Abstract
In this review, we describe the key molecular entities involved in the process of infection by SARS-CoV-2, while also detailing how those key entities influence the spread of the disease. We further introduce the molecular mechanisms of preventive and treatment strategies including drugs, antibodies, and vaccines.
Keywords: Enzymes, Proteins, Peptides: Nucleic acids, DNA, RNA, Drugs, Vaccines, SARS-COV-2, COVID-19, CORONA virus, Epidemiology, Antibody, Transmission
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), also known as “the novel coronavirus” due to genome variation relative to previously identified coronaviruses, is a positive sense RNA virus and the etiological agent of COVID-19. SARS-CoV-2 is a member of the viral family, Coronaviridae, and subfamily, Coronavirinae, which are large, enveloped, single-stranded RNA viruses between 65 and 125 nm in diameter. Under electron microscopy, virions take on a roughly spherical shape and possess finger like extensions (glycoproteins), commonly referred to as spikes, on their surface (Fig. 1 ) [1], [2], [3], [4]. These spikes resemble a crown (or a “corona” in Latin) that is the basis for the name of this family of viruses [5]. Members of the subfamily Coronavirinae infect a variety of mammals, causing diverse clinical syndromes. They are grouped into four sub-groups based on sequence homology: alpha, beta, gamma and delta [6]. To date, only seven coronaviruses infect humans. Four (two α-CoVs and two β-CoVs) cause mild cold-like illnesses but three, including severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV) and the recently discovered SARS-CoV-2 lead to serious or fatal disease. SARS-CoV and MERS-CoV emerged in the human population in 2002 and 2012, respectively, while SARS-CoV-2 emerged in 2019, leading to the current global pandemic [7], [8]. The 2002 SARS-CoV infected approximately 8096 people, with a mortality rate of ~10%, whereas MERS-CoV infected over 2500 people with a mortality rate of ~36% [9], [10], [11], [12]. As of August 20, 2021, the COVID-19 pandemic caused by the SARS-CoV-2 coronavirus has infected over 210 million people and has killed close to 4.5 million people worldwide [13], [14], [15]. Genomic analysis shows patterns of molecular divergence between SARS-CoV-2 and other coronaviruses, leading to a disease that is distinct from that of both SARS-CoV and MERS-CoV [5], [16]. In this article, we will focus on describing the molecular basis of SARS-CoV-2 infection and antiviral drugs that target the virus. More detailed medical, epidemiological, and technologies used to identify and combat SARS-CoV-2 have been reviewed elsewhere [17], [18], [19], [20], [21], [22], [23], [24], [25], [26].
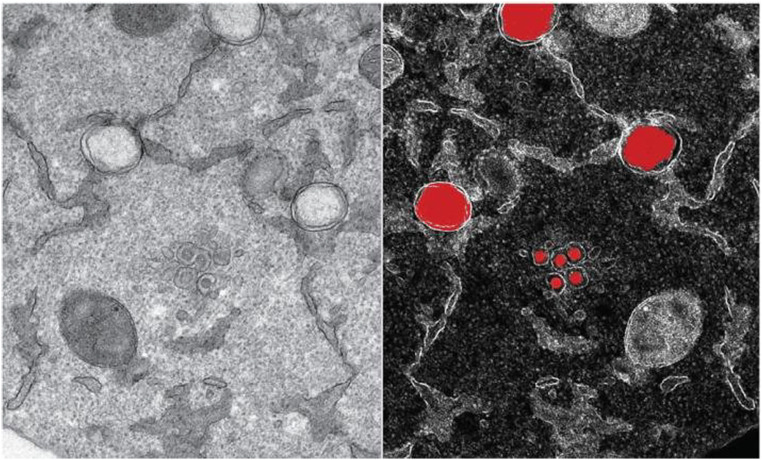
Fig. 1.
Transmission electron micrograph of SARS-CoV-2 viral particle, isolated from a patient. Large red circles represent newly formed genome copies and the small red balloons are new virions formed by budding at the interface of Golgi apparatus and endoplasmic reticulum. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Credit: Yannick Schwab/EMBL in Ref. [4].
2. Pathogenicity and transmission
Like other coronaviruses, transmission of SARS-CoV-2 can occur primarily via infected respiratory droplets through direct, indirect, or close contact with infected individuals [27]. The virus targets and infects cells at the nasal, conjunctival, or oral mucosa once respiratory particles are inhaled or deposited on these locations. Receptors for SARS-CoV-2, which are found mainly in the human respiratory tract epithelium, facilitate virus entry and allow the virus to efficiently replicate and spread to various organs. Approximately five days after infection, the host is contagious and can transmit the virus easily through exhalation to potentially infect others. Symptoms associated with the virus infection are due to transient damage to susceptible cells in various organs, including the lung, intestines, kidneys, and blood vessels [28].
Infection by the virus triggers an initial inflammatory immune response during which white blood cells originating from the lymph nodes, such as helper T-cells and cytotoxic T-cells, infiltrate to the site of infection to eliminate virus-infected cells [29]. In addition, body temperature is raised, causing fever in an attempt to kill (i.e., denature) the virus while facilitating the immune response [30], [31]. In patients who develop more severe disease, SARS-CoV-2 induces an aberrant host immune response. Overproduction of proinflammatory cytokines leads to a so-called “cytokine storm” which, if not remedied with the help of immunomodulators that regulate cytokine production or drugs like dexamethasone or blood thinners, can lead to death [32], [33], [34], [35]. This “cytokine storm”, can adversely affect several organs, including the liver, kidneys, and lungs [32], [36]. The latter leads to breakdown of lung epithelial cells that line the air sacs in lungs, causing the air sacs to fill with fluid. This induces pneumonia and, in severe cases, acute respiratory distress syndrome (ARDS) that can cause lung failure and death. Studies have shown that levels of certain cytokines can be correlated with the severity of COVID-19 symptoms [32].
SARS-CoV-2 has an enhanced rate of transmission compared to the 2002 SARS-CoV. This is due to several characteristics, both of the virus (i.e., SARS-CoV-2) and its associated disease (i.e., COVID-19). For instance, active virus can remain viable on some surfaces for up to 9 days and in the indoor air for several hours, thus increasing the chances of exposure [37]. Likewise, differences in the severity and onset of symptoms of COVID-19 compared to SARS may lead to increased contact with infected individuals. For example, during the SARS-CoV epidemic of 2002, mortality rates were nearly twice that currently observed for COVID-19 pandemic (9.6% vs. 5.4%, respectively) [10], [12], and infected individuals exhibited clear symptoms of respiratory distress almost immediately [38], [39], [40]. In contrast, people infected with SARS-CoV-2 may not show signs of infection or illness for a week or longer, despite being contagious and shedding virus [41]. Furthermore, asymptomatic individuals and people with mild symptoms may carry large amounts of virus in the upper respiratory tract, thus contributing to the rapid spread of SARS-CoV-2. In fact, in some situations, even after symptoms disappear, SARS-CoV-2-infected individuals may be shedding virus. In addition, re-infection may occur in specific cases, especially given that human coronaviruses are well equipped to subvert host immunity. For instance, the emergence of new variants of the virus that are more contagious and may carry an increased risk of death, such as the South African variant (B.1.351), the UK variant (B.1.1.7), the Indian variant (B.1.617.2, also commonly known as the delta variant), or the Brazil/Japan variant (P.1), suggest that SARS-CoV-2 is rapidly evolving mechanisms to increase contagiousness and subvert existing immune defenses [42], [43]. Clearly, additional information is needed to understand the global risks these new variants pose.
2.1. Genome structure
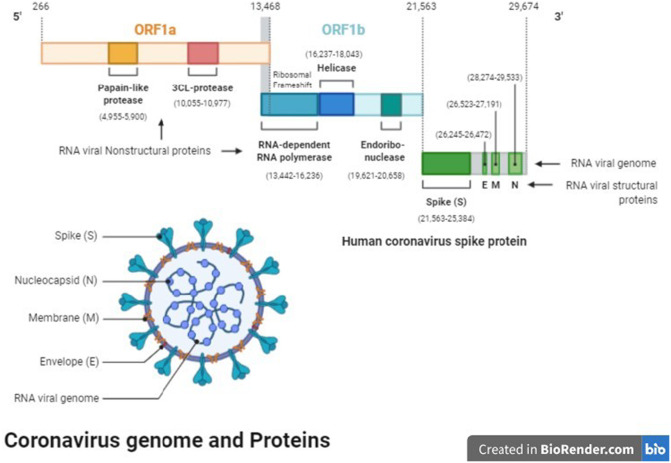
After the initial sequencing of the SARS-CoV-2 genome in December 2019 [16], the viral genome has been sequenced numerous times in order to detect mutations and identify variants [44], [45], [46]. The genome of SARS-CoV-2 is comprised of a single-stranded RNA molecule housed inside a fatty acid membrane (Fig. 2 ), known as the envelope [47]. The viral RNA, which is capped and polyadenylated, contains a 5′ leader sequence of 70 bases, with 7–10 of those bases being transcription-regulatory sequences (TRS-L), and encodes 13–15 open reading frames (ORFs). ORF1a is the longest region of the genome, comprising ~13,200 of the 29,674 bases in the sequence and encoding two genes, the papain-like protease and the 3CL protease. ORF1b, which overlaps with ORF1a (Fig. 2) [47], encompasses bases 14,442–21,563 and encodes enzymes important for viral replication, such as a RNA-dependent RNA polymerase (RdRp), a helicase, and an endonuclease. Subsequent regions spanning from nucleotides 21,562–29,674 encode the structural proteins: the spike (S) protein, membrane (M) protein, envelope (E) protein, nucleocapsid (N) protein, and accessory proteins. Each ORF also has its own transcriptional-regulatory sequences (TRSs), which are located immediately adjacent to the ORFs. The RdRp pauses when it crosses a TRS in the body (TRS-B) and switches the template to the TRS in the leader (TRS-L) during replication. This leads to discontinuous transcription and the fusion of negative (or complementary) strand RNAs that serve as templates for generation of genomic strand RNAs.
Fig. 2.
The genome of SARS-CoV-2 contains over 29,000 bases and codes for 29 proteins. SARS-CoV-2 has four structural proteins: The proteins E and M that form the viral envelope, protein N binds to the RNA genome of SARS-CoV-2 and S protein, which binds to host ACE2 receptor to initiate infection. The nonstructural proteins (NsPs) initially form as two long polypeptides, which by the action of the virus' main protease (MP), Nsp5, aside from autocatalytically releasing itself, also releases the RNA-dependent RNA polymerase (RdRp), Nsp12. Because of its important role in the replication and transcription of the virus, MP can be targeted by antiviral drugs against SARS-CoV-2. Ref: [47].
It is important to note that while most living organisms possess DNA as their fundamental genetic material, using RNA as the basis of the genome is unique to viruses [16], [48]. There are several important differences between DNA and RNA that can affect their cellular function. For instance, RNA uses the nitrogenous base uracil in place of the thymine found in DNA. Likewise, RNA nucleotides are composed of ribose sugar molecules that contain a hydroxyl moiety in the 2′ position. In contrast, DNA contains a hydrogen atom at the 2′ position, making it a deoxyribose. The absence of a hydroxyl at the 2′ position causes the ribose sugar to adopt a C3′-endo sugar pucker vs. the C2′-endo pucker typically found in B-form DNA. As a consequence, rather than aligning neatly with the helical axis like in B-DNA, the nucleobases of RNA are tilted relative to the helical axis. This reduces the stability of complementary base pairs in RNA molecules, preventing RNA from forming long double helices. Instead, RNA tends to exhibit more single-stranded character than DNA, which is predominantly double-stranded. In DNA-based organisms, gene expression involves the transcription of DNA to RNA using RNA polymerase. RNA viruses, on the other hand, use RNA as templates for the transcription of additional copies of RNA. Due to this unique transcriptional requirement, virally encoded RdRps facilitate this process.
The genomic structure and content of SARS-CoV-2 has been shown to promote its survival in host cells and may contribute to the progression of disease. Coronaviruses, including SARS-CoV-2, contain cytosine-phosphate-guanine (CpG) nucleotides as part of their genome. CpG sequences allow cellular detection of viruses through association with toll-like receptor 9 (TLR9) in endosomes to trigger a type I interferon (IFN) antiviral immune response in the host. This antiviral mechanism may be driven by the zinc-finger antiviral protein (ZAP), which restricts numerous viral pathogens by targeting CpG-rich RNA sequences. Interestingly, studies have shown that during replication of SARS-CoV-2, there is a selective pressure to lower CpG content as a means of evading the host immune response [49]. Interestingly, as another survival mechanism, the SARS-CoV-2 viral genome contains large-scale internal RNA base pairing, or genome-scale ordered RNA structures (GORS), that are associated with shielding viral RNA recognition in the host cell and may contribute to persistent mechanisms [45].
Many secondary structures (SS) are also built into the genome. These structures are generated once the viral RNA has been synthesized and released into the cytoplasm of the host cell. The various forms of SS include hairpin-forming inverted repeats (IR), quadruplex sites, and a slippery sequence with a downstream pseudoknot, all of which are needed to equip the virus for replication. For instance, a pseudoknot structure in ORF1ab controls the frameshift during the overlapped transition of ORF1a and ORF1ab. This is known as a programmed −1 ribosomal frameshift (−1 PRF) signal. The −1 PRF signal also contains a slippery sequence and linker region that play a role in protein translation [50], [51].
2.2. Viral replication
2.2.1. Entry into host cells
All viruses, including SARS-CoV-2, enter host cells and efficiently modulate cellular factors to facilitate replication (Fig. 3 ) [52]. SARS-CoV-2 has been shown to infect cells in two ways: entry through endocytic mechanisms or through fusion of the viral envelope with the host cell plasma membrane [46]. In both cases, the spike protein subunits, S1 and S2, mediate attachment and entry by binding with the cell surface protein, angiotensin-converting enzyme 2 (ACE2). Recent structural data obtained via high resolution cryo-electron microscopy has revealed the simultaneous binding of two S glycoprotein trimers to an ACE2 dimer [53], [54]. Binding of the viral glycoproteins to ACE2 is further facilitated by heparan sulfate (HS). Finally, cleavage by the serine protease, TMPRSS2, promotes fusion with the host membrane. Other cellular proteases (e.g., furin) facilitate pH-dependent entry through the endocytic pathway. Studies have shown that the main entry routes are dependent on the presence of select proteases in specific cell types [55]. Once the virus has entered the host cell, the RNA genome is released into the cytoplasm in a process known as uncoating [56].
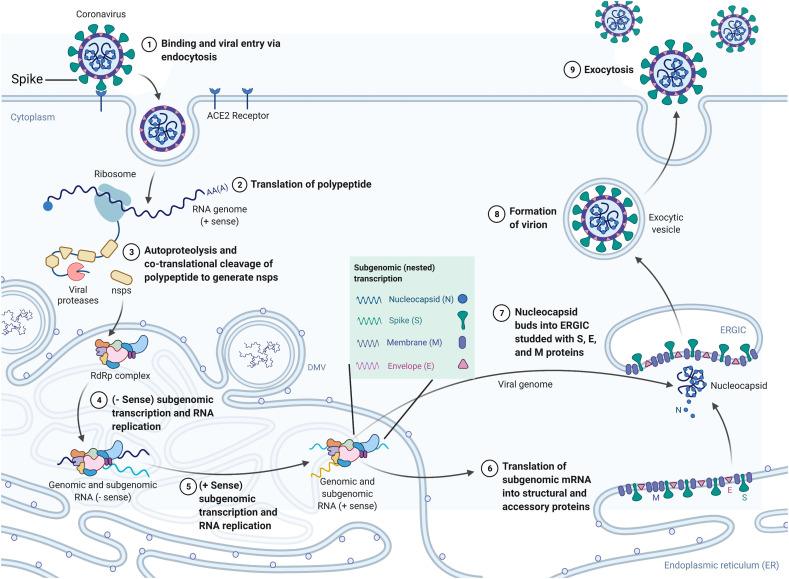
Fig. 3.
SARS-CoV-2 life cycle. SARS-CoV-2 spike (S) glycoprotein binds to the ACE2 receptor on the host cell surface and the virus enters the cell via endocytosis. Viral genomic RNA is released into the cytoplasm of the host cell, and the single-stranded positive-sense genome is transcribed and translated to produce nonstructural proteins (nsps) including replicase polyproteins (RNA-dependent RNA polymerase and helicase) to create an RdRp complex. Subgenomic transcription and RNA replication occur within the RdRp complex to synthesize negative-strand guide RNA (gRNA) and a set of subgenomic RNAs for viral replication and transcription. The newly produced subgenomic RNAs are translated into viral structural proteins such as the spike (S), nucleocapsid (N), membrane (M), and envelope (E) proteins. These proteins are inserted into the membrane of the rough endoplasmic reticulum (ER) and then transported to the ER-Golgi intermediate compartment (ERGIC) to assemble with the N protein-encapsidated RNA to form viral particles. Virions are then released from the cell through exocytosis. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2.2. Translation of viral proteins
The release of the coronavirus genome into the host cytoplasm triggers a complex and highly regulated program of viral gene expression [46]. The two polyproteins (pp1a and pp1ab) are produced through translation of ORF1a and ORF1b from the genomic RNA. As mentioned above, pp1ab is expressed from a programmed-1 ribosome frameshift between ORF1a and ORF1b. The efficiency of the frameshift can be measured through ribosomal profiling and, in the case of SARS-CoV-2, pp1a is expressed at levels approximately 1.4–2.2 times greater than pp1ab [57]. Through co-translational and post-translation mechanisms, including processing by viral proteases nsp3 and nsp5, sixteen non-structural proteins are released from pp1a (nsp1–11) and pp1ab (nsp1–10, nsp12–16), of which fifteen compose the viral replication and transcription complex (RTC). The RTC includes RNA-processing and RNA-modifying enzymes as well as an RNA proofreading function. Of these enzymes, RNA synthesis is performed by the viral RdRp (nsp12) together with its two cofactors, nsp7 and nsp8, which has a 3′-terminal adenylyltransferase activity. The bifunctional nsp14 contains both a 3′ to 5′ exoribonuclease (ExoN) domain and a guanine-N7-methyltransferase (N7-MTase) domain. Previous studies have shown that the ExoN function of nsp14 is critical for boosting longer-term replication fidelity for SARS-CoV. However, recently, it was revealed through biochemical evaluation of MERS-CoV and SARS-CoV-2 mutants that, for these viruses, ExoN plays a more direct role in RNA synthesis in addition to safeguarding the long-term fidelity of the viral genome [58]. In addition to these enzymes, non-structural proteins also provide important support functions to accommodate the RTC, such as modulating intracellular membranes, host immune evasion, and providing cofactors to facilitate replication. The 3′ one third of the SARS-CoV-2 genome expresses genes for viral structural proteins and accessory proteins, including the spike (S), nucleocapsid (N), membrane (M) and envelope (E) proteins. The S1 subunit of S protein mediates attachment to ACE2 while S2 promotes membrane fusion. Likewise, the N, M and E proteins are critical for different aspects of viral functioning, maturation and assembly and will be described in more detail in the sections below [59].
2.2.3. Transcription of the new genetic material
The early expression of viral genes leads to the biogenesis of viral replication organelles in the cytoplasm of infected cells that create a protected microenvironment for viral genomic RNA expression and the transcription of subgenomic mRNAs [46]. Viral genome replication is initiated by the transcription of full-length negative-sense genomic copies that serve as templates for the production of positive-sense genomic RNAs. These newly synthesized genomes also generate more nsps and RTCs or are packaged into progeny virions during the assembly process. In addition, all coronaviruses, including SARS-CoV-2, have a conserved leader sequence in their 5′-ends that function as cis-acting elements for the transcription of subgenomic mRNAs directed by viral RdRP. The TRS-L sequence described previously constitutes a signal for the transcription of subgenomic mRNA while each transcriptional unit on the genomic RNA are preceded by TRS-B, additional transcription regulatory elements [60]. Studies have also demonstrated that trans-acting factors encoded by the virus as well as cellular proteins play a role in subgenomic mRNA synthesis [61].
Once subgenomic mRNAs are expressed, nsp16 in conjunction with nsp10, methylates the 5′-end of viral mRNAs to create a 5′-methyl-cap [62]. This mechanism serves to not only promote translation of virally encoded messages by the host translational machinery, but it is also an essential strategy to protect the virus from the host immune response. RNA capping in coronaviruses involves several nonstructural proteins, including nsp13, nsp14, and nsp16. Nsp16, a cap ribose 2′O methyltransferase, is known to form a complex with nsp10 to convert mRNA species from the Cap-O (me7GopppA1) to the Cap-1 form (me7GopppA1m) by methylation of the 2′O ribose of the first nucleotide. Mass spectrometry studies have revealed that an essential antiviral factor, IFT1, has high affinity for unmethylated Cap-O forms of RNA and impairs binding of eukaryotic translation initiation factors to 2′O-unmethylated RNA templates, resulting in the inhibition of translation [63]. Therefore, the specificity of IFIT1 for the Cap-O form of mRNA serves as a potent antiviral mechanism against viruses that lack 2′-O methyltransferase activity. Since coronaviruses actively generate Cap-1 forms of viral mRNAs, this not only promotes translation, but also serves as a viral strategy to avoid detection by the innate immune response.
3. Viral proteins
The SARS-CoV-2 genomes contains 29,811 nucleotides and codes for a total of 29 different viral proteins (see Fig. 2) [64]. For comparison, the human genome contains 3.2 billion DNA nucleotides, which contains ~20,000 protein-coding genes [65]. Each of the SARS-COV-2 viral proteins is classified into one of three groups: 1) structural proteins, 2) non-structural proteins, or 3) accessory proteins (Table 1 ). Many structures of SARS-CoV-2 proteins have been described in the literature and have been compared with those of other coronaviruses [66]. The variation between these proteins accounts for the observed differences in each virus's contagiousness and infectivity in human cells.
Table 1.
List of names, functions, and length of proteins.a
| Name | Name-function | Length (aa) | Name | Name-function | Length (aa) |
|---|---|---|---|---|---|
| ORF1ab | ORF1ab polyprotein | 7096 | nsp14 | 3′-to-5′ exonuclease, proof reading during RNA synthesis | 527 |
| ORF1a | ORF1a polyprotein | 4405 | nsp15 | endoRNAse, cleaves RNA at polyuridylate sites | 346 |
| nsp1 | leader protein, inhibits host protein translation | 180 | nsp16 | 2′-o-ribose methyltransferase, in RNA capping | 298 |
| nsp2 | nsp2, disrupts host cell cycle | 638 | nsp11 | nsp11 | 13 |
| nsp3 | Protease, lowers host immune response, and host translation | 1945 | S | Surface glycoprotein | 1273 |
| nsp4 | nsp4 | 500 | ORF3a | ORF3a | 275 |
| nsp5 | 3C-like proteinase | 306 | E | Envelope protein | 75 |
| nsp6 | nsp6 | 290 | M | Membrane glycoprotein | 222 |
| nsp7 | nsp7, primer synthesis and RNA replication | 83 | ORF6 | ORF6 | 61 |
| nsp8 | nsp8, primer synthesis and RNA replication | 198 | ORF7a | ORF7a | 121 |
| nsp9 | nsp9, interacts with DDX5 protein to facilitate replication of virus | 113 | ORF7b | ORF7b | 43 |
| nsp10 | nsp10, mRNA cap methylation | 139 | ORF8 | ORF8 | 121 |
| RdRp | RNA-dependent RNA polymerase, RNA replication | 932 | N | Nucleocapsid phosphoprotein | 419 |
| nsp13 | Helicase, activity during RNA replication | 601 | ORF10 | ORF10 | 38 |
Adapted from Kadam et al. [64].
3.1. Structure and function of the SARS-CoV-2 proteins
Structural analysis of proteins and protein complexes can provide key insights into the molecular basis for their observed function. Therefore, researchers have examined the structure of several SARS-CoV-2 proteins and related complexes. Traditionally, X-ray crystallography has been used to determine high resolution three-dimensional protein structures important for determining biological function and drug design [67], [68], [69]. Though X-ray crystallography is a very detailed and powerful method, it is often limited by difficulties associated with protein crystalization caused by size and flexibility constraints. In addition, due to their flexibility, it is difficult to observe glycosylation patterns on the surface of crystallized proteins. Finally, since crystals require static positioning within a crystal lattice, it is very difficult to observe and predict conformational changes and protein dynamics without assuming movement between crystals in different static conformational states. Therefore, as an alternative technique, many structural biologists have begun to use cryogenic electron microscopy (cryo-EM) to determine protein structure [70], [71]. During a cryo-EM experiment, the protein is deposited frozen onto a metal grid in a single molecule layer. The deposited layer is then irradiated with low energy electron beams and a 2-D image of the protein is obtained. By adding additional layers and using a computer to collect and sort the data, a 3D image of the protein is obtained. With cryo-EM, conformational changes and protein dynamics can also be recorded. In recent years, the quality of the images from cryo-EM experiments has improved to resolutions that are comparable to those obtained using crystallography [72], [73], [74]. Because of these technologies, several structures of SARS-CoV-2 S-protein in a complex with the ACE2 receptor have been solved [8], [53], [75], [76].
3.1.1. The S-protein
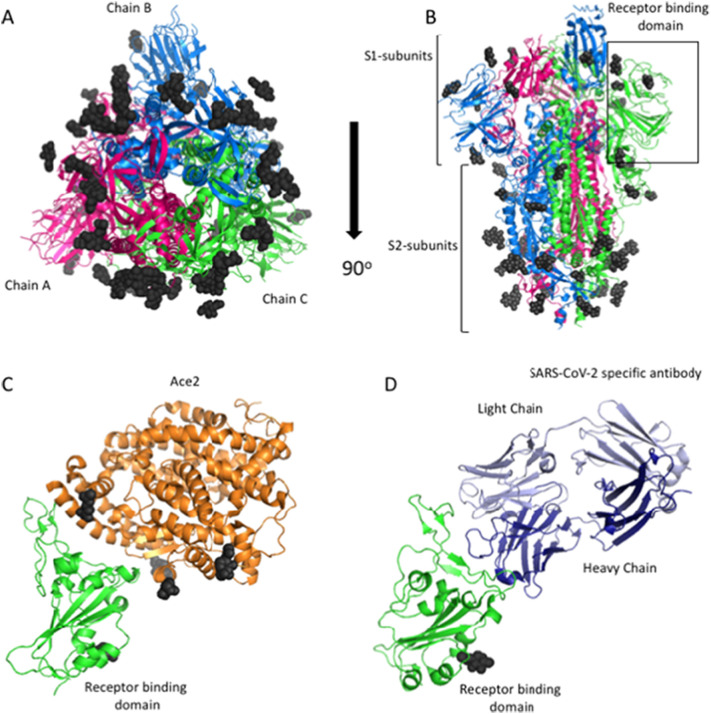
The S-protein of SARS-CoV-2 is a 150 kDa transmembrane protein embedded into the surface of the viral envelope. As mentioned previously, S-protein binds to ACE2 receptors on the surface of host cells to facilitate viral entry [8], [77]. Several groups have solved the three-dimensional structure of the S-protein, thus providing valuable insights into its overall shape, plasticity, and specific target interactions [8], [53], [54], [72], [75]. The S-protein consists of an S1 subunit that binds the host cell receptor and an S2 subunit that is responsible for fusion of the viral envelope with host-cell membranes. In addition, S-protein has a receptor binding domain (RBD), composed of approximately 230 amino acids, that binds to specific sequences of the ACE2 receptor. The protein is fully glycosylated and forms a homotrimer made of A, B and C chains embedded within the viral envelope (in Fig. 9A and B, glycosylation sites are shown in dark grey). Interestingly, the S1 subunit has several domains that have been shown to vary in function depending on the specific viral strain. This variation leads to its ability to use alternative host cell recognition targets.
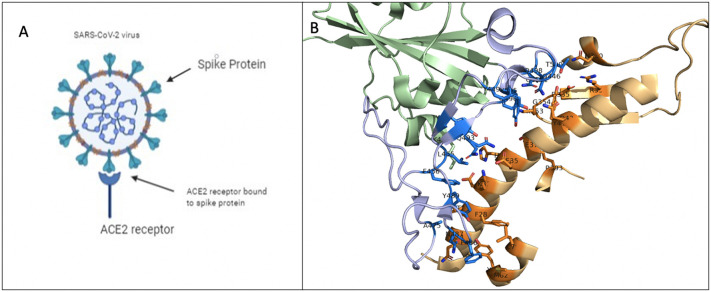
Fig. 9.
Depiction of SARS-CoV-2 using its spike protein (S-protein) to bind to angiotensin converting enzyme 2 (ACE2) receptor to initiate infection. A) Schematic diagram depicting interactions between SARS-CoV-2 S-protein and ACE2. B) Co-crystal structure highlighting the binding interface between S-protein (purple) and ACE2 (orange). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Recently a cryo-EM structure of the SARS-CoV-2 S-protein ectodomain trimer was solved [8], [54]. The result suggests that the SARS-CoV-2 S-protein adopts conformations similar to those reported for both the SARS-CoV and MERS-CoV S-proteins. As observed in other β-coronavirus S-glycoproteins, the SARS-CoV-2 S1 subunit takes on a V-shape and harbors three human ACE2-recognition motifs in its closed state (known as the receptor binding domain or RBD shown in Fig. 9C). Structural information shows that these motifs are buried, and therefore an opening of the structure is likely required for interactions with ACE2. This interaction would initiate a conformational change that would subsequently allow S2 protease cleavage, leading to membrane fusions and viral entry. In comparison to SARS-CoV, which uses S-protein residues Y422, L472, N479, D480, T487, and Y4911 for binding to ACE2, the SARS-CoV-2 S-protein may use amino acids L455, F486, Q493, S494, N501, Y505 for binding to ACE2 [78], [79], [80], [81]. The differences in the amino acids promote stronger binding interactions between the SARS-CoV-2 S1 RBD and ACE2 receptors on the host cell. While cryo-EM is a beneficial strategy to elucidate the overall protein structure of S1, it is not an optimal method to identify the structure of the sugars that are covalently linked to the surface of the protein [82], [83], [84]. The sugars play important roles in stabilizing the proteins to aid in folding. In addition, they have been shown to contribute to viral host immune evasion tactics. Because of their importance, the molecular composition and the structure of these sugars have been analyzed through mass spectrometry [85].
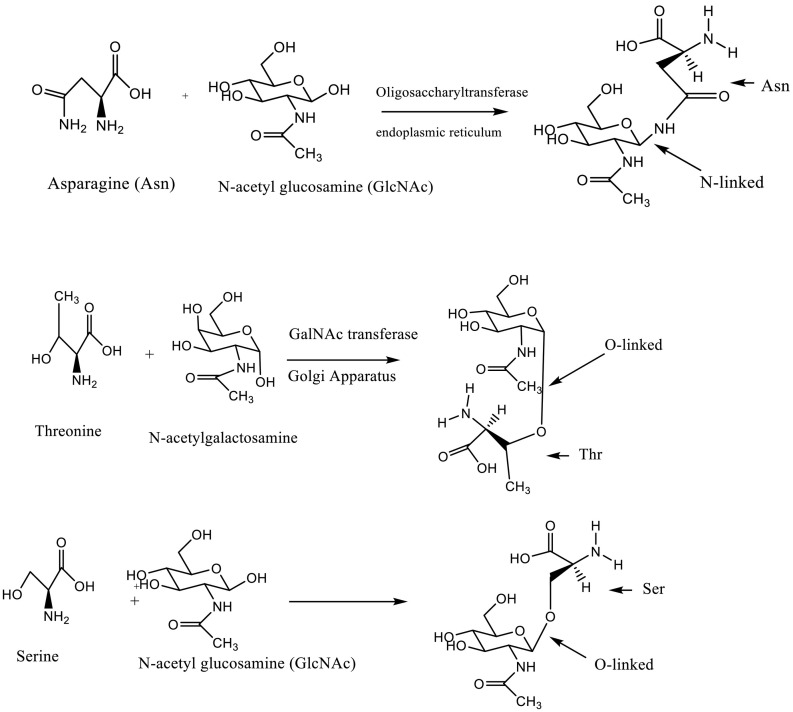
S-protein is heavily glycosylated, undergoing glycosylation on multiple asparagine (Asn) or serine (Ser) residues via N- and O-glycosylation, respectively. The formation of the N-glycan, which involves conjugation of N-acetylglucosamine to asparagine residues, is catalyzed by oligosaccharyltransferases present in the endoplasmic reticulum (ER) while the formation of O-glycans is mediated by O-GlcNAc transferase (OGT), likely in the ER or the Golgi apparatus [86] (Fig. 4).
Fig. 4.
The amine functional group of asparagine (Asn) functions as a nucleophile to promote N-linked glycosylation, in which a carbohydrate chain (e.g., N-acetyl glucosamine (GlcNAc)) is added to the protein chain. In general, a carbohydrate chain is added to an Asn residue when it is flanked C-terminally by X-serine or X-threonine, where X is any amino acid other than proline. In the case of O-linked glycosylation, the hydroxyl group of threonine or serine acts as the nucleophile to promote conjugation of GlcNAc or N-acetyl galacosamine (GalNAc). See ref. [277].
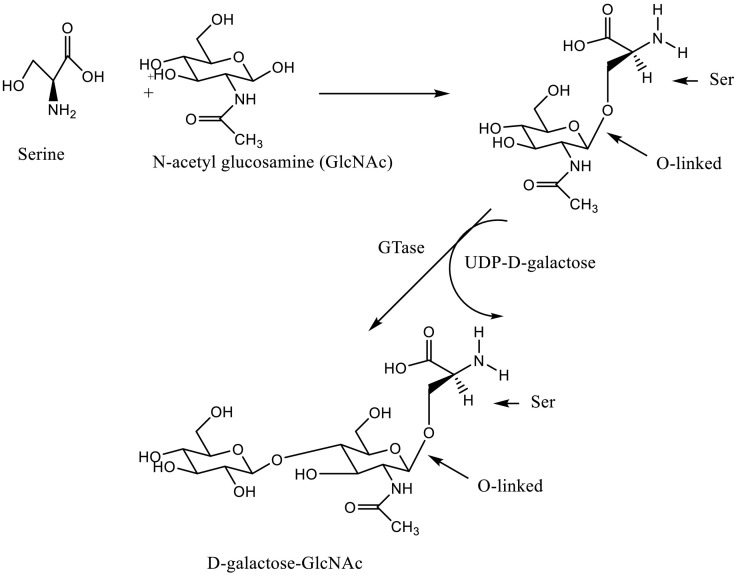
After being conjugated to Asn or Ser, N-acetyl glucosamine (GlcNAc) is further glycosylated by the enzyme GTase to produce D-galactose-GlcNAc [87] (Fig. 5, Fig. 6 ). After this initial reaction, more sugars are added by other glycosyltransferases, eventually leading to the formation of long chains of various polysaccharides.
Fig. 5.
Addition of D-galactose to GlcNAc-serine. A similar reaction may be observed with threonine-N-acetylgalactosamine.
Fig. 6.
Components of O-linked glycans.
The SARS-CoV-2 S-protein has 22 putative N-linked glycosylation sites and 4 possible O-linked glycosylation sites (Fig. 7A and B). Interestingly, mass spectral analysis shows differential glycosylation patterns in different cell types [88], [89], [90], [91]. It appears that, depending on the tissue type, the overall level of glycosylation, as well as the relative ratios of N- to O-glycosylation, is different. In O-glycans, the sugar molecule is attached to the oxygen atom of serine (Ser) or threonine (Thr) of the protein (Fig. 5, Fig. 6) after the protein is synthesized. O-glycosylation occurs in the ER, Golgi apparatus, and even the cytoplasm. In humans, the S1 protein has been shown to be O-glycosylated on Thr323, Ser325, Ser673, Thr678 and Ser686 [92]. The presence of O-glycosylation at Thr323 and Ser325 of the S1 subunit of SARS-COV-2 has been correlated with the active structure for the protein, which leads to the biological function [93]. These differences can have a bearing on vaccine production, as they can influence how the S-protein interacts with ACE2 receptor [94], [95].
Fig. 7.
Structures of the SARS-CoV-2 S-glycoprotein. A) Cartoon diagram of the 3D structure of the S-glycoprotein solved using cryo-EM to 3.46 Å (PDB ID: 6VSB). The S-glycoprotein forms a trimer composed of an S1-subunit (pink), an S2-subunit B (green) and an S3-subunit (blue). This structure represents the prefusion confirmation and was found to have 22 glycosylation sites (dark grey). B) 90° rotation of A. Here, the receptor binding domain (RBD) is located at the top of the structure. The RBD of the subunit C is boxed in black. C) Cartoon diagram of the RBD of the S-glycoprotein (green) bound to the host receptor ACE2 (orange) (PDB ID: 6M0J). This structure was solved to 2.45 Å using X-ray protein crystallography. D) Cartoon diagram of the SARS-CoV-2 RBD (green) bound to a SARS-CoV-2 specific antibody (the antibody heavy and light chains are colored dark blue and light blue, respectively). This structure was solved using X-ray crystallography to 2.85 Å (PDB ID: 7BWJ). It is important to note that the antibody binding site is at the same position as the ACE2 binding site, thereby blocking interactions between the virus and the host receptor. All figures were made using PyMol. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1.2. Changes in S-Protein due to mutations
Mutations are commonplace in viruses. As viruses replicate, mutations cause changes in the genomes of the newly synthesized viral particles, which carry the mutation(s) to hosts during subsequent infections. Only some mutations produce more aggressive, more deadly diseases. Changes in the genome causes changes in the viral proteins and since S-protein is the instrument of infection for SARS-COV-2, changes in the S-protein determines how easily virus can infect its host, or how rapidly infection spreads among populations. Similarly, mutations can impact how effective an antibody is to the S-protein. Antibodies bind to the N-terminal domain of the S-protein while the receptor binding domain (RBD) associates with the ACE-2 receptor at the surface of host cells. In the B.1.1.7 Variant that was first detected in the UK, nine mutations in the S-protein, including N501Y in the RBD and deletion of 3 amino acids in positions 69, 70, and 144 in the N-terminal domain. Tighter binding of the S-protein with ACE-2 due to these mutations makes the virus 50–70% more infectious. In the B.1.351 Variant, which first appeared in South Africa, of the nine amino acid changes, three were involved in N501Y as in the B.1.1.7 Variant, the other two were K417N (lysine to asparagine) and E484K (switching glutamic acid to lysine). These changes made the virus 50% more infectious than the original variant. In P.1 Variant initially detected in Brazil, 11 mutations occur in the S-protein, which include N501Y and E484K seen in other variants and K417T, which involves changing lysine to threonine. These mutations also increase the binding between the RBD with the ACE2 receptor on the host cells. In the B.1.427 and B.1.429 Variants, which were first detected in California, there are four mutations in the spike protein, including L452R in the RBD. This variant, which was about 20% more aggressive, is disappearing [96]. Delta and Kappa Variants first identified in India both have two mutations, E484Q and L452R. In addition, the Delta Variant contains a T478K mutation. These mutations in S1 increase the binding affinity to ACE2, causing higher transmissibility and virulence [44].
3.1.3. E-protein
The SARS-CoV-2 envelope protein (E-protein) is a 10 kDa protein with an N-terminal transmembrane domain and a C-terminal cytoplasmic tail [97]. In SARS-CoV, E-protein mediates the budding and release of progeny viruses and also activates the host inflammasome. Studies have shown that the E-protein of SARS-CoV-2 is structurally and functionally similar to that of SARS-CoV and is highly conserved in their N-terminal regions [98], [99]. E-protein oligomerizes to form viroporins, which are small membrane-embedded proteins present on different viruses with ion-conducting properties. They form cation selective channels across the ER-Golgi intermediate compartment to aid in the viral production, maturation and release processes, which are regulated by ion homeostasis of cellular organelles [99], [100], [101]. This protein has also been found to influence folding of other proteins, which has an effect on the survivability of the host cells [101], [102], [103], [104], [105].
3.1.4. M-protein
The most abundant structural protein of coronaviruses is the membrane (M)-glycoprotein. M-protein is a membrane bound glycoprotein with protease activity and three transmembrane domains. The M-protein can interact with viral RNA and bind to the other structural proteins, including S-, E- and N-proteins [106]. Binding with M-protein helps to stabilize N-proteins to promote viral assembly. In addition, M-protein also aids the S-protein during the attachment phase and may be involved in immune evasion and pathogenicity functions of the virus [107], [108], [109]. The C-terminus of the M-protein resides inside the cell with the N-terminus on the outside. The third transmembrane domain, common to all coronaviruses, contains both polar and non-polar ends. The presence of an N-terminal Ser at position 70 distinguishes the M-protein of SARS-CoV-2 from M-proteins of other CoVs and may have allowed SARS-CoV-2 to jump from bats to humans [110]. An additional change is substitution of Ala for Thr at position 285 in SARS-CoV-2 M-protein, which may have increased the protease activity of the SARS-CoV-2 enzyme [84].
3.1.5. N-protein (nucleocapsid protein)
The N-protein has a molecular weight of 47 kDa and binds to gRNA. It has three domains, termed the N-terminal domain (NTD), central linker (CL), and C-tail. This protein is involved in the replication and transcription of viral RNA and formation of the ribonucleoprotein (RNP) complex [108]. It also influences the host cell cycle to enhance viral multiplication [111]. The CTD domain of SARS-CoV-2 N-protein, which is required for dimerization and oligomerization, interacts with the M-protein through the CL region [112].
The NTD binds to RNA and contains many positively-charged amino acids residues, which aid in binding the negatively-charged RNA. The amino acid sequence of SARS-CoV-2 N-protein is 90% similar to that of SARS-CoV [113]. Most of the differences are due to the presence of these positively-charged amino acids in the SARS-COV-2, which are absent in other CoVs.
3.2. Viral non-structural proteins
In addition to the structural proteins, there are also 16 non-structural proteins (Nsps) in SARS-CoV-2, including polyproteins, nucleoproteins, and membrane proteins, each with varying functions (Fig. 8 ). For instance, Nsp1 and Nsp2 are involved in modulating the immune response [114]. Nsp1 has 180 amino acids and binds to the small subunit of the ribosome, stopping synthesis of antiviral proteins. In addition, Nsp1 halts all host protein synthesis by cleaving host mRNA [47], [115]. As a result, production of type-1 interferons (which provide innate immunity) is halted, disabling the host defense mechanisms against the virus. Nsp2, on the other hand, influences cell division by altering the host cell cycle (Fig. 3) [116].
Fig. 8.
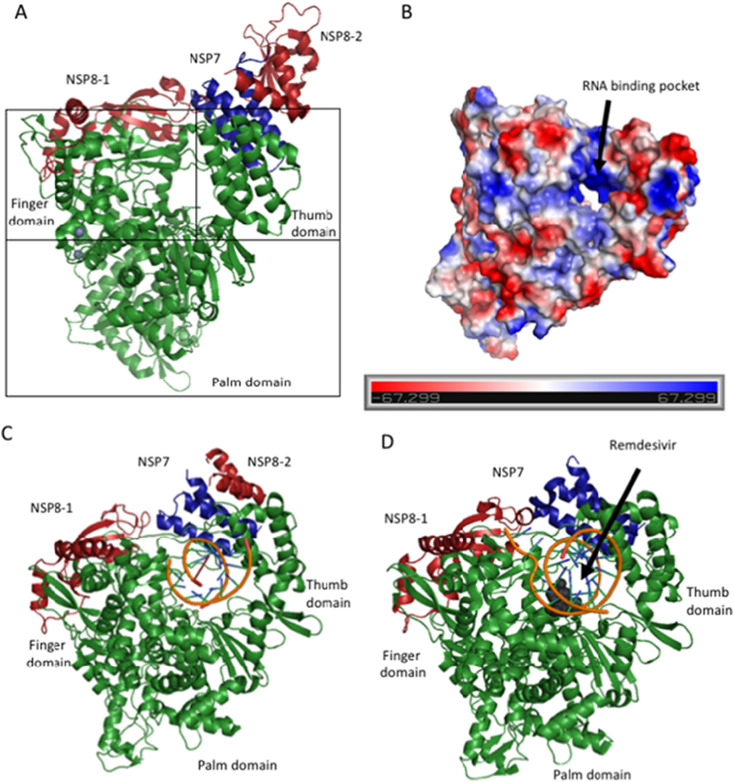
The SARS-CoV-2 RNA dependent RNA polymerase (RdRp). The structure of the RdRP was solved to 2.95 Å using cryo-EM. A) Cartoon diagram of the RdRP (green) in complex with its required co-factors, NSP7 (blue), Nsp8-1 (light red) and Nsp8-2 (dark red). In addition, there are two molecules of Zn bound to the RdRP (PDB ID: 7BTF). The structure takes on a typical polymerase fold, with a thumb, palm and finger domain that cradles the RNA substrate during synthesis. B) Electrostatic surface diagram of Nsp12 alone. Here, the hole traversing between the domains is highly positively charged in order to provide a complementary binding site for the negatively charged RNA substrate. C) SARS-CoV-2 Nsp12/RNA complex solved using Cryo-EM to 3.53 Å (PDB ID: 6X2G). D) Cryo-EM structure solved to 2.5 Å of Nsp12, its cofactors, template RNA, and the drug Remdesivir. When Remdesivir is covalently incorporated into the RNA primer, it terminates chain elongation and therefore inhibits the polymerase activity of the enzyme (PDB ID: 7BV2). Figures made using PyMol. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Nsp3, or papain-like protease (PLpro), is the largest of all Nsps and plays many roles. Along with Nsp5, it is responsible for cleavage of viral polyproteins into functional units [117], [118]. Nsp3 also plays a role in the onset of cytokine storm, assembly of viral particles, and, via interactions with Nsp4, in formation of double membrane vesicles [119]. Finally, Nsp3 both inhibits the host enzyme poly-[ADP-ribose] polymerase (PARP), whose role is to block viral replication, and suppresses the expression of interferon genes (required for inhibition of viral infections).
Nsp9 is a single-stranded RNA-binding protein that plays a role in viral replication. Nsp12 is an RNA-dependent RNA polymerase (RdRp) and nsp13 is a helicase [80], [118]. Both are essential for RNA replication. Nsp12, along with Nsp7 and Nsp8, recognize the RNA template and facilitate processivity [120] (Fig. 8A [121]. In SARS-CoV-2, it has been shown that the hetero-dimer Nsp7–Nsp8 and individual Nsp8 can bind with the RdRp [82].
The nsp13 helicase contributes to RNA synthesis and 5′-RNA capping by unwinding double stranded RNA in a 5′–3′ direction to enable elongation by the RdRp. The RdRp (Nsp12) from SARS-COV-2 has the structure of a typical polymerase, which resembles a right hand with finger, thumb, and palm domains [122]. When overlayed with the RdRp of SARS-CoV, there is only a root mean squared distance (rmsd) of 0.82 for 1078 of the C-alpha for SARS-COV-2, indicating that there are very few structural differences between the two proteins [82]. Despite these small differences, the amino acid sequence and the active sites of the RdRps from the two viruses are nearly identical [123]. This has enabled the rapid development of inhibitors against SARS-CoV-2 using the lessons learned from the previous inhibition studies on SARS-CoV.
Nsp14 is an exoribonuclease protein that acts as a methyl transferase for N7 guanine in mRNA cap synthesis. For activation, Nsp14 requires Nsp10 to form a heterodimer [124]. Nsp15 is a nidoviral RNA uridylate specific endoribonuclease (NendoU) that protects from attacks by the host immune system [125]. Meanwhile, Nsp16 is a 2′-O-methyltransferase, which uses SAM for its methyl source. It complexes with nsp10 but has two S-adenosyl-L-methionine (SAM) binding sites of its own. Binding between SAM and nsp16 is mediated by both hydrophobic and ionic interactions [126], [127], [123], [128], [129].
3.3. Accessory proteins
Accessory proteins are encoded primarily on the 3′-end of the viral RNA. They, like many of the other SARS-CoV-2 proteins, help to protect the virus against the host immune system. For instance, accessory proteins 3a and 7a are thought to be involved in induction of inflammation. The six accessory proteins, 3a, 6, 7a, 8a and 9b, are similar to those in SARS-COV. The only exception is 8b, which has an additional 37 amino acids not observed in SARS-COV proteins [5], [130], [131], [132]. Because the structure of most SARS-CoV-2 proteins is very similar to the structure of the proteins of coronaviruses studied previously [133], similar techniques can be applied to the new virus during the development of the best inhibitors for target enzymes [71], [72], [134].
3.4. Target receptors
As alluded to above, respiratory droplets and aerosols carrying the SARS-CoV-2 virus produced by an infected person target the host nasal goblet epithelial cells, type II pneumocytes, and enterocytes cells [64], [135], [136] via interactions between ACE2 and the viral S-protein, which is primed by transmembrane serine protease 2 (TMPRSS2) located on the surface of the cells (Fig. 2) [36], [137].
The receptor binding motif (RBM) is the specific region within the RBD of the S-protein that binds to the ACE2 receptor. The RBM is made up of residues 438–506 and is located in an extended insertion between β4 and β7. The concave surface of the RBD contacts the N-terminal helix of ACE2. There is significant sequence variation in this region of the S-protein compared to SARS-CoV, with 17 total resides of SARS-CoV-2 S-protein making contact with 20 residues of ACE2 (vs. 16 of SARS-CoV S-protein to 20 of ACE2). Of the 20 ACE2 residues making contact, 17 are shared between the proteins. These changes could account for the observed differences in binding affinity for SARS-CoV-2 (4.7 nM) and SARS-CoV (31 nM), despite their overall structural similarities (rmsd of 1.3 Å) [108], [138].
The TMPRSS2 protein, which is also expressed in the prostate, salivary glands, colon, and stomach, is a chymotrypsin family serine protease (492 aa) composed of three domains [64], [139], [140]. It mediates the cleavage of S-protein within the S1–S2 linker (R685) and at S2′ (R815). The S1-S2 cleavage site is facilitated by the presence of several repeated basic arginine residues [64], [137], [141]. Additional help is provided to the virus by Furin, a host cell protease that cleaves at the polybasic furin cleavage site (e.g., with the PQRESRRKK/GLF sequence of amino acids) in S-protein, promoting cell fusion and entry of the virus into human cells [64], [79], [135], [136], [141]. After cleavage at the S2′ site, the fusion peptide is then inserted into the host membrane. The two HR regions, HR1 and HR2, in the S2 domain form antiparallel six-helix bundles (6-HB). Once inside the host cell, the virus can effectively replicate and initiate the process of infection and disease development [136], [142]. The RBD from SARS-CoV-2 S-protein has higher binding affinity (~5 kcal/mol) towards human ACE2 than the corresponding RBD from SARS-CoV S-protein, which is higher than its affinity for ACE2 from other animals [64], [143]. This higher affinity, according to contact map analysis, is due to greater electrostatic interactions between the virus's S-protein and ACE2 receptor [144]. Computational alanine scanning analysis has identified key residues responsible for binding of the three RBDs of SARS-COV-2 with ACE2 [145]. Accordingly, SARS-CoV-2 uses almost all (90%) of the tyrosine (Tyr) and glycine (Gly) residues that are present at its ACE2 interface for binding. Molecular dynamics simulation on membrane-bound ACE2 supports the presence of about seven additional hydrogen-bonded contacts between the SARS-CoV-2 RBD and ACE2 [146]. Compared to human ATR1, preferential electrostatic interactions between ACE2 and the RBD of SARS-COV-2 are due to 7 additional hydrogen bonds, which is equivalent to ~3.4 kCal/mol of binding energy. According to mutagenesis analysis, residues 484 and 498 of the S-protein in SARS-CoV-2 are involved in recognizing human ACE2 [147].
As stated above, in addition to the Type II alveolar cells of lungs, ACE2 and TMPRSS2 are also expressed in the outer layer of the nasal and esophagus epithelium, the absorptive enterocytes from ileum and colon, and cornea [36], [148], [149]. In fact, single-cell RNA sequencing has shown high ACE2 mRNA expression in Type II alveolar cells of lungs, myocardial cells, esophagus upper and stratified epithelial cells, and digestive tract and kidneys [53], [150], [151], [152]. The initial contact may also occur on the skin, throat, or in the gastrointestinal (GI) tract [36], [153], [154], [155], [156], [157].
3.5. Comparing SARS-CoV-2, SARS-CoV, and MERS-CoV receptor binding
The SARS-CoV-2 genome is different from all previously identified coronaviruses, including SARS-CoV and MERS-CoV. These differences likely account for differences in cellular targets, as SARS-COV-2 targets host nasal epithelium in addition to many other organs, in contrast SARS-CoV, which does not target the nasal epithelium but targets lung, trachea/bronchus, stomach, small intestine, distal convoluted renal tubule, sweat gland, parathyroid, pituitary, pancreas, adrenal gland, liver and cerebrum [158]. The SARS-CoV-2 S-protein exhibits even less homology with the MERS-CoV S-protein, which recognizes dipeptidyl peptidase-4 (DPP-4) as a specific receptor for host cell entry (as opposed to ACE2) [79], [141], [159], [160].
Aside from molecular factors, other factors such as age, smoking, and gender may also influence the expression of ACE2 and TMPRSS2 in the organs and tissues and thus influence the severity and rate of infection of SARS-CoV-2 [137], [161], [162]. There is evidence that by directly infecting macrophages and T cells, SARS-CoV represses the body's immunity. But it is not yet clear if this is also the case for SARS-CoV-2, as macrophages and monocytes in the lung do not express a large number of the ACE2 receptors [163].
The SARS-CoV-2 genome sequence was first published in January 2020 and is 79.5% similar to the genome of SARS-CoV [66], [80], [164], [165]. Both SARS-CoV and SARS-CoV-2 have S-proteins that are nearly 80% identical in composition [166], [167], [168], [169]. Interestingly, the relatively small number of differences between the two S-proteins has a dramatic influence on parameters such as affinity between the S-protein and its receptor (ACE2) [170], [171]. The increased affinity of its S-protein for ACE2 allows SARS-CoV-2 to attach to human cells much more strongly than SARS-CoV. This characteristic of SARS-CoV-2 enhances both the contagiousness and infectivity of the virus [54], [168]. Importantly, for infection, viral S-protein binds to host ACE2, therefore people with increased numbers of ACE2 sites in their lungs are more susceptible to the infection by this virus [172]. One additional characteristic that sets SARS-CoV-2 apart from other coronaviruses is the presence of a furin cleavage site within the S-protein. Furin is a host cell protease and has been found to be important for S-protein–mediated cell-cell fusion and entry into human lung cells [79], [141].
4. Strategies to fight SARS-COV-2
Strategies to fight SARS-CoV-2 include vaccines, drug repurposing, novel antivirals, and therapeutic antibodies [171], [173], [174], [175], [176]. For effective development, all these strategies require knowledge of the viral genome and/or proteins, along with an understanding of the basic biology of its replication cycle. To this end, interdisciplinary teams of scientists use computer modeling, structure determination, metabolism/pharmacokinetics, biochemical assays, and synthetic chemistry protocols to design and optimize effective therapeutics. As a result, some drugs have been designed to inhibit capsid formation through formation of van der Waals forces, hydrogen bonds, and electrostatic interactions with the capsid directly. Meanwhile, others change the forces that hold the capsid together, disrupting their ability to attach and enter host cells. Many of the non-structural proteins, including the S-protein, M-protein, and RdRp, are also targeted by small molecule inhibitors, which bind to the enzyme active site. In some cases, these drugs are already available. For instance, several in silico screening efforts have shown similarities between SARS-CoV-2 and other non-coronaviruses, enabling the use of a number of available antivirals and drugs that are currently on the market [177], [178], [179], [180].
4.1. Spike protein as target for inhibitors (potential drugs)
Much of the current efforts in combating SARS-CoV-2 is focused on finding ways to block the interactions between ACE2 and the viral S-protein, sometimes through specialized maneuvering that stabilizes the spike [181]. The difficulty in doing this is that ACE2 is also present on the cells in other parts of the body that are not targets of the virus. Therefore, non-tissue specific inhibition of ACE2 could lead to unwanted side effects [182], [183]. ACE2's physiological role in the cell is in the maturation of angiotensin, which is a hormone that controls blood pressure by regulating blood vessel constriction. Consequently, the inhibition of ACE2 can lead to several complications, including cardiovascular disease and hypertension [53]. As a result, alternative strategies are being explored to block the interaction between the SARS-CoV-2 S-protein and ACE2 directly using therapeutic antibodies. The ideal therapeutic antibody would specifically bind the S-protein and prevent it from interacting with the ACE2 receptor, achieving a similar result as blocking ACE2 directly without the negative side effects [77]. Alternatively, designing small peptides or small molecules that bind to different subunits of the SARS-CoV-2 S-protein will prevent the subunits from interacting with ACE2 and viral entry into the cell. As previously stated, the S-protein is initially cleaved into S1 and S2 subunits by the host cell protease, furin. S1 facilitates receptor binding by engaging with the host cell membrane while S2 provides structural support and regulates the fusion with the host's epithelial cell membrane. For this reason, blocking the cleavage of the S-protein could also be a potential strategy for inhibiting viral entry [8], [54], [77], [184], [185], [186].
Using knowledge gained from HKU1, a common cold virus, and MERS virus, scientists found two small loops of amino acids that held two α-helices in the spike together. When the S-protein binds to human cells, the coil-like structure is released and the two helices and one loop are elongated to bring the human cell and the virus close together for fusion. By adding up to six proline residues to the SARS-CoV-2 S-protein, scientists believe that they can prevent the “spring” from being released, which would prevent the fusion of SARS-CoV-2 with human cells [181].
4.2. Chemistry of selected antiviral compounds
Due to the relatively small size of the SARS-CoV-2 genome and recent rapid advances in whole genome sequencing technologies, scientists were able to sequence the genome of the SARS-CoV-2 rather quickly once it was isolated [80], [165]. As a result, the sequence has been used to determine the primary structure (i.e., linear amino acid sequence) of the viral proteins. Once a protein's primary sequence has been determined, computational modeling programs can be used to deduce its secondary and tertiary structures. These programs are based largely on homology modeling (which uses the structures of closely related proteins to model the structure of the protein-of-interest). This is either in lieu of 3D structure determination, as discussed previously, or as a complement to confirm what was observed in vitro. Once a putative structure is determined, different therapies can be designed to target the protein's active site and/or other protein surfaces important for enzyme function [118]. Of course, similar strategies can be used once a bona fide structure of the enzyme-of-interest is solved experimentally using structural biology methods, such as X-ray crystallography, NMR spectroscopy or, more recently, cryo-EM. The structural information can, in turn, be used to develop drugs that target those proteins involved in infection, viral replication, or transmission of the disease. In the process, computer models of the viral proteins are constructed and their interactions with human proteins are studied at the molecular level [74], [187]. At the same time, high-throughput screening may be performed for the compounds that look promising based on in silico docking studies [83], [179].
At the onset of the COVID-19 pandemic, a list of 50,000 compounds with potential antiviral properties was provided by the American Chemical Society (ACS) Central Science. The compounds were extracted from the CAS registry, which contains 160 million compounds [188]. The identified compounds were anti-infective, enzyme inhibitors, or have shown an effect on the respiratory system [189].
Recently, the cryo-EM structure of the RdRP was determined (Fig. 8) [74], [82], [190]. The structural information led to computational simulations (in silico) of the interactions between RdRp and potential drug candidates. Similarly, knowledge of the structure of the S-protein and main protease (M-protein or Mpro) has enabled similar studies [82], [170], [191], [192]. To make these studies possible, computational resources around the world have been made readily available to COVID-19 researchers by major international computer centers inside and outside the U.S. during the COVID-19 pandemic [193].
In addition to in silico analysis, animal models have been used to model infection, including mice that express the necessary receptors and succumb to similar symptoms as humans [189], [194]. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. This affords the opportunity to devise appropriate treatments through docking studies [83], [135].
Researchers at Novartis had quickly discovered that the Mpro from SARS-CoV-2 (cysteine-main protease) was very similar to the protease from the previous corona viruses. This includes the active site cysteine, which serves as a target for identifying inhibitors. As a result, sixteen potential inhibitors were identified using computational methods [195], [196], [197]. A select number of identified inhibitors were synthesized and tested in models. Chemical structures were then further refined based on the results of the inhibition studies. A new smaller set of inhibitors were then obtained and some of the final compounds have entered randomized clinical trials [198], [199]. Computational studies have suggested that lopinavir, oseltamivir, and ritonavir may bind to SARS-CoV-2 Mpro [198], [199]. A similar approach could be used to develop inhibitors for all other viral proteins, including RdRp and the S-protein. More recently, it was discovered that drug repurposing studies for COVID-19 provided compounds that were harmful to the body as their mechanism of action was misunderstood in the initial studies [200]. As a result, some of such drugs like hydroxychloroquine, Amiodarone, and Sertraline, cause phospholipidosis in the infected host cells which mistakenly in the studies was correlated with antiviral efficacy.
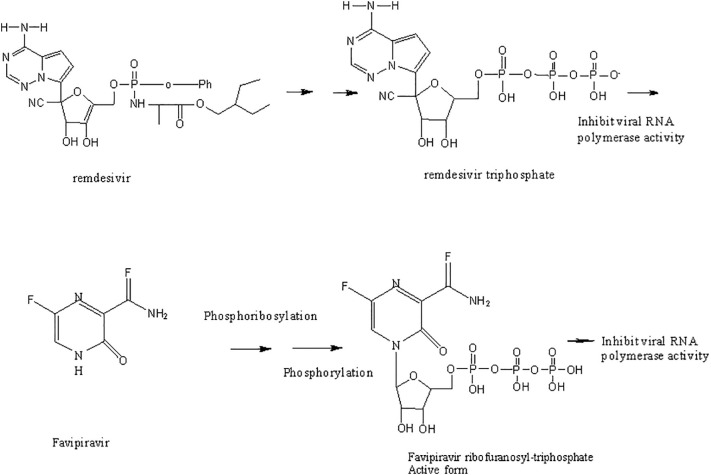
As previously discussed, RdRp is responsible for replication of the viral RNA (Fig. 8) [201]. Remdesivir (See Table 2 ) (an inhibitor of this enzyme), was originally developed as anti-Ebola drug [202], [203], [204], [205],and soon entered the Phase I clinical trial against COVID-19 [205], [206]. In addition, remdesivir was tested in combination with chloroquine and the combination compared with chloroquine alone [194], [207], [208]. Remdesivir in combination with interferon beta was also shown to be effective against SARS-CoV-2 [208], [209]. Based on clinical trial results, and the compassionate use of this drug [18], [203], [210], remdesivir received Emergency Use Authorization by the FDA [205], [211], [212], [213], [214]. However, similar studies conducted in China suggested that remdesivir did not offer any benefit compared to placebo [205], [215], [216], [217]. Considering all the results, clinicians think of remdesivir as a helpful treatment for shortening recovery time but not a cure [215], [218]. Phase III Clinical trials of RdRp inhibitors, are currently underway. Remdesivir was recently approved by the FDA [205], [206], [213].
Table 2.
List of compounds tested against SARS-CoV-2 and their chemical structures.a
| Name of the compound | Chemical structure | Function | Reference |
|---|---|---|---|
| Camostat mesylate | Protease inhibitor | [137] | |
| PB28 | 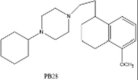 |
Replication of SARS-CoV-2 by inhibiting the translation of viral RNA, or by modulation cell signaling | [232], [233] |
| Zotatifin | 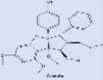 |
Replication of SARS-CoV-2 by inhibiting the translation of viral RNA, or by modulation cell signaling | [232], [233] |
| Cloperastine | 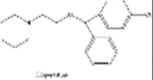 |
Antihistamines showed good antiviral properties | [233] |
| Clemastine | 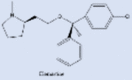 |
Antihistamines showed good antiviral properties | [233] |
| Famotidine (Pepcid) | 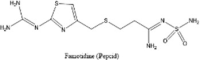 |
Targets histamine H2 receptors which are abundant in all parts of the body release of histamine by the mast cells is blocked and the overreaction of immune system dampened |
[234] [235] |
| Remdesivir | 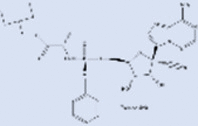 |
Inhibitors of viral RdRp's | |
| EIDD-2801 (Molnupiravir) | 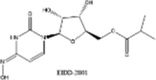 |
Inhibitors of viral RdRps | [219], [220], [221] |
| Favipiravir | 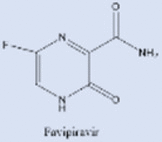 |
Inhibitors of viral RdRp's | [219], [220], [221] |
| Tautomers of EIDD-1931 | 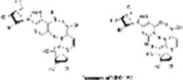 |
Antiviral against RNA viruses | [189] |
| Dexamethasone |  |
Suppresses the overreaction of the immune system | [236] |
| Hydroxychloroquine |  |
Anti-malarial drug. Inhibits viral replication in vitro Now not recommended for COVID-19 treatment |
[237], [238], [239], [240] |
| Baricitinib |  |
Anti-inflammation rheumatoid arthritis drug Could prevent the cytokine storm |
[241], [242], [243] |
| Ruxolittinib |  |
A cancer drug Could prevent the cytokine storm |
[241], [242], [243] |
| Brensocatib |  |
[231] | |
| Abu-Bip-CN |  |
[231] | |
| IcatCxpz41 |  |
[231] |
Structures created with ChemDraw.
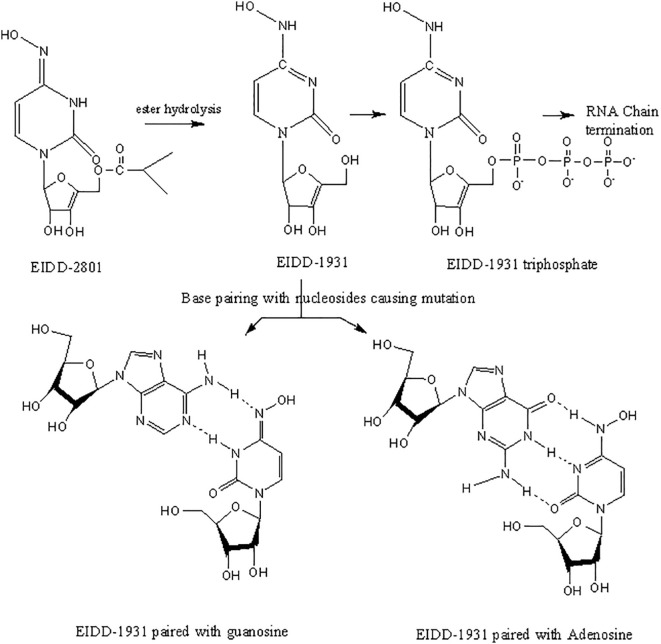
Two other inhibitors of viral RdRp's, favipiravir and the ribonucleoside analog, EIDD-2801 (Molnupiravir, See Table 2), were originally developed against HIV and hepatitis B [219], [220], [221]. In animals infected with SARS-CoV and MERS-CoV, EIDD significantly reduced viral titer and loss of body weight. Also, in vitro studies using primary human airway epithelial cells (HAECs) suggested that EIDD reduced SARS-CoV-2 reproduction in a dose-dependent manner [189], [222]. Remdesivir (administered through IV) and EIDD-2801 (administered orally) both hamper viral replication by targeting RdRp [223]. Interestingly, EIDD, unlike other antiviral compounds, cannot be overcome by resistance through mutations in the virus. EIDD-2801 has a pyrimidine as its base structure altered with a N-hydroxycytidine (Fig. 10). As a result, the RdRp incorporates EIDD-2801 into its genome. However, since EIDD-2801 can tautomerize, the RdRp erroneously reads EIDD-2801 as uridine instead of cytidine when it replicates. Incorporation of adenosine instead of guanosine is a mutation that becomes part of the viral genome and affects all future generations of the virus by dictating how viral genomes are synthesized and copied. Since all RNA-based viruses have RdRp enzyme, this drug should work on SARS-COV-2 and other similar viruses [218].
Fig. 10.
The oxime form of EIDD-1931 mimics uridine, base pairing with adenosine (left), while the other tautomer mimics cytidine and base pairs with guanosine (right).
The drug EIDD-1931 has two tautomers (Fig. 10). One tautomer is an oxime that mimics uridine and, upon incorporation into the genome (Fig. 11), base pairs with adenosine, while the other tautomer mimics cytidine and base pairs with guanosine [219]. As a result, the RdRp incorporates EIDD-2801 into the SARS-CoV-2 genome. However, since EIDD-2801 can tautomerize, the RdRp erroneously reads EIDD-2801 as uridine instead of cytidine when it replicates. Incorporation of adenosine instead of guanosine is a mutation that becomes part of the viral genome and affects all future generations of the virus by dictating how viral genomes are synthesized and copied. Since all RNA-based viruses have RdRp enzyme, this drug should work on SARS-COV-2 and other similar viruses [224]. However, in practice, initially it did not work, when monkeys were dosed with this drug in their GI, the CH2OH group in the molecule of EIDD-2801 was phosphorylated and the phosphorylated drug was readily excreted without being absorbed to work against the virus. To prevent this, the CH2OH group on the sugar moiety of the molecule was first converted to 2-methyl propionate (prodrug formation) before dosing. This protected the CH2OH group against pre-mature phosphorylation. The 2-methyl propionate moiety of the prodrug is broken back into the CH2OH upon absorption and then along with two other hydroxyl groups is phosphorylated to make the active drug. EIDD-2801 is developed in the United Kingdom (UK) by Merck. Phase I clinical trials of EIDD-2801 began on April 10, 2020 in the UK [225]. EIDD-2801 is recommended to be given to patients as soon as infection is detected. Because of its mechanism of action, this drug is likely to work against many RNA viruses, including MERS-CoV, HIV, Ebola, H1N1, and SARS-CoV. However, since other compounds that are similar to EIDD-2801 have been linked to birth defects in animals, expecting mothers or those who intend to become pregnant should avoid this drug [226]. The mechanism of incorporation of these drugs into RNA is shown in Fig. 11 [227].
Fig. 11.
Comparison of the mechanism of actions of antivirus Remdesivir, favipiravir, EIDD-2801, and EIDD-1931 through incorporation into DNA [227].
Other potential drugs under study against COVID-19 include interferon, ribavirin, tocilizumab, sarilumab, lopinavir, and chloroquine. To date, no benefit has been observed with lopinavir-ritonavir treatment of patients [118], [228], [229].
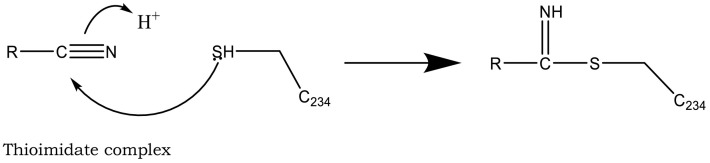
As additional potentially useful drug target, protein kinases also have a crucial role in entry, spread, and replication of the virus. AK-1 inhibitors, caffeic acid and its ester, propolis, ketorolac, and triptolide may help patients by inhibition of these crucial stages of virus life cycle. However, these compounds are not easily available to the body due to their solubility problems [176], [230]. Cathepsin C inhibition has also been pursued as an option to save overburdened lungs in serious cases [231]. Structures of three such inhibitors, Brensocatib, Abu-Bip-CN, IcatCxpz41 are included in Table 2. In terms of mechanism, in the inhibitor compound containing nitrile group, the nitrile group is said to interact with the cysteine 234 of Cathepsin C, resulting in a thioimidate complex and inhibition (Fig. 12) [231].
Fig. 12.
Interaction of Cysteine 234 and nitrile leads to the formation of a thioimidate complex, which inhibits cathepsin C. See Ref. [230].
4.3. Blood pressure drugs
Blood pressure drugs are thought to increase the expression of ACE2 receptors (though this idea has been disputed by more recent studies) [244], [245], [246]. Therefore, initially, it was thought that people taking these drugs would have an increased number of ACE2 receptors on their cell surfaces, thus providing more sites for the virus to attach and making the patient more susceptible to infection. However, more recent evidence suggests that blood pressure drugs actually protect patients against the induced lung injury, as described in more detail below [18], [244], [247], [248].
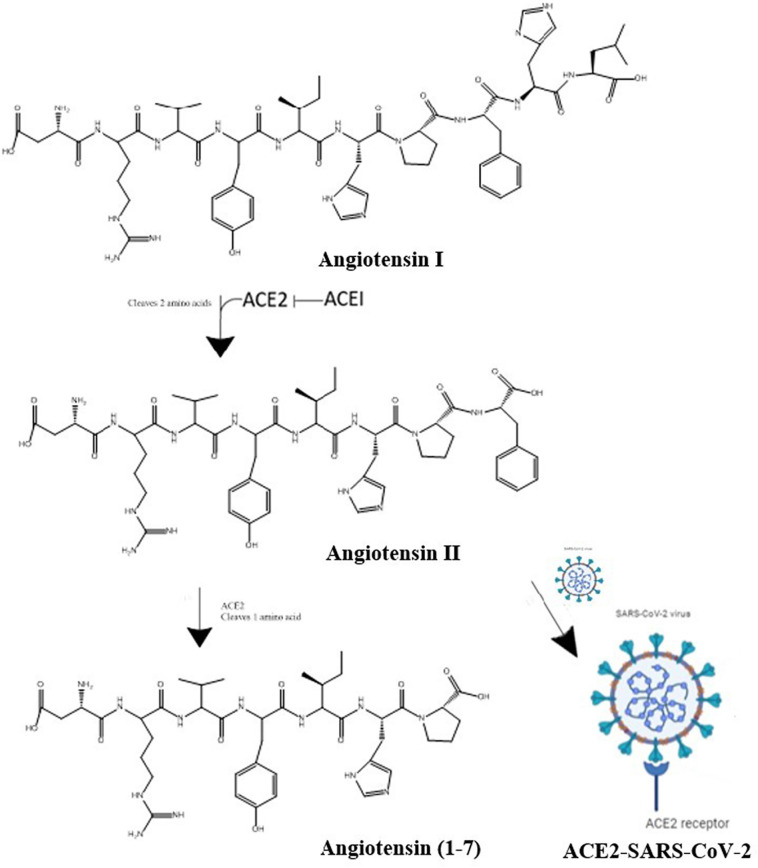
Since COVID-19 is more serious disease in older patients, they are also more likely to take ACE inhibitors (ACEIs) and angiotensin II receptor blockers (ARBs) for hypertension, heart disease, and cardiovascular disease. These drugs regulate the renin-angiotensin equilibrium. ACEIs prevent ACE1 from converting angiotensin I to angiotensin II. Angiotensin II controls lung injury and severe inflammation, preventing angiotensin II from binding to its receptors. To lower blood pressure, these drugs keep the level of angiotensin II at a desired level by effecting the renin-angiotensin equilibrium (Fig. 13) [244], [247]. In the lungs, heart, kidneys, nose, and GI tract, ACE2 helps regulate the angiotensin II levels by shifting this equilibrium to the right or the left, as needed. For instance, when angiotensin II levels are too high, ACE2 converts angiotensin II to angiotensin 1–7, which is inactive. When angiotensin II levels are too low, more angiotensin II is produced [53]. If the level of angiotensin II gets too high, fibrous tissue will form in the heart, kidney, and lungs, which can lead to severe fibrosis in these organs. Antihypertensive drugs like captopril, Lisinopril, and losartan either stop the production of angiotensin II or make it inactive. Towards the beginning of COVID-19 pandemic, many scientists warned against using ACEIs and ARBs in COVID-19 patients [249], [even though a prolonged absence of these drugs could lead to serious health problems or even death in the patients by increasing their hypertension]. Subsequently, it was discovered that using these drugs does not, in fact, harm COVID-19 patients [244]. In fact, they may actually help. This is because, during COVID-19 infection, the virus binds to host ACE2 and occupies host ACE2 sites. As a result, the ACE2 needed to convert angiotensin II to angiotensin 1–7 is no longer available, causing angiotensin II levels to go up. The increase in angiotensin II levels causes cell death and injury in lung and heart tissue. The injury in lung causes the release of cytokines (small proteins), which enhance the inflammation reaction and leads to more cell death [33], [35]. Fibroblast's form scar tissues and pneumonia ensues, filling the lung with fluid. The blood pressure drugs stop all these negative consequences of angiotensin build-up by increasing the number of ACE2 sites, which help the patient by converting excess angiotensin II to angiotensin 1–7 through the removal of one amino acid.
Fig. 13.
Blood pressure is regulated by the renin-angiotensin system. To enter target cells, SARS-CoV-2 binds to ACE2, preventing it from breaking down angiotensin II into angiotensin 1–7.
In a study on mice infected with SARS-CoV-2 virus, levels of angiotensin II increased while levels of ACE2 decreased. Giving the blood pressure drug, losartan, to the animals reduced injury. The effect of blood pressure drugs is under study about in 30 clinical trials. In related studies, to study the effect of ACE2 on viral infection, by making changes in the translation, ACE2 levels are lowered [250].
4.4. Antibody-based therapies
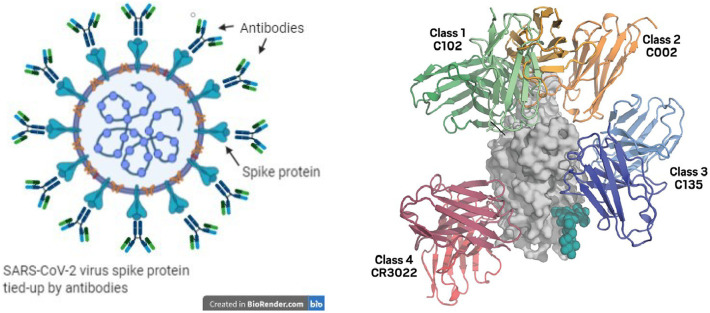
Antibodies are proteins of the immune system that bind to a specific antigen (such as virus before it infects a host cell). The process involves both the innate and adaptive immune responses [171]. Antibodies bind to the antigens on the surface of infected cells (see Fig. 14) and mark them for destruction by killer T cells. Among the SARS-CoV-2 and SARS-CoV proteins, the S-protein elicits an antibody response, as it resides on the external surface of the virus, while the structural proteins, M-, E-, and N-protein (in SARS-CoV) do not. As a result, over 90% of antibodies are directed against S-protein [188], [251]. Antibodies are made of a constant region (which interacts with other host immune proteins) and a variable region (that targets the specific antigen). The variable region is constructed to match the shape and chemical properties of their target antigen with high specificity. Construction of the variable region is generated through a process called VDJ recombination. One type of specific antibodies, “neutralizing antibodies” can be used as therapies where they capture antigens like bacteria and viruses to prevent them from infecting our cells. Neutralizing antibodies for therapy have been developed against COVID-19 [252], [253], [254].
Fig. 14.
Depiction of how antibodies block the spike protein of SARS-CoV-2, preventing the virus from attaching its spike protein to ACE2 receptors to initiate infection (left) [252], [254]. Four different ways that antibodies bind to receptor binding domain of spike protein from SARS-COV-2 virus (right).
(From Ref [74]).
Due to similarity of viruses, the antibody raised against the SARS-CoV S-protein can also prevent infection by binding S-proteins from SARS-CoV-2 and MERS-CoV [254], [255], [256], [257], [251]. However, because the S-protein in SARS-CoV has fewer sugar moieties on its surface, antibody binding is weaker to the SARS-CoV-2 S-protein. As a result, for treatment purposes, antibodies are isolated from convalescent plasma collected from patients who recently recovered from COVID-19. Currently, scientists are working to identify the gene in the B cells that code for the SARS-CoV-2 antibody to then recombinantly express those genes as therapeutics. This approach has already worked in the case of SARS [26], [188].
In another approach used by Regeneron, genetically modified mice were injected with the S-protein. B cells were then isolated from the mouse and among the B cells, those that produced the best monoclonal antibody were isolated, compared with best antibody from patients, and then mass produced through cell culture [251]. Palivizumab, a monoclonal antibody produced in this manner, is effective against respiratory syncytial virus. With financial support from the U.S. government, using VelociMouse technology, a mouse genetically modified to have human immune system, Regeneron developed monoclonal antibody against the S-protein of SARS-CoV-2, which received EUP from FDA approval [74], [258]. Tychan, a company from Singapore, started its clinical trial in China. Working with AbCellera, Eli Lilly also developed its monoclonal antibody (Ly-CoV555), a neutralizing antibody, designed to prevent SARS-CoV-2 from entering the cells. The antibody targets the S-protein of the virus and thus prevents infection by clearing the virus from the body. One out of three doses were given to people with mild symptoms of COVID-19 or placebo. In these studies, the rate of hospitalization in those patients who received the real dose was reduced to 1.7% compared to 6% for the placebo [259]. Another antibody against S-protein (JS016) is a human monoclonal neutralizing antibody developed by Junshi Biosciences. When combined with Ly-CoV555, this product makes a strong antibody [260]. Some of the difficulty for these techniques lies in scaling production. As a result, with the immediate threat of these viruses, it is important to note that these companies have been able to cut the time of the development and evaluation of these therapies from 10 to 12 months to possibly 5–6 months [261].
4.5. SARS-CoV-2 vaccines
Vaccines trick our bodies to perceive the vaccine as an antigen, causing our immune system to mount an attack to destroy the antigen [262]. The body's defense is presented in the form of antibodies which are proteins created to match the structure of the antigen in addition to secondary defensive mechanisms involving T-cells. Close to 200 vaccine development programs have been working to create vaccines against SARS-CoV-2 virus. Some, like Moderna and Pfizer, use mRNA as vaccine to generate antibodies against the S-protein inside the human cells [263]. Other strategies use the S-protein or the inactivated virus itself as the vaccine. Vaccination of the populations against SARS-CoV-2 has been successful in preventing the infection and death in many countries. However, some people refuse the vaccines while many do not have access to the vaccines. Initially, the major challenge for the vaccination, especially in the U.S., was the creation and production of the vaccines. However, after vaccination of about 50% of the population, the challenge has shifted to convincing the unvaccinated people to accept the vaccine. In the meantime, the virus has sufficient hosts to mutate to new variants including alpha, beta, gamma, and delta. The emergence of newer variants resistant to current vaccines may require the development and production of booster vaccines [264].
5. Summary and conclusions
The fight against SARS-CoV-2 has been a major challenge for our best scientists, massive investments of pharmaceutical companies and governments, and our available technology. In a very short period of time, significant advances were made in finding ways to lessen the spread of the virus and to prevent and treat the viral infection through therapeutics and vaccines. Understanding the mechanism of interaction between the viral S-protein and ACE2, along with the structure and function of the RdRp, Mpro, viral RNA, and various proteases, have been instrumental in discovering novel treatment methods [171], [265]. We have seen that biochemically, SARS-CoV-2 shows higher affinity towards the ACE2 receptor compared to SAR-CoV. Despite these differences, fortunately there are also many similarities between these two viruses, which has facilitated the discovery and development of antibody therapies and vaccines to help control the COVID-19 pandemic [256]. Vaccine development used many different techniques, some very new, but all focused on inducing the production of neutralizing antibodies against the spike protein [266], [267], [268], [269]. In addition to developing vaccines, scientists also explored other potential strategies that have not been discussed here. They include targeting the viral proteases, therapies based on cytokines, and nucleic acids. Knowledge of these and other relevant technologies is essential, not only for fighting SARS-CoV-2 today, but also to combat future viruses and pathogens.
Despite development of so many prevention and treatment tools in a short time period, mutation in SARS-CoV-2 RNA may lead to drastic changes in the viral genome and render all or some of these strategies useless [5], [16]. Fighting a mutating virus is possible, but as experience has shown, may be challenging. Thus, the most effective way of combating the virus is to vaccinate the whole population and follow safety guidelines, such as social distancing, wearing masks, sanitization, and contact tracing. However, experience has shown that for political or other reasons, these are not fully achivable [270], [271], [272], [273], [274], [275], [276].
Just as in the case of the Spanish flu of 1918, SARS-CoV-2 will continue to dominate the news for years to come. In fact, it is expected that this virus, like other viruses such as flu virus, will remain within the human population for the foreseeable future. As younger people acquire immunity, older adults will remain susceptible until heard immunity is established within the population. The potential for childhood vaccines for long-term protection, as in the case of other childhood vaccines, is still uncertain as we continue to understand the immune response of all segments of the population including children. In the meantime, new experimental treatments, along with social distancing and proper hygiene will be necessary in conjunction with vaccines to protect susceptible populations. In the process, cooperation of national and international leaders is important for fighting the current pandemic.
Abbreviations
- 3CLpro
3C-like protease or main protease
- ACE2
angiotensin-converting enzyme II
- Ang
angiotensin
- ARDS
acute respiratory distress syndrome
- CatC
cathepsin C
- CCR2
CC chemokine receptor 2
- CCL2
CC chemokine ligand 2
- CCR5
CC chemokine receptor 5
- COVID-19
coronavirus disease 2019
- CPG
CpG is shorthand for 5′—C—phosphate—G—3′ [cytosine and guanine separated by one phosphate group]
- E protein
envelope protein
- ELISA
enzyme-linked immunosorbent assay
- FP
fusion peptide
- HA
hemagglutinin
- HCoV
human coronavirus
- HCV
hepatic C virus
- HIV
human immune deficiency virus
- HTS
high-throughput screen
- IFA
immunofluorescence assay
- IFN-β
interferon β
- M protein
membrane protein
- MERS-CoV
Middle East respiratory syndrome coronavirus
- N protein
nucleocapsid protein
- nsp
non-structural proteins
- PCR
polymerase chain reaction
- PLpro
papain-like protease
- RAS
renin-angiotensin system
- RdRp
RNA-dependent RNA polymerase
- S protein
spike protein
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- SERMs
selective estrogen receptor modulators
- SFKs
Src-family of tyrosine kinases
- TCM
traditional Chinese medicine
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. ‡These authors contributed equally.
Funding sources
Any funds used to support the research of the manuscript should be placed here (per journal style).
Declaration of competing interest
The authors declare no competing financial interest.
References
- 1.National Institute of Allergy and Infectious Diseases (NIAID) Integrated research facility (IRF)., Novel coronavirus SARS-CoV-2, flickr. 2020. https://flic.kr/p/2iDwzAJ s.
- 2.Cortese M., Lee J.-Y., Hennies J., Mizzon G., Laketa V., Ruggieri A., Schwab Y., Bartenschlager R. Mendeley Data. V1. Elsevier; 2020. Integrative imaging reveals SARS-CoV-2 induced reshaping of subcellular morphologies. Cortese et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortese M., Lee J.-Y., Cerikan B., Neufeldt C.J., Oorschot V.M.J., Köhrer S., Hennies J., Schieber N.L., Ronchi P., Mizzon G., Romero-Brey I., Santarella-Mellwig R., Schorb M., Boermel M., Mocaer K., Beckwith M.S., Templin R.M., Gross V., Pape C., Tischer C., Frankish J., Horvat N.K., Laketa V., Stanifer M., Boulant S., Ruggieri A., Chatel-Chaix L., Schwab Y., Bartenschlager R. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28(6):853–866. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Molecular Biology Laboratory Replication cycle of SARS-CoV-2 in 3D, Healthcare-In-Europe.com. November 26, 2020. https://healthcare-in-europe.com/en/news/replication-cycle-of-sars-cov-2-in-3d.html s.
- 5.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24(July):91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Mil. Med. Res. 2020;7 doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e1–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katsnelson A. What we know about the novel coronavirus’s proteins. C&EN Glob. Enterp. 2020;98(15):19–21. doi: 10.1021/cen-09815-feature2. [DOI] [Google Scholar]
- 10.Park S.E. Epidemiology, virology, and clinical features of severe acute respiratory syndrome -coronavirus-2 (SARS-CoV-2; coronavirus disease-19) Clin. Exp. Pediatr. 2020;63(4):119–124. doi: 10.3345/cep.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peiris J.S.M., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10(12):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003, emergencies preparedness, response. December 31, 2003. https://www.who.int/csr/sars/country/table2004_04_21/en/ s.
- 13.Johns Hopkins University & Medicine Home, Coronavirus Resource Center. 2021. https://coronavirus.jhu.edu/ s.
- 14.World Health Organization Coronavirus disease (COVID-19): weekly epidemiological update and weekly operational update and situation reports, Coronavirus Disease (COVID-19) pandemic: situation reports. 2021. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ s.
- 15.World Health Organization., Overview, WHO coronavirus (COVID-19) dashboard. https://covid19.who.int/ (2929)s (Accessed 20 August 2021).
- 16.Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7(6):1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neerukonda S.N., Katneni U. A review on SARS-CoV-2 virology, pathophysiology, animal models, and anti-viral interventions. Pathogens. 2020;9(6) doi: 10.3390/pathogens9060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 19.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16(1) doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoeman D., Fielding B.C. Is there a link between the pathogenic human coronavirus envelope protein and immunopathology? A review of the literature. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.02086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., Ng O.-T., Marimuthu K., Ang L.W., Mak T.M., Lau S.K., Anderson D.E., Chan K.S., Tan T.Y., Ng T.Y., Cui L., Said Z., Kurupatham L., Chen M.I.-C., Chan M., Vasoo S., Wang L.-F., Tan B.H., Lin R.T.P., Lee V.J.M., Leo Y.-S., Lye D.C., Singapore Novel Coronavirus Outbreak Research Team Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323(15):1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J., Xie B., Hashimoto K. Current status of potential therapeutic candidates for the COVID-19 crisis. Brain Behav. Immun. 2020;87(July):59–73. doi: 10.1016/j.bbi.2020.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richman D.D., Whitley R.J., Hayden F.G., editors. Clinical virology. 4th ed. ASM Press; Washington, DC: 2017. [DOI] [Google Scholar]
- 24.Carroll K.C., Pfaller M.A., Landry M.L., McAdam A.J., Patel R.M.D., Richter S.S., Warnock D.W. 12th ed. 1-2. ASM Press; Washington, DC: 2019. Manual of clinical microbiology. [Google Scholar]
- 25.Lostroh P. Taylor & Francis; Boca Raton, FL: 2019. Molecular and Cellular Biology of Viruses. [Google Scholar]
- 26.Moharana S., Khuntia P., Das S.S., Das R., Samantaray P., Mohanty S. Evolution of experimental techniques & control measures in COVID-19 reservoir studies: A systematic review. Int. J. Innov. Sci. Res. Technol. 2020;5(5):918–934. https://www.ijisrt.com/assets/upload/files/IJISRT20MAY713.pdf [Google Scholar]
- 27.Muralidar S., Ambi S.V., Sekaran S., Krishnan U.M. The emergence of COVID-19 as a global pandemic: understanding the epidemiology, immune response and potential therapeutic targets of SARS-CoV-2. Biochimie. 2020;179(December):85–100. doi: 10.1016/j.biochi.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu J., Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 2007;170(4):1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry B.M., de Oliveira M.H.S., Benoit S., Plebani M., Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin. Chem. Lab. Med. 2020;58(7):1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 30.Cartmell T., Mitchell D. In: Techniques in the Behavioral and Neural Sciences: Handbook of Stress and the Brain, Part 2, Stress: Integrative and Clinical Aspects. Steckler T., Kalin N.H., Reul J.M.H.M., editors. Vol. 15. Elsevier; Amsterdam: 2005. The molecular basis of fever (Ch. 2.4) pp. 193–227. Pt. 2. [DOI] [Google Scholar]
- 31.Roth J., Blatteis C.M. Mechanisms of fever production and lysis: lessons from experimental LPS fever. Compr. Physiol. 2014;4(4):1563–1604. doi: 10.1002/cphy.c130033. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y., Zhang C., Huang F., Yang Y., Wang F., Yuan J., Zhang Z., Qin Y., Li X., Zhao D., Li S., Tan S., Wang Z., Li J., Shen C., Li J., Peng L., Wu W., Cao M., Xing L., Xu Z., Chen L., Zhou C., Liu W.J., Liu L., Jiang C. Elevated plasma level of selective cytokines in COVID-19 patients reflect viral load and lung injury. Natl. Sci. Rev. 2020;7(6):1003–1011. doi: 10.1093/nsr/nwaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 2020;8(6):e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye Q., Wang B., Mao J. The pathogenesis and treatment of the `Cytokine Storm' in COVID-19. J. Infect. 2020;80(6):607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyk W., Janik M.K., Portincasa P., Milkiewicz P., Lammert F., Krawczyk M. COVID-19: focus on the lungs but do not forget the gastrointestinal tract. Eur. J. Clin. Investig. 2020;50 doi: 10.1111/eci.13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020;104(3):246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abd El-Aziz T.M., Stockand J.D. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2) - an update on the status. Infect. Genet. Evol. 2020;83(September) doi: 10.1016/j.meegid.2020.104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B.M., Yang Q.Q., Zhao L.Y., Xie W., Si X.Y. Epidemiological characteristics of COVID-19 patients in convalescence period. Epidemiol. Infect. 2020;148 doi: 10.1017/S0950268820001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheervalilou R., Shirvaliloo M., Dadashzadeh N., Shirvalilou S., Shahraki O., Pilehvar-Soltanahmadi Y., Ghaznavi H., Khoei S., Nazarlou Z. COVID-19 under spotlight: A close look at the origin, transmission, diagnosis, and treatment of the 2019-nCoV disease. J. Cell. Physiol. 2020;235(12):8873–8924. doi: 10.1002/jcp.29735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 42.Centers for Disease Control and Prevention, National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases SARS-CoV-2 variant classifications and definitions, Coronavirus Disease 2019 (COVID-19) August 17, 2021. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html s.
- 43.Hacisuleyman E., Hale C., Saito Y., Blachere N.E., Bergh M., Conlon E.G., Schaefer-Babajew D.J., DaSilva J., Muecksch F., Gaebler C., Lifton R., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D., Darnell R.B. Vaccine breakthrough infections with SARS-CoV-2 variants. N. Engl. J. Med. 2021;384(23):2212–2218. doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khateeb J., Li Y., Zhang H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit. Care. 2021;25(1) doi: 10.1186/s13054-021-03662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simmonds P., Griffin D.E. Pervasive RNA secondary structure in the genomes of SARS-CoV-2 and Other Coronaviruses. mBio. 2020;11(6) doi: 10.1128/mBio.01661-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.V'kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Straub J.H., Stürzel C.M., Fröhlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369(6508):1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krokan H.E., Kavli B., Slupphaug G., Drablos F. In: Genomic uracil: Evolution, biology, immunology and disease. Slupphaug G., Krokan H.E., editors. World Scientific; New Jersey: 2018. The Earth, life, and genomic uracil (Ch. 2) [Google Scholar]
- 49.Di Gioacchino A., Šulc P., Komarova A.V., Greenbaum B.D., Monasson R., Cocco S. The heterogeneous landscape and early evolution of pathogen-associated CpG dinucleotides in SARS-CoV-2. Mol. Biol. Evol. 2021;38(6):2428–2445. doi: 10.1093/molbev/msab036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartas M., Brázda V., Bohálová N., Cantara A., Volná A., Stachurová T., Malachová K., Jagelská E.B., Porubiaková O., Červeň J., Pečinka P. In-depth bioinformatic analyses of Nidovirales including human SARS-CoV-2, SARS-CoV, MERS-CoV viruses suggest important roles of non-canonical nucleic acid structures in their lifecycles. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelly J.A., Olson A.N., Neupane K., Munshi S., San Emeterio J., Pollack L., Woodside M.T., Dinman J.D. Structural and functional conservation of the programmed −1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2) J. Biol. Chem. 2020;295(31):10741–10748. doi: 10.1074/jbc.AC120.013449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choudhry N., Zhao X., Xu D., Zanin M., Chen W., Yang Z., Chen J. Chinese therapeutic strategy for fighting COVID-19 and potential small-molecule inhibitors against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) J. Med. Chem. 2020;63(22):13205–13227. doi: 10.1021/acs.jmedchem.0c00626. [DOI] [PubMed] [Google Scholar]
- 53.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bergmann C.C., Silverman R.H. COVID-19: coronavirus replication, pathogenesis, and therapeutic strategies. Cleve. Clin. J. Med. 2020;87(6):321–327. doi: 10.3949/ccjm.87a.20047. [DOI] [PubMed] [Google Scholar]
- 56.Fehr A.R., Perlman S. In: Maier H.J., Bickerton E., Britton P., editors. Vol. 1282. Humana Press; New York: 2015. Coronaviruses: An overview of their replication and pathogenesis (Ch. 1) pp. 1–23. (Coronaviruses: Methods and Protocols. Methods in Molecular Biology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth-Din A., Melamed S., Weiss S., Israely T., Paran N., Schwartz M., Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature. 2021;589(7840):125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 58.Ogando N.S., Zevenhoven-Dobbe J.C., van der Meer Y., Bredenbeek P.J., Posthuma C.C., Snijder E.J., Gallagher T. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020;94(23) doi: 10.1128/JVI.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Satarker S., Nampoothiri M. Structural proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Arch. Med. Res. 2020;51(6):482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Budzilowicz C.J., Wilczynski S.P., Weiss S.R. Three intergenic regions of coronavirus mouse hepatitis virus strain A59 genome RNA contain a common nucleotide sequence that is homologous to the 3' end of the viral mRNA leader sequence. J. Virol. 1985;53(3):834–840. doi: 10.1128/JVI.53.3.834-840.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sawicki S.G., Sawicki D.L. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 1986;57(1):328–334. doi: 10.1128/JVI.57.1.328-334.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viswanathan T., Arya S., Chan S.-H., Qi S., Dai N., Misra A., Park J.-G., Oladunni F., Kovalskyy D., Hromas R.A., Martinez-Sobrido L., Gupta Y.K. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020;11(1):3718. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collier A.M., Lyytinen O.L., Guo Y.R., Toh Y., Poranen M.M., Tao Y.J. Initiation of RNA polymerization and polymerase encapsidation by a small dsRNA virus. PLoS Pathog. 2016;12(4) doi: 10.1371/journal.ppat.1005523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kadam S.B., Sukhramani G.S., Bishnoi P., Pable A.A., Barvkar V.T. SARS-CoV-2, the pandemic coronavirus: molecular and structural insights. J. Basic Microbiol. 2021;61(3):180–202. doi: 10.1002/jobm.202000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health SARS-CoV-2 proteins – NCBI datasets, NCBI datasets. 2020. https://www.ncbi.nlm.nih.gov/datasets/coronavirus/proteins/ s.
- 66.Rabaan A.A., Al-Ahmed S.H., Haque S., Sah R., Tiwari R., Malik Y.S., Dhama K., Yatoo M.I., Bonilla-Aldana D.K., Rodriguez-Morales A.J. SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview. Infez. Med. 2020;28(2):174–184. https://www.infezmed.it/index.php/article?Anno=2020&numero=2&ArticoloDaVisualizzare=Vol_28_2_2020_174 s. [PubMed] [Google Scholar]
- 67.Chasman D.I. Marcel Dekker; New York: 2003. Protein Structure: Determination, Analysis and Applications for Drug Discovery. [Google Scholar]
- 68.Ilari A., Savino C. In: Bioinformatics: Data, Sequence Analysis and Evolution. Keith J.M., editor. Humana Press; Totowa, NJ: 2008. Protein structure determination by X-ray crystallography; pp. 63–87. [DOI] [Google Scholar]
- 69.Ramesh V., Baslé A., Lewis R.J. In: Biomolecular and Bioanalytical Techniques: Theory, Methodology and Applications. Ramesh V., editor. Wiley; Hoboken, NJ: 2019. Principles and practice in macromolecular X-ray crystallography; pp. 385–419. [DOI] [Google Scholar]
- 70.Yang Y.-J., Wang S., Zhang B., Shen H.-B. Resolution measurement from a single reconstructed cryo-em density map with multiscale spectral analysis. J. Chem. Inf. Model. 2018;58(6):1303–1311. doi: 10.1021/acs.jcim.8b00149. [DOI] [PubMed] [Google Scholar]
- 71.Bartesaghi A., Matthies D., Banerjee S., Merk A., Subramaniam S. Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy. Proc. Natl. Acad. Sci. 2014;111(32):11709–11714. doi: 10.1073/pnas.1402809111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Howes L. The race to reveal the new coronavirus’s structure. C&EN Glob. Enterp. 2020;98(17):16–19. doi: 10.1021/cen-09817-feature1. [DOI] [Google Scholar]
- 73.Steven A., Belnap D. Electron microscopy and image processing: An essential tool for structural analysis of macromolecules. Curr. Protoc. Protein Sci. 2005;42(1) doi: 10.1002/0471140864.ps1702s42. 17.2.1-17.2.39. [DOI] [PubMed] [Google Scholar]
- 74.Cross R., Howes L., Satyanarayana M. 8 tools that helped us tackle the coronavirus. C&EN Glob. Enterp. 2021;99(3):38–43. doi: 10.1021/cen-09903-feature3. [DOI] [Google Scholar]
- 75.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. e1–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176(April) doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020;92(6):595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 84.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu H., Zhang Y., Yang P. Advancements in mass spectrometry-based glycoproteomics and glycomics. Natl. Sci. Rev. 2016;3(3):345–364. doi: 10.1093/nsr/nww019. [DOI] [Google Scholar]
- 86.Burda P., Aebi M. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta. 1999;1426(2):239–257. doi: 10.1016/S0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- 87.Brooker R.J., Widmaier E.P., Graham L.E., Stiling P.D. Biology. McGraw-Hill Higher Education; Boston: 2008. Systems biology of cell organization (Ch. 6) [Google Scholar]
- 88.Arnaud C.H. Adding the missing sugars to coronavirus protein structures. C&EN Glob. Enterp. 2020;98(16):24–25. doi: 10.1021/cen-09816-feature2. [DOI] [Google Scholar]
- 89.Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30(12):981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Y., Zhao W., Mao Y., Chen Y., Wang S., Zhong Y., Su T., Gong M., Du D., Lu X., Cheng J., Yang H. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins. Mol. Cell. Proteomics. 2021;20 doi: 10.1074/mcp.RA120.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bagdonaite I., Thompson A.J., Wang X., Søgaard M., Fougeroux C., Frank M., Diedrich J.K., Yates J.R., Salanti A., Vakhrushev S.Y., Paulson J.C., Wandall H.H. Site-specific O-glycosylation analysis of SARS-CoV-2spike protein produced in insect and human cells. Viruses. 2021;13(4):551. doi: 10.3390/v13040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Imperiali B., O’Connor S.E. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr. Opin. Chem. Biol. 1999;3(6):643–649. doi: 10.1016/S1367-5931(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 94.Hioe C.E., Jan M., Feyznezhad R., Itri V., Liu X., Upadhyay C. HIV-1 envelope glycan composition influences virus-host interaction. J. Immunol. 2019;202(1):197.17. http://www.jimmunol.org/content/202/1_Supplement/197.17.abstract [Google Scholar]
- 95.Hioe C.E., Jan M., Upadhyay C. HIV-1 envelope glycan composition is a key determinant of efficient virus transmission via DC-SIGN and resistance to antibodies and inhibitory lectins. J. Immunol. 2020;204(1):247.8. doi: 10.1016/j.isci.2019.10.030. http://www.jimmunol.org/content/204/1_Supplement/247.8.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Howes L. COVID-19: What you need to know about SARS-CoV-2 variants. C&EN Glob. Enterp. 2021;99(20):18–19. doi: 10.1021/cen-09920-feature1. [DOI] [Google Scholar]
- 97.Alam I., Kamau A.A., Kulmanov M., Jaremko Ł., Arold S.T., Pain A., Gojobori T., Duarte C.M. Functional pangenome analysis shows key features of E protein are preserved in SARS and SARS-CoV-2. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilson L., McKinlay C., Gage P., Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330(1):322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nieva J.L., Madan V., Carrasco L. Viroporins: structure and biological functions. Nat. Rev. Microbiol. 2012;10(8):563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang R., Wang K., Lv W., Yu W., Xie S., Xu K., Schwarz W., Xiong S., Sun B. The ORF4a protein of human coronavirus 229E functions as a viroporin that regulates viral production. Biochim. Biophys. Acta. 2014;1838(4):1088–1095. doi: 10.1016/j.bbamem.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pham T., Perry J.L., Dosey T.L., Delcour A.H., Hyser J.M. The rotavirus NSP4 viroporin domain is a calcium-conducting ion channel. Sci. Rep. 2017;7 doi: 10.1038/srep43487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Duart G., García-Murria M.J., Grau B., Acosta-Cáceres J.M., Martínez-Gil L., Mingarro I. SARS-CoV-2 envelope protein topology in eukaryotic membranes. Open Biol. 2020;10(9) doi: 10.1098/rsob.200209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liao Y., Tam J.P., Liu D.X. Viroporin activity of SARS-CoV E protein. Adv. Exp. Med. Biol. 2006;581:199–202. doi: 10.1007/978-0-387-33012-9_34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu Q., Zhang Y., Lü H., Wang J., He X., Liu Y., Ye C., Lin W., Hu J., Ji J., Xu J., Ye J., Hu Y., Chen W., Li S., Wang J., Wang J., Bi S., Yang H. The E protein is a multifunctional membrane protein of SARS-CoV. Genomics Proteomics Bioinformatics. 2003;1(2):131–144. doi: 10.1016/S1672-0229(03)01017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bianchi M., Benvenuto D., Giovanetti M., Angeletti S., Ciccozzi M., Pascarella S. SARS-CoV-2 envelope and membrane proteins: structural differences linked to virus characteristics? Biomed. Res. Int. 2020;2020 doi: 10.1155/2020/4389089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.de Haan C.A.M., Vennema H., Rottier P.J.M. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74(11):4967–4978. doi: 10.1128/JVI.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 109.Hu Y., Wen J., Tang L., Zhang H., Zhang X., Li Y., Wang J., Han Y., Li G., Shi J., Tian X., Jiang F., Zhao X., Wang J., Liu S., Zeng C., Wang J., Yang H. The M protein of SARS-CoV: Basic structural and immunological properties. Genomics Proteomics Bioinformatics. 2003;1(2):118–130. doi: 10.1016/S1672-0229(03)01016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arndt A.L., Larson B.J., Hogue B.G. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J. Virol. 2010;84(21):11418–11428. doi: 10.1128/JVI.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McBride R., Van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Surjit M., Lal S.K. The SARS-CoV nucleocapsid protein: a protein with multifarious activities. Infect. Genet. Evol. 2008;8(4):397–405. doi: 10.1016/j.meegid.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10(7):1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vandelli A., Monti M., Milanetti E., Armaos A., Rupert J., Zacco E., Bechara E., Delli Ponti R., Tartaglia Gian G. Structural analysis of SARS-CoV-2 genome and predictions of the human interactome. Nucleic Acids Res. 2020;48(20):11270–11283. doi: 10.1093/nar/gkaa864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.-A., Leibundgut M., Thiel V., Mühlemann O., Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27(10):959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 116.Cornillez-Ty C.T., Liao L., Yates J.R., Kuhn P., Buchmeier M.J. Severe Acute Respiratory Syndrome Coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 2009;83(19):10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149(January):58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 119.Claverie J.-M. A putative role of de-mono-ADP-ribosylation of STAT1 by the SARS-CoV-2 nsp3 protein in the cytokine storm syndrome of COVID-19. Viruses. 2020;12(6) doi: 10.3390/v12060646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M.C., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43(17):8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.-C., Tian G., Jiang H.-W., Tao S.-C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Howes L. Another coronavirus drug target imaged. C&EN Glob. Enterp. 2020;98(15):4. doi: 10.1021/cen-09815-scicon1. [DOI] [Google Scholar]
- 123.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc. Natl. Acad. Sci. 2015;112(30):9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29(7):1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Wiersum G., Kim Y., Jedrzejczak R., Maltseva N.I., Endres M., Jaroszewski L., Godzik A., Joachimiak A., Satchell K.J.F. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. bioRxiv. 2020 doi: 10.1101/2020.04.17.047498. (Preprint) [DOI] [Google Scholar]
- 127.Konkolova E., Klima M., Nencka R., Boura E. Structural analysis of the putative SARS-CoV-2 primase complex. J. Struct. Biol. 2020;211(2) doi: 10.1016/j.jsb.2020.107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47(12):6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Frieman M., Yount B., Agnihothram S., Page C., Donaldson E., Roberts A., Vogel L., Woodruff B., Scorpio D., Subbarao K., Baric R.S. Molecular determinants of Severe Acute Respiratory Syndrome Coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J. Virol. 2012;86(2):884–897. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.de Haan C.A.M., Kuo L., Masters P.S., Vennema H., Rottier P.J.M. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72(8):6838–6850. doi: 10.1128/JVI.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133(1):113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Das A., Bhattacharya M., Roy R., Ghosh P., Mondal N., Das T., Behera B.K., Patra B.C., Acharya S. Research Square. 2020. Genetic diversity and structural characterization of spike glycoprotein of newly emerged SARS-CoV-2. (Preprint), PPR167372. [DOI] [Google Scholar]
- 134.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat. Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., Hennig B.P., Kreuter M., Conrad C., Eils R. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39(10) doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.-E., Barbry P., Leslie A., Kiem H.-P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J., Banovich N., Barbry P., Brazma A., Desai T., Duong T.E., Eickelberg O., Falk C., Farzan M., Glass I., Haniffa M., Horvath P., Hung D., Kaminski N., Krasnow M., Kropski J.A., Kuhnemund M., Lafyatis R., Lee H., Leroy S., Linnarson S., Lundeberg J., Meyer K., Misharin A., Nawijn M., Nikolic M.Z., Ordovas-Montanes J., Pe’er D., Powell J., Quake S., Rajagopal J., Tata P.R., Rawlins E.L., Regev A., Reyfman P.A., Rojas M., Rosen O., Saeb-Parsy K., Samakovlis C., Schiller H., Schultze J.L., Seibold M.A., Shalek A.K., Shepherd D., Spence J., Spira A., Sun X., Teichmann S., Theis F., Tsankov A., van den Berge M., von Papen M., Whitsett J., Xavier R., Xu Y., Zaragosi L.-E., Zhang K. SARS-CoV-2 receptor ACE2 Is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035. doi: 10.1016/j.cell.2020.04.035. e1-e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e1–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., Aziz B. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020;92(9):1580–1586. doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hussain M., Jabeen N., Amanullah A., Baig A.A., Aziz B., Shabbir S., Raza F., Uddin N. Molecular docking between human TMPRSS2 and SARS-CoV-2 spike protein: conformation and intermolecular interactions. AIMS Microbiol. 2020;6(3):350–360. doi: 10.3934/microbiol.2020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78(4):779–784. doi: 10.1016/j.molcel.2020.04.022. e1–e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L., Banovich N.E., Barbry P., Brazma A., Collin J., Desai T.J., Duong T.E., Eickelberg O., Falk C., Farzan M., Glass I., Gupta R.K., Haniffa M., Horvath P., Hubner N., Hung D., Kaminski N., Krasnow M., Kropski J.A., Kuhnemund M., Lako M., Lee H., Leroy S., Linnarson S., Lundeberg J., Meyer K.B., Miao Z., Misharin A.V., Nawijn M.C., Nikolic M.Z., Noseda M., Ordovas-Montanes J., Oudit G.Y., Pe’er D., Powell J., Quake S., Rajagopal J., Tata P.R., Rawlins E.L., Regev A., Reyfman P.A., Rozenblatt-Rosen O., Saeb-Parsy K., Samakovlis C., Schiller H.B., Schultze J.L., Seibold M.A., Seidman C.E., Seidman J.G., Shalek A.K., Shepherd D., Spence J., Spira A., Sun X., Teichmann S.A., Theis F.J., Tsankov A.M., Vallier L., van den Berge M., Whitsett J., Xavier R., Xu Y., Zaragosi L.-E., Zerti D., Zhang H., Zhang K., Rojas M., Figueiredo F. HCA Lung Biological Network, SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lam S.D., Bordin N., Waman V.P., Scholes H.M., Ashford P., Sen N., van Dorp L., Rauer C., Dawson N.L., Pang C.S.M., Abbasian M., Sillitoe I., Edwards S.J.L., Fraternali F., Lees J.G., Santini J.M., Orengo C.A. SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals. Sci. Rep. 2020;10(1):16471. doi: 10.1038/s41598-020-71936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chowdhury R., Boorla V.S., Maranas C.D. Computational biophysical characterization of the SARS-CoV-2 spike protein binding with the ACE2 receptor and implications for infectivity. Comput. Struct. Biotechnol. J. 2020;18:2573–2582. doi: 10.1016/j.csbj.2020.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Laurini E., Marson D., Aulic S., Fermeglia M., Pricl S. Computational Alanine Scanning and Structural Analysis of the SARS-CoV-2 Spike Protein/Angiotensin-Converting Enzyme 2 Complex. ACS Nano. 2020;14(9):11821–11830. doi: 10.1021/acsnano.0c04674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang Y., Liu M., Gao J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc. Natl. Acad. Sci. 2020;117(25):13967–13974. doi: 10.1073/pnas.2008209117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hatmal M.M., Alshaer W., Al-Hatamleh M.A.I., Hatmal M., Smadi O., Taha M.O., Oweida A.J., Boer J.C., Mohamud R., Plebanski M. Comprehensive Structural and Molecular Comparison of Spike Proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and their interactions with ACE2. Cells. 2020;9(12) doi: 10.3390/cells9122638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Baxter B.D., Larson E.D., Merle L., Feinstein P., Polese A.G., Bubak A.N., Niemeyer C.S., Hassell J., Shepherd D., Ramakrishnan V.R., Nagel M.A., Restrepo D. Transcriptional profiling reveals potential involvement of microvillous TRPM5-expressing cells in viral infection of the olfactory epithelium. BMC Genomics. 2021;22(1) doi: 10.1186/s12864-021-07528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hikmet F., Méar L., Edvinsson Å., Micke P., Uhlén M., Lindskog C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020;16(7) doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Guo J., Huang Z., Lin L., Lv J. Coronavirus disease 2019 (COVID-19) and cardiovascular disease: a viewpoint on the potential influence of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers on onset and severity of Severe Acute Respiratory Syndrome Coronavirus 2 infection. J. Am. Heart Assoc. 2020;9(7) doi: 10.1161/JAHA.120.016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12(1) doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang Y., Geng X., Tan Y., Li Q., Xu C., Xu J., Hao L., Zeng Z., Luo X., Liu F., Wang H. New understanding of the damage of SARS-CoV-2 infection outside the respiratory system. Biomed. Pharmacother. 2020;127(July) doi: 10.1016/j.biopha.2020.110195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Seah I., Su X., Lingam G. Revisiting the dangers of the coronavirus in the ophthalmology practice. Eye (London) 2020;34:1155–1157. doi: 10.1038/s41433-020-0790-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang J., Zhao S., Liu M., Zhao Z., Xu Y., Wang P., Lin M., Xu Y., Huang B., Zuo X., Chen Z., Bai F., Cui J., Lew A.M., Zhao J., Zhang Y., Luo H., Zhang Y. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. medRxiv. 2020 doi: 10.1101/2020.02.05.20020545. Preprint. [DOI] [Google Scholar]
- 155.Xia J., Tong J., Liu M., Shen Y., Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020;92(6):589–594. doi: 10.1002/jmv.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. e1–e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Z., Cui X., Xiao J., Meng T., Zhou W., Liu J., Xu H. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxic. 2020 doi: 10.1101/2020.01.30.927806. Preprint 01.30.927806. [DOI] [Google Scholar]
- 158.Ding Y., He L., Zhang Q., Huang Z., Che X., Hou J., Wang H., Shen H., Qiu L., Li Z., Geng J., Cai J., Han H., Li X., Kang W., Weng D., Liang P., Jiang S. Organ distribution of Severe Acute Respiratory Syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol. 2004;203(2):622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., Arledge K.C., Chen Y.-H., Zhang L., Wang X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23(8):986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Brake S.J., Barnsley K., Lu W., McAlinden K.D., Eapen M.S., Sohal S.S. Smoking upregulates angiotensin-converting enzyme-2 receptor: a potential adhesion site for novel coronavirus SARS-CoV-2 (Covid-19) J. Clin. Med. 2020;9(3) doi: 10.3390/jcm9030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Muus C., Luecken M.D., Eraslan G., Waghray A., Heimberg G., Sikkema L., Kobayashi Y., Vaishnav E.D., Subramanian A., Smilie C., Jagadeesh K., Duong E.T., Fiskin E., Triglia E.T., Ansari M., Cai P., Lin B., Buchanan J., Chen S., Shu J., Haber A.L., Chung H., Montoro D.T., Adams T., Aliee H., Samuel J., Andrusivova A.Z., Angelidis I., Ashenberg O., Bassler K., Bécavin C., Benhar I., Bergenstråhle J., Bergenstråhle L., Bolt L., Braun E., Bui L.T., Chaffin M., Chichelnitskiy E., Chiou J., Conlon T.M., Cuoco M.S., Deprez M., Fischer D.S., Gillich A., Gould J., Guo M., Gutierrez A.J., Habermann A.C., Harvey T., He P., Hou X., Hu L., Jaiswal A., Jiang P., Kapellos T., Kuo C.S., Larsson L., Leney-Greene M.A., Lim K., Litviňuková M., Lu J., Ludwig L.S., Luo W., Maatz H., Madissoon E., Mamanova L., Manakongtreecheep K., Marquette C.-H., Mbano I., McAdams A.M., Metzger R.J., Nabhan A.N., Nyquist S.K., Penland L., Poirion O.B., Poli S., Qi C., Queen R., Reichart D., Rosas I., Schupp J., Sinha R., Sit R.V., Slowikowski K., Slyper M., Smith N., Sountoulidis A., Strunz M., Sun D., Talavera-López C., Tan P., Tantivit J., Travaglini K.J., Tucker N.R., Vernon K., Wadsworth M.H., Waldman J., Wang X., Yan W., Zhao W., Ziegler C.G.K. Integrated analyses of single-cell atlases reveal age, gender, and smoking status associations with cell type-specific expression of mediators of SARS-CoV-2 viral entry and highlights inflammatory programs in putative target cells. bioRxiv. 2020 doi: 10.1101/2020.04.19.049254. Preprint. [DOI] [Google Scholar]
- 163.Dandekar A.A., Perlman S. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5(12):917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rangan R., Zheludev I.N., Hagey R.J., Pham E.A., Wayment-Steele H.K., Glenn J.S., Das R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA. 2020;26(8):937–959. doi: 10.1261/rna.076141.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A Novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., Tan T.K., Rijal P., Coombes N., Bewley K.R., Tree J.A., Radecke J., Paterson N.G., Supasa P., Mongkolsapaya J., Screaton G.R., Carroll M., Townsend A., Fry E.E., Owens R.J., Stuart D.I. Neutralization of SARS-CoV-2 by destruction of the prefusion spike. Cell Host Microbe. 2020;28(3):445–454. doi: 10.1016/j.chom.2020.06.010. e1–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1) doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cellular & Molecular Immunology. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., Pia L., Risson E., Saffern M., Salomé B., Selvan M.E., Spindler M.P., Tan J., van der Heide V., Gregory J.K., Alexandropoulos K., Bhardwaj N., Brown B.D., Greenbaum B., Gümüş Z.H., Homann D., Horowitz A., Kamphorst A.O., de Lafaille M.A.C., Mehandru S., Merad M., Samstein R.M., The Sinai Immunology Review Project Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pinto B.G.G., Oliveira A.E.R., Singh Y., Jimenez L., Gonçalves A.N.A., Ogava R.L.T., Creighton R., Peron J.P.S., Nakaya H.I. ACE2 expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. J. Infect. Dis. 2020;222(4):556–563. doi: 10.1093/infdis/jiaa332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rameshrad M., Ghafoori M., Mohammadpour A.H., Nayeri M.J.D., Hosseinzadeh H. A comprehensive review on drug repositioning against coronavirus disease 2019 (COVID19) Naunyn Schmiedeberg's Arch. Pharmacol. 2020;393:1137–1152. doi: 10.1007/s00210-020-01901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wu R., Wang L., Kuo H.-C.D., Shannar A., Peter R., Chou P.J., Li S., Hudlikar R., Liu X., Liu Z., Poiani G.J., Amorosa L., Brunetti L., Kong A.-N. An update on current therapeutic drugs treating COVID-19. Curr. Pharmacol. Rep. 2020;6:56–70. doi: 10.1007/s40495-020-00216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Subramanian K., Nalli A., Senthil V., Jain S., Nayak A., Bhat A. Perspectives on the early quality of evidence guiding the therapeutic management of SARS-CoV-2: A systematic literature review. Adv. Ther. 2020;37(10):4107–4131. doi: 10.1007/s12325-020-01460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Uddin M., Mustafa F., Rizvi T.A., Loney T., Al Suwaidi H., Al-Marzouqi A.H.H., Kamal Eldin A., Alsabeeha N., Adrian T.E., Stefanini C., Nowotny N., Alsheikh-Ali A., Senok A.C. SARS-CoV-2/COVID-19: Viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses. 2020;12(5) doi: 10.3390/v12050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Al-Bari M.A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017;5(1) doi: 10.1002/prp2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ancy I., Sivanandam M., Kumaradhas P. Possibility of HIV-1 protease inhibitors-clinical trial drugs as repurposed drugs for SARS-CoV-2 main protease: a molecular docking, molecular dynamics and binding free energy simulation study. J. Biomol. Struct. Dyn. 2020;39(15):5368–5375. doi: 10.1080/07391102.2020.1786459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gül Ş., Özcan O., Aşar S., Okyar A., Barış İ., Kavakli I.H. In silico identification of widely used and well-tolerated drugs as potential SARS-CoV-2 3C-like protease and viral RNA-dependent RNA polymerase inhibitors for direct use in clinical trials. J. Biomol. Struct. Dyn. 2020;39(17):6772–6791. doi: 10.1080/07391102.2020.1802346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Panda G., Mishra N., Ray A. OSF Preprints; 2020. Genetic variations and drug repurposing provides key insights into the disruption of the SARS COV2. (Preprint), b7y2c. [DOI] [Google Scholar]
- 181.Hsieh C.-L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.-C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., Chou C.-W., Byrne P.O., Hjorth C.K., Johnson N.V., Ludes-Meyers J., Nguyen A.W., Park J., Wang N., Amengor D., Lavinder J.J., Ippolito G.C., Maynard J.A., Finkelstein I.J., McLellan J.S. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369(6510):1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Osman E.E.A., Toogood P.L., Neamati N. COVID-19: Living through another pandemic. ACS Infect. Dis. 2020;6(7):1548–1552. doi: 10.1021/acsinfecdis.0c00224. [DOI] [PubMed] [Google Scholar]
- 183.Wadman M., Couzin-Frankel J., Kaiser J., Matacic C. A rampage through the body. Science. 2020;368(6489):356–360. doi: 10.1126/science.368.6489.356. [DOI] [PubMed] [Google Scholar]
- 184.Luytjes W., Sturman L.S., Bredenbee P.J., Charite J., van der Zeijst B.A.M., Horzinek M.C., Spaan W.J.M. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology. 1987;161(2):479–487. doi: 10.1016/0042-6822(87)90142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Abraham S., Kienzle T.E., Lapps W., Brian D.A. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology. 1990;176(1):296–301. doi: 10.1016/0042-6822(90)90257-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.de Groot R.J., Luytjes W., Horzinek M.C., van der Zeijst B.A.M., Spaan W.J.M., Lenstra J.A. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 1987;196(4):963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395(10223):e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Central Science. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12(541):eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J., Sun Q., Yang X., Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182(2):417–428. doi: 10.1016/j.cell.2020.05.034. e1–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020;526(1):165–169. doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Amaro R.E., Mulholland A.J. A community letter regarding sharing biomolecular simulation data for COVID-19. J. Chem. Inf. Model. 2020;60(6):2653–2656. doi: 10.1021/acs.jcim.0c00319. [DOI] [PubMed] [Google Scholar]
- 194.Gao J., Tian Z., Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. BioSci. Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 195.Kaeppler U., Stiefl N., Schiller M., Vicik R., Breuning A., Schmitz W., Rupprecht D., Schmuck C., Baumann K., Ziebuhr J., Schirmeister T. A new lead for nonpeptidic active-site-directed inhibitors of the severe acute respiratory syndrome coronavirus main protease discovered by a combination of screening and docking methods. J. Med. Chem. 2005;48(22):6832–6842. doi: 10.1021/jm0501782. [DOI] [PubMed] [Google Scholar]
- 196.Martina E., Stiefl N., Degel B., Schulz F., Breuning A., Schiller M., Vicik R., Baumann K., Ziebuhr J., Schirmeister T. Screening of electrophilic compounds yields an aziridinyl peptide as new active-site directed SARS-CoV main protease inhibitor. Bioorg. Med. Chem. Lett. 2005;15(24):5365–5369. doi: 10.1016/j.bmcl.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shah F., Mukherjee P., Desai P., Avery M. Computational approaches for the discovery of cysteine protease inhibitors against malaria and SARS. Current Computer-Aided Drug Design. 2010;6(1):1–23. doi: 10.2174/157340910790980142. [DOI] [PubMed] [Google Scholar]
- 198.Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J. Biomol. Struct. Dyn. 2020;39(7):2673–2678. doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 199.Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tummino T.A., Rezelj V.V., Fischer B., Fischer A., O’Meara M.J., Monel B., Vallet T., White K.M., Zhang Z., Alon A., Schadt H., O’Donnell H.R., Lyu J., Rosales R., McGovern B.L., Rathnasinghe R., Jangra S., Schotsaert M., Galarneau J.-R., Krogan N.J., Urban L., Shokat K.M., Kruse A.C., García-Sastre A., Schwartz O., Moretti F., Vignuzzi M., Pognan F., Shoichet B.K. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science. 2021;373(6554):541–547. doi: 10.1126/science.abi4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Chu C.M., Cheng V.C.C., Hung I.F.N., Wong M.M.L., Chan K.H., Chan K.S., Kao R.Y.T., Poon L.L.M., Wong C.L.P., Guan Y., Peiris J.S.M., Yuen K.Y. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2) doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X., Nicastri E., Oda R., Yo K., Quiros-Roldan E., Studemeister A., Redinski J., Ahmed S., Bernett J., Chelliah D., Chen D., Chihara S., Cohen S.H., Cunningham J., D’Arminio Monforte A., Ismail S., Kato H., Lapadula G., L’Her E., Maeno T., Majumder S., Massari M., Mora-Rillo M., Mutoh Y., Nguyen D., Verweij E., Zoufaly A., Osinusi A.O., DeZure A., Zhao Y., Zhong L., Chokkalingam A., Elboudwarej E., Telep L., Timbs L., Henne I., Sellers S., Cao H., Tan S.K., Winterbourne L., Desai P., Mera R., Gaggar A., Myers R.P., Brainard D.M., Childs R., Flanigan T. Compassionate use of remdesivir for patients with severe COVID-19. N. Engl. J. Med. 2020;382(24):2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jarvis L.M. Scaling up remdesivir amid the coronavirus crisis. C&EN Glob. Enterp. 2020;98(17):23–24. doi: 10.1021/cen-09817-feature2. [DOI] [Google Scholar]
- 205.Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent. Sci. 2020;6(5):672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.U.S. National Library of Medicine Studies found for COVID-19 and inhibitors, ClinicalTrials.gov. 2020. https://www.clinicaltrials.gov/ct2/results?cond=COVID-19&term=inhibitors s.
- 207.Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care. 2020;57(June):279–283. doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hendaus M.A. J. Biomol. Struct. Dyn. 2019;39(10):3787–3792. doi: 10.1080/07391102.2020.1767691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.O’Day D., Gilead Sciences, Inc An open letter from Daniel O'Day, Chairman & CEO, Gilead Sciences, GILEAD: newsroom - press releases. June 29, 2020. https://www.gilead.com/news-and-press/press-room/press-releases/2020/6/an-open-letter-from-daniel-oday-chairman--ceo-gilead-sciences s.
- 213.Srinivas P., Sacha G.L., Koval C., Antivirals for COVID-19, Cleve. Clin. J. Med. (COVID-19 Curbside Consults) (September 29, 2020) 1-5, doi:10.3949/ccjm.87a.ccc030. [DOI] [PubMed]
- 214.U.S. Food and Drug Administration., Emergency use authorization. https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization, (Accessed July 2020).
- 215.Jarvis L.M. Remdesivir data offer hope, with caveats. C&EN Glob. Enterp. 2020;98(17):3. doi: 10.1021/cen-09817-leadcon. [DOI] [Google Scholar]
- 216.Matsuoka K. Fujifilm tests favipiravir as COVID-19 treatment. C&EN Glob. Enterp. 2020;98(15):11. doi: 10.1021/cen-09815-buscon4. [DOI] [Google Scholar]
- 217.Wang Y., Zhou F., Zhang D., Zhao J., Du R., Hu Y., Cheng Z., Gao L., Jin Y., Luo G., Fu S., Lu Q., Du G., Wang K., Lu Y., Fan G., Zhang Y., Liu Y., Ruan S., Liu W., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Evaluation of the efficacy and safety of intravenous remdesivir in adult patients with severe COVID-19: study protocol for a phase 3 randomized, double-blind, placebo-controlled, multicentre trial. Trials. 2020;21(1) doi: 10.1186/s13063-020-04352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Brophy J.M. U.S. purchases world stocks of remdesivir: why the rest of the world should be glad to be at the back of the queue. BMJ. 2020;370 doi: 10.1136/bmj.m2797. [DOI] [PubMed] [Google Scholar]
- 219.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y., Shen C., Li X., Peng L., Huang D., Zhang J., Zhang S., Wang F., Liu J., Chen L., Chen S., Wang Z., Zhang Z., Cao R., Zhong W., Liu Y., Liu L. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering (Beijing) 2020;6(10):1192–1198. doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mullard A. FDA approves Eli Lilly's baricitinib. Nat. Rev. Drug Discov. 2018;17(7):460. doi: 10.1038/nrd.2018.112. [DOI] [PubMed] [Google Scholar]
- 222.National Institute of Allergy and Infectious Diseases (NIAID) Office of Communications NIH clinical trial shows remdesivir accelerates recovery from advanced COVID-19, NIAID News Releases. April 29, 2020. https://www.niaid.nih.gov/news-events/nih-clinical-trial-shows-remdesivir-accelerates-recovery-advanced-covid-19 s.
- 223.Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11(4) doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Halford B. An emerging antiviral takes aim at COVID-19: EIDD-2801’s wily chemistry might make it an ideal weapon in this pandemic and the next. C&EN Glob. Enterp. 2020;98(20):22–23. doi: 10.1021/cen-09820-feature2. [DOI] [Google Scholar]
- 225.U.S. National Library of Medicine Studies found for COVID-19 and EIDD-2801, ClinicalTrials.gov. 2020. https://www.clinicaltrials.gov/ct2/results?cond=COVID-19&term=EIDD-2801 s.
- 226.Cross R. Merck & Co. joins race for COVID-19 vaccines and therapies. C&EN Glob. Enterp. 2020;98(21):12. doi: 10.1021/cen-09821-buscon1. [DOI] [Google Scholar]
- 227.Jena N.R. Drug targets, mechanisms of drug action, and therapeutics against SARS-CoV-2. Chem. Phys. Impact. 2021;2 doi: 10.1016/j.chphi.2021.100011. [DOI] [Google Scholar]
- 228.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sarma P., Prajapat M., Avti P., Kaur H., Kumar S., Medhi B. Therapeutic options for the treatment of 2019-novel coronavirus: an evidence-based approach. Indian Journal of Pharmacology. 2020;52(1):1–5. doi: 10.4103/ijp.IJP_119_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Nguyen B.C.Q., Yoshimura K., Kumazawa S., Tawata S., Maruta H. Frondoside A from sea cucumber and nymphaeols from Okinawa propolis: Natural anti-cancer agents that selectively inhibit PAK1 in vitro. Drug Discov. Ther. 2017;11(2):110–114. doi: 10.5582/ddt.2017.01011. [DOI] [PubMed] [Google Scholar]
- 231.Korkmaz B., Lesner A., Marchand-Adam S., Moss C., Jenne D.E. Lung protection by cathepsin C inhibition: a new hope for COVID-19 and ARDS? J. Med. Chem. 2020;63(22):13258–13265. doi: 10.1021/acs.jmedchem.0c00776. [DOI] [PubMed] [Google Scholar]
- 232.Boerner L.K. Cell studies follow up on previous SARS-CoV-2 study: Earlier study identified 69 compounds that could disrupt viral and human protein interactions. C&EN Glob. Enterp. 2020;98(18):5. doi: 10.1021/cen-09818-leadcon. [DOI] [Google Scholar]
- 233.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Freedberg D.E., Conigliaro J., Wang T.C., Tracey K.J., Callahan M.V., Abrams J.A., Sobieszczyk M.E., Markowitz D.D., Gupta A., O’Donnell M.R., Li J., Tuveson D.A., Jin Z., Turner W.C., Landry D.W. Famotidine use is associated with improved clinical outcomes in hospitalized COVID-19 patients: a propensity score matched retrospective cohort study. Gastroenterology. 2020;159(3):1129–1131. doi: 10.1053/j.gastro.2020.05.053. e1–e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Malone R., Tisdall P., Fremont-Smith P., Liu Y., Huang X.-P., White K., Miorin L., Moreno Del Olmo E., Alon A., Delaforge E., Hennecker C., Wang G., Pottel J., Bona R., Smith N., Hall J., Shapiro G., Clark H., Mittermaier A., Kruse A., García-Sastre A., Roth B., Glasspool-Malone J., Francone V., Hertzog N., Fremont-Smith M., Ricke D. Research Square; 2020. COVID-19: Famotidine, histamine, mast cells, and mechanisms. Preprint PPR179097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Horby P., Landray M., University of Oxford, Nuffield Department of Public Health Low-cost dexamethasone reduces death by up to one third in hospitalised patients with severe respiratory complications of COVID-19, randomised evaluation of COVid-19 thERapY (RECOVERY): news. June 16, 2020. https://www.recoverytrial.net/news/low-cost-dexamethasone-reduces-death-by-up-to-one-third-in-hospitalised-patients-with-severe-respiratory-complications-of-covid-19 s.
- 237.Borba M.G.S., Val F.F.A., Sampaio V.S., Alexandre M.A.A., Melo G.C., Brito M., Mourão M.P.G., Brito-Sousa J.D., Baía-da-Silva D., Guerra M.V.F., Hajjar L.A., Pinto R.C., Balieiro A.A.S., Pacheco A.G.F., Santos J.D.O., Jr., Naveca F.G., Jr., Xavier M.S., Jr., Siqueira A.M., Jr., Schwarzbold A., Jr., Croda J., Jr., Nogueira M.L., Jr., Romero G.A.S., Jr., Bassat Q., Jr., Fontes C.J., Jr., Albuquerque B.C., Jr., Daniel-Ribeiro C.-T., Jr., Monteiro W.M., Jr., Lacerda M.V.G., Jr. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA Network Open. 2020;3(4) doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PubMed] [Google Scholar]
- 238.Fiolet T., Guihur A., Rebeaud M.E., Mulot M., Peiffer-Smadja N., Mahamat-Saleh Y. Effect of hydroxychloroquine with or without azithromycin on the mortality of coronavirus disease 2019 (COVID-19) patients: a systematic review and meta-analysis. Clin. Microbiol. Infect. 2021;27(1):19–27. doi: 10.1016/j.cmi.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Magagnoli J., Narendran S., Pereira F., Cummings T.H., Hardin J.W., Sutton S.S., Ambati J. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with COVID-19. Med. 2020;1(1):114–127. doi: 10.1016/j.medj.2020.06.001. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Singh S., Khan A., Chowdhry M., Chatterjee A. Outcomes of hydroxychloroquine treatment among hospitalized COVID-19 patients in the United States-real-world evidence from a federated electronic medical record network. medRxiv. 2020 doi: 10.1101/2020.05.12.20099028. Preprint. [DOI] [Google Scholar]
- 241.McCoy M. Lilly to test baricitinib against COVID-19. C&EN Glob. Enterp. 2020;98(15):13. doi: 10.1021/cen-09815-buscon17. [DOI] [Google Scholar]
- 242.Halford B. Ruxolitinib to be tested in fight against COVID-19. C&EN Glob. Enterp. 2020;98(14):11. doi: 10.1021/cen-09814-buscon3. [DOI] [Google Scholar]
- 243.Zhang C., Wu Z., Li J.-W., Zhao H., Wang G.-Q. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Boerner L.K. Rethinking the role of blood pressure drugs in COVID-19. C&EN Glob. Enterp. 2020;98(20):29–33. doi: 10.1021/cen-09820-cover. [DOI] [Google Scholar]
- 245.Lo K.B., McCullough P.A., Rangaswami J. Antihypertensive drugs and risk of COVID-19? Lancet Respir. Med. 2020;8(5) doi: 10.1016/S2213-2600(20)30156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Perrotta F., Matera M.G., Cazzola M., Bianco A. Severe respiratory SARS-CoV2 infection: does ACE2 receptor matter? Respir. Med. 2020;168(July) doi: 10.1016/j.rmed.2020.105996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.-C., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436(7047):112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Götz V., Magar L., Dornfeld D., Giese S., Pohlmann A., Höper D., Kong B.-W., Jans D.A., Beer M., Haller O., Schwemmle M. Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci. Rep. 2016;6(1):23138. doi: 10.1038/srep23138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.U.S. National Library of Medicine Studies found for COVID-19 and ACE2, ClinicalTrials.gov. 2020. https://www.clinicaltrials.gov/ct2/results?cond=COVID-19&term=ACE2 s.
- 251.Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H.W., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 253.Sesterhenn F., Yang C., Bonet J., Cramer J.T., Wen X., Wang Y., Chiang C.-I., Abriata L.A., Kucharska I., Castoro G., Vollers S.S., Galloux M., Dheilly E., Rosset S., Corthésy P., Georgeon S., Villard M., Richard C.-A., Descamps D., Delgado T., Oricchio E., Rameix-Welti M.-A., Más V., Ervin S., Eléouët J.-F., Riffault S., Bates J.T., Julien J.-P., Li Y., Jardetzky T., Krey T., Correia B.E. De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science. 2020;368(6492) doi: 10.1126/science.aay5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Whittaker G.R., Daniel S. Going back in time for an antibody to fight COVID-19. Nature. 2020;583(7815):203–204. doi: 10.1038/d41586-020-01816-5. [DOI] [PubMed] [Google Scholar]
- 255.Kasem S., Qasim I., Al-Hufofi A., Hashim O., Alkarar A., Abu-Obeida A., Gaafer A., Elfadil A., Zaki A., Al-Romaihi A., Babekr N., El-Harby N., Hussien R., Al-Sahaf A., Al-Doweriej A., Bayoumi F., Poon L.L.M., Chu D.K.W., Peiris M., Perera R.A.P.M. Cross-sectional study of MERS-CoV-specific RNA and antibodies in animals that have had contact with MERS patients in Saudi Arabia. J. Infect. Public Health. 2018;11(3):331–338. doi: 10.1016/j.jiph.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/ap-200220-0772. [DOI] [PubMed] [Google Scholar]
- 257.Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Cross R. U.S. preorders COVID-19 vaccine and antibody therapy. C&EN Glob. Enterp. 2020;98(27):11. doi: 10.1021/cen-09827-buscon2. [DOI] [Google Scholar]
- 259.Jarvis L.M. Lilly antibody shows promise in COVID-19. C&EN Glob. Enterp. 2020;98(36):15. doi: 10.1021/cen-09836-buscon14. [DOI] [Google Scholar]
- 260.Antibody Society., Archives for COVID-19. Welcome to the Antibody Society: An international non-profit supporting antibody-related research and development. https://www.antibodysociety.org/covid-19/ (Accessed August 2020).
- 261.Kelley B. Developing therapeutic monoclonal antibodies at pandemic pace. Nat. Biotechnol. 2020;38(5):540–545. doi: 10.1038/s41587-020-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Iwasaki A., Omer S.B. Why and how vaccines work. Cell. 2020;183(2):290–295. doi: 10.1016/j.cell.2020.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lin F., Xu J., Shi J., Li H., Li B. Molecular cloning and characterization of a novel glyoxalase I gene TaGly I in wheat (Triticum aestivum L.) Mol. Biol. Rep. 2010;37(2):729–735. doi: 10.1007/s11033-009-9578-3. [DOI] [PubMed] [Google Scholar]
- 264.Callaway E. COVID vaccine boosters: the most important questions. Nature. 2021;596(7871):178–180. doi: 10.1038/d41586-021-02158-6. [DOI] [PubMed] [Google Scholar]
- 265.Boukhatem M.N., Setzer W.N. Aromatic herbs, medicinal plant-derived essential oils, and phytochemical extracts as potential therapies for coronaviruses: Future perspectives. Plants (Basel) 2020;9(6) doi: 10.3390/plants9060800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Le T.T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19(5):305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 267.McKeever A., National Geographic Staff Here’s the latest on COVID-19 vaccines: everything you need to know about the COVID-19 vaccines—from their safety and efficacy to the global vaccine rollout. National Geographic Science: Coronavirus Coverage. August 20, 2021. https://www.nationalgeographic.com/science/health-and-human-body/human-diseases/coronavirus-vaccine-tracker-how-they-work-latest-developments-cvd/ s.
- 268.O’Callaghan K.P., Blatz A.M., Offit P.A. Developing a SARS-CoV-2 vaccine at warp speed. JAMA. 2020;324(5):437–438. doi: 10.1001/jama.2020.12190. [DOI] [PubMed] [Google Scholar]
- 269.World Health Organization., COVID-19 vaccine tracker and landscape. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (Accessed October 28, 2021).
- 270.Nishiura H., Kobayashi T., Yang Y., Hayashi K., Miyama T., Kinoshita R., Linton N.M., Jung S.-M., Yuan B., Suzuki A., Akhmetzhanov A.R. The rate of underascertainment of novel coronavirus (2019-nCoV) infection: estimation using Japanese passengers data on evacuation flights. J. Clin. Med. 2020;9(2) doi: 10.3390/jcm9020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Eikenberry S.E., Mancuso M., Iboi E., Phan T., Eikenberry K., Kuang Y., Kostelich E., Gumel A.B. To mask or not to mask: modeling the potential for face mask use by the general public to curtail the COVID-19 pandemic. Infect. Dis. Modell. 2020;5:293–308. doi: 10.1016/j.idm.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Zhao M., Liao L., Xiao W., Yu X., Wang H., Wang Q., Lin Y.L., Kilinc-Balci F.S., Price A., Chu L., Chu M.C., Chu S., Cui Y. Household materials selection for homemade cloth face coverings and their filtration efficiency enhancement with triboelectric charging. Nano Lett. 2020;20(7):5544–5552. doi: 10.1021/acs.nanolett.0c02211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lin Q., Lim J.Y.C., Xue K., Yew P.Y.M., Owh C., Chee P.L., Loh X.J. Sanitizing agents for virus inactivation and disinfection. View. 2020;1(2) doi: 10.1002/viw2.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Konda A., Prakash A., Moss G.A., Schmoldt M., Grant G.D., Guha S. Aerosol filtration efficiency of common fabrics used in respiratory cloth masks. ACS Nano. 2020;14(5):6339–6347. doi: 10.1021/acsnano.0c03252. [DOI] [PubMed] [Google Scholar]
- 275.Wood-Black F., Lewin J., Blayney M.B., Galindo L., Foreman R., Zelivyanskaya M., Reid M. Highlights: reusing masks, face covering efficacy, plant restarts, and more. ACS Chem. Health Saf. 2020;27(4):204–208. doi: 10.1021/acs.chas.0c00069. [DOI] [PubMed] [Google Scholar]
- 276.Bettenhausen C.A. A chemist’s guide to disinfectants. C&EN Glob. Enterp. 2020;98(20):24–25. doi: 10.1021/cen-09820-feature3. [DOI] [Google Scholar]
- 277.Protein glycosylation in: Lodish H, Berk A, Zipursky SL, et al.New York: W. H. Freeman; 2000. found at : https://www.ncbi.nlm.nih.gov/books/NBK21744/.