ABSTRACT
Nontyphoidal Salmonella bacteria are the causative agent of salmonellosis, which accounts for the majority of foodborne illness of bacterial etiology in humans. Here, we demonstrate the safety and efficacy of the prophylactic administration of a bacteriophage preparation termed FOP (foodborne outbreak pill), which contains lytic phages targeting Salmonella (SalmoFresh phage cocktail), Shiga toxin-producing Escherichia coli (STEC), and Listeria monocytogenes, for lowering Salmonella burdens in OMM12 gnotobiotic mice. Prophylactic administration of FOP significantly reduced the levels of Salmonella in feces and in intestinal sections compared to the levels in controls. Moreover, the overall symptoms of the disease were also considerably lessened. Dose-dependent administration of FOP showed that phage amplification reached similarly high levels in less than 48 h independent of dose. In addition, 16S rRNA gene analysis showed that FOP did not alter the intestinal microbiota of healthy OMM12 mice and reduced microbiota perturbations induced by Salmonella. FOP maintained its full potency against Salmonella in comparison to that of SalmoFresh, its Salmonella-targeting component phages alone. Altogether, the data support that preventive administration of FOP may offer a safe and effective approach for reducing the risk of foodborne infections caused by Salmonella and, potentially, other foodborne bacteria (namely, STEC and L. monocytogenes) targeted by the FOP preparation.
IMPORTANCE Foodborne bacterial infections cause worldwide economic loss. During an epidemic, the use of antibiotics to slow down the spread of the disease is not recommended because of their side effects on the resident microbiota and the selection of antibiotic-resistant bacteria. Here, we investigated the potential for the prophylactic administration of bacteriophages (viruses infecting bacteria) to reduce the burden of Salmonella in vivo using mice colonized by a synthetic microbiota. We found that the repeated administration of bacteriophages was safe and efficient in lowering the Salmonella burden. Perturbations of the microbiota by the Salmonella infection were also reduced when mice received bacteriophages. Altogether, these data support the use of bacteriophages as a prophylactic intervention to lower the spread of foodborne epidemics.
KEYWORDS: foodborne disease, gnotobiotic model, epidemic, prevention, food-borne disease
INTRODUCTION
Foodborne illnesses are a major cause of morbidity and mortality, with an estimated 600 million foodborne infections and >400,000 deaths worldwide in 2010 (1, 2). The majority of foodborne disease (FBD) is caused by bacterial pathogens, where nontyphoidal Salmonella (NTS) strains are among those most commonly associated with invasive disease (3) and gastroenteritis (4). NTS infections are consistently the leading cause of an estimated 80 million cases of FBD worldwide, and approximately 1 million cases of FBD occur in the United States each year. NTS strains are Gram-negative, facultative, anaerobic bacteria consisting of multiple serovars of Salmonella enterica subsp. enterica. Salmonella infections (i.e., salmonellosis) in humans typically are acquired during transmission from the consumption of contaminated food or water, often from animal sources (5, 6).
Salmonellosis, a form of gastroenteritis, is often self-limiting, resulting in nausea, abdominal cramps, vomiting, and diarrhea that, while often lasting several days in healthy individuals, can lead to serious and deadly complications in vulnerable populations, such as the immunocompromised, elderly, or very young (7, 8). Salmonellosis may be treated using broad-spectrum antibiotics, but recent developments, including antibiotic resistance, have exposed limitations and risks with this approach (9–13). Thus, considerable effort is increasingly directed at finding prevention strategies to lower the dissemination of foodborne pathogens in a more specific manner (14).
Bacteriophages (or phages) offer one such approach. Phages are bacterial viruses that, unlike antibiotics, are highly specific and effective at killing their targeted host bacteria. In addition, the mechanisms by which antibiotics and phages kill bacteria and, conversely, the mechanisms of bacterial resistance to antibiotics or phages are fundamentally different (15–17). There is a long history of using phages to treat bacterial infections in humans, namely, phage therapy (18, 19), and currently, the number of successful compassionate treatments is increasing worldwide (20, 21). In contrast, the prophylactic use of phages remains underevaluated, in particular in the field of FBD.
Here, we utilize gnotobiotic mice (OMM12) to examine the tolerability and efficacy of the prophylactic administration of a phage preparation to target an intestinal foodborne pathogen. The OMM12 mice harbor a defined and stable intestinal community composed of 12 bacterial strains of mouse origin, which represent the five most prevalent and abundant phyla of the native laboratory mouse intestine (22). Importantly, when challenged by the human foodborne pathogen Salmonella enterica subsp. enterica serovar Typhimurium, OMM12 mice exhibited symptoms similar to those of salmonellosis in humans, without the need to use an antibiotic treatment to favor intestinal colonization, making these mice a relevant model for assessing the prophylactic impact of phages (22, 23).
We evaluated two phage preparations, SalmoFresh and FOP (foodborne outbreak pill), that target Salmonella Typhimurium. SalmoFresh is a cocktail of 6 phages targeting multiple Salmonella strains, and FOP is a cocktail of 15 phages that includes those from SalmoFresh, 6 phages targeting Listeria monocytogenes strains, and 3 phages targeting Shiga toxin-producing Escherichia coli (STEC) strains, including E. coli O157:H7 (24). FOP was equally as effective as SalmoFresh in reducing the Salmonella burden in feces and intestinal sections and in reducing symptoms of the disease. The prophylactic administration of FOP to healthy mice did not perturb the native gut microbiome, and FOP alone did not induce a gut inflammatory response as measured by the biomarker lipocalin-2. Altogether, these observations show that the prophylactic administration of FOP is both safe and effective to reduce Salmonella burdens in vivo.
RESULTS
SalmoFresh delays the burden of Salmonella in OMM12 mice.
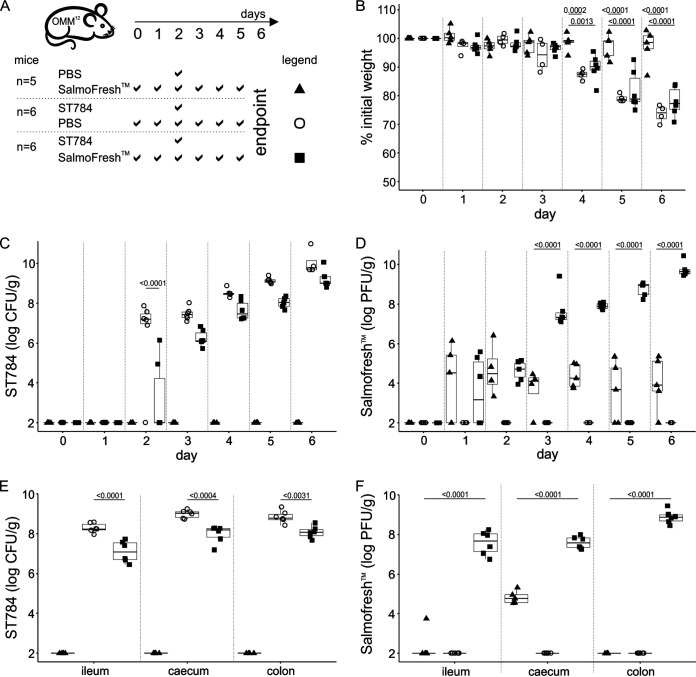
To assess the impact of the prophylactic administration of phage products on the burden of Salmonella in vivo, we first evaluated SalmoFresh, a product that contains six lytic phages (Fig. 1A and Table 1) (24, 25). Animals (3 groups) received for 2 days (days 0 and 1) either phosphate-buffered saline (PBS) (1 group) or SalmoFresh (2 groups) administered twice a day by oral gavage to reduce variability between animals and to ensure appropriate dosing and efficient delivery of the treatments and control. On day 2, the PBS group and one of the SalmoFresh groups received a single administration of Salmonella Typhimurium strain ST784 (1 × 108 CFU by oral gavage) (referred to as the ST784 PBS and ST784 SalmoFresh groups), while the other SalmoFresh group received PBS as a SalmoFresh control (referred to as the PBS SalmoFresh group). SalmoFresh (1 × 109 PFU/dose, quantified on strain ST784 prior to use) and PBS were then administered to the respective groups by oral gavage twice a day for an additional 4 days (from day 2 to day 5). Of note, four of the six phages included in SalmoFresh infect strain ST784 with similar efficiencies in vitro (see Materials and Methods). The single oral administration of ST784 in OMM12 mice receiving PBS (ST784 PBS group) led to a significant loss of weight compared to the weight loss in uninfected mice (PBS SalmoFresh group), starting 48 h post-ST784 administration and lasting two more days before animals reached the experimental endpoint (loss of 25% of their initial weight) (Fig. 1B and Table S1). Infected animals that received SalmoFresh twice a day (ST784 SalmoFresh group) displayed significant weight loss compared to the weight loss in uninfected mice (PBS SalmoFresh group). No significant weight variations were observed between ST784 PBS and ST784 SalmoFresh groups post-ST784 administration (Fig. 1B and Table S1). Additionally, symptoms of the disease (fur appearance, mobility, weight loss, and fecal consistency) were delayed in ST784 SalmoFresh animals compared to the onset in the ST784 PBS controls (Table 1). Concomitant to these observations, the levels of Salmonella in fecal samples increased steadily over time in both infected groups (Fig. 1C and Table S1). However, this increase was delayed by approximately 24 h in the ST784 SalmoFresh group and trended lower than in the ST784 PBS group (Fig. 1C and Table S1). This suggests that phages were able to infect ST784 in vivo, which was confirmed by the highly significant difference in the fecal phage levels when comparing uninfected and infected groups receiving SalmoFresh, which culminated with a 4-log difference by day 6 (Fig. 1D). Intestinal gut sections (ileum, cecum, and colon) were collected at day 6, and the Salmonella and phage levels were measured (Fig. 1E and F). Significant differences in Salmonella counts between the ST784 SalmoFresh and ST784 PBS groups were confirmed in all gut sections (Fig. 1E and Table S1). Phage levels in all three gut sections from ST784 SalmoFresh mice reached 108 PFU/g of feces, while in PBS SalmoFresh mice, the levels only reached 104 PFU/g in the cecum and were below the threshold of detection in both the ileum and colon (Fig. 1F). Phages were undetectable in the gut sections of the ST784 PBS group. These data demonstrate that the prophylactic administration of phages delays the burden of Salmonella in OMM12 mice. They are congruent with results previously obtained when testing SalmoFresh to reduce Salmonella contamination on fresh products (25). Additionally, the repeated administration of SalmoFresh in uninfected mice (PBS SalmoFresh group) did not impact their overall health (i.e., behavior and weight), showing that in the absence of their bacterial target, phages are innocuous.
FIG 1.
SalmoFresh delays the burden of Salmonella after challenge in OMM12 mice. (A) Experimental design. Groups of 4 to 6 OMM12 mice were orally administered PBS or SalmoFresh (1 × 109 PFU) on the indicated days. On day 2, mice were challenged with ST784 (1 × 108 CFU) or a PBS control as indicated. (B) Mice were weighed daily. Shown are the percentages of weight loss compared to starting weights of OMM12 mice over time. (C and D) The amounts of ST784 (CFU) (C) and phages (PFU) (D) in OMM12 mouse feces were quantitated daily (day 2 corresponds to 3 h after ST784 challenge). (E and F) Intestinal organs were collected on day 6 and homogenized, and the amounts of ST784 (CFU) (E) and phage (PFU) (F) were quantitated. Statistical analyses are described in Materials and Methods and reported in Table S1.
TABLE 1.
SalmoFresh reduces disease symptoms associated with the burden of Salmonella in OMM12 mice
| Treatment groupa | No. of mice that wereb: |
Treatment |
Challenge |
Clinical signs and symptoms on dayd: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | SFc | PBS | ST784 | PBS | 0 | 1 | 2 | 3 | 4 | 5 | 6 | |
| ST784 SalmoFresh | 2 | − | + | − | + | − | None | None | None | None | None | Mild | Mod |
| − | 4 | + | − | + | − | None | None | None | None | None | None | Mod | |
| PBS SalmoFresh | 2 | − | + | − | − | + | None | None | None | None | None | None | None |
| − | 3 | + | − | − | + | None | None | None | None | None | None | None | |
| ST784 PBS | 2 | − | − | + | + | − | None | None | None | None | None | Mod | Mod |
| − | 4 | − | + | + | − | None | None | None | None | Mod | Mod | Sev | |
Treatment groups were as shown in Fig. 1A.
M, male; F, female.
SF, SalmoFresh.
Clinical signs and symptoms over time (days 0 to 6) are scored based on animal behavior (e.g., activity, hunching) and consistency of fecal pellets (e.g., formed/no liquid, formed/liquid, liquid) as follows: None, no signs of disease; Mild, 50% of animals in group exhibited mild signs of disease; Mod, all animals in group exhibited moderate disease; Sev, all animals in group exhibited severe disease.
FOP strongly reduces Salmonella burden in vivo.
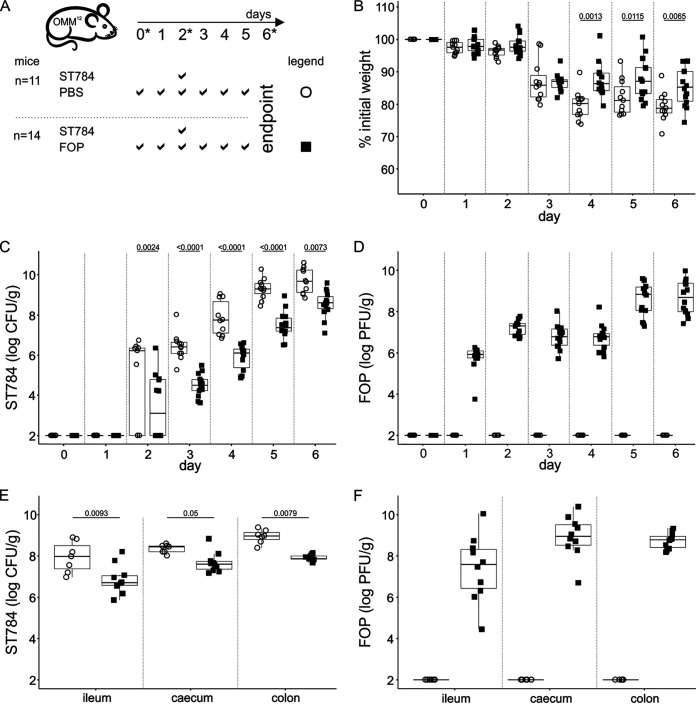
Using an experiment similar to the one described above, we next assessed the impact of FOP, which contains a combination of products including SalmoFresh (Fig. 2A and Table 2). In two independent experiments, two groups of mice were administered by oral gavage either FOP (1 × 109 PFU/dose; n = 14) or PBS (n = 11) twice a day for 6 days. Both groups were challenged with ST784 (1 × 108 CFU) on day 2 (ST784 PBS and ST784 FOP groups). We observed in both groups a median weight loss of approximately 10% of their initial weights within 24 h after Salmonella administration (Fig. 2B and Table S2). During the following 3 days (days 3 to 5), the median weight loss increased to 20% in the ST784 PBS group, while it stayed roughly stable in the ST784 FOP group (Fig. 2B and Table S2). Fecal pellets collected 3 h after Salmonella administration showed that in the ST784 PBS group, Salmonella levels reached 106 CFU/g of feces in all but two mice (Fig. 2C and Table S2). This was in sharp contrast to the ST784 FOP group, in which only 6 of 14 mice had Salmonella levels above the detection threshold (Fig. 2C and Table S2). During the next 4 days (days 3 to 6), the initial impact of the FOP product was significantly maintained, despite a progressive increase of fecal Salmonella levels, where the Salmonella burden in the ST784 FOP group was >1-log lower than in the ST784 PBS group (Fig. 2C and Table S2). Additionally, clinical signs and symptoms were significantly reduced and delayed in the ST784 FOP group compared to the clinical signs and symptoms in the ST784 PBS group (Table 2). Fecal phage levels were approximately 107 PFU/g 3 h after Salmonella administration, remained stable during the next 2 days (days 3 and 4), and increased up to 108 to 1010 PFU/g during the last 2 days (days 5 and 6), which is a pattern expected from continuous infection of increasing levels of Salmonella in the intestine (Fig. 2D). The levels of both Salmonella and phages in gut sections examined at day 6 were in agreement with the observations presented in Fig. 2C and D, where smaller amounts of Salmonella were measured in the ST784 FOP group than in the ST784 PBS group (Fig. 2E and Table S2), together with concomitant elevated levels of phages (Fig. 2F).
FIG 2.
FOP reduces Salmonella burden in vivo. (A) Experimental design. OMM12 mice were orally administered PBS or FOP (1 × 109 PFU) on the indicated days. On day 2, all mice were challenged with strain ST784 (1 × 108 CFU). Stars indicate the time points at which samples were taken for the 16S rRNA gene analysis reported in Fig. 3. (B) Mice were weighed daily. Shown are the percentages of weight loss compared to starting weights of OMM12 mice over time. (C and D) The amounts of ST784 (CFU) (C) and phages (PFU) (D) in OMM12 mouse feces were quantitated daily (day 2 corresponds to 3 h after ST784 challenge). (E and F) Intestinal organs were collected on day 6, and the amounts of ST784 (CFU) (E) and phages (PFU) quantitated. Statistical analyses are described in Materials and Methods and reported in Table S2.
TABLE 2.
FOP reduces disease symptoms associated with the burden of Salmonella in OMM12 mice
| Treatment groupa | No. of mice that wereb: |
Treatment |
Clinical signs and symptoms on dayc: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | FOP | PBS | 0 | 1 | 2 | 3 | 4 | 5 | 6 | |
| ST784 FOP | 6 | − | + | − | None | None | None | None | None | None | Mild |
| − | 8 | + | − | None | None | None | None | None | None | Mild | |
| ST784 PBS | 6 | − | − | + | None | None | None | None | None | Mod | Sev |
| − | 5 | − | + | None | None | None | None | Mild | Mod | Mod | |
Treatment groups were as shown in Fig. 2.
M, male; F, female.
Clinical signs and symptoms over time (days 0 to 6) are scored based on animal behavior (e.g., activity, hunching) and consistency of fecal pellets (e.g., formed/no liquid, formed/liquid, liquid) as follows: None, indicates no signs of disease; Mild, indicates 50% of animals in group exhibited mild signs of disease; Mod, indicates all animals in group exhibited moderate disease; Sev, indicates 50% of animals in group exhibited severe disease.
An independent experiment was performed with uninfected OMM12 mice to assess the safety of repeated administration of FOP, evaluated by observation of mouse behavior and measures of weight and fecal levels of lipocalin-2, a marker of intestinal inflammation (26), over time (Fig. S1A). When comparing groups of mice that received either FOP or PBS, we observed that all animals lost a slight amount of weight in both groups, which was attributed to repeated handling (i.e., twice-daily gavage) (Fig. S1B). No changes in overall health were observed (i.e., fur appearance, mobility, and fecal consistency). Fecal levels of phages were high and remained stable over time (Fig. S1C). Fecal levels of lipocalin-2 were quantitated before and 24 h and 96 h after FOP or PBS administration (Fig. S1D). Overall, lipocalin-2 levels remained below the threshold of inflammation (5 ng/g of feces [26]), and no increases were observed in the FOP group compared to the PBS group, indicating that FOP does not induce gastrointestinal tract inflammation (Fig. S1D).
The FOP treatment lowers Salmonella-induced perturbation of the bacterial consortium of the OMM12 mice.
We next assessed whether the prophylactic administration of the FOP product to Salmonella-infected OMM12 mice would affect the relative abundances of the resident bacteria of the gut. Fecal DNA was extracted from samples (Fig. 2C) at three time points: before FOP or PBS administrations (day 0), before ST784 challenge (day 2), and before sacrifice on day 6. Samples were subjected to 16S rRNA gene amplification and sequencing (Fig. 3A). No significant differences in bacterial abundance were seen between day 0 and day 2 for each of the PBS and FOP groups of animals. Additionally, no significant differences in bacterial abundance were seen between the FOP or PBS groups on day 2 prior to the bacterial challenge (Fig. 3B and Table S3). As expected, Salmonella bacteria were substantially more abundant in day 6 samples but significantly lower in ST784 FOP than in ST784 PBS samples, further demonstrating the efficacy of FOP in reducing Salmonella burdens in vivo (Fig. 3B). In the ST784 PBS group (day 6 versus day 0), the relative abundances of four strains (Clostridium clostridioforme, Muribaculum intestinale, Lactobacillus reuteri, and Enterococcus faecalis) were altered by the presence of Salmonella (Tables S3). Interestingly, in ST784 FOP samples, the abundances of C. clostridioforme and M. intestinale were not significantly altered, while the abundances of L. reuteri and E. faecalis were. These results indicate that the latter two strains were strongly associated with the burden of Salmonella, while the other two were not, suggesting that FOP prevented their variation (Table S3). These data are consistent with those previously obtained with an in vitro human model system (24) showing that FOP administration does not alter the resident gastrointestinal microbial community.
FIG 3.
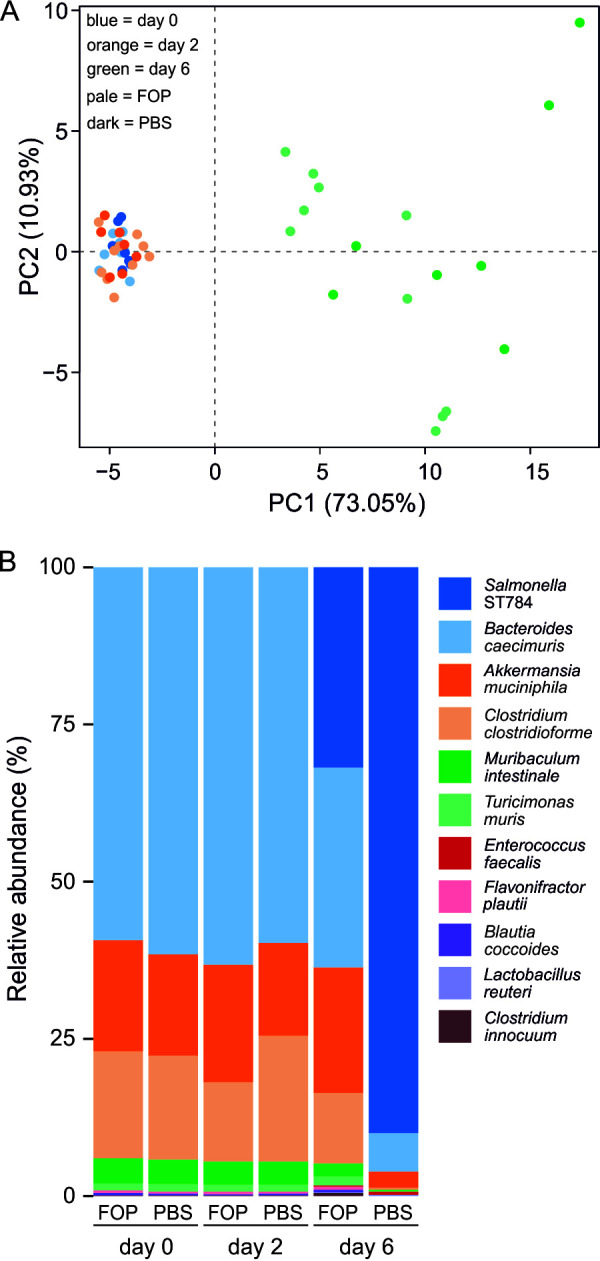
Salmonella infection disturbs the microbiota in OMM12 mice but FOP does not. (A) Shown is a principal-component analysis (PCA) plot of 16S rRNA gene data obtained from fecal pellets taken at days 0 (baseline), 2 (just before the ST784 challenge), and 6 (before sacrifice) from OMM12 mice that received FOP (n = 10) or PBS (n = 7). All animals were infected with strain ST784 on day 2 (see Fig. 2A for the experimental design). (B) The relative abundances of the ST784 challenge strain and OMM12 mouse origin bacteria from the fecal samples are shown. A comparison of log2-fold changes of 16S rRNA gene read abundances between days and conditions with statistical values is given in Table S3. Note that only 10 of the 12 mouse origin bacteria are detectable by either 16S rRNA gene or quantitative PCR (qPCR), as reported previously (31).
Decreasing the FOP dose does not reduce its in vivo efficacy.
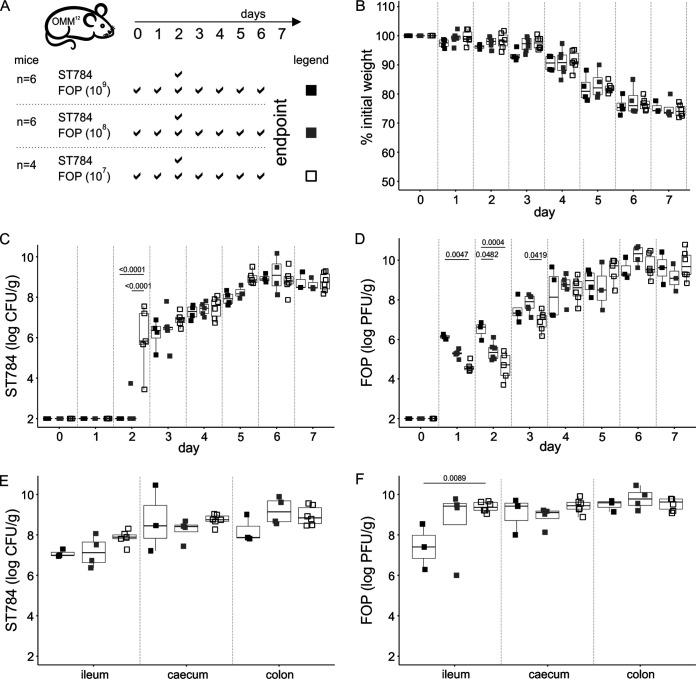
To assess whether there was a dose-dependent effect on the ability of FOP to reduce Salmonella in vivo, three groups of 4 to 6 OMM12 mice were orally administered FOP twice a day for 7 days at different doses with 10-fold reductions between doses, ranging from 1 × 109 to 1 × 107 PFU (Fig. 4A). All animals received a single dose of ST784 (1 × 108 CFU) on day 2. No differences in mouse behavior or weight were observed between the three groups (Fig. 4B and Table S4). At 3 h post-Salmonella administration, all mice that received the lowest dose of FOP (1 × 107 PFU) had detectable levels of Salmonella in feces, while only one mouse in the group receiving the middle dose (1 × 108 PFU) and none receiving the highest dose (1 × 109 PFU) had detectable ST784 counts (Fig. 4C and Table S4). This indicates that the highest prophylactic dose of FOP had a stronger impact on the ST784 inoculum. During the next 3 days (days 3 to 5), fecal levels of Salmonella rose in all three groups with no significant differences; however, the levels trended inversely proportional to the FOP dose (Fig. 4C and Table S4). During the last 2 days (days 5 and 6), Salmonella levels reached a median value of 108 CFU/g in feces and the difference between the groups vanished. Fecal levels of phages showed initial differences between the three groups on days 1 and 2 in agreement with the administered doses. Then, over the next several days, phage levels progressively increased, mirroring Salmonella fecal levels, with diminishing differences between the dosing groups until reaching a plateau where the median phage level was approximately 109 PFU/g (Fig. 4D and Table S4). We measured Salmonella (Fig. 4E) and phage (Fig. 4F) levels from gut sections on day 7 (1 day later than the experiments whose results are shown in Fig. 1 and 2). No significant differences in levels of ST784 between the three gut compartments were discerned, as expected from fecal contents (Fig. 4C), while a significant difference in phage counts could only be detected in the ileum between the highest and lowest dose groups (Fig. 4F and Table S4).
FIG 4.
Dose dependence of effect of FOP on ST784 burden in OMM12 mice. (A) Experimental design. OMM12 mice (n = 4 to 6) were orally administered the indicated doses of FOP (107 PFU, white squares; 108 PFU, gray squares; 109 PFU, black squares) on the indicated days. On day 2, all mice were challenged with strain ST784 (1 × 108 CFU). (B) Mice were weighed daily. Shown are the percentages of weight loss compared to starting weights of OMM12 mice over time. (C and D) The amounts of ST784 (CFU) (C) and phages (PFU) (D) in OMM12 mouse feces were quantitated daily (day 2 corresponds to 3 h after ST784 challenge). (E and F) Intestinal organs were collected on day 6, and the amounts of ST784 (CFU) (E) and phages (PFU) quantitated. Statistical analyses are described in Materials and Methods and reported in Table S4.
DISCUSSION
Several phage products have been granted generally recognized as safe (GRAS) status by the U.S. Food and Drug Administration (FDA), which is in agreement with a large body of literature that has shown that phages are innocuous when ingested by humans (27–29). This is not surprising, given the large and diverse amount of phages already present in the human gut (1015) (30). Nevertheless, the characterization of any new phage product for human applications must include information on safety as well as efficacy. Here, we evaluated the prophylactic application of FOP, a novel phage cocktail designed to target FBDs, including gut infections of Salmonella, which remains one of the most common causes of FBD worldwide. We attempted to mimic a real-life situation where individuals at risk, residing in the close proximity of an ongoing foodborne epidemic, would prophylactically take a phage product to lower the probability of contracting a disease. For this purpose, we used OMM12 mice that do not provide colonization resistance to Salmonella and therefore do not require antibiotic treatment (22, 23), in order to perform our study with an in vivo controlled-microbiota environment.
Given the expected rate of passive transit of phages in the guts of mice (31), we administered phages twice a day to maintain a high level of phages in order to maximize their impact on incoming Salmonella bacteria. The safety of such repetitive treatment was confirmed by the lack of any abnormal behavior or clinical symptoms, as well as the quantitation of an intestinal inflammatory marker (lipocalin-2), which remained below the threshold of inflammation. In addition, no impact on the microbiota structure was observed following 48 h of such treatment. Altogether, these data confirm that FOP is equally as safe as any of the three GRAS phage cocktails that compose it (i.e., EcoShield, ListShield, and SalmoFresh) (32).
We show that the prophylactic administration of SalmoFresh or FOP delayed the onset and lessened the most severe symptoms of Salmonella infection compared to the time of onset and severity of symptoms in PBS controls. Remarkably, the nearly 2-log decrease in CFU in the feces of mice that received FOP was also observed in each gut section, showing that phages can kill Salmonella with the same efficacy throughout the intestine. When comparing three different doses of FOP, we observed that 3 h after the ST784 challenge, the highest dose was sufficient to reduce the shedding of the inoculum to below the limit of detection. This rapid efficacy is congruent with data obtained with an in vitro human gut simulation model, which showed that FOP treatment killed ≥90% of ST784 after 5 h while simultaneously inhibiting the ability of ST784 to invade human intestinal cells (24). This strong impact on the Salmonella inoculum is encouraging for the prophylactic application of phages. Indeed, the Salmonella dose used in mice (1 × 108 CFU for a 20-g mouse) would correspond to a dose of 3.5 × 1011 CFU for a 70-kg human, which is several log higher than the infective dose for nontyphoidal salmonellosis (<103 CFU) (33, 34), as well as the most probable numbers (MPNs) of contaminating bacteria in retail settings, which are estimated to be even lower (35–37). Therefore, an FOP dose of 1 × 109 PFU would still remain several log higher than the target dose of Salmonella.
During all experiments, and more strikingly during the FOP dose experiments, the fecal levels of phages rose rapidly following ST784 challenge, demonstrating that orally administered FOP is actively replicating in the gut at the expense of ST784. Interestingly, during the dose experiment, within 24 h there was already no difference between the phage levels in mice that received the middle and the high doses, and within another 24 h, the phage levels resulting from the three doses were undistinguishable, while all three doses were significantly different before the challenge. These data showed that within 48 h, a 100-fold-lower dose of FOP reached a similar density in the gut that was associated with an equivalent impact on the Salmonella burden.
The relative abundances of 4 of the 12 intestinal bacteria of OMM12 mice were altered by increased levels of ST784 Salmonella. However, the FOP treatment limited these variations to increased abundances of L. reuteri and E. faecalis, suggesting that these two strains benefit from the presence of Salmonella. These observations illustrate that the growth of a pathogen has a direct impact on microbiota structure and that a phage intervention can limit such impact. They also demonstrate that murine gnotobiotic models, despite their limitations, are suitable to study mechanisms involved in the role of the gut microbiota during infection by Salmonella (22) and, potentially, other intestinal pathogens.
Finally, we showed that the prophylactic administration of phage products like FOP lowers the burden of a foodborne intestinal pathogen and could represent a strategy to reduce the dissemination of FBD during an outbreak. Lowering the spread of the contamination until the identification of its source, which can last several weeks, would have direct economic impact (38). In addition, the use of phages instead of antibiotics will be beneficial to the resident microbiota and will not increase the selection for antibiotic-resistant pathogens.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were approved by the committee on animal experimentation at the Institut Pasteur (Paris, France) and by the French Ministry of Research. A total of 68 OMM12 (C57BL/6J) mice from 7 to 9 weeks old, bred at Institut Pasteur, were used.
Bacterial strains and phage products.
S. enterica subsp. enterica serovar Typhimurium strain ST784 was obtained from Intralytix, Inc. (Columbia, MD), and routinely cultured in lysogeny broth (LB), on LB agar, or on Drigalski agar (Bio-Rad, Hercules, CA) at 37°C. For oral administration of ST784, an overnight liquid culture in LB was diluted 1/10 in sucrose bicarbonate buffer (20% sucrose and 2.6% sodium bicarbonate, pH 8). SalmoFresh (25, 39) and FOP (24) phage preparations were obtained from Intralytix. SalmoFresh is a commercially available phage cocktail consisting of 6 lytic phages at approximately equivalent titers targeting Salmonella spp., 4 of which infect strain ST784 with similar efficiencies. The FOP phage cocktail, which includes 15 phages at approximately equivalent titers, was prepared as described previously (24). It is a combination of three previously described commercial FDA-affirmed generally recognized as safe (GRAS) phage cocktails (i.e., ListShield, EcoShield PX, and SalmoFresh) that, combined, target Listeria monocytogenes, E. coli (STEC), and Salmonella, respectively (24). Before use, the phage cocktails were diluted in PBS to the indicated concentrations, which were calculated from the titration of these products on strain ST784. Salmonella-specific-phage quantification was performed by spotting serial dilutions of fecal or intestinal-section samples on LB plates overlaid with strain ST784 (24).
Murine model.
In all experiments, mice were randomly assigned to a group, and each group included approximately the same numbers of male and female animals kept in separate cages. Every day, mice were observed and weighed and feces were collected. Fecal pellets were transferred into preweighed, sterile, 2 ml tubes, weighed, and then resuspended in 1 ml of PBS. Serial dilutions in PBS were performed and plated onto Drigalski plates for Salmonella quantitation (CFU) and onto LB plates overlaid with ST784 for phage titration (PFU).
SalmoFresh, FOP, or PBS was administered by oral gavage in 200 μl twice daily, 6 h apart. The dose of phages administered through each gavage was 1 × 109 PFU, or lower as specified. Strain ST784 (1 × 108 CFU) was administered by oral gavage in 200 μl on day 2, 3 h after the first phage gavage on that day. Control mice not receiving ST784 received PBS instead. At the end of all experiments, mice were sacrificed by cervical dislocation, and intestinal sections (i.e., ileum, cecum, and colon) were collected without the separation of luminal and mucosal contents. These sections were homogenized in PBS using the gentleMACS OctoDissociator (Miltenyi Biotec), serially diluted in PBS, and plated on both Drigalski plates and LB plates overlaid with strain ST784.
Lipocalin-2 assay.
Frozen supernatants of fecal samples resuspended in PBS were thawed before lipocalin-2 quantification using a commercial enzyme-linked immunosorbent assay (ELISA) kit (catalog number DY1857; R&D Systems) according to the manufacturer’s instructions. The threshold of inflammation (5 ng/g of feces) was defined according to the method in reference 26.
16S rRNA gene analysis.
Resuspended fecal samples (500 μl) were centrifuged at 8,000 × g for 10 min, and the supernatant removed. Pellets were resuspended in 500 μl of lysis buffer (500 mM NaCl, 50 mM Tris-HCl, pH 8.0, 50 mM EDTA, 4% sodium dodecyl sulfate) and incubated for 15 min at 50°C (40). Then, 100 μl of lysozyme (25 mg/ml) was added and samples were incubated at 37°C for 2 h. DNA extraction was performed using the Maxwell 16 tissue DNA purification kit (Promega). Amplicon libraries targeting the V3-V4 16S rRNA gene region were then constructed using Illumina primers (forward primer, 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′, and reverse primer, 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′) and amplified by PCR for 25 cycles. The libraries were sequenced on an Illumina MiSeq instrument (2 × 300 bp). FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used for the quality control of the reads. Read filtering, operational taxonomic unit (OTU) clustering, and annotation were performed with the MASQUE pipeline (https://github.com/aghozlane/masque). All statistical analyses were performed with SHAMAN (http://shaman.c3bi.pasteur.fr) as previously described (41).
Statistical analysis.
Statistical analysis on the numbers of bacteria (CFU) and phages (PFU), as well as mouse weights, were carried out using the lme4 and lmerTest packages of R (42, 43). Both CFU and PFU were log10 transformed prior to analysis. Given the nonlinearity of responses, the day at which a measure was performed was considered a categorical variable. Linear mixed models were used to account for random experimental effects (i.e., individuals, experiments, and cage effects). Overall effects were assessed with analysis of variance (ANOVA) and post hoc Tukey’s comparisons and were performed using the lsmeans R package (44). A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Thierry Angélique and Eddie Maranghi, members of the Centre for Gnotobiology Platform of the Institut Pasteur, for their help with the animal work. We thank Amine Ghozlane from the Bioinformatics and Biostatistics Hub, Department of Computational Biology, Institut Pasteur, for help with 16S rRNA gene analysis. We also thank Joelle Woolston (Intralytix) for editorial contributions.
Q.L.-B. was funded by École Doctorale FIRE—Program Bettencourt. L.C. was funded by a Ph.D. fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche, France, Ecole Doctorale 394. M.L. belongs to the Pasteur—Paris University (PPU) international Ph.D. program funded by Institut Carnot Pasteur Maladies Infectieuses (grant number ANR 11-CARN 017-01).
The study was funded in part by Intralytix, Inc. (Columbia, MD, USA). A.S. holds an equity stake in Intralytix, Inc., a Maryland-based corporation specializing in developing and commercializing phage-based preparations, including FOP. A.S., J.A.S., J.T.T., and R.B.P. are employees of Intralytix, Inc.
A.S., J.A.S., and L.D. designed the study. Q.L.-B., L.C., and M.L. performed experiments. Q.L.-B., L.C., J.A.S., J.T.T., and L.D. analyzed the data. J.A.S., R.B.P., and L.D. drafted the paper, and all the authors contributed to and approved the submitted version.
Footnotes
Supplemental material is available online only.
Contributor Information
Laurent Debarbieux, Email: laurent.debarbieux@pasteur.fr.
Daria Van Tyne, University of Pittsburgh School of Medicine.
REFERENCES
- 1.Ashida H, Mimuro H, Sasakawa C. 2015. Shigella manipulates host immune responses by delivering effector proteins with specific roles. Front Immunol 6:219. doi: 10.3389/fimmu.2015.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, Praet N, Bellinger DC, de Silva NR, Gargouri N, Speybroeck N, Cawthorne A, Mathers C, Stein C, Angulo FJ, Devleesschauwer B, World Health Organization Foodborne Disease Burden Epidemiology Reference Group . 2015. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med 12:e1001923. doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vugia DJ, Samuel M, Farley MM, Marcus R, Shiferaw B, Shallow S, Smith K, Angulo FJ, Emerging Infections Program FoodNet Working Group . 2004. Invasive Salmonella infections in the United States, FoodNet, 1996–1999: incidence, serotype distribution, and outcome. Clin Infect Dis 38(Suppl 3):S149–S156. doi: 10.1086/381581. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2010. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. MMWR Morb Mortal Wkly Rep 59:418–422. [PubMed] [Google Scholar]
- 5.Greig J, Rajić A, Young I, Mascarenhas M, Waddell L, LeJeune J. 2015. A scoping review of the role of wildlife in the transmission of bacterial pathogens and antimicrobial resistance to the food chain. Zoonoses Public Health 62:269–284. doi: 10.1111/zph.12147. [DOI] [PubMed] [Google Scholar]
- 6.Levantesi C, Bonadonna L, Briancesco R, Grohmann E, Toze S, Tandoi V. 2012. Salmonella in surface and drinking water: occurrence and water-mediated transmission. Food Res Int 45:587–602. doi: 10.1016/j.foodres.2011.06.037. [DOI] [Google Scholar]
- 7.Havelaar AH, Garssen J, Takumi K, Koedam MA, Dufrenne JB, Van Leusden FM, De La Fonteyne L, Bousema JT, Vos JG. 2001. A rat model for dose–response relationships of Salmonella Enteritidis infection. J Appl Microbiol 91:442–452. doi: 10.1046/j.1365-2672.2001.01399.x. [DOI] [PubMed] [Google Scholar]
- 8.Scallan E, Fitzgerald M, Collins C, Crowley D, Daly L, Devine M, Igoe D, Quigley T, Robinson T, Smyth B. 2004. Acute gastroenteritis in northern Ireland and the Republic of Ireland: a telephone survey. Commun Dis Public Health 7:61–67. [PubMed] [Google Scholar]
- 9.Asperilla MO, Smego RA, Scott LK. 1990. Quinolone antibiotics in the treatment of Salmonella infections. Rev Infect Dis 12:873–889. doi: 10.1093/clinids/12.5.873. [DOI] [PubMed] [Google Scholar]
- 10.Bartlett JG. 1992. Antibiotic-associated diarrhea. Clin Infect Dis 15:573–581. doi: 10.1093/clind/15.4.573. [DOI] [PubMed] [Google Scholar]
- 11.Francino MP. 2016. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol 6:1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huddleston JR. 2014. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist 7:167–176. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S, Bhutta ZA, Coates A, Bergstrom R, Wright GD, Brown ED, Cars O. 2013. Antibiotic resistance—the need for global solutions. Lancet Infect Dis 13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh C, Sarkar P, Issa R, Haldar J. 2019. Alternatives to conventional antibiotics in the era of antimicrobial resistance. Trends Microbiol 27:323–338. doi: 10.1016/j.tim.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Lu TK, Koeris MS. 2011. The next generation of bacteriophage therapy. Curr Opin Microbiol 14:524–531. doi: 10.1016/j.mib.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 16.Merril CR, Scholl D, Adhya SL. 2003. The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov 2:489–497. doi: 10.1038/nrd1111. [DOI] [PubMed] [Google Scholar]
- 17.Sulakvelidze A, Alavidze Z, Morris JG, Jr.. 2001. Bacteriophage therapy. Antimicrob Agents Chemother 45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chanishvili N. 2012. Phage therapy–history from Twort and d’Herelle through Soviet experience to current approaches. Adv Virus Res 83:3–40. doi: 10.1016/B978-0-12-394438-2.00001-3. [DOI] [PubMed] [Google Scholar]
- 19.Kortright KE, Chan BK, Koff JL, Turner PE. 2019. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25:219–232. doi: 10.1016/j.chom.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 20.Aslam S, Lampley E, Wooten D, Karris M, Benson C, Strathdee S, Schooley RT. 2020. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infect Dis 7:ofaa389. doi: 10.1093/ofid/ofaa389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Djebara S, Maussen C, De Vos D, Merabishvili M, Damanet B, Pang KW, De Leenheer P, Strachinaru I, Soentjens P, Pirnay JP. 2019. Processing phage therapy requests in a Brussels military hospital: lessons identified. Viruses 11:265. doi: 10.3390/v11030265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brugiroux S, Beutler M, Pfann C, Garzetti D, Ruscheweyh HJ, Ring D, Diehl M, Herp S, Lötscher Y, Hussain S, Bunk B, Pukall R, Huson DH, Münch PC, McHardy AC, McCoy KD, Macpherson AJ, Loy A, Clavel T, Berry D, Stecher B. 2016. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat Microbiol 2:16215. doi: 10.1038/nmicrobiol.2016.215. [DOI] [PubMed] [Google Scholar]
- 23.Herp S, Brugiroux S, Garzetti D, Ring D, Jochum LM, Beutler M, Eberl C, Hussain S, Walter S, Gerlach RG, Ruscheweyh HJ, Huson D, Sellin ME, Slack E, Hanson B, Loy A, Baines JF, Rausch P, Basic M, Bleich A, Berry D, Stecher B. 2019. Mucispirillum schaedleri antagonizes Salmonella virulence to protect mice against colitis. Cell Host Microbe 25:681–694.e688. doi: 10.1016/j.chom.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Moye ZD, Woolston J, Abbeele PVD, Duysburgh C, Verstrepen L, Das CR, Marzorati M, Sulakvelidze A. 2019. A bacteriophage cocktail eliminates Salmonella Typhimurium from the human colonic microbiome while preserving cytokine signaling and preventing attachment to and invasion of human cells by Salmonella in vitro. J Food Prot 82:1336–1349. doi: 10.4315/0362-028X.JFP-18-587. [DOI] [PubMed] [Google Scholar]
- 25.Woolston J, Parks AR, Abuladze T, Anderson B, Li M, Carter C, Hanna LF, Heyse S, Charbonneau D, Sulakvelidze A. 2013. Bacteriophages lytic for Salmonella rapidly reduce Salmonella contamination on glass and stainless steel surfaces. Bacteriophage 3:e25697. doi: 10.4161/bact.25697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, Vijay-Kumar M. 2012. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One 7:e44328. doi: 10.1371/journal.pone.0044328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarker SA, Brüssow H. 2016. From bench to bed and back again: phage therapy of childhood Escherichia coli diarrhea. Ann N Y Acad Sci 1372:42–52. doi: 10.1111/nyas.13087. [DOI] [PubMed] [Google Scholar]
- 28.Vandenheuvel D, Lavigne R, Brüssow H. 2015. Bacteriophage therapy: advances in formulation strategies and human clinical trials. Annu Rev Virol 2:599–618. doi: 10.1146/annurev-virology-100114-054915. [DOI] [PubMed] [Google Scholar]
- 29.Weiss M, Denou E, Bruttin A, Serra-Moreno R, Dillmann ML, Brüssow H. 2009. In vivo replication of T4 and T7 bacteriophages in germ-free mice colonized with Escherichia coli. Virology 393:16–23. doi: 10.1016/j.virol.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 30.Dalmasso M, Hill C, Ross RP. 2014. Exploiting gut bacteriophages for human health. Trends Microbiol 22:399–405. doi: 10.1016/j.tim.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Lourenço M, Chaffringeon L, Lamy-Besnier Q, Pédron T, Campagne P, Eberl C, Bérard M, Stecher B, Debarbieux L, De Sordi L. 2020. The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28:390–401.e395. doi: 10.1016/j.chom.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Dissanayake U, Ukhanova M, Moye ZD, Sulakvelidze A, Mai V. 2019. Bacteriophages reduce pathogenic Escherichia coli counts in mice without distorting gut microbiota. Front Microbiol 10:1984. doi: 10.3389/fmicb.2019.01984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan KJ, Ray CG, Sherris JC. 2004. Sherris medical microbiology: an introduction to infectious disease, 4th ed, McGraw-Hill, New York, NY. [Google Scholar]
- 34.Bronze MS, Greenfield RA (ed). 2005. Biodefense: principles and pathogens. Horizon Bioscience, Wymondham, Norfolk, England. [Google Scholar]
- 35.Boughton C, Leonard FC, Egan J, Kelly G, O’Mahony P, Markey BK, Griffin M. 2004. Prevalence and number of Salmonella in Irish retail pork sausages. J Food Prot 67:1834–1839. doi: 10.4315/0362-028x-67.9.1834. [DOI] [PubMed] [Google Scholar]
- 36.Dufrenne J, Ritmeester W, Delfgou-van Asch E, van Leusden F, de Jonge R. 2001. Quantification of the contamination of chicken and chicken products in The Netherlands with Salmonella and Campylobacter. J Food Prot 64:538–541. doi: 10.4315/0362-028X-64.4.538. [DOI] [PubMed] [Google Scholar]
- 37.Fegan N, Vanderlinde P, Higgs G, Desmarchelier P. 2004. Quantification and prevalence of Salmonella in beef cattle presenting at slaughter. J Appl Microbiol 97:892–898. doi: 10.1111/j.1365-2672.2004.02380.x. [DOI] [PubMed] [Google Scholar]
- 38.Van Duynhoven YT, Isken LD, Borgen K, Besselse M, Soethoudt K, Haitsma O, Mulder B, Notermans DW, De Jonge R, Kock P, Van Pelt W, Stenvers O, Van Steenbergen J, Outbreak Investigation Team . 2009. A prolonged outbreak of Salmonella Typhimurium infection related to an uncommon vehicle: hard cheese made from raw milk. Epidemiol Infect 137:1548–1557. doi: 10.1017/S0950268809002337. [DOI] [PubMed] [Google Scholar]
- 39.Moye ZD, Woolston J, Sulakvelidze A. 2018. Bacteriophage applications for food production and processing. Viruses 10:205. doi: 10.3390/v10040205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Z, Morrison M. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808–812. doi: 10.2144/04365ST04. [DOI] [PubMed] [Google Scholar]
- 41.Volant S, Lechat P, Woringer P, Motreff L, Campagne P, Malabat C, Kennedy S, Ghozlane A. 2020. SHAMAN: a user-friendly website for metataxonomic analysis from raw reads to statistical analysis. BMC Bioinformatics 21:345. doi: 10.1186/s12859-020-03666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 43.Kuznetsova A, Brockhoff PB, Christensen RHB. 2017. lmerTest package: tests in linear mixed effects models. J Stat Softw 82:1–26. doi: 10.18637/jss.v082.i13. [DOI] [Google Scholar]
- 44.Lenth RV. 2016. Least-squares means: the R package lsmeans. J Stat Softw 69:1–33. doi: 10.18637/jss.v069.i01. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00497-21_Supp_1_seq10.xlsx, XLSX file, 0.02 MB (18.1KB, xlsx)
Supplemental material. Download SPECTRUM00497-21_Supp_2_seq11.xlsx, XLSX file, 0.1 MB (67.1KB, xlsx)
Supplemental material. Download SPECTRUM00497-21_Supp_3_seq8.xlsx, XLSX file, 0.02 MB (17.5KB, xlsx)
Supplemental material. Download SPECTRUM00497-21_Supp_4_seq9.xlsx, XLSX file, 0.02 MB (21.9KB, xlsx)
Supplemental material. Download SPECTRUM00497-21_Supp_5_seq12.pdf, PDF file, 0.1 MB (136.3KB, pdf)