Abstract
Lung microbiota (LM) is an interesting new way to consider and redesign pathogenesis and possible therapeutic approach to many lung diseases, such as idiopathic pulmonary fibrosis (IPF), which is an interstitial pneumonia with bad prognosis. Chronic inflammation is the basis but probably not the only cause of lung fibrosis and although the risk factors are not completely clear, endogenous factors (e.g. gastroesophageal reflux) and environmental factors like cigarette smoking, industrial dusts, and precisely microbial agents could contribute to the IPF development. It is well demonstrated that many bacteria can cause epithelial cell injuries in the airways through induction of a host immune response or by activating flogosis mediators following a chronic, low-level antigenic stimulus. This persistent host response could influence fibroblast responsiveness suggesting that LM may play a role in repetitive alveolar injury in IPF. We reviewed literature regarding not only bacteria but also the role of virome and mycobiome in IPF. In fact, some viruses such as hepatitis C virus or certain fungi could be etiological agents or co-factors in the IPF progress. We aim to illustrate how the cross-talk between different local microbiotas throughout specific axis and immune modulation governed by microorganisms could be at the basis of lung dysfunctions and IPF development. Finally, since the future direction of medicine will be personalized, we suggest that the analysis of LM could be a goal to research new therapies also in IPF.
Keywords: Idiopathic pulmonary fibrosis, Lung microbiota, Immune Response
Idiopathic pulmonary fibrosis: a complex world between immune dysregulation and chronic inflammation
Idiopathic pulmonary fibrosis (IPF) is an heterogeneous interstitial pneumonia of unknown cause with a highly variable evolution and response to therapies, which complicate patient management and prognostic evaluation (1). IPF primarily occurs in older adults between 50 and 80 years, predominantly males; throughout Europe and North America, the estimated IPF incidence has been reported to range between 2.8 and 19 cases per 100 000 people per year (2,3). The highest rates of IPF in Europe are reported in the UK, with incidence rates between 4.6 and 8.65 per 100 000 people per year, and 6000 people diagnosed annually (3). Even if chronic inflammation is commonly believed to be the pathological basis of fibrosis (such happens in other diseases), the etiology of Idiopathic pulmonary fibrosis remain unknown (4). IPF has a poor prognosis yet and a mortality rate higher than that of numerous cancers with an estimated 5-year survival rate less than 30% (5). Commonly approved guidelines recommend multidisciplinary discussion as the diagnostic gold standard (6). IPF high-resolution thin section computed tomographic (HRCT) scan images is characterized by an usual interstitial pneumonia, including honeycombing pattern localized especially in lower lobes and subpleural regions (7,8). When clinical and/or radiologic evaluation is nondiagnostic, histopathologic samples has the major weight on the final diagnosis (9). To obtain adequate lung specimens, surgical lung biopsy (10) or a less invasive bronchoscopic lung cryobiopsy can be used (11). Aging, the major risk factor, can be defined as “a degenerative process due to accumulation of extrinsic and intrinsic damages that results in cellular dysfunction, altered tissue response and finally death” (12), and there are interesting data on how the aging biology may influence the susceptibility to lung fibrosis (13). Several studies have shown a close correlation between IPF and different risk factors such as gastroesophageal reflux (GER), cigarette smoking, some viral infections, environmental factors and occupational exposure (14–17). In addition genetic can contribute to the risk of developing familiar IPF (especially with surfactant protein C mutation) (18,19). Although more than a few limitations, bleomycin-induced lung fibrosis is a widely utilized animal model of Idiopathic pulmonary fibrosis (20). Various proinflammatory immune cells, such as macrophages, neutrophils, and T helper 17 (Th17) cells, have been reported to play important roles in pulmonary fibrosis (4) and histological analysis documented: i) myofibroblast foci located in the fibrotic areas, ii) increased hyperplastic type II alveolar epithelial cells, and ii) the reduction of type I alveolar epithelial cells (21). As it happens in other organs, the main effector cells for fibrosis are the mesenchymal cells, which is identified in three distinct but interrelated cell types: fibroblast, myofibroblast, and smooth muscle cell (22).
The inflammation seems have as a crucial role in IPF being responsible for the lung fibrosis evolution; in particular the function of macrophages, the loss of T-cell and B-cell tolerance leading auto-immune responses, and the interaction of immune cells with myofibroblasts could be key mechanisms in IPF (23). Many authors reported that repetitive damage to the alveolar epithelium can disrupts the balance between injury and repair mechanisms resulting in exacerbated inflammatory response and profibrotic signaling (1,6,24). Intriguingly, recent studies found in mitochondria dysfunction and metabolic reprogramming a crucial way in lung fibrosis evolution; mitochondria could promote low resilience and increase susceptibility to activation profibrotic responses in the lungs (25). To date, although numerous clinical studies are ongoing, two anti-fibrotic therapies are available to slow down the disease evolution: Nintedanib, a tyrosine kinase inhibitor and Pirfenidone, a pyridine whit anti-inflammatory, anti-oxidant, and anti-fibrotic actions (26,27). A promising approach for developing new therapies could be researching new biomarkers and molecular endotypes to personalized treatment (5,28).
The human lung microbiome: a world in continuous discovery
From birth humans have a deep relationship with microorganisms and their products due to the co-evolution in the environment and each body district has a unique set of microorganisms in its microbiota (29). The microbiota of one habitat at a specific time consists of bacteria, viruses, fungi, and protozoans living and sharing our body in commensal, symbiotic, and pathogenic conditions (30). We are more bacteria than human cells and our genome is associated with ‘another genome’: the microbiome. Differences between the term microbiota and microbiome and other useful definitions are explained in Table 1 (31). The knowledge that each human body district hosts its own microbiota comes from “The Human Microbiome Project” where analyzed more than 11000 primary specimens from 18 body sites in 300 healthy volunteers (32). The most studied human microbiota is the intestinal microbiota that is the most densely populated by microbial communities and plays some important metabolic and protective roles in human health. The gut microbiota (GM) contains predominantly obligate anaerobes from the phyla Actinobacteria, Verrucomicrobia, Firmicutes, Fusobacteria, Proteobacteria,and Bacteroidetes (33). But gut isn’t the unique organ harboring a own microbiota, and from few years we know the existence of others microbiotas (34–38). During normal breathing respiratory tract is a site of microbial exchanges with the environment and it is believed that between 1,500 and 14,000 microbes are inhaled each hour; the dynamic composition of lung microbiota (LM) can reflect relative immigration, elimination through mechanisms such as coughing and microbial growth (39). So, the knowledge of its composition seems to be important both in lung health and in the development of respiratory diseases (40). Like a tide pool is subject to immigration from the ocean’s fauna, the lungs are subject to continuous microorganism’s immigration from the oropharynx (41). In healthy conditions, lungs are relatively low-nutrient environment for microbes (42): the epithelial layers are coated with a thin mucosa with only <100 mL of mucus produced per day and rich in surfactant to prevent alveolar collapse (43). Therefore, as estimated by culture methods the mouse LM mice shows a low density (about 10^3–10^5 CFU/g of lung tissue) (44) while it is assessed that human lungs harbor about 2.2 x 10^3 bacterial genomes per cm^2 (45)and that microbial population of the lung is comparable to that of the duodenum (46). Firmicutes, Bacteriodetes, Proteobacteria, Fusobacteria and Actinobacteria are generally the five major phyla in healthy lung and the preservation of a small bacterial community seems to be an health hallmark (47,48). Lung epithelial cells can interact with microbes through various innate sensors on their membranes and in their cytoplasm like toll-like receptors and NOD-like receptors (49) activating molecular cascades and triggering the induction of tolerance or inflammation (50). Differences in lung environments like temperature, pressure, mucus and surfactant presence may have an impact on the installation and location of bacterial communities, particularly if they advantage certain bacteria being selected in lung diseases (Figure 1)(51).
Table 1.
Most common definitions
| Microbiota | the microbes (including bacteria, viruses, fungi, and protozoans) in a given population (103) |
| Microbiome | microbiota found in a particular habitat at a specific time and consists of the ecological community of commensal, symbiotic, and pathogenic microorganisms that share our body(103); “microbiome” and “microbiota” are often used interchangeably but they refer to different ecological principles. |
| Microbial community | microbes that interact functionally and metabolically (40) |
| Holobiont | the host organism together with its microbiome constitutes the “holobiont” (Greek, holos, whole/entire) and the totality of the genome is the “hologenome” (104) |
| 16S ribosomal RNA gene | Characterization of the microbiome can be done by sequencing regions of a conserved gene, such as the hypervariable regions of the 16S ribosomal RNA gene (105). The gene comprises nine constant regions (C) and nine hypervariable regions (V1–V9). The variable regions enable sequence-specific discrimination between different bacteria. (106). 16S ribosomal RNA gene sequencing has been described to identify bacterial DNA in 95.7% of bronchoalveolar lavage (BAL) specimens compared with conventional culture techniques,which detected bacteria in 39.1% of BAL samples (107). |
| OTU | Operational taxonomic units: is an operational definition used to classify groups of closely related individuals |
Figure 1.
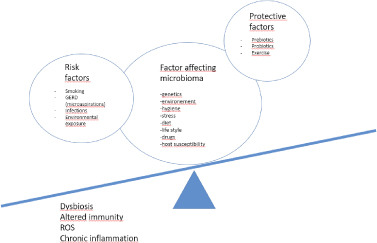
Microbiota imbalance in lung disease. When factors inducing lung dysbiosis have more weight compared to protective factors, can induce disease state. In detail, ROS activation and chronic inflammation could have a crucial role in the pathogenesis of IPF.
All microorganisms of different organs not only can influence “their” sites (or have systemic influences modulating immune responses), but are not close that we think; in fact there is a clear cross-talk between the gut and the lungs, also called gut–lung axis, that is vital for maintaining homeostasis and educating the host immune system (Figure 2) (52).
Figure 2.
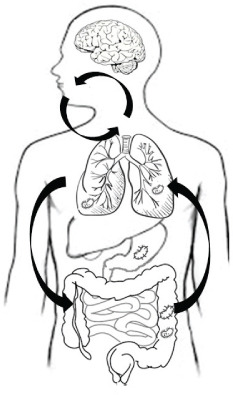
Schematic representation of gut-lung microbiome interactions. Bidirectional Gut-Lung axis: production of bacterial metabolites and immunity modulation influences lung microbiome. (A) Bacterial migration via inhalations. (B) Bacterial elimination. (C) Gastrointestinal-lung microaspirations. (D) Diet influence bacterial reproduction in the gut.
Gut dysbiosis can be associated with lung disease or infections (53). This strength correlation between gut and lung could be also responsible for perpetuating inflammatory damage. When intestinal dysbiosis occurs, for example, during infection or antibiotic use, the microbiota-derived signals are altered too, leading to changes in the immune response against pathogens. A similar situation could happen in the lung with a modulation of the microbiota composition, which, in turn, could induce an altered immune response against pathogens. The existence of a gut–lung axis could perpetuates this phenomena establishing a vicious circle (54).
LM is altered in numerous respiratory disorders such as obstructive airway diseases, infections, interstitial lung disease and lung cancer (55). In Chronic Obstructive Pulmonary Disease (COPD) patients bacterial colonization correlates with the severity of inflammatory response, radiological changes, pathological variations of the local immune response, and increased daily symptoms (56). In asthma there is an interesting correlation between a lower numbers of some lung bacteria genera such as Bifdobacteria, Akkermansia and Faecalibacterium, and higher risk of developing atopy and/or early life asthma (57). Studies in patients with bronchiectasis, showed that lung bacterial communities are dominated by Pseudomonas, Haemophilus and Streptococcus, while exhibiting intraindividual stability and large interindividual variability (58). Pseudomonas- and Haemophilus-dominated LM have been shown to be linked to severity of bronchiectasis and the frequent exacerbations (58).
In addition, many data are available about the role of LM in carcinogenesis of lung cancers (59–63).
Notably, in addition to LM, also the gut microbiota seems linked with lung disease as recently demonstrated on cystic fibrosis patients where gastrointestinal tract dysbiosis is associated with pulmonary outcomes (64).
Finally, the majority of our knowledge about the LM impact on interstitial lung diseases mainly relates to the IPF (55).
Recent data suggest that the LM is molded in large part by silent micro-aspiration of bacteria from the oropharynx (42) (Figure 2). Interestingly although the risk factors are not completely clear, environmental factors like cigarette smoking, industrial dusts, microbial agents and endogenous factors like GER could contribute to the development to IPF (6). Increased bacterial burden in the oropharynx due to GER and successive lungs immigration via micro-aspiration, is one probable explanation for the presence of increased bacterial colonies in IPF lungs compared to controls (65). Increased micro-organisms’ colonies and/or richness of potentially pathogenic bacteria may drive disease progression, acute exacerbations and mortality (66). In addition, as numerous studies hypothesize that antibiotic administration or immunization against pathogenic organisms may improve IPF outcomes, seems interestingly evaluate the LM composition as a potentially therapeutic target (67). As other respiratory diseases, a microbial selection and a diversity loss along with dysbiosis seems to be present in IPF (68). In fact, recent LM characterization of IPF patients, showed an over-representation of specific genera such as Streptococcus, Prevotella and Staphylococcus, compared to healthy controls (66, 69) (Figure 3).
Figure 3.
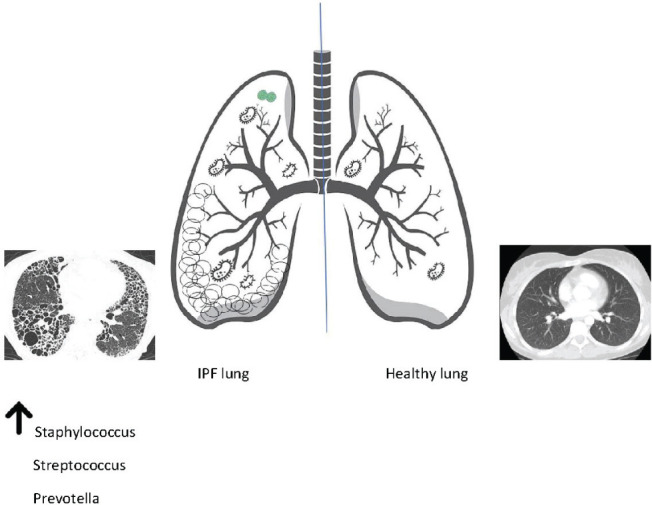
Microbial community diversity in IPF lung and in healthy lung
Could we consider microorganisms in the pathogenesis and management of IPF?
Bacteria can directly cause epithelial cell injuries in the airways or indirectly, by induction of an host immune response or activating flogosis’ mediators following a chronic, low-level antigenic stimulus (70). Bacteria can activate a persistent host response and influence fibroblast responsiveness (71) suggesting that LM may play a role in repetitive alveolar injury in IPF (72). As previously mentioned, some evidence suggest that mitochondria are crucial in maintaining the innate immune system and chronic inflammation (73), especially in chronic disease such as COPD or lung cancer (74). The intriguingly relationship between mitochondria and microbiota is strengthened by the probable prokaryotic origin of mitochondria and some studies demonstrated that the latter targets mitochondria by modulating the Reactive Oxygen Species (ROS) production and the mitochondrial activity through interactions with toxins, proteins or other metabolites released by gut microbiota (75). In addition, the cross-talk between gut and lung microbiota (the named gut-lung axis (76,77)) and mitochondria could modulate the increased systemic ROS level that leads to the proliferation of stem cells in target organs like the lungs followed by cell differentiation (75).
Interestingly, O’Dwyer and coll. (78) validated the finding that lung bacterial burden predicts disease progression in IPF patients while different LM strains correlate with increased alveolar profibrotic cytokines, contributing to lung inflammation. Knippenberg et all (79), in two mouse models, demonstrated that Streptococcus pneumoniae producing pneumolysin, a toxin which mediates fibrotic progression via injury of the alveolar epithelium, triggered the progression of pulmonary fibrosis. Interestingly, antibiotic treatment stopped infection and induced fibrosis progression. Evaluating the LM in 17 IPF patients (80), a recent study found in bronchoalveolar lavage (BAL) samples, bacteria often present in the oropharynx, as well as uncultured bacterial sequences corresponding to the Neisseria, Streptococcus and Actinobacterium sp. genera. Takahashi and coll. (81) documented in the BAL of thirty-four IPF patients a loss of diversity of the lung microbiota correlated with the progression of IPF. Increase in Firmicutes and Bacteroidetes phyla and decrease in the phylum Proteobacteria were involved in the reduction of diversity and consequent lung dysbiosis. The latter correlated with IPF negative prognostic factors, including decreased 6 minutes walking test (6MWT), low forced lung capacity (FVC) and high serum surfactant protein D (SP-D) and LDH. In addition, the same authors observed that the increased Streptococcaceae correlated with a reduction in 6MWT suggesting that LM and lung fibrosis progression could be closely related and potentially involved in the IPF pathogenesis (81). The COMET-IPF study revealed that Prevotella, Veillonella, and Cronobacter species (spp.) were the most prevalent and abundant bacteria across IPF patients. After adjusting for age, sex, smoking status, GER, baseline pulmonary function and 6MWT desaturation status, samples enriched by Streptococcus or Staphylococcus were associated with a clinically significant reduction in progression-free survival time (82). These data provide additional support to the hypothesis that at least microorganisms could be potential biomarkers of disease progression and may be involved in disease pathogenesis (82). If LM dysbiosis can drive disease progression is a hypothesis that merits future research (69). To that proposal, interestingly Molyneaux and coll. demonstrated in BAL IPF samples, that increased bacterial load at the time of diagnosis identified patients with more rapidly progressive fibrosis and a higher risk of mortality (65). However, they did not find a specific correlation with specific bacteria genera. In addition, the authors compared LM in IPF and chronic obstructive pulmonary disease (COPD) patients, with healthy subjects. IPF patients had significant higher bacterial burden compared to COPD and healthy controls (65). IPF patients were more likely to harbor potentially pathogenic bacteria such as Haemophilus, Neisseria and Streptococcus spp. compared to controls (65). Therefore, in agreement with the authors, an early LM characterization could be helpful to stratify IPF patients according to bacterial burden to predict the mortality risk.
Since specific viruses enhance fibrosis in animal models (69), some studies investigated the role of hepatitis C virus as etiologic agent or co-factor in the IPF development (83). Despite high prevalence found in independent studies, the association of HCV with IPF has not been constantly observed (84). Others IPF studies were performed on human herpes viruses including herpes simplex virus, cytomegalovirus, Epstein-Barr virus and human herpes virus-7 and -8 (85,86). Surprisingly herpes viruses have been identified in the lung tissue of a greater proportion of IPF patients as compared to controls (86). These data may be confounded by immunosuppressive drugs used in IPF patients but could suggest a role of viruses as co-factors driving fibrosis progression. In addition, other authors have evaluated the human herpesvirus infection in IPF acute exacerbations founding in the airways an epithelial damage that could be linked to a viral–mediated pathogenesis of acute exacerbations (85). Seems possible that viruses could reprogram epithelial and mesenchymal cells and may drive fibrotic processes over time (85). However, there are only few studies on the relationship between human virome and IPF.
Finally, regarding mycobiome in IPF only few data are available in the literature. Molyneaux and coll. (87) found in a small cohort of patients that mycobiome in IPF was dominated by Candida species despite the presence of other respiratory pathogens including Aspergillus, Cryptococcus, Malassezia and Exophiala species more abundant in IPF patients rather than in healthy subjects (87). Furthermore, the presence of certain fungi could interact with the bacterial pool. Interestingly, bacterial DNA was not detected most of patients colonized with Pneumocystis jirovecii, suggesting this fungus could impair lungs bacterial colonization (80). Different authors (65,88,89) are interested to better understand if an altered lung microbiota is the cause or result of destruction of the normal lung structure and induce fibrosis.
However, despite growing evidence of association, the cause-effect correlation between lung dysbiosis and IPF evolution remains elusive and more and detailed studies are required.
Could be the microbiota modulation a potential therapeutic target in IPF exacerbations?
The progression of IPF is correlated with frequencies and severity of exacerbations. Acute exacerbation of IPF (AE-IPF) was first defined by Kondo and colleagues as “an acute, clinically significant respiratory worsening of unidentifiable cause in a patient with underlying IPF” (90). AE-IPF occurs in about 10% of patients per year, and is associated with a particularly poor prognosis (91). Among patients with acute exacerbations, non-survivors had shorter durations of dyspnea, higher flogosis indexes such as C reactive protein levels, lower arterial oxygen tension /inspiratory oxygen fraction ratios (P/F), higher percentages of neutrophils and lower percentages of lymphocytes in BAL compared with survivors (92). Only C reactive protein was found to be an independent predictor of survival, suggesting that infection and/or inflammation could be a pathogenic mechanisms contributing to AE-IPF (91).
American, European and Asian guidelines suggest that AE-IPF should be treated with corticosteroids’ therapy, including pulse administrations although the latter may often be complicated by opportunistic infections such as pneumocystis pneumonia or viral infections (6, 91). Undoubtedly, the corticosteroids immunomodulatory and anti-inflammatory role could modulate gut and lung microbiota composition, and this must be considered. Others important findings about the use of prednisone, azathioprine, and N-acetylcysteine was elucidated in PANTHER-IPF study (93); IPF patients treated with a combination of those drugs showed an increased risk of death and hospitalization as compared with placebo. This may suggests that immunosuppression is associated with an increased rate of acute exacerbations (93).
Despite few data on the role of bacterial infection in IPF patients, recent studies suggest the importance of infections in the development of AE-IPF(69). Anti-microbial therapy include antibiotics to reduce bacterial burden or target pathogenic identified organisms, vaccination to reduce the risk of infection with specific pathogens, or interventions aimed to reduce immigration of bacteria to the airways (i.e. reducing aspiration from the oropharynx) (67). Fastrès and coll. (94) demonstrated that a high bacterial burden at the time of IPF diagnosis seem to be a biomarker for a more-rapidly progressive disease with an increased mortality risk. Despite evidence supporting an infectious hypothesis as a trigger of AE-IPF (95), the use of antibiotics in stable IPF and AE-IPF remains controversial. For example, Oda and coll. (96) suggested that IPF patients with AE-IPF and rapid development of respiratory failure treated with cotrimoxazole and macrolides were significantly associated with a good prognosis expecially when administered in combination with high-dose corticosteroids. On the other hand a randomized controlled trial by Sulgina and coll. in 181 patients with interstitial pneumonia who receives cotrimoxazole 960 mg twice daily or placebo showed that the addition of cotrimoxazole therapy to standard treatment had no effect on lung function but resulted in improved quality of life and a reduction in mortality (97). Lung dysbiosis and the resulting dysregulated local and systemic immune response seem to be new and promising search fields; recently was published the study CleanUP-IPF, a clinical trial where will be randomized approximately 500 IPF participants. Patients will be treated with antimicrobial treatment strategy (trimethoprim 160 mg/ sulfamethoxazole 800 mg twice a day plus folic acid 5 mg daily or doxycycline 100 mg once daily if body weight is < 50 kg or 100 mg twice daily if ≥50 kg) and blood, oral and fecal samples for DNA sequencing and genome wide transcriptomics will be collected (98).
Finally, as mentioned above, many authors claim a correlation between GER and IPF. The intimate relationship between the bacterial flora of the upper digestive tract and the airways has also been demonstrated in IPF. Molyneaux and coll (99) enrolled twenty AE-IPF patients and 15 stable IPF patients as a control and BAL was performed comparing microbial composition. AE-IPF patients showed a notable microbiota change, with an increase in two potentially pathogenic Proteobacterial; Campylobacter sp. and Stenotrophomonas sp., coupled with a significant decrease in Veillonella sp. The apparent translocation of bacteria usually confined to the gastrointestinal tract suggests a role for aspiration in the development of IPF acute exacerbations(99). From this evidence, modulating digestive bacterial flora would seem possible to influence the lung microbiota.
Conclusion
To date although the management of a complex disease like IPF is personalized using precision medicine (5), there are some convincing evidence of the potential role of the lung microbiota influencing natural history of this disease. The IPF genesis is multifactorial and crucial are environment factors (24). A potential role for infections, both as a cofactor of initial development and/ or of fibrosis progression, has been widely postulated (65). IPF is often treated with immunosuppressive therapy and this may significantly modulate lung microbiota. On the one hand, this makes patients more susceptible to the development of bacterial infections. On the other hand, the reduced immune reserve can alter as the composition as the functionality of the microbiota (100). Retrospective data suggest that IPF patients receiving invasive ventilation and corticosteroids in addition to co-trimoxazole or a macrolide antibiotic have a better prognosis compared to not receiving these drugs (96). The microbiota modulation could influence immune system, systemic inflammation, ROS productions and consequently activation pathways like organ fibrosis. The cross talk between different microbiotas (especially gut and lung) throughout specific axis could be at the basis of organ dysfunctions and so promoting many diseases. The future medicine will be personalized, and the quality, quantity and diversity of microbiota could be a goal to research new therapies (101,102). Based on this consideration, we could better understand the pathogenesis and the evolution of some idiopathic diseases such as IPF and preventive or therapeutic strategies to shaping the microbiota (e.g prebiotics and/or probiotics, diet and lifestyle modification, fecal transplantation) could be administrated to patients. Despite much evidence from described studies, how the altered lung microbiota can contribute to IPF pathogenesis is not well understand and more specific studies are required.
Supported by:
the study was founded by University of Florence Amedeiex60%
Conflict of interest:
the authors declare no confict of interest related to the present manuscript.
References
- 1.Richeldi L, Collard HR, Jones MG. Idiopathic pulmonary fibrosis. Lancet. 2017;389(10082):1941–52. doi: 10.1016/S0140-6736(17)30866-8. [DOI] [PubMed] [Google Scholar]
- 2.Richeldi L, Rubin AS, Avdeev S, Udwadia ZF, Xu ZJ. Idiopathic pulmonary fibrosis in BRIC countries: the cases of Brazil, Russia, India, and China. BMC Med. September 2015;13:237. doi: 10.1186/s12916-015-0495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duchemann B, Annesi-Maesano I, Jacobe de Naurois C, et al. Prevalence and incidence of interstitial lung diseases in a multi-ethnic county of Greater Paris. Eur Respir J. Augustus 2017;50(2):1602419. doi: 10.1183/13993003.02419-2016. [DOI] [PubMed] [Google Scholar]
- 4.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18(7):1028–40. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magnini D, Montemurro G, Iovene B, et al. Idiopathic Pulmonary Fibrosis: Molecular Endotypes of Fibrosis Stratifying Existing and Emerging Therapies. Respiration. 2017 doi: 10.1159/000475780. [DOI] [PubMed] [Google Scholar]
- 6.Raghu G, Collard HR, Egan JJ, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. Maart 2011;183(6):788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz DA. Idiopathic Pulmonary Fibrosis Is a Genetic Disease Involving Mucus and the Peripheral Airways. Ann Am Thorac Soc. November 2018;15(Supplement_3):S192–7. doi: 10.1513/AnnalsATS.201802-144AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Invernizzi R, Wu BG, Barnett J, et al. The Respiratory Microbiome in Chronic Hypersensitivity Pneumonitis Is Distinct from That of Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. Julie 2020;203(3):339–47. doi: 10.1164/rccm.202002-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monaghan H, Wells AU, Colby T V, du Bois RM, Hansell DM, Nicholson AG. Prognostic Implications of Histologic Patterns in Multiple Surgical Lung Biopsies From Patients With Idiopathic Interstitial Pneumonias. Chest. Februarie 2004;125(2):522–6. doi: 10.1378/chest.125.2.522. [DOI] [PubMed] [Google Scholar]
- 10.Cottin V. Lung biopsy in interstitial lung disease: balancing the risk of surgery and diagnostic uncertainty. Eur Respir J. November 2016;48(5):1274 LP–1277. doi: 10.1183/13993003.01633-2016. [DOI] [PubMed] [Google Scholar]
- 11.Tomassetti S, Wells AU, Costabel U, et al. Bronchoscopic Lung Cryobiopsy Increases Diagnostic Confidence in the Multidisciplinary Diagnosis of Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. November 2015;193(7):745–52. doi: 10.1164/rccm.201504-0711OC. [DOI] [PubMed] [Google Scholar]
- 12.Phillip JM, Aifuwa I, Walston J, Wirtz D. The Mechanobiology of Aging. Annu Rev Biomed Eng. Desember 2015;17(1):113–41. doi: 10.1146/annurev-bioeng-071114-040829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulati S, Thannickal VJ. The Aging Lung and Idiopathic Pulmonary Fibrosis. Am J Med Sci. Mei 2019;357(5):384–9. doi: 10.1016/j.amjms.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Lee JS. The Role of Gastroesophageal Reflux and Microaspiration in Idiopathic Pulmonary Fibrosis. Clin Pulm Med. Maart 2014;21(2):81–5. doi: 10.1097/cpm.0000000000000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell PD, Das JP, Murphy DJ, et al. Idiopathic Pulmonary Fibrosis With Emphysema: Evidence of Synergy Among Emphysema and Idiopathic Pulmonary Fibrosis in Smokers. Respir Care. Februarie 2015;60(2):259 LP–268. doi: 10.4187/respcare.03389. [DOI] [PubMed] [Google Scholar]
- 16.Harari S, Caminati A. IPF: new insight on pathogenesis and treatment. Allergy. Mei 2010;65(5):537–53. doi: 10.1111/j.1398-9995.2009.02305.x. [DOI] [PubMed] [Google Scholar]
- 17.Tanjore H, Blackwell TS, Lawson WE. Emerging evidence for endoplasmic reticulum stress in the pathogenesis of idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012/01/27. April 2012;302(8):L721–9. doi: 10.1152/ajplung.00410.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaur A, Mathai SK, Schwartz DA. Genetics in Idiopathic Pulmonary Fibrosis Pathogenesis, Prognosis, and Treatment. Front Med. September 2017;4:154. doi: 10.3389/fmed.2017.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogee LM, Dunbar AE, Wert SE, Askin F, Hamvas A, Whitsett JA. A Mutation in the Surfactant Protein C Gene Associated with Familial Interstitial Lung Disease. N Engl J Med. Februarie 2001;344(8):573–9. doi: 10.1056/NEJM200102223440805. [DOI] [PubMed] [Google Scholar]
- 20.Moeller A, Ask K, Warburton D, Gauldie J, Kolb M. The bleomycin animal model: a useful tool to investigate treatment options for idiopathic pulmonary fibrosis? Int J Biochem Cell Biol. 2007/08/30. 2008;40(3):362–82. doi: 10.1016/j.biocel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mora AL, Rojas M, Pardo A, Selman M. Erratum: Emerging therapies for idiopathic pulmonary fibrosis, a progressive age-related disease. Nat Rev Drug Discov. 2017;16(11):810. doi: 10.1038/nrd.2017.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman SL, Sheppard D, Duffield JS, Violette S. Therapy for Fibrotic Diseases: Nearing the Starting Line. Sci Transl Med. Januarie 2013;5(167):167sr1 LP–167sr1. doi: 10.1126/scitranslmed.3004700. [DOI] [PubMed] [Google Scholar]
- 23.Heukels P, Moor CC, von der Thüsen JH, Wijsenbeek MS, Kool M. Inflammation and immunity in IPF pathogenesis and treatment. Respir Med. Februarie 2019;147:79–91. doi: 10.1016/j.rmed.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 24.Maher TM, Wells AU, Laurent GJ. Idiopathic pulmonary fibrosis: multiple causes and multiple mechanisms? Eur Respir J. November 2007;30(5):835 LP–839. doi: 10.1183/09031936.00069307. [DOI] [PubMed] [Google Scholar]
- 25.Bueno M, Calyeca J, Rojas M, Mora AL. Mitochondria dysfunction and metabolic reprogramming as drivers of idiopathic pulmonary fibrosis. Redox Biol. 2020;(February):101509. doi: 10.1016/j.redox.2020.101509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noble PW, Albera C, Bradford WZ, et al. Pirfenidone in patients with idiopathic pulmonary fibrosis (CAPACITY): two randomised trials. Lancet. Mei. 2011;377(9779):1760–9. doi: 10.1016/S0140-6736(11)60405-4. [DOI] [PubMed] [Google Scholar]
- 27.Richeldi L, Costabel U, Selman M, et al. Efficacy of a Tyrosine Kinase Inhibitor in Idiopathic Pulmonary Fibrosis. N Engl J Med. September 2011;365(12):1079–87. doi: 10.1056/NEJMoa1103690. [DOI] [PubMed] [Google Scholar]
- 28.Inoue Y, Kaner RJ, Guiot J, et al. Diagnostic and Prognostic Biomarkers for Chronic Fibrosing Interstitial Lung Diseases with a Progressive Phenotype. Chest. 2020 doi: 10.1016/j.chest.2020.03.037. [DOI] [PubMed] [Google Scholar]
- 29.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009/11/05. Desember 2009;326(5960):1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao L. The tale of our other genome. Nature. Junie 2010;465:879. doi: 10.1038/465879a. [DOI] [PubMed] [Google Scholar]
- 31.Lederberg BJ, McCray AT. ’ Ome Sweet ’ Omics-A Genealogical Treasury of Words. Sci. 2001;15(7):8. [Google Scholar]
- 32.Proctor LM. The human microbiome project in 2011 and beyond. Cell Host Microbe. 2011;10(4):287–91. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011/04/20. Mei 2011;473(7346):174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The Placenta Harbors a Unique Microbiome. Sci Transl Med. Mei 2014;6(237):237ra65–237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearce MM, Hilt EE, Rosenfeld AB, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio. Julie 2014;5(4):e01283. doi: 10.1128/mBio.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. The Human Nasal Microbiota and Staphylococcus aureus Carriage. PLoS One. Mei 2010;5(5):e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grice EA, Kong HH, Renaud G, et al. A diversity profile of the human skin microbiota. Genome Res. Julie 2008;18(7):1043–50. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Traykova D, Schneider B, Chojkier M, Buck M. Blood Microbiome Quantity and the Hyperdynamic Circulation in Decompensated Cirrhotic Patients. PLoS One. Februarie 2017;12(2):e0169310–e0169310. doi: 10.1371/journal.pone.0169310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The Microbiome and the Respiratory Tract. Annu Rev Physiol. Februarie 2016;78(1):481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui L, Morris A, Huang L, et al. The microbiome and the lung. Annals of the American Thoracic Society. 2014 doi: 10.1513/AnnalsATS.201402-052PL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dickson RP, Erb-Downward JR, Huffnagle GB. Homeostasis and its Disruption in the Lung Microbiome. Am J Physiol - Lung Cell Mol Physiol. 2015 doi: 10.1152/ajplung.00279.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bassis CM, Erb-Downward JR, Dickson RP, et al. Analysis of the Upper Respiratory Tract Microbiotas as the Source of the Lung and Gastric Microbiotas in Healthy Individuals. Ravel J, redakteur. MBio. Mei 2015;6(2) doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H, Kuzmenko A, Wan S, et al. Surfactant proteins A and D inhibit the growth of Gram-negative bacteria by increasing membrane permeability. J Clin Invest. Mei 2003;111(10):1589–602. doi: 10.1172/JCI16889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remot A, Descamps D, Noordine M-L, et al. Bacteria isolated from lung modulate asthma susceptibility in mice. Isme J. Januarie 2017;11:1061. doi: 10.1038/ismej.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One. Januarie 2010;5(1):e8578–e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aron-Wisnewsky J, Doré J, Clement K. The importance of the gut microbiota after bariatric surgery. Nat Rev Gastroenterol &Amp; Hepatol. Augustus 2012;9:590. doi: 10.1038/nrgastro.2012.161. [DOI] [PubMed] [Google Scholar]
- 47.Dickson RP, Erb-Downward JR, Freeman CM, et al. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann Am Thorac Soc. Junie 2015;12(6):821–30. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segal LN, Alekseyenko A V, Clemente JC, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. Julie 2013;1(1):19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung J-W, Choi J-C, Shin J-W, et al. Lung Microbiome Analysis in Steroid-Naїve Asthma Patients by Using Whole Sputum. Tuberc Respir Dis. Julie 2016;79(3):165–78. doi: 10.4046/trd.2016.79.3.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Stefano A, Ricciardolo FLM, Caramori G, et al. Bronchial inflammation and bacterial load in stable COPD is associated with TLR4 overexpression. Eur Respir J. Mei 2017;49(5) doi: 10.1183/13993003.02006-2016. [DOI] [PubMed] [Google Scholar]
- 51.Mathieu E, Escribano-Vazquez U, Descamps D, et al. Paradigms of lung microbiota functions in health and disease, particularly, in asthma. Front Physiol. 2018;9(AUG):1–11. doi: 10.3389/fphys.2018.01168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dang AT, Marsland BJ. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019;12(4):843–50. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 53.Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol. 2019 doi: 10.1038/s41590-019-0451-9. [DOI] [PubMed] [Google Scholar]
- 54.Spagnolo P, Molyneaux PL, Bernardinello N, et al. The Role of the Lung’s Microbiome in the Pathogenesis and Progression of Idiopathic Pulmonary Fibrosis. Int J Mol Sci. November 2019;20(22):5618. doi: 10.3390/ijms20225618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fabbrizzi A, Amedei A, Lavorini F, Renda T, Fontana G. The lung microbiome: clinical and therapeutic implications. Intern Emerg Med. November 2019;14(8):1241–50. doi: 10.1007/s11739-019-02208-y. [DOI] [PubMed] [Google Scholar]
- 56.Langille MGI, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013/08/25. September 2013;31(9):814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. September 2016;22:1187. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richardson H, Dicker AJ, Barclay H, Chalmers JD. The microbiome in bronchiectasis. Eur Respir Rev. September 2019;28(153):190048. doi: 10.1183/16000617.0048-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mur LAJ, Huws SA, Cameron SJS, Lewis PD, Lewis KE. Lung cancer: A new frontier for microbiome research and clinical translation. Ecancermedicalscience. 2018;12:1–9. doi: 10.3332/ecancer.2018.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou H, Suo J, Zhu J. Therapeutic Relevance of Human Microbiota and Lung Cancer. Zhongguo Fei Ai Za Zhi. Julie 2019;22:464–9. doi: 10.3779/j.issn.1009-3419.2019.07.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagano T, Otoshi T, Hazama D, et al. Novel cancer therapy targeting microbiome. Onco Targets Ther. Mei 2019;12:3619–24. doi: 10.2147/OTT.S207546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahn HJ, Lee DS. Helicobacter pylori in gastric carcinogenesis. World J Gastrointest Oncol. 2015/12/15. Desember 2015;7(12):455–65. doi: 10.4251/wjgo.v7.i12.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng Y, Fang Z, Xue Y, et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes. April 2020:1–13. doi: 10.1080/19490976.2020.1737487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Budden KF, Shukla SD, Rehman SF, et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med. 2019;2600(18):1–14. doi: 10.1016/S2213-2600(18)30510-1. [DOI] [PubMed] [Google Scholar]
- 65.Molyneaux PL, Cox MJ, Willis-Owen SAG, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. Oktober 2014;190(8):906–13. doi: 10.1164/rccm.201403-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han MLK, Zhou Y, Murray S, et al. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: An analysis of the COMET study. Lancet Respir Med. 2014 doi: 10.1016/S2213-2600(14)70069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salisbury ML, Han MK, Dickson RP, Molyneaux PL. Microbiome in interstitial lung disease: from pathogenesis to treatment target. Curr Opin Pulm Med. September 2017;23(5):404–10. doi: 10.1097/MCP.0000000000000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Faner R, Sibila O, Agustí A, et al. The microbiome in respiratory medicine: Current challenges and future perspectives. Eur Respir J. 2017;49(4):1–12. doi: 10.1183/13993003.02086-2016. [DOI] [PubMed] [Google Scholar]
- 69.Molyneaux PL, Maher TM. The role of infection in the pathogenesis of idiopathic pulmonary fibrosis. Eur Respir Rev. September 2013;22(129):376 LP–381. doi: 10.1183/09059180.00000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Molyneaux PL, Maher TM. Respiratory microbiome in IPF: cause, effect, or biomarker? Lancet Respir Med. Julie 2014;2(7):511–3. doi: 10.1016/S2213-2600(14)70088-8. [DOI] [PubMed] [Google Scholar]
- 71.Huang Y, Ma S-F, Espindola MS, et al. Microbes Are Associated with Host Innate Immune Response in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. Julie 2017;196(2):208–19. doi: 10.1164/rccm.201607-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Molyneaux PL, Willis-Owen SAG, Cox MJ, et al. Host-Microbial Interactions in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. Junie 2017;195(12):1640–50. doi: 10.1164/rccm.201607-1408OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hill S, Van Remmen H. Mitochondrial stress signaling in longevity: a new role for mitochondrial function in aging. Redox Biol. Julie 2014;2:936–44. doi: 10.1016/j.redox.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nam H-S, Izumchenko E, Dasgupta S, Hoque MO. Mitochondria in chronic obstructive pulmonary disease and lung cancer: where are we now? Biomark Med. 2017/06/09. Mei 2017;11(6):475–89. doi: 10.2217/bmm-2016-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saint-Georges-Chaumet Y, Attaf D, Pelletier E, Edeas M. Targeting microbiota-mitochondria inter-talk: Microbiota control mitochondria metabolism. Cell Mol Biol. 2015;61(4):121–4. [PubMed] [Google Scholar]
- 76.He Y, Wen Q, Yao F, Xu D, Huang Y, Wang J. Gut–lung axis: The microbial contributions and clinical implications. Crit Rev Microbiol. Januarie 2017;43(1):81–95. doi: 10.1080/1040841X.2016.1176988. [DOI] [PubMed] [Google Scholar]
- 77.Marsland BJ, Trompette A, Gollwitzer ES. The gut-lung axis in respiratory disease. Ann Am Thorac Soc. 2015;12(November):S150–6. doi: 10.1513/AnnalsATS.201503-133AW. [DOI] [PubMed] [Google Scholar]
- 78.O’Dwyer DN, Ashley SL, Gurczynski SJ, et al. Lung Microbiota Contribute to Pulmonary Inflammation and Disease Progression in Pulmonary Fibrosis. Am J Respir Crit Care Med. Februarie 2019;199(9):1127–38. doi: 10.1164/rccm.201809-1650OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Knippenberg S, Ueberberg B, Maus R, et al. <em>Streptococcus pneumoniae</em> triggers progression of pulmonary fibrosis through pneumolysin. Thorax. Julie 2015;70(7):636 LP–646. doi: 10.1136/thoraxjnl-2014-206420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Friaza V, la Horra C de, Rodríguez-Domínguez MJ, et al. Metagenomic analysis of bronchoalveolar lavage samples from patients with idiopathic interstitial pneumonia and its antagonic relation with Pneumocystis jirovecii colonization. J Microbiol Methods. 2010;82(1):98–101. doi: 10.1016/j.mimet.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 81.Takahashi Y, Saito A, Chiba H, et al. Impaired diversity of the lung microbiome predicts progression of idiopathic pulmonary fibrosis. Respir Res. 2018;19(1):34. doi: 10.1186/s12931-018-0736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han MK, Zhou Y, Murray S, et al. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: an analysis of the COMET study. Lancet Respir Med. 2014/04/21. Julie 2014;2(7):548–56. doi: 10.1016/S2213-2600(14)70069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ueda T, Ohta K, Suzuki N, et al. Idiopathic Pulmonary Fibrosis and High Prevalence of Serum Antibodies to Hepatitis C Virus. Am Rev Respir Dis. Julie 1992;146(1):266–8. doi: 10.1164/ajrccm/146.1.266. [DOI] [PubMed] [Google Scholar]
- 84.Irving WL, Day S, Johnston IDA. Idiopathic Pulmonary Fibrosis and Hepatitis C Virus Infection. Am Rev Respir Dis. Desember 1993;148(6_pt_1):1683–4. doi: 10.1164/ajrccm/148.6_Pt_1.1683. [DOI] [PubMed] [Google Scholar]
- 85.Moore BB, Moore TA. Viruses in Idiopathic Pulmonary Fibrosis. Etiology and Exacerbation. Ann Am Thorac Soc. November 2015;12(Suppl 2(Suppl 2)):S186–92. doi: 10.1513/AnnalsATS.201502-088AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tang Y-W, Johnson JE, Browning PJ, et al. Herpesvirus DNA is consistently detected in lungs of patients with idiopathic pulmonary fibrosis. J Clin Microbiol. Junie 2003;41(6):2633–40. doi: 10.1128/JCM.41.6.2633-2640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Molyneaux P, James P, Cookson W, Moffatt M, Maher T. P062 The Role of the Fungal Microbiome in IPF. QJM An Int J Med. September 2016;109(suppl_1):S42–S42. [Google Scholar]
- 88.Varone F, Gibiino G, Gasbarrini A, Richeldi L. Evaluation of the lung microbiome as a therapeutic target in the management of idiopathic pulmonary fibrosis: Role of antioxidant/antibiotic combination therapy. Eur Rev Med Pharmacol Sci. 2019;23(14):6379–86. doi: 10.26355/eurrev_201907_18463. [DOI] [PubMed] [Google Scholar]
- 89.Wang J, Lesko M, Badri MH, et al. Lung microbiome and host immune tone in subjects with idiopathic pulmonary fibrosis treated with inhaled interferon-γ. ERJ Open Res. 2017;3(3):00008–2017. doi: 10.1183/23120541.00008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kondoh Y, Cottin V, Brown KK. Recent lessons learned in the management of acute exacerbation of idiopathic pulmonary fibrosis. Eur Respir Rev. September 2017;26(145):170050. doi: 10.1183/16000617.0050-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song JW, Hong S-B, Lim C-M, Koh Y, Kim DS. Acute exacerbation of idiopathic pulmonary fibrosis: incidence, risk factors and outcome. Eur Respir J. Februarie 2011;37(2):356 LP–363. doi: 10.1183/09031936.00159709. [DOI] [PubMed] [Google Scholar]
- 92.Juarez MM, Chan AL, Norris AG, Morrissey BM, Albertson TE. Acute exacerbation of idiopathic pulmonary fibrosis-a review of current and novel pharmacotherapies. J Thorac Dis. Maart 2015;7(3):499–519. doi: 10.3978/j.issn.2072-1439.2015.01.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Network IPFCR, Raghu G, Anstrom KJ, King Jr TE, Lasky JA, Martinez FJ. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. 2012/05/20. Mei 2012;366(21):1968–77. doi: 10.1056/NEJMoa1113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fastrès A, Felice F, Roels E, et al. The Lung Microbiome in Idiopathic Pulmonary Fibrosis: A Promising Approach for Targeted Therapies. Int J Mol Sci. Desember 2017;18(12):2735. doi: 10.3390/ijms18122735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huie TJ, Olson AL, Cosgrove GP, et al. A detailed evaluation of acute respiratory decline in patients with fibrotic lung disease: Aetiology and outcomes. Respirology. Augustus 2010;15(6):909–17. doi: 10.1111/j.1440-1843.2010.01774.x. [DOI] [PubMed] [Google Scholar]
- 96.Oda K, Yatera K, Fujino Y, et al. Efficacy of concurrent treatments in idiopathic pulmonary fibrosis patients with a rapid progression of respiratory failure: an analysis of a national administrative database in Japan. BMC Pulm Med. 2016;16(1):91. doi: 10.1186/s12890-016-0253-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shulgina L, Cahn AP, Chilvers ER, et al. Treating idiopathic pulmonary fibrosis with the addition of co-trimoxazole: a randomised controlled trial. Thorax. Februarie 2013;68(2):155 LP–162. doi: 10.1136/thoraxjnl-2012-202403. [DOI] [PubMed] [Google Scholar]
- 98.Anstrom KJ, Noth I, Flaherty KR, et al. Design and rationale of a multi-center, pragmatic, open-label randomized trial of antimicrobial therapy - the study of clinical efficacy of antimicrobial therapy strategy using pragmatic design in Idiopathic Pulmonary Fibrosis (CleanUP-IPF) clinical trial. Respir Res. Maart 2020;21(1):68. doi: 10.1186/s12931-020-1326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Molyneaux PL, Cox MJ, Wells AU, et al. Changes in the respiratory microbiome during acute exacerbations of idiopathic pulmonary fibrosis. Respir Res. Februarie. 2017;18(1):29. doi: 10.1186/s12931-017-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park H, Shin JW, Park S-G, Kim W. Microbial Communities in the Upper Respiratory Tract of Patients with Asthma and Chronic Obstructive Pulmonary Disease. PLoS One. Oktober 2014;9(10):e109710. doi: 10.1371/journal.pone.0109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boem F, Amedei A. Healthy axis: Towards an integrated view of the gut-brain health. World J Gastroenterol [Internet] 07 Augustus 2019;25(29):3838–41. doi: 10.3748/wjg.v25.i29.3838. Available at: https://pubmed.ncbi.nlm.nih.gov/31413521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boem F, Nannini G, Amedei A. Not just ‘immunity’: how the microbiota can reshape our approach to cancer immunotherapy. Immunotherapy [Internet] 01 April 2020;12(6):407–16. doi: 10.2217/imt-2019-0192. Available at: https://www.futuremedicine.com/doi/abs/10.2217/imt-2019-0192. [DOI] [PubMed] [Google Scholar]
- 103.Group NIHHMPW, Peterson J, Garges S, et al. The NIH Human Microbiome Project. Genome Res. Desember 2009;19(12):2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bordenstein SR, Theis KR. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PLoS Biol. Augustus 2015;13(8):e1002226–e1002226. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huse SM, Ye Y, Zhou Y, Fodor AA. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One. Junie 2012;7(6):e34242–e34242. doi: 10.1371/journal.pone.0034242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007/02/22. Mei 2007;69(2):330–9. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dickson RP, Erb-Downward JR, Prescott HC, et al. Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid. J Clin Microbiol. Oktober 2014;52(10):3605–13. doi: 10.1128/JCM.01028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


