonged agonist bioactivity. Peptides 1996, 17, 995–1002. [DOI] [PubMed] [Google Scholar]
- 37.Castrucci AM; Hadley ME; Sawyer TK; Wilkes BC; Al-Obeidi F; Staples DJ; de Vaux AE; Dym O; Hintz MF; Riehm JP; Rao KR; Hruby V, α-Melanotropin: The minimal active sequence in the lizard skin bioassay. Gen. Comp. Endocrinol 1989, 73, 157–163. [DOI] [PubMed] [Google Scholar]
- 38.Hruby VJ; Wilkes BC; Hadley ME; Al-Obeidi F; Sawyer TK; Staples DJ; Devaux AE; Dym O; Castrucci AMD; Hintz MF; Riehm JP; Rao KR, α-Melanotropin: The minimal active sequence in the frog-skin bioassay. J. Med. Chem 1987, 30, 2126–2130. [DOI] [PubMed] [Google Scholar]
- 39.Hruby VJ; Lu DS; Sharma SD; Castrucci AD; Kesterson RA; Al-Obeidi FA; Hadley ME; Cone RD, Cyclic lactam α-melanotropin analogs of Ac-Nle4-cyclo[Asp5,D-Phe7,Lys10] α-melanocyte-stimulating hormone-(4-10)-NH2 with bulky aromatic amino acids at position 7 show high antagonist potency and selectivity at specific melanocortin receptors. J. Med. Chem 1995, 38, 3454–3461. [DOI] [PubMed] [Google Scholar]
- 40.Ericson MD; Wilczynski A; Sorensen NB; Xiang ZM; Haskell-Luevano C, Discovery of a β-hairpin octapeptide, c[Pro-Arg-Phe-Phe-Dap-Ala-Phe-DPro], mimetic of agouti-related protein(87-132) [AGRP(87-132)] with equipotent mouse melanocortin-4 receptor (mMC4R) antagonist pharmacology. J. Med. Chem 2015, 58, 4638–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheung AWH; Danho W; Swistok J; Qi LD; Kurylko G; Rowan K; Yeon M; Franco L; Chu XJ; Chen L; Yagaloff K, Structure-activity relationship of linear peptide Bu-His-DPhe-Arg-Trp-Gly-NH2 at the human melanocortin-1 and-4 receptors: Histidine substitution. Bioorg. Med. Chem. Lett 2003, 13, 133–137. [DOI] [PubMed] [Google Scholar]
- 42.Doering SR; Freeman KT; Schnell SM; Haslach EM; Dirain M; Debevec G; Geer P; Santos RG; Giulianotti MA; Pinilla C; Appel JR; Speth RC; Houghten RA; Haskell-Luevano C, Discovery of mixed pharmacology melanocortin-3 agonists and melanocortin-4 receptor tetrapeptide antagonist compounds (TACOs) based on the sequence Ac-Xaa1-Arg-(pI)DPhe-Xaa4-NH2. J. Med. Chem 2017, 60, 4342–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleming KA; Freeman KT; Powers MD; Santos RG; Debevec G; Giulianotti MA; Houghten RA; Doering SR; Pinilla C; Haskell-Luevano C, Discovery of polypharmacological melanocortin-3 and -4 receptor probes and identification of a 100-fold selective nM MC3R agonist versus a μM MC4R partial agonist. J. Med. Chem 2019, 62, 2738–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haslach EM; Huang H; Dirain M; Debevec G; Geer P; Santos RG; Giulianotti MA; Pinilla C; Appel JR; Doering SR; Walters MA; Houghten RA; Haskell-Luevano C, Identification of tetrapeptides from a mixture based positional scanning library that can restore nM full agonist function of the L106P, I69T, I102S, A219V, C271Y, and C271R human melanocortin-4 polymorphic receptors (hMC4Rs). J. Med. Chem 2014, 57, 4615–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fosgerau K; Hoffmann T, Peptide therapeutics: current status and future directions. Drug Discovery Today 2015, 20, 122–128. [DOI] [PubMed] [Google Scholar]
- 46.Todorovic A; Haskell-Luevano C, A review of melanocortin receptor small molecule ligands. Peptides 2005, 26, 2026–2036. [DOI] [PubMed] [Google Scholar]
- 47.Ericson MD; Lensing CJ; Fleming KA; Schlasner KN; Doering SR; Haskell-Luevano C, Bench-top to clinical therapies: A review of melanocortin ligands from 1954 to 2016. Biochim. Biophys. Acta, Mol. Basis Dis 2017, 1863, 2414–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ujjainwalla F; Sebhat IK, Small molecule ligands of the human melanocortin-4 receptor. Curr. Top. Med. Chem 2007, 7, 1068–1084. [DOI] [PubMed] [Google Scholar]
- 49.Haskell-Luevano C; Rosenquist A; Souers A; Khong KC; Ellman JA; Cone RD, Compounds that activate the mouse melanocortin-1 receptor identified by screening a small molecule library based upon the β-turn. J. Med. Chem 1999, 42, 4380–4387. [DOI] [PubMed] [Google Scholar]
- 50.Bondebjerg J; Xiang ZM; Bauzo RM; Haskell-Luevano C; Meldal M, A solid-phase approach to mouse melanocortin receptor agonists derived from a novel thioether cyclized peptidomimetic scaffold. J. Am. Chem. Soc 2002, 124, 11046–11055. [DOI] [PubMed] [Google Scholar]
- 51.Sebhat IK; Martin WJ; Ye ZX; Barakat K; Mosley RT; Johnston DBR; Bakshi R; Palucki B; Weinberg DH; MacNeil T; Kalyani RN; Tang R; Stearns RA; Miller RR; Tamvakopoulos C; Strack AM; McGowan E; Cashen DE; Drisko JE; Hom GJ; Howard AD; MacIntyre DE; van der Ploeg LHT; Patchett AA; Nargund RP, Design and pharmacology of N-[(3R)-1,2,3,4-tetrahydroisoquinolinium-3-ylcarbonyl]-(1R)-1-(4-chlorobenzyl)2-[4-cyclohexyl-4-(1H-1,2,4-triazol-1-ylmethyl)piperidin-1-yl]-2-oxoethylamine (1), a potent, selective, melanocortin subtype-4 receptor agonist. J. Med. Chem 2002, 45, 4589–4593. [DOI] [PubMed] [Google Scholar]
- 52.Palucki BL; Park MK; Nargund RP; Ye ZX; Sebhat IK; Pollard PG; Kalyani RN; Tang R; MacNeil T; Weinberg DH; Vongs A; Rosenblum CI; Doss GA; Miller RR; Stearns RA; Peng QP; Tamvakopoulos C; McGowan E; Martin WJ; Metzger JM; Shepherd CA; Strack AM; MacIntyre DE; Van der Ploeg LHT; Patchett AA, Discovery of (2S)-N-[(1R)-2-[4-cyclohexyl-4-[[(1,1-dimethylethyl)amino]carbonyl]-1-piperidinyl]-1-[(4-fluorophenyl)methyl]-2-oxoethyl]-4-methyl-2-piperazinecarboxamide (MB243), a potent and selective melanocortin subtype-4 receptor agonist. Bioorg. Med. Chem. Lett 2005, 15, 171–175. [DOI] [PubMed] [Google Scholar]
- 53.Ujjainwalla F, Design and syntheses of human melanocortin subtype-4 receptor (hMC4R) agonists: Discovery of the tert-butylpyrrolidine archetype. Abstr. Pap. Am. Chem. S 2005, 230, U2659–U2660. [Google Scholar]
- 54.Conde-Frieboes K; Ankersen M; Breinholt J; Hansen BS; Raun K; Thogersen H; Wulff BS, Serendipitous discovery of a new class of agonists for the melanocortin 1 and 4 receptors and a new class of cyclophanes. Bioorg. Med. Chem. Lett 2011, 21, 1459–1463. [DOI] [PubMed] [Google Scholar]
- 55.Joseph CG; Bauzo RM; Xiang ZM; Haskell-Luevano C, Urea small molecule agonists on mouse melanocortin receptors. Bioorg. Med. Chem. Lett 2003, 13, 2079–2082. [DOI] [PubMed] [Google Scholar]
- 56.Singh A; Kast J; Dirain MLS; Huang HS; Haskell-Luevano C, Synthesis and structure-activity relationships of substituted urea derivatives on mouse melanocortin receptors. ACS Chem. Neurosci 2016, 7, 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joseph CG; Wilson KR; Wood MS; Sorenson NB; Phan DV; Xiang ZM; Witek RM; Haskell-Luevano C, The 1,4-benzodiazepine-2,5-dione small molecule template results in melanocortin receptor agonists with nanomolar potencies. J. Med. Chem 2008, 51, 1423–1431. [DOI] [PubMed] [Google Scholar]
- 58.Szewczyk JR; Laudeman CP; Sammond DM; Villeneuve M; Minick DJ; Grizzle MK; Daniels AJ; Andrews JL; Ignar DM, A concise synthesis of 1,4-dihydro-[1,4]diazepine-5,7-dione, a novel 7-TM receptor ligand core structure with melanocortin receptor agonist activity. Bioorg. Med. Chem 2010, 18, 1822–1833. [DOI] [PubMed] [Google Scholar]
- 59.Houghten RA; Pinilla C; Appel JR; Blondelle SE; Dooley CT; Eichler J; Nefzi A; Ostresh JM, Mixture-based synthetic combinatorial libraries. J. Med. Chem 1999, 42, 3743–3778. [DOI] [PubMed] [Google Scholar]
- 60.Pinilla C; Appel JR; Borras E; Houghten RA, Advances in the use of synthetic combinatorial chemistry: mixture-based libraries. Nat. Med 2003, 9, 118–122. [DOI] [PubMed] [Google Scholar]
- 61.Houghten RA; Pinilla C; Giulianotti MA; Appel JR; Dooley CT; Nefzi A; Ostresh JM; Yu Y; Maggiora GM; Medina-Franco JL; Brunner D; Schneider J, Strategies for the use of mixture-based synthetic combinatorial libraries: scaffold ranking, direct testing in vivo, and enhanced deconvolution by computational methods. J. Comb. Chem 2008, 10, 3–19. [DOI] [PubMed] [Google Scholar]
- 62.Chen WB; Shields TS; Stork PJS; Cone RD, A colorimetric assay for measuring activation of Gs- and Gq-coupled signaling pathways. Anal. Biochem 1995, 226, 349–354. [DOI] [PubMed] [Google Scholar]
- 63.Reilley KJ; Giulianotti M; Dooley CT; Nefzi A; McLaughlin JP; Houghten RA, Identification of two novel, potent, low-liability antinociceptive compounds from the direct in vivo screening of a large mixture-based combinatorial library. AAPS J. 2010, 12, 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hensler ME; Bernstein G; Nizet V; Nefzi A, Pyrrolidine bis-cyclic guanidines with antimicrobial activity against drug-resistant Gram-positive pathogens identified from a mixture-based combinatorial library. Bioorg. Med. Chem. Lett 2006, 16, 5073–5079. [DOI] [PubMed] [Google Scholar]
- 65.Nefzi A; Ostresh JM; Appel JR; Bidlack J; Dooley CT; Houghten RA, Identification of potent and highly selective chiral tri-amine and tetra-amine μ opioid receptors ligands: An example of lead optimization using mixture-based libraries. Bioorg. Med. Chem. Lett 2006, 16, 4331–4338. [DOI] [PubMed] [Google Scholar]
- 66.Baell J; Walters MA, Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [DOI] [PubMed] [Google Scholar]
- 67.Nefzi A; Appel J; Arutyunyan S; Houghten RA, Parallel synthesis of chiral pentaamines and pyrrolidine containing bis-heterocyclic libraries. Multiple scaffolds with multiple building blocks: A double diversity for the identification of new antitubercular compounds. Bioorg. Med. Chem. Lett 2009, 19, 5169–5175. [DOI] [PubMed] [Google Scholar]
- 68.Manku S; Laplante C; Kopac D; Chan T; Hall DG, A mild and general solid-phase method for the synthesis of chiral polyamines. Solution studies on the cleavage of borane-amine intermediates from the reduction of secondary amides. J. Org. Chem 2001, 66, 874–885. [DOI] [PubMed] [Google Scholar]
- 69.Ostresh JM; Schoner CC; Hamashin VT; Nefzi A; Meyer JP; Houghten RA, Solid-phase synthesis of trisubstituted bicyclic guanidines via cyclization of reduced N-acylated dipeptides. J. Org. Chem 1998, 63, 8622–8623. [Google Scholar]
- 70.Ericson MD; Schnell SM; Freeman KT; Haskell-Luevano C, A fragment of the Escherichia coli ClpB heat-shock protein is a micromolar melanocortin 1 receptor agonist. Bioorg. Med. Chem. Lett 2015, 25, 5306–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tala SR; Schnell SM; Haskell-Luevano C, Microwave-assisted solid-phase synthesis of side-chain to side-chain lactam-bridge cyclic peptides. Bioorg. Med. Chem. Lett 2015, 25, 5708–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh A; Tala SR; Flores V; Freeman K; Haskell-Luevano C, Synthesis and pharmacology of α/β3-peptides based on the melanocortin agonist Ac-His-DPhe-Arg-Trp-NH2 sequence. ACS Med. Chem. Lett 2015, 6, 568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schild HO, pA, a new scale for the measurement of drug antagonism. Br. J. Pharmacol 1947, 2, 189–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Enriori PJ; Chen W; Garcia-Rudaz MC; Grayson BE; Evans AE; Comstock SM; Gebhardt U; Muller HL; Reinehr T; Henry BA; Brown RD; Bruce CR; Simonds SE; Litwak SA; McGee SL; Luquet S; Martinez S; Jastroch M; Tschop MH; Watt MJ; Clarke IJ; Roth CL; Grove KL; Cowley MA, α-Melanocyte stimulating hormone promotes muscle glucose uptake via melanocortin 5 receptors. Mol. Metab 2016, 5, 807–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang L; Li WH; Anthonavage M; Pappas A; Rossetti D; Cavender D; Seiberg M; Eisinger M, Melanocortin-5 receptor and sebogenesis. Eur. J. Pharmacol 2011, 660, 202–206. [DOI] [PubMed] [Google Scholar]
- 76.Rideout MC; Boldt JL; Vahi-Ferguson G; Salamon P; Nefzi A; Ostresh JM; Giulianotti M; Pinilla C; Segall AM, Potent antimicrobial small molecules screened as inhibitors of tyrosine recombinases and Holliday junction-resolving enzymes. Mol. Diversity 2011, 15, 989–1005. [DOI] [PubMed] [Google Scholar]
- 77.Fleeman R; LaVoi TM; Santos RG; Morales A; Nefzi A; Welmaker GS; Medina-Franco JL; Giulianotti MA; Houghten RA; Shaw LN, Combinatorial libraries as a tool for the discovery of novel, broad-spectrum antibacterial agents targeting the ESKAPE pathogens. J. Med. Chem 2015, 58, 3340–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doering SR; Todorovic A; Haskell-Luevano C, Melanocortin antagonist tetrapeptides with minimal agonist activity at the mouse melanocortin-3 receptor. ACS Med. Chem. Lett 2015, 6, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen CA; Okayama H, Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques 1988, 6, 632–638. [PubMed] [Google Scholar]
- 80.Houghten RA, General method for the rapid solid-phase synthesis of large numbers of peptides: specificity of antigen-antibody interaction at the level of individual amino acids. Proc. Natl. Acad. Sci. U. S. A 1985, 82, 5131–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


