Summary
High-resolution and rapid imaging of host cell ultrastructure can generate insights toward viral disease mechanism, for example for a severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection. Here, we employ full-rotation soft X-ray tomography (SXT) to examine organelle remodeling induced by SARS-CoV-2 at the whole-cell level with high spatial resolution and throughput. Most of the current SXT systems suffer from a restricted field of view due to use of flat sample supports and artifacts due to missing data. In this approach using cylindrical sample holders, a full-rotation tomogram of human lung epithelial cells is performed in less than 10 min. We demonstrate the potential of SXT imaging by visualizing aggregates of SARS-CoV-2 virions and virus-induced intracellular alterations. This rapid whole-cell imaging approach allows us to visualize the spatiotemporal changes of cellular organelles upon viral infection in a quantitative manner.
Keywords: soft X-ray, cryo-tomography, viral replication organelles, SARS-CoV-2, cells, tomography, organelles, synchrotron
Graphical abstract
Highlights
-
•
Soft X-ray tomography enables label-free imaging of organelle morphology
-
•
Cylindrical sample holders enable capture of a full-rotation tomogram of large cells
-
•
Tomogram obtained rapidly within 10 min
-
•
Whole-cell quantitative phenotypic analysis of SARS-CoV-2-induced alterations
Motivation
The analysis of 3D architecture of infected cells is critical for understanding the mechanisms of viral diseases. Fluorescence and electron microscopy techniques, in particular, focused ion beam scanning electron microscopy (FIB-SEM) have been extensively used to image the 3D structure of cells. However, although FIB-SEM achieves superior spatial resolution, the method remains low throughput: an entire mammalian cell at 8 nm isotropic voxel size takes about a week to image and generates an enormous amount of data that is daunting to analyze. To overcome these limitations, a whole-cell imaging technique with minimal sample preparation and rapid image acquisition is required for quantitative analysis of structural changes induced in virus-infected cells.
High-resolution, rapid imaging techniques are needed to analyze 3D cell architecture for understanding viral disease mechanisms. Here, Loconte et al. use soft X-ray tomography to rapidly image SARS-CoV-2-infected whole cells, opening avenues to analyze virus-cell interactions and efficacy of antiviral drugs in statistically significant numbers.
Introduction
Soft X-ray tomography (SXT) is a technology that can image whole intact cells in 3D under normal and pathological conditions (Carzaniga et al., 2014a; Guo and Larabell, 2019; Harkiolaki et al., 2018; Schneider et al., 2010; Weiß et al. 2000). SXT has been applied to study yeast (Larabell and Le Gros, 2003; Liu et al., 2018; Uchida et al., 2009; Walters et al., 2019; Yang et al., 2010), algae (Hummel et al., 2012; Roth et al., 2019), and primary and immortalized cell lines (Darrow et al., 2016; Duke et al., 2014; Elgass et al., 2015; Le Gros et al., 2016; Smith et al., 2014; White et al., 2020). SXT imaging is done on cells without labeling or chemical fixation at high throughput (about 5 min for a whole 3D volume of 15 × 15 × 15 μm3) and with a spatial resolution of a few tens of nanometers (Larabell and Nugent, 2010; Le Gros et al., 2014; Parkinson et al., 2012). Moreover, SXT uses a specific energy range of soft X-rays (from about 282 eV to 533 eV or λ = 4.4 nm to λ = 2.3 nm) with superior and intrinsically quantitative contrast of molecular building blocks, such as carbon and nitrogen (Jacobsen and Kirz, 1998). The absorption of soft X-rays linearly depends on the concentration of biomolecules and thus can be used to differentiate not only cellular organelles (Le Gros et al., 2016; McDermott et al., 2009; Uchida et al., 2011), but also the metabolic state of organelles and proteins of similar composition (Boeynaems et al., 2019; Hanssen et al., 2012). This natural contrast averts the need for staining and the possible misinterpretation of cellular structures (Kellenberger et al., 1992; Mielanczyk et al., 2014; Miller and Goldsmith, 2020).
In the past, SXT has successfully been used to visualize single virions and associated changes in cells infected with vaccinia virus (Carrascosa et al., 2009; Chichón et al., 2012), herpes simplex virus 1 (Aho et al., 2017, 2019; Myllys et al., 2016), hepatitis C virus (Pérez-Berná et al., 2016), and reovirus (Kounatidis et al., 2020). Recently, a correlative imaging approach was used to visualize SARS-CoV-2 replication in Vero cells, combining cryo-electron tomography and SXT (Mendonça et al., 2021); however, quantitative whole-cell analyses of virus-associated changes are still lacking.
SXT has the ability to image whole cells. However, many of the recent SXT studies of virus-infected cells have been done using microscopes that have adopted the same sample holders as used in electron tomography–flat grids (Groen et al., 2019; Harkiolaki et al., 2018). When tilting such flat specimen holders, there is a progressive increase in specimen thickness and decrease in angular tilt capability. This leads to “missing wedge” artifacts and, with high-resolution SXT optics, some out-of-focus cell structures (Ekman et al., 2019, Li et al., 2017, Otón et al., 2017, Selin et al., 2015). In addition, cells adhere and spread out on grids, making it necessary to stitch together multiple tomograms to image an entire cell. As a result, only local information is typically obtained, and a small sampling of the cell is imaged instead of all organelles in the whole cell; this limits statistical comparison of cellular changes associated with viral infection.
To ensure safe handling of viruses at the reduced biosafety level of the synchrotron, infected cells must be neutralized or inactivated with fixatives prior to imaging. As SXT does not require chemical fixation of cells, previous studies focused on characterizing the achievable contrast and quantitative nature of the X-ray absorption coefficient used to discriminate cellular organelles in unfixed cells (Ekman et al., 2019; Uchida et al., 2011). The quantitative nature of the soft X-ray absorption contrast in fixed cells has not been characterized to date.
To address this discrepancy, we characterized the soft X-ray linear absorption coefficient in native compared with fixed human B cells. We were able to discriminate heterogeneous organelles in the cytoplasm and changes in molecular composition, mainly carbon and nitrogen, in fixed cells that were comparable to those in the native state. Our data show that aldehyde fixation as performed in this study does not interfere with quantitative X-ray imaging and, as with other microscopy techniques, many insights into virus-induced cell remodeling can be studied on fixed cells with SXT.
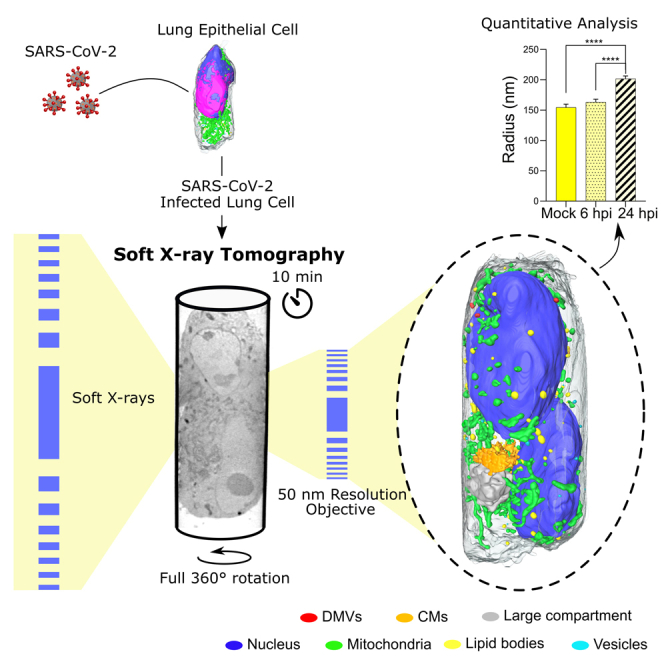
Here, we illustrate the potential of SXT to characterize and quantify structural changes induced in cells infected by SARS-CoV-2. Unlike previous studies, we mounted cells in thin-wall glass capillaries; this enabled us to visualize whole mammalian cells in one tomogram. Furthermore, we used human lung epithelial cells, showing that SXT can visualize individual clusters of SARS-CoV-2 virions on the cell surface and double-membrane vesicles, which are the sites of viral RNA replication in the cytoplasm of infected cells. In addition, we performed a quantitative comparison of cellular organelles at 6 and 24 hours post infection (hpi) compared with mock infected cells, revealing profound alterations of the subcellular architecture induced by viral infection in a time-dependent manner. Our data demonstrate the power of SXT to both visualize and quantify changes in whole cells. The ongoing development of tabletop SXT systems (Hertz et al., 2012; Kim et al., 2006; McEnroe et al., 2017; Takman et al., 2007) will make this technology available to any laboratory and will provide the opportunity to visualize subcellular structures in whole-cell volumes without the need for complex and demanding imaging equipment.
Results
Absorption coefficient of soft X-rays in fixed cells
The physical process resulting in a soft X-ray image is that of X-ray absorption, with a linear X-ray absorption coefficient associated with each voxel of the reconstructed 3D image. This is similar to medical computed tomography (CT) in which a linear attenuation coefficient, measured in Hounsfield units (HU), is associated with each voxel of the patient’s image. This is a quantitative way to distinguish density differences between different tissues. For example, bone has a very different HU than lung tissue, and a tumor has a different HU compared with normal tissue. For SXT, the linear absorption coefficient (LAC) is well characterized in human B lymphocytes in hydrated and cryo-preserved cells (Larabell and Nugent, 2010; Ma et al., 2020; Myllys et al., 2016; Weinhardt et al., 2020). However, when working with infectious pathogens cells must be neutralized with fixatives prior to imaging. In these cases, fixation protocols are designed to ensure full inactivation of viruses and not optimized in terms of ultrastructural preservation.
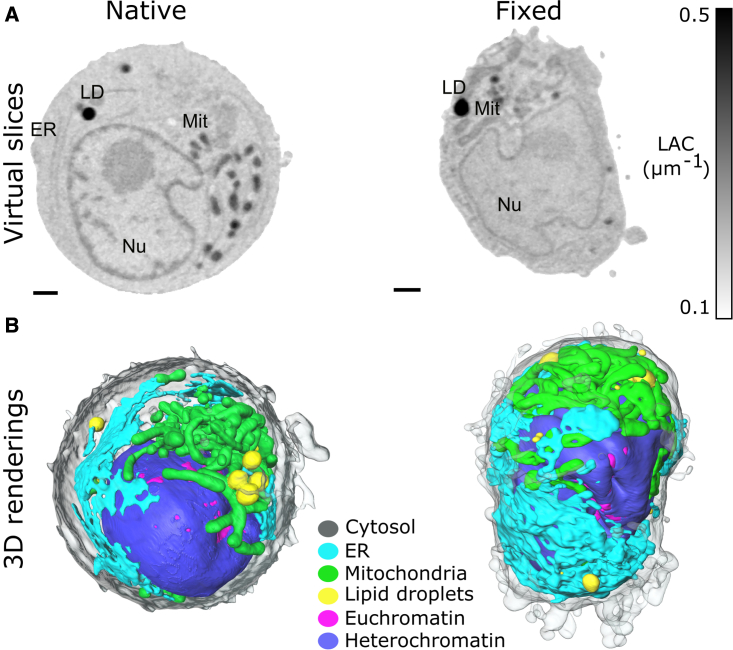
To investigate whether the LAC of soft X-rays can be used to differentiate organelles in fixed cells, we cultured, fixed with glutaraldehyde, then plunge froze the human B cells (cell line GM12878). The virtual slices of fixed and “native-state” cells are shown in Figure 1A and Video S1. To the naked eye, X-ray absorption contrast between organelles in native and fixed cells is comparable. In terms of gross morphology, fixed human B cells show a convoluted shape with distorted cytoplasm. This cell blebbing is a well-known effect of aldehydes on cell structure (Zhao et al., 2014). In addition, we found distorted lipid droplets in some fixed cells (n = 2/10, Figure S1), potentially due to known challenges in fixation and permeabilization of lipid droplets (Ohsaki et al., 2005). At the level of SXT resolution, we did not observe significant differences in organelle morphology.
Figure 1.
Comparison of soft X-ray LAC of organelles in native and fixed cells
(A) Virtual slices and (B) 3D renderings of native and fixed human B lymphocytes of cell line GM12878. Virtual slices are scaled as depicted with colorbar from 0.1 to 0.5 μm−1 LAC values. Nu, nucleus; LD, lipid droplets; Mit, mitochondria. Scale bars, 1 μm.
Virtual slices are scaled as depicted with colorbar from 0.1 to 0.5 μm-1 LAC values.
To quantitatively compare X-ray LAC, we segmented the major organelles in native (n = 7) and fixed cells (n = 10), see Table S1. Although aldehyde-based fixatives have been shown to alter biochemical content of cells (Hobro and Smith, 2017; Jearanaikoon and Abraham-Peskir, 2005), we found no significant difference in measured (mean) LAC values between fixed and native human B cells for hetero- and euchromatin, mitochondria, lipid droplets, endoplasmic reticulum (ER), and cytoplasm. Furthermore, the LAC values for organelles in fixed human B cells fall in a similar range as previously reported for Candida albicans cells (McDermott et al., 2009), mouse lymphoma cells (Smith et al., 2014), and human stem cells (Niclis et al., 2015). Thus, we conclude that the LAC of soft X-rays can be applied as a quantitative parameter to identify and unambiguously segment organelles in SXT datasets for cells fixed as described in this study (see STAR Methods).
Full-rotation imaging
Most mammalian cells are between 10 and 20 μm in diameter. Cell types used to characterize entry and kinetics of SARS-CoV-2 replication have an average diameter of about 15 to 20 μm (Dietmair et al., 2012). On flat sample holders, these cells adhere and spread out, extending large lamellipodia and filopodia that can cause them to increase in size to about 100 μm, and with changing thickness that can reach 6 μm at the nucleus, see Figure 2A. With previously reported penetration depth of 10 μm (Duke et al., 2014; Hagen et al., 2012) and field of view (depending on the objective) of 16 μm × 16 μm (depicted in yellow), SXT imaging of whole cells on flat sample holders is limited to the fraction (region of interest) of the whole cell. In addition, due to the progressive increase in specimen thickness during rotation (Cinquin et al., 2014), visualizing cells from all angles is not possible and results in “missing wedge” artifacts.
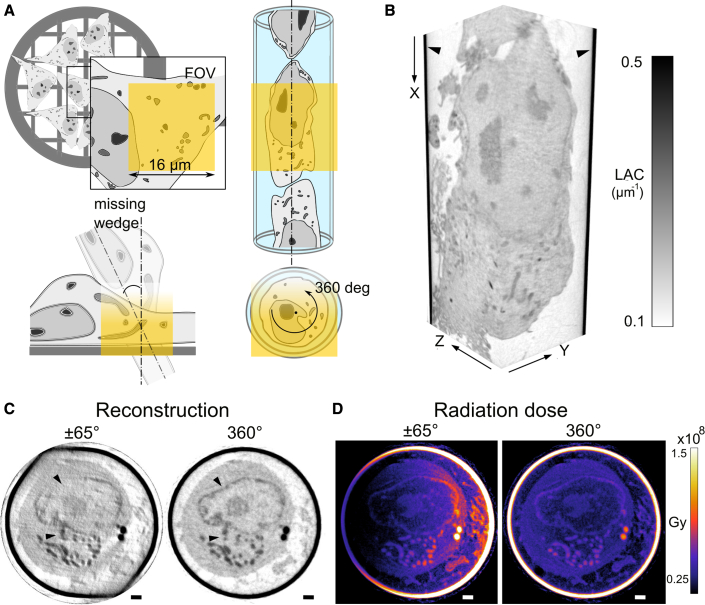
Figure 2.
Cylindrical capillary versus flat grid for single-cell mounting in SXT
(A) Schematics of flat grid and glass capillary cell mounting for SXT. Depicted in yellow: SXT field of view is about 16 μm × 16 μm with 50-nm objective lens. X-ray progressive absorption (from yellow to white) limits depth of imaging and maximum rotation angle for flat specimens, resulting in a “missing wedge.”
(B) In a cylindrical sample holder (black arrows), the full cell volume of a single cell is imaged, seen in both axial (XY) and sagittal (XZ) view. The gray values show the different level of absorption of each type of organelle with a threshold set from 0.1 to 0.5 μm−1.
(C) The effect of limited ±65° rotation, there is “shadowing” with the sample holder and decrease in X-ray transmission in comparison with full-rotation tomography. Arrows show streak artifacts. (D) Calculated X-ray radiation dose for limited angle and full-rotation tomography in Gy. The colorbar is from 1.5 × 107 to 1.5 × 108 Gy. Scale bars are 1 μm.
To investigate the difference between flat and cylindrical sample holders, we reconstructed a native human B cell, depicted in Figure 1A, using those projection images corresponding to the typical tilt angles of ±65° rotation (Harkiolaki et al., 2018), typically used for flat grids, see Figures 2C and S2. The missing angular data result in streak artifacts over the virtual plane and, at the regions where X-ray projections are missing, leads to distortions and decreased contrast between organelles. This simulation has the advantage of examining the exact same cell but excludes the interference from the flat specimen holder itself that is typically encountered. This “shadowing” of a sample holder shifts X-ray LAC values used for segmentation and identification of organelles in SXT, see Figures S2A and S2B. The complex and potentially varying geometry of flat supports makes reconstructions of 3D data particularly prone to ambiguity, undermining the quantitative nature of LAC values in SXT.
We employ an alternative method of sample handling and place cells in thin-wall glass capillaries, see Figure 2A. This approach helps to match the SXT field of view with the diameter of a cell, ensures artifact-free imaging enabled by full 360° range of rotation and similar transmission for all angles, and generates a reconstruction of the cell volume unobscured from the sample holder; this approach ensures quantitative LAC values. As projections are acquired over more rotation angles than in limited angle tomography, SXT imaging in cylindrical sample holders could potentially introduce more radiation dose. Thus, we have simulated X-ray radiation dose in SXT reconstructions for 360° rotation and limited ±65° angle tomography acquired with the same sampling (1 projection image per 1° angle), see Figure 2D. As X-ray radiation dose is linearly proportional to the number of X-ray photons per reconstructed voxel, the X-ray radiation dose of 360° rotation tomography does not increase in comparison with limited angle tomography. On the contrary, the progressive increase of exposure time required to compensate for the lower transmission of X-rays at higher rotation angles results in higher radiation dose, which is distributed unevenly throughout the sample. The distribution of X-ray dose in full 360° rotation tomography is more homogeneous for a cell than tomography acquisitions with 180° rotation, and ±65° angle tomography, see Figures S2B and S2B′. Moreover, with the increasing rotation angle θ, the thickness of cells on flat specimen holders increases as cos(θ), such that a 5-μm-thick region at 0° becomes 11.8 μm thick at ±65°. The gradual increase in cell thickness results in lower transmission of X-rays and is typically compensated by a comparable increase in exposure time (Chiappi et al., 2016), resulting in even higher X-ray dose in limited angle tomography (Figure S2B). However, if positioned correctly (toward incoming X-ray beam) a flat support will act as a “shield” with more homogeneous distribution of X-ray dose in ±65° angle tomography as well. For full-rotation tomography, the X-ray dose is about 0.5 × 108 Gy within the cell, which is below the previously reported limit of 108 to 109 Gy for X-ray radiation dose of cryogenically preserved specimens (Schneider et al., 1995).
Taken together, 3D tomography of single cells on flat holders is not optimal in terms of throughput and data quality, and should be used only when cells cannot be placed in cylindrical sample holders or the biological question requires imaging of adherent cells.
Whole-cell architecture of human epithelial cells by SXT
To demonstrate whole-cell imaging of virus-infected cells with SXT, we selected the well-established human lung epithelial cell lines Calu-3 and A549-ACE2 and human embryonic kidney cells HEK293T-ACE2 cells, the latter two stably expressing the SARS-CoV-2 entry factor. These cell lines have been recently used to study subcellular alterations induced by SARS-CoV-2 with light and electron microscopy (e.g., Cortese et al., 2020; Harcourt et al., 2020; Klein et al., 2020; Ou et al., 2020). Using cylindrical sample holders, we obtained the structural organization of whole cells in a single tomographic acquisition of about 5 min, see Figure 3B. Larger cells were imaged with two tomographic acquisitions by translating the capillary vertically (in Figure 2B X direction), resulting in a volume of more than 16 μm × 16 μm × 32 μm. The datasets were reconstructed in real time within 5 min per tomogram by the automatic alignment and reconstruction software for SXT: AREC3D (Parkinson et al., 2012; Ekman et al., 2018).
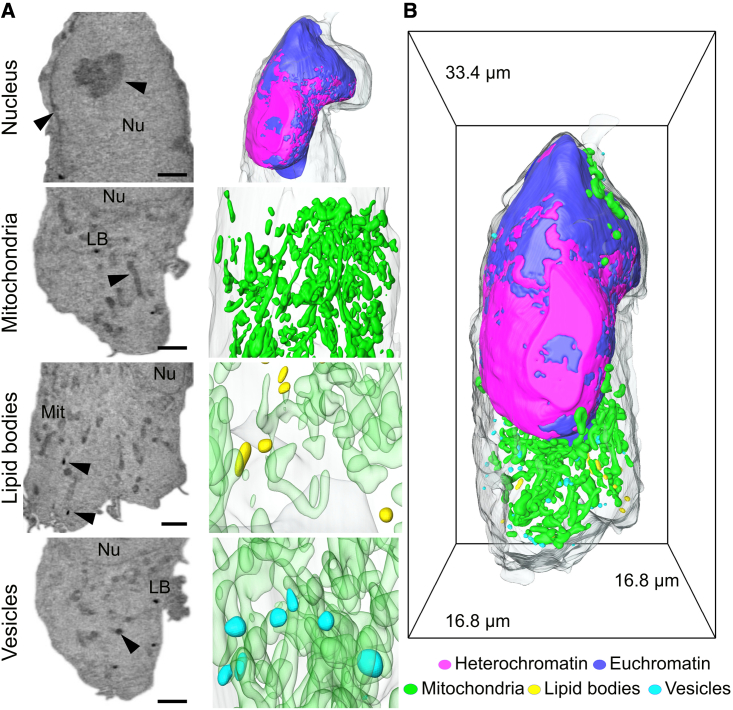
Figure 3.
SXT of uninfected HEK293T-ACE2 cells and 3D rendering of individual organelles
(A) Raw orthoslices showing respectively: Nu, nucleus (with distinct euchromatin and heterochromatin); Mit, mitochondria; LB, lipid bodies; and vesicles. Each type of organelle is highlighted by a black arrowhead. Scale bar, 2 μm.
(B) Rendering of the whole cell showing the segmented organelles labeled according to their LAC and morphology. Heterochromatin is represented in magenta; euchromatin in blue; mitochondria in green; lipid bodies in yellow; and vesicles in cyan.
Based on the established X-ray absorption coefficient of cellular organelles with SXT, we identified different subcellular compartments in uninfected cells. First, we semiautomatically segmented the nucleus in each cell (see STAR Methods) and separated chromatin into hetero- and euchromatin regions identified by double-peak fit of X-ray LAC values (Le Gros et al., 2016). In the remaining cytoplasm volume, we used automatic thresholding based on the X-ray LAC observed in human B cells to identify lipid bodies, mitochondria, and other organelles.
An example of an HEK293T-ACE2 cell is shown in Figures 3A and 3B. All three cell lines had similar gross morphology (Figure S3). Based on such rapid, whole, 3D cell volume acquisition, we established the morphology of these cells in normal (mock) conditions (n = 3 for each cell line). The cells are approximately the same size (Calu-3 2272 ± 74.5μm3, A549-ACE2 2280 ± 625μm3, HEK293T-ACE2 1738 ± 458μm3) and possess evident cytoplasmic protrusions. Nuclei of these cells occupy 32.5% ± 0.04% of total cell volume and contain mainly a loosely packed form of chromatin, euchromatin (57.2 ± 23.4 μm3, 9.97% ± 2.31% of the total nuclear volume; LAC value 0.29 ± 0.02 μm−1; see Figures 3A and 3B), with a thin rim of heterochromatin beneath the nuclear membrane and isolated nucleoli. Based on the recent established association of such nuclear morphology with aging (Lee et al., 2020), we assume the nuclear morphology seen here is a sign of extensive passaging of these cell types. Nevertheless, access to genome editing to both improve viral infectivity of cell cultures and to create virus-specific reporter cell lines, together with wide availability of a broad range of cell types, makes cell culture one of the important characterization approaches currently available (Dolskiy et al., 2020).
Using SXT, we identified mitochondria, lipid bodies (lipid droplets and lamellar bodies), and vesicles in the cytoplasm, potentially involved in the endosomal pathway. Based on the heterogeneous size and specific X-ray absorption value, these vesicles are potentially the class of lipid-bound vesicles that might be related to the endocytic cycle in the cell (Klumperman and Raposo, 2014).
Interestingly, although SXT has been used to visualize networks of ER and Golgi apparatus in many cell types, see Figure S1 and (Elgass et al., 2015; Groen et al., 2019; Hagen et al., 2012; Weinhardt et al., 2020), these organelles were not apparent in Calu-3, A549-ACE2, and HEK293T-ACE2 cells in the current study (see Figure S3). Previous ultrastructural characterization by electron microscopy showed that the Golgi apparatus is rare in Calu-3 cells (Finkbeiner et al., 1995); however, more recent studies with electron microscopy showed a network of ER spanning throughout almost the whole cell and multiple Golgi-like structures in the same cell type (Cortese et al., 2020). Consistent with this observation, SXT imaging of ER and other organelles has been successfully detected in A549 cells upon uptake of gold nanoparticles with higher resolution (30 nm versus 50 nm in the current study) SXT microscopes (Kepsutlu et al., 2020). The architecture of the ER has been shown to have two different configurations, tubules, and sheets. While ER sheets are rough and highly packed with ribosomes, serving as the main site for synthesis, folding, and post-translational modifications (Shibata et al., 2010; West et al., 2011), fewer ribosomes are present on the smooth ER tubules (Schwarz and Blower, 2016). Proportions of these two ER configurations reflect functions of different cell types. While secreting cells like B cells are largely made up of ER sheets, cells involved in the processes of lipid signaling and calcium metabolism are composed primarily of tubules of ER (Baumann and Walz, 2001). As seen in human B cells, 50 nm spatial resolution is sufficient to differentiate ER, even though the X-ray absorption coefficient (LACER = 0.24 ± 0.01 μm−1) is very similar to that of cytosol (LACCyt = 0.22 ± 0.01 μm−1). Thus, we hypothesize that the ER network of these particular cell lines is in a different configuration from the ER of B cells, and that there is an insufficient difference between the X-ray LAC of ER and cytoplasm for unambiguous identification, at this resolution.
The mitochondria, however, are clearly visible and appear as a large tubular network, occupying 4.70% ± 0.95% of the cytoplasm with rare, isolated regions that underwent fission. Interestingly, we found a small number of lipid bodies (0.019% ± 0.012% of total cell volume) with a volume of 0.20 ± 0.08 μm3 dispersed throughout cytoplasm.
In Calu-3 and HEK293T cells, we found vesicles previously not seen under SXT (Figure 3A and Video S2). The X-ray absorption coefficient of 0.37 ± 0.04 μm−1 together with radius of 143 ± 43 nm clearly separates these vesicles from mitochondria and lipid bodies or recently reported insulin secretory granules (White et al., 2020). Other potential cellular structures, such as endosomes, lysosomes, and multi-vesicle bodies in these cell lines have been shown to have a dense granular matrix filled with small heterogeneous vesicles and/or lamellar material (Cortese et al., 2020); such structures are not visible in our data. Given the homogeneity of such vesicles, their radius size, and their presence in mock cells, we suggest that they might be endosome-like vesicles involved in the late stage of the endocytic cycle of the cell (Huotari and Helenius, 2011; Longatti et al., 2010).
Heterochromatin is represented in magenta; euchromatin in blue; mitochondria in green; lipid bodies in yellow; and vesicles in cyan.
Hallmarks of SARS-CoV-2 infection at a whole-cell level
To demonstrate the potential of volumetric imaging with SXT, we imaged the same cell lines at 6 and 24 hpi with SARS-CoV-2. Infectious viral particles were detectable at high efficiency starting at 6 hpi, as confirmed by immunofluorescent imaging (Figure S4A). Since the precise time of infection for individual cells is not known, imaged datasets effectively included cells infected at any given time between 0 and 24 hpi. To ensure correct identification of virus-associated organelles, we performed a systematic comparison of the SXT data with transmission electron microscopy (TEM) images on thin chemically fixed sections of the same cells (Figure S4B). With the spatial resolution in SXT of 50 nm, we could detect individual virus particles at the cell surface and organelles associated with SARS-CoV-2 pathology (Figure 4). We found one cell (n = 1/17, 6 hpi) with individual SARS-CoV-2 virions and clusters of virions extracellularly, adhering to the cell surface, and no structural changes intracellularly (Figures 4A and S4B). These clusters consisted of multiple SARS-CoV-2 particles (diameter of whole clusters 239 ± 54 nm) with an X-ray absorption coefficient of 0.35 ± 0.04 μm−1. Interestingly, aggregations of virions were found predominantly on one side of the cell surface (Figure 4A). While not explicitly discussed, a similar localization of virus particles was visible in the recent ultrastructural characterization by cryoFIB/SEM (Mendonca et al., 2020). This localization of viral entry might be due to the polarization of epithelial cells in cultures and respective polarization of ACE localization (Lambert, 2009).
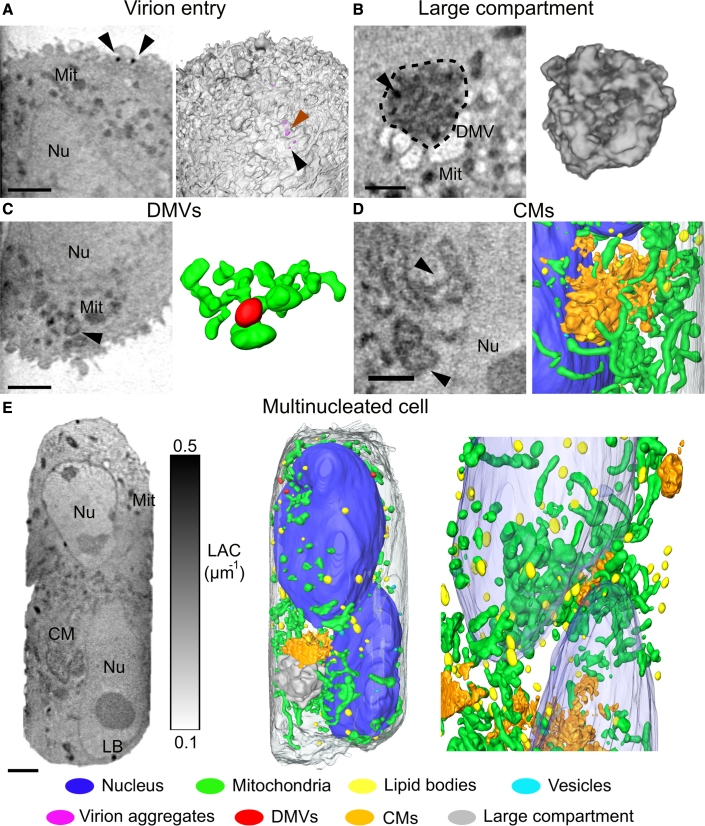
Figure 4.
The hallmarks of SARS-CoV-2 infection
(A) Orthoslice showing the entry of viral particles at the surface of the cell (n = 1/17). Individual SARS-CoV-2 virions (black arrows) and aggregates (orange arrows) are characterized by 0.35 ± 0.043 μm−1 of LAC and 239 ± 54 nm of diameter. On the right, cell surface and 3D rendering of virions.
(B) Orthoslices and 3D rendering of the large compartment found in three cells, showing high density and heterogeneous structure of the compartment.
(C) Orthoslices identifying DMVs as hollowed structures in proximity of the mitochondrial network with 3D rendering of the interacting organelles.
(D) Single orthoslice showing the position of convoluted membranes and the 3D rendering of the CMs in proximity with nucleus and mitochondrial network.
(E) Orthoslice (left) of a whole multinucleated cell and 3D rendering (right) of identified organelles. Each organelle is color coded as follows: nucleus - blue, mitochondria - green, lipid bodies - yellow, vesicles - cyan, virions - magenta, DMVs - red, CMs - orange, and large compartment - gray. Scale bars, 2 μm.
Many viruses induce modifications of cellular organelles to create an environment conducive to viral replication. At 24 hpi, a prominent organelle generated in infected cells is the membranous replication organelle, which in the case of SARS-CoV-2 is composed of double-membrane vesicles (DMVs) occasionally connected to convoluted membranes (CMs) (Knoops et al., 2008; Snijder et al., 2020). Cytosolic DMVs are associated with coronavirus genome replication (Klein et al., 2020; Snijder et al., 2020; Wolff et al., 2020a). With SXT, we found both DMVs and CMs in the cytoplasm of infected cells (Figures 4C and 4D). Although double membranes are not resolved, the DMVs in SXT are clearly visible due to the higher LAC (0.33 ± 0.03 μm−1) than that of mitochondria (LAC: 0.31 to 0.32 μm−1) and typical “ring-like” morphology (Figures 4C and S4B). The DMVs were often found with physical contacts to mitochondria, suggesting their involvement in the cross-talk between mitochondria and ER, as suggested by others (Singh et al., 2020). An example of a cell with highly convoluted CMs that are easily distinguishable from DMVs can be seen in Figures 4C and 4D.
The DMVs are about 366 ± 37 nm in diameter, which is in good agreement with previously reported sizes of DMVs induced by SARS-CoV-2 (Cortese et al., 2020). The CMs form a large structure, altogether occupying 11.8 μm3 of the cell volume. Although continuously interconnected, CMs were localized as clusters close to the nucleus (Figure 4D). Notably, the inner lumen of both DMVs and CMs has a similar X-ray absorption coefficient to that of cytosol, indicating that these organelles are filled with similar components found in cytosol or that their contents have been already released (Figure 4D). Recent publications (Klein et al., 2020; Wolff et al., 2020b) have shown luminal filaments inside the DMVs.
Furthermore, we found that all HEK293T-ACE2 cells at 24 hpi show one large membrane compartment within the cytosol (Figures 4B and S6, Video S3). These compartments are packed with highly dense material (LAC = 0.29 ± 0.05 μm−1). Based on their large diameter (3.20 ± 0.36 μm), nonconvoluted morphology, and LAC values distinctly different from that of DMV and CM X-ray absorption values, we suggest these structures as distinct hallmarks of SARS-CoV-2 infection. Based on results from blob detection (Figure S4C), we suggest that these compartments consist of an assembly of DMVs and CMs, or that they might be involved in the degradation of other chemically dense material. Indeed, similar compartments of DMVs and other organelles were validated with TEM imaging (Figure S4B). Based on previous characterization of endocytic pathway and autophagy in CoV infections (Yang and Shen, 2020), we hypothesize that these compartments are large autophagosomes (1–5 μm in diameter) or autolysosomes involved in the disposal of the viral replication machinery.
Among cells at 24 hpi, we found one cell containing a double nucleus, see Figure 4E and Video S4. In this single giant cell, we found both DMVs and CMs often located at the periphery of nuclei. Such multinucleated cells, also known as syncytium, are a result of cell-to-cell cytoplasmic fusion to spread the virus (Sattentau, 2008), recently reported for SARS-CoV-2 infected cells as well (Sanders et al., 2020).
Each organelle is color coded as: nucleus - blue, mitochondria - green, lipid bodies - yellow, vesicles - cyan, virions - magenta, DMVs - red, CMs - orange, and large compartment - gray.
Quantitative analysis of SARS-CoV-2-induced cellular remodeling
Taking advantage of rapid whole-cell imaging with SXT, we performed a quantitative analysis of the morphological remodeling of SARS-CoV-2-infected HEK293T-ACE2 cells at 6 and 24 hpi, see Figure 5. Compared with mock cells, infected cells display alterations in morphology and organization of mitochondria, lipid bodies, vesicles, and virus-associated organelles. The total volume of mitochondria networks increased at 6 hpi by 21% and remained unchanged at 24 hpi. Recent observations via cryoFIB/SEM have reported that SARS-CoV-2 leads to disruption of the mitochondria network (Mendonca et al., 2020). We observed only local alterations of mitochondria in proximity to viral replication organelles (see Figure 4D), consistent with a recent report (Cortese et al., 2020). On a whole-cell level (see Figure 5A), we did not find any significant difference in the volume of mitochondria (mock: 3.81% ± 1.52%, 6 hpi: 4.61% ± 0.94%, 24 hpi: 3.78% ± 1.01%), X-ray LAC (mock: 0.30 ± 0.01 μm−1, 6 hpi: 0.32 ± 0.01 μm−1, 24 hpi: 0.32 ± 0.02 μm−1), number of isolated mitochondria per cell volume (mock: 5.36 ± 1.47∗10−6, 6 hpi: 5.97 ± 1.24∗10−6, 24 hpi: 4.08 ± 2.29∗10−6) or changes in shape, measured as surface to volume ratio (mock: 0.29% ± 0.06%, 6 hpi: 0.39% ± 0.13%, 24 hpi: 0.21% ± 0.06%). Similarly, we did not observe differences in the nucleus volume or DNA packing within the nucleus in infected cells (see Figure S5).
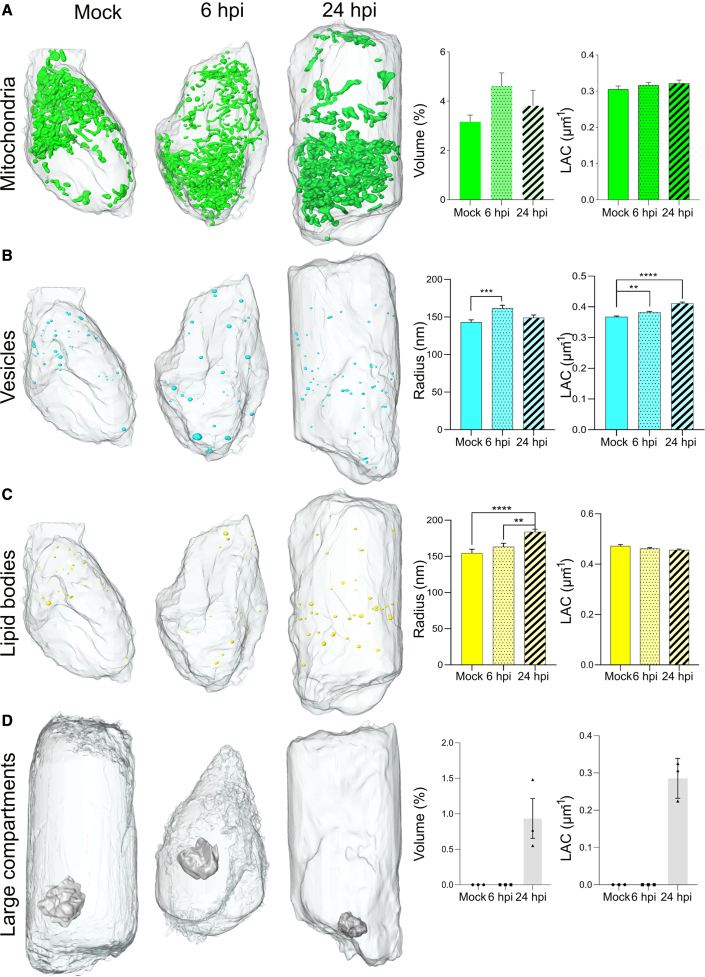
Figure 5.
Quantitative analysis of changes in organelle morphology and composition at 6 and 24 hpi as compared with uninfected HEK293T-ACE2 cells
(A–D) Exemplarily 3D renderings of (A) mitochondria, (B) lipid bodies, (C) vesicles, and (D) apoptotic bodies, and their corresponding quantitative analysis of total volume (normalized to total cell volume) and soft X-ray absorption coefficient (LAC). Each organelle is color coded as follows: mitochondria - green, lipid bodies - yellow, vesicles - cyan, large apoptotic bodies - lavender. Statistical significance is reported for lipid droplet radius in mock cells compared with 24 hpi (∗∗∗∗p < 0.0001) and 6 hpi cells compared with 24 hpi (∗∗p = 0.0042). Similarly, we report a significance difference in the vesicle radius at 6 hpi compared with mock cells (∗∗∗p = 0.0006) and for the X-ray absorption of the vesicles in mock cells compared with 24 hpi (∗∗∗∗p < 0.001) and compared with 6 hpi (∗∗p < 0.0020) (Dunnett’s multiple comparison test). The statistical analysis for all organelles is performed based on Tukey’s multiple comparison test.
We observed an increase in the total volume of lipid bodies in cells at 24 hpi (0.044% ± 0.070%, total volume normalized by cell volume) compared with 6 hpi (0.010% ± 0.004%) and mock (0.012% ± 0.007%) cells with up to n = 75 lipid bodies in individual cell. Together with total volume, we observed an increase in average radius of the bodies (mock: 154 ± 38 nm; 6 hpi: 155 ± 33 nm; 184 ± 45 nm), but no changes in the chemical composition are detected (X-ray absorption coefficient in mock: 0.48 ± 0.06 μm−1; 6 hpi: 0.50 ± 0.04 μm−1; 24 hpi: 0.50 ± 0.05 μm−1; Figure 5B). Such an increased accumulation of lipid bodies at later infection stages are signs of a possible inflammatory response, which was recently described in SARS-CoV-2-infected monocytes (Dias et al., 2020).
Compared with lipid bodies, the total volume of vesicles in infected cells at first (6 hpi) increased and then dropped at 24 hpi as compared with mock cells (mock: 1.01% ± 0.51∗10−3%, 6 hpi: 1.88% ± 1.22∗10−3%, 24 hpi: 1.26% ± 1.22∗10−3%, total vesicle volume normalized by cell volume), see Figure 5B. In addition, we observed a gradual increase in chemical density (mock: 0.36 ± 0.04 μm−1, 6 hpi: 0.37 ± 0.04 μm−1, 24 hpi: 0.40 ± 0.04 μm−1 LAC values), indicating a denser biochemical content toward 24 hpi. The behavior can be related to the release of the virus to neighbor cells via endocytic pathway and has been previously described in relation to SARS-CoV-2 (Ghosh et al., 2020).
Finally, we found single large compartments in every segmented cell at 24 hpi (n = 3), see Figure 5D. Though no membrane blebbing or membrane protrusion was visible, we suspect that presence of large compartments (Caruso and Poon, 2018), release of vesicles, accumulation of lipids, and convoluted membrane structures visualized by SXT in SARS-CoV-2-infected cells are signs of apoptosis at 24 hpi.
Discussion
Emerging viruses, for example SARS-CoV-2, present challenges for the scientific community at multiple levels and call for development and adaptation of new methods, including high-throughput imaging techniques. Because viruses can highjack different cell organelles, high-throughput imaging of all organelles in whole cells is needed to understand the pathological changes induced by viral infection and to devise novel concepts for antiviral therapy. Although super-resolution fluorescence and FIB/SEM have proven to be powerful for 3D imaging of viruses in cells and tissues (Castelletto and Boretti, 2021; Grove, 2014; Hoffman et al., 2020; Richert-Pöggeler et al., 2019; Romero-Brey and Bartenschlager, 2015; Weiner and Enninga, 2019), these methods typically require extensive sample preparation and time-consuming imaging experiments (a 100-μm3 sample at 10-nm spatial resolution would take about 8 years to complete [Xu et al., 2017]) plus data interpretation.
Our group and other labs are attempting to overcome these limitations through the development of a single-cell 3D imaging technique, based on SXT, that can examine intact cells without labeling or chemical fixation, at high throughput and spatial resolution. While SXT has been successfully applied to study various cell types (Guo and Larabell, 2019; Harkiolaki et al., 2018), imaging of infected cells has been limited due to potential changes of X-ray absorption by fixatives that are required for neutralization of viruses and small partial volumetric imaging of cells on flat substrates.
Here, we demonstrate that soft X-ray microscopy is a rapid imaging technique that can visualize SARS-CoV-2–induced cellular remodeling at the whole-cell level. We show that SXT is capable of imaging chemically (aldehyde)-fixed cells in a quantitative manner and, similar to native-state cells, enables identification of organelles based on their chemical density. To ensure the 3D reconstruction and visualization of the whole cell with isotropic resolution, we have imaged cells in glass capillaries with full rotation of the specimen during data collection. Thanks to the large-volume imaging of 16 μm × 16 μm × 16 μm, we could visualize the cell composition (nucleus, mitochondria, lipid bodies and intracellular vesicles) of three different cell types (Calu-3, A549-ACE2, HEK293T-ACE2). In addition, we were able to visualize virus-induced alterations, including spherical membrane vesicles and highly convoluted structures. The combination of rapid imaging (5 min per tomogram), large volume, and quantitative X-ray absorption coefficients, we conducted a quantitative analysis of cellular organelles at 6 and 24 hpi as compared with uninfected cells.
Limitations of the study
Currently, the main drawback of SXT is limited access provided by only five synchrotron facilities around the world (Ekman et al., 2019; Harkiolaki et al., 2018). Recent developments of laboratory-based SXT (Hertz et al., 2012; Kördel et al., 2020; McEnroe et al., 2017) and its first commercial prototypes (Fahy et al., 2021) will open up the possibility for systematic day-to-day imaging experiments. Another limitation is a need to enzymatically detach cells and place them inside glass capillaries for optimal full-cell tomography. Although the absolute number of organelles will barely change within such a short time, organelles might be displaced with respect to each other, limiting colocalization analysis to the use of specialized cryo-light microscopes (Le Gros et al., 2009). Recently, many of the features (such as insulin granule clusters) visible by electron microscopy have been reproducibly found in cells after enzymatic detachment and imaging inside glass capillaries by SXT (White et al., 2020). The effect of enzymatic detachment will be further investigated by comparing SXT data of adherent cells on flat (Carzaniga et al., 2014a; Ekman et al., 2019) and cylindrical holders. In addition, in our current workflow, segmentation is primarily a manual task, which is tedious, time-consuming, and subject to human errors or bias. This issue can be alleviated by use of a semiautomatic segmentation pipeline (Lösel and Heuveline, 2016; Lösel et al., 2020) or development of fully automatic image analysis pipelines for SXT data (Ekman et al., 2020; Pelt and Sethian, 2018; Plautz et al., 2017), such that whole-cell phenotypic changes can be analyzed in statistically significant numbers to allow analysis of virus-cell interactions and efficacy of antiviral drugs.
To conclude, we believe that the combination of minimal sample preparation, quantitative identification of cellular organelles in native and fixed cells, and rapid imaging of whole cells will meet the need for systematic analysis of virus-induced cellular remodeling with statistically significant sample sizes. Moreover, the cells for imaging with SXT can be pre-sorted based on different stages of viral infection (Aho et al., 2019) and visualized with fluorescence microscopy (Carzaniga et al., 2014b; Hagen et al., 2012; Pereiro, 2019; Smith et al., 2014), including super-resolution techniques (Kounatidis et al., 2020). Such combinatorial and correlative studies with other imaging modalities will help to visualize single virions or individual biomolecules within the 3D structure of cells. This approach also will be useful for statistical analyses of the structural consequences of potential pharmaceutical agents being developed for treatment of viral infections.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SARS-CoV-2 (isolate Bavpat1/2020) | European Virology Archive | Ref-SKU: 026V-03883 |
| Chemicals, peptides, and recombinant proteins | ||
| PBS | Thermo Scientific | Cat.#10010023 |
| Paraformaldehyde | Thermo Scientific | Cat.#J61899-AP |
| Formaldehyde | Thermo Scientific | Cat.#28906 |
| Deposited data | ||
| Raw data | This paper | Mendeley Data: https://data.mendeley.com/datasets/gkc557ssvp/2 |
| TEM data | Cortese et al. (2020) | EMPIAR: 10490 |
| Experimental models: Cell lines | ||
| Calu-3 | ATCC | Cat.#HTB-55 |
| A549-ACE2 | Klein et al. (2020) | N/A |
| HEK293T-ACE2 | Gift by Prof. Becker | N/A |
| Human B lymphocytes | Coriell Institute of Medical Research | GM12878 |
| Software and algorithms | ||
| ImageJ – Fiji 1.53 c | Schneider et al. (2012) | https://imagej.nih.gov/ij/ |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Amira 2019.1 | Thermo Scientific | https://thermofisher.com/amira-avizo |
| IMOD – 4.10.42 | Kremer et al. (1996) | https://bio3d.colorado.edu/imod/ |
| AREC reconstruction | Parkinson et al. (2012) | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Venera Weinhardt (venera.weinhardt@cos.uni-heidelberg.de).
Materials availability
This study did not generate new reagents.
Experimental model and subject details
Cell culture
The human B lymphocytes (GM12878) were purchased from the NGIMS Human Genetics Cell Repository, Cornell Institute of Medical Research (Camden, NJ). The B cells were maintained as suspension cultures at 37°C in Advanced RPMI-1640 medium supplemented with 15% of fetal bovine serum (FBS), L-glutamine, (Gibco™, Carlsbad, CA), and 5% CO2. The medium was refreshed every two to three days to maintain a cell density of 106 ml−1.
A549 cells were obtained from ATCC (CRL-1586 and CCL-185, respectively) and were grown in Dulbecco’s modified Eagle medium (DMEM) containing 2 mM L-glutamine, non-essential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin and 10% fetal calf serum (DMEM complete). Calu-3 cells (ATCC, HTB-55) were a kind gift from Dr. Manfred Frey, Mannheim, and were cultured in DMEM complete with 20% fetal calf serum. HEK293-ACE2 cells were a kind gift from Prof. Stephan Becker at University of Marburg. A549 cells stably expressing ACE2 (A549-ACE2) were generated as described recently (Klein et al., 2020). SARS-CoV-2 isolate Bavpat1/2020 was kindly provided by Prof. Christian Drosten (Charité Berlin, Germany) through the European Virology Archive (Ref-SKU: 026V-03883) at passage 2. Working virus stocks were produced as described previously (Klein et al., 2020) by passaging the virus two times in VeroE6 cells.
Method details
SARS-CoV-2 infection
Cells were seeded in 6-well plates to achieve 80% confluency on the day of infection. For identification of virus-related organelles, cells were infected with SARS-CoV-2 at multiplicity of infection (MOI) of 0.5 in a final volume of 1 ml per well (Figure S4A) and imaged at 6 and 26hpi. For the kinetic study, infection was synchronized by using MOI = 10 or 20 for HEK293-ACE2 and Calu-3 cells, respectively. At different time post-infection, cells were detached by trypsinization, washed twice in PBS(1X) and then fixed as described below.
Fixation and freezing
Human B cell culture was pelleted at 300 × g for 10 min and washed in PBS(1X) and then fixed. They were firstly fixed in a solution of 2.5% of glutaraldehyde and later resuspend in a solution of 6% of formaldehyde. Cells were incubated at room temperature for 30 min in both cases and finally washed in PBS(1X) before the freezing. 1.5 μl of cells was loaded into thin-wall (200 nm) glass capillary and frozen by plunging in liquid propane. Native human B cells were spun down by centrifugation at 300 × g for 10 min and resuspended in 20 μl of PBS(1X). After checking the density, cells were loaded in thin-walled glass capillaries and rapidly plunged-frozen.
HEK-293T-ACE2, A549-ACE2, and Calu-3 cells were washed and later resuspended in fresh PBS(1X). 1.5 μl of each batch of cells was then loaded into thin-walled glass capillaries and rapidly plunge-frozen. All cells were stored in liquid nitrogen until data collection.
Imaging
Projection images were collected at 517 eV using XM-2, the National Center for X-ray Tomography soft X-ray microscope at the Advanced Light Source of Lawrence Berkeley National Laboratory; the microscope was equipped with a 50-nm resolution defining objective lens. During data collection, the cells were maintained in a stream of helium gas cooled to liquid nitrogen temperatures (McDermott et al., 2009), which allows the collection of projection images while reducing the effects of exposure to radiation. Projection images were collected sequentially around a rotation axis of 180°, with 2° increments. Depending on the synchrotron ring current, an exposure time of 250 and 350 ms was used.
Quantification and statistical analysis
All statistical analyses were undertaken using Prism (version 8.0, GraphPad Software). t test and analysis of variance (ANOVA) analyses were performed to determine statistical significance among two or three conditions, respectively. Post hoc analysis was performed to identify difference in means for specific conditions.
3D reconstructions
2D projections images were normalized by the reference image (without a sample), automatically aligned by image cross-correlation and reconstructed in 3D volumes by the software package AREC3D (Parkinson et al., 2012). The reconstructions do not require fiducial markers or manual interaction with the software and gives virtually real-time initial 3D results with the final 3D volume available within 5 minutes. Selected datasets (Figures 2 and 3) were additionally reconstructed by incorporating point spread function (PSF) of the microscope (Ekman et al., 2018). For further analysis and quantitative comparison for all conditions, all data were normalized and transferred to linear absorption coefficient by accounting for pixel size.
Simulations
To simulate the effect of limited rotation angle on SXT data, we have used part of 2D projection images obtained for the B cell shown in Figure 1A, corresponding to the ±65° (total 130°) tilt angles. To include a “shadowing effect” of the flat support, each 2D projection was multiplied by the transmission function of a flat grid: Img_plate(θ) = Img(θ)∗exp[-μt(θ)]. Here, μ is the absorption coefficient of a support grid (formvar with carbon coating), and t is the thickness of the support for each projection angle θ.
The X-ray dose calculations are done as in Weiß et al. (2000) by cumulatively adding local dose from each illumination angle and by propagating the intensity obtained from the flat field image through the reconstructed sample. We used X-rays with an energy of 543 eV, and a Fano factor of 0.11 for the CCD. The calculated photon count is scaled by the optical transmission of the microscope to account for the reduction of dose between the sample and the CCD. For this, an X-ray objective lens, with transmission of 0.125 and a Si3N4 membrane with transmission of 0.67863 have been considered. Similar to 3D reconstructions, X-ray dose calculations were performed for different rotation angles, that is full rotation of 360°, 180°, ±65° and ±65° rotation with gradual increase in exposure time, see Figure S2C.
Organelle segmentation
All cells were manually segmented using the software Amira 2019.1. The identification of each type of organelles was based on previously published data (McDermott et al., 2009; White et al., 2020). Nucleus was firstly manually segmented at first in every 10 orthoslices; each orthoslice was consequently interpolated to reconstruct the 3D label field. Heterochromatin, mitochondria, lipids, and structures involved in the viral replication were both manually and semi-automatic segmented, setting the threshold for the absorption at different values as reported in literature (McDermott et al., 2009). Specifically, lipid bodies were identified setting the threshold between 0.48 and 0.90 μm−1, with average value set at about 0.58 μm−1 and by thanks to their elongated shape; vesicles were labeled according to the lower X-ray absorption, with threshold sets between 0.25 and 0.48 μm−1, and average value around 0.38 μm−1. The large apoptotic body was identified by its natural contrast and the presence of an external rim in contact with the cytosol; whereas the CMs were recognized due to their curved and hollow shape. DMVs were outlined based on X-ray absorption values (typically higher than mitochondria) and typical “donut”-like shape. Aggregates of virions were found at the cell surface and were segmented based on X-ray absorption values. Finally, the ER was manually segmented by outlining features in each virtual slice.
Blob analysis
To visualize ultrastructure of large compartments, we applied series of Laplacian filters on the images. We used the Laplace of the Gaussian susceptible to spherical structures between 100 and 200 nm in radius. The maximum of the scale-normalized Laplacian operator t(Lxx + Lyy + Lzz), where t is the standard deviations of the Gaussian filter was used to visualize the substructure of the compartment, as shown in Figure S4C.
Acknowledgments
We thank Dr. Manfred Frey, Mannheim; Prof. Stephan Becker, University of Marburg; and Prof. Christian Drosten, Charité Berlin, Germany, for their kind gift of cell cultures used in this study. V.W. is supported by German Research Foundation research fellowship WE 6221/2-1. The National Center for X-ray Tomography is supported by NIH NIGMS (P41GM103445, P30GM138441) and the DOE’s Office of Biological and Environmental Research (DE-AC02-5CH11231). V.W. was supported by the ERC Synergy Grant IndiGene (no. 810172). V.W and R.B are supported by the CoCID project (no. 101017116) funded within EU Research and Innovation Act.
Author contributions
V.L. and J-H.C. performed experiments and formal analysis. M.C. provided samples. A.E. and M A.L.G. provided methodology, software, and data curation. R.B. and M.C. provided expertise and feedback. V.W., R.B., and C.L. conceived experiments and secured funding. All authors wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 28, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100117.
Supplemental information
Data and code availability
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
References
- Aho V., Myllys M., Ruokolainen V., Hakanen S., Mäntylä E., Virtanen J., Hukkanen V., Kühn T., Timonen J., Mattila K., et al. Chromatin organization regulates viral egress dynamics. Sci. Rep. 2017;7:3692. doi: 10.1038/s41598-017-03630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aho V., Mäntylä E., Ekman A., Hakanen S., Mattola S., Chen J.-H., Weinhardt V., Ruokolainen V., Sodeik B., Larabell C., et al. Quantitative microscopy reveals stepwise alteration of chromatin structure during herpesvirus infection. Viruses. 2019;11:935. doi: 10.3390/v11100935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann O., Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- Boeynaems S., Holehouse A.S., Weinhardt V., Kovacs D., Lindt J.V., Larabell C., Bosch L.V.D., Das R., Tompa P.S., Pappu R.V., et al. Spontaneous driving forces give rise to protein−RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. 2019;116:7889–7898. doi: 10.1073/pnas.1821038116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrascosa J.L., Chichón F.J., Pereiro E., Rodríguez M.J., Fernández J.J., Esteban M., Heim S., Guttmann P., Schneider G. Cryo-X-ray tomography of vaccinia virus membranes and inner compartments. J. Struct. Biol. 2009;168:234–239. doi: 10.1016/j.jsb.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Caruso S., Poon I.K.H. Apoptotic cell-derived extracellular vesicles: more than just debris. Front. Immunol. 2018;9:1486. doi: 10.3389/fimmu.2018.01486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carzaniga R., Domart M.-C., Collinson L.M., Duke E. Cryo-soft X-ray tomography: a journey into the world of the native-state cell. Protoplasma. 2014;251:449–458. doi: 10.1007/s00709-013-0583-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carzaniga R., Domart M.-C., Duke E., Collinson L.M. Correlative cryo-fluorescence and cryo-soft X-ray tomography of adherent cells at European synchrotrons. Methods Cell Biol. 2014;124:151–178. doi: 10.1016/B978-0-12-801075-4.00008-2. [DOI] [PubMed] [Google Scholar]
- Castelletto S., Boretti A. Viral particle imaging by super-resolution fluorescence microscopy. Chem. Phys. Impact. 2021;2:100013. [Google Scholar]
- Chiappi M., Conesa J.J., Pereiro E., Sorzano C.O.S., Rodríguez M.J., Henzler K., Schneider G., Chichón F.J., Carrascosa J.L. Cryo-soft X-ray tomography as a quantitative three-dimensional tool to model nanoparticle:cell interaction. J. Nanobiotechnol. 2016;14:15. doi: 10.1186/s12951-016-0170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichón F.J., Rodríguez M.J., Pereiro E., Chiappi M., Perdiguero B., Guttmann P., Werner S., Rehbein S., Schneider G., Esteban M., et al. Cryo X-ray nano-tomography of vaccinia virus infected cells. J. Struct. Biol. 2012;177:202–211. doi: 10.1016/j.jsb.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquin B.P., Do M., McDermott G., Walters A.D., Myllys M., Smith E.A., Cohen-Fix O., Gros M.A.L., Larabell C.A. Putting molecules in their place. J. Cell. Biochem. 2014;115:209–216. doi: 10.1002/jcb.24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M., Lee J.-Y., Cerikan B., Neufeldt C.J., Oorschot V.M.J., Köhrer S., Hennies J., Schieber N.L., Ronchi P., Mizzon G., et al. Integrative imaging reveals SARS-CoV-2 induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28:853–866. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow M.C., Zhang Y., Cinquin B.P., Smith E.A., Boudreau R., Rochat R.H., Schmid M.F., Xia Y., Larabell C.A., Chiu W. Visualizing red blood cell sickling and the effects of inhibition of sphingosine kinase 1 using soft X-ray tomography. J. Cell Sci. 2016;129:3511–3517. doi: 10.1242/jcs.189225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S.da S.G., Soares V.C., Ferreira A.C., Sacramento C.Q., Fintelman-Rodrigues N., Temerozo J.R., Teixeira L., Barreto E., Mattos M., Freitas C.S.de, et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. BioRxiv. 2020 doi: 10.1101/2020.08.22.262733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmair S., Hodson M.P., Quek L.-E., Timmins N.E., Gray P., Nielsen L.K. A multi-omics analysis of recombinant protein production in Hek293 cells. PLoS One. 2012;7:e43394. doi: 10.1371/journal.pone.0043394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolskiy A.A., Grishchenko I.V., Yudkin D.V. Cell cultures for virology: usability, advantages, and prospects. Int. J. Mol. Sci. 2020;21:7978. doi: 10.3390/ijms21217978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke E.M.H., Razi M., Weston A., Guttmann P., Werner S., Henzler K., Schneider G., Tooze S.A., Collinson L.M. Imaging endosomes and autophagosomes in whole mammalian cells using correlative cryo-fluorescence and cryo-soft X-ray microscopy (cryo-CLXM) Ultramicroscopy. 2014;143:77–87. doi: 10.1016/j.ultramic.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman A., Weinhardt V., Chen J.-H., McDermott G., Le Gros M.A., Larabell C. PSF correction in soft X-ray tomography. J. Struct. Biol. 2018;204:9–18. doi: 10.1016/j.jsb.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman A., Chen J.-H., Weinhardt V., Do M., McDermott G., Gros M.A.L., Larabell C.A. In: Synchrotron Light Sources and Free-Electron Lasers: Accelerator Physics, Instrumentation and Science Applications. Jaeschke E., Khan S., Schneider J.R., Hastings J.B., editors. Springer International Publishing; 2019. Putting molecules in the picture: using correlated light microscopy and soft X-ray tomography to study cells; pp. 1–32. [Google Scholar]
- Ekman A., Chen J.-H., Dermott G.M., Gros M.A.L., Larabell C. Task based semantic segmentation of soft X-ray CT images using 3D convolutional neural networks. Microsc. Microanal. 2020;26:3152–3154. [Google Scholar]
- Elgass K.D., Smith E.A., LeGros M.A., Larabell C.A., Ryan M.T. Analysis of ER-mitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. J. Cell Sci. 2015;128:2795–2804. doi: 10.1242/jcs.169136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy K., Weinhardt V., Vihinen-Ranta M., Fletcher N., Skoko D., Pereiro E., Gastaminza P., Bartenschlager R., Scholz D., Ekman A., et al. Compact cell imaging device (CoCID) to provide insights into the cellular origins of viral infections. J. Phys. Photonics. 2021;3:031002. [Google Scholar]
- Finkbeiner W.E., Zlock L.T., Carrier S.D., Chun S.Y., Watt L., Chow A. Expression of airway secretory epithelial functions by lung carcinoma cells. in vitro cell. Dev. Biol. Anim. 1995;31:379–386. doi: 10.1007/BF02634287. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Dellibovi-Ragheb T.A., Kerviel A., Pak E., Qiu Q., Fisher M., Takvorian P.M., Bleck C., Hsu V.W., Fehr A.R., et al. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell. 2020;183:1520–1535.e14. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen J., Conesa J.J., Valcárcel R., Pereiro E. The cellular landscape by cryo soft X-ray tomography. Biophys. Rev. 2019;11:611–619. doi: 10.1007/s12551-019-00567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J. Super-resolution microscopy: a virus’ eye view of the cell. Viruses. 2014;6:1365–1378. doi: 10.3390/v6031365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Larabell C.A. Soft X-ray tomography: virtual sculptures from cell cultures. Curr. Opin. Struct. Biol. 2019;58:324–332. doi: 10.1016/j.sbi.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen C., Guttmann P., Klupp B., Werner S., Rehbein S., Mettenleiter T.C., Schneider G., Grünewald K. Correlative VIS-fluorescence and soft X-ray cryo-microscopy/tomography of adherent cells. J. Struct. Biol. 2012;177:193–201. doi: 10.1016/j.jsb.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen E., Knoechel C., Dearnley M., Dixon M.W.A., Le Gros M., Larabell C., Tilley L. Soft X-ray microscopy analysis of cell volume and hemoglobin content in erythrocytes infected with asexual and sexual stages of Plasmodium falciparum. J. Struct. Biol. 2012;177:224–232. doi: 10.1016/j.jsb.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., et al. Isolation and characterization of SARS-CoV-2 from the first US COVID-19 patient. BioRxiv. 2020 doi: 10.1101/2020.03.02.972935. [DOI] [Google Scholar]
- Harkiolaki M., Darrow M.C., Spink M.C., Kosior E., Dent K., Duke E. Cryo-soft X-ray tomography: using soft X-rays to explore the ultrastructure of whole cells. Emerg. Top. Life Sci. 2018;2:81–92. doi: 10.1042/ETLS20170086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz H.M., von Hofsten O., Bertilson M., Vogt U., Holmberg A., Reinspach J., Martz D., Selin M., Christakou A.E., Jerlström-Hultqvist J., et al. Laboratory cryo soft X-ray microscopy. J. Struct. Biol. 2012;177:267–272. doi: 10.1016/j.jsb.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Hobro A.J., Smith N.I. An evaluation of fixation methods: spatial and compositional cellular changes observed by Raman imaging. Vib. Spectrosc. 2017;91:31–45. [Google Scholar]
- Hoffman D.P., Shtengel G., Xu C.S., Campbell K.R., Freeman M., Wang L., Milkie D.E., Pasolli H.A., Iyer N., Bogovic J.A., et al. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science. 2020;367:eaaz5357. doi: 10.1126/science.aaz5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel E., Guttmann P., Werner S., Tarek B., Schneider G., Kunz M., Frangakis A.S., Westermann B. 3D ultrastructural organization of whole chlamydomonas reinhardtii cells studied by nanoscale soft X-ray tomography. PLoS One. 2012;7:e53293. doi: 10.1371/journal.pone.0053293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J., Helenius A. Endosome maturation. EMBO J. 2011;30:3481. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen C., Kirz J. X-ray microscopy with synchrotron radiation. Nat. Struct. Biol. 1998;5:650–653. doi: 10.1038/1341. [DOI] [PubMed] [Google Scholar]
- Jearanaikoon S., Abraham-Peskir J.V. An X-ray microscopy perspective on the effect of glutaraldehyde fixation on cells. J. Microsc. 2005;218:185–192. doi: 10.1111/j.1365-2818.2005.01472.x. [DOI] [PubMed] [Google Scholar]
- Kellenberger E., Johansen R., Maeder M., Bohrmann B., Stauffer E., Villiger W. Artefacts and morphological changes during chemical fixation. J. Microsc. 1992;168:181–201. doi: 10.1111/j.1365-2818.1992.tb03260.x. [DOI] [PubMed] [Google Scholar]
- Kepsutlu B., Wycisk V., Achazi K., Kapishnikov S., Pérez-Berná A.J., Guttmann P., Cossmer A., Pereiro E., Ewers H., Ballauff M., et al. Cells undergo major changes in the quantity of cytoplasmic organelles after uptake of gold nanoparticles with biologically relevant surface coatings. ACS Nano. 2020;14:2248–2264. doi: 10.1021/acsnano.9b09264. [DOI] [PubMed] [Google Scholar]
- Kim K.W., Kwon Y., Nam K.-Y., Lim J.-H., Kim K.-G., Chon K.S., Kim B.H., Kim D.E., Kim J., Ahn B.N., et al. Compact soft X-ray transmission microscopy with sub-50 nm spatial resolution. Phys. Med. Biol. 2006;51:N99. doi: 10.1088/0031-9155/51/6/N01. [DOI] [PubMed] [Google Scholar]
- Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J., Raposo G. The complex ultrastructure of the Endolysosomal system. Cold Spring Harb. Perspect. Biol. 2014;6:a016857. doi: 10.1101/cshperspect.a016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., van den Worm S.H.E., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kördel M., Kördel M., Dehlinger A., Dehlinger A., Dehlinger A., Seim C., Seim C., Vogt U., Fogelqvist E., Fogelqvist E., et al. Laboratory water-window X-ray microscopy. Optica. 2020;7:658–674. [Google Scholar]
- Kounatidis I., Stanifer M.L., Phillips M.A., Paul-Gilloteaux P., Heiligenstein X., Wang H., Okolo C.A., Fish T.M., Spink M.C., Stuart D.I., et al. 3D correlative cryo-structured illumination fluorescence and soft X-ray microscopy elucidates reovirus intracellular release pathway. Cell. 2020;182:515–530.e17. doi: 10.1016/j.cell.2020.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lambert D.W. In: Molecular Biology of the SARS-Coronavirus. Lal S., editor. Springer; 2009. The cell biology of the sars coronavirus receptor, angiotensin-converting enzyme 2; pp. 23–30. [Google Scholar]
- Larabell C.A., Le Gros M.A. X-ray tomography generates 3-D reconstructions of the yeast, Saccharomyces cerevisiae, at 60-nm resolution. Mol. Biol. Cell. 2003;15:957–962. doi: 10.1091/mbc.E03-07-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larabell C.A., Nugent K.A. Imaging cellular architecture with X-rays. Curr. Opin. Struct. Biol. 2010;20:623–631. doi: 10.1016/j.sbi.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gros M.A., McDermott G., Uchida M., Knoechel C.G., Larabell C.A. High-aperture cryogenic light microscopy. J. Microsc. 2009;235:1–8. doi: 10.1111/j.1365-2818.2009.03184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gros M.A., McDermott G., Cinquin B.P., Smith E.A., Do M., Chao W.L., Naulleau P.P., Larabell C.A. Biological soft X-ray tomography on beamline 2.1 at the advanced light source. J. Synchrotron Radiat. 2014;21:1370–1377. doi: 10.1107/S1600577514015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gros M.A., Clowney E.J., Magklara A., Yen A., Markenscoff-Papadimitriou E., Colquitt B., Myllys M., Kellis M., Lomvardas S., Larabell C.A. Soft X-ray tomography reveals gradual chromatin compaction and reorganization during neurogenesis in vivo. Cell Rep. 2016;17:2125–2136. doi: 10.1016/j.celrep.2016.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Kim E.W., Croteau D.L., Bohr V.A. Heterochromatin: an epigenetic point of view in aging. Exp. Mol. Med. 2020;52:1–9. doi: 10.1038/s12276-020-00497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Guan Y., Xiong Y., Zhang X., Liu G., Tian Y. Method for extending the depth of focus in X-ray microscopy. Opt. Express. 2017;25:7657–7667. doi: 10.1364/OE.25.007657. [DOI] [PubMed] [Google Scholar]
- Liu J., Li F., Chen L., Guan Y., Tian L., Xiong Y., Liu G., Tian Y. Quantitative imaging of Candida utilis and its organelles by soft X-ray Nano-CT. J. Microsc. 2018;270:64–70. doi: 10.1111/jmi.12650. [DOI] [PubMed] [Google Scholar]
- Longatti A., Orsi A., Tooze S.A. Autophagosome formation: not necessarily an inside job. Cell Res. 2010;20:1181–1184. doi: 10.1038/cr.2010.132. [DOI] [PubMed] [Google Scholar]
- Lösel P., Heuveline V. Medical Imaging 2016: Image Processing. International Society for Optics and Photonics; 2016. Enhancing a diffusion algorithm for 4D image segmentation using local information; p. 97842L. [Google Scholar]
- Lösel P.D., van de Kamp T., Jayme A., Ershov A., Faragó T., Pichler O., Tan Jerome N., Aadepu N., Bremer S., Chilingaryan S.A., et al. Introducing Biomedisa as an open-source online platform for biomedical image segmentation. Nat. Commun. 2020;11:5577. doi: 10.1038/s41467-020-19303-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Do M., Gros M.A.L., Peskin C.S., Larabell C.A., Mori Y., Isaacson S.A. Strong intracellular signal inactivation produces sharper and more robust signaling from cell membrane to nucleus. PLoS Comput. Biol. 2020;16:e1008356. doi: 10.1371/journal.pcbi.1008356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott G., Le Gros M.A., Knoechel C.G., Uchida M., Larabell C.A. Soft X-ray tomography and cryogenic light microscopy: the cool combination in cellular imaging. Trends Cell Biol. 2009;19:587–595. doi: 10.1016/j.tcb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEnroe T., O’Reilly F., Sheridan P., Howard J., Byrne R., Rogers D., Rogers C., McCarthy C., Manzoni A., Skoko D., et al. Development of a commercial laboratory scale soft X-ray microscope. Microsc. Microanal. 2017;23:982–983. [Google Scholar]
- Mendonca L., Howe A., Gilchrist J.B., Sun D., Knight M., Zanetti-Domingues L.C., Bateman B., Krebs A.-S., Chen L., Radecke J., et al. SARS-CoV-2 assembly and egress pathway revealed by correlative multi-modal multi-scale cryo-imaging. BioRxiv. 2020 doi: 10.1101/2020.11.05.370239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonça L., Howe A., Gilchrist J.B., Sheng Y., Sun D., Knight M.L., Zanetti-Domingues L.C., Bateman B., Krebs A.-S., Chen L., et al. Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress. Nat. Commun. 2021;12:4629. doi: 10.1038/s41467-021-24887-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielanczyk L., Matysiak N., Michalski M., Buldak R., Wojnicz R. Closer to the native state. Critical evaluation of cryo-techniques for transmission electron microscopy: preparation of biological samples. Folia Histochem. Cytobiol. 2014;52:1–17. doi: 10.5603/FHC.2014.0001. [DOI] [PubMed] [Google Scholar]
- Miller S.E., Goldsmith C.S. Caution in identifying coronaviruses by electron microscopy. J. Am. Soc. Nephrol. 2020;31:2223–2224. doi: 10.1681/ASN.2020050755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllys M., Ruokolainen V., Aho V., Smith E.A., Hakanen S., Peri P., Salvetti A., Timonen J., Hukkanen V., Larabell C.A., et al. Herpes simplex virus 1 induces egress channels through marginalized host chromatin. Sci. Rep. 2016;6:28844. doi: 10.1038/srep28844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niclis J.C., Murphy S.V., Parkinson D.Y., Zedan A., Sathananthan A.H., Cram D.S., Heraud P. Three-dimensional imaging of human stem cells using soft X-ray tomography. J. R. Soc. Interfaces. 2015;12:20150252. doi: 10.1098/rsif.2015.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y., Maeda T., Fujimoto T. Fixation and permeabilization protocol is critical for the immunolabeling of lipid droplet proteins. Histochem. Cell Biol. 2005;124:445–452. doi: 10.1007/s00418-005-0061-5. [DOI] [PubMed] [Google Scholar]
- Otón J., Pereiro E., Conesa J.J., Chichón F.J., Luque D., Rodríguez J.M., Pérez-Berná A.J., Sorzano C.O.S., Klukowska J., Herman G.T., et al. XTEND: Extending the depth of field in cryo soft X-ray tomography. Sci. Rep. 2017;7:45808. doi: 10.1038/srep45808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson D.Y., Knoechel C., Yang C., Larabell C.A., Le Gros M.A. Automatic alignment and reconstruction of images for soft X-ray tomography. J. Struct. Biol. 2012;177:259–266. doi: 10.1016/j.jsb.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelt D.M., Sethian J.A. A mixed-scale dense convolutional neural network for image analysis. Proc. Natl. Acad. Sci. U S A. 2018;115:254–259. doi: 10.1073/pnas.1715832114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereiro E. Correlative cryo-soft X-ray tomography of cells. Biophys. Rev. 2019;11:529–530. doi: 10.1007/s12551-019-00560-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Berná A.J., Rodríguez M.J., Chichón F.J., Friesland M.F., Sorrentino A., Carrascosa J.L., Pereiro E., Gastaminza P. Structural changes in cells imaged by soft X-ray cryo-tomography during hepatitis C virus infection. ACS Nano. 2016;10:6597–6611. doi: 10.1021/acsnano.6b01374. [DOI] [PubMed] [Google Scholar]
- Plautz T., Boudreau R., Chen J.-H., Ekman A., LeGros M., McDermott G., Larabell C. Progress toward automatic segmentation of soft X-ray tomograms using convolutional neural networks. Microsc. Microanal. 2017;23:984–985. [Google Scholar]
- Richert-Pöggeler K.R., Franzke K., Hipp K., Kleespies R.G. Electron microscopy methods for virus diagnosis and high resolution analysis of viruses. Front. Microbiol. 2019;9:3255. doi: 10.3389/fmicb.2018.03255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I., Bartenschlager R. Viral infection at high magnification: 3D electron microscopy methods to analyze the architecture of infected cells. Viruses. 2015;7:6316–6345. doi: 10.3390/v7122940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M.S., Gallaher S.D., Westcott D.J., Iwai M., Louie K.B., Mueller M., Walter A., Foflonker F., Bowen B.P., Ataii N.N., et al. Regulation of oxygenic photosynthesis during trophic transitions in the green alga Chromochloris zofingiensis. Plant Cell. 2019;31:579–601. doi: 10.1105/tpc.18.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D.W., Jumper C.C., Ackerman P.J., Bracha D., Donlic A., Kim H., Kenney D., Castello-Serrano I., Suzuki S., Tamura T., et al. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. BioRxiv. 2020 doi: 10.1101/2020.12.14.422737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau Q. Avoiding the void: cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 2008;6:815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G., Niemann B., Guttmann P., Rudolph D., Schmahl G. Cryo X-ray microscopy. Synchrotron Radiat. News. 1995;8:19–28. [Google Scholar]
- Schneider G., Guttmann P., Heim S., Rehbein S., Mueller F., Nagashima K., Heymann J.B., Müller W.G., McNally J.G. Three-dimensional cellular ultrastructure resolved by X-ray microscopy. Nat. Methods. 2010;7:985–987. doi: 10.1038/nmeth.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D.S., Blower M.D. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell. Mol. Life Sci. 2016;73:79–94. doi: 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selin M., Fogelqvist E., Werner S., Hertz H.M. Tomographic reconstruction in soft x-ray microscopy using focus-stack back-projection. Opt. Lett. 2015;40:2201–2204. doi: 10.1364/OL.40.002201. [DOI] [PubMed] [Google Scholar]
- Shibata Y., Shemesh T., Prinz W.A., Palazzo A.F., Kozlov M.M., Rapoport T.A. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K.K., Chaubey G., Chen J.Y., Suravajhala P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol.-Cell Physiol. 2020;319:C258–C267. doi: 10.1152/ajpcell.00224.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.A., McDermott G., Do M., Leung K., Panning B., Le Gros M.A., Larabell C.A. Quantitatively imaging chromosomes by correlated cryo-fluorescence and soft X-ray tomographies. Biophys. J. 2014;107:1988–1996. doi: 10.1016/j.bpj.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Limpens R.W.A.L., Wilde A.H.de, Jong A.W.M.de, Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takman P.a.C., Stollberg H., Johansson G.A., Holmberg A., Lindblom M., Hertz H.M. High-resolution compact X-ray microscopy. J. Microsc. 2007;226:175–181. doi: 10.1111/j.1365-2818.2007.01765.x. [DOI] [PubMed] [Google Scholar]
- Uchida M., McDermott G., Wetzler M., Le Gros M.A., Myllys M., Knoechel C., Barron A.E., Larabell C.A. Soft X-ray tomography of phenotypic switching and the cellular response to antifungal peptoids in Candida albicans. Proc. Natl. Acad. Sci. U S A. 2009;106:19375–19380. doi: 10.1073/pnas.0906145106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida M., Sun Y., McDermott G., Knoechel C., Le Gros M.A., Parkinson D., Drubin D.G., Larabell C.A. Quantitative analysis of yeast internal architecture using soft X-ray tomography. Yeast. 2011;28:227–236. doi: 10.1002/yea.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters A.D., Amoateng K., Wang R., Chen J.-H., McDermott G., Larabell C.A., Gadal O., Cohen-Fix O. Nuclear envelope expansion in budding yeast is independent of cell growth and does not determine nuclear volume. Mol. Biol. Cell. 2019;30:131–145. doi: 10.1091/mbc.E18-04-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A., Enninga J. The pathogen–host interface in three dimensions: correlative FIB/SEM applications. Trends Microbiol. 2019;27:426–439. doi: 10.1016/j.tim.2018.11.011. [DOI] [PubMed] [Google Scholar]
- Weinhardt V., Chen J.-H., Ekman A.A., Guo J., Remesh S.G., Hammel M., McDermott G., Chao W., Oh S., Gros M.A.L., et al. Switchable resolution in soft X-ray tomography of single cells. PLoS One. 2020;15:e0227601. doi: 10.1371/journal.pone.0227601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiß D., Schneider G., Niemann B., Guttmann P., Rudolph D., Schmahl G. Computed tomography of cryogenic biological specimens based on X-ray microscopic images. Ultramicroscopy. 2000;84:185–197. doi: 10.1016/s0304-3991(00)00034-6. [DOI] [PubMed] [Google Scholar]
- West M., Zurek N., Hoenger A., Voeltz G.K. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K.L., Singla J., Loconte V., Chen J.-H., Ekman A., Sun L., Zhang X., Francis J.P., Li A., Lin W., et al. Visualizing subcellular rearrangements in intact β cells using soft X-ray tomography. Sci. Adv. 2020;6:eabc8262. doi: 10.1126/sciadv.abc8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G., Melia C.E., Snijder E.J., Bárcena M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020;28:1022–1033. doi: 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G., Limpens R.W.A.L., Zevenhoven-Dobbe J.C., Laugks U., Zheng S., Jong A.W.M., de Koning R.I., Agard D.A., Grünewald K., Koster A.J., et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395–1398. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C.S., Hayworth K.J., Lu Z., Grob P., Hassan A.M., García-Cerdán J.G., Niyogi K.K., Nogales E., Weinberg R.J., Hess H.F. Enhanced FIB-SEM systems for large-volume 3D imaging. Elife. 2017;6:e25916. doi: 10.7554/eLife.25916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Shen H.-M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16:1724–1731. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Li W., Liu G., Zhang X., Chen J., Wu W., Guan Y., Xiong Y., Tian Y., Wu Z. 3D visualization of subcellular structures of Schizosaccharomyces pombe by hard X-ray tomography. J. Microsc. 2010;240:14–20. doi: 10.1111/j.1365-2818.2010.03379.x. [DOI] [PubMed] [Google Scholar]
- Zhao S., Liao H., Ao M., Wu L., Zhang X., Chen Y. Fixation-induced cell blebbing on spread cells inversely correlates with phosphatidylinositol 4,5-bisphosphate level in the plasma membrane. FEBS Open Bio. 2014;4:190–199. doi: 10.1016/j.fob.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Virtual slices are scaled as depicted with colorbar from 0.1 to 0.5 μm-1 LAC values.
Heterochromatin is represented in magenta; euchromatin in blue; mitochondria in green; lipid bodies in yellow; and vesicles in cyan.
Each organelle is color coded as: nucleus - blue, mitochondria - green, lipid bodies - yellow, vesicles - cyan, virions - magenta, DMVs - red, CMs - orange, and large compartment - gray.
Data Availability Statement
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request