ABSTRACT
The Burkholderia cepacia complex (Bcc) comprises several species of closely related, versatile bacteria. Some Bcc strains produce 4-hydroxy-3-methyl-2-alkylquinolines (HMAQs), analogous to the 4-hydroxy-2-alkylquinolines of Pseudomonas aeruginosa. Using in silico analyses, we previously estimated that the hmqABCDEFG operon, which encodes enzymes involved in the biosynthesis of HMAQs, is carried by about one-third of Bcc strains, with considerable inter- and intraspecies variability. In the present study, we investigated by PCR, using consensus primers, the distribution of hmqABCDEFG in a collection of 312 Bcc strains (222 of clinical and 90 of environmental origins) belonging to 18 Bcc species. We confirmed that this operon is not distributed evenly among Bcc species. Among the 30% of strains bearing the hmqABCDEFG operon, we found that 92% of environmental isolates and 82% of clinically isolated Bcc strains produce levels of HMAQs detectable by liquid chromatography-mass spectrometry in at least one of the tested culture conditions. Among the hmqABCDEFG-positive but HMAQ-negative strains, none expressed the hmqA gene under the specified culture conditions. Interestingly, the hmqABCDEFG operon is more prevalent among plant root environment species (e.g., Burkholderia ambifaria and Burkholderia cepacia) and absent in species commonly found in chronically colonized individuals with cystic fibrosis (e.g., Burkholderia cenocepacia and Burkholderia multivorans), suggesting a role for the Hmq system in niche adaptation. We investigated the impact of the Hmq system on plant growth promotion and found that Pisum sativum root development by B. ambifaria required a functional HMAQ system.
IMPORTANCE Environmental bacteria belonging to the various closely related species forming the Burkholderia cepacia complex (Bcc) can infect plants and animals, including humans. Their pathogenicity is regulated by intercellular communication, or quorum sensing, allowing them to collaborate instead of acting individually. Bcc organisms generally exploit interacting quorum sensing systems based on N-acyl-homoserine lactones as signaling molecules. Several Bcc strains also carry an hmqABCDEFG operon responsible for the biosynthesis of 4-hydroxy-3-methyl-2-alkylquinolines (HMAQs), molecules analogous to the Pseudomonas quinolone signal (PQS) system of P. aeruginosa. Our finding that the prevalences of the Hmq system and HMAQ production are very different between various Bcc species suggests a key role in niche adaptation or pathogenicity. This is supported by a significant reduction in plant growth promotion in the absence of HMAQ production for a beneficial Bcc strain.
KEYWORDS: hmqABCDEFG operon, 4-hydroxy-2-alkylquinolines, pqsABCDE, Pseudomonas quinolone signal, plant growth promotion, quorum sensing, secondary metabolites
INTRODUCTION
The environmental species of Burkholderia can be divided into two phylogenetic groups: (i) pathogenic species and (ii) plant-beneficial species (1). Based on this still-controversial separation (2), the latter group was reclassified as Paraburkholderia, based on lower GC content and lack of virulence in Caenorhabditis elegans (3, 4). The pathogenic Burkholderia group comprises (i) plant pathogens (e.g., Burkholderia andropogonis, which causes leaf streak on sorghum [5], and B. glumae, which causes bacterial panicle blight on rice [6–8]); (ii) the pseudomallei group, composed of B. pseudomallei (the causative agent of melioidosis), B. thailandensis (closely related to B. pseudomallei but avirulent), and B. mallei (causing glanders in equids) (9–11); and (iii) the Burkholderia cepacia complex (Bcc), comprising at least 26 different species (12–14; reviewed in reference 1), most considered opportunistic pathogens.
Bcc bacteria have been used (i) in agriculture for the biocontrol of phytopathogens and for their plant growth-promoting properties (e.g., pea protection by B. ambifaria against Pythium and Aphanomyces [15, 16]) and (ii) in bioremediation (e.g., B. vietnamiensis with its trichloroethylene degradation abilities [17, 18; reviewed in reference 19]). Bcc bacteria are also well known for secondary-metabolite production, including antibiotics (reviewed in reference [20]). However, since the 1980s, Bcc opportunistic pathogens have emerged as a serious threat among certain immunocompromised individuals (e.g., those with chronic granulomatous disease [CGD]) and people with cystic fibrosis (CF), causing the cepacia syndrome and pushing authorities to prohibit their use in biotechnological applications. It is now generally accepted that their high transmissibility and intrinsic resistance to clinically relevant antibiotics make them particularly problematic (21–23).
Cell-to-cell communication mechanisms, e.g., quorum sensing, act by (i) controlling gene transcription at the population level (24), (ii) promoting colonization, and (iii) optimizing interaction with hosts and increasing resistance to stress (25, 26). Depending on the Bcc species, at least two Cep-like quorum sensing systems may be present, synthesizing different acyl-homoserine lactones (AHLs) as ligands and regulating each other and genes implicated in virulence (27–31).
The bacterium Pseudomonas aeruginosa carries a quorum sensing system based on signaling using 4-hydroxy-2-alkylquinolines (HAQs), such as the Pseudomonas quinolone signal (PQS) and 4-hydroxy-2-heptylquinoline (HHQ) (32–34). Interestingly, some species of the Bcc (B. ambifaria and B. cepacia), as well as B. pseudomallei and B. thailandensis, produce analogous molecules called 4-hydroxy-3-methyl-2-alkylquinolines (HMAQs) (35–37) that are synthesized by enzymes encoded by the hmqABCDEFG operon (36). In contrast with P. aeruginosa HAQs, the main HMAQs produced by Burkholderia bear a methyl group at the 3′ position and lack saturation of their alkyl side chain. Although an increasing number of Bcc strains are being reported to produce some HMAQ congeners (36, 38–44), this remains mostly anecdotal. In contrast with the HAQ/PQS quorum sensing system of P. aeruginosa, the Burkholderia Hmq system does not appear to form a quorum sensing system per se (specific transcriptional regulation of target genes in response to concentration of signaling molecules), and instead, we and Le Guillouzer have shown that it is closely interrelated with the Cep quorum sensing system in B. ambifaria HSJ1 and with the three Bta quorum sensing systems in B. thailandensis E264 (36, 45, 46), as HMAQs impact AHL production in both species (36, 45, 46).
Functions of P. aeruginosa HHQ and PQS as quorum sensing autoinducers, immunomodulators, and antimicrobials have been described (47). Several studies report novel molecules belonging to the HAQ family and various bacterial species having antimicrobial activities (48–52). Apart from some intra- and interspecies activity as signals (36, 45, 46) and as low-activity antimicrobials, the primary function of HMAQs remains enigmatic (38–41, 53, 54). Nonetheless, given the demonstrated role of HAQs and PQS in P. aeruginosa, HMAQs may also play roles in niche competition and virulence of producing Burkholderia strains (36, 46). We previously characterized a series of clinical B. ambifaria strains able to produce HMAQs (36, 43). Their respective colony morphotype variant had lost the ability to produce several secondary metabolites, including HMAQs, similarly to a set of environmental B. ambifaria isolates (36, 43). Therefore, to better understand HMAQs, we postulated that (i) the hmqABCDEFG operon is more frequently found among clinical Bcc strains and (ii) among HMAQ-producing Bcc isolates, clinical strains produce higher concentrations than environmental ones. Our previous bioinformatic study on the distribution of the hmqABCDEFG operon in the Bcc, based on available whole-genome sequences for 1,257 strains belonging to 21 Bcc species, showed that B. ambifaria, B. cepacia, B. contaminans, B. pyrrocinia, B. stagnalis, B. territorii, and B. ubonensis strains carry the hmqABCDEFG operon, although not all strains within a species do (55). On the other hand, all sequenced genomes of B. anthina, B. arboris, B. cenocepacia, B. diffusa, B. latens, B. metallica, B. multivorans, B. pseudomultivorans, B. seminalis, B. stabilis, and B. vietnamiensis—species mainly isolates from clinical cases—that have been investigated are lacking the hmqABCDEFG operon (55). To experimentally validate our in silico study and verify the ability of Bcc isolates carrying the hmqABCDEFG operon to actually produce HMAQs, a collection of 312 Bcc strains, comprising 222 clinical and 90 environmental isolates belonging to 18 different Bcc species, was analyzed. We first assessed the presence of the hmqABCDEFG operon in their genomes and then directly determined, by liquid chromatography coupled to mass spectrometry (LC-MS), the ability of all the strains bearing the hmqABCDEFG operon to produce HMAQs. Our data show that the Hmq system is heterogeneously distributed between Bcc species, with high prevalence in some species (e.g., B. cepacia) and near absence in others (e.g., B. cenocepacia and B. multivorans). Finally, we investigated the impact of the Hmq system on plant growth using hmqA and hmqG mutants and found that Pisum sativum root growth promotion by a B. ambifaria strain was lost in the absence of HMAQ production.
RESULTS
The hmqABCDEFG operon is heterogeneously distributed across and within Bcc species.
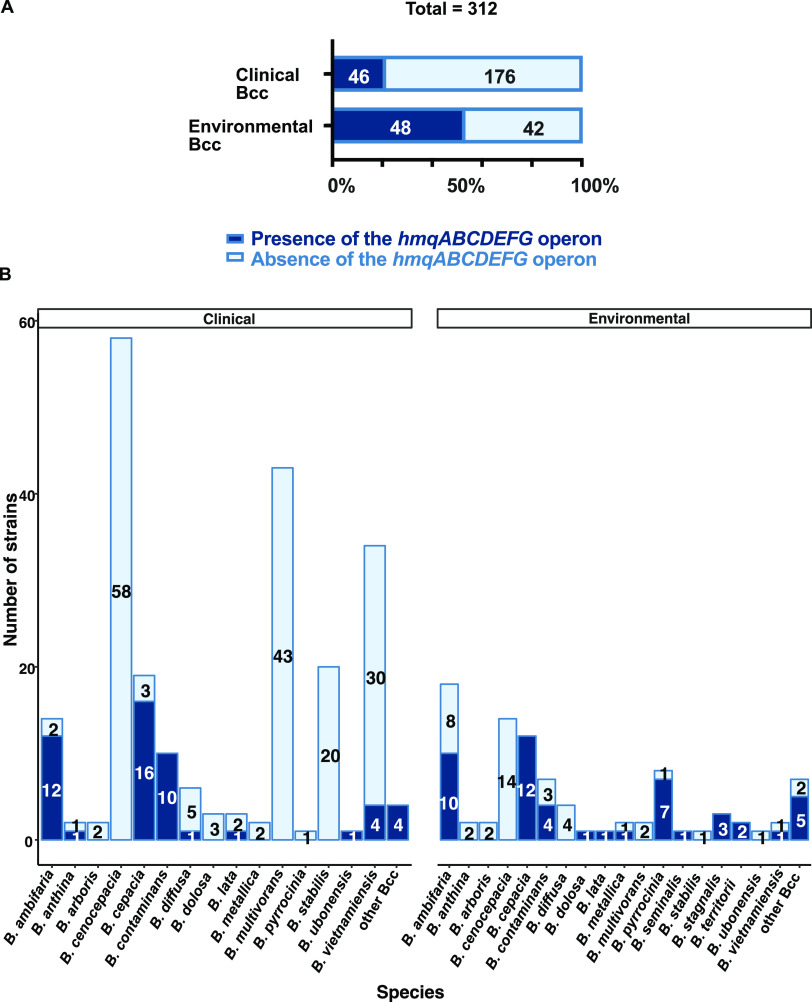
We previously examined 1,257 whole-genome sequences belonging to 21 different Bcc species to assess the distribution of the hmqABCDEFG operon by orthology and homology analyses (55). We found that at least one sequenced strain belonging to 7 of the 21 Bcc species carries the hmqABCDEFG operon (B. ambifaria, B. cepacia, B. contaminans B. pyrrocinia, B. stagnalis. B. territorii, and B. ubonensis); one striking initial finding was that prevalence of the hmqABCDEFG operon within a species appeared highly variable (55). Here, to validate our in silico analyses (55) and to globally determine the ability of Bcc to actually produce HMAQs, we screened a collection of 312 Bcc strains (222 of clinical and 90 of environmental origins; listed in Table S1), belonging to 18 Bcc species: B. ambifaria, B. anthina, B. arboris, B. cenocepacia, B. cepacia, B. contaminans, B. diffusa, B. dolosa, B. lata, B. metallica, B. multivorans, B. pyrrocinia, B. seminalis, B. stabilis, B. stagnalis. B. territorii, B. ubonensis, and B. vietnamiensis, plus a few more classified in the “other Bcc” group (PubMLST database [https://pubmlst.org/organisms/burkholderia-cepacia-complex]). The presence of hmqABCDEFG was determined by PCR using consensus primers targeting hmqA and hmqG (based on highly conserved regions [Fig. S1]). We had previously shown that presence of a hmqG orthologue correlates with the presence of a complete hmqABCDEFG operon (55). Here, we found that 30% of the 312 tested strains possess an hmqABCDEFG operon, including 53% of the environmental strains but only 21% of clinical strains (Fig. 1A). A Fisher test with a P value of 2.13 × 10−10 indicates that the prevalence of hmqABCDEFG is significantly different depending on the strains’ sampling origin (environmental versus clinical). Among the 18 different Bcc species investigated, 14 species comprise at least one strain carrying the operon (Fig. 1B and Fig. S2). However, B. cenocepacia, B. multivorans, and B. vietnamiensis are overrepresented among clinical strains compared to the other species. We thus decided to subsample these three species with only 20 clinical strains, increasing the prevalence of the hmqABCDEFG operon in clinical Bcc strains to 30% (46 strains carrying the operon among the 151 subsampled clinical strains), which is still statistically significantly lower than the prevalence of the hmqABCDEFG operon within environmental strains (Fisher test with a P value of 1.324 × 10−6).
FIG 1.
Bcc strain screening for presence of the hmqABCDEFG operon in their genomes. (A) Distribution of the operon between environmental and clinical Bcc strains investigated in this study (B) Distribution of the hmqABCDEFG operon within tested Bcc species.
More precisely, the hmqABCDEFG operon is more prevalent among clinical strains of B. ambifaria, B. anthina, B. contaminans, B. diffusa, B. ubonensis, and B. vietnamiensis, while it is more prevalent among environmental strains of B. cepacia, B. contaminans, B. dolosa, B. lata, B. metallica, B. pyrrocinia, and the “other Bcc” group. Clinical B. seminalis and environmental B. stagnalis and B. territorii species carry the hmqABCDEFG operon.
We found isolates of B. anthina, B. dolosa, and B. vietnamiensis carrying the hmqABCDEFG operon, which was not predicted in our previous analysis of available genomic data (55); we confirmed these PCR results by sequencing the amplicons using primers listed in Table S2.
Considering that horizontal gene transfer and pc3 chromosomal rearrangement could explain the heterogeneous distribution of the hmqABCDEFG operon in the Bcc.
Since not all Bcc species carry hmqABCDEFG, we asked whether the distribution of the operon could be related to a loss of the third chromosome in Bcc or to horizontal gene transfer. Bcc bacteria can lose their third chromosome, also described as a virulence megaplasmid (pc3) containing a few core genes (56, 57). The hmqABCDEFG operon being generally located on the pc3 replicon (except in B. ubonensis, which carries this operon on its second chromosome like B. pseudomallei and B. thailandensis), and because the synteny of the hmqABCDEFG operon is conserved within a species (55), we determined whether the strains missing the operon were also missing their pc3 replicon. Our analysis showed that the absence of the hmqABCDEFG operon does not correlate with the loss of pc3 (Table S3).
By comparing the phylogenetic tree based on multilocus sequence typing (MLST) sequences and the distribution of strains possessing the hmqABCDEFG operon, we found that the resulting MLST and hmqABCDEFG phylogenies mostly match, except for two groups, which are inverted (Fig. 2). Hence, the Hmq system originates from a common ancestor, with possible chromosomic rearrangement for some strains.
FIG 2.
Phylogeny indicates that acquisition of the hmqABCDEFG operon results from horizontal gene transfer within Bcc species. (A) Phylogeny of Bcc strains carrying the hmqABCDEFG operon, based on MLST genes (atpD, gltB, gyrB, recA, lepA, phaC, and trpB) (8). (B) Phylogeny of the Bcc species based on their hmqABCDEFG operon. Trees were generated by RAxML using the GTRGAMMA model and 1,000 bootstraps. The branches are labeled where bootstrap values are >50%. Strains in black were chosen as models and were not a part of our study. Strains used in this study are in red.
Most Bcc strains carrying the hmqABCDEFG operon produce HMAQs.
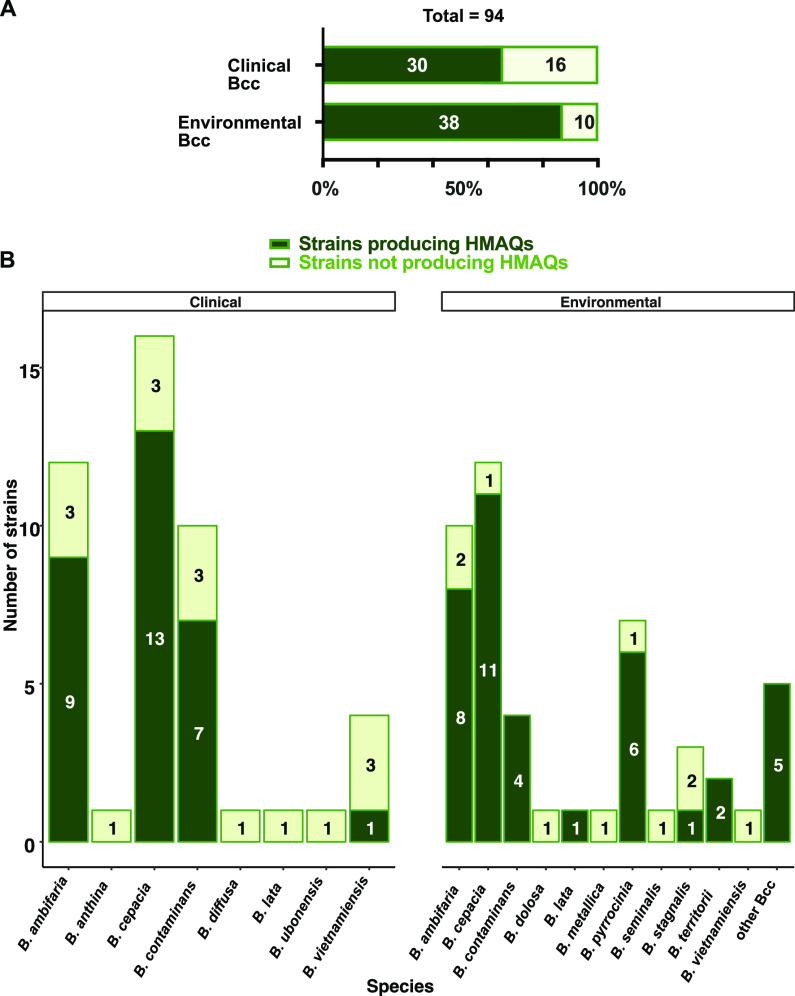
We then verified whether the presence of the biosynthetic genes is indicative of known HMAQ production. We cultured the 94 strains that were PCR-positive for hmqA and hmqG in tryptic soy broth (TSB) medium at 30°C and 60 rpm overnight and detected the production of HMAQ in 72% of the PCR-positive strains—65% clinical and 79% environmental—carrying the hmqABCDEFG operon (Table S4). None of the PCR-positive B. anthina, B. diffusa, B. dolosa, B. metallica, B. seminalis, and B. ubonensis strains produced detectable HMAQ under these culture conditions.
Assuming that the absence of HMAQ production in 26 of the 94 PCR-positive strains could simply be the result of unfavorable culture conditions, we assayed the production of these metabolites in TSB but at 37°C, in artificial sputum medium (ASM; at 30°C at 60 rpm, overnight), and on tryptic soy agar plates (TSA; incubated at 30°C for 4 days). As shown in Table 1, 4 of 26 previously negative strains produced HMAQs when grown in TSB at 37°C, while ASM growth induced production levels allowing detection of HMAQs in 10 of the 26 Bcc strains—seven environmental and three clinical strains. Surface growth on TSA plates induced the detectable production of HMAQs for five environmental and four clinical strains. Among the strains already producing HMAQs in TSB at 30°C, most of them also produced HMAQs in ASM and TSA (Table S4). Interestingly, two B. contaminans strains that were negative in both TSB and ASM produced detectable HMAQ when cultured on TSA plates. These additional culture conditions reduced the number of HMAQ-negative strains from 26 to 15, increasing the number of Bcc isolates able to produce HMAQs to a total of 82, that is, 87% of strains carrying the hmqABCDEFG operon, including 92% environmental and 82% clinical isolates (Fig. 3 and Fig. S3).
TABLE 1.
Production of HMAQs under alternative culture conditions for hmqABCDEFG-positive strains not producing HMAQs in tryptic soy broth at 30°C
| Strain | Type | HMAQ production |
||
|---|---|---|---|---|
| TSB (37°C) | ASM (30°C) | TSA (30°C) | ||
| B. ambifaria AMMD | Environmental | − | + | + |
| B. ambifaria AU7994 | Clinical | − | − | − |
| B. ambifaria CEP1231 | Clinical | − | + | − |
| B. ambifaria HI2425 | Environmental | − | + | + |
| B. ambifaria VC11631 | Clinical | − | − | − |
| B. anthina VC15382 | Clinical | − | − | − |
| B. cepacia ATCC 25416 | Environmental | − | − | − |
| B. cepacia VC13196 | Clinical | − | − | − |
| B. cepacia VC13575 | Clinical | + | − | − |
| B. cepacia VC19225 | Clinical | + | + | + |
| B. contaminans VC15406 | Clinical | − | − | + |
| B. contaminans VC16897 | Clinical | + | − | − |
| B. contaminans VC16948 | Clinical | + | − | + |
| B. diffusa VC14008 | Clinical | − | − | − |
| B. dolosa LMG21443 | Environmental | − | − | − |
| B. lata VC6377 | Clinical | − | + | + |
| B. metallica ES0559 | Environmental | − | − | − |
| B. pyrrocinia Bcc indeterminate 9 ES0209 | Environmental | + | + | + |
| B. seminalis HI2490 | Environmental | − | + | − |
| B. stagnalis Bcc indeterminate 6 HI3537 | Environmental | − | + | + |
| B. stagnalis HI2720 | Environmental | − | + | − |
| B. ubonensis LMG24263 | Clinical | − | − | − |
| B. vietnamiensis CEP0040 | Clinical | − | − | − |
| B. vietnamiensis VC17180 | Clinical | + | − | − |
| B. vietnamiensis VC9237 | Clinical | − | − | − |
FIG 3.
Distribution of HMAQ production among Bcc strains carrying the hmqABCDEFG operon. (A) Distribution of environmental and clinical Bcc strains regarding their ability to produce HMAQs in at least one of the tested culture conditions. (B) Distribution of HMAQ-producing Bcc species. HMAQs were detected and quantified by LC-MS with a limit of detection of 50 μg/liter for each molecule in the total culture.
To validate our strategy, we assessed the production of HMAQs by 31 Bcc strains determined by PCR not to carry the hmqABCDEFG operon; all 31 strains—belonging to B. ambifaria, B. anthina, B. arboris, B. cenocepacia, B. multivorans, B. pyrrocinia, B. stabilis, B. ubonensis, and B. vietnamiensis—indeed did not produce detectable HMAQs (Table S5).
All HMAQ-producing Bcc strains mainly produce the HMAQ-C7:2′ and HMAQ-C9:2′ congeners.
To verify which HMAQ congeners are produced by the various Bcc, we scanned by LC-MS for the 14 congeners of HAQs and HMAQs we had previously identified (Table S6) (36). We found that the main congeners produced were HMAQ-C7:2′ and HMAQ-C9:2′, as previously observed for B. ambifaria HSJ1 (36), followed by HMAQ-C8 and HMAQ-C6 (Fig. 4; Table S6). Most of the strains produced other HMAQs, such as HMAQ-C7 and HMAQ-C8:2′ (also known as burkholone).
FIG 4.
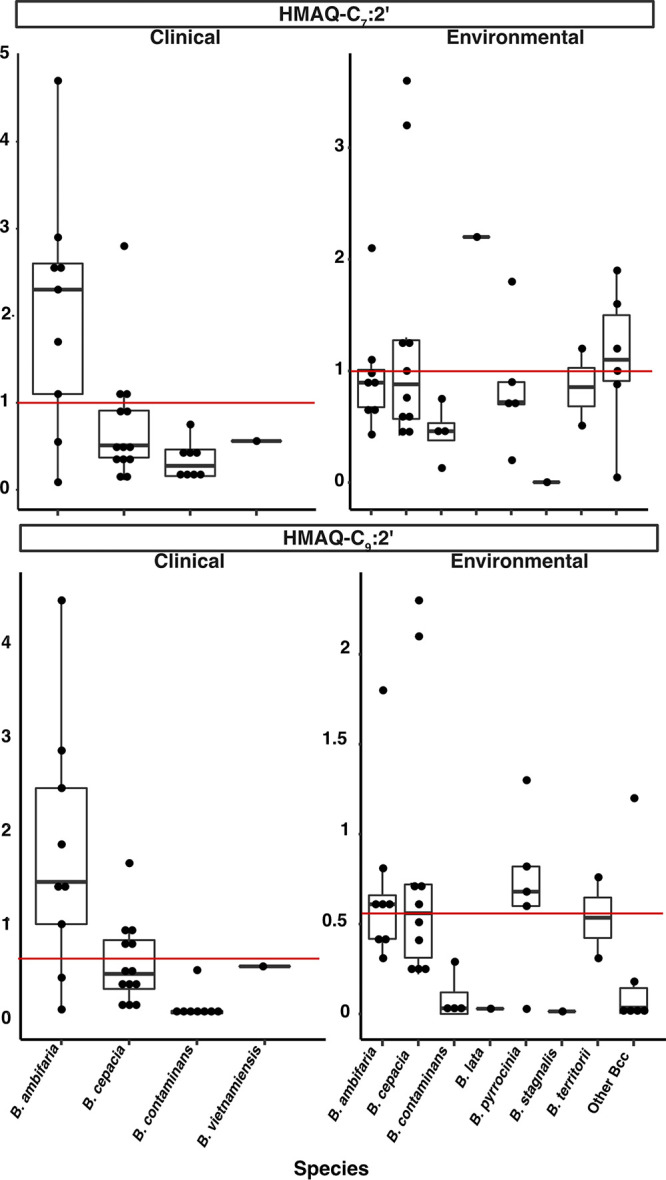
Concentration of the main produced HMAQs among Bcc species when cultured in tryptic soy broth. HMAQ-C7:2′ and HMAQ-C9:2′ were quantified by LC-MS with a lower limit of detection of 50 μg/liter for each molecule in the total culture. Tested strains are grouped by species. The production of HMAQs was quantified in three biological replicate cultures for each strain, and each dot represents the average HMAQ production for each strain, in milligrams per liter. Lines represent the production average for the clinical and environmental strains.
We also found that the concentration of HMAQs produced was variable among the various species when grown in TSB at 30°C. A Kruskal-Wallis test confirmed the absence of statistical difference in the concentrations produced between the clinical and environmental strains (P value of 0.19 for HMAQ-C7:2′ and 0.22 for HMAQ-C9:2′). However, clinical strains of B. ambifaria produced more HMAQs than environmental ones. The opposite was observed for B. cepacia and B. contaminans strains (Fig. 4).
Presence of the hmqABCDEFG operon and ability to produce HMAQs do not correlate with P. aeruginosa coisolation or the origin of the strain.
Since the hmqABCDEFG operon is homologous to the pqsABCDE operon in P. aeruginosa, and HMAQs might interact with the PQS system, we asked if HMAQ production of clinical Bcc is correlated with a coisolation or a colocalization with P. aeruginosa from the patient at some point as well as with the origin of the sample (sputum, throat, sinus, etc.). Information was available for 53 of 222 clinical strains (Table S7). Using Fisher's exact test for count data, we did not find a correlation between the presence of the hmqABCDEFG operon and the presence of P. aeruginosa at the sampling time or within the previous year or with the origin of the sample (Table 2).
TABLE 2.
Correlation between the presence of the hmqABCDEFG operon and the production of HMAQs in clinical Bcc, along with sampling data
| Characteristic | Correlation (P) witha: |
||
|---|---|---|---|
| Coisolation with P. aeruginosa | Colocalization with P. aeruginosa (previous yr) | Origin of sampleb | |
| Presence of the hmqABCDEFG operon | 0.34 | 0.41 | 0.11 |
| Production of HMAQs | 0.57 | 1 | 0.59 |
Raw data are from Table S7. P values are representative of the correlation (tested by Fisher’s exact test for count data) and are considered significant under 0.05.
Possible origins: sputum, respiratory sample, throat, or sinus.
Lack of expression of the hmqABCDEFG operon explains the absence of HMAQs detection in some Bcc cultures.
Our screening revealed that 26 strains carrying the hmqABCDEFG operon do not produce HMAQs in TSB at 30°C. To investigate the possibility that too low transcription of the biosynthetic genes would explain this absence of detectable production, which is compatible with the induction seen when the culture conditions were changed, we measured the transcription of the hmqABCDEFG operon for one HMAQ-negative and one HMAQ-positive strain per species of B. ambifaria, B. cepacia, B. contaminans, and B. vietnamiensis by targeting the hmqA gene by reverse transcription-PCR (RT-PCR).
Results show that the hmqABCDEFG-positive but HMAQ-negative strains B. ambifaria AMMD, B. cepacia ATCC 25416, B. contaminans VC15406, and B. vietnamiensis VC9237 do not express the hmqA gene when grown in TSB at 30°C, while B. ambifaria HSJ1, B. cepacia VC13394, B. contaminans FFH2055, and B. vietnamiensis VC8245, which produce HMAQs under these conditions, produce a clear hmqA transcript (Fig. S5). Thus, we decided to investigate if a mutation in the promoter region could explain the lack of expression of the hmqABCDEFG operon, but there was no difference between B. ambifaria HSJ1 and AMMD promoter region sequences. The lack of expression of hmqABCDEFG is a matter of regulation, especially since strain AMMD produced HMAQs in ASM and TSA. Extending these results, we hypothesize that the other strains which carry the hmqABCDEFG operon and do not produce HMAQs do not express the hmqA gene under the specified culture conditions. This is supported by the finding that several of these strains could eventually produce measurable levels of HMAQs when culture conditions were changed (as described above) (Table 1).
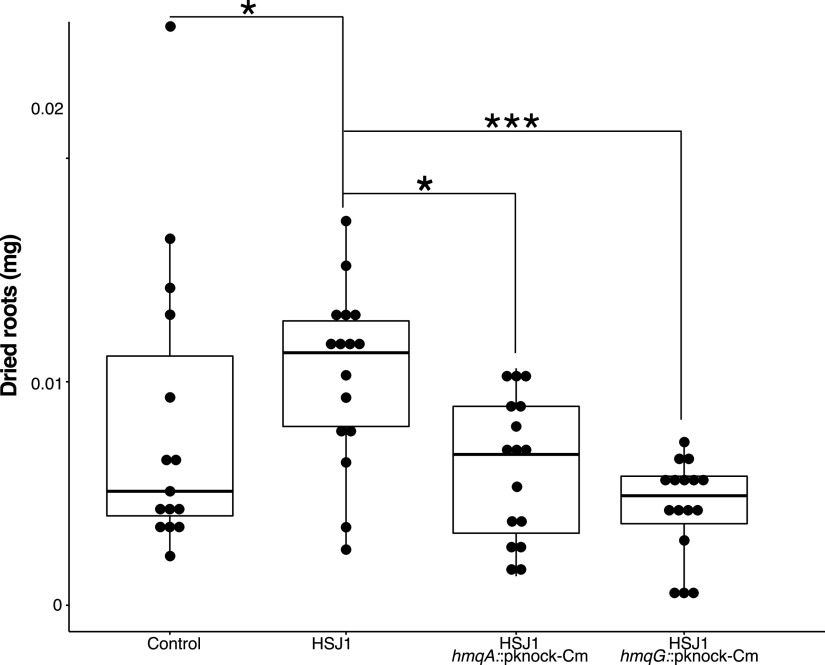
The capacity to produce HMAQs is required for root growth promotion.
Since B. ambifaria is a good plant growth promoter (15, 16; reviewed in reference 19) and the Hmq system is more prevalent among environmental strains, we investigated the impact of the Hmq system on growth of the common pea (P. sativum). While B. ambifaria HSJ1 promoted P. sativum roots’ development, the isogenic hmqA::pKnock-Cm and hmqG::pKnock-Cm mutants lost this effect (Fig. 5).
FIG 5.
Presence of the intact hmqABCDEFG operon in B. ambifaria promotes the development of roots in a P. sativum plant model. The impact of hmqA and hmqG mutants of B. ambifaria HSJ1 on root development of P. sativum compared to that of the wild-type strain was measured. Weight of dried roots was measured after 5 days of cultivation. A Dunn test was performed, and P values are represented as follows: *, between 0.5 and 0.01; **, between 0.01 and 0.001; ***, between 0.001 and 0.0001.
DISCUSSION
This study aimed at understanding the prevalence of the Hmq system and corresponding HMAQ production among the various species belonging to the Bcc to experimentally validate and extend our previous in silico analyses (55). Besides the non-Bcc species B. pseudomallei and B. thailandensis, only a few strains of B. ambifaria and B. cepacia were already known to carry the hmqABCDEFG operon and to produce HMAQs (16, 36, 38–44). Indeed, the presence of the hmqABCDEFG operon is well conserved in B. pseudomallei and B. thailandensis, but its prevalence within the Bcc remains unclear (55). To understand the ecological role of the Hmq system, we first needed to evaluate its distribution and prevalence. Since available genomes are not equally distributed between Bcc species, we screened a panel of 312 Bcc strains for the presence of the hmqABCDEFG operon by PCR. Since the hmqG gene is detectable only when the complete hmqABCDEFG operon is present and no significant variation of the hmqABCDEFG operon was observed in the 447 positive genomes analyzed by bioinformatics (55), we considered the operon to be present when both hmqA and hmqG genes were amplified. While the possibility that that some strains carry variations in the hmqABCDEFG operon cannot be excluded, we did not detect any in our previous bioinformatic analysis comparing whole genomes (55). The prevalence of the hmqABCDEFG operon found here for the Bcc followed the same distribution as that found by in silico analysis. Our PCR-based screening could miss some strains carrying the operon due to the limited availability of whole-genome sequences for some Bcc species (e.g., B. arboris, B. metallica, and B. stabilis). Nevertheless, our primers could amplify hmqA and hmqG targets in species not previously known to carry a hmqABCDEFG operon (e.g., B. vietnamiensis), which was expected due to the high level of identity (>88%) of hmqA and hmqG among identified and sequenced Bcc species (55). Furthermore, testing the production of HMAQs in a group of 31 PCR-negative strains confirmed the absence of production of these metabolites. Taking all the results together, we are confident that we identified the strains carrying the genetic ability to synthesize HMAQs.
Based on our previous results obtained with a few B. ambifaria isolates, we had hypothesized that the Hmq system would be more prevalent among clinical isolates and produce more HMAQs than environmental ones (36, 43, 46). Unexpectedly, we discovered that B. cenocepacia, B. multivorans, and B. vietnamiensis (the prominent Bcc species colonizing immunosuppressed individuals and those with CF) transmitted between patients (22, 23, 58) never or rarely (0 of 72 B. cenocepacia, 0 of 35 B. multivorans, and 5 of 35 B. vietnamiensis strains) carry the hmqABCDEFG operon. As this operon in Bcc results from a common ancestor, its absence could result from chromosomic rearrangement (by deletion or replacement) (55). In contrast, our data suggest that the Hmq system could play a beneficial role in niche adaptation to the rhizosphere microbial community due to the large prevalence of the hmqABCDEFG operon among environmental strains of B. ambifaria, B. cepacia, B. contaminans, B. pyrrocinia, and B. ubonensis, species known for their preference for the plant root environment (19, 59–62). The presence of the Hmq system in clinical strains of these five common environmental species is compatible with a recent or direct environmental acquisition in CF patients (60, 63, 64).
Among the strains carrying the hmqABCDEFG operon, not all produced HMAQs under the tested conditions. Since the limit of detection for these molecules was 50 μg/liter, we cannot exclude the possibility that some of these strains actually produced lower levels. For those strains producing HMAQs in TSB, growth in ASM did not inhibit HMAQ production and stimulated a detectable production in 10 additional strains, which could be explained by differences in regulation of the biosynthetic operon. Further, an increase in the number of HMAQ-positive strains when a third culture condition (4 days on TSA plates) was tested highlights the need to use nutritionally appropriate media for investigating the production of secondary metabolites (53). For this purpose, we tried to optimize a medium for HMAQ production. Nevertheless, our results show that the production of HMAQs in the Bcc is dependent on the conditions of growth and that hmqABCDEFG-positive strains not producing detectable HMAQs under the conditions tested here could produce enough HMAQs under different ones.
Lack of transcription of the hmqABCDEFG operon seemed to explain the observation that 28% of Bcc strains carrying the genes did not produce HMAQs when grown in TSB at 30°C. This result may be observed due to nonpromoter effects (e.g., posttranscriptional or posttranslational control). Indeed, production could be detected simply by changing the culture conditions, suggesting that the promoter of the hmqABCDEFG was not expressed, although we cannot exclude the possibility that it could be nonfunctional in some strains. We thus believe the same explanation (poor hmqABCDEFG transcription) could apply to the remaining 13% strains for which we could not yet identify appropriate culture conditions. Future work will involve developing a better understanding of the nutritional and regulatory elements (e.g., posttranscriptional or posttranslational regulation) controlling the expression of the hmqABCDEFG operon and production of HMAQs.
Because the hmqABCDEFG operon is more prevalent in environmental strains, which are often isolated from the rhizosphere, we investigated the impact of the Hmq system on the growth promotion ability of a B. ambifaria strain of a model plant, P. sativum. The inactivation of hmqA or hmqG prevented root growth promotion, presumably because of the inability to produce HMAQs. As is the case for cepacin and pyrrolnitrin, HMAQs could influence the rhizosphere microbial community, for instance, due to their antimicrobial activity (16, 36, 39–41, 45, 46, 50, 54, 65) or other properties. As we have previously reported that the HMAQs influence the Cep regulatory system, the molecules could indirectly impact the production of other plant-beneficial metabolites and functions regulated by quorum sensing (e.g., enacyloxin, pyrrolnitrin, cepacin, and antifungal cluster (afc) lipopeptide) (16, 30, 36, 39–41, 45, 46, 50, 54, 65–67). Further experiments with other HMAQ producers and pure HMAQs will provide exciting insights.
MATERIALS AND METHODS
Strains and culture conditions.
A total of 312 strains isolated from either clinical or environmental settings were used in this study (Table S1). Uncertain identification was confirmed by amplifying and sequencing the recA and gyrB genes at the IRCM Sequencing Platform (Montreal) following the directions on the PubMLST database (https://pubmlst.org/organisms/burkholderia-cepacia-complex) (Table S8).
Strains were cultured in borosilicate tubes containing 3 ml tryptic soy broth (TSB; Difco) from stocks frozen at −80°C in 15% glycerol and incubated at 30°C with 60 rpm rotative shaking overnight (∼16 h).
Detection of the presence of hmqABCDEFG operon by PCR.
Genomic DNA was extracted following a previously described method (68). Briefly, cells were resuspended in lysis buffer (50 mM Tris-HCl [pH 8], 5 mM EDTA-2Na [pH 8], 3% SDS) and transferred to a tube containing glass beads. The cells were lysed in a Fast-Prep-24 instrument (MP Biomedicals). Then the lysates were centrifuged at 8,000 × g for 5 min, the supernatants were transferred to a new tube, and 2.5 N ammonium acetate was added. The supernatant was transferred to a new tube, and 1 volume of isopropanol was added. The pellets were washed with 75% ethanol and dried before resuspension in 50 μl water.
For the strains acquired from the Burkholderia cepacia Research Laboratory and Repository and from the Canadian Burkholderia cepacia complex Research and Referral Repository, genomic DNA was extracted using a 96-well plate gDNA extraction kit (Favorgen, Canada).
The hmqA and hmqG genes were amplified by PCR using EasyTaq polymerase (Transgen, Canada). Primers were designed based on a consensus sequence of 11 complete Bcc sequences available in 2017 (Table S8). A strain was considered to have a complete hmqABCDEFG operon when amplification of both hmqA and hmqG targets was attained, based on our previous analyses (55).
Detection of the presence of the pc3 chromosome and the hmqABCDEFG operon in Bcc strains.
Assembled genomes are available on DB Burkholderia (http://www.Burkholderia.com). We selected only complete assembled genomes and searched for the presence of the hmqABCDEFG operon by orthology and homology directly on DB Burkholderia. Results are listed in Table S3.
Phylogeny of the Bcc based on MLST and hmqABCDEFG sequences.
First, atpD, gltB, gyrB, recA, lepA, phaC, and trpB gene sequences were used to generate a Bcc phylogeny based on MLST. Second, the hmqABCDEFG operon sequence was used to determine the coevolution or horizontal transfer of the Hmq system within Bcc species. Sequences were found on DB Burkholderia (http://www.Burkholderia.com) and concatenated in the order atpD, gyrB, recA, gltB, lepA, phaC, and trpB, with hmqABCDEFG in parallel. The resulting concatenated sequences were aligned using Clustal Omega (69), and trees were generated by RAxML using the GTRGAMMA model and 1,000 bootstraps. The branches are labeled where bootstrap values are >50.
Quantification of HMAQ production by LC-MS/MS.
Bcc strains were cultured in 5 ml TSB at a starting optical density at 600 nm (OD600) of 0.05 and incubated at 30°C with shaking for an overnight. 5,6,7,8-Tetradeutero-4-hydroxy-2-heptylquinoline (HHQ-d4) was used as an internal standard (70). The total HMAQs were extracted from 4 ml culture with 1 volume of ethyl acetate. After nitrogen evaporation, the residues were dissolved in 400 μl high-performance liquid chromatography (HPLC)-grade acetonitrile. Samples were analyzed by liquid chromatography coupled with mass spectrometry (LC-MS) in positive electrospray ionization using a Kinetex 5-μm EVO C18 100-Å 100- by 3-mm reverse-phase column as previously described (70). A Quattro Premier XE triple quadrupole was used as a detector (Waters). A multiple reaction monitoring (MRM) program was used to detect HMAQ families (36). This experiment was conducted with three independent biological replicates.
ASM, TSB, and TSA medium assays.
The strains were grown in ASM liquid medium (71) and TSA agar plates out from overnight cultures and incubated at 30°C for 24 h and 4 days, respectively, or in grown TSB and incubated at 37°C for 24 h.
One milliliter of ASM culture was extracted as described for the HMAQ production method. For each TSA plate, 5 ml water was added to extract the HMAQs from the agar. For each sample, 1 ml was extracted with 1 volume ethyl acetate containing 4 ppm HHQ-D4, concentrated 10 times, dissolved in HPLC-grade acetonitrile, and analyzed as described above for the HMAQ extraction method.
The experiments were performed in two independent biological replicates.
Detection of the expression of the hmqABCDEFG operon by RT-PCR.
Total RNA was extracted from cultures grown in TSB to an OD600 of 3.0 using TransZol (Transgene, Canada) by following the manufacturer’s instructions. Residual DNA was removed using the Turbo DNase (Thermo Fisher, Canada). Reverse transcription was performed using the iScript kit (Bio-Rad, Canada). The expression of the hmqABCDEFG operon was determined by PCR targeting hmqA. The ndh gene served as a reference gene (Table S8) (72).
Correlation between the presence of the hmqABCDEFG operon and the production of HMAQs in Bcc with different characteristics.
Based on our qualitative data (Table 2 and Table S7), we studied the correlation by Fisher’s exact test for count data using R software (http://www.R-project.org) (73).
Pisum sativum growth promotion.
Pisum sativum seeds were decontaminated using successive bleach and 70% ethanol treatments (10 min each, under shaking [60 rpm], 3 times) and then germinated for 4 days at room temperature on 0.8% agar plates. B. ambifaria HSJ1 and isogenic hmqA::pKnock-Cm, and hmqG::pKnock-Cm mutants, previously respectively generated by integrating a 757-bp internal fragment of hmqA and a 525-bp fragment of hmqG from HSJ1 into the suicide vector pKnock-Cm and by selecting single-crossover insertion mutants obtained following mating Escherichia coli SM10 (pKnock-Cm-hmqA or pKnock-Cm-hmqG) with B. ambifaria HSJ1 (36), were incubated at 30°C overnight in TSB with shaking at 60 rpm. Seeds were exposed by adding 105 bacteria/ml in 5 ml Murashige and Skoog (MS) basal medium (Sigma) (43). Plants were grown at room temperature for 5 days. Roots were then cut, dried at 52°C overnight, and weighed. A Dunn test was used for statistical analyses. This experiment was repeated twice, with the same conclusions.
ACKNOWLEDGMENTS
This work was supported by grant MOP-142466 from the Canadian Institutes of Health Research (CIHR). E.D. held the Canada Research Chair in Sociomicrobiology.
We thank Silvia Cardona (University of Manitoba, Canada) for providing B. contaminans FFH2055; Paola Cescutti (University of Trieste, Italy) for providing B. cepacia BTS13 strain; David Wagner (Menzies School of Health Research and Northern Arizona University, USA) for providing B. stagnalis MSMB1956WGS, B. territorii MSMB1301WGS, and B. territorii MSMB1502WGS; and John LiPuma (University of Michigan, USA) for providing the 75 environmental strains listed in Table S1. We also thank Phillipe Constant (INRS) for help with statistics and Marie-Christine Groleau for achieving experimental wonders and for her great comments on reviewing the article draft.
P.M.L.C.: conception and design of the manuscript and acquisition of data; J.E.A.Z.: provision and identification of 200 clinical Bcc; E.D.: conception of the project, resources, and funding; P.M.L.C., J.E.A.Z., and E.D.: analysis and interpretation of data, revision of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Eric Déziel, Email: eric.deziel@inrs.ca.
Jeffrey A Gralnick, University of Minnesota.
REFERENCES
- 1.Eberl L, Vandamme P. 2016. Members of the genus Burkholderia: good and bad guys. F1000Res 5:1007. doi: 10.12688/f1000research.8221.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vandamme P, Peeters C, Smet BD, Price EP, Sarovich DS, Henry DA, Hird TJ, Zlosnik JEA, Mayo M, Warner J, Baker A, Currie BJ, Carlier A. 2017. Comparative genomics of Burkholderia singularis sp. nov., a low G+C content, free-living bacterium that defies taxonomic dissection of the genus Burkholderia. Front Microbiol 8:1679. doi: 10.3389/fmicb.2017.01679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angus AA, Agapakis CM, Fong S, Yerrapragada S, los Santos PE, Yang P, Song N, Kano S, Caballero-Mellado J, de Faria SM, Dakora FD, Weinstock G, Hirsch AM. 2014. Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS One 9:e83779. doi: 10.1371/journal.pone.0083779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawana A, Adeolu M, Gupta RS. 2014. Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet 5:429. doi: 10.3389/fgene.2014.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramundo BA, Claflin LE. 2005. Identification of Burkholderia andropogonis with a repetitive sequence BOX element and PCR. Curr Microbiol 50:52–56. doi: 10.1007/s00284-004-4354-z. [DOI] [PubMed] [Google Scholar]
- 6.Azegami K, Nishiyama K, Watanabe Y, Suzuki T, Yoshida M, Nose K, Toda S. 1985. Tropolone as a root growth-inhibitor produced by a plant pathogenic Pseudomonas sp. causing seedling blight of rice. Jpn J Phytopathol 51:315–317. doi: 10.3186/jjphytopath.51.315. [DOI] [Google Scholar]
- 7.Nandakumar R, Shahjahan AKM, Yuan XL, Dickstein ER, Groth DE, Clark CA, Cartwright RD, Rush MC. 2009. Burkholderia glumae and B. gladioli cause bacterial panicle blight in rice in the southern United States. Plant Dis 93:896–905. doi: 10.1094/PDIS-93-9-0896. [DOI] [PubMed] [Google Scholar]
- 8.Ham JH, Melanson RA, Rush MC. 2011. Burkholderia glumae: next major pathogen of rice? Mol Plant Pathol 12:329–339. doi: 10.1111/j.1364-3703.2010.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanford JP. 1990. Pseudomonas species (including melioidosis and glanders), p 1692–1696. In Mandell GL, Douglas RG, Bennett JE (ed), Principles and practice of infectious disease. Elsevier, New York, NY. [Google Scholar]
- 10.Chaowagul W, White NJ, Dance DAB, Wattanagoon Y, Naigowit P, Davis TME, Looareesuwan S, Pitakwatchara N. 1989. Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand. J Infect Dis 159:890–899. doi: 10.1093/infdis/159.5.890. [DOI] [PubMed] [Google Scholar]
- 11.Howe C. 1950. Glanders, p 185–202. In Christian HA (ed), The Oxford medicine. Oxford University Press, New York, NY. [Google Scholar]
- 12.Bach E, Sant’Anna FH, Passos JFMD, Balsanelli E, de Baura VA, de Pedrosa F O, de Souza EM, Passaglia LMP. 2017. Detection of misidentifications of species from the Burkholderia cepacia complex and description of a new member, the soil bacterium Burkholderia catarinensis sp. nov. Pathog Dis 75:ftx076. doi: 10.1093/femspd/ftx076. [DOI] [PubMed] [Google Scholar]
- 13.Martina P, Leguizamon M, Prieto CI, Sousa SA, Montanaro P, Draghi WO, Stämmler M, Bettiol M, de Carvalho CCCR, Palau J, Figoli C, Alvarez F, Benetti S, Lejona S, Vescina C, Ferreras J, Lasch P, Lagares A, Zorreguieta A, Leitão JH, Yantorno OM, Bosch A. 2018. Burkholderia puraquae sp. nov., a novel species of the Burkholderia cepacia complex isolated from hospital settings and agricultural soils. Int J Syst Evol Micr 68:14–20. doi: 10.1099/ijsem.0.002293. [DOI] [PubMed] [Google Scholar]
- 14.Weber CF, King GM. 2017. Volcanic soils as sources of novel CO-oxidizing Paraburkholderia and Burkholderia: Paraburkholderia hiiakae sp. nov., Paraburkholderia metrosideri sp. nov., Paraburkholderia paradisi sp. nov., Paraburkholderia peleae sp. nov., and Burkholderia alpina sp. nov. a member of the Burkholderia cepacia complex. Front Microbiol 8:207. doi: 10.3389/fmicb.2017.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parke JL. 1991. Biological control of pythium damping-off and aphanomyces root rot of peas by application of Pseudomonas cepacia or P fluorescens to seed. Plant Dis 75:987. doi: 10.1094/PD-75-0987. [DOI] [Google Scholar]
- 16.Mullins AJ, Murray JAH, Bull MJ, Jenner M, Jones C, Webster G, Green AE, Neill DR, Connor TR, Parkhill J, Challis GL, Mahenthiralingam E. 2019. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria. Nat Microbiol 4:996–1005. doi: 10.1038/s41564-019-0383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillis M, Van TV, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez MP. 1995. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Evol Micr 45:274–289. doi: 10.1099/00207713-45-2-274. [DOI] [Google Scholar]
- 18.O'Sullivan LA, Mahenthiralingam E. 2005. Biotechnological potential within the genus Burkholderia. Lett Appl Microbiol 41:8–11. doi: 10.1111/j.1472-765X.2005.01758.x. [DOI] [PubMed] [Google Scholar]
- 19.Vial L, Chapalain A, Groleau M-C, Déziel E. 2011. The various lifestyles of the Burkholderia cepacia complex species: a tribute to adaptation. Environ Microbiol 13:1–12. doi: 10.1111/j.1462-2920.2010.02343.x. [DOI] [PubMed] [Google Scholar]
- 20.Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P, Mahenthiralingam E. 2016. Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl Microbiol Biotechnol 100:5215–5229. doi: 10.1007/s00253-016-7520-x. [DOI] [PubMed] [Google Scholar]
- 21.Gilligan PH, Downey DG, Elborn JS, Flume PA, Funk S, Gilpin D, Kidd TJ, McCaughan J, Millar BC, Murphy PG, Rendall JC, Tunney MM, Moore JE. 2018. “Pathogen eradication” and “emerging pathogens”: difficult definitions in cystic fibrosis. J Clin Microbiol 56:e00193-18. doi: 10.1128/JCM.00193-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speert DP, Bond M, Woodman RC, Curnutte JT. 1994. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J Infect Dis 170:1524–1531. doi: 10.1093/infdis/170.6.1524. [DOI] [PubMed] [Google Scholar]
- 23.Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60:539–574. doi: 10.1128/MR.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuqua C, Greenberg EP. 1998. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr Opin Microbiol 1:183–189. doi: 10.1016/S1369-5274(98)80009-X. [DOI] [PubMed] [Google Scholar]
- 25.Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 26.Juhas M, Eberl L, Tümmler B. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ Microbiol 7:459–471. doi: 10.1111/j.1462-2920.2005.00769.x. [DOI] [PubMed] [Google Scholar]
- 27.McKenney D, Brown KE, Allison DG. 1995. Influence of Pseudomonas aeruginosa exoproducts on virulence factor production in Burkholderia cepacia: evidence of interspecies communication. J Bacteriol 177:6989–6992. doi: 10.1128/jb.177.23.6989-6992.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewenza S, Conway B, Greenberg EP, Sokol PA. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J Bacteriol 181:748–756. doi: 10.1128/JB.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewenza S, Sokol PA. 2001. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J Bacteriol 183:2212–2218. doi: 10.1128/JB.183.7.2212-2218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapalain A, Vial L, Laprade N, Dekimpe V, Perreault J, Déziel E. 2013. Identification of quorum sensing‐controlled genes in Burkholderia ambifaria. MicrobiologyOpen 2:226–242. doi: 10.1002/mbo3.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choudhary KS, Hudaiberdiev S, Gelencsér Z, Coutinho BG, Venturi V, Pongor S. 2013. The organization of the quorum sensing luxI/R family genes in Burkholderia. Int J Mol Sci 14:13727–13747. doi: 10.3390/ijms140713727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pesci EC, Milbank JBJ, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Déziel E, Lépine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Cámara M. 2011. Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev 35:247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diggle SP, Lumjiaktase P, Dipilato F, Winzer K, Kunakorn M, Barrett DA, Chhabra SR, Cámara M, Williams P. 2006. Functional genetic analysis reveals a 2-alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem Biol 13:701–710. doi: 10.1016/j.chembiol.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Vial L, Lépine Fois, Milot S, Groleau M-C, Dekimpe V, Woods DE, Déziel E, 2008. Burkholderia pseudomallei, B. thailandensis, and B. ambifaria produce 4-hydroxy-2-alkylquinoline analogues with a methyl group at the 3 position that is required for quorum-sensing regulation. J Bacteriol 190:5339–5352. doi: 10.1128/JB.00400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritzmann NH, Mährlein A, Ernst S, Hennecke U, Drees SL, Fetzner S. 2019. Bromination of alkyl quinolones by Microbulbifer sp. HZ11, a marine Gammaproteobacterium, modulates their antibacterial activity. Environ Microbiol 21:2595–2609. doi: 10.1111/1462-2920.14654. [DOI] [PubMed] [Google Scholar]
- 38.Mori T, Yamashita T, Furihata K, Nagai K, Suzuki K, Hayakawa Y, Shin-Ya K. 2007. Burkholone, A new cytotoxic antibiotic against IGF-I dependent cells from Burkholderia sp. J Antibiot 60:713–716. doi: 10.1038/ja.2007.92. [DOI] [PubMed] [Google Scholar]
- 39.Kilani-Feki O, Zouari I, Culioli G, Ortalo-Magné A, Zouari N, Blache Y, Jaoua S. 2012. Correlation between synthesis variation of 2-alkylquinolones and the antifungal activity of a Burkholderia cepacia strain collection. World J Microbiol Biotechnol 28:275–281. doi: 10.1007/s11274-011-0817-0. [DOI] [PubMed] [Google Scholar]
- 40.Kilani-Feki O, Culioli G, Ortalo-Magné A, Zouari N, Blache Y, Jaoua S. 2011. Environmental Burkholderia cepacia strain Cs5 acting by two analogous alkyl-quinolones and a didecyl-phthalate against a broad spectrum of phytopathogens fungi. Curr Microbiol 62:1490–1495. doi: 10.1007/s00284-011-9892-6. [DOI] [PubMed] [Google Scholar]
- 41.Mahenthiralingam E, Song L, Sass A, White J, Wilmot C, Marchbank A, Boaisha O, Paine J, Knight D, Challis GL. 2011. Enacyloxins are products of an unusual hybrid modular polyketide synthase encoded by a cryptic Burkholderia ambifaria genomic island. Chem Biol 18:665–677. doi: 10.1016/j.chembiol.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 42.Li D, Oku N, Hasada A, Shimizu M, Igarashi Y. 2018. Two new 2-alkylquinolones, inhibitory to the fish skin ulcer pathogen Tenacibaculum maritimum, produced by a rhizobacterium of the genus Burkholderia sp. Beilstein J Org Chem 14:1446–1451. doi: 10.3762/bjoc.14.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vial L, Groleau M-C, Lamarche MG, Filion G, Castonguay-Vanier J, Dekimpe V, Daigle F, Charette SJ, Déziel E. 2010. Phase variation has a role in Burkholderia ambifaria niche adaptation. ISME J 4:49–60. doi: 10.1038/ismej.2009.95. [DOI] [PubMed] [Google Scholar]
- 44.Li D, Oku N, Shinozaki Y, Kurokawa Y, Igarashi Y. 2020. 4-Hydroxy-3-methyl-2(1H)-quinolone, originally discovered from a Brassicaceae plant, produced by a soil bacterium of the genus Burkholderia sp.: determination of a preferred tautomer and antioxidant activity. Beilstein J Org Chem 16:1489–1494. doi: 10.3762/bjoc.16.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Guillouzer S. 2018. Étude de la régulation des systèmes de communication intercellulaire chez la bactérie Burkholderia thailandensis. PhD thesis. Institut National de la Recherche Scientifique, Laval, Quebec, Canada. [Google Scholar]
- 46.Chapalain A, Groleau M-C, Guillouzer SL, Miomandre A, Vial L, Milot S, Déziel E. 2017. Interplay between 4-hydroxy-3-methyl-2-alkylquinoline and N-acyl-homoserine lactone signaling in a Burkholderia cepacia complex clinical strain. Front Microbiol 8:1021. doi: 10.3389/fmicb.2017.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin J, Cheng J, Wang Y, Shen X. 2018. The Pseudomonas quinolone signal (PQS): not just for quorum sensing anymore. Front Cell Infect Microbiol 8:230. doi: 10.3389/fcimb.2018.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wratten SJ, Wolfe MS, Andersen RJ, Faulkner DJ. 1977. Antibiotic metabolites from a marine Pseudomonad. Antimicrob Agents Chemother 11:411–414. doi: 10.1128/aac.11.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamasaki N, Ishii E, Tominaga K, Tezuka Y, Nagaoka T, Kadota S, Kuroki T, Yano I. 2000. Highly selective antibacterial activity of novel alkyl quinolone alkaloids from a chinese herbal medicine, gosyuyu (wu-chu-yu), against Helicobacter pylori in vitro. Microbiol Immunol 44:9–15. doi: 10.1111/j.1348-0421.2000.tb01240.x. [DOI] [PubMed] [Google Scholar]
- 50.Whalen KE, Becker JW, Schrecengost AM, Gao Y, Giannetti N, Harvey EL. 2019. Bacterial alkylquinolone signaling contributes to structuring microbial communities in the ocean. Microbiome 7:93. doi: 10.1186/s40168-019-0711-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer N, Bigalke A, Kaulfuß A, Pohnert G. 2017. Strategies and ecological roles of algicidal bacteria. FEMS Microbiol Rev 41:880–889. doi: 10.1093/femsre/fux029. [DOI] [PubMed] [Google Scholar]
- 52.Dow L, Stock F, Peltekis A, Szamosvári D, Prothiwa M, Lapointe A, Böttcher T, Bailleul B, Vyverman W, Kroth PG, Lepetit B. 2020. The multifaceted inhibitory effects of an alkyl quinolone on the diatom Phaeodactylum tricornutum. Chembiochem 21:1206–1216. doi: 10.1002/cbic.201900612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klaus JR, Coulon PML, Koirala P, Seyedsayamdost MR, Déziel E, Chandler JR. 2020. Secondary metabolites from the Burkholderia pseudomallei complex: structure, ecology, and evolution. J Ind Microbiol Biot 47:877–887. doi: 10.1007/s10295-020-02317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piochon M, Coulon PML, Caulet A, Groleau M-C, Déziel E, Gauthier C. 2020. Synthesis and antimicrobial activity of Burkholderia-related 4-hydroxy-3-methyl-2-alkenylquinolines (HMAQs) and their N-oxide counterparts. J Nat Prod 83:2145–2154. doi: 10.1021/acs.jnatprod.0c00171. [DOI] [PubMed] [Google Scholar]
- 55.Coulon PML, Groleau M-C, Déziel E. 2019. Potential of the Burkholderia cepacia Complex to produce 4-hydroxy-3-methyl-2-alkyquinolines. Front Cell Infect Microbiol 9:33. doi: 10.3389/fcimb.2019.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.diCenzo GC, Mengoni A, Perrin E. 2019. Chromids aid genome expansion and functional diversification in the family Burkholderiaceae. Mol Biol Evol 36:562–574. doi: 10.1093/molbev/msy248. [DOI] [PubMed] [Google Scholar]
- 57.Agnoli K, Schwager S, Uehlinger S, Vergunst A, Viteri DF, Nguyen DT, Sokol PA, Carlier A, Eberl L. 2012. Exposing the third chromosome of Burkholderia cepacia complex strains as a virulence plasmid. Mol Microbiol 83:362–378. doi: 10.1111/j.1365-2958.2011.07937.x. [DOI] [PubMed] [Google Scholar]
- 58.Gilligan PH. 1991. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev 4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balandreau J, Viallard V, Cournoyer B, Coenye T, Laevens S, Vandamme P. 2001. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl Environ Microbiol 67:982–985. doi: 10.1128/AEM.67.2.982-985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 61.Vidal-Quist JC, O'Sullivan LA, Desert A, Fivian-Hughes AS, Millet C, Jones TH, Weightman AJ, Rogers HJ, Berry C, Mahenthiralingam E. 2014. Arabidopsis thaliana and Pisum sativum models demonstrate that root colonization is an intrinsic trait of Burkholderia cepacia complex bacteria. Microbiology (Reading) 160:373–384. doi: 10.1099/mic.0.074351-0. [DOI] [PubMed] [Google Scholar]
- 62.Cipolla L, Rocca F, Martinez C, Aguerre L, Barrios R, Prieto M. 2018. Prevalence of Burkholderia cepacia complex species in cystic fibrosis patients in Argentina during the period 2011–2015. Enferm Infecc Microbiol Clin 36:431–434. doi: 10.1016/j.eimc.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 63.Huang CH, Jang TN, Liu CY, Fung CP, Yu KW, Wong WW. 2001. Characteristics of patients with Burkholderia cepacia bacteremia. J Microbiol Immunol Infect 34:215–219. [PubMed] [Google Scholar]
- 64.Loutet SA, Valvano MA. 2010. A decade of Burkholderia cenocepacia virulence determinant research. Infect Immun 78:4088–4100. doi: 10.1128/IAI.00212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patten CL, Blakney AJC, Coulson TJD. 2013. Activity, distribution and function of indole-3-acetic acid biosynthetic pathways in bacteria. Crit Rev Microbiol 39:395–415. doi: 10.3109/1040841X.2012.716819. [DOI] [PubMed] [Google Scholar]
- 66.Gomes MC, Tasrini Y, Subramoni S, Agnoli K, Feliciano JR, Eberl L, Sokol P, O'Callaghan D, Vergunst AC. 2018. The afc antifungal activity cluster, which is under tight regulatory control of ShvR, is essential for transition from intracellular persistence of Burkholderia cenocepacia to acute pro-inflammatory infection. PLoS Pathog 14:e1007473. doi: 10.1371/journal.ppat.1007473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jung BK, Hong S-J, Park G-S, Kim M-C, Shin J-H. 2018. Isolation of Burkholderia cepacia JBK9 with plant growth-promoting activity while producing pyrrolnitrin antagonistic to plant fungal diseases. Appl Biol Chem 61:173–180. doi: 10.1007/s13765-018-0345-9. [DOI] [Google Scholar]
- 68.Durand A-A, Bergeron A, Constant P, Buffet J-P, Déziel E, Guertin C. 2015. Surveying the endomicrobiome and ectomicrobiome of bark beetles: the case of Dendroctonus simplex. Sci Rep 5:17190. doi: 10.1038/srep17190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sievers F, Higgins DG. 2014. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol 1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- 70.Lépine F, Déziel E. 2011. Liquid chromatography/mass spectirometry for the detection and quantification of N-acyl-l-homoserine lactone and 4-hydroxy-2-alkylquinolines. Methods Mol Biol 692:61–69. doi: 10.1007/978-1-60761-971-0_5. [DOI] [PubMed] [Google Scholar]
- 71.Price EP, Viberg LT, Kidd TJ, Bell SC, Currie BJ, Sarovich DS. 2018. Transcriptomic analysis of longitudinal Burkholderia pseudomallei infecting the cystic fibrosis lung. Microb Genom 4:e000194. doi: 10.1099/mgen.0.000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Subsin B, Chambers CE, Visser MB, Sokol PA. 2007. Identification of genes regulated by the cepIR quorum-sensing system in Burkholderia cenocepacia by high-throughput screening of a random promoter library. J Bacteriol 189:968–979. doi: 10.1128/JB.01201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00127-21_Supp_1_seq10.docx, DOCX file, 1.1 MB (1.2MB, docx)