ABSTRACT
In this study, we characterize the response of the free-living oligotrophic alphaproteobacterium Caulobacter crescentus to low temperatures by global transcriptomic analysis. Our results showed that 656 genes were upregulated and 619 were downregulated at least 2-fold after a temperature downshift. The identified differentially expressed genes (DEG) belong to several functional categories, notably inorganic ion transport and metabolism, and a subset of these genes had their expression confirmed by reverse transcription quantitative real-time PCR (RT-qPCR). Several genes belonging to the ferric uptake regulator (Fur) regulon were downregulated, indicating that iron homeostasis is relevant for adaptation to cold. Several upregulated genes encode proteins that interact with nucleic acids, particularly RNA: cspA, cspB, and the DEAD box RNA helicases rhlE, dbpA, and rhlB. Moreover, 31 small regulatory RNAs (sRNAs), including the cell cycle-regulated noncoding RNA (ncRNA) CcnA, were upregulated, indicating that posttranscriptional regulation is important for the cold stress response. Interestingly, several genes related to transport were upregulated under cold stress, including three AcrB-like cation/multidrug efflux pumps, the nitrate/nitrite transport system, and the potassium transport genes kdpFABC. Further characterization showed that kdpA is upregulated in a potassium-limited medium and at a low temperature in a SigT-independent way. kdpA mRNA is less stable in rho and rhlE mutant strains, but while the expression is positively regulated by RhlE, it is negatively regulated by Rho. A kdpA-deleted strain was generated, and its viability in response to osmotic, acidic, or cold stresses was determined. The implications of such variation in the gene expression for cold adaptation are discussed.
IMPORTANCE Low-temperature stress is an important factor for nucleic acid stability and must be circumvented in order to maintain the basic cell processes, such as transcription and translation. The oligotrophic lifestyle presents further challenges to ensure the proper nutrient uptake and osmotic balance in an environment of slow nutrient flow. Here, we show that in Caulobacter crescentus, the expression of the genes involved in cation transport and homeostasis is altered in response to cold, which could lead to a decrease in iron uptake and an increase in nitrogen and high-affinity potassium transport by the Kdp system. This previously uncharacterized regulation of the Kdp transporter has revealed a new mechanism for adaptation to low temperatures that may be relevant for oligotrophic bacteria.
KEYWORDS: cold adaptation, Caulobacter crescentus, transcriptomic analysis, cation homeostasis, potassium transport
INTRODUCTION
Bacteria must quickly adapt to constant changes in the environment, such as nutrient starvation, oxygen availability, and abiotic stresses such as a temperature shift, which is particularly important for free-living organisms. A sudden drop in temperature causes global changes in the gene expression of mesophilic bacteria, resulting in physiological alterations that lead to a lower growth rate, including changes in the membrane lipid composition, a decrease in protein synthesis, and a change in the metabolic pathways (reviewed in references 1 and 2), which are also observed as a strategy of the psychrophiles (3). As temperature decreases, the main consequences are a decrease in membrane fluidity and increased stabilization of the secondary structures in nucleic acids, affecting important processes such as transcription and translation. Triggering the expression of a specific set of proteins, in turn, helps the cell to adjust to these unfavorable changes, mainly by increasing the fluidity of the membrane and destabilizing the secondary structures in nucleic acids (1, 2, 4).
The bacterial response to low temperatures was comprehensively studied initially in the model bacteria Escherichia coli (5) and Bacillus subtilis (6, 7). Ever since, several other species have been investigated in that context. Many of the cold-induced proteins have been identified and shown to be essential for cellular adaptation to low temperatures (8). The importance of keeping the functionality of RNA molecules can be demonstrated by the ubiquitous cold induction of RNA-modifying enzymes in all domains of life. Small cold shock proteins (CSPs), DEAD box RNA helicases, and RNases have been shown to play a critical role in the cold shock response of Gram-negative (1, 9–11) and Gram-positive (12–15) bacteria, a conserved feature in Archaea (16) and plants (17). CSPs possess a conserved cold shock domain, composed of two nucleic acid-binding motifs in tandem that bind to RNA and prevent the formation of secondary structures (18–21). These proteins have been shown to participate in many cellular processes, such as transcription antitermination and the initiation of translation (22, 23). The DEAD box RNA helicases unwind the RNA:RNA bonds in the RNA secondary structures and are frequently found associated with the RNA degradosome complex, where they play a role in RNA decay and are also important for the correct assembly of the rRNA-protein complexes (24–28).
The response to temperature downshift frequently overlaps with the response to other stresses, indicating a more universal stress response. In cyanobacteria, it has been reported that cold-induced genes also respond to other stresses, such as high osmolarity, oxidative and acidic stresses, or redox perturbations (29). The pre-exposure to low temperature can increase the tolerance to other stresses, which indicates that the alteration in gene expression induced by cold may prepare the cells to withstand different severe situations (30, 31). Moreover, an interesting overlap between cold and hyperosmotic shock responses has been reported, showing that the accumulation of compatible solutes can increase cryoprotection (32, 33).
The cold shock genes are mainly regulated posttranscriptionally; mRNA stabilization and translation efficiency are differentially affected according to temperature. These mechanisms have been mostly studied for the CSP genes, in which the long 5′ untranslated region (UTR) was shown to have a role in cspA and cspB mRNA stability and translation (34–36). The CSP mRNAs contain cis elements that favor translation initiation at low temperatures, while other mRNAs are transiently inefficiently translated under these conditions, until the ribosomes become cold adapted by association with other factors (1). The CSPs, in turn, have a role in facilitating the transcription and translation of other highly structured mRNAs.
The alphaproteobacterium genus Caulobacter comprises free-living mesophiles and oligotrophs, widespread in humid soils associated with plants and in aquatic environments that can freeze for long periods of time (37, 38). The remarkable ability of Caulobacter crescentus NA1000 to withstand freezing is stimulated by nonfreezing temperatures, indicating that its cryotolerance can be induced by low temperatures (39). The genome of C. crescentus NA1000 encodes four paralogs of CSPs, two cold-induced (cspA and cspB) and the other two induced at stationary phase (cspC and cspD) (40). Three DEAD box RNA helicases are present: RhlE, DbpA, and RhlB, the latter being part of the RNA degradosome complex (41). The involvement of several of these proteins with cold stress resistance has been reported by previous studies (36, 39, 42). The cspA and cspA/cspB mutants present a more severe growth phenotype at 10°C than that of the cspB null mutant, suggesting that CspA may be more important to cold adaptation than CspB (36). The C. crescentus rhlE null mutant is deficient in growth at low temperatures and survival to freezing (39), and rhlE is upregulated at low temperatures due to both transcriptional and posttranscriptional mechanisms (42). Moreover, a change in the RNA degradosome composition was observed at low temperatures, when other proteins become attached to the main complex, including RhlE and Rho (42). These studies have identified a few genes important for cold adaptation, but they were mostly related to the physiological response toward the increase of RNA secondary structure stability. The study hereby presented aimed to identify other cold-regulated genes that could unveil a comprehensive picture of physiological adaptations to low temperatures in the oligotroph C. crescentus.
Although much is known about the cold stress response in many genera, scarce information is available regarding the strategies utilized by the alphaproteobacteria group. Therefore, a comprehensive analysis of the C. crescentus cold-induced genes could provide a picture of how this oligotrophic alphaproteobacterium responds to low-temperature stress. In this study, we used a global transcriptomic analysis (RNA-Seq) approach to characterize the C. crescentus response to low temperatures. This strategy allowed us to identify the differentially expressed genes (DEG) encoding proteins involved in a wide range of biological processes. In particular, a predominant category is the transmembrane transport of cations, with an expressive downregulation of the iron transporters and upregulation of the assimilatory nitrate reduction pathway and of the Kdp potassium uptake system, indicating that Fe2+, NH4+, and K+ homeostasis is relevant for this response. The basis for the cold induction of the kdp operon was investigated, showing that the DEAD box RNA helicase RhlE and the transcription termination factor Rho have opposite roles in maintaining the levels of kdp mRNA. Moreover, several small regulatory RNAs (sRNAs) were upregulated, indicating that posttranscriptional control is relevant for the regulation of cold stress response.
RESULTS
Global expression analysis.
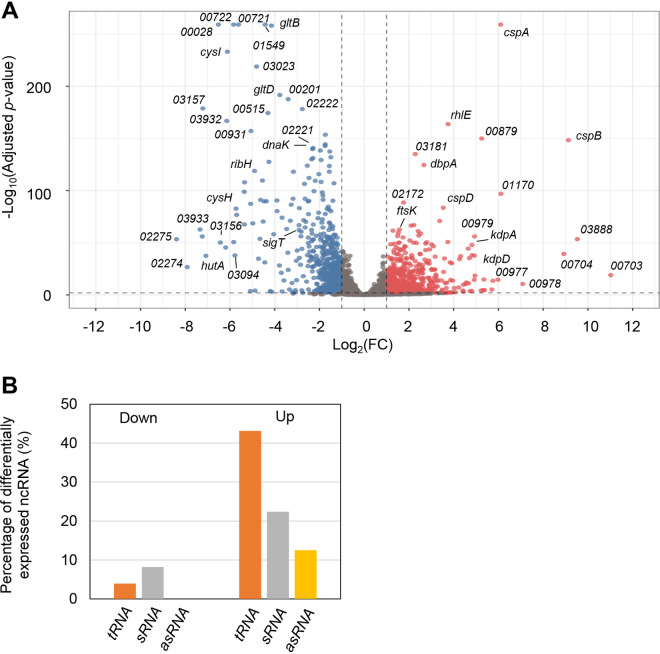
The response to temperature decrease in bacteria was previously characterized and develops in a two-step manner: an immediate response (called the acclimation phase), when there is a transient arrest of cell growth, followed by an adaptation phase, when cells resume growth (9). It was previously determined that in C. crescentus NA1000, the growth lag period following cold shock is about 4 h (39), but after 2 h the synthesis of most cold shock proteins is back to their initial levels (36). Therefore, to identify the genes differentially expressed in response to cold stress, we compared the transcriptome of the wild-type C. crescentus strain NA1000 under optimal growth conditions (30°C) and after 2 h under cold stress (10°C). Our results showed that 656 genes were upregulated and 619 were downregulated with a log2 fold change of ≥1 and ≤–1, respectively (Fig. 1A, Table S1 in the supplemental material). Of these, 66 genes for noncoding RNAs (ncRNAs) were differentially expressed, encoding tRNAs, small noncoding RNAs (sRNAs), and antisense RNA (asRNAs) (Fig. 1B). Interestingly, a higher percentage of the tRNAs and sRNAs was found among the upregulated genes.
FIG 1.
Genes that are differentially expressed during cold stress. (A) Volcano plot for the transcriptomic analysis of C. crescentus under cold stress. The genes represented by red dots are those that met the cutoff criteria of a log2 fold change of ≥1 and those by blue dots of ≤-1 and an adjusted P value of <0.01, while those represented by the gray dots did not meet the cutoff criteria. Dots corresponding to several of the most DEG, or the genes of interest, were labeled with the respective gene name as indicated. Genes satisfying the condition of adjusted P value = 0 had these values replaced by the smallest nonzero adjusted P value multiplied by 10−2. (B) Percentage of differentially expressed noncoding RNAs (ncRNA) that were decreased (Down) or increased (Up) in abundance (10°C/30°C). The percentage is relative to the total number of ncRNA in each category.
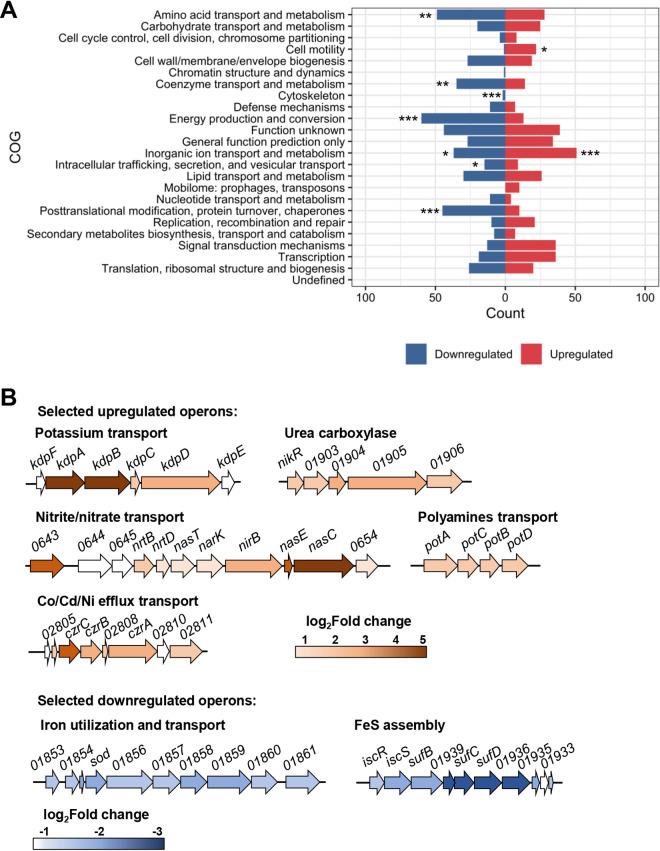
The Clusters of Orthologous Groups of proteins (COG) were used to identify the functional categories of the DEG (Fig. 2A, Table S1). An analysis of the enrichment of categories within the up- and downregulated groups was carried out, and the results showed that the inorganic ion transport and metabolism category was significantly enriched in both groups (Fig. 2A). Furthermore, a few other categories were enriched among the downregulated genes: amino acid transport and metabolism; coenzyme transport and metabolism; energy production and conversion; intracellular trafficking, secretion, and vesicular transport; and posttranslational modification, protein turnover, and chaperones. Regarding the set of upregulated genes, the cell motility category was the only one to be overrepresented besides inorganic ion transport and metabolism.
FIG 2.
Functional characterization of the DEG. (A) Enrichment of functional categories among DEG. The total number of genes belonging to distinct orthologous groups of encoded proteins (COG) was analyzed to verify whether the representation of each COG within the up- or downregulated groups was significant against the total gene background. The categories defined as being prominent in each group are those with an adjusted P value of <0.05, as indicated: *adjusted P value < 0.05; **adjusted P value < 0.01; ***adjusted P value < 0.001. (B) Graphic representation of selected up- and downregulated operons. Most of the operons encode transport systems. The levels of expression are color-coded approximately to the log2 fold change of each gene.
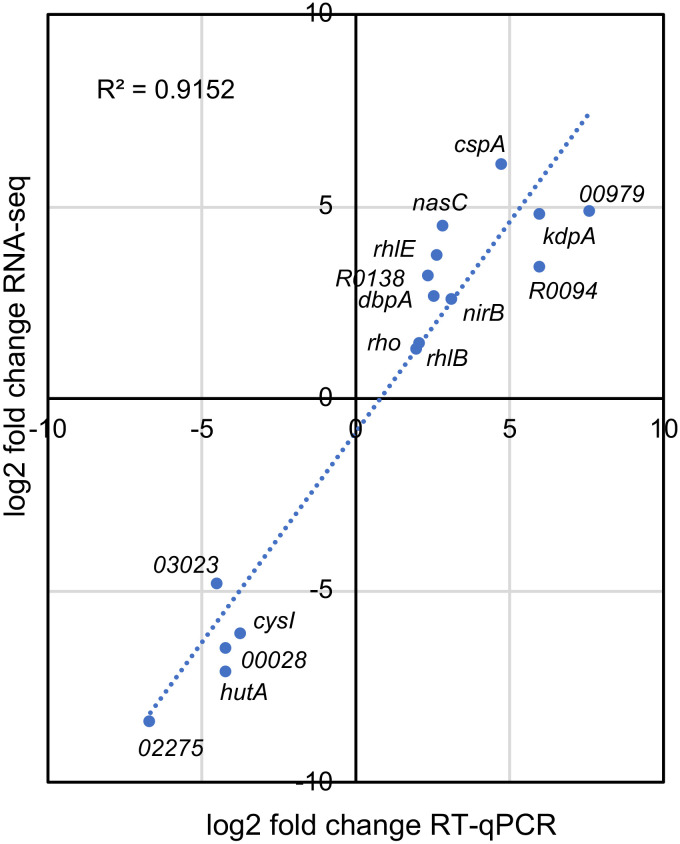
A more detailed analysis of the genes belonging to the transport category showed that the DEG were organized into large operons that were coregulated (Fig. 2B). We measured the expression of 11 upregulated and 5 downregulated genes by reverse transcription quantitative real-time PCR (RT-qPCR), and the results were consistent with the pattern of expression observed in the RNA-Seq experiments (Fig. 3), validating the transcriptome analysis. Some of the most relevant DEG will be discussed below.
FIG 3.
Validation of RNA-Seq data by RT-qPCR analysis. The relative expression was determined by RT-qPCR from the total RNA of cells incubated at 10°C for 2 h, relative to each gene expression at 30°C. The log2 fold change in the RT-qPCR data was plotted against the log2 fold change obtained from the RNA-Seq data. The expression levels for selected genes were plotted, and the identification of each gene is indicated.
Iron and sulfur uptake and central metabolism genes are downregulated under cold stress.
As discussed above, the genes encoding enzymes for the central energy metabolism (belonging to the energy production and conversion category) were downregulated at low temperatures, leading the cells to enter a slow-growth phase compatible with the new condition. All the genes encoding enzymes of the glycolysis/gluconeogenesis, citrate cycle, pentose phosphate, and oxidative phosphorylation pathways were downregulated (Table S1). Furthermore, the genes encoding assimilatory sulfate reduction and cysteine and methionine synthesis were strongly downregulated.
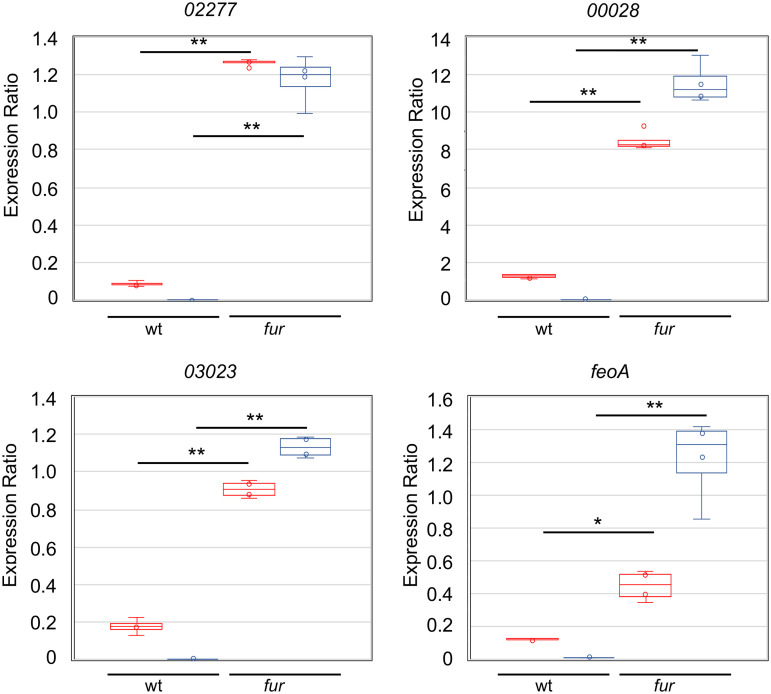
Interestingly, many of the most downregulated genes are involved in iron metabolism and are regulated by the ferric uptake regulator (Fur) (43). This was observed mostly for those negatively regulated, such as the iron transporters feoAB, hutA, CCNA_00028, CCNA_00138, and CCNA_03023; the bacterioferritin-associated ferredoxin CCNA_03372; the riboflavin synthesis operon CCNA_00929-0932; and the Fe-S cluster biogenesis operon CCNA_03156-3159 (Fig. 2B) (43, 44). In order to verify whether downregulation of these genes was Fur-dependent, RT-qPCR was carried out for four genes directly repressed by Fur (43, 44) in the wt and fur mutant, at 30°C and 10°C (Fig. 4). The results showed that while the relative expression was much higher in the fur mutant, as expected, downregulation was no longer observed at 10°C, and in fact expression increased (CCNA_00028, CCNA_03023, and feoA) or did not change (CCNA_02277). These results suggest that Fur is mediating the cold-induced downregulation of its regulon, perhaps due to changes in its affinity for iron or for its operator. Other Fur-independent genes were also severely downregulated, such as the groESL operon, which is upregulated in response to iron limitation (43), indicating that this effect could be related mostly to iron levels. The downregulation of the protein chaperones GroELS and DnaK also indicates that protein homeostasis is maintained at cold temperatures and that the sigma32-dependent heat shock response is turned off, agreeing with the lower protein synthesis activity.
FIG 4.
mRNA levels of Fur-regulated genes in C. crescentus. The expression was determined by RT-qPCR from the total RNA isolated from wild-type strain NA1000 (wt) or from the fur strain cultures growing in M2 medium, at 30°C (red) and after 2h at 10°C (blue). The dots indicate the individual results; the central bar indicates the mean, and the vertical bars indicate the standard error. The significance was calculated using Student’s t test (*P < 0.05, **P < 0.001).
The genes encoding the general stress extracellular function (ECF) sigma factors sigT and sigU were also downregulated, along with their coregulators NepR, PhyK, PhyR, and the regulatory RNA GsrN, indicating that the adaptation to low temperatures does not trigger a general stress response (45).
Upregulation of genes encoding RNA-binding proteins aids in coping with RNA secondary structures.
The most upregulated genes in response to cold stress encode proteins that interact with nucleic acids, notably RNA (Table S1). In particular, a pronounced increase in the expression of cspA, cspB, and rhlE was observed. C. crescentus NA1000 has four cold shock proteins (CSPs), CspA, CspB, CspC, and CspD. The cold-induced CspA and CspB have been reported to be essential for cold stress survival, while the stationary-phase-induced CspC and CspD are involved in adaptation to long starvation periods (36, 40, 46). While cspA, cspB, and cspD were among the cold-induced genes in our analysis, the levels of induction were distinct, with cspB being the most highly differentially expressed of all genes (560-fold), cspA presenting a 68-fold increase and cspD only an 11-fold increase. This high level of cspB expression was also observed for the genes downstream (CCNA_03888, CCNA_00703, and CCNA_00704) (see Fig. S1 in the supplemental material). These genes are transcribed independently from cspB and also encode small proteins (65 to 119 amino acids) with no sequence similarity. Interestingly, CCNA_03888 encodes a conserved basic protein (predicted pI = 10.6) and is syntenic with cspB in several genera of the alphaproteobacteria group (data not shown). The role of these small proteins in the cold-stress response is still unknown.
The genes for the DEAD box RNA helicases RhlE, DbpA, and RhlB and the transcription terminator factor Rho were also induced. The RNA helicase RhlE was already described as important for cold adaptation in C. crescentus, since the mutant rhlE is extremely sensitive to low temperatures (39, 42). RhlB is the main helicase associated with the RNA degradosome, but RhlE and DbpA were shown to also be associated with it at low temperatures (42). The DEAD box RNA helicases’ role in unwinding short RNA secondary structures may provide more effective degradation of highly structured mRNAs and therefore have an important role in mRNA stabilization under cold stress.
Upregulated noncoding RNAs indicate a posttranscriptional layer of regulation.
While 11 noncoding small regulatory RNAs (sRNAs) were downregulated, 31 sRNAs were upregulated at low temperatures at least 2-fold (Fig. 1B, Table S1). sRNAs act as posttranscriptional regulators, and several studies on their roles in environmental stress responses have been performed over the last decades (47–49). Although the gene for the RNA chaperone Hfq was not differentially expressed, previous studies have shown that it participates in this type of regulation, acting as an intermediate between the sRNA and its target (50, 51). Among the most upregulated sRNAs is the cell cycle-regulated CcnA (R0094) (11-fold), which is responsible for controlling two master regulators of the cell cycle, CtrA and GcrA (52). As the overexpression of CcnA leads to a slow growth phenotype (52), it is tempting to speculate that it contributes to the decrease in growth rates observed at low temperatures.
While 22 tRNAs were upregulated under our conditions, only two were downregulated (Fig. 1B). The complex regulation of protein synthesis during adaptation to stress is also associated with changes in the tRNA pool of the cell and the utilization of unusual codons (53, 54). However, the altered tRNAs in our analysis predominantly correspond to the expected codon usage preference of Caulobacter.
Upregulated genes for transport systems for detoxification and cation homeostasis.
Several genes related to transport and membrane components were upregulated at low temperatures, belonging to the secretion systems, lipid metabolism, TonB-dependent receptors, and flagellum assembly categories. Under cold stress, the membrane suffers from rigidification of its structure, and the cell needs to adapt its components to maintain its membrane fluidity and functions (2). Interestingly, however, very few DEG related to lipid metabolism and transport seem to be specifically associated with processes involved in countermeasures to the effects of cold on the membrane. In fact, only two genes encoding a fatty acid desaturase (CCNA_03535) and a sterol desaturase (CCNA_01743) were upregulated. Among those encoding transport systems, three genes encoding proteins belonging to the AcrB-like cation/multidrug efflux pump (CCNA_00850, CCNA_02809, and CCNA_03219) were upregulated. These proteins are part of the resistance/nodulation/division (RND) family and are involved in the export of cations or drugs. In fact, the upregulated czrCBA (CCNA_02806-09) operon (Fig. 2B) encodes an RND efflux system for cadmium/zinc/cobalt (55), suggesting that the detoxification of cations is an important response to low temperature conditions.
Nitrogen is one of the most important nutrients and also performs a regulatory role in modulating cell cycle progression via the regulation of (p)ppGpp accumulation (56), and C. crescentus cells use nitrate (NO3-) and ammonium (NH4+) as nitrogen sources (57). The upregulation of the genes encoding nitrogen regulatory proteins, the sigma factor RpoN, histidine kinase NtrB, and the nitrogen assimilation response regulator NtrC, agrees with the upregulation of many genes belonging to the ammonium limitation stimulon (57). The operon encoding the nitrate/nitrite transport systems nrtABD and narK-nirB-nasE-nasC was upregulated in our analysis (Fig. 2B). The Nrt ABC transporter and the NarK protein transport NO3- and NO2-, respectively, from the extracellular medium, and subsequently NasC converts NO3- into NO2-. Nitrite is then converted into NH4+ by NirB in the assimilatory nitrate reduction pathway. However, only one glutamine synthetase gene, glnA2, was upregulated, and the glutamate synthase genes (gltB and gltD) were highly downregulated, indicating that not all NH4+ may be incorporated into amino acids. Since the M2 medium provides sufficient ammonium chloride and the cultures are not under nitrogen deprivation, these data suggest that the upregulation of nitrogen metabolism could be providing an increase in the NH4+ concentration in the cytosol. In fact, other expression profiles of genes that could be involved in this response agree with this idea: the operon encoding the polyamine uptake system (putrescine/spermidine transport CCNA_03235-03239) was also upregulated, indicating the need to increase the uptake of NH4+; and the operon encoding urea-carboxylase (CCNA_01902-06), which regulates intracellular nickel concentration and was implicated in acid response in Helicobacter (58), was also upregulated (Fig. 2B). The expression of this operon could also be regulated by nickel ions, since urease is a nickel-binding protein, agreeing with the upregulation of the RND cation export system.
The Kdp potassium transporter is highly upregulated during cold stress.
The kdp operon, encoding a high-affinity potassium transporter system, was also upregulated at low temperatures, except for kdpE, which was not differentially expressed. However, a closer inspection identified a nonannotated gene upstream of kdpA that was also upregulated. This gene codes for a protein with high similarity to E. coli KdpF (Fig. S2A, B), so it is likely that in C. crescentus the operon kdpFABCDE is organized similarly as in other bacteria.
Upstream of kdpF, there is a σT-dependent promoter, and SigT is important for the induction of the operon under osmotic stress (45). Although the transcription start site (TSS) upstream of kdpF has not been determined, a putative TSS upstream of kdpC was identified, and the levels of expression of kdpCDE were higher than those for kdpFAB (59), suggesting that they are also independently regulated. Transcription from the σT-dependent promoter to the start codon of KdpF would generate a 5′ UTR comprising about 100 nucleotides that is predicted to be highly structured (Fig. S2C). This could reflect in posttranscriptional regulation of the kdp operon, altering the mRNA half-life or its translation, depending on the secondary structure.
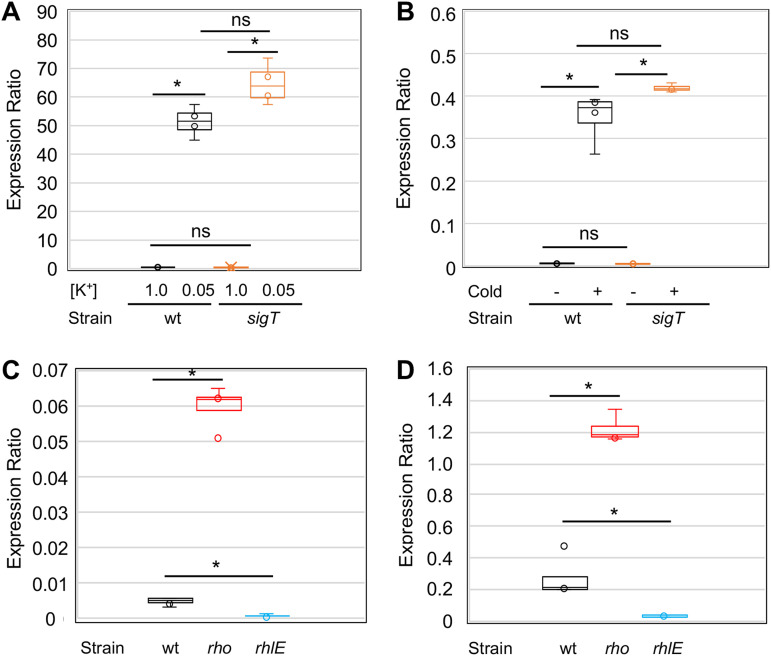
The upregulation of the kdp operon at low temperatures was intriguing, so we further analyzed its regulation in order to understand the molecular basis for the cold induction. Since this operon should respond to low K+ levels, we determined its expression in response to low [K+] concentrations and low temperatures, both in the wild-type strain NA1000 and in the sigT mutant strain. For the response to low [K+], kdpA expression was determined by RT-qPCR, using RNA isolated from cells grown under different [K+] in a Na+ phosphate-buffered M2 medium with increasing concentrations of KCl added. The results showed that the gene is highly induced by low [K+] both in the wt strain and in the sigT mutant (Fig. 5A), indicating that upregulation in this case does not require transcription from the SigT promoter. The same experiment was carried out using RNA from exponentially growing cells in M2 medium that were either incubated at 30°C or after 2 h at 10°C, and kdpA cold induction was also SigT-independent (Fig. 5B). In order to confirm that the observed cold induction was not restricted to the minimal medium condition, we determined the kdpA expression in rich peptone-yeast extract (PYE) medium at both temperatures (Fig. S3), and the results showed that it was induced to the same extent as in minimal medium. Therefore, the cold induction of kdpA might be regulated by a putative second promoter and/or by posttranscriptional mechanisms such as transcript stabilization.
FIG 5.
kdpA mRNA levels in C. crescentus. The expression was determined by RT-qPCR from the total RNA isolated from cultures growing in M2 medium, calculated as 2-ΔΔCT. (A) Expression was measured either in the wild-type (wt) strain NA1000 or in the sigT strain using total RNA from cultures incubated for 2 h in sodium phosphate-based M2 medium (M2-K) with sufficient (1 mM KCl) or deficient (50 μM KCl) potassium levels. The expression was determined relative to the respective culture in the K+-sufficient condition. (B) Expression was measured either in the wild-type strain NA1000 (wt) or in the sigT strain using the total RNA from cultures incubated in M2 medium at 30°C (Cold -) and after 2 h at 10°C (Cold +). The expression was determined relative to the wt strain at 30°C. (C) Expression from cells grown in M2 medium at 30°C from the wild-type strain (wt) and in the rho mutant strain (NA1000 rho::Tn5) and rhlE mutant strain (NA1000 rhlE::Tn5) (Table S2). (D) The same as in part C, but cultures were grown at 10°C. The results shown are from 4 independent experiments using RNA from 2 biological replicas. The significance was calculated using Student’s t test (*P < 0.001).
The RNA helicase RhlE and the transcription terminator Rho are induced during cold stress and probably have a role in the cold-adapted RNA degradosome (42). To investigate the role of Rho and RhlE in kdp expression, the expression of kdpA was determined in the C. crescentus rhlE and rho mutant strains by RT-qPCR (Fig. 5C and D). We found that, although the levels of kdpA mRNA were still higher at low temperatures in both strains (about 20-fold and 30-fold in the rho and rhlE mutant strains, respectively), the steady-state transcript levels were very different from those in the wt strain. The kdpA transcript levels were 5- to 10-fold higher in the rho mutant and lower to the same extent in the rhlE mutant at either temperature.
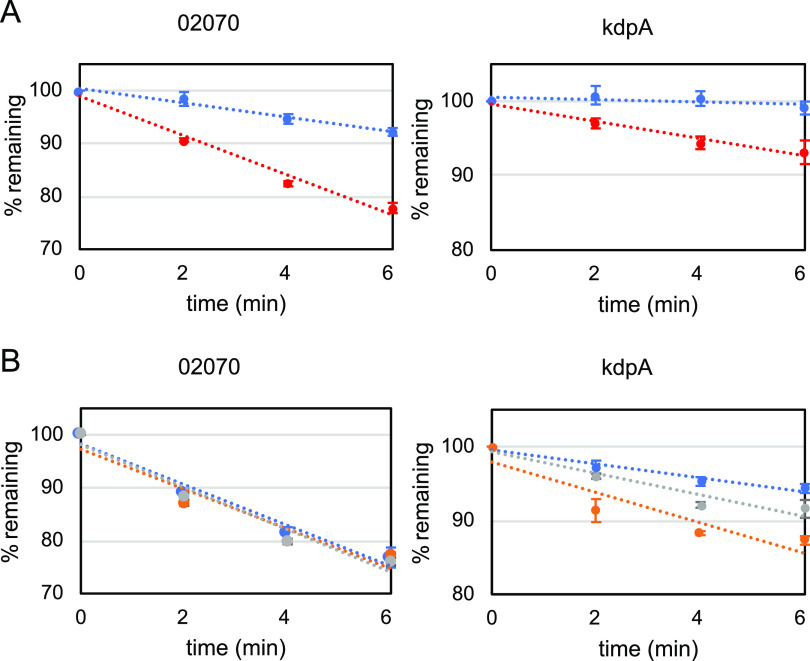
To investigate whether different mRNA stability could account for these differences, an mRNA decay assay was carried out in the wt strain grown at 30°C and 10°C (Fig. 6A). The results showed that both the control CCNA_02070 mRNA, which is not cold-induced, and the kdpA mRNA were stabilized at 10°C. However, the steady-state mRNA levels (defined by the expression ratio) of kdpA were much higher at 10°C (Fig. 5B), while those of CCNA_02070 were the same under both conditions. Moreover, mRNA decay was assessed in the wt, rho, and rhlE strains at 30°C (Fig. 6B), and the results showed that while the control mRNA decayed at a constant rate in all strains, the kdpA mRNA had a more pronounced decay rate in the rho and rhlE strains. These results indicate that mRNA stabilization could partly account for the increase in the expression of kdpA at 10°C. RhlE is required for the stabilization of the kdp mRNA and therefore for maximal levels of expression, perhaps by interacting with the highly structured 5′ region of the kdp operon.
FIG 6.
mRNA decay of kdpA. (A) C. crescentus NA1000 (wt) was grown in M2 medium at 30°C (red) or after 2 h at 10°C (blue), followed by treatment with 200 μg/ml rifampin. At the indicated times before (t = 0) and after the addition of rifampin, the cells were collected and the total RNA was extracted from 2 biological replicas. (B) C. crescentus NA1000 (blue), rho (orange), and rhlE (gray) strains were grown in M2 medium at 30°C, followed by treatment with 200 μg/ml rifampin. At the indicated times before (t = 0) and after the addition of rifampin, the cells were collected and the total RNA was extracted from 2 biological replicas. The mRNA levels of CCNA_02070 (control) and kdpA were determined relative to t = 0 by RT-qPCR using primer pairs for the amplification of each gene. The results shown are from 4 independent experiments using RNA from 2 biological replicas. The dotted lines indicate the tendency lines. The vertical bars indicate the standard deviation.
Surprisingly, the kdpA mRNA was also less stable in the rho mutant, despite the fact that Rho negatively regulates kdpA expression. One explanation for this effect on kdpA expression would be Rho-mediated transcription termination, since the kdpA transcript levels are much higher in the rho mutant. Interestingly, analysis of the RNA-Seq data for the kdp operon showed an abrupt decrease of the reads immediately after the kdpF gene (more visible at 10°C) (Fig. S4). This could be explained either by premature transcription termination or by cleavage followed by the destabilization of the downstream transcript. Although this region of the mRNA is highly structured, we did not find any putative intrinsic terminator, suggesting that if transcription attenuation is occurring, it might be mediated by Rho.
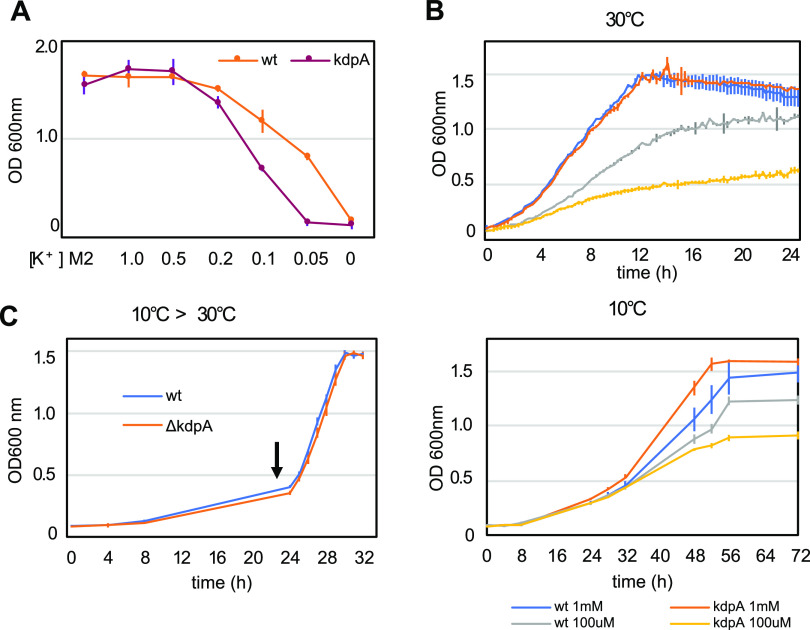
In order to investigate whether the potassium transport mediated by KdpA is important for response to low temperatures and other stresses, a strain with an in-frame deletion of kdpA was generated (Fig. S5) and phenotypically characterized. As expected, the kdpA strain showed a growth phenotype only in low [K+] (Fig. 7A). This difference in growth can be observed both at 30°C and at 15°C (Fig. 7B), indicating that this phenotype is not aggravated at low temperature. To verify whether the mutant could have a defect in the resumption of growth after cold stress, the cultures were grown at 10°C for 24 h and then transferred to 30°C (Fig. 7C), and no difference was observed in the growth rate. Moreover, the kdpA mutant phenotype was similar to that of the wt strain in response to osmotic stress generated by the addition of either 85 mM NaCl or 150 mM sucrose and to acidic pH (4.0) (Fig. S6). The resistance to low temperature was assessed by incubating the plates at 15°C for 5 days or freezing at –20°C before plating, and the results showed that the kdpA mutant presented no phenotypic alteration compared to the wt strain (Fig. S6). These results indicate that despite the kdp operon being highly induced at low temperatures, K+ uptake mediated by KdpA is not essential for growth at low temperatures, but it may contribute to the overall osmotic balance of the cell under these conditions.
FIG 7.
Phenotypic characterization of the kdpA mutant. (A) Growth of the wild-type strain and kdpA mutant in different potassium concentrations. Cells from ON cultures were washed with M2-K and inoculated into M2 medium or M2-K with increasing concentrations of KCl added as indicated. The growth was determined by measuring the OD600 of the cultures after 24 h at 30°C with agitation. [K+] is indicated in mM; M2, standard M2 minimal medium. (B) Growth of the wild-type strain and kdpA mutant in M2-K with either 100 μM or 1 mM KCl, incubated at 30°C or 10°C with agitation. (C) Cultures of the wild-type strain and kdpA mutant were grown in M2 medium at 10°C for 24 h and then transferred to 30°C at the time indicated (arrow). The lines indicate the averages and the vertical bars the standard deviation for each time point. The growth at 30°C was measured every 15 min and at 10°C every 4 h at the indicated time points.
DISCUSSION
The study of how bacteria cope with the physiological alterations caused by low temperature stress is of significance for understanding the different strategies used for each group. Although responses are common to several bacteria, such as those regarding RNA metabolism, other responses are particular to each group and are related to their way of life. C. crescentus is unique in this respect. It is a free-living alphaproteobacterium that thrives in oligotrophic aquatic environments and therefore severely suffers the effects of temperature change. This study has unveiled the strategies C. crescentus uses to maintain cell homeostasis and showed that the regulation of cation homeostasis is important for this response.
Bacteria have diverse mechanisms to overcome the cold stress effect in membranes, including incorporating desaturated fatty acids (2, 60). In E. coli, this adaptation occurs mainly by the incorporation of a monounsaturated fatty acid, via the cold-induced acyltransferase LpxP (61). B. subtilis and cyanobacteria seem to utilize similar approaches, but instead of introducing unsaturated fatty acids, they desaturate the current ones in the membrane by upregulating desKR and desABD, respectively (29, 62). It was proposed that C. crescentus can incorporate exogenous fatty acids into phospholipids and utilizes a similar synthesis pathway of unsaturated fatty acids as E. coli, via the β-hydroxydecanoyl thioester dehydrase (63). Moreover, genes encoding a fatty acid desaturase (CCNA_03535) and a sterol desaturase (CCNA_01743) were upregulated and perhaps have a role in adjusting the membrane lipid saturation in response to cold.
As expected, the cold shock genes cspA and cspB were highly induced, but surprisingly the stationary-phase-induced cspD gene was also upregulated. C. crescentus cspD is induced by carbon starvation via ppGpp but also responds to a decline in the growth rate, which is also observed after cold stress (64). The genes encoding components of the RNA degradosome, rlhB, and the genes for PNPase and RNase D were upregulated, as well as the DEAD box RNA helicases RhlE and DbpA and the transcription terminator Rho, as described for other bacteria (1).
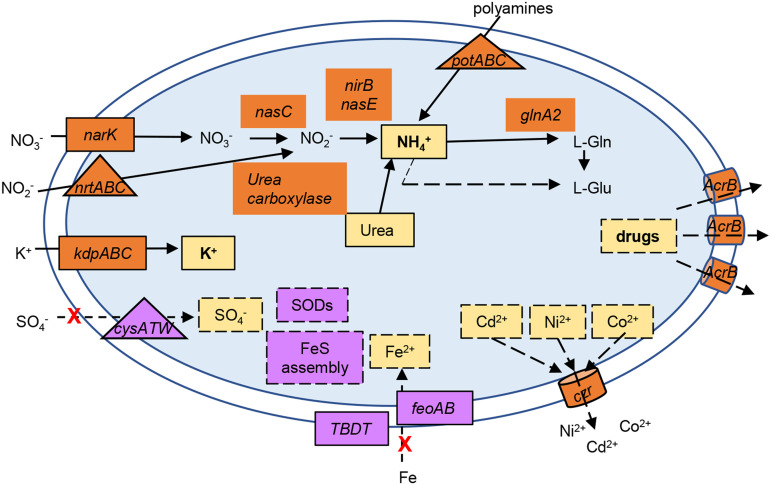
The most conspicuous alteration in gene expression was noted in the genes encoding transport systems (Fig. 8), indicating that cation homeostasis in oligotrophic environments is even more important at low temperatures. The downregulation of uptake systems and upregulation of efflux systems may lead to a decrease in the concentration of divalent cations, namely, Fe2+, SO42−, Ni2+, and Co2+, as well as toxic species such as Cd2+ and harmful molecules. Our results showed that the cold-induced downregulation of the Fur-regulated genes involved in iron homeostasis is mediated by Fur, perhaps to prevent oxidative stress. Several transport systems and enzymes for the assimilation and interconversion of nitrogen species were upregulated, suggesting an increase in the accumulation of NH4+. The preferred inorganic nitrogen source of the cell is NH4+, which can also be obtained by nitrate/nitrite reduction and in turn incorporated into glutamate by glutamine synthase (GlnA), generating glutamine. The C. crescentus NA1000 genome has three annotated glnA genes, glnA2 (CCNA_03230) being 2-fold upregulated. Moreover, the glutamate synthase gltDB genes were downregulated, suggesting that this increase in intracellular [NH4+] could result in the accumulation of free ammonium ions in the cell, probably to keep pH homeostasis. This, in turn, could inhibit the PTSNtr relay, limiting (p)ppGpp and delaying the progression of the cell cycle (56, 65, 66). This is also in agreement with the upregulation of the cell-cycle-regulated sRNA CcnA, which also leads to a decrease in the growth rate (52).
FIG 8.
Overview of the putative alterations in ion homeostasis upon cold stress. The ion transport systems or converting enzymes whose expression was upregulated (in orange) or downregulated (in purple) at 10°C compared to 30°C are indicated. The solid lines indicate a putative increase in the intracellular concentration, while the dashed lines indicate a putative decrease in the intracellular concentration of a given ion or molecule. The red X indicates a decrease of transport; the triangles indicate ABC-type transport systems, and the cylinders indicate RND efflux systems. Probably as a consequence of a lower intracellular concentration of Fe2+ and S, the genes encoding superoxide dismutases (SODs) and FeS assembly were also downregulated. Please refer to Fig. 2B for details of the genes involved.
Potassium is the main cation present within the bacterial cell, acting together with the compatible solute group, such as trehalose, to regulate osmolarity and other processes such as enzymatic activity and antifreezing resistance (67). C. crescentus does not synthesize trehalose and therefore must rely on a more restricted set of osmoregulatory compounds. The upregulation of the kdpFABCDE operon at low temperature indicates that intracellular K+ concentration is adjusted in response to cold. In K+-deficient environments, the high-affinity KdpFABC complex uses ATP to pump K+ into the cell. Interestingly, the low-affinity K+ transporters KefB and KefC were not differentially expressed under our conditions.
While the levels of K+ respond mainly to a shift in osmolarity, previous studies have reported that in Pseudomonas putida, kdpE is induced during cold adaptation, and in Salmonella enterica a kdpA mutant strain showed reduced long-term survival at low temperature after dehydration, suggesting that the importance of potassium homeostasis in low-temperature adaptation is spread among bacteria (68, 69). The kdpFABCDE operon is regulated at several levels, being activated by the two-component system KdpD/KdpE in response to low K+ levels and other signals (70, 71). A second regulatory system described in E. coli is via the nitrogen phosphotransferase system (PTSNtr), which is able to sense the cellular levels of nitrogen; under N-rich conditions, the nonphosphorylated PtsN binds to KdpD, which activates the expression of the kdp operon (72). The use of an ATP-driven high-affinity potassium transporter can be an adaptation to the oligotrophic condition of the environment but may cause a lower energetic efficiency, which leads to several alterations in the cell. Interestingly, the Kdp system also transports NH4+ when it is at a high concentration in the medium, leading to a futile cycle that leads to the acidification of the cytoplasm (73). The induction of these systems at low temperatures suggests that under these conditions, nutrient availability is perceived differently, perhaps as a result of the osmotic changes in the surrounding medium.
In C. crescentus, the kdp operon is regulated by the ECF-type sigma factor σT in response to osmotic stress (45), but the induction of kdpA both in response to cold as well as to low [K+] is not dependent on SigT (Fig. 4A and B), suggesting that there is a second promoter yet to be identified that could be regulated by KdpDE. Moreover, kdpA, and probably the whole kdpFABCDE operon, are posttranscriptionally regulated via transcript stability, which is affected by RhlE. The increase in transcript stability at low temperatures has been proposed as the main regulatory mechanism of cold-induced genes (23, 36, 74, 75), but transcript stabilization is observed for many genes under this condition. A second putative mechanism that could explain the kdpA transcript levels being much higher in the rho mutant even when the mRNA is less stable is transcription termination at a Rho-dependent terminator, which could also explain the abrupt decrease in RNA-Seq reads at the end of kdpF. In fact, the kdp operon is upregulated in bicyclomycin-treated E. coli cells, suggesting that Rho-mediated attenuation is a more general mechanism for the regulation of this operon (76).
In this study, we have characterized the cold-stress response in an oligotrophic free-living bacterium. Our results showed that there is a downshift in the central metabolism, leading to a slow growth mode, together with a decrease in the iron metabolism and the assimilatory sulfate reduction. On the other hand, the upregulation of transport systems, leading to an increase in cytoplasmic NH4+ and K+, suggests the need for adjusting the pH and osmotic homeostasis of the cytoplasm. The expressive number of transporters and efflux systems that were induced suggests that the response to cold stress is also increasing the resistance to other stresses, such as toxic compounds and drugs. The upregulation of genes for both transcriptional and posttranscriptional regulatory networks showed that the fine tuning of the cold stress response is complex and involves several layers of regulation.
MATERIALS AND METHODS
Strains and growth conditions.
The C. crescentus and E. coli strains are described in Table S2. C. crescentus NA1000 (77) was used as the wild-type strain for all experiments. C. crescentus cultures were grown at 30°C with agitation at 250 rpm in M2 minimal medium (78). E. coli strains were grown in LB complex medium. When necessary, kanamycin was then added to the cultures: 5 μg/ml for C. crescentus and 50 μg/ml for E. coli.
Growth at low [K+] was carried out in a sodium phosphate-buffered M2 medium (M2-K), where sodium phosphate substituted for potassium phosphate, and different final K+ concentrations were obtained by adding KCl back to the medium. To establish the [K+] concentration at which the mutant phenotype was most pronounced, the inoculum was grown in M2 medium overnight, washed twice in M2-K, and the cells were diluted to an optical density at 600 nm (OD600) of 0.1 in 15-ml tubes in 2 ml M2-K with different [K+] concentrations. The growth of NA1000 and kdpA strains was determined after incubation for 24 h at 30°C with agitation. Once the best [K+] concentration was determined, the growth was measured every 15 min in M2-K containing either 1 mM or 100 μM KCl in 2-ml cultures in 12-well plates incubated in the SpectraMax Paradigm device (Molecular Devices) with agitation at 30°C. Growth at 10°C was carried out the same way, except that the plates were incubated in an orbital shaker at 10°C and the OD600 value was measured in the device every 4 h.
For the RNA-Seq and RT-qPCR experiments, six 15-ml NA1000 cultures were grown in M2 in Erlenmeyer flasks at 30°C with agitation at 250 rpm until the OD600 value reached 0.5. Three cultures were collected and maintained at –80°C until RNA extraction, and three cultures were transferred to a shaker at 10°C and incubated with agitation at 250 rpm for 2 h before harvesting. The OD600 values of the cultures after this period at 10°C were very similar (0.6). The RNAs were obtained from three independent biological replicates at each temperature as described below. For RT-qPCR, the NA1000, sigT, and kdpA cultures were grown in M2 medium until the OD600 value equalled 0.3 to 0.4 and washed twice in M2-K; the cells were resuspended in the same volume of M2-K containing either 1 mM or 50 μM KCl. After incubation for 2 h at 30°C with agitation, the cells were collected by centrifugation for RNA extraction.
Total RNA extraction and sequencing (RNA-Seq).
Total RNA was extracted from exponential phase (OD600 = 0.5) cultures grown in the same batch of M2 minimal medium at 30°C and then transferred or not to 10°C for 2 h. For RT-qPCR experiments, total RNA was extracted from 1-ml cultures from two independent biological replicates using the TRIzol Reagent (Invitrogen Life Technologies), following the manufacturer’s instructions. The RNA concentration after extraction was determined using the OD260/280 ratio, and the integrity was confirmed by electrophoresis. The samples were maintained at –80°C until use.
For the RNA-Seq experiments, total RNA was extracted from 2-ml cultures from three independent biological replicates using an RNeasy minikit (Qiagen), and the RNA concentration and integrity were verified using a 2100 Bioanalyzer instrument (Agilent Technologies, Waldbronn, Germany). RNA-Seq libraries were prepared using the Illumina stranded total RNA ligation with a RiboZero Plus kit (Illumina) and sequenced using the NextSeq 500/550 midoutput kit v2.0 (150 cycles) (Illumina) on the Illumina NextSeq 500 platform.
RNA-Seq and functional enrichment analyses.
We processed the RNA-Seq raw data using the frtc pipeline (available at https://github.com/alanlorenzetti/frtc/) (43). Then, we performed differential expression analysis using DESeq2 (79) (available at https://github.com/alanlorenzetti/ccrescentus_RNASeq_analysis). A detailed description of the whole analysis is available in the supplemental material (RNASeq data analysis).
We carried out COG category enrichment analysis by checking for the overrepresentation of categories within the set of upregulated and downregulated genes. In summary, for each category, we performed the hypergeometric test implemented in the stats::phyper function of R and adjusted the computed P values using the Benjamini-Hochberg correction method implemented in the stats::p.adjust function of the same statistical suite.
Reverse transcription quantitative real-time PCR.
Two micrograms of total RNA from each sample was treated with one unit of DNase I (Invitrogen) and tested for the absence of genomic DNA by PCR using primers for the rho gene (Table S3). Single-strand cDNA synthesis was performed using the SuperScript III first-strand synthesis kit for RT-PCR (Invitrogen). Quantitative real-time PCR experiments were performed using Power SYBR green and PCR master mix (Applied Biosystems) with the respective primer pairs (Table S3) and the CCNA_02070 gene as a reference control. The reactions were performed in duplicate for each biological replicate in a StepOnePlus real-time PCR system (Thermo Fisher Scientific). The relative change in the expression of each gene was calculated using the 2-ΔΔCt relative expression quantification method (80).
mRNA decay experiments.
Cultures from strains NA1000 (wt), rho::Tn5, and rhlE::Tn5 were grown in M2 medium at 30°C with agitation until the OD600 value reached 0.5. Samples (1 ml) were centrifuged for 1 min; the pellets were resuspended in 0.5 ml of TRIzol Reagent (Invitrogen Life Technologies) and frozen in a dry ice/ethanol bath (t0). Then, 200 μg/ml rifampin (Sigma-Aldrich) was added to each culture kept at 30°C with agitation; samples were taken at several time points and immediately treated and frozen in the same manner. Determination of the mRNA decay at 10°C was carried out in the same way, except that the cultures were incubated for 2 h at 10°C before taking the t0 aliquot and adding rifampin.
Total RNA was isolated and converted to cDNA as described above. The mRNA decay was assessed by RT-qPCR using equal amounts of cDNA for each point, with primer pairs for the CCNA_02070 and kdpA genes (Table S3).
Construction of the ΔkdpA strain.
The kdpA gene was removed from the C. crescentus chromosome by an in-frame deletion generated by double homologous recombination. Fragments corresponding to the flanking regions of the kdpA gene were obtained via PCR amplification using the primer pairs kdpA IF1/kdpA IF2 and kdpA IF3/kdpA IF4 (Table S3). These fragments were then cloned into the pNPTS138 suicide vector (gift of M.R.K. Alley) using the In-Fusion HD cloning kit (TaKaRa Bio) and introduced into competent E. coli DH10B cells. The confirmed recombinant vector was then transformed into the conjugative strain E. coli S17-1 (81) and transferred to C. crescentus NA1000 through conjugation. The resulting colonies, containing the vector integrated into their chromosomes via homologous recombination, were selected for kanamycin resistance. Enrichment for the second recombination was obtained by growth in PYE medium for 48 h, followed by plating in PYE with 3% sucrose. The Kan-sensitive clones were analyzed using PCR with the primers del-Fw/del-Rv (Table S3), and a confirmed clone presenting a 0.65-kb fragment (Fig. S5) was the kdpA-deleted mutant selected for further characterization.
Statistical analyses.
The graphs show the average of the number of experiments indicated in each figure, and the bars indicate the standard deviation. The statistical significance was calculated pairwise using Student’s t test.
Data availability.
The raw RNA-Seq data generated in this study are publicly available at NCBI’s Sequence Read Archive (SRA) under accession number SRP310957. Detailed information about the samples and a project overview are available at NCBI under BioProject accession number PRJNA714975.
ACKNOWLEDGMENTS
This study was supported by the São Paulo Research Foundation (FAPESP, grant 2019/08514-0), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-Brasil, grant 307974/2017-0), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES) (finance code 001). During the course of this study, H.L.D.A., A.P.R.L., and A.M.V. were supported by FAPESP fellowships (grant 2018/17309-8 to H.L.D.A., grants 2017/03052-2 and 2019/13440-5 to A.P.R.L., and grant 2014/13552-4 to A.M.V.). H.L.D.A. and B.P.M. were supported by fellowships from CNPq-Brasil (grants 142391/2018-2 and 135489/2019-9, respectively).
Footnotes
Supplemental material is available online only.
Contributor Information
Marilis V. Marques, Email: mvmarque@usp.br.
Jeffrey A. Gralnick, University of Minnesota
REFERENCES
- 1.Phadtare S, Severinov K. 2010. RNA remodeling and gene regulation by cold shock proteins. RNA Biol 7:788–795. doi: 10.4161/rna.7.6.13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barria C, Malecki M, Arraiano CM. 2013. Bacterial adaptation to cold. Microbiology (Reading) 159:2437–2443. doi: 10.1099/mic.0.052209-0. [DOI] [PubMed] [Google Scholar]
- 3.Tribelli PM, López NI. 2018. Reporting key features in cold-adapted bacteria. Life 8:8. doi: 10.3390/life8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber MHW, Klein W, Müller L, Niess UM, Marahiel MA. 2001. Role of the Bacillus subtilis fatty acid desaturase in membrane adaptation during cold shock. Mol Microbiol 39:1321–1329. doi: 10.1111/j.1365-2958.2001.02322.x. [DOI] [PubMed] [Google Scholar]
- 5.Jones PG, VanBogelen RA, Neidhardt FC. 1987. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol 169:2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lottering EA, Streips UN. 1995. Induction of cold shock proteins in Bacillus subtilis. Curr Microbiol 30:193–199. doi: 10.1007/BF00293633. [DOI] [PubMed] [Google Scholar]
- 7.Graumann P, Schröder K, Schmid R, Marahiel MA. 1996. Cold shock stress-induced proteins in Bacillus subtilis. J Bacteriol 178:4611–4619. doi: 10.1128/jb.178.15.4611-4619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamanaka K. 1999. Cold shock response in Escherichia coli. J Mol Microbiol Biotechnol 1:193–202. [PubMed] [Google Scholar]
- 9.Phadtare S, Inouye M. 2004. Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J Bacteriol 186:7007–7014. doi: 10.1128/JB.186.20.7007-7014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langklotz S, Narberhaus F. 2011. The Escherichia coli replication inhibitor CspD is subject to growth-regulated degradation by the Lon protease. Mol Microbiol 80:1313–1325. doi: 10.1111/j.1365-2958.2011.07646.x. [DOI] [PubMed] [Google Scholar]
- 11.Awano N, Xu C, Ke H, Inoue K, Inouye M, Phadtare S. 2007. Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J Bacteriol 189:5808–5815. doi: 10.1128/JB.00655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunger K, Beckering CL, Wiegeshoff F, Graumann PL, Marahiel MA. 2006. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J Bacteriol 188:240–248. doi: 10.1128/JB.188.1.240-248.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loepfe C, Raimann E, Stephan R, Tasara T. 2010. Reduced host cell invasiveness and oxidative stress tolerance in double and triple csp gene family deletion mutants of Listeria monocytogenes. Foodborne Pathog Dis 7:775–783. doi: 10.1089/fpd.2009.0458. [DOI] [PubMed] [Google Scholar]
- 14.Söderholm H, Lindström M, Somervuo P, Heap J, Minton N, Lindén J, Korkeala H. 2011. cspB encodes a major cold shock protein in Clostridium botulinum ATCC 3502. Int J Food Microbiol 146:23–30. doi: 10.1016/j.ijfoodmicro.2011.01.033. [DOI] [PubMed] [Google Scholar]
- 15.Salze M, Muller C, Bernay B, Hartke A, Clamens T, Lesouhaitier O, Rincé A. 2020. Study of key RNA metabolism proteins in Enterococcus faecalis. RNA Biol 17:794–804. doi: 10.1080/15476286.2020.1728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giaquinto L, Curmi PMG, Siddiqui KS, Poljak A, DeLong E, DasSarma S, Cavicchioli R. 2007. Structure and function of cold shock proteins in archaea. J Bacteriol 189:5738–5748. doi: 10.1128/JB.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JS, Park SJ, Kwak KJ, Kim YO, Kim JY, Song J, Jang B, Jung C-H, Kang H. 2007. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res 35:506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang W, Hou Y, Inouye M. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem 272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 19.Schröder K, Graumann P, Schnuchel A, Holak TA, Marahiel MA. 1995. Mutational analysis of the putative nucleic acid‐binding surface of the cold‐shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single‐stranded DNA containing the Y‐box motif. Mol Microbiol 16:699–708. doi: 10.1111/j.1365-2958.1995.tb02431.x. [DOI] [PubMed] [Google Scholar]
- 20.Phadtare S, Tyagi S, Inouye M, Severinov K. 2002. Three amino acids in Escherichia coli CspE surface-exposed aromatic patch are critical for nucleic acid melting activity leading to transcription antitermination and cold acclimation of cells. J Biol Chem 277:46706–46711. doi: 10.1074/jbc.M208118200. [DOI] [PubMed] [Google Scholar]
- 21.Woodson SA, Panja S, Santiago-Frangos A. 2018. Proteins that chaperone RNA regulation. Microbiol Spectr 6:1–13. doi: 10.1128/microbiolspec.RWR-0026-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phadtare S, Inouye M, Severinov K. 2002. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J Biol Chem 277:7239–7245. doi: 10.1074/jbc.M111496200. [DOI] [PubMed] [Google Scholar]
- 23.Horn G, Hofweber R, Kremer W, Kalbitzer HR. 2007. Structure and function of bacterial cold shock proteins. Cell Mol Life Sci 64:1457–1470. doi: 10.1007/s00018-007-6388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Py B, Higgins CF, Krisch HM, Carpousis AJ. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 25.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol 48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 26.Charollais J, Dreyfus M, Iost I. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res 32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peil L, Virumäe K, Remme J. 2008. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J 275:3772–3782. doi: 10.1111/j.1742-4658.2008.06523.x. [DOI] [PubMed] [Google Scholar]
- 28.Phadtare S. 2012. Escherichia coli cold-shock gene profiles in response to over-expression/deletion of CsdA, RNase R and PNPase and relevance to low-temperature RNA metabolism. Genes Cells 17:850–874. doi: 10.1111/gtc.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinetova MA, Los DA. 2016. Systemic analysis of stress transcriptomics of Synechocystis reveals common stress genes and their universal triggers. Mol Biosyst 12:3254–3258. doi: 10.1039/c6mb00551a. [DOI] [PubMed] [Google Scholar]
- 30.Shah J, Desai PT, Chen D, Stevens JR, Weimer BC. 2013. Preadaptation to cold stress in Salmonella enterica serovar Typhimurium increases survival during subsequent acid stress exposure. Appl Environ Microbiol 79:7281–7289. doi: 10.1128/AEM.02621-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pittman JR, Buntyn JO, Posadas G, Nanduri B, Pendarvis K, Donaldson JR. 2014. Proteomic analysis of cross protection provided between cold and osmotic stress in Listeria monocytogenes. J Proteome Res 13:1896–1904. doi: 10.1021/pr401004a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wemekamp-Kamphuis HH, Sleator RD, Wouters JA, Hill C, Abee T. 2004. Molecular and physiological analysis of the role of osmolyte transporters BetL, Gbu, and OpuC in growth of Listeria monocytogenes at low temperatures. Appl Environ Microbiol 70:2912–2918. doi: 10.1128/AEM.70.5.2912-2918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann T, Bremer E. 2011. Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J Bacteriol 193:1552–1562. doi: 10.1128/JB.01319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gualerzi CO, Giuliodori AM, Pon CL. 2003. Transcriptional and post-transcriptional control of cold-shock genes. J Mol Biol 331:527–539. doi: 10.1016/S0022-2836(03)00732-0. [DOI] [PubMed] [Google Scholar]
- 35.Giuliodori AM, Fabbretti A, Gualerzi C. 2019. Cold-responsive regions of paradigm cold-shock and non-cold-shock mRNAs responsible for cold shock translational bias. Int J Mol Sci 20:457. doi: 10.3390/ijms20030457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazzon RR, Lang EAS, Silva CAPT, Marques MV. 2012. Cold shock genes CspA and CspB from Caulobacter crescentus are posttranscriptionally regulated and important for cold adaptation. J Bacteriol 194:6507–6517. doi: 10.1128/JB.01422-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang S, Wen X, Zhao L, Shi Y, Jin H. 2014. Crude oil treatment leads to shift of bacterial communities in soils from the deep active layer and upper permafrost along the China-Russia Crude Oil Pipeline route. PLoS One 9:e96552. doi: 10.1371/journal.pone.0096552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosoi-Tanabe S, Zhang H, Zhu D, Nagata S, Ban S, Imura S. 2010. Comprehensive analysis of an Antarctic bacterial community with the adaptability of growth at higher temperatures than those in Antarctica. Biocontrol Sci 15:57–62. doi: 10.4265/bio.15.57. [DOI] [PubMed] [Google Scholar]
- 39.Mazzon RR, Lang EAS, Braz VS, Marques MV. 2008. Characterization of Caulobacter crescentus response to low temperature and identification of genes involved in freezing resistance. FEMS Microbiol Lett 288:178–185. doi: 10.1111/j.1574-6968.2008.01337.x. [DOI] [PubMed] [Google Scholar]
- 40.Lang EAS, Marques MV. 2004. Identification and transcriptional control of Caulobacter crescentus genes encoding proteins containing a cold shock domain. J Bacteriol 186:5603–5613. doi: 10.1128/JB.186.17.5603-5613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardwick SW, Chan VSY, Broadhurst RW, Luisi BF. 2011. An RNA degradosome assembly in Caulobacter crescentus. Nucleic Acids Res 39:1449–1459. doi: 10.1093/nar/gkq928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguirre AA, Vicente AM, Hardwick SW, Alvelos DM, Mazzon RR, Luisi BF, Marques MV. 2017. Association of the cold shock DEAD-box RNA helicase RhlE to the RNA degradosome in Caulobacter crescentus. J Bacteriol 199:e00135-17. doi: 10.1128/JB.00135-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.da Silva Neto JF, Lourenço RF, Marques MV. 2013. Global transcriptional response of Caulobacter crescentus to iron availability. BMC Genomics 14:549. doi: 10.1186/1471-2164-14-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.da Silva Neto JF, Braz VS, Italiani VCS, Marques MV. 2009. Fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Res 37:4812–4825. doi: 10.1093/nar/gkp509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lourenço RF, Kohler C, Gomes SL. 2011. A two-component system, an anti-sigma factor and two paralogous ECF sigma factors are involved in the control of general stress response in Caulobacter crescentus. Mol Microbiol 80:1598–1612. doi: 10.1111/j.1365-2958.2011.07668.x. [DOI] [PubMed] [Google Scholar]
- 46.Balhesteros H, Mazzon RR, da Silva CAPT, Lang EAS, Marques MV. 2010. CspC and CspD are essential for Caulobacter crescentus stationary phase survival. Arch Microbiol 192:747–758. doi: 10.1007/s00203-010-0602-8. [DOI] [PubMed] [Google Scholar]
- 47.Sledjeski DD, Gupta A, Gottesman S. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J 15:3993–4000. doi: 10.1002/j.1460-2075.1996.tb00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoe C-H, Raabe CA, Rozhdestvensky TS, Tang T-H. 2013. Bacterial sRNAs: regulation in stress. Int J Med Microbiol 303:217–229. doi: 10.1016/j.ijmm.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Robledo M, Schlüter J-P, Loehr LO, Linne U, Albaum SP, Jiménez-Zurdo JI, Becker A. 2018. An sRNA and cold shock protein homolog-based feedforward loop post-transcriptionally controls cell cycle master regulator ctrA. Front Microbiol 9:763. doi: 10.3389/fmicb.2018.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santiago-Frangos A, Fröhlich KS, Jeliazkov JR, Małecka EM, Marino G, Gray JJ, Luisi BF, Woodson SA, Hardwick SW. 2019. Caulobacter crescentus Hfq structure reveals a conserved mechanism of RNA annealing regulation. Proc Natl Acad Sci USA 116:10978–10987. doi: 10.1073/pnas.1814428116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Assis NG, Ribeiro RA, da Silva LG, Vicente AM, Hug I, Marques MV. 2019. Identification of Hfq-binding RNAs in Caulobacter crescentus. RNA Biol 16:719–726. doi: 10.1080/15476286.2019.1593091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beroual W, Prévost K, Lalaouna D, Ben Zaina N, Valette O, Denis Y, Djendli M, Brasseur G, Brilli M, Robledo Garrido M, Jimenez-Zurdo J-I, Massé E, Biondi EG. 2019. Pulling the strings of cell cycle: a non-coding RNA, CcnA, modulates the master regulators CtrA and GcrA in Caulobacter crescentus. bioRxiv. doi: 10.1101/756452. [DOI] [PMC free article] [PubMed]
- 53.Torrent M, Chalancon G, de Groot NS, Wuster A, Madan Babu M. 2018. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci Signal 11:eaat6409. doi: 10.1126/scisignal.aat6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gingold H, Dahan O, Pilpel Y. 2012. Dynamic changes in translational efficiency are deduced from codon usage of the transcriptome. Nucleic Acids Res 40:10053–10063. doi: 10.1093/nar/gks772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valencia EY, Braz VS, Guzzo C, Marques MV. 2013. Two RND proteins involved in heavy metal efflux in Caulobacter crescentus belong to separate clusters within proteobacteria. BMC Microbiol 13:79. doi: 10.1186/1471-2180-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ronneau S, Petit K, De Bolle X, Hallez R. 2016. Phosphotransferase-dependent accumulation of (p)ppGpp in response to glutamine deprivation in Caulobacter crescentus. Nat Commun 7:11423. doi: 10.1038/ncomms11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.England JC, Perchuk BS, Laub MT, Gober JW. 2010. Global regulation of gene expression and cell differentiation in Caulobacter crescentus in response to nutrient availability. J Bacteriol 192:819–833. doi: 10.1128/JB.01240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones MD, Li Y, Zamble DB. 2018. Acid-responsive activity of the Helicobacter pylori metalloregulator NikR. Proc Natl Acad Sci USA 115:8966–8971. doi: 10.1073/pnas.1808393115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou B, Schrader JM, Kalogeraki VS, Abeliuk E, Dinh CB, Pham JQ, Cui ZZ, Dill DL, McAdams HH, Shapiro L. 2015. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet 11:e1004831. doi: 10.1371/journal.pgen.1004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber MHW, Marahiel MA. 2003. Bacterial cold shock responses. Sci Prog 86:9–75. doi: 10.3184/003685003783238707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vorachek-Warren MK, Carty SM, Lin S, Cotter RJ, Raetz CRH. 2002. An Escherichia coli mutant lacking the cold shock-induced palmitoleoyltransferase of lipid A biosynthesis: absence of unsaturated acyl chains and antibiotic hypersensitivity at 12°C. J Biol Chem 277:14186–14193. doi: 10.1074/jbc.M200408200. [DOI] [PubMed] [Google Scholar]
- 62.Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, de Mendoza D. 2001. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J 20:1681–1691. doi: 10.1093/emboj/20.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Letts V, Shaw P, Shapiro L, Henry S. 1982. Synthesis and utilization of fatty acids by wild-type and fatty acid auxotrophs of Caulobacter crescentus. J Bacteriol 151:1269–1278. doi: 10.1128/jb.151.3.1269-1278.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Da Silva CAPT, Lourenço RF, Mazzon RR, Ribeiro RA, Marques MV. 2016. Transcriptomic analysis of the stationary phase response regulator SpdR in Caulobacter crescentus. BMC Microbiol 16:66. doi: 10.1186/s12866-016-0682-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boutte CC, Crosson S. 2011. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment. Mol Microbiol 80:695–714. doi: 10.1111/j.1365-2958.2011.07602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonzalez D, Collier J. 2014. Effects of (p)ppGpp on the progression of the cell cycle of Caulobacter crescentus. J Bacteriol 196:2514–2525. doi: 10.1128/JB.01575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Welsh DT. 2000. Ecological significance of compatible solute accumulation by micro- organisms: from single cells to global climate. FEMS Microbiol Rev 24:263–290. doi: 10.1111/j.1574-6976.2000.tb00542.x. [DOI] [PubMed] [Google Scholar]
- 68.Frank S, Schmidt F, Klockgether J, Davenport CF, Gesell Salazar M, Völker U, Tümmler B. 2011. Functional genomics of the initial phase of cold adaptation of Pseudomonas putida KT2440. FEMS Microbiol Lett 318:47–54. doi: 10.1111/j.1574-6968.2011.02237.x. [DOI] [PubMed] [Google Scholar]
- 69.Gruzdev N, McClelland M, Porwollik S, Ofaim S, Pinto R, Saldinger-Sela S. 2012. Global transcriptional analysis of dehydrated Salmonella enterica serovar Typhimurium. Appl Environ Microbiol 78:7866–7875. doi: 10.1128/AEM.01822-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ballal A, Basu B, Apte SK. 2007. The Kdp-ATPase system and its regulation. J Biosci 32:559–568. doi: 10.1007/s12038-007-0055-7. [DOI] [PubMed] [Google Scholar]
- 71.Ali MK, Li X, Tang Q, Liu X, Chen F, Xiao J, Ali M, Chou S-H, He J. 2017. Regulation of inducible potassium transporter KdpFABC by the KdpD/KdpE two-component system in Mycobacterium smegmatis. Front Microbiol 8:570. doi: 10.3389/fmicb.2017.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sánchez-Cañizares C, Prell J, Pini F, Rutten P, Kraxner K, Wynands B, Karunakaran R, Poole PS. 2020. Global control of bacterial nitrogen and carbon metabolism by a PTSNtr-regulated switch. Proc Natl Acad Sci USA 117:10234–10245. doi: 10.1073/pnas.1917471117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teixeira De Mattos MJ, Neijssel OM. 1997. Bioenergetic consequences of microbial adaptation to low-nutrient environments. J Biotechnol 59:117–126. doi: 10.1016/s0168-1656(97)00174-0. [DOI] [PubMed] [Google Scholar]
- 74.Fang L, Jiang W, Bae W, Inouye M. 1997. Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol Microbiol 23:355–364. doi: 10.1046/j.1365-2958.1997.2351592.x. [DOI] [PubMed] [Google Scholar]
- 75.Ermolenko DN, Makhatadze GI. 2002. Bacterial cold-shock proteins. Cell Mol Life Sci 59:1902–1913. doi: 10.1007/pl00012513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. 2012. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev 26:2621–2633. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Evinger M, Agabian N. 1977. Envelope associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol 132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol 204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 79.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 81.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00710-21_Supp_1_seq10.xlsx, XLSX file, 0.1 MB (113.3KB, xlsx)
Supplemental material. Download SPECTRUM00710-21_Supp_2_seq11.pdf, PDF file, 0.5 MB (548.1KB, pdf)
Data Availability Statement
The raw RNA-Seq data generated in this study are publicly available at NCBI’s Sequence Read Archive (SRA) under accession number SRP310957. Detailed information about the samples and a project overview are available at NCBI under BioProject accession number PRJNA714975.