ABSTRACT
Antibiotic resistance is a global challenge for tuberculosis control, and accelerating its diagnosis is critical for therapy decisions and controlling transmission. Genotype-based molecular diagnostics now play an increasing role in accelerating the detection of such antibiotic resistance, but their accuracy depends on the instructed detection of genetic variations. Genetic mobile elements such as IS6110 are established sources of genetic variation in Mycobacterium tuberculosis, but their implication in clinical antibiotic resistance has thus far been unclear. Here, we describe the discovery of an intragenic IS6110 insertion into Rv0678 that caused antibiotic resistance in an in vitro-selected M. tuberculosis isolate. The subsequent development of bioinformatics scripts allowed genome-wide analysis of intragenic IS6110 insertions causing gene disruptions in 6,426 clinical M. tuberculosis strains. This analysis identified 10,070 intragenic IS6110 insertions distributed among 333 different genes. Focusing on genes whose disruption leads to antibiotic resistance, 12 clinical isolates were identified with high confidence to be resistant to bedaquiline, clofazimine, pyrazinamide, ethionamide, and para-aminosalicylic acid because of an IS6110-mediated gene disruption event. A number of these IS6110-mediated resistant strains had identical genomic distributions of IS6110 elements and likely represent transmission events of a single resistant isolate. These data provide strong evidence that IS6110-mediated gene disruption is a clinically relevant mechanism of antibiotic resistance in M. tuberculosis that should be considered for molecular diagnostics. Concomitantly, this analysis provides a list of 333 IS6110-disrupted genes in clinical tuberculosis isolates that can be deemed nonessential for human infection.
IMPORTANCE To help control the spread of drug-resistant tuberculosis and to guide treatment choices, it is important that rapid and accurate molecular diagnostic tools are used. Current molecular diagnostic tools detect the most common antibiotic-resistance-conferring mutations in the form of single nucleotide changes, small deletions, or insertions. Mobile genetic elements, named IS6110, are also known to move within the M. tuberculosis genome and cause significant genetic variations, although the role of this variation in clinical drug resistance remains unclear. In this work, we show that both in vitro and in data analyzed from 6,426 clinical M. tuberculosis strains, IS6110 elements are found that disrupt specific genes essential for the function of a number of pivotal antituberculosis drugs. By providing ample evidence of clinically relevant IS6110-mediated drug resistance, we believe that this shows that this form of genetic variation must not be overlooked in molecular diagnostics of drug resistance.
KEYWORDS: IS6110, Mycobacterium tuberculosis, antibiotic resistance, diagnostics, genetic polymorphisms
INTRODUCTION
With more than 400,000 annual infections by multidrug-resistant (MDR) and extensively drug-resistant (XDR) Mycobacterium tuberculosis (World Health Organization [WHO] Global Tuberculosis Report 2020 [1]), the WHO recommends M. tuberculosis drug susceptibility testing to guide treatment choices, improve therapy outcomes, and minimize the spread of drug-resistant infections. Culture-based drug susceptibility testing remains a gold-standard diagnostic test; however, the sluggish growth of M. tuberculosis makes this too slow for quick decision-making and tackling drug-resistant tuberculosis (TB). Molecular genotyping tests such as Xpert or Truenat (1) allow rapid initial diagnostics for antibiotic resistance by detecting the most prevalent mutations linked to isoniazid and rifampicin resistance but have as a limitation that they cannot detect less-frequent mutations or mutations that confer resistance to other antibiotics. To achieve better diagnostics, next-generation sequencing (NGS) approaches such as deep sequencing of multiple targeted amplicons (Deeplex technology) (2) or whole-genome sequencing (WGS) are used to increase the coverage of multiple genomic regions. To extract the most out of these next-generation technologies, it is vital that the bioinformatics pipeline can detect diverse genetic variations that could lead to antibiotic resistance, such as single nucleotide polymorphisms (SNPs) and short insertions-deletions (indels) but also genome locus duplications and insertions.
An important source of genetic variation in M. tuberculosis is mobile genetic elements (MGEs), in particular the Mycobacterium complex-specific IS6110 element, that occasionally transpose within the bacterial genome. Such events of transposition into genes largely result in a loss of gene function (akin to saturating transposon mutagenesis used to define essential genes [3–5]), although the presence of an outward-directed promoter at the 3′ end of IS6110 can occasionally allow for gene overexpression when the insertion is upstream of genes (6–8). IS6110 carries two out-of-frame open reading frames (ORFs) of a split transposase that normally do not allow the translation of an active transposase. Occasionally, however, this −1 ribosomal frameshift can be overcome by ribosome stalling and backtracking, a feature aided by an RNA pseudoknot (9), and increased under adverse bacterial growth conditions and during infection (9). In general, M. tuberculosis strains carry a high number of genomic IS6110 copies, whose detection has served as a valuable diagnostic marker for M. tuberculosis infection (10) and where IS6110 restriction fragment length polymorphism (RFLP) insertion mapping has traditionally allowed for strain epidemiology (11, 12). The IS6110 distribution is thought to play a major role in the genetic variation and characteristics of M. tuberculosis strains, with suggestions that the particularly large number of IS6110 copies in M. tuberculosis Beijing/W lineage strains may be associated with their higher virulence (13). While IS6110 insertions are frequently found in M. tuberculosis strains, they do not always result in gene inactivation. As an example, H37Rv carries 16 copies of IS6110 (see Table S1 in the supplemental material), 10 of which are within intergenic regions (with potential polar effects) and 6 of which appear to have caused insertion sequence (IS)-mediated gene inactivation. To date, it remains unclear how frequently genes are pseudogenized by IS6110 elements in clinical M. tuberculosis strains and if this has repercussions on antibiotic resistance.
As the inactivation of certain genes (such as by premature stop codons) is known to cause drug resistance to particular antibiotics, it would be expected that IS6110-mediated gene inactivation could achieve the same result. Indeed, genome-wide analysis of MGEs in other bacteria found evidence of transposon-mediated antibiotic resistance through the inactivation of porins (ompF) and transcriptional regulators of efflux pumps (acrR) (14). With respect to M. tuberculosis, however, despite the increasing availability of next-generation sequencing of clinical stains, to the best of our knowledge, no clear evidence has been reported showing IS6110-mediated antibiotic resistance. Unique evidence from the “pregenomic era,” however, clearly showed in an isolated clinical case, that IS6110-mediated inactivation of pncA, the pyrazinamidase responsible for activating pyrazinamide, conferred resistance to this prodrug (15). Considering this, we postulated that IS6110-mediated drug resistance may be underdiagnosed, particularly considering the large number of prodrugs used for the treatment of tuberculosis, whose bacterium-specific bioactivation is often through nonessential genes.
In this study, we describe the finding of IS6110-mediated antibiotic resistance in an in vitro-selected M. tuberculosis-resistant isolate. We then develop bioinformatics tools to detect intragenic IS6110 insertions from whole-genome sequencing data and use this script to define the intragenic IS6110 landscape in 6,426 clinical isolates, finding strong evidence of IS6110-mediated resistance to bedaquiline, clofazimine, pyrazinamide, ethionamide, and para-aminosalicylic acid (PAS).
RESULTS
Selection and characterization of OH190-resistant H37Rv isolates.
OH190 is a synthetic analogue of pyridomycin whose antituberculosis activity appeared unchanged by episomal overexpression of the pyridomycin target, the enoyl-reductase InhA (16). In search of alternative/additional protein targets, OH190-resistant H37Rv isolates were selected on plates containing 10 μg/ml OH190 (giving 121 colonies, a frequency of resistance of 5 × 10−6) and 20 μg/ml OH190 (only 1 colony appeared). As a comparison, selection on 10 and 20 μg/ml pyridomycin resulted in 4 and 3 colonies, respectively (frequency of resistance of 1.5 × 10−8). For both antibiotics, no colonies were isolated at 40 μg/ml. The 3 OH190-resistant clones (randomly selected from 122 resistant clones) were confirmed to have low-level resistance (2-fold) to OH190 and pyridomycin (Table 1), while the 3 pyridomycin-resistant isolates were >16-fold more resistant to pyridomycin and 8-fold more resistant to OH190.
TABLE 1.
Genotypes and antibiotic susceptibilities of selected H37Rv pyridomycin- and OH190-resistant isolates
| Isolate | Genotypea | MIC (μg/ml) |
||
|---|---|---|---|---|
| Pyridomycin | OH190 | Isoniazid | ||
| Parental H37Rv | WT | 0.8 | 1.6 | 0.08 |
| PYR RC10.1 | InhA(M161L) | 25 | 12.5 | 0.08 |
| PYR RC10.2 | InhA(M161L) | 12.5 | 6.3 | 0.04 |
| PYR RC20.1 | InhA(M161L) | 25 | 12.5 | 0.04 |
| OH190 RC10.4 | Rv0678(A71D) | 1.6 | 3.1 | 0.08 |
| OH190 RC10.5 | Rv0678::IS6110 | 1.6 | 3.1 | 0.08 |
| OH190 RC20.1 | Rv0678(N70D) | 1.6 | 3.1 | 0.08 |
WT, wild type.
Sanger sequencing of inhA and its promoter.
Sanger sequencing of InhA and its promoter confirmed that all three OH190-resistant isolates were wild type for this DNA region, while all three pyridomycin-resistant strains carried a previously undescribed mutation in InhA, namely, A481C (M161L). Structural data (17) show that M161 interacts through hydrophobic interactions with pyridomycin, and the isolated mutation likely eliminates this binding interaction. The pyridomycin-resistant isolates were also partially resistant to OH190, which suggests that this analogue still interacts with InhA but perhaps has gained a second mechanism of action.
Whole-genome sequencing and variant analysis of the 3 OH190-resistant isolates and the parental H37Rv wild-type strain found that 2 of the resistant isolates (named OH190 RC10.4 and OH190 RC20.1) carried a unique nonsynonymous SNP in Rv0678 (C212A [A71D] and A208G [N70D], respectively, located in the helix-turn-helix DNA binding domain). No variants (SNPs or small indels) were found in the third OH190-resistant isolate (named OH190 RC10.5). Due to the mutations identified in Rv0678, the sequence reads of OH190 RC10.5 that mapped to this region were scrutinized. In doing so, a short, 5-base stretch (bases 28 to 32 of Rv0678) was found to have a doubled read depth, a hallmark of a transposon insertion event (9). The reads that mapped to this 5′ region of Rv0678 were also found to only partially map (soft-clipped sequences) (Fig. 1), with the unmapped portion aligning to the extremities of IS6110, indicating its insertion into Rv0678 in this resistant clone. This finding was confirmed by an increase in the size of a PCR product for Rv0678 (see Fig. S1 in the supplemental material) and subsequent Sanger sequencing. Rv0678 codes for a transcriptional repressor whose loss of function leads to the overexpression of the efflux pump mmpL5-mmpS5, which in turn results in low-level resistance to the antituberculosis drugs bedaquiline and clofazimine (18). The identification of this IS6110-mediated inactivation of Rv0678 and the consequent resistance to OH190 led us to question if such IS6110-mediated resistance is also observed clinically.
FIG 1.
(A and B) Representation of the sequencing reads mapped onto the H37Rv reference genome for parental H37Rv (A) and OH190-resistant clone 10.5 (B). Green and red sequencing reads represent mapping in the forward and reverse directions, respectively. While reads from the parental strain map fully to the reference genome, for the resistant strain, there is a 5-bp duplication of mapped sequencing reads (highlighted in the inset of panel B), a hallmark of a transposon insertion. In addition, sequencing reads of the OH190-resistant clone only partially map to this genomic region (soft clipping), with the nonaligned end of the reads (lighter color) aligning with IS6110. (C) Sequencing reads of the OH190-resistant clone mapped onto a “corrected” genome containing the intragenic IS6110 element in Rv0678 showing the now full alignment of the sequencing reads across the border of the IS6110 insertion sites.
Identification of IS6110-disrupted genes in genomic data from clinical M. tuberculosis isolates.
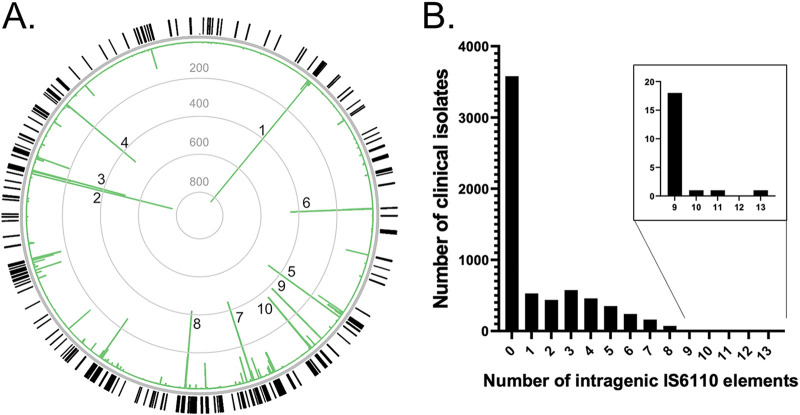
To determine if IS6110 insertions could mediate antibiotic drug resistance in clinical isolates of M. tuberculosis, we decided to analyze IS6110 elements in the genomes of 6,426 clinical M. tuberculosis isolates available from the NCBI genome repository (https://www.ncbi.nlm.nih.gov/genome/166). As these data are derived from clinical isolates, some of these isolates may present a single M. tuberculosis strain with dendrograms (based on BLAST) of their genetic variation presented on the NCBI website. These genomic data were available as preassembled contigs for all 6,426 isolates as well as associated raw sequencing reads (but not for all isolates). As it is challenging to predict the impact of IS6110 insertions in noncoding regions on antibiotic susceptibility, we decided to analyze only the distribution of intragenic IS6110 insertions that likely lead to a gene disruption and loss of function. To achieve this, a contig-based bioinformatics analysis was performed on the genomic data of all 6,426 isolates, identifying 10,070 intragenic IS6110 insertion events distributed among 333 different genes (the full list of insertions per genome is available upon request). Among the most frequently found intragenic IS6110 insertions were mmpS1 (in 828/6,426 genomes), Rv3113, and Rv3128c (Fig. 2A; Table S2). Interestingly, upon determining the frequency of the top 10 IS6110 intragenic insertions with the M. tuberculosis strain lineage of the clinical isolates, a clear lineage bias distribution was observed (Table S3), with, for example, 80% of mmpS1 insertions being lineage 4.1 strains, 99.5% of Rv3113 insertions being lineage 4.3 strains, and 77% of Rv3128c insertions being lineage 2 strains. The majority (56%) of clinical isolates carried no intragenic insertions relative to H37Rv (which has 16 IS6110 elements) (Table S1), with the remaining isolates being found to harbor up to as many as 13 insertions per clinical isolate (Fig. 2B; Table S4).
FIG 2.
Genome-wide analysis of intragenic IS6110 insertions found in 6,426 clinical M. tuberculosis isolates using a contig-based bioinformatics approach. (A) Representation of the genomic distribution of identified intragenic IS6110 insertions (outer circle), with the relative frequency for each intragenic insertion shown in the bar chart within the circle (linear scale). The top 10 most frequent genes found to be disrupted by IS6110 insertion are marked (1, mmpS1; 2, Rv3113; 3, Rv3128c; 4, idsB; 5, Rv1371; 6, Rv963c; 7, Rv1754c; 8, Rv2016; 9, ctpD; 10, mmpL12). The image was generated using Circos (39). (B) Bar chart showing the frequency of clinical isolates with different numbers of identified intragenic IS6110 insertions. The inset is a zoomed-in section of the main bar chart. Note that while all the isolates come from different patients, some may be classified as the same strain.
Evidence of IS6110-mediated antibiotic resistance.
To evaluate if any of the 333 identified IS6110 gene disruption events in the clinical isolates could mediate antibiotic resistance, it was important to define a list of genes whose inactivation is expected, with high confidence, to result in antibiotic resistance. This was particularly important as the clinical strains examined were not readily available for antibiotic testing and validation. In M. tuberculosis, Rv0678 is one such high-confidence resistance gene whose inactivation is known to cause both in vitro and clinical resistance to bedaquiline and clofazimine through the upregulation of mmpL5-mmpS5 (18, 19). Similarly, inactivation of the thyA gene encoding folate-dependent thymidylate synthase causes resistance to para-aminosalicylic acid (PAS) (20). Finally, the unique abundance of bacterially activated prodrugs in tuberculosis therapy makes the inactivation of the bioactivation enzymes a high-confidence marker for associated antibiotic resistance. To this extent, the absence of a functional copy of katG, pncA, or ethA will result in resistance to pyrazinamide, isoniazid, or ethionamide, respectively. Similarly, the inactivation of ddn or any of the genes involved in F420 cofactor biosynthesis (fbiA, fbiB, fbiC, fbiD, and fdg) will result in resistance to the newly approved nitroimidazole prodrugs delamanid and pretomanid. Of the 333 identified intragenic IS6110 elements identified in the 6,426 M. tuberculosis genomes using the contig-based analysis, 12 clinical strains were identified to carry an IS6110 insertion in Rv0678, ethA, pncA, or thyA (Table 2) but not in katG, ddnI, fbiA, fbiB, fbiC, fbiD, and fdg.
TABLE 2.
Clinical M. tuberculosis strains identified to carry an IS6110 insertion element in genes whose disruption confers antibiotic resistancea
| M. tuberculosis genome | Gene with IS6110 insertion | Associated resistance to clinical antibiotic | Insertion position/gene length (bp) | Confirmation by read-based analysis | Geographic location |
|---|---|---|---|---|---|
| TB_RSA126 | Rv0678 | Bedaquiline/clofazimine | 71/498b | Yes | South Africa |
| TB_RSA64 | Rv0678 | Bedaquiline/clofazimine | 71/498b | Yes | South Africa |
| TB_RSA63 | Rv0678 | Bedaquiline/clofazimine | 71/498b | Yes | South Africa |
| TKK-01-0074 | Rv0678 | Bedaquiline/clofazimine | 76/498 | Yes | South Africa |
| KT-0084 | Rv0678 | Bedaquiline/clofazimine | 71/498 | Yes | South Korea |
| Strain 02-R0861 | ethA | Ethionamide | 1,267/1,470c | Yes | Lima, Peru |
| Strain 99-09115 | ethA | Ethionamide | 1,267/1,470c | Reads not available | Lima, Peru |
| Strain 02-R0014 | pncA | Pyrazinamide | 159/561c | Reads not available | Lima, Peru |
| Strain 01-R0774 | pncA | Pyrazinamide | 159/561c | Yes | Lima, Peru |
| XTB13-251 | pncA | Pyrazinamide | 462/561 | Yes | Belarus |
| KT-0109 | thyA | PAS | 84/792 | Yes | South Korea |
| KT-0077 | thyA | PAS | 150/792 | Yes | South Korea |
Mapped reads can be visualized in Fig. S2 in the supplemental material. PAS, para-aminosalicylic acid.
Identical by IS6110 barcode: different clinical isolate, same M. tuberculosis strain.
Identical by contig-based genotype comparison: different clinical isolate, same M. tuberculosis strain.
As a means of validating the contig-based IS6110 insertion analysis and to gain greater resolution of the overall distribution of IS6110 elements, a sequence-read-based bioinformatics analysis was established to identify both intergenic and intragenic IS6110 insertions from the original raw sequence reads. Using this sequence read-based analysis, the predicted IS6110 disruption of Rv0678, ethA, pncA, and thyA in the clinical isolates by the contig-based method was confirmed (mapped sequence reads are also shown in Fig. S2). In addition, it was verified that no wild-type gene copy was present that could have originated from a gene duplication event. Together, these results leave it unlikely that these specific clinical isolates have a functional Rv0678, ethA, pncA, or thyA gene and are therefore most certainly resistant to the associated antibiotics (Table 2).
The higher resolution of the read-based analysis also allowed the generation of an IS6110 “barcode” that illustrates the exact location of IS6110 insertions on an H37Rv reference genome. This IS6110 barcode therefore acts as a high-resolution version of the IS6110 mapping performed by RFLP approaches for tuberculosis epidemiology studies. As 4 of the 5 clinical isolates with IS6110-disrupted Rv0678 carried this insertion in an identical location (base 71 of Rv0678) (Table 2), the developed IS6110 barcode was used to determine if they were closely related and possibly originated from the transmission of a single isolate. This analysis indeed found that 3 of the 4 clinical strains have identical IS6110 barcodes (TB_RSA126, TB_RSA64, and TB_RSA63) (Fig. 3). As these three strains are also documented to come from the same geographic location (South Africa, with no further detailed information available), it is likely that they represent a single transmitted isolate resistant to bedaquiline due to an IS6110-mediated Rv0678 disruption. For the two clinical isolates with IS6110 insertions in either ethA or pncA, IS6110 barcodes could not be generated (as the original sequencing reads were not available), but pairwise analysis of their genotype found these isolates to be identical and hence also represent single M. tuberculosis strains.
FIG 3.
The IS6110 “barcode” of four identified clinical strains carrying an IS6110 insertion at base 71 of Rv0678 (marked with asterisks). This barcode was generated from the sequencing-read-based analysis and shows the distribution of IS6110 insertions along the H37Rv reference genome (x axis) against the coverage of each insertion event (y axis). The data clearly indicate that TB_RSA126, TB_RSA64, and TB_RSA63 have identical IS6110 barcodes, likely suggesting high similarity of the isolates and the potential transmission of this clinical strain, while the KT_0084 IS6110 barcode is different and likely not related.
Previously identified essential genes.
As well as its repercussions for antibiotic susceptibility, the intragenic IS6110 insertion landscape in clinical isolates also provides insight into gene essentiality in human infection. IS6110 elements are a natural surrogate of the saturating transposon mutagenesis analysis performed to define the essentiality of M. tuberculosis genes under in vitro and ex vivo conditions (3, 4, 21, 22). Of the 333 genes found with intragenic IS6110 elements using our contig-based analysis, only Rv2017, a reported transcriptional regulator, was previously described as essential by transposon-site hybridization (TraSH) analysis (3) (Fig. S3). This IS6110-disrupted Rv2017 gene was found in seven clinical strains, all at the same genomic location and all originating from Lima, Peru. Additional analysis also confirmed the absence of an intact copy of Rv2017 in these genomes; however, an IS6110 barcode could not be generated, as raw sequencing reads were unavailable for these isolates. In addition to essential genes, IS6110 insertions were also identified in genes whose inactivation was previously found to cause a bacterial growth defect by DeJesus and colleagues (3), namely, Rv2491 and Rv2764c (thyA). In the case of thyA, this selection could have been driven by para-aminosalicylic acid as discussed above for antibiotic resistance. While developing the script to identify intragenic IS6110 insertions, it was also found that a few clinical isolates carried IS6110 elements in core essential genes (dnaA, dnaN, pyrH, purF, and sodA) (Fig. S4), but these genomes were also found to carry a second uninterrupted gene copy indicative of a previous gene duplication event. Similar genomic plasticity and gene duplications have been observed in griselimycin-resistant M. tuberculosis isolates where multiple copies of dnaN were identified (23).
DISCUSSION
Considering the described mobility of IS6110 elements in the M. tuberculosis genome; the substantial number of genes whose inactivation leads to antibiotic resistance; a previous, pregenomic-era report of IS6110-mediated pyrazinamide resistance in tuberculosis (15); and the current regularity of whole-genome sequencing of M. tuberculosis, it was remarkable that gene inactivation by IS6110 has not been reported as a source of clinical antibiotic resistance from genome sequencing data. Analysis of such genomic data in Enterobacteriaceae found evidence of likely MGEs mediating antibiotic resistance (14), but despite efforts, this has not previously been shown in M. tuberculosis (14, 24). The custom analysis performed here on 6,426 clinical M. tuberculosis strains, however, provides clear evidence that intragenic IS6110 insertions play a vital role in defining clinical M. tuberculosis drug susceptibility and that such resistance is likely transmitted. In addition, as the identified IS6110 insertions lead to the full inactivation of Rv0678, ethA, thyA, and pncA, these strains are likely fully clinically resistant to bedaquiline, ethionamide, PAS, and pyrazinamide and not just less susceptible.
The identified frequency of 12/6,426 clinical isolates with likely antibiotic resistance due to IS6110 gene disruption suggests that such a source of antibiotic resistance is rare. Nonetheless, the bioinformatics analysis performed here was designed to define high-probability antibiotic-resistant cases with high confidence and not the absolute number of such events, hence likely leading to a conservative evaluation. For one, this analysis relied on contig data, and it may be that some information was lost because contigs carrying IS6110 insertions did not correctly assemble. In addition, this work focused only on intragenic IS6110 insertions, while it is known that intergenic insertions can also affect local gene expression (6–8), which could lead to antibiotic resistance. Finally, M. tuberculosis also contains additional, non-IS6110 mobile elements that can equally play a role in gene inactivation and antibiotic resistance (25). Despite this, it is likely that IS6110 remains a low-frequency but definite source of antibiotic resistance that needs to be factored into molecular genotyping diagnostics and analysis.
While the genomes of the clinical strains analyzed in this study predate the clinical introduction of bedaquiline (as far as can be determined from NCBI data), it is interesting to note that intragenic insertions of IS6110 into Rv0678 were observed. Loss-of-function mutations in Rv0678 cause in vitro and clinical resistance to bedaquiline and clofazimine, and finding these prebedaquiline strains with inactivated Rv0678 (and, thus, likely bedaquiline resistance) is in accordance with findings demonstrating preexisting SNP-based Rv0678 mutations (19, 26). This finding further illustrates the importance of considering IS6110-mediated genetic variations in molecular diagnostic predictions of bedaquiline resistance.
Having found evidence of IS6110 insertions mediating resistance to the antituberculosis prodrugs ethionamide and pyrazinamide, it is worth noting that despite many decades of selective pressure from the first-line antibiotic isoniazid, no IS6110 insertions were observed in the nonessential catalase gene katG, vital for isoniazid bioactivation. A likely explanation for this is that katG inactivation has been shown to confer a considerable fitness cost (27) that may limit its competitive advantage over more classical point mutations in katG. In addition to this lack of isoniazid resistance, no clinical isolates were identified with IS6110-mediated resistance to the new class of antituberculosis nitroimidazole prodrugs, pretomanid and delamanid, despite the many genes potentially being involved in their bioactivation (ddn, fbiA, fbiB, fbiC, fbiD, and fdg). As all samples predate the clinical use of pretomanid and delamanid to treat tuberculosis, there appears to be no evidence of preexisting resistance through IS6110 insertions. Nonetheless, due to the large number of genes involved in the activation of nitroimidazoles, it would be prudent not to ignore such possibilities now that they are approved for clinical use.
This work clearly illustrates the role of IS6110 elements in mediating antibiotic resistance, and current molecular genotyping diagnostics are not set up to detect such events. To better detect such IS6110 insertions from WGS data, it is important that de novo contig assembly is performed, rather than mapping against a reference genome. Alternatively, sequencing technology based on deep sequencing of targeted amplicons (such as Deeplex) may allow the indirect detection of such events based on the failure to generate targeted amplicons, although this remains to be assessed. In addition to intragenic IS6110 insertions, the potential role of intergenic insertions will also need to be evaluated, although such studies will necessitate access to the clinical strains themselves for validation experiments. In addition to the bioinformatics approaches described here to define intragenic IS6110 elements, similar bioinformatics tools have been reported that can detect sequence-specific IS element insertions (ISMapper [28]) or sequence-independent IS insertion events (panISa [29] and MGEfinder [14]). In our opinion, it is necessary that these bioinformatics approaches act as blueprints for the development of easily accessible molecular diagnostic tools that can be used in a clinical setting for the detection of antibiotic drug resistance and molecular epidemiology.
Overall, this work puts into perspective the importance of bioinformatics analysis for NGS-based diagnostics of M. tuberculosis clinical resistance and provides evidence for the role of IS6110-mediated gene inactivation in antibiotic resistance. Improving the detection and understanding of these mobile elements will allow more accurate diagnostics and treatment decisions and help further increase our understanding of gene essentiality in human tuberculosis infection.
MATERIALS AND METHODS
Chemicals.
Pyridomycin was purified from the supernatant of Dactylosporangium fulvum cultures as described previously (17). OH190, a C2-cyclohexyl pyridomycin analogue, was synthesized by de novo chemistry methods as described previously (30, 31) and kindly provided by Karl-Heinz Altmann (ETH Zurich, Switzerland).
In vitro selection of OH190-resistant H37Rv.
Following reports that InhA overexpression does not induce resistance to OH190 (30), we decided to define the mechanism of resistance to OH190 in H37Rv, with the aim of defining a new mechanism of action. H37Rv was grown to mid-log phase (optical density at 600 nm [OD600] of 0.4 to 0.8) in Middlebrook 7H9 medium supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC), 0.2% glycerol, and 0.05% Tween 80. Log-phase cultures were then pelleted (3,200 × g for 6 min) and resuspended to an OD600 of 100. One hundred microliters (∼6 × 108 cells) of this concentrated suspension was then plated onto Middlebrook 7H11 medium supplemented with 10% OADC, 0.5% glycerol, and 0.05% Tween 80 and containing either pyridomycin (10, 20, or 40 μg/ml) or OH190 (10, 20, or 40 μg/ml) and incubated for 4 weeks (37°C) to allow the growth of resistant colonies.
Whole-genome sequencing of OH190-resistant strains.
Genomic DNA was isolated from three OH190-resistant isolates and the parental H37Rv wild-type strain and submitted for library preparation (Nextera XT for 2- by 250-bp paired-end reads) and whole-genome sequencing on an Illumina HiSeq 2500 platform (Genoscreen, France). Sequencing reads were mapped onto the M. tuberculosis H37Rv reference genome (GenBank accession number NC_000962.3; 4,411,532 bp) using Bowtie 2.0. The stringent mapping procedure used allowed coverage of 94.5% of the reference genome on average that will be used for subsequent variant analyses (average read depth of >40×). Based on the mapping obtained, SNPs and indels were identified against the accessible portion (>94.4%) of the M. tuberculosis reference genome (GenBank accession number NC_000962.3), independently for the four samples, and compared.
Contig-based bioinformatics analysis of IS6110 gene disruption in clinical M. tuberculosis strains.
A total of 6,426 genome sequences of M. tuberculosis with various levels of assembly were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/genome/166). M. tuberculosis lineage attribution was performed according to the classification of SNPs as previously described (32). All sequence files were concatenated to build a single nucleotide BLAST sequence database, which was searched (BLASTN [33]) using the sequences of all 3,998 annotated genes of M. tuberculosis strain H37Rv (GenBank accession number AL123456), and saved in XML format. A custom python script was subsequently used to parse the BLASTN result files, providing a list of all mutations (SNPs and short indels) detected relative to the H37Rv reference genes but also a list of “incomplete” alignments. These incomplete alignments occurred where a gene aligned to two separate contigs or because the gene alignment on a single contig was split into two high-scoring pairs (HSPs), suggesting a large insertion. When two HSPs were separated by 1,000 to 3,000 bp (space for one or two IS6110 insertion elements), the inter-HSP sequences were aligned to the IS6110 sequence (BLASTN) to confirm that the insertion was mediated by an IS6110 element. In all cases where IS6110 insertions were predicted to mediate antibiotic resistance, this contig-based analysis was validated using the raw sequencing reads where available.
Sequence-read-based bioinformatics analysis of IS6110 gene disruption in clinical M. tuberculosis strains.
The sequencing reads from the studied clinical isolates were downloaded and mapped to the IS6110 sequence using Bowtie 2 (34) in local mode (using the –local and –no-unal parameters). This allowed the selection of all the sequencing reads that mapped fully or partially to IS6110. The output SAM alignment files were converted to BAM files, sorted using SAMtools (35) (default parameters), and converted into a fastq file using bamTofastq (36) (default parameters).
A synthetic M. tuberculosis reference genome (named H37Rv_Del_IS6110) was constructed by deleting the 16 IS6110 copies (see Table S1 in the supplemental material) in the H37Rv genome (GenBank accession number NC_000962.3) as well as a repeated region loosely related in sequence to IS6110 (H37Rv coordinates 3119185 to 3123576). As the IS6110 elements in H37Rv are not considered to cause antibiotic resistance, we decided to avoid reporting these insertion sites by further deleting the 150-bp flanking regions of the H37Rv IS6110 elements. Together, the coordinates (H37Rv based) of these 17 deleted regions are as follows: 888871 to 890526, 1541802 to 1543457, 1987553 to 1989208, 1995951 to 1997606, 2365264 to 2366919, 2429967 to 2431622, 2549864 to 2551519, 2635427 to 2637082, 2784464 to 2786121, 2971959 to 2973614, 3119185 to 3123576, 3120373 to 3122048, 3551080 to 3552735, 3552563 to 3554218, 3710232 to 3711887, 3794908 to 3796563, and 3890629 to 3892284. The extracted IS6110 mapped reads (in fastq format) were mapped onto the H37Rv_Del_IS6110 synthetic genome using Bowtie 2 (in local mode using the –local and –no-unal parameters). The SAM alignment files were then filtered to remove alignments with a quality of mapping (MAPQ) parameter of <2 (option -q 2) using SAMtools view (default parameters). The SAM alignment files were converted to BAM files and sorted using SAMtools view with default parameters.
The coverage depth at each position of the H37Rv_Del_IS6110 synthetic genome sequence was calculated using SAMtools depth (default parameters). The depth text file was converted into CSV (comma-separated values) file format using a custom python script. Using a custom python script, the sequencing depth was plotted using matplotlib.pyplot (37) along the H37Rv_Del_IS6110 synthetic genome, forming an IS6110 barcode (png file exported), and the peaks were detected using the find_peaks function of the scipy.signal package (38) with the following parameters: distance of 1,500 and prominence of 20. This script also provided the coordinates of peak summits detected in a text file. Finally, a custom script defined the IS6110 insertions as intragenic (insertions in the coding DNA sequence [CDS]) or an intergenic region (IGR).
Data availability.
Whole-genome sequencing reads for the OH190-resistant M. tuberculosis H37Rv isolates (fastq format) have been deposited in the NCBI database (BioProject accession number PRJNA739842).
ACKNOWLEDGMENTS
This work was funded by Foundation pour la Recherche Médicale (FRM) grant number DBF20160635729.
We thank Maryline Dong (Kienle) and Oliver Horlacher from the group of Karl-Heinz Altmann at ETH Zürich for the synthesis of OH190. We thank Alain Baulard for scientific discussion and proofreading.
R.C.H. performed research on H37Rv; R.A., C.G., and R.C.H. were involved in the identification of IS6110 elements in H37Rv; R.A. wrote the script to analyze genomic data from clinical M. tuberculosis isolates; and R.A. and R.C.H. designed the study and wrote the paper.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Ruben C. Hartkoorn, Email: ruben.hartkoorn@inserm.fr.
William Lainhart, University of Arizona/Banner Health.
REFERENCES
- 1.World Health Organization. 2020. Global tuberculosis report 2020. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller PP, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO. 2017. BUSTER version 2.10.3. Global Phasing Ltd, Cambridge, United Kingdom. [Google Scholar]
- 3.DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, Ioerger TR. 2017. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8:e02133-16. doi: 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 5.Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. 2012. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol 19:218–227. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McEvoy CRE, Falmer AA, van Pittius NCG, Victor TC, van Helden PD, Warren RM. 2007. The role of IS6110 in the evolution of Mycobacterium tuberculosis. Tuberculosis (Edinb) 87:393–404. doi: 10.1016/j.tube.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Soto CY, Menéndez MC, Pérez E, Samper S, Gómez AB, García MJ, Martín C. 2004. IS6110 mediates increased transcription of the phoP virulence gene in a multidrug-resistant clinical isolate responsible for tuberculosis outbreaks. J Clin Microbiol 42:212–219. doi: 10.1128/JCM.42.1.212-219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beggs ML, Eisenach KD, Cave MD. 2000. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J Clin Microbiol 38:2923–2928. doi: 10.1128/JCM.38.8.2923-2928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalo-Asensio J, Pérez I, Aguiló N, Uranga S, Picó A, Lampreave C, Cebollada A, Otal I, Samper S, Martín C. 2018. New insights into the transposition mechanisms of IS6110 and its dynamic distribution between Mycobacterium tuberculosis complex lineages. PLoS Genet 14:e1007282. doi: 10.1371/journal.pgen.1007282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brisson-Noël A, Aznar C, Chureau C, Nguyen S, Pierre C, Bartoli M, Bonete R, Pialoux G, Gicquel B, Garrigue G. 1991. Diagnosis of tuberculosis by DNA amplification in clinical practice evaluation. Lancet 338:364–366. doi: 10.1016/0140-6736(91)90492-8. [DOI] [PubMed] [Google Scholar]
- 11.Mendiola MV, Martin C, Otal I, Gicquel B. 1992. Analysis of the regions responsible for IS6110 RFLP in a single Mycobacterium tuberculosis strain. Res Microbiol 143:767–772. doi: 10.1016/0923-2508(92)90104-v. [DOI] [PubMed] [Google Scholar]
- 12.Van Embden JDA, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM, Small PM. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alonso H, Samper S, Martín C, Otal I. 2013. Mapping IS6110 in high-copy number Mycobacterium tuberculosis strains shows specific insertion points in the Beijing genotype. BMC Genomics 14:422. doi: 10.1186/1471-2164-14-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durrant MG, Li MM, Siranosian BA, Montgomery SB, Bhatt AS. 2020. A bioinformatic analysis of integrative mobile genetic elements highlights their role in bacterial adaptation. Cell Host Microbe 27:140–153.e9. doi: 10.1016/j.chom.2019.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemaitre N, Sougakoff W, Truffot-Pernot C, Jarlier V. 1999. Characterization of new mutations in pyrazinamide-resistant strains of Mycobacterium tuberculosis and identification of conserved regions important for the catalytic activity of the pyrazinamidase PncA. Antimicrob Agents Chemother 43:1761–1763. doi: 10.1128/AAC.43.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartkoorn RC, Sala C, Neres J, Pojer F, Magnet S, Mukherjee R, Uplekar S, Boy-Röttger S, Altmann KH, Cole ST. 2012. Towards a new tuberculosis drug: pyridomycin—nature’s isoniazid. EMBO Mol Med 4:1032–1042. doi: 10.1002/emmm.201201689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartkoorn RC, Pojer F, Read JA, Gingell H, Neres J, Horlacher OP, Altmann KH, Cole ST. 2014. Pyridomycin bridges the NADH- and substrate-binding pockets of the enoyl reductase InhA. Nat Chem Biol 10:96–98. doi: 10.1038/nchembio.1405. [DOI] [PubMed] [Google Scholar]
- 18.Hartkoorn RC, Uplekar S, Cole ST. 2014. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:2979–2981. doi: 10.1128/AAC.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J, Wang B, Hu M, Huo F, Guo S, Jing W, Nuermberger E, Lu Y. 2017. Primary clofazimine and bedaquiline resistance among isolates from patients with multidrug-resistant tuberculosis. Antimicrob Agents Chemother 61:e00239-17. doi: 10.1128/AAC.00239-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minato Y, Thiede JM, Kordus SL, McKlveen EJ, Turman BJ, Baughn AD. 2015. Mycobacterium tuberculosis folate metabolism and the mechanistic basis for para-aminosalicylic acid susceptibility and resistance. Antimicrob Agents Chemother 59:5097–5106. doi: 10.1128/AAC.00647-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin JE, Gawronski JD, DeJesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinich A, Zaveri A, DeJesus MA, Flores-Bautista E, Smith CM, Sassetti FCM, Rock CJM, Ehrt S, Schnappinger D, Ioerger GTR, Rhee K. 2021. The Mycobacterium tuberculosis transposon sequencing database (MtbTnDB): a large-scale guide to genetic conditional essentiality. bioRxiv. doi: 10.1101/2021.03.05.434127. [DOI]
- 23.Kling A, Lukat P, Almeida DV, Bauer A, Fontaine E, Sordello S, Zaburannyi N, Herrmann J, Wenzel SC, Konig C, Ammerman NC, Barrio MB, Borchers K, Bordon-Pallier F, Bronstrup M, Courtemanche G, Gerlitz M, Geslin M, Hammann P, Heinz DW, Hoffmann H, Klieber S, Kohlmann M, Kurz M, Lair C, Matter H, Nuermberger E, Tyagi S, Fraisse L, Grosset JH, Lagrange S, Muller R. 2015. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science 348:1106–1112. doi: 10.1126/science.aaa4690. [DOI] [PubMed] [Google Scholar]
- 24.Roychowdhury T, Mandal S, Bhattacharya A. 2015. Analysis of IS6110 insertion sites provide a glimpse into genome evolution of Mycobacterium tuberculosis. Sci Rep 5:12567. doi: 10.1038/srep12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon SV, Heym B, Parkhill J, Barrell B, Cole ST. 1999. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology 145:881–892. doi: 10.1099/13500872-145-4-881. [DOI] [PubMed] [Google Scholar]
- 26.Torrea G, Coeck N, Desmaretz C, Van De Parre T, Van Poucke T, Lounis N, de Jong BC, Rigouts L. 2015. Bedaquiline susceptibility testing of Mycobacterium tuberculosis in an automated liquid culture system. J Antimicrob Chemother 70:2300–2305. doi: 10.1093/jac/dkv117. [DOI] [PubMed] [Google Scholar]
- 27.Pym AS, Saint-Joanis B, Cole ST. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect Immun 70:4955–4960. doi: 10.1128/IAI.70.9.4955-4960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawkey J, Hamidian M, Wick RR, Edwards DJ, Billman-Jacobe H, Hall RM, Holt KE. 2015. ISMapper: identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genomics 16:667. doi: 10.1186/s12864-015-1860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Treepong P, Guyeux C, Meunier A, Couchoud C, Hocquet D, Valot B. 2018. PanISa: ab initio detection of insertion sequences in bacterial genomes from short read sequence data. Bioinformatics 34:3795–3800. doi: 10.1093/bioinformatics/bty479. [DOI] [PubMed] [Google Scholar]
- 30.Kienle M, Eisenring P, Stoessel B, Horlacher OP, Hasler S, Van Colen G, Hartkoorn RC, Vocat A, Cole ST, Altmann KH. 2020. Synthesis and structure-activity relationship studies of C2-modified analogs of the antimycobacterial natural product pyridomycin. J Med Chem 63:1105–1131. doi: 10.1021/acs.jmedchem.9b01457. [DOI] [PubMed] [Google Scholar]
- 31.Horlacher OP, Hartkoorn RC, Cole ST, Altmann K-H. 2013. Synthesis and antimycobacterial activity of 2,1′-dihydropyridomycins. ACS Med Chem Lett 4:264–268. doi: 10.1021/ml300385q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, Viveiros M, Portugal I, Pain A, Martin N, Clark TG. 2014. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng 9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 38.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat İ, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, SciPy 1.0 Contributors. 2020. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download Spectrum.00019-21-sf0001.pdf, PDF file, 2.3 MB (2.3MB, pdf)
Supplemental material. Download Spectrum.00019-21-sf0002.xlsx, XLSX file, 0.2 MB (251.8KB, xlsx)
Data Availability Statement
Whole-genome sequencing reads for the OH190-resistant M. tuberculosis H37Rv isolates (fastq format) have been deposited in the NCBI database (BioProject accession number PRJNA739842).