Abstract
Objectives
Patients hospitalised abroad can become colonised with multidrug-resistant bacteria and import them to their home countries. In this study, we characterised an OXA-484 carbapenemase-producing Escherichia coli strain from a Swiss patient infected by SARS-CoV-2 and repatriated from India.
Methods
At admission to Switzerland (April 2021), the patient undertook a nasopharyngeal swab to search for SARS-CoV-2 and a rectal swab to detect multidrug-resistant bacteria. Both SARS-CoV-2 and E. coli isolates were whole-genome sequenced and analysed for phylogenetic relatedness.
Results
The patient was infected with the SARS-CoV-2 B.1.617.2 lineage (VOC Delta), a lineage that began to be reported across Switzerland at that time. He was also colonised with a sequence type 410 (ST410) E. coli strain (L3452210II) producing OXA-484, a single amino acid variant of OXA-181. The blaOXA-484 gene was carried by a 51.5 kb IncX3 plasmid identical to those described in blaOXA-181-harbouring ST410 E. coli strains. Core genome analysis showed that L3452210II was identical (ΔSNV ≤23) to two ST410 OXA-484 producers recently reported in Qatar and Germany, but differed from other ST410 OXA-181 producers reported worldwide.
Conclusion
The patient was infected by an emerging SARS-CoV-2 variant and also imported an E. coli producing OXA-484, an OXA-48-like carbapenemase not yet reported in Switzerland. The genetic background of L3452210II indicated that blaOXA-484 shared the same plasmid as blaOXA-181, but its bacterial host differed from most of the pandemic OXA-181-producing ST410 strains reported previously. This case description underlines that the COVID-19 crisis can contribute to the worldwide spread of emerging carbapenemase producers.
Keywords: Carbapenemase, OXA-48, OXA-484, Plasmid, SARS-CoV-2, IncX3
1. Introduction
The spread of carbapenemase-producing Enterobacterales (CPE) represents a worldwide public-health concern. In particular, the prevalence of OXA-48-type producers is increasing as their numerous OXA-48-like variants (e.g. OXA-181, OXA-232, OXA-244) emerge [1]. For instance, OXA-181 is the second most common OXA-48 derivative, which differs from OXA-48 by four amino acid substitutions. Its bla gene is commonly located on a 51.5-kb IncX3 plasmid that is frequently harboured by high-risk sequence type 410 (ST410) Escherichia coli strains. Such bla OXA-181-possessing ST410 E. coli (bla OXA-181-ST410-Ec) strains originated on the India subcontinent and are nowadays reported worldwide both in human and non-human settings [1,2].
One of the ways in which CPE spread throughout the world is by the transfer of patients between countries. In fact, it is well known that hospitalised subjects can become colonised at the intestinal level with multidrug-resistant Enterobacterales (MDR-E). Importation of these pathogens into the country of origin (i.e. after patient transfer) can lead to difficult-to-treat infections; importantly, their dissemination can contribute to the spread to other people and/or settings (e.g. animals, food chain and the environment) [3,4].
In this context, the current COVID-19 (coronavirus disease 2019) pandemic crisis may amplify this phenomenon. Constant hygiene precautions of healthcare workers and pre-emptive isolation of patients repatriated from abroad are important elements to prevent the dissemination of MDR-E. However, the current COVID-19 pandemic has placed pressure on these precaution standards because healthcare workers are overworked and there is a lack of training of supplementary staff working in rooms with multiple beds and in intensive care units (ICUs) [5], [6], [7]. This may result in an uneven focus of hygiene precautions for COVID-19, while infection prevention and control measures linked to MDR-E are involuntarily (and unknowingly) relaxed.
In this study, we report a case of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) infection that was associated with intestinal colonisation of a rare OXA-48 variant in a Swiss patient transferred from India. State-of-the-art molecular and genomic techniques were implemented to characterise both the viral and bacterial pathogens.
2. Materials and methods
2.1. Routine laboratory tests
In April 2021, an adult patient hospitalised in India was transferred back to Switzerland. At admission, the patient underwent a nasopharyngeal swab to search for SARS-CoV-2 by implementing both the Cobas® SARS-CoV-2 and Influenza A/B assays on a Cobas® Liat® System (Roche), and the Xpert® Xpress SARS-CoV-2/Flu/RSV assay (Cepheid) was used as a confirmatory test to estimate the viral load. A rectal swab was also obtained to screen for MDR-E as part of the routine hygiene guidelines of the hospital. No further clinical data are available. The bacterial strain was named L3452210II to assure anonymity of the patient.
The rectal swab was plated on CHROMagarTM Orientation ESBL, mSuperCARBA and vancomycin-resistant enterococci (VRE) selective media and was incubated overnight at 36°C. Species identification was performed by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF/MS) (Bruker). Antimicrobial susceptibility testing was performed using broth microdilution ESB1F and GNX2F Sensititre panels (Thermo Fisher). Minimum inhibitory concentrations (MICs) for antibiotics were interpreted according to the 2021 European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria (https://www.eucast.org/; version 11.0). eazyplex® SuperBug complete B assay was used to screen for the presence of the main bla carbapenemase genes (Amplex Diagnostics; https://www.eazyplex.com/).
2.2. Sequencing of the SARS-CoV-2 genome
The SARS-CoV-2 RNA genome was reverse transcribed and amplified following the ARTIC V3 protocol. Nanopore library preparation was performed with the sequencing kit SQK-LSK109, and whole-genome sequencing (WGS) was performed on a GridION X5 (Oxford Nanopore Technologies). Reagents, quality control and flow cell preparation were done as previously described [8,9]. Bioinformatic analyses followed the ARTIC workflow (v.1.1.3). Consensus sequences were generated using medaka and bcftools. All methods are described in detail in Supplementary File 1.
Overall, 59 291 Nanopore reads were generated (coverage 250 ×). The final assembly (29 775 bases, 98.67% covered bases) contained 8.92% unknown bases and a 7-nucleotide gap compared with the reference sequence of Wuhan-Hu-1 (GenBank MN908947.3). The resulting consensus sequence was deposited in GISAID (https://www.gisaid.org/) with accession no. EPI_ISL_1916510.
2.3. Characterisation of the Escherichia coli strain
Escherichia coli strain L3452210II underwent WGS implementing both NovaSeq 6000 (Illumina Inc.) and MinION (SQK-RBK004 library, FLO-MIN 106D R9 flow-cell; Oxford Nanopore Technologies) technologies as previously described [10], [11], [12]. The hybrid assembly was generated using Unicycler v.0.4.8, and annotation was performed with the NCBI pipeline. The final genome was analysed using tools from the Center for Genomic Epidemiology (www.genomicepidemiology.org/). Species identification was confirmed implementing the Type (Strain) Genome Server (https://tygs.dsmz.de/). Genome and plasmid assemblies were deposited in GenBank (CP076527–CP076531) under BioProject PRJNA735932.
2.4. Phylogenetic analyses for SARS-CoV-2 and Escherichia coli
All genomic sequences of SARS-CoV-2 B.1.617.2 lineages available for Switzerland (n = 180 as of 28 June 2021) were retrieved from GISAID. The multiple sequence alignment generated with MAFFT v.7.407 was further subjected to maximum likelihood phylogenetic reconstruction with IQ-TREE 2 using the TIM2+F+I model and 1000 ultrafast bootstrap replicates. All methods are described in detail in Supplementary File 1.
To study the link between E. coli L3452210II and other bla OXA-181/bla OXA-484-ST410-Ec strains, a core genome single nucleotide variant (SNV) analysis was performed as done previously [10,12]. Briefly, the genome assemblies of bla OXA-181 / OXA-484-ST410-Ec strains were retrieved from Enterobase and the NCBI genome database (access date 15 June 2021). Core genome alignment and calculation of the phylogenetic tree was performed with Parsnp v.1.2. Variants with no flags (PASS) were determined as reliable and were used for downstream SNV analysis with a custom R v.3.6.2 script.
3. Results and discussion
3.1. SARS-CoV-2
The routine nasopharyngeal swab resulted positive for the SARS-CoV-2 target (cycle threshold value of 25.9). According to WGS, the patient's case was classified as Pango lineage B.1.617.2 or VOC Delta (World Health Organization), which was first detected in India (https://covariants.org/variants/21A.Delta). The B.1.617.2 lineage harbours the L452R and P681R spike substitutions that impact antibody binding; moreover, it is associated with a more rapid transmission than other circulating SARS-CoV-2 variants [13].
The present COVID-19 case occurred in calendar Week 17, 3 weeks after the first Swiss case of VOC Delta was reported in GISAID from the Canton of Aargau (7 April 2021). Noticeably, during Weeks 14 to 17, a total of 3, 5, 12 and 21 Delta genome sequences were reported in Switzerland (GISAID), respectively. Phylogenetic comparison of the 180 VOC Delta genomes available in GISAID for Switzerland indicated that our case belonged to the initial wave of introduction of VOC Delta and suggests a multiple-origin introduction scenario (Supplementary Fig. S1).
3.2. Resistome and plasmidome of the OXA-484-producing Escherichia coli
An E. coli strain (L3452210II) was isolated from selective agar plates and was resistant to various antibiotics, including fluoroquinolones, cephalosporins, β-lactam/β-lactamase inhibitory combinations and ertapenem (Supplementary Table S1). The eazyplex® SuperBug assay indicated that the strain possessed bla OXA-181, although this was not confirmed by WGS (see below).
According to the WGS analysis, strain L3452210II was of ST410 and its chromosome (4.7 Mb) possessed the mdf(A) antimicrobial resistance gene along with the GyrA Ser83Leu/Asp87Asn, ParC Ser80Ile and ParE Ser458Ala amino acid substitutions.
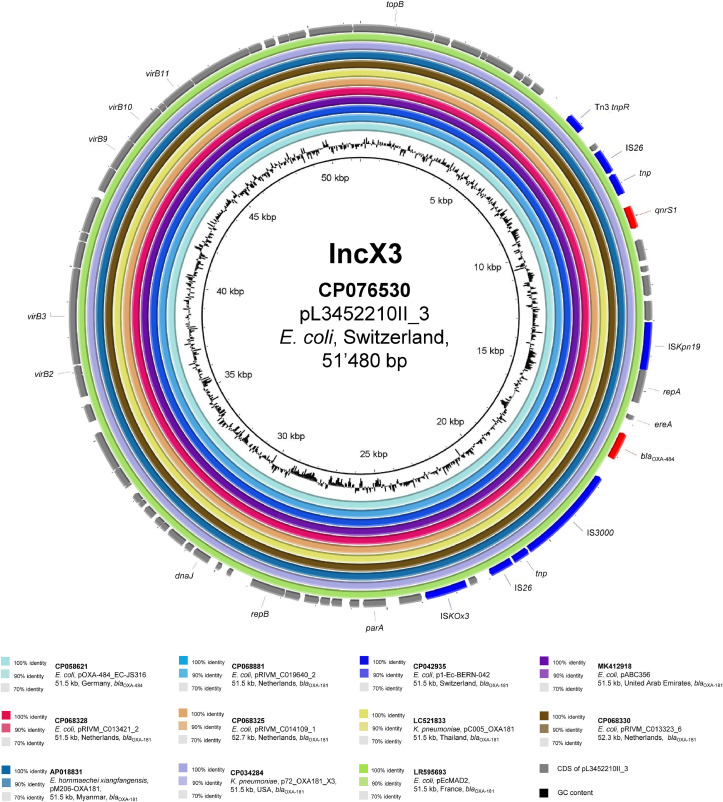
Strain L3452210II also possessed bla OXA-484 and qnrS1 within a composite transposon (IS26–tnp–IS3000–ΔISEcp1–bla OXA-484–ΔereA–ΔrepA1–ISKpn19–tnp–qnrS1–tnp–IS26) carried by a 51.5 kb IncX3 plasmid (pL3452210II_3). As shown in Fig. 1 , this mobile genetic element (MGE) was indistinguishable (coverage 100%, identity 100%) from another bla OXA-484-harbouring plasmid recently described in a ST410 E. coli (EC-JS316) isolated in Germany [14]. Moreover, with the exception of the expected single nucleotide substitution (responsible for Arg214Gly) in bla OXA-484, this MGE was identical (coverage 100%, identity 99.99%) to the numerous bla OXA-181-carrying plasmids reported worldwide, including those isolated in E. coli strains from human and non-human Swiss settings [2,11,15,16].
Fig. 1.
BLASTn comparison of plasmid pL3452210II_3 (GenBank accession no. CP076530) against similar sequences. Plasmid sequences were selected based on high homology to pL3452210II_3 in a BLASTn search against the NCBI non-redundant nucleotide database (date of access 9 June 2021). Rings were constructed using BRIG (BLAST Ring Image Generator) v.0.95. Coloured rings represent similarities to the reference sequence. For each sequence we report GenBank accession, species of isolation, sequence name, plasmid size and carbapenemase gene. Coding sequences (CDS) are represented as arrows in the outermost circle. Mobile genetic elements are depicted in blue, antimicrobial resistance genes in red, and other CDS in grey.
Strain L3452210II carried three additional plasmids: pL3452210II_1 (82.8 kb, IncY) without any antimicrobial resistance genes; pL3452210II_2 (67.3 kb, IncFIA/FII) carrying bla TEM-1B, aac(3)-IId, aph(3′′)-Ib, aph(6)-Id, tet(B) and sul2; and pL3452210II_4 (34.2 kb; IncIγ) harbouring the bla CMY-42 plasmid-mediated AmpC β-lactamase [17]. We note that most of the bla OXA-181/OXA-484-ST410-Ec strains previously reported carry bla CMY-42 in IncI1γ plasmids with a size of 34–51 kb. All of them shared (identity >99.62%) an ∼30 kb region that included bla CMY-42 (Supplementary Fig. S2). The bla CMY-42 gene was located upstream of the outer membrane lipoprotein gene blc and downstream of an IS1 family transposase. The same genetic context has been described in the IncIγ plasmid pISV_IncI_CMY-42 (GenBank MN242251) that was carried by an E. coli isolated from the stool of an Italian infant with recent travel history to India [18].
3.3. Global comparison of the ST410 blaOXA-181/OXA-484-positive Escherichia coli strains
As shown in Fig. 2 , core genome analysis indicated that L3452210II was identical (8–23 ΔSNVs) to the bla OXA-484-ST410-Ec German isolate EC-JS316 [14], but also to one (QU-31) recently found in Qatar. In contrast, a third bla OXA-484-ST410-Ec strain (C214) reported in Thailand was not clonally related. Nevertheless, analysis of the WGS assembly of C214 (GenBank GCA_017902335) with PlasmidFinder 2.0 indicated that the strain contained an IncX3 plasmid. Furthermore, the contigs containing the IncX3 replicon sequence (34 kb, GenBank DADVDF010000036) and the bla OXA-484 gene (6.7 kb; GenBank DADVDF010000052) aligned with a coverage of 100% (identity 99.99%) to pL3452210II_4, indicating that C214 carried the same IncX3 plasmid (data not shown).
Fig. 2.
Core genome analysis for Escherichia coli L3452210II (in red) and 75 E. coli ST410 possessing blaOXA-484 or blaOXA-181 obtained from the NCBI genome collection and Enterobase (date of access 15 June 2021). Core genome analysis of the whole-genome sequencing (WGS) assemblies is presented in a single nucleotide variant (SNV) hierarchical clustering dendrogram tree. The ΔSNV values correspond to the number of non-identical SNVs between two strains within a clade. We indicate association of the isolate with previous stay abroad (Link), country of origin, source of isolation and year of isolation. IND, India; QAT, Qatar; HKG, Hong Kong; NED, the Netherlands; PAK, Pakistan; DEN, Denmark; AUS, Australia; CAN, Canada; USA, United States of America; GBR, United Kingdom; SUI, Switzerland; GER, Germany; NGR, Nigeria; THA, Thailand; MYA, Myanmar; CRO, Croatia; LBN, Lebanon; SWE, Sweden; EGY, Egypt; ICU, intensive care unit; env., environment; na, not available.
We also note that the majority of bla OXA-181-ST410-Ec strains were not clonally related to the three identical bla OXA-484-ST410-Ec isolates. Only several bla OXA-181-ST410-Ec strains showed a certain homology (226 to 254 ΔSNVs) with those harbouring bla OXA-484 (Fig. 2). Among them, we emphasise the presence of E. coli AR24.2b and Ec-042, both representative strains responsible for a large outbreak that occurred in the Swiss veterinary setting involving both pets and employees [2,11].
4. Conclusions
Numerous COVID-19 patients hospitalised in ICUs were found to have a further infection and/or colonisation due to CPE. However, all of these subjects acquired the CPE after their admission [5], [6], [7]. In contrast, our patient was already colonised with a rare OXA-484-producing E. coli originating from India and later imported to Switzerland where it had not yet been observed.
The bla OXA-484 sequence was first reported (2017) in five Klebsiella pneumoniae isolates from the UK [19]. However, only recently the genetic background of this OXA-48-like carbapenemase has been described in EC-JS316, an E. coli strain obtained from a rectal swab of a patient hospitalised in Germany (December 2019) due to acute dengue fever following a stay in India [14]. The bla OXA-484-ST410-Ec strain isolated in the present work also originated from India and possessed both an IncX3 bla OXA-48-like-carrying plasmid and a core genome identical to EC-JS316 (23 ΔSNVs). Therefore, bla OXA-484-ST410-Ec strains are probably endemic on the Indian subcontinent, as for the bla OXA-181-ST410-Ec strains [1]. However, these two groups of bacteria (OXA-181 vs. OXA-484 producers) share only the same IncX3 plasmid (Fig. 1), whereas the E. coli host is different (Fig. 2). An exception could be noted for the bla OXA-484-positive strain C214 (detected in Thailand in 2016) that harboured the IncX3 plasmid in a ST410 identical to those usually possessing bla OXA-181. This unique finding suggests that both bla OXA-484 and bla OXA-181 may have evolved from a common ancestor and spread to different hosts. Importantly, the presence of these genes on a common IncX3 plasmid underlines the promiscuous ability of this MGE.
In conclusion, the COVID-19 crisis is contributing to the further expansion of life-threatening MDR bacteria endemic in some countries. This situation could also amplify the importation of these pathogens to low-prevalence nations [20], favouring their uncontrolled spread and the occurrence of outbreaks [5], [6], [7]. Therefore, in COVID-19 wards, infection prevention and control programmes must be rigorously maintained, or rapidly restored, to mitigate any future clinical impact due to CPE.
Acknowledgments
Acknowledgments
The authors thank Mrs Angela Vallone and the IFIK sequencing centre team for excellent technical assistance, especially Christian Baumann, Miguel A. Terrazos Miani and Stefan Neuenschwander.
Funding
This work was supported by the Swiss National Science Foundation (SNF) [grant no. 192514 to AE].
Competing interests
None declared.
Ethical approval
Not required.
Editor: Prof Wen-Chien Ko
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jgar.2021.10.012.
Appendix. Supplementary materials
References
- 1.Pitout JDD, Peirano G, Kock MM, Strydom KA, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2019;33:e00102. doi: 10.1128/CMR.00102-19. -19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nigg A, Brilhante M, Dazio V, Clement M, Collaud A, Gobeli Brawand S, et al. Shedding of OXA-181 carbapenemase-producing Escherichia coli from companion animals after hospitalisation in Switzerland: an outbreak in 2018. Euro Surveill. 2019;24 doi: 10.2807/1560-7917.ES.2019.24.39.1900071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leblebicioglu H, Rodriguez-Morales AJ, Rossolini GM, Lopez-Velez R, Zahar JR, Rello J, et al. Management of infections in critically ill returning travellers in the intensive care unit–I: considerations on infection control and transmission of resistance. Int J Infect Dis. 2016;48:113–117. doi: 10.1016/j.ijid.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Bij AK, Pitout JD. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother. 2012;67:2090–2100. doi: 10.1093/jac/dks214. [DOI] [PubMed] [Google Scholar]
- 5.Belvisi V, Del Borgo C, Vita S, Redaelli P, Dolce P, Pacella D, et al. Impact of SARS CoV-2 pandemic on carbapenemase-producing Klebsiella pneumoniae prevention and control programme: convergent or divergent action? J Hosp Infect. 2021;109:29–31. doi: 10.1016/j.jhin.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez-Simmonds A, Annavajhala MK, McConville TH, Dietz DE, Shoucri SM, Laracy JC, et al. Carbapenemase-producing Enterobacterales causing secondary infections during the COVID-19 crisis at a New York City hospital. J Antimicrob Chemother. 2021;76:380–384. doi: 10.1093/jac/dkaa466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farfour E, Lecuru M, Dortet L, Le Guen M, Cerf C, Karnycheff F, et al. Carbapenemase-producing Enterobacterales outbreak: another dark side of COVID-19. Am J Infect Control. 2020;48:1533–1536. doi: 10.1016/j.ajic.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neuenschwander SM, Terrazos Miani MA, Amlang H, Perroulaz C, Bittel P, Casanova C, et al. A sample-to-report solution for taxonomic identification of cultured bacteria in the clinical setting based on Nanopore sequencing. J Clin Microbiol. 2020;58:e00060. doi: 10.1128/JCM.00060-20. -20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gradel C, Terrazos Miani MA, Barbani MT, Leib SL, Suter-Riniker F, Ramette A. Rapid and cost-efficient enterovirus genotyping from clinical samples using Flongle flow cells. Genes (Basel) 2019;10:659. doi: 10.3390/genes10090659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budel T, Kuenzli E, Campos-Madueno EI, Mohammed AH, Hassan NK, Zinsstag J, et al. On the island of Zanzibar people in the community are frequently colonized with the same MDR Enterobacterales found in poultry and retailed chicken meat. J Antimicrob Chemother. 2020;75:2432–2441. doi: 10.1093/jac/dkaa198. [DOI] [PubMed] [Google Scholar]
- 11.Endimiani A, Brilhante M, Bernasconi OJ, Perreten V, Schmidt JS, Dazio V, et al. Employees of Swiss veterinary clinics colonized with epidemic clones of carbapenemase-producing Escherichia coli. J Antimicrob Chemother. 2020;75:766–768. doi: 10.1093/jac/dkz470. [DOI] [PubMed] [Google Scholar]
- 12.Moser AI, Kuenzli E, Budel T, Campos-Madueno EI, Bernasconi OJ, DeCrom-Beer S, et al. Travellers returning from the island of Zanzibar colonized with MDR Escherichia coli strains: assessing the impact of local people and other sources. J Antimicrob Chemother. 2021;76:330–337. doi: 10.1093/jac/dkaa457. [DOI] [PubMed] [Google Scholar]
- 13.Lazarevic I, Pravica V, Miljanovic D, Cupic M. Immune evasion of SARS-CoV-2 emerging variants: what have we learnt so far? Viruses. 2021;13:1192. doi: 10.3390/v13071192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommer J, Gerbracht KM, Krause FF, Wild F, Tietgen M, Riedel-Christ S, et al. OXA-484, an OXA-48-type carbapenem-hydrolyzing class D β-lactamase from Escherichia coli. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.660094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bleichenbacher S, Stevens MJA, Zurfluh K, Perreten V, Endimiani A, Stephan R, et al. Environmental dissemination of carbapenemase-producing Enterobacteriaceae in rivers in Switzerland. Environ Pollut. 2020;265 doi: 10.1016/j.envpol.2020.115081. [DOI] [PubMed] [Google Scholar]
- 16.Zurfluh K, Poirel L, Nordmann P, Klumpp J, Stephan R. First detection of Klebsiella variicola producing OXA-181 carbapenemase in fresh vegetable imported from Asia to Switzerland. Antimicrob Resist Infect Control. 2015;4:38. doi: 10.1186/s13756-015-0080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carattoli A, Villa L, Fortini D, Garcia-Fernandez A. Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid. 2018 Dec 5 doi: 10.1016/j.plasmid.2018.12.001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Marchetti VM, Bitar I, Mercato A, Nucleo E, Bonomini A, Pedroni P, et al. Complete nucleotide sequence of plasmids of two Escherichia coli strains carrying blaNDM-5 and blaNDM-5 and blaOXA-181 from the same patient. Front Microbiol. 2019;10:3095. doi: 10.3389/fmicb.2019.03095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Findlay J, Hopkins KL, Loy R, Doumith M, Meunier D, Hill R, et al. OXA-48-like carbapenemases in the UK: an analysis of isolates and cases from 2007 to 2014. J Antimicrob Chemother. 2017;72:1340–1349. doi: 10.1093/jac/dkx012. [DOI] [PubMed] [Google Scholar]
- 20.Albrecht R, Knapp J, Theiler L, Eder M, Pietsch U. Transport of COVID-19 and other highly contagious patients by helicopter and fixed-wing air ambulance: a narrative review and experience of the Swiss air rescue Rega. Scand J Trauma Resusc Emerg Med. 2020;28:40. doi: 10.1186/s13049-020-00734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.