ABSTRACT
Ceftazidime-avibactam (CZA) has emerged as a promising solution to the lack of new antibiotics against Pseudomonas aeruginosa infections. Data from in vitro assays of CZA combinations, however, are scarce. The objective of our study was to perform a time-kill analysis of the effectiveness of CZA alone and in combination with other antibiotics against a collection of extensively drug-resistant (XDR) P. aeruginosa isolates. Twenty-one previously characterized representative XDR P. aeruginosa isolates were selected. Antibiotic susceptibility was tested by broth microdilution, and results were interpreted using CLSI criteria. The time-kill experiments were performed in duplicate for each isolate. Antibiotics were tested at clinically achievable free-drug concentrations. Different treatment options, including CZA alone and combined with amikacin, aztreonam, meropenem, and colistin, were evaluated to identify the most effective combinations. Seven isolates were resistant to CZA (MIC ≥ 16/4 mg/liter), including four metallo-β-lactamase (MBL)-carrying isolates and two class A carbapenemases. Five of them were resistant or intermediate to aztreonam (MIC ≥ 16 mg/liter). Three isolates were resistant to amikacin (MIC ≥ 64 mg/liter) and one to colistin (MIC ≥ 4 mg/liter). CZA monotherapy had a bactericidal effect in 100% (14/14) of the CZA-susceptible isolates. Combination therapies achieved a greater overall reduction in bacterial load than monotherapy for the CZA-resistant isolates. CZA plus colistin was additive or synergistic in 100% (7/7) of the CZA-resistant isolates, while CZA plus amikacin and CZA plus aztreonam were additive or synergistic in 85%. CZA combined with colistin, amikacin, or aztreonam was more effective than monotherapy against XDR P. aeruginosa isolates. A CZA combination could be useful for treating XDR P. aeruginosa infections, including those caused by CZA-resistant isolates.
IMPORTANCE The emergence of resistance to antibiotics is a serious public health problem worldwide and can be a cause of mortality. For this reason, antibiotic treatment is compromised, and we have few therapeutic options to treat infections. The main goal of our study is to search for new treatment options for infections caused by difficult-to-treat resistant germs. Pseudomonas aeruginosa is a Gram-negative bacterium distributed throughout the world with the ability to become resistant to most available antibiotics. Ceftazidime-avibactam (CZA) emerged as a promising solution to the lack of new antibiotics against infections caused by P. aeruginosa strains. This study intended to analyze the effect of CZA alone or in combination with other available antibiotics against P. aeruginosa strains. The combination of CZA with other antibiotics could be more effective than monotherapy against extensively drug-resistant P. aeruginosa strains.
KEYWORDS: ceftazidime-avibactam, colistin, aztreonam, amikacin, combination therapy, Pseudomonas aeruginosa
INTRODUCTION
New therapeutic options for multidrug-resistant (MDR) and extensively drug-resistant (XDR) Pseudomonas aeruginosa infections are required to overcome the growing problem of antimicrobial resistance. According to the U.S. Centers for Disease Control and Prevention, XDR P. aeruginosa is a “serious threat” to human health, and resistance is on the rise (1). This bacterium has a nonclonal epidemic population structure (2) and can develop antibiotic resistance through several mechanisms. XDR P. aeruginosa high-risk clones are disseminated in hospitals around the world (2) and pose a major public health problem because of limited treatment options and rising costs. Sequence type 111 (ST111) and ST235 are the predominant high-risk clones worldwide, but in Spain, the predominant clone is ST175 (2). High-risk clones are frequently responsible for nosocomial infections and are associated with the acquisition of horizontally transferable beta-lactamases and resistance mechanisms through chromosomal mutations (2, 3).
The problem of increasing antimicrobial resistance is compounded by a dwindling supply of new drugs. Given the few antibiotics in the clinical pipeline before 2010, the treatment options for XDR P. aeruginosa infections were suboptimal and consisted largely of antibiotics with a narrow therapeutic window and high toxicity (aminoglycosides, polymyxins) or unpredictable pharmacokinetics (colistin), yielding poor patient outcomes (4–7).
Ceftazidime-avibactam (CZA) was approved by the U.S. Food and Drug Administration in 2015 and was the first β-lactam combination to provide broad coverage against XDR Gram-negative pathogens, including P. aeruginosa (8). Few studies, however, have examined the effectiveness of CZA against infections caused by XDR P. aeruginosa high-risk clones. An in vitro study of a large collection of P. aeruginosa strains reported a CZA resistance rate of 2.9% (9). Most studies, however, have reported higher rates, up to 18% in some cases (10) and over 50% when XDR strains are involved (11, 12). Strains carrying metallo-β-lactamases (MBLs) have the highest resistance rates (>95%) as they are resistant to CZA, and CZA is not expected to be efficacious against these strains (13).
The use of CZA to treat P. aeruginosa infections caused by XDR high-risk clones may be clinically more effective and less toxic than colistin, which is often the only option available (14). However, given the high risk for the emergence of CZA-resistant mutants, it is paramount to monitor their selection during treatment and to evaluate associated risk factors. Combination therapy is a useful strategy for achieving maximum antimicrobial activity against various resistant organisms and for preventing antibiotic resistance (15). In vitro experiments have shown synergy for certain antipseudomonal antibiotics against MDR P. aeruginosa (5, 15–20). In vitro studies evaluating the activity of CZA combined with other antibiotics against P. aeruginosa, however, are lacking, and only few reports covering a small number of isolates have been published (21).
The aim of this study was to perform a comprehensive time-kill analysis of CZA alone or in combination with standard antipseudomonal antibiotics against a representative collection of the most common resistance mechanisms and XDR P. aeruginosa clones, including high-risk clones.
RESULTS
Antimicrobial susceptibility testing.
The antibiotic susceptibility profiles and previously characterized antibiotic resistance mechanisms of the 21 XDR P. aeruginosa isolates are shown in Table 1. Seven isolates were resistant to both CZA (MIC ≥ 16/4 mg/liter) and meropenem (MIC ≥ 8 mg/liter), and of these, four were resistant and one was intermediate to aztreonam (MIC ≥ 16 mg/liter), three were resistant to amikacin (MIC ≥ 64 mg/liter), and one was resistant to colistin (MIC ≥ 4 mg/liter). Six of the seven CZA-resistant isolates harbored carbapenemases belonging to Ambler class A or B and had OprD deficiency, except for one, and two of them showed AmpC hyperproduction.
TABLE 1.
Antibiotic susceptibility profile and resistance mechanisms of the 21 XDR P. aeruginosa isolatesa
| Isolate | ST | Acquired β- lactamase(s) | AmpC hyperproduction | OprD deficiency | MIC (mg/liter) |
||||
|---|---|---|---|---|---|---|---|---|---|
| AMK | ATM | MEM | CST | CZA | |||||
| 04-017 | 111 | OXA-46 | Yes | No | 4 | 64 | 32 | 2 | 8 |
| 04-025 | 175 | Yes | Yes | 4 | 16 | 16 | 1 | 4 | |
| 10-023 | 175 | Yes | Yes | 4 | 16 | 16 | 2 | 4 | |
| 06-014 | 179 | OXA-10 | Yes | Yes | 8 | 16 | 32 | 2 | 4 |
| 12-003 | 244 | Yes | Yes | 8 | 32 | 32 | 2 | 4 | |
| 09-011 | 274 | Yes | Yes | 128 | 64 | 32 | 1 | 4 | |
| 09-007 | 313 | Yes | Yes | 8 | 32 | 16 | 2 | 4 | |
| 10-017 | 395 | Yes | No | 4 | 32 | 8 | 2 | 4 | |
| 06-035 | 455 | Yes | No | <2 | 64 | >32 | 0.5 | 8 | |
| 10-019 | 2221 | Yes | Yes | <2 | 64 | 32 | 2 | 8 | |
| 06-025 | 2534 | Yes | Yes | <2 | 64 | 8 | 2 | 8 | |
| 06-027 | 2535 | Yes | No | 8 | 32 | 8 | 2 | 4 | |
| 06-001 | 2536 | Yes | Yes | 8 | 64 | 32 | 2 | 8 | |
| 09-012 | 175 | Yes | Yes | 8 | 64 | 16 | 2 | 8 | |
| 10-009 | 111 | VIM-2 | Yes | Yes | 32 | >128 | >32 | 4 | >32 |
| 07-016 | 175 | GES-5 | No | Yes | 16 | 16 | >32 | 2 | 32 |
| 12-012 | 175 | VIM-20, OXA-2 | No | Yes | 16 | 8 | >32 | 2 | 32 |
| 07-004 | 235 | GES-19, OXA-2 | No | Yes | 128 | 128 | >32 | 2 | >32 |
| 06-042 | 235 | VIM-47 | No | No | 64 | 32 | >32 | 2 | 32 |
| 01-008 | 253 | VIM-1 | No | Yes | 8 | 4 | >32 | 2 | >32 |
| 10-021 | 2533 | Yes | Yes | <2 | 64 | 32 | 1 | 16 | |
MICs (mg/liter) of the following antibiotics tested in this study are shown: amikacin (AMK), aztreonam (ATM), meropenem (MEM),colistin (CST), and ceftazidime-avibactam (CZA). CZA-resistant isolates are highlighted in gray.
Time-kill studies.
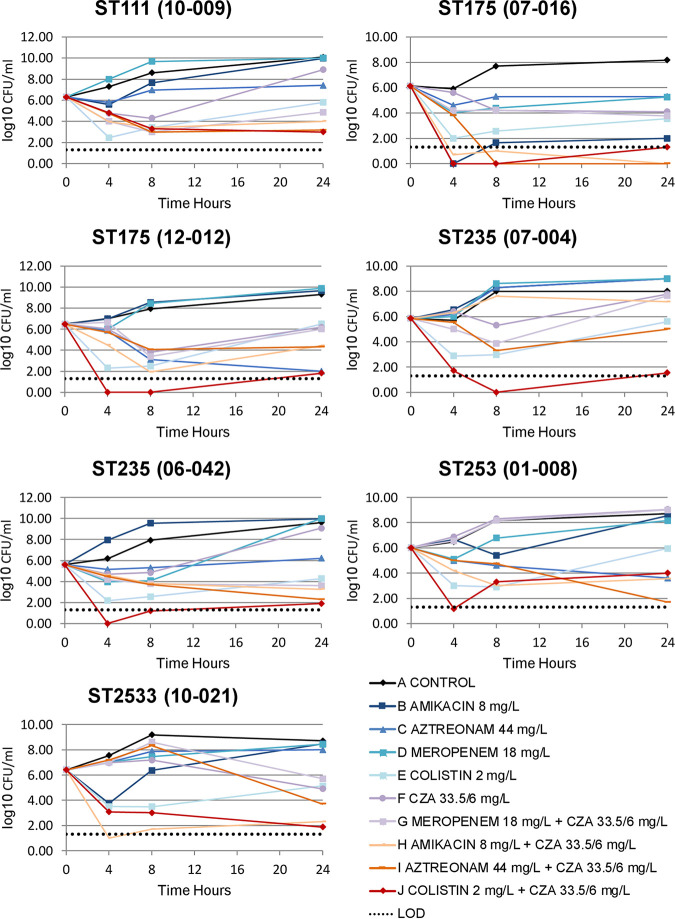
Bacterial growth without antibiotic reached 8 to 9 log10 CFU/ml at 24 h for all isolates. The results of the time-kill experiments for the 21 XDR P. aeruginosa isolates are shown in Table S1 in the supplemental material. The mean bacterial loads (log10 CFU/ml) over 24 h for the seven CZA-resistant XDR P. aeruginosa isolates treated with each antibiotic regimen are shown in Fig. 1. Table 2 shows the synergistic and additive effects of each combination against CZA-susceptible and CZA-resistant isolates. Table S2 shows the time-kill results (log difference at 24 h) for each antibiotic compared with the control and for each antibiotic combination compared with each antibiotic.
FIG 1.
Bacterial load (log10 CFU/ml) over 24 h in the seven CZA-resistant XDR P. aeruginosa isolates for each antibiotic regimen. LOD, lower limit of detection.
TABLE 2.
Synergistic and additive effects of each antibiotic combination against CZA-susceptible and CZA-resistant P. aeruginosa isolatesa
| Antibiotic combination | % of isolates |
|||||
|---|---|---|---|---|---|---|
| CZA susceptible |
CZA resistant |
|||||
| Synergy | Additivity | Total | Synergy | Additivity | Total | |
| AMK+CZA | 8 | 3 | 78.6 | 5 | 1 | 85.7 |
| ATM+CZA | 2 | 4 | 42.9 | 4 | 2 | 85.7 |
| MEM+CZA | 1 | 1 | 14.3 | 2 | 0 | 28.6 |
| CST+CZA | 6 | 2 | 57.1 | 6 | 1 | 100.0 |
AMK, amikacin; ATM, aztreonam; MEM, meropenem; CST, colistin; CZA, ceftazidime-avibactam.
Single antibiotics (aztreonam, meropenem, colistin, amikacin) were not bactericidal against any of the isolates at 24 h. Despite this, when compared with the control, all single antibiotics resulted in fewer bacteria than the control (F4, 64 = 8.7, P < 0.001; amikacin dif = −1.34, t = −2.5, P = 0.02; aztreonam dif = −1.43, t = −2.63, P = 0.01; meropenem dif = −1.42, t = −2.62, P = 0.01; colistin dif = −3.18, t = −5.87, P < 0.001).
CZA monotherapy was bactericidal against all the CZA-susceptible isolates, with a mean reduction of 3.19 log10 CFU/ml. In a comparison of the effects of the combination of CZA with other antibiotics, we found differences (F4, 65 = 11.08, P < 0.001). CZA plus amikacin (dif = −1.74, t = −3.58, P < 0.001) and CZA plus colistin (dif = −1.59, t = −3.25, P = 0.001) achieved a mean reduction of >4 log10 CFU/ml in the same isolates. The best combination against the CZA-susceptible isolates was CZA plus amikacin, which was synergistic or additive in approximately 80% of cases. On the other hand, no differences between CZA alone and CZA with aztreonam were found (dif = −0.48, t = −0.99, P = 0.33). Furthermore, combining CZA with meropenem increased the number of bacteria in comparison with CZA alone (dif = 1.02, t = 1.09, P = 0.04).
CZA combination therapies achieved a higher overall reduction in bacterial load than any of the treatments in isolation for the seven CZA-resistant isolates (F1, 61 = 33.92, P < 0.001). The log10 CFU/ml mean for the treatments in isolation was 0.94, and combining treatments reduced that mean to 3.44 (t = −5.82, P < 0.001). Hence, the mean reduction was 4.4 log10 CFU/ml for CZA plus colistin, amikacin, or aztreonam. As can be seen in Table 2, CZA plus colistin was either additive or synergistic in 100% of cases, while CZA plus amikacin or aztreonam was additive or synergistic in 85% of cases. The combination of CZA with aztreonam was effective against three of the four MBL-carrying isolates and against the two isolates that harbored class A carbapenemases.
DISCUSSION
We investigated the use of CZA alone or in combination with four antibiotics to assess the potential synergistic effects against XDR P. aeruginosa. As expected, a bactericidal effect was observed for CZA monotherapy in all the CZA-susceptible P. aeruginosa isolates, which had AmpC hyperproduction and/or OprD deficiency. To preserve the effectiveness of CZA, its clinical use should be avoided in naturally resistant strains and in those carrying MBLs and certain class D β-carbapenemases (22). Combination therapy has an important role in these clinical scenarios, and CZA combined with other antibacterial agents should be considered.
CZA resistance has already been described in Gram-negative bacilli. β-Lactamase-related mutations are the main mechanism behind CZA resistance in Enterobacterales. Recent reports suggest that the development of different resistance mechanisms within the course of treatment (e.g., mutations in KPC-encoding genes) might threaten the effectiveness of CZA (23, 24), a phenomenon that could be further complicated by horizontal spread (25). The development of CZA resistance during treatment of P. aeruginosa infections is frequently due to the selection of mutations in the AmpC β-lactamase structure, which are associated with coresistance with ceftolozane-tazobactam (16). Other contributory factors might be diminished outer membrane permeability and/or overexpression of efflux pumps (26). High-level resistance to CZA might also be due to MBL acquisition (27). Overall, six of the seven CZA-resistant isolates in our study harbored acquired β-lactamases, including several MBLs (VIM type) and a serine carbapenemase.
Little has been published on antibiotic combinations containing CZA, especially in the context of XDR P. aeruginosa isolates. Combination therapy with CZA plus aztreonam, amikacin, colistin, fosfomycin, and meropenem was recently evaluated in MDR Klebsiella pneumoniae and P. aeruginosa strains, but none of the isolates carried MBLs and few time-kill curves were analyzed (28). A synergistic effect was also reported for the combined use of CZA and colistin against MDR P. aeruginosa strains, including those resistant to colistin (29). In the present study, the combination of CZA with colistin showed a synergistic or additive effect against all the CZA-resistant P. aeruginosa isolates, including a colistin-resistant strain. Synergy was also observed against 85% of these isolates when CZA was combined with amikacin or aztreonam. In the combination of CZA with colistin, several bacterial isolates reached bacterial eradication at 4 and 8 h but then showed a little regrowth at 24 h. The phenomenon of bacterial regrowth could be due to either a loss of functionality of these antibiotics or selection of resistant isolates. Presumably, the latter could include selection of preexisting resistant subpopulations, de novo mutations, adaptive resistance, or formation of persistent cells (30). Further studies are required in order to evaluate these possibilities.
A double β-lactam strategy has been tested against carbapenemase-producing enterobacterial isolates in which CZA combined with meropenem or imipenem showed synergy against certain KPC-producing K. pneumoniae strains (31). In our study, however, CZA plus meropenem was the only combination to show no synergistic or additive activity against most XDR P. aeruginosa isolates. This could be because nonenzymatic mechanisms, alongside acquired β-lactamases, may have contributed to high meropenem MICs in the CZA-resistant isolates.
As mentioned, CZA is not active against MBL-bearing strains (22). The addition of aztreonam might overcome this resistance, as MBLs are known to have a weak hydrolysis capacity against aztreonam (32, 33). Combination therapy with ceftazidime and aztreonam may also be beneficial due to the simultaneous inhibition of multiple penicillin-binding proteins (34). Additionally, CZA plus aztreonam could exert an independent effect by acting on the “divisome” of Gram-negative bacteria (27). A recent report based on time-kill experiments with five P. aeruginosa isolates resistant to both CZA and aztreonam found that the combined use of the antibiotics had a synergistic effect and restored bactericidal activity in four of the isolates (21). In our study, this combination was effective against three of the four MBL-carrying isolates.
This study had some limitations. Our results are based on short in vitro assays with minimal antibiotic exposure compared with other pharmacokinetic/pharmacodynamic studies. Since these results are not representative of clinical guidelines for the administration of most antibiotics, they must be validated in in vivo experiments (35). The experimental design of this type of study does not allow identification of mechanisms of interactions or taking the emergence of resistance into consideration. A strength of our study is that our results are based on a large number of time-kill assays and show evidence of synergistic or additive effects in a considerable proportion of cases.
In conclusion, CZA is effective against XDR P. aeruginosa isolates both alone and in combination with other antibiotics. Combination regimens featuring CZA may be a good option against infections caused by these difficult-to-treat bacteria. Our data support the potential use of CZA in combination with amikacin, aztreonam, and colistin against XDR P. aeruginosa isolates, including CZA-resistant isolates and prevalent high-risk clones. These findings may help identify strategies to improve the clinical management of XDR P. aeruginosa infections using currently available drugs.
MATERIALS AND METHODS
Bacterial isolates and resistance mechanisms.
We studied 21 XDR P. aeruginosa clinical isolates which had been previously collected by our group as a part of the COLIMERO trial, a multicenter Spanish trial involving the molecular characterization of 150 XDR P. aeruginosa isolates from nine Spanish hospitals using pulsed-field gel electrophoresis, multilocus sequence typing, and whole-genome sequencing (3). The 21 isolates were representative of the clones and the most prevalent and relevant resistance mechanisms detected in the trial, namely, chromosomal mutations (AmpC hyperproduction and OprD inactivation) and horizontally acquired enzymes, including several MBLs and class A carbapenemases.
Antibiotics.
The antipseudomonal antibiotics used in the experiments were amikacin, aztreonam, colistin, meropenem (Sigma-Aldrich), and CZA (Pfizer). The antibiotics were chosen based on the mechanism of action and availability in the hospital’s pharmacy. Antibiotic solutions were prepared according to CLSI guidelines (36). Antibiotic concentrations for time-kill experiments were based on area-under-the-curve (AUC) serum levels: for amikacin, 1 g every 24 h (q24h), with an area under the concentration-time curve for 24 h (AUC24) of 196 μg · h/ml (37, 38); for aztreonam, 2 g q8h, with an AUC24 of 1,050 μg · h/ml (39); for meropenem, 2 g q8h, with an AUC24 of 425 μg · h/ml (40); for colistin, 4.5 MIU (million International units) q12h, with an AUC24 of 50 μg · h/ml (41, 42); for CZA, 2 g q8h, with an AUC24 of 800 μg · h/ml (43); and for avibactam, 2 g q8h, with an AUC24 of 147 μg · h/ml (43).
Antibiotic susceptibility testing.
The susceptibility profiles of the XDR isolates were obtained from the COLIMERO trial (3). Antimicrobial susceptibility was tested using broth microdilution and agar dilution methods with cation-adjusted Mueller-Hinton II broth (CAMHB) and Mueller-Hinton (MH) agar media, according to the CLSI guidelines (36). Ceftazidime susceptibility testing was conducted alone and in combination with a fixed avibactam concentration (4 mg/liter).
Time-kill experiments.
Time-kill studies were performed to analyze the activity of the selected antibiotics alone and in combination with CZA at clinically achievable free-drug concentrations. All experiments were performed in duplicate. An overnight culture of isolate was diluted with CAMHB and further incubated at 37°C for an hour to reach early log-phase growth. The bacterial suspension was diluted with CAMHB according to the absorbance at 630 nm. The magnitudes of absorbance ranged from 0.2 to 0.4. Sterile 50-ml conical flasks were used with 30 ml of CAMHB supplemented with the corresponding antibiotics. The final bacterial inoculum was approximately 6 to 7 log10 CFU/ml per flask. Flasks were incubated at 37°C in a shaker water bath for 24 h. Samples were collected at 0, 4, 8, and 24 h to measure bacterial growth. A 1-ml aliquot was obtained from each flask at each time point, centrifuged at 13,000 rpm for 3 min, and reconstituted with sterile saline solution to its original volume to minimize drug carryover. Serial decimal dilutions in CAMHB were performed; MH agar plates were inoculated (200 μl per plate) and incubated in a humidified incubator (37°C) for 18 to 24 h. Bacterial colonies for each sample were counted after overnight incubation. The bacterial density from the original sample was calculated based on the dilution factor. The limit of detection (LOD) was 1.3 log10 CFU/ml.
Apart from describing the results, in order to assess the effect of monotherapy and of the antibiotic combinations, we performed a series of regression analyses in which we entered the log difference in 24 h as dependent variable and each antibiotic regimen as independent variable. We checked for the application conditions of the regression, and all the conditions were met (normality of the residuals [assessed with Shapiro-Wilk’s test] and homoscedasticity [assessed with the Breusch-Pagan test]).
Pharmacodynamic time-kill parameters.
The results of the time-kill experiments were read at the different time points (0, 4, 8, and 24 h). Bactericidal activity was defined as a ≥3-log10 CFU/ml reduction, synergy as a ≥2-log10 CFU/ml reduction for a given combination compared with the most active single agent, additivity as a 1- to 2-log10 CFU/ml reduction in the final colony count for the combination compared with the most active single agent, and antagonism as a regrowth to ≥1-log10 CFU/ml for the combination compared with the least active single agent (44, 45). In addition to the aforementioned relevance criteria, we applied regression analysis to determine if the difference in log10 was statistically significant.
ACKNOWLEDGMENTS
We thank the Institute for Clinical Pharmacodynamics (ICPD), Schenectady, NY, and the Infectious Pathology and Antimicrobials Research Group (IPAR), Institute Hospital del Mar d’Investigacions Mèdiques (IMIM), for their support.
This study was supported by a medical grant from Pfizer Spain and was partially supported by the Ministerio de Economía y Competitividad of Spain, Instituto de Salud Carlos III, FEDER PI16/00669, PI17/00251, and PI18/0076, and the Spanish Network for Research in Infectious Diseases (REIPI RD16/0016).
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
María M. Montero, Email: 95422@parcdesalutmar.cat.
Juan P. Horcajada, Email: jhorcajada@psmar.cat.
William Lainhart, University of Arizona/Banner Health.
REFERENCES
- 1.Noval M, Banoub M, Claeys KC, Heil E. 2020. The battle is on: new beta-lactams for the treatment of multidrug-resistant Gram-negative organisms. Curr Infect Dis Rep 22:1. doi: 10.1007/s11908-020-0710-9. [DOI] [PubMed] [Google Scholar]
- 2.Oliver A, Mulet X, López-Causapé C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21–22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 3.del Barrio-Tofiño E, López-Causapé C, Cabot G, Rivera A, Benito N, Segura C, Montero MM, Sorlí L, Tubau F, Gómez-Zorrilla S, Tormo N, Durá-Navarro R, Viedma E, Resino-Foz E, Fernández-Martínez M, González-Rico C, Alejo-Cancho I, Martínez JA, Labayru-Echverria C, Dueñas C, Ayestarán I, Zamorano L, Martinez-Martinez L, Horcajada JP, Oliver A. 2017. Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob Agents Chemother 61:e01589-17. doi: 10.1128/AAC.01589-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horcajada JP, Sorlí L, Luque S, Benito N, Segura C, Campillo N, Montero M, Esteve E, Mirelis B, Pomar V, Cuquet J, Martí C, Garro P, Grau S. 2016. Validation of a colistin plasma concentration breakpoint as a predictor of nephrotoxicity in patients treated with colistin methanesulfonate. Int J Antimicrob Agents 48:725–727. doi: 10.1016/j.ijantimicag.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 5.Montero MM, Domene Ochoa S, López-Causapé C, VanScoy B, Luque S, Sorlí L, Campillo N, Padilla E, Prim N, Segura C, Pomar V, Rivera A, Grau S, Ambrose PG, Oliver A, Horcajada JP. 2019. Colistin plus meropenem combination is synergistic in vitro against extensively drug-resistant Pseudomonas aeruginosa, including high-risk clones. J Glob Antimicrob Resist 18:37–44. doi: 10.1016/j.jgar.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Zusman O, Avni T, Leibovici L, Adler A, Friberg L, Stergiopoulou T, Carmeli Y, Paul M. 2013. Systematic review and meta-analysis of in vitro synergy of polymyxins and carbapenems. Antimicrob Agents Chemother 57:5104–5111. doi: 10.1128/AAC.01230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorlí L, Luque S, Grau S, Berenguer N, Segura C, Montero MM, Alvarez-Lerma F, Knobel H, Benito N, Horcajada JP. 2013. Trough colistin plasma level is an independent risk factor for nephrotoxicity: a prospective observational cohort study. BMC Infect Dis 13:380. doi: 10.1186/1471-2334-13-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.U.S. Food and Drug Administration. 2016. Novel drugs summary 2015. U.S. Food and Drug Administration, Washington, DC. [Google Scholar]
- 9.Sader HS, Castanheira M, Shortridge D, Mendes RE, Flamm RK. 2017. Antimicrobial activity of ceftazidime-avibactam tested against multidrug-resistant Enterobacteriaceae and Pseudomonas aeruginosa isolates from U.S. medical centers, 2013 to 2016. Antimicrob Agents Chemother 61:e01045-17. doi: 10.1128/AAC.01045-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez MD, Mcmullen AR, Wallace MA, Crotty MP, Ritchie DJ, Burnham C-AD. 2017. Susceptibility of ceftolozane-tazobactam and ceftazidime-avibactam against a collection of β-lactam-resistant Gram-negative bacteria. Ann Lab Med 37:174–176. doi: 10.3343/alm.2017.37.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaumburg F, Bletz S, Mellmann A, Becker K, Idelevich EA. 2019. Comparison of methods to analyse susceptibility of German MDR/XDR Pseudomonas aeruginosa to ceftazidime/avibactam. Int J Antimicrob Agents 54:255–260. doi: 10.1016/j.ijantimicag.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Humphries RM, Hindler JA, Wong-Beringer A, Miller SA. 2017. Activity of ceftolozane-tazobactam and ceftazidime-avibactam against beta-lactam-resistant Pseudomonas aeruginosa isolates. Antimicrob Agents Chemother 61:e01858-17. doi: 10.1128/AAC.01858-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlowsky JA, Kazmierczak KM, Bouchillon SK, De Jonge BLM, Stone GG, Sahm DF. 2018. In vitro activity of ceftazidime-avibactam against clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa collected in Asia-Pacific countries: results from the INFORM global surveillance program, 2012 to 2015. Antimicrob Agents Chemother 62:e02569-17. doi: 10.1128/AAC.02569-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montero M, Horcajada JP, Sorlí L, Alvarez-Lerma F, Grau S, Riu M, Sala M, Knobel H. 2009. Effectiveness and safety of colistin for the treatment of multidrug-resistant Pseudomonas aeruginosa infections. Infection 37:461–465. doi: 10.1007/s15010-009-8342-x. [DOI] [PubMed] [Google Scholar]
- 15.Montero M, VanScoy BD, López-Causapé C, Conde H, Adams J, Segura C, Zamorano L, Oliver A, Horcajada JP, Ambrose PG. 2018. Evaluation of ceftolozane-tazobactam in combination with meropenem against Pseudomonas aeruginosa sequence type 175 in a hollow-fiber infection model. Antimicrob Agents Chemother 62:e00026-18. doi: 10.1128/AAC.00026-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraile-Ribot PA, Cabot G, Mulet X, Periañez L, Luisa Martín-Pena M, Juan C, Pérez JL, Oliver A. 2018. Mechanisms leading to in vivo ceftolozane/tazobactam resistance development during the treatment of infections caused by MDR Pseudomonas aeruginosa. J Antimicrob Chemother 73:658–663. doi: 10.1093/jac/dkx424. [DOI] [PubMed] [Google Scholar]
- 17.Montero M, Ochoa SD, López-Causapé C, VanScoy B, Luque S, Sorlí L, Campillo N, Angulo-Brunet A, Padilla E, Prim N, Pomar V, Rivera A, Grau S, Ambrose PG, Oliver A, Horcajada JP. 2020. Efficacy of ceftolozane-tazobactam in combination with colistin against extensively drug-resistant Pseudomonas aeruginosa, including high-risk clones, in an in vitro pharmacodynamic model. Antimicrob Agents Chemother 64:e02542-19. doi: 10.1128/AAC.02542-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunderson BW, Ibrahim KH, Hovde LB, Fromm TL, Reed MD, Rotschafer JC. 2003. Synergistic activity of colistin and ceftazidime against multiantibiotic-resistant Pseudomonas aeruginosa in an in vitro pharmacodynamic model. Antimicrob Agents Chemother 47:905–909. doi: 10.1128/AAC.47.3.905-909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Y, Yang D, Wang J, Wang R. 2018. Activity of colistin alone or in combination with rifampicin or meropenem in a carbapenem-resistant bioluminescent Pseudomonas aeruginosa intraperitoneal murine infection model. J Antimicrob Chemother 73:456–461. doi: 10.1093/jac/dkx399. [DOI] [PubMed] [Google Scholar]
- 20.Louie A, Grasso C, Bahniuk N, B VS, Brown DL, Kulawy R, Drusano GL. 2010. The combination of meropenem and levofloxacin is synergistic with respect to both Pseudomonas aeruginosa kill rate and resistance suppression. Antimicrob Agents Chemother 54:2646–2654. doi: 10.1128/AAC.00065-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee M, Abbey T, Biagi M, Wenzler E. 2021. Activity of aztreonam in combination with ceftazidime-avibactam against serine- and metallo-β-lactamase–producing Pseudomonas aeruginosa. Diagn Microbiol Infect Dis 99:115227. doi: 10.1016/j.diagmicrobio.2020.115227. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Wang J, Wang R, Cai Y. 2020. Resistance to ceftazidime-avibactam and underlying mechanisms. J Glob Antimicrob Resist 22:18–27. doi: 10.1016/j.jgar.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Compain F, Arthur M. 2017. Impaired inhibition by avibactam and resistance to the ceftazidime-avibactam combination due to the D179Y substitution in the KPC-2 β-lactamase. Antimicrob Agents Chemother 61:e00451-17. doi: 10.1128/AAC.00451-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, Gregson A. 2015. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59:6605–6607. doi: 10.1128/AAC.01165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haidar G, Clancy CJ, Shields RK, Hao B, Cheng S, Nguyen MH. 2017. Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum β-lactamases. Antimicrob Agents Chemother 61:e02534-16. doi: 10.1128/AAC.02534-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winkler ML, Papp-Wallace KM, Hujer AM, Domitrovic TN, Hujer KM, Hurless KN, Tuohy M, Hall G, Bonomo RA. 2015. Unexpected challenges in treating multidrug-resistant Gram-negative bacteria: resistance to ceftazidime-avibactam in archived isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 59:1020–1029. doi: 10.1128/AAC.04238-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall S, Hujer AM, Rojas LJ, Papp-Wallace KM, Humphries RM, Spellberg B, Hujer KM, Marshall EK, Rudin SD, Perez F, Wilson BM, Wasserman RB, Chikowski L, Paterson DL, Vila AJ, Van Duin D, Kreiswirth BN, Chambers HF, Fowler VG, Jacobs MR, Pulse ME, Weiss WJ, Bonomo RA. 2017. Can ceftazidime-avibactam and aztreonam overcome β-lactam resistance conferred by metallo-β-lactamases in Enterobacteriaceae? Antimicrob Agents Chemother 61:e02243-16. doi: 10.1128/AAC.02243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikhail S, Singh NB, Kebriaei R, Rice SA, Stamper KC, Castanheira M, Rybak MJ. 2019. Evaluation of the synergy of ceftazidime-avibactam in combination with meropenem, amikacin, aztreonam, colistin, or fosfomycin against well-characterized multidrug-resistant Klebsiella pneumoniae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 63:e00779-19. doi: 10.1128/AAC.00779-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mataraci Kara E, Yilmaz M, İstanbullu Tosun A, Özbek Çelik B. 2020. Synergistic activities of ceftazidime-avibactam in combination with different antibiotics against colistin-nonsusceptible clinical strains of Pseudomonas aeruginosa. Infect Dis (Lond) 52:616–624. doi: 10.1080/23744235.2020.1767803. [DOI] [PubMed] [Google Scholar]
- 30.Fernández L, Breidenstein EBM, Hancock REW. 2011. Creeping baselines and adaptive resistance to antibiotics. Drug Resist Updat 14:1–21. doi: 10.1016/j.drup.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Gaibani P, Lewis RE, Volpe SL, Giannella M, Campoli C, Landini MP, Viale PL, Re MC, Ambretti S. 2017. In vitro interaction of ceftazidime-avibactam in combination with different antimicrobials against KPC-producing Klebsiella pneumoniae clinical isolates. Int J Infect Dis 65:1–3. doi: 10.1016/j.ijid.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Sy S, Zhuang L, Xia H, Beaudoin M-E, Schuck VJ, Derendorf H. 2017. Prediction of in vivo and in vitro infection model results using a semimechanistic model of avibactam and aztreonam combination against multidrug resistant organisms. CPT Pharmacometrics Syst Pharmacol 6:197–207. doi: 10.1002/psp4.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livermore DM, Mushtaq S, Barker K, Hope R, Warner M, Woodford N. 2012. Characterization of β-lactamase and porin mutants of Enterobacteriaceae selected with ceftaroline1avibactam (NXL104). J Antimicrob Chemother 67:1354–1358. doi: 10.1093/jac/dks079. [DOI] [PubMed] [Google Scholar]
- 34.Asli A, Brouillette E, Krause KM, Nichols WW, Malouin F. 2016. Distinctive binding of avibactam to penicillin-binding proteins of gram-negative and gram-positive bacteria. Antimicrob Agents Chemother 60:752–756. doi: 10.1128/AAC.02102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tängdén T, Karvanen M, Friberg LE, Odenholt I, Cars O. 2017. Assessment of early combination effects of colistin and meropenem against Pseudomonas aeruginosa and Acinetobacter baumannii in dynamic time-kill experiments. Infect Dis (Lond) 49:521–527. doi: 10.1080/23744235.2017.1296183. [DOI] [PubMed] [Google Scholar]
- 36.CLSI. 2020. Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI, Wayne, PA. [Google Scholar]
- 37.Mahmoudi L, Mohammadpour AH, Ahmadi A, Niknam R, Mojtahedzadeh M. 2013. Influence of sepsis on higher daily dose of amikacin pharmacokinetics in critically ill patients. Eur Rev Med Pharmacol Sci 17:285–291. [PubMed] [Google Scholar]
- 38.de Montmollin E, Bouadma L, Gault N, Mourvillier B, Mariotte E, Chemam S, Massias L, Papy E, Tubach F, Wolff M, Sonneville R. 2014. Predictors of insufficient amikacin peak concentration in critically ill patients receiving a 25 mg/kg total body weight regimen. Intensive Care Med 40:998–1005. doi: 10.1007/s00134-014-3276-x. [DOI] [PubMed] [Google Scholar]
- 39.Smith PF, Ballow CH, Booker BM, Forrest A, Schentag JJ. 2001. Pharmacokinetics and pharmacodynamics of aztreonam and tobramycin in hospitalized patients. Clin Ther 23:1231–1244. doi: 10.1016/S0149-2918(01)80103-X. [DOI] [PubMed] [Google Scholar]
- 40.Tam VH, Nikolaou M. 2011. A novel approach to pharmacodynamic assessment of antimicrobial agents: new insights to dosing regimen design. PLoS Comput Biol 7:e1001043. doi: 10.1371/journal.pcbi.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plachouras D, Karvanen M, Friberg LE, Papadomichelakis E, Antoniadou A, Tsangaris I, Karaiskos I, Poulakou G, Kontopidou F, Armaganidis A, Cars O, Giamarellou H. 2009. Population pharmacokinetic analysis of colistin methanesulfonate and colistin after intravenous administration in critically ill patients with infections caused by Gram-negative bacteria. Antimicrob Agents Chemother 53:3430–3436. doi: 10.1128/AAC.01361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garonzik SM, Li J, Thamlikitkul V, Paterson DL, Shoham S, Jacob J, Silveira FP, Forrest A, Nation RL. 2011. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob Agents Chemother 55:3284–3294. doi: 10.1128/AAC.01733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das S, Zhou D, Nichols WW, Townsend A, Newell P, Li J. 2020. Selecting the dosage of ceftazidime-avibactam in the perfect storm of nosocomial pneumonia. Eur J Clin Pharmacol 76:349–361. doi: 10.1007/s00228-019-02804-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gómez-Junyent J, Benavent E, Sierra Y, El Haj C, Soldevila L, Torrejón B, Rigo-Bonnin R, Tubau F, Ariza J, Murillo O. 2019. Efficacy of ceftolozane/tazobactam, alone and in combination with colistin, against multidrug-resistant Pseudomonas aeruginosa in an in vitro biofilm pharmacodynamic model. Int J Antimicrob Agents 53:612–619. doi: 10.1016/j.ijantimicag.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Lim TP, Cai Y, Hong Y, Chan ECY, Suranthran S, Teo JQM, Lee WH, Tan TY, Hsu LY, Koh TH, Tan TT, Kwa ALH. 2015. In vitro pharmacodynamics of various antibiotics in combination against extensively drug-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother 59:2515–2524. doi: 10.1128/AAC.03639-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00585-21_Supp_1_seq4.pdf, PDF file, 0.3 MB (296.4KB, pdf) .