ABSTRACT
Morphological transition is an important adaptive mechanism in the host invasion process. Wor1 is a conserved fungal regulatory protein that controls the phenotypic switching and pathogenicity of Candida albicans. By modulating growth conditions, we simulated three models of Beauveria bassiana morphological transitions, including CTH (conidia to hyphae), HTC (hyphae to conidia), and BTB (blastospore to blastospore). Disruption of BbWor1 (an ortholog of Wor1) resulted in a distinct reduction in the time required for conidial germination (CTH), a significant increase in hyphal growth, and a decrease in the yield of conidia (HTC), indicating that BbWor1 positively controls conidium production and negatively regulates hyphal growth in conidium-hypha switching. Moreover, ΔBbWor1 prominently decreased blastospore yield, shortened the G0/G1 phase, and prolonged the G2/M phase under the BTB model. Importantly, BbWor1 contributed to conidium-hypha switching and blastospore propagation via different genetic pathways, and yeast one-hybrid testing demonstrated the necessity of BbWor1 to control the transcription of an allergen-like protein gene (BBA_02580) and a conidial wall protein gene (BBA_09998). Moreover, the dramatically weakened virulence of ΔBbWor1 was examined by immersion and injection methods. Our findings indicate that BbWor1 is a vital participant in morphological transition and pathogenicity in entomopathogenic fungi.
IMPORTANCE As a well-known entomopathogenic fungus, Beauveria bassiana has a complex life cycle and involves transformations among single-cell conidia, blastospores, and filamentous hyphae. This study provides new insight into the regulation of the fungal cell morphological transitions by simulating three models. Our research identified BbWor1 as a core transcription factor of morphological differentiation that positively regulates the production of conidia and blastospores but negatively regulates hyphal growth. More importantly, BbWor1 affects fungal pathogenicity and the global transcription profiles within three models of growth stage transformation. The present study lays a foundation for the exploration of the transition mechanism of entomopathogenic fungi and provides material for the morphological study of fungi.
KEYWORDS: Beauveria bassiana, Wor1, morphological transition, virulence, germination, conidia, blastospore
INTRODUCTION
The capacity of pathogenic fungi for morphological changes extends through the entire life cycle (1). The morphological transitions are usually controlled by transcriptional regulators, which initiate expression of a series of genes based on time, space, or environmental signals (2, 3). Candida albicans can transform among multiple phenotypic forms to adapt to and reside on a variety of hosts (4). The transcription factor Wor1 is considered to be the master regulator of cell morphological transformation of fungal white-opaque states in this fungal pathogen (5, 6). Wor1 controls its own level of expression, forming a stable autoregulatory feedback loop to induce and maintain the opaque state (7–9). The disruption of Wor1 prevents opaque cell formation, while ectopic expression of the gene translates all fungal cells into stable opaque cells (7).
Although the morphological transformation of white-opaque cells only occurs in C. albicans and in closely related fungi, the Wor1 transcription factor is conserved throughout the fungal kingdom (10). Ryp1, a homologue of the Wor1 protein in the human pathogenic fungus Histoplasma capsulatum, is crucial for temperature-dependent mycelia-to-yeast transition and virulence (11). In plant pathogenic fungi, the ros1 (a homologue of Wor1) deletion strain locks Ustilago maydis development in the filamentous stage and inhibits spore formation (12). ZtWor1 in Zymoseptoria tritici is pivotal for the production of spores and pathogenesis, and the gene disruption strain produces an extensive, dense mycelial network accompanied by a large number of abnormally swollen cell structures (13). Deletion of CfWor1 in Cladosporium fulvum results in loss of virulence and damage to the formation of sclerotium-like structures and conidia (14). These results demonstrate that homologous genes of Wor1 have evolved divergently in various human and plant fungal pathogens.
As a well-known entomopathogenic fungus, Beauveria bassiana is not only widely used as a fungal insecticide in controlling pests but also serves as a model system to investigate the interaction between fungal development and the host (15, 16). Under natural conditions, B. bassiana undergoes a transition among three cell morphologies, including hyphae, conidia, and blastospores (3). The conidia germinate, generate hyphae, and then directly invade host cuticles to infect insects (17). After intruding into the host hemocoel, B. bassiana experiences a dimorphic change to generate blastospores, which consume the nutrients existing in the hemolymph (15, 18). Finally, B. bassiana penetrates host tissues and cuticles and forms conidia to begin a new cycle of infection (19). For B. bassiana, both BbMbp1 (a component of the MluI cell cycle box-binding complex) and BbGEL1 (a gelsolin) play crucial roles in changing morphology to affect the development of conidia and blastospores (2, 3). Moreover, BbMbp1 mediates different transcriptomes and directly controls the expression of one cell wall protein gene and integral membrane protein gene to adapt to aerial and submerged conditions (2).
In this study, the role of BbWor1 was characterized in three models of the morphological transition process, including CTH (conidia to hyphae), HTC (hyphae to conidia), and BTB (blastospore to blastospore) in B. bassiana by gene disruption, complementation methods, and comparative transcriptomics analysis.
RESULTS
Bioinformatics description of BbWor1 and generation of its mutant strains.
Wor1 is considered the master regulator of cell morphological transformation in C. albicans (5). Based on a BLAST search with the CaWor1 protein sequence, BBA_06411 (identity 57.14%; E value 5e-27) was characterized and named BbWor1. Phylogenetic tree analysis revealed that BbWor1 shared 40% to ∼61% identity with homologues in human pathogenic fungi, plant pathogenic fungi, and yeast (Fig. S1A in the supplemental material). BbWor1 encodes a 469-amino acid protein with a GTI1_PAC2 domain (amino acids +11 to +180) in its N-terminal region (Fig. S1B). According to quantitative real-time PCR (qRT-PCR), the transcript level of BbWor1 increased by 5.4- and 1.8-fold in the BTB and HTC models, respectively, in contrast to the CTH model (Fig. S1C).
To reveal the functions of BbWor1 in B. bassiana, a partial gene fragment was substituted by the phosphinothricin resistance gene to generate the ΔBbWor1 disruption strain. The BbWor1 open reading frame and the corresponding promoter regions were integrated into the deletion mutant strain to select the ΔBbWor1/BbWor1 complementation mutant strain. The mutant strains were successively confirmed via PCR and qRT-PCR (Fig. S1D and E).
Influence of BbWor1 on fungal development in the three models.
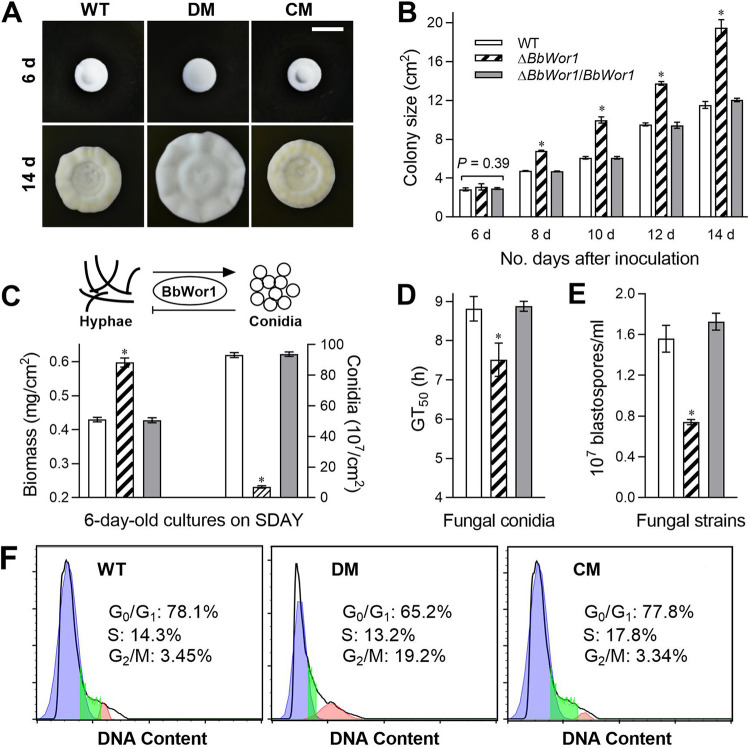
The growth ability of fungal hyphae was measured by colony area comparison and biomass measurement. Colony area assays revealed no significant difference on day 6 postincubation on Sabouraud dextrose agar plates (SDAY) cultures between mutant strains and wild-type (WT) strains (P > 0.05; Fig. 1A and B). However, the ΔBbWor1 strain displayed obvious superiority in colony growth compared with the WT strain at 14 days (Fig. 1A) and was 1.4 to 1.7 times larger than the control strains (WT and complementation mutant) from 8 to 14 days (Fig. 1B).
FIG 1.
Disruption of BbWor1 affects fungal vegetative growth and asexual development of B. bassiana. (A) Images of fungal colonies at 6 days and 14 days of growth under the temperature condition of 25°C on SDAY medium spotted with 1-μl aliquots of conidial suspension (scale bar: 20 mm). (B) Colony size of fungal colonies from 6 days to 14 days of culture on SDAY at 25°C. (C) Biomass and conidial yield of the fungal strains on SDAY plates for 6 days at 25°C in the HTC model (bottom). In the model of conidium-hypha switching by BbWor1 regulation, arrows and bars represent positive regulation and negative regulation, respectively (top). (D) Time until 50% conidial germination (GT50) in GB broth. (E) Blastospore yields after 4 days of culture in NLB medium at 25°C. (F) Cell cycle (G0/G1, S and G2/M phases) of unicellular blastospores determined by DNA content profiles with FACS analysis. The asterisk (*) denotes significant differences (Tukey’s HSD, P < 0.05). Error bars, standard deviations (SDs) of three replicates.
Although no difference in colony area was observed at 6 days, the ΔBbWor1 mutation biomass was increased by 18% compared with that of the WT in the HTC model (Fig. 1C). Moreover, the deletion strain produced 0.69 ± 0.11 × 108 conidia/cm2 with an approximate 93% reduction compared with 9.32 ± 0.23 × 108 conidia/cm2 for the WT strain and 9.38 ± 0.21 × 108 conidia/cm2 for the complementation strain (Fig. 1C). In the CTH model, the ΔBbWor1 strain showed an acceleration of approximately 15% in germination, with time to 50% germination (GT50) = 7.52 ± 0.24 h for the ΔBbWor1 mutant compared with that of the control strains (Fig. 1D). In the CTH and HTC models, disruption of BbWor1 significantly promoted the growth of hyphae and inhibited the production of conidia, indicating that BbWor1 positively regulates conidium production and negatively controls hyphal growth in conidium-hypha switching (Fig. 1C).
Blastospore incubation in nitrogen-limited broth (NLB) (BTB model) imitates the growth of B. bassiana in the hemolymph of insects. BbWor1 disruption resulted in an approximately 55% decrease in blastospore yield. The ΔBbWor1 mutant generated only 0.77 ± 0.02 × 108 spores/ml, whereas the WT and complementation strains produced 1.68 ± 0.04 and 1.72 ± 0.05 × 108 spores/ml, respectively (Fig. 1E). To explore the effect of BbWor1 on the cell cycle in the BTB model, the DNA concentration from fluorescence-activated cell sorting (FACS) analysis was used to differentiate blastospores containing DNA-specific dye for the unduplicated (G1), duplicated (G2), and intermediate DNA concentration (S) profiles. The disruption mutant exhibited an altered cell cycle characterized by a shorter G0/G1 phase and longer G2/M phase compared with those of the control strains (Fig. 1F). Overall, the ΔBbWor1 strain displayed significantly reduced spore production and a shortened G0/G1 phase in blastospores, but a prolonged G2/M phase.
Effect of BbWor1 on fungal virulence.
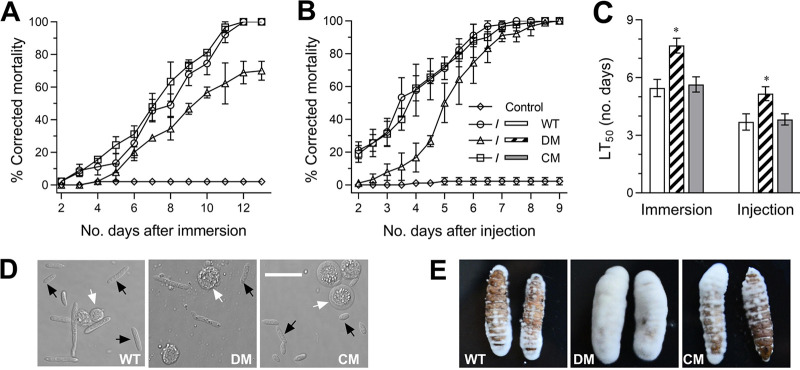
To explore how BbWor1 loss affects virulence, the lethality of B. bassiana against the moth Galleria mellonella was assayed to evaluate virulence as the median lethal time (LT50) by immersion or direct injection. Compared with the control strains, ΔBbWor1 showed a dramatic decrease in mortality of insects infected by fungi (Fig. 2A and B). In immersion bioassays, the ΔBbWor1 mutant strain displayed an LT50 of 7.67 ± 0.39 days, with LT50 = 5.46 ± 0.45 days for WT and LT50 = 5.64 ± 0.39 days for the complementation strain (Fig. 2C), demonstrating that disruption mutants require more time to kill insects than control strains. Similarly, direct injection bioassays displayed LT50 values of 5.16 ± 0.36 days for ΔBbWor1, 3.69 ± 0.43 days for WT, and 3.82 ± 0.28 days for the complementation strain (Fig. 2C). To further understand the cause of delayed lethal action, we examined hemolymph samples of live larvae that were cultured for 3 days following injection. Abundant blastospores were observed in the control strains; however, this number of individual cells was obviously reduced in the larvae infected with ΔBbWor1 (Fig. 2D). In addition, after 3 days of maintenance under humid conditions, a large number of mycelia was observed outside of the cadavers killed by the ΔBbWor1 mutant through the two infection modes, and the surfaces of cadavers killed by control strains were largely exposed (Fig. 2E). This phenomenon indicated that BbWor1 deletion promoted the accumulation of B. bassiana hyphae on larval cadavers.
FIG 2.
Effects of BbWor1 loss on the virulence of B. bassiana. (A and B) Mortalities in immersion and intrahemoceol injection assays of ΔBbWor1 (DM) and WT or ΔBbWor1/BbWor1 (CM) were recorded. (C) The mean lethal time (LT50) for immersion and intrahemoceol injection tests were analyzed with Probit analysis. (D) Microscopy images (scale: 20 μm) obtained by LSCM for the blastospores in the hemolymph samples of surviving larvae at 3 days postinjection. Black arrows indicate blastospores, and white arrows indicate host hemocytes. (E) Images of fungal outgrowths at the surface of cadavers 3 days post-death through immersion (left) or injection (right) bioassays. The asterisk (*) denotes significant differences (Tukey’s HSD, P < 0.05). Error bars, SDs of three replicates.
Requirement of BbWor1 for global expression of the three models.
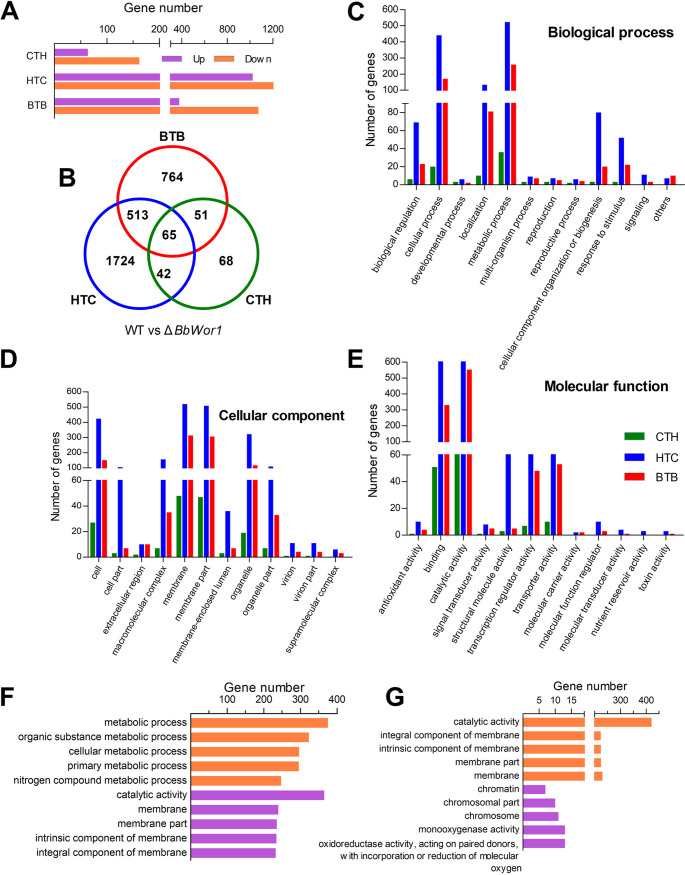
According to the Cuffdiff method (20), differentially expressed genes (DEGs) (Q < 0.05, |log2FC| > 1) were screened between the WT and ΔBbWor1 groups within three models of stage transformation simulated by modulated growth conditions. Compared with the BTB (1,393 DEGs) and HTC (2,344 DEGs) models, the CTH model had the fewest DEGs (226 genes) (Fig. 3A), indicating that disruption of BbWor1 had the weakest effect on the number of genes in the CTH model. Further comparison analyses showed that most DEGs (578 genes) overlapped between the BTB and HTC libraries. Moreover, 107 DEGs overlapped between CTH and HTC, and 116 DEGs overlapped between CTH and BTB (Fig. 3B). Moreover, 65 DEGs overlapped among the three databases of the CTH, HTC and BTB models (Fig. 3B and Table S3).
FIG 3.
Disruption of BbWor1 affects the transcription profile of B. bassiana in the three models. (A) DEGs whose expression was upregulated and downregulated were determined in the ΔBbWor1 strain via comparison with WT in the CTH, HTC, and BTB models. (B) Venn diagram showing the number of DEGs between the WT and ΔBbWor1 strains in the three models. (C to E) GO classification of DEGs into three main categories: biological processes, cellular components, and molecular functions. (F and G) GO enrichment analysis for DEGs whose expression was upregulated and downregulated between WT and ΔBbWor1 in the HTC and BTB models. The top 5 terms are displayed (Q < 0.05).
Following gene ontology (GO) functional annotation analysis, the DEGs were concentrated in the three functional groups of biological processes, cell components, and molecular functions. As the GO terms were arranged in accordance with the number of genes, this suggested that the following top eight terms of the three models were the same: (i) cellular process and (ii) metabolic process in biological processes (Fig. 3C); (iii) cell, (iv) membrane, (v) membrane part, and (vi) organelle in cell components (Fig. 3D); and (vii) catalytic activity and (viii) binding in molecular functions (Fig. 3E).
However, despite that the top eight GO terms of DEGs were similar in the three models, gene expression regulated by BbWor1 within the three models was significantly different after GO enrichment. The DEGs in the CTH model were not significantly enriched in GO terms, suggesting that disruption of BbWor1 does not significantly alter the type of genes expressed during germination. In the HTC model, genes whose expression was repressed were most enriched in metabolic processes (375 genes), and genes whose expression was upregulated were mostly enriched in catalytic activity (365 genes) (Fig. 3F; Table S4). Under the BTB model, catalytic activity (421 genes) was the enrichment term with the largest number of genes whose expression was repressed (Fig. 3G and Table S5). In addition, both the genes whose expression was upregulated in the HTC model and genes whose expression was repressed in the BTB model were collectively enriched in some terms, such as catalytic activity, integral component of membrane, intrinsic component of membrane, membrane part, and membrane (Fig. 3G). Such differences in GO enrichment indicated that BbWor1 plays different roles in the three models.
Interaction of BbWor1 with downstream genes.
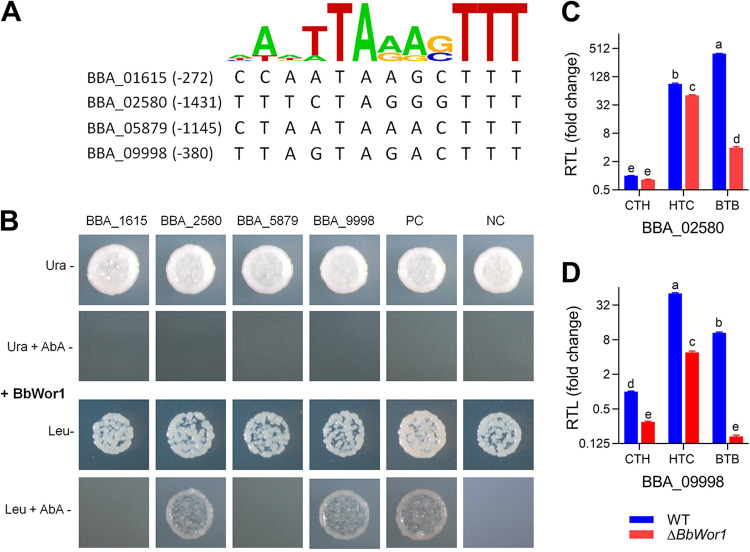
To identify the potential BbWor1-mediated target genes, approximately 1.5 kbp upstream of each start codon (i.e., the promoter regions) of 37 genes that were repressed in all three models (Table S3) was screened in accordance with the binding motif of Wor1 (21, 22). Four genes (BBA_01615, BBA_02580, BBA_05879, and BBA_09998) were captured (Fig. 4A) and were considered potential targets of the BbWor1 protein for yeast one-hybrid experiments, in which it was demonstrated that BbWor1 directly mediated transcriptional activation of the BBA_02580 and BBA_09998 motif constructs incorporated into the reporter gene (Fig. 4B). BBA_02580 and BBA_09998 are allergen-like protein genes and conidial wall protein genes, respectively, on the basis of the content within the NCBI database. Compared with those in the WT under the CTH model, BBA_02580 and BBA_09998 transcript levels of the ΔBbWor1 mutant were verified to be downregulated in all three models by qRT-PCR, especially in the BTB model, in which the levels decreased by 99% and 98%, respectively (Fig. 4C and D). The results from the qRT-PCR and the transcriptome sequencing (RNA-seq) data were consistent (Table 1), confirming the accuracy and quality of the transcriptome experiment.
FIG 4.
Identification of BbWor1 downstream genes and determination of the expression of target genes. (A) The binding site data of Wor1 (21, 22) were used to identify consensus binding sites in promoter regions of 4 genes selected from 37 common repressed DEGs in three models. (B) Yeast one-hybrid assay of the interaction of BbWor1 and 4 gene motifs. All motifs were introduced to autoactivation testing with 250 ng/ml aureobasidin A (AbA) on SD/−Ura medium, and physical interactions were measured on SD/−Leu medium with 250 ng/ml AbA. PC, positive control (yeast cells transformed with the pGADT7-Rec-p53 vector and p53-AbAi). NC, negative control (yeast cells transformed with pGADT7-Rec-BbWor1 and a blank vector [pAbAi]). (C and D) qRT-PCR validation of BBA_02580 and BBA_0998 expression. The relative transcript levels (RTLs) of the two genes are presented as the ratio of WT in the CTH model. Different lowercase letters denote significant differences (Tukey’s HSD, P < 0.05).
TABLE 1.
BbWor1-related genes described in this study and from the analysis of transcriptomes between ΔBbWor1 and WTa
| Locus tag | Annotation | CTH |
HTC |
BTB |
|||
|---|---|---|---|---|---|---|---|
| Log2 FC | Q value | Log2 FC | Q value | Log2 FC | Q value | ||
| BBA_02580 | allergen-like protein | −1.09 | 0.00 | −5.46 | 0.00 | −6.05 | 0.00 |
| BBA_09998 | conidial wall protein | −5.96 | 0.00 | −3.87 | 0.00 | −6.02 | 0.00 |
| BBA_03082 | β-1,3-glucanosyltransferase | 1.08 | 0.00 | 2.34 | 0.00 | −1.38 | 0.00 |
| BBA_09307 | acidic chitinase | 1.43 | 0.00 | 0.11 | 0.00 | −3.64 | 0.00 |
| BBA_00047 | Cel5b-like protein | 1.52 | 0.00 | −0.62 | 0.00 | 0.05 | 0.13 |
| BBA_00048 | hypothetical protein | 1.14 | 0.00 | −0.69 | 0.00 | 0.00 | 0.19 |
| BBA_04942 | fluG protein | 0.34 | 0.28 | −6.70 | 0.00 | −1.39 | 0.00 |
| BBA_02348 | hsp70-like protein | −0.06 | 0.00 | −1.29 | 0.00 | 0.11 | 0.00 |
| BBA_00257 | hsp70-like protein | −0.09 | 0.00 | −1.12 | 0.00 | 0.06 | 0.00 |
CTH, condia to hyphae; HTC, hyphae to condia; BTB, blastospore to blastospore; FC, fold change.
DISCUSSION
As a well-known insect pathogenic filamentous fungus, B. bassiana is becoming a new typical fungus with which to investigate fungal development and pathogen-host interactions (15, 16). In the infection process, B. bassiana undergoes a series of morphological changes throughout the infection cycle, including filamentous hyphae, single-cell conidia, and blastospores. In the present study, we simulated three models of the morphological transition process of B. bassiana in vitro, including CTH, HTC, and BTB. After RNA-seq analysis of the three models with the WT strain was performed, the largest number of DEGs and the weakest correlation were found in the CTH and HTC models, indicating that the performance of B. bassiana in conidium-hypha switching differs more during gene transcription.
Wor1, a conserved fungal transcription factor, has been thoroughly studied in both human and plant pathogenic fungi. Wor1 is the master regulator of cell morphological transformation, especially in C. albicans (5, 7, 23). The change in fungal cellular states is accompanied by a change in the gene expression level of Wor1 (6). The qRT-PCR results exhibited a notable discrepancy in BbWor1 gene expression among the three simulation models, indicating that BbWor1 may be related to the morphological transition of B. bassiana. In the present study, BbWor1 was highly influential in the control of vegetative growth and asexual development. Moreover, comparative transcriptomics demonstrated that BbWor1 participated in controlling the morphological transitions in the three models of B. bassiana via different genetic pathways.
Disruption of BbWor1 reduced conidial germination time and promoted hyphal growth, possibly due to cell wall remodeling. Fungal β-1,3-glucanosyltransferases are glycosyl phosphatidylinositol-anchored proteins that affect cell wall biogenesis of fungi (24). In C. albicans, β-1,3-glucanosyltransferase (Phr1p) is a fungal cell wall remodeling enzyme conducive to hyphal plasticity and extension (25). The increased expression of BBA_03082 (which encodes a β-1,3-glucanosyltransferase) may accelerate the extension of the cell wall during conidial germination and hyphal growth (Table 1). The degradation by hydrolytic enzymes causes the cell wall to continue to relax and expand to release constraints on the cell wall to meet cell growth requirements (26–28). ChiE1 (chitinase) in Coprinopsis cinerea has been proven to participate in the extension and growth of stipe cell walls through breaking tethers and allowing chitin microfibrils to unclasp, thus increasing the space to add more polymers of chitin units and β-glucan subject to in vivo turgor pressure (29). In addition to upregulation of BBA_09307 (the homologous gene of ChiE1), the expression levels of two endoglucanase hydrolases (BBA_00047 and BBA_00048) were upregulated in CTH, which might account for the accelerated germination in the ΔBbWor1 mutant, to some extent.
Conidia are important fungal cells for dispersal and persistence in the environment. B. bassiana grow saprophytically and produce conidia on cadavers and then enter the next stage of the infection cycle (2). In the HTC model, disruption of BbWor1 significantly reduced (93%) conidial yield. FluG, as a conserved protein, plays vital roles in the conidiation process of filamentous fungi and is an upstream developmental activator in Aspergillus nidulans (30, 31). The expression level of BBA_04942 (the FluG homologous gene) significantly decreased (log2FC = −6.7) in ΔBbWor1 mutant strains, suggesting there is an interaction between the Wor1-mediated pathway and the central regulatory pathway for conidiation. As a molecular chaperone in the endoplasmic reticulum (ER), LHS1 (from the heat shock protein 70 family) functions during protein translocation and protein folding in the ER, and loss of LHS1 severely impairs the conidiation of Magnaporthe oryzae (32). Expression of Lhs1 homologous genes (BBA_02348 and BBA_00257) was repressed in the ΔBbWor1 mutant, which might lead to a decrease in conidiation. The CTH and HTC models showed that BbWor1 positively controlled conidial production and negatively controlled hyphal growth by regulating different transcriptional maps in conidium-hypha switching.
Morphological transformation is critical for the pathogenesis of mycopathogens. The switch between opaque and white cells remarkably influences the interaction between C. albicans and the host. Opaque cells thrive in skin infections, while white cells are more frequently observed in internal infections (33). The disruption of Wor1 maintains cells in the white state, which weakens the adaptability of C. albicans on the skin, thus affecting pathogenicity (5). In Fusarium oxysporum, deletion of Sge1 not only is quantitatively involved in conidiogenesis, but also influences the expression of six effector proteins and reduces secondary metabolites, thereby reducing virulence (34). Disruption of Fgp1 in Fusarium graminearum results in a lack of trichothecene toxin accumulation and greatly reduces virulence to wheat plants (35). ZtWor1, a transcriptional regulator of another wheat pathogen, plays a decisive role in the expression level of many genes encoding small secreted proteins and contributes to pathogenicity (13). In B. bassiana, blastospores consume insect hemolymph nutrients and/or result in symptoms caused by toxic metabolites from the pathogen (36). Therefore, the remarkable deterioration ability of the ΔBbWor1 mutant for blastospore propagation might lead to significantly weakened fungal pathogenicity.
MATERIALS AND METHODS
Strains and growth conditions.
WT B. bassiana strain ARSEF 2860 was routinely fostered under a temperature of 25°C on Sabouraud dextrose agar plates (SDAY; 4% glucose, 1% yeast extract, 1% peptone, plus 1.5% agar) for 12 h in light and 12 h in dark. Escherichia coli strain DH5α (Shanghai, China) was amplified in Luria-Bertani (LB) broth. Agrobacterium tumefaciens strain AGL-1 was amplified in yeast extract broth (YEB; wt/vol: 0.5% sucrose, 0.1% yeast extract, 1% peptone, and 0.05% MgSO4) (37).
Sequence analysis and generation of BbWor1.
The Wor1 protein sequence in C. albicans (XP_723567.2) was used as a query to search the B. bassiana genome (38) and a homologous protein was identified and appointed to BbWor1. BbWor1 was structurally compared with the protein sequences of human pathogens, phytopathogens, and yeasts, and the alignment was displayed in line with the ClustalW algorithm (39). A phylogenetic tree was constructed by MEGA 7 software (https://www.megasoftware.net/) with a neighbor-joining approach (40).
Generation of gene deletion and complementation strains.
The BbWor1 gene deletion and complementation vectors were constructed as described previously (41, 42). The primers used in this study are shown in Table S1 in the supplemental material. Briefly, a phosphinothricin resistance gene (bar) was used to replace a partial gene fragment (from −77 to +405) to form a gene deletion vector, and the entire BbWor1 open reading frame, together with its corresponding promoter, was cloned into the complementation vector. The deletion and complementation vectors were transformed into WT and gene deletion mutant strains by the Agrobacterium-mediated transformation procedure. PCR and qRT-PCR were performed on potential transformants with primer pairs (Table S1) to verify correct recombination events.
Assessments of fungal development in three models.
For the ΔBbWor1 mutant, WT, and complementation strains, the diameters of fungal colonies inoculated by spotting of a 104 conidial suspension (1 μl) were measured from 6 days to 14 days, and the colony areas on SDAY plates at 25°C were used as growth indices.
To explore morphological transitions in entomopathogenic fungi, we simulated the morphological transition of B. bassiana by three model sets of growth conditions. For the HTC model, aliquots (100 μl) of conidial suspension (107 cells/ml) were plated on SDAY medium and cultured at 25°C for 6 days. The growth of mycelia was measured by drying fungal cultures overnight at 50°C and weighing them. The number of conidia per square centimeter was used to quantify the production ability of conidia. For the CTH model, fungal conidia cultured on SDAY were collected and added to 20 ml (106 conidia/ml) of germination broth (GB; 2% sucrose and 0.5% peptone). The rate of germinating conidia was counted hourly after incubation for 6 h at 25°C (with aeration, 200 rpm). The median germination time (GT50, h), a viability indicator of 50% conidial germination, was computed following the fitted germination tendency of fungal conidia. For the BTB model, hyphae collected from SDAY medium were inoculated into NLB (4% glucose, 0.4% NH4NO3, 0.3% KH2PO4 and 0.3% MgSO4) to collect blastospores. As initial inocula, the resultant blastospores were adjusted to a final concentration of 104/ml and then cultured for 4 days at 25°C in NLB to simulate a BTB model.
Determination of the cell cycle.
Examination of the cell cycle for the ΔBbWor1 mutant and control strains was carried out as mentioned above (43). Specifically, WT and mutant strain blastospore suspensions were treated with propidium iodide (DNA-specific stain) at a final concentration of 50 μg/ml at 4°C for 30 min. FACS analysis was performed on three 500-μl aliquots of every stained suspension (105 conidia/ml) using a Beckman Coulter CytoFLEX LX flow cytometer for determination of DNA concentration. The G0/G1, G2/M, and S phases of the cell cycle were evaluated with the DNA concentrations.
Insect bioassays.
The immersion and hemocoel injection methods were adopted to determine fungal virulence to G. mellonella (44). Specifically, a total of 30 larvae (∼300 mg each) were immersed in 30 ml of a conidial suspension (107 conidia/ml) for approximately 10 s or injected into the abdomen with 1 μl of conidial suspension (5 × 105 conidia/ml; 500 conidia per larva). The same volume of Tween 80 solution (0.02%) was used as a blank control. All batches of larvae were incubated in petri dishes (20 cm diameter) at room temperature at 25°C, and the mortality was examined at 12-h intervals. Probit analysis revealed the distinction between sigmoid time-mortality trends and median lethal time (LT50) estimates. The blastospores were observed in G. mellonella blood (45) using a laser scanning confocal microscope (Leica DMi8, Germany). The fungal hyphal growth on the humid cadaveric surface was recorded by taking photos.
Extraction of RNA and comparative transcriptomics analysis of the three models.
To explore the relationship among the three models of WT and the role of BbWor1 in morphological transitions, comparative transcriptomics analysis was carried out among six treatments under the CTH (9 h), HTC (6 day), and BTB (4 day) models using WT and the ΔBbWor1 mutant. Total RNA was extracted from fungal cultures of each mode with TRIzol-A+ reagent. Each treatment included three independent biological replicates. Sequencing of RNA samples was performed on the BGISEQ-500 platform at BGI-Shenzhen, China. Entire clean reads were mapped to the reference genome with HISAT2 (v2.0.4) (46). Bowtie2 (v2.2.5) (47) was applied to align the clean reads to the reference coding gene set, and the gene expression level was then evaluated by RSEM (v1.2.12) (48). The Cuffdiff method was used to analyze the differentially expressed genes (DEGs; Q value < 0.05, |log2 fold change (FC)| > 1) between the WT and disruption mutant strains (20). According to the gene ontology (GO) annotation results following the functional classification of DEGs, the phyper function in R software was used for enrichment analysis. Significant levels of terms and pathways were corrected by Q value (Q value < 0.05) with a Bonferroni correction (49).
Screening of downstream genes and yeast one-hybrid test.
The yeast one-hybrid test was performed as previously described (2). The primer pair (Table S2) corresponding to different motifs was mixed, heated, and annealed by PCR to form double strands, which were then inserted into the pAbAi vector (Clontech) digested by HindIII and XhoI to generate the pBait-AbAi construct. The resulting construct was confirmed by AflII and XmaI digestion. Full-length BbWor1 was amplified from cDNA with the p9/p10 primers (Table S2) and then cloned into the NdeI and EcoRI sites to construct the pGADT7-Rec-BbWor1 vector. The pGADT7-Rec-BbWor1 plasmid was transformed to a bait-specific reporter strain and then selected on appropriate selection plates (leucine-free SD medium with 250 ng/ml aureobasidin A; AbA). Transformation of yeast cells with the pGADT7-Rec-BbWor1 vector and one blank vector (pAbAi) was used as the negative control, while transformation of yeast cells with the p53-AbAi vector and pGADT7-Rec-p53 (Clontech) was used as the positive control. The positive colonies displayed an interaction of BbWor1 and the tested gene motifs.
Gene expression analysis using qRT-PCR.
Total RNA samples were extracted and reverse transcribed into cDNA with TRIzol-A+ reagent (Tiangen Biotech, Beijing, China) and a FastKing RT kit (with gDNase) (Tiangen Biotech, Beijing, China). The 96 RT-PCR system (Thermo Fisher Scientific, USA) was used for quantitative real-time PCR (qRT-PCR) analysis with 2 × M5 HiPer SYBR Premix EsTaq (with Tli RNaseH) (Mei5 Biotechnology, Beijing, China) and the primers listed in Table S1. Fungal 18S rRNA was considered an endogenous standard, and the relative transcript levels of target genes were computed with the threshold cycle (2-ΔΔCt) method (50).
Statistical analysis.
Entire phenotypic estimates from triplicate tests were acquired with one-way ANOVA. Tukey's honestly significantly difference (HSD) test was applied to determine the notable differences among fungal strains.
Data availability.
RNA-seq data for this study are available at the NCBI GEO database (accession number GSE178974).
ACKNOWLEDGMENTS
Funding of this work was provided by the National Natural Science Foundation of China (grants 31800062 and 31600063) and the Foundation (no. 202013) of Qilu University of Technology of Cultivating Subject for Biology and Biochemistry, Qilu University of Technology, Shandong Academy of Sciences.
Footnotes
Supplemental material is available online only.
Contributor Information
Lei Qiu, Email: qiulei.2005@163.com.
Juan-Juan Wang, Email: wjj880414@163.com.
Christina A. Cuomo, Broad Institute
REFERENCES
- 1.Gauthier GM. 2015. Dimorphism in fungal pathogens of mammals, plants, and insects. PLoS Pathog 11:e1004608. doi: 10.1371/journal.ppat.1004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding JL, Lin HY, Feng MG, Ying SH. 2020. Mbp1, a component of the MluI cell cycle box‐binding complex, contributes to morphological transition and virulence in the filamentous entomopathogenic fungus Beauveria bassiana. Environ Microbiol 22:584–597. doi: 10.1111/1462-2920.14868. [DOI] [PubMed] [Google Scholar]
- 3.He PH, Dong WX, Chu XL, Feng MG, Ying SH. 2016. The cellular proteome is affected by a gelsolin (BbGEL1) during morphological transitions in aerobic surface versus liquid growth in the entomopathogenic fungus Beauveria bassiana. Environ Microbiol 18:4153–4169. doi: 10.1111/1462-2920.13500. [DOI] [PubMed] [Google Scholar]
- 4.Noble SM, Gianetti BA, Witchley JN. 2017. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol 15:96–108. doi: 10.1038/nrmicro.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alkafeef SS, Yu C, Huang L, Liu H. 2018. Wor1 establishes opaque cell fate through inhibition of the general co-repressor Tup1 in Candida albicans. PLoS Genet 14:e1007176. doi: 10.1371/journal.pgen.1007176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao L, Du H, Guan GB, Dai Y, Nobile CJ, Liang WH, Cao CJ, Zhang QY, Zhong J, Huang GH. 2014. Discovery of a “white-gray-opaque” tristable phenotypic switching system in Candida albicans: roles of non-genetic diversity in host adaptation. PLoS Biol 12:e1001830. doi: 10.1371/journal.pbio.1001830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. 2006. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A 103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zordan RE, Galgoczy DJ, Johnson AD. 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A 103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Q, Zhang Q, Bing J, Ding X, Huang G. 2017. Environmental and genetic regulation of white-opaque switching in Candida tropicalis. Mol Microbiol 106:999–1017. doi: 10.1111/mmi.13862. [DOI] [PubMed] [Google Scholar]
- 10.Porman AM, Alby K, Hirakawa MP, Bennett RJ. 2011. Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc Natl Acad Sci U S A 108:21158–21163. doi: 10.1073/pnas.1112076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen VQ, Sil A. 2008. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci U S A 105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tollot M, Assmann D, Becker C, Altmüller J, Dutheil JY, Wegner CE, Kahmann R. 2016. The WOPR protein Ros1 is a master regulator of sporogenesis and late effector gene expression in the maize pathogen Ustilago maydis. PLoS Pathog 12:e1005697. doi: 10.1371/journal.ppat.1005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirzadi Gohari A, Mehrabi R, Robert O, Ince IA, Boeren S, Schuster M, Steinberg G, de Wit PJGM, Kema GHJ. 2014. Molecular characterization and functional analyses of ZtWor1, a transcriptional regulator of the fungal wheat pathogen Zymoseptoria tritici. Mol Plant Pathol 15:394–405. doi: 10.1111/mpp.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okmen B, Collemare J, Griffiths S, van der Burgt A, Cox R, De Wit P. 2014. Functional analysis of the conserved transcriptional regulator CfWor1 in Cladosporium fulvum reveals diverse roles in the virulence of plant pathogenic fungi. Mol Microbiol 92:10–27. doi: 10.1111/mmi.12535. [DOI] [PubMed] [Google Scholar]
- 15.Ying SH, Ji XP, Wang XX, Feng MG, Keyhani NO. 2014. The transcriptional co‐activator multiprotein bridging factor 1 from the fungal insect pathogen, Beauveria bassiana, mediates regulation of hyphal morphogenesis, stress tolerance and virulence. Environ Microbiol 16:1879–1897. doi: 10.1111/1462-2920.12450. [DOI] [PubMed] [Google Scholar]
- 16.Wang C, Wang S. 2017. Insect pathogenic fungi: genomics, molecular interactions, and genetic improvements. Annu Rev Entomol 62:73–90. doi: 10.1146/annurev-ento-031616-035509. [DOI] [PubMed] [Google Scholar]
- 17.Ortiz-Urquiza A, Keyhani NO. 2013. Action on the surface: entomopathogenic fungi versus the insect cuticle. Insects 4:357–374. doi: 10.3390/insects4030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wanchoo A, Lewis MW, Keyhani NO. 2009. Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and haemolymph-derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiology (Reading) 155:3121–3133. doi: 10.1099/mic.0.029157-0. [DOI] [PubMed] [Google Scholar]
- 19.He PH, Wang XX, Chu XL, Feng MG, Ying SH. 2015. RNA sequencing analysis identifies the metabolic and developmental genes regulated by BbSNF1 during conidiation of the entomopathogenic fungus Beauveria bassiana. Curr Genet 61:143–152. doi: 10.1007/s00294-014-0462-x. [DOI] [PubMed] [Google Scholar]
- 20.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohse MB, Zordan RE, Cain CW, Johnson AD. 2010. Distinct class of DNA-binding domains is exemplified by a master regulator of phenotypic switching in Candida albicans. Proc Natl Acad Sci U S A 107:14105–14110. doi: 10.1073/pnas.1005911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cain CW, Lohse MB, Homann OR, Sil A, Johnson AD. 2012. A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics 190:511–521. doi: 10.1534/genetics.111.134080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohse MB, Hernday AD, Fordyce PM, Noiman L, Sorrells TR, Hanson-Smith V, Nobile CJ, DeRisi JL, Johnson AD. 2013. Identification and characterization of a previously undescribed family of sequence-specific DNA-binding domains. Proc Natl Acad Sci U S A 110:7660–7665. doi: 10.1073/pnas.1221734110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazáň M, Ragni E, Popolo L, Farkaš V. 2011. Catalytic properties of the Gas family β-(1,3)-glucanosyltransferases active in fungal cell-wall biogenesis as determined by a novel fluorescent assay. Biochem J 438:275–282. doi: 10.1042/BJ20110405. [DOI] [PubMed] [Google Scholar]
- 25.Ragni E, Calderon J, Fascio U, Sipiczki M, Fonzi WA, Popolo L. 2011. Phr1p, a glycosylphosphatidylinsitol-anchored β(1,3)-glucanosyltransferase critical for hyphal wall formation, localizes to the apical growth sites and septa in Candida albicans. Fungal Genet Biol 48:793–805. doi: 10.1016/j.fgb.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Rast DM, Baumgartner D, Mayer C, Hollenstein G. 2003. Cell wall-associated enzymes in fungi. Phytochemistry 64:339–366. doi: 10.1016/S0031-9422(03)00350-9. [DOI] [PubMed] [Google Scholar]
- 27.Riquelme M, Aguirre J, Bartnicki-García S, Braus GH, Feldbrügge M, Fleig U, Hansberg W, Herrera-Estrella A, Kämper J, Kück U, Mouriño-Pérez RR, Takeshita N, Fischer R. 2018. Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures. Microbiol Mol Biol Rev 82:e00068-17. doi: 10.1128/MMBR.00068-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinberg G, Peñalva MA, Riquelme M, Wösten HA, Harris SD. 2017. Cell biology of hyphal growth, p 231–265. In The fungal kingdom. American Society of Microbiology, Washington, DC. doi: 10.1128/9781555819583.ch11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou JS, Kang LQ, Liu CC, Niu X, Wang XJ, Liu HL, Zhang WM, Liu ZH, Latge JP, Yuan S. 2019. Chitinases play a key role in stipe cell wall extension in the mushroom Coprinopsis cinerea. Appl Environ Microbiol 85:e00532-19. doi: 10.1128/AEM.00532-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park HS, Yu JH. 2016. Developmental regulators in Aspergillus fumigatus. J Microbiol 54:223–231. doi: 10.1007/s12275-016-5619-5. [DOI] [PubMed] [Google Scholar]
- 31.Park HS, Yu JH. 2012. Genetic control of asexual sporulation in filamentous fungi. Curr Opin Microbiol 15:669–677. doi: 10.1016/j.mib.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Yi M, Chi MH, Khang CH, Park SY, Kang S, Valent B, Lee YH. 2009. The ER chaperone LHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae. Plant Cell 21:681–695. doi: 10.1105/tpc.107.055988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lachke SA, Lockhart SR, Daniels KJ, Soll DR. 2003. Skin facilitates Candida albicans mating. Infect Immun 71:4970–4976. doi: 10.1128/IAI.71.9.4970-4976.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michielse CB, van Wijk R, Reijnen L, Manders EM, Boas S, Olivain C, Alabouvette C, Rep M. 2009. The nuclear protein Sge1 of Fusarium oxysporum is required for parasitic growth. PLoS Pathog 5:e1000637. doi: 10.1371/journal.ppat.1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonkers W, Dong Y, Broz K, Kistler HC. 2012. The Wor1-like protein Fgp1 regulates pathogenicity, toxin synthesis and reproduction in the phytopathogenic fungus Fusarium graminearum. PLoS Pathog 8:e1002724. doi: 10.1371/journal.ppat.1002724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng M, Poprawski T, Khachatourians GG. 1994. Production, formulation and application of the entomopathogenic fungus Beauveria bassiana for insect control: current status. Biocontrol Sci Techn 4:3–34. doi: 10.1080/09583159409355309. [DOI] [Google Scholar]
- 37.Ding JL, Peng YJ, Chu XL, Feng MG, Ying SH. 2018. Autophagy‐related gene BbATG11 is indispensable for pexophagy and mitophagy, and contributes to stress response, conidiation and virulence in the insect mycopathogen Beauveria bassiana. Environ Microbiol 20:3309–3324. doi: 10.1111/1462-2920.14329. [DOI] [PubMed] [Google Scholar]
- 38.Xiao G, Ying SH, Zheng P, Wang ZL, Zhang S, Xie XQ, Shang YF, St Leger RJ, Zhao GP, Wang CS, Feng MG. 2012. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci Rep 2:483. doi: 10.1038/srep00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace M, Wilm A, Lopez R, Thompson JD, Gibson TL, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 40.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu L, Wei XY, Wang SJ, Wang JJ. 2020. Characterization of trehalose-6-phosphate phosphatase in trehalose biosynthesis, asexual development, stress resistance and virulence of an insect mycopathogen. Pestic Biochem Physiol 163:185–192. doi: 10.1016/j.pestbp.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Qiu L, Zhang J, Song JZ, Hu SJ, Zhang TS, Li Z, Wang JJ, Cheng W. 2021. Involvement of BbTpc1, an important Zn(II)(2)Cys(6) transcriptional regulator, in chitin biosynthesis, fungal development and virulence of an insect mycopathogen. Int J Biol Macromol 166:1162–1172. doi: 10.1016/j.ijbiomac.2020.10.271. [DOI] [PubMed] [Google Scholar]
- 43.Qiu L, Wang JJ, Ying SH, Feng MG. 2015. Wee1 and Cdc25 control morphogenesis, virulence and multistress tolerance of Beauveria bassiana by balancing cell cycle‐required cyclin‐dependent kinase 1 activity. Environ Microbiol 17:1119–1133. doi: 10.1111/1462-2920.12530. [DOI] [PubMed] [Google Scholar]
- 44.Wang JJ, Yin YP, Song JZ, Hu SJ, Cheng W, Qiu L. 2021. A p53-like transcription factor, BbTFO1, contributes to virulence and oxidative and thermal stress tolerances in the insect pathogenic fungus, Beauveria bassiana. PLoS One 16:e0249350. doi: 10.1371/journal.pone.0249350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He Z, Luo L, Keyhani NO, Yu X, Ying S, Zhang Y. 2017. The C-terminal MIR-containing region in the Pmt1 O-mannosyltransferase restrains sporulation and is dispensable for virulence in Beauveria bassiana. Appl Microbiol Biotechnol 101:1143–1161. doi: 10.1007/s00253-016-7894-9. [DOI] [PubMed] [Google Scholar]
- 46.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdi H. 2007. The Bonferonni and Sidak Corrections for multiple comparisons, p 1–9. In Salkind N (ed). Encylopedia of measurement and statistics. Sage, Thousand Oaks, CA. [Google Scholar]
- 50.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00203-21_Supp_1_seq7.pdf, PDF file, 0.5 MB (521.2KB, pdf)
Supplemental material. Download SPECTRUM00203-21_Supp_2_seq8.xls, XLS file, 0.04 MB (41.5KB, xls)
Supplemental material. Download SPECTRUM00203-21_Supp_3_seq9.xls, XLS file, 0.04 MB (40.5KB, xls)
Supplemental material. Download SPECTRUM00203-21_Supp_4_seq10.xls, XLS file, 0.03 MB (33KB, xls)
Data Availability Statement
RNA-seq data for this study are available at the NCBI GEO database (accession number GSE178974).