Abstract
To date, the time of acquisition of a Cryptococcus neoformans infectious strain has never been studied. We selected a primer, (GACA)4, and a probe, CNRE-1, that by randomly amplified polymorphic DNA (RAPD) analysis and restriction fragment length polymorphism (RFLP), respectively, regrouped strains from control samples of C. neoformans var. grubii environmental isolates according to their geographical origins. The two typing techniques were then used to analyze 103 isolates from 29 patients diagnosed with cryptococcosis in France. Nine of the 29 patients lived in Africa a median of 110 months prior to moving to France; 17 of the patients originated from Europe. Results showed a statistically significant clustering of isolate subtypes from patients originating from Africa compared to those from Europe. We conclude that the patients had acquired the C. neoformans infectious strain long before their clinical diagnoses were made.
Cryptococcus neoformans is a ubiquitous and opportunistic yeast that causes life-threatening meningoencephalitis in 3 to 30% of patients with AIDS (24). This encapsulated basidiomycete exists in three varieties: C. neoformans var. grubii (serotype A) (14) and C. neoformans var. neoformans (serotype D), both with worldwide distributions, as well as C. neoformans var. gattii (serotypes B and C), which is limited to tropical and subtropical regions (21). Cryptococcosis, like many other fungal infections, is thought to begin with inhalation of airborne fungi from an environmental source. Basidiospores, which are smaller, more easily aerosolized, and much more resistant to desiccation than yeast cells, are most likely to be the infectious particles (22, 35). It has been reported that this yeast’s most important natural source is weathered pigeon droppings or soil contaminated with avian guano (11, 12). Data reported in the literature indicate that C. neoformans can be found as a transient commensal organism on humans or as an incidental colonizer in the respiratory tract or on the skin of healthy subjects or even patients with bronchopulmonary disorders (19, 26).
Several observations converge toward the hypothesis that the infectious particles can be acquired long before the infection develops and is diagnosed. First, a high percentage of healthy subjects have anticryptococcal antibodies, which suggests prior contact with the fungus (7, 18). Second, patients coming from tropical areas can be diagnosed with C. neoformans var. gattii cryptococcosis long after they have left their countries of origin (8). Finally, unlike French patients, African patients living in France and diagnosed with cryptococcosis are rarely infected with C. neoformans var. neoformans strains (10). To verify this hypothesis, a well-characterized group of patients and a molecular method able to distinguish between isolates of the same serotype but from different geographical regions should be selected. The molecular typing methods currently available are reported to be unable to regroup Cryptococcus neoformans var. grubii isolates by their geographical origins (4, 5, 33). On the other hand, Kwon-Chung and Bennett, using standard immunological methods of serotyping, have described a nonrandom distribution of serotypes around the world (21). Although serotyping is not sufficiently discriminative to determine the geographical origin of a cryptococcal isolate, it provides good evidence that a technique capable of clustering strains from the same geographical region might exist.
In this study, we addressed the question of the time of acquisition of the infecting organism, an issue that has never before been raised. Using control samples of environmental isolates and two typing methods capable of clustering strains based on their geographical origins, we were able to demonstrate that patients diagnosed with cryptococcosis in France but born in Africa had acquired their infectious strains a long time ago, prior to emigrating from their countries of origin.
MATERIALS AND METHODS
Patients and strains.
Twenty environmental isolates of C. neoformans var. grubii from different geographical regions were used in this study: Japanese isolates J1, J2, J3, J4, and J5 were kindly provided by S. Kohno (Nagasaki University School of Medicine, Nagasaki, Japan) as M12, SH1311, MT11, SUMO1, and SASO1, respectively (36); African isolates AF1 (Morocco), AF2 (Togo), AF3 (Ivory Coast), AF4 (Burundi), and AF5 (Zimbabwe) were provided by D. Swinne (Institute of Tropical Medicine, Antwerp, Belgium) as RV45718, RV45880, RV46288, RV67312, and RV70273, respectively; American isolates US1, US2, and US3 (Kentucky) as well as US4 (New York) were provided by J. M. Clauson (Western Kentucky University, Bowling Green) and A. Casadevall (Albert Einstein College of Medicine, Bronx, N.Y.) as FE-1, PE-1, SSE-1, and B5 respectively (6); and French isolates F1 through F6 were provided by S. Mathoulin and B. Couprie (Centre Hospitalo-Universitaire, Bordeaux, France) as 115A, 57B, 109B, 13A, 110B, and 122A, respectively (16).
A total of 103 clinical C. neoformans var. grubii isolates were recovered from 29 patients who had been diagnosed with cryptococcosis in France and whose infections had been reported to the National Reference Center for Mycoses during the first year (1997) of a multicentric clinical study, étude Crypto A/D (Direction Générale de la Santé no. 970089). Detailed information on clinical and epidemiological issues (particularly the patients’ trips and stays since childhood) and on all of the isolates recovered at the time of diagnosis and during the course of the infection were collected. Among the 29 patients, 17 had been born in Europe and 9 had been born in Africa (see Table 1). The African patients had been living in France for a median of 110 months before cryptococcosis was diagnosed. The last trip back to Africa had occurred as long as 13 years ago (patient P17).
TABLE 1.
RAPD profiles, generated with primer (GACA)4, among patients of different origins
| Patient no. | Sexa | HIV statusb | Country of birth | No. of isolates | (GACA)4 profile | Time since emigration (mo) |
|---|---|---|---|---|---|---|
| P1 | M | + | France | 8 | I | |
| P2 | M | + | France | 6 | I | |
| P3 | M | + | France | 11 | I | |
| P4 | M | + | Haiti | 4 | I | 189 |
| P5 | F | + | France | 1 | I | |
| P6 | M | + | France | 9 | I | |
| P7 | M | + | Algeria | 3 | I | 205 |
| P8 | M | + | France | 7 | I | |
| P9 | M | + | France | 14 | I | |
| P10 | M | + | Tunisia | 2 | I | 109 |
| P11 | M | + | Ivory Coast | 2 | I | 314 |
| P12 | M | + | France | 1 | I | |
| P13 | M | + | Italy | 4 | I | 34 |
| P14 | M | + | France | 3 | I | |
| P15 | M | + | France | 1 | I | |
| P16 | M | + | Ivory Coast | 1 | II | 0 |
| P17 | M | + | DRCc | 1 | II | 110 |
| P18 | M | + | Colombia | 5 | II | 41 |
| P19 | M | + | Gambia | 3 | II | 139 |
| P20 | F | + | DRC | 2 | II | 11 |
| P21 | M | + | Ivory Coast | 1 | II | 48 |
| P22 | M | + | Cambodia | 3 | II | 0 |
| P23 | F | + | France | 2 | III | |
| P24 | M | + | France | 4 | III | |
| P25 | F | − | France | 1 | III | |
| P26 | F | − | France | 1 | III | |
| P27 | M | + | France | 1 | III | |
| P28 | M | + | France | 1 | III | |
| P29 | M | − | DRC | 1 | IV | 239 |
M, male; F, female.
+, HIV positive; −, HIV negative.
DRC, Democratic Republic of Congo.
The identification of all cultured organisms as C. neoformans was confirmed by standard biochemical methods. Isolates were identified as C. neoformans var. grubii by the use of canavanine-glycine-bromothymol medium, d-proline assimilation, and a direct immunofluorescence assay using a monoclonal antibody (9). All strains were stored frozen in 40% glycerol at −80°C and were grown overnight in YPD medium (5 g of yeast extract, 10 g of Bacto Peptone, and 10 g of glucose per liter) at 30°C.
Randomly amplified polymorphic DNA (RAPD) analysis.
C. neoformans DNA was extracted as previously described (31). The following primers were chosen from the literature and tested for their discriminatory power on 20 environmental isolates under various annealing temperatures: (CA)8 RY (25), (GTG)5, (GACA)4 (23), the phage M13 core sequence (27), and two enterobacterial repetitive intergenic consensus sequences, ERIC1 and ERIC2 (2). PCR was carried out in a thermal cycler (Omnigene; Hybaid, Teddington, United Kingdom) in 100-μl reaction volumes, each containing 50 ng of genomic DNA, 50 pmol of primer, 200 μM deoxynucleoside triphosphates, and 2 U of recombinant Taq polymerase (Pharmacia Biotech, Uppsala, Sweden), with the manufacturer’s recommended buffers.
Two primers, ERIC1 and (GACA)4, were then selected and used to study clinical isolates under the following optimized conditions: reactions were cycled 35 times, with a 4-min denaturation at 94°C, 1 min of annealing each at 28 and 48°C, and a 2-min primer extension at 74°C. Amplification products were analyzed by electrophoresis through a 2% agarose gel and visualized under UV light after being stained with ethidium bromide. The reproducibility of this method was confirmed by reanalyzing another DNA preparation from five clinical isolates. In every case, identical profiles were obtained (data not shown).
RFLP analysis.
Restriction fragment length polymorphisms (RFLPs) were detected by Southern blot hybridization after restriction enzyme SstI digestion of total-DNA samples. The restriction fragments obtained were then separated by electrophoresis through a 0.8% agarose gel and transferred onto positively charged nylon membranes (Boehringer Mannheim, Mannheim, Germany). The DNA probe (C. neoformans repetitive element CNRE-1), generously provided by E. Spitzer and S. Spitzer (28), was labeled with DIG digoxigenin–11-dUTP by using a DIG-High Prime Kit (Boehringer Mannheim). After an overnight hybridization at 68°C and stringent washes, bands were detected and exposed according to the manufacturer’s instructions. Reproducibility was confirmed by reanalyzing another DNA preparation from five clinical isolates (data not shown).
Data analysis.
DNA fingerprint patterns were analyzed by using the software Taxotron, developed by P. D. Grimont (Institut Pasteur, Paris, France) (17), which automatically identified band positions and compared two profiles by calculating the Dice coefficient complement (number of different bands per total number of fragments in the two profiles). Dendograms were then generated by using the unweighted pair group method of average linkage (17).
Statistical analysis.
The distributions of the patients’ isolates by subtype according to the European or African origin were analyzed by using Fisher’s exact test.
RESULTS
Selection of a typing method for environmental isolates.
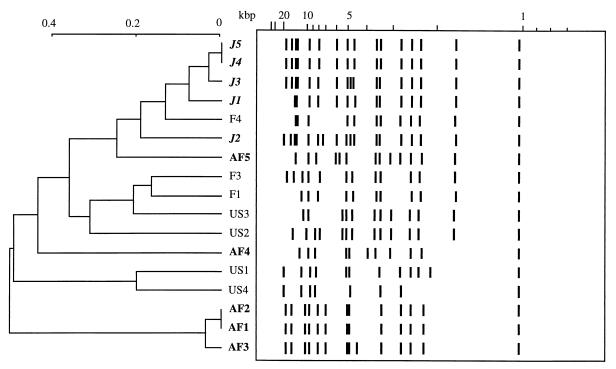
We tested different typing methods to evaluate their abilities to cluster environmental C. neoformans var. grubii strains according to their geographical origins. Because RFLP is known to be reproducible and easy to perform, we first tested the ability of CNRE-1 to geographically classify the isolates. Figure 1 shows the Taxotron-derived schematic representation of the RFLP profiles obtained after hybridization of CNRE-1 to SstI-digested genomic DNA. Fifteen different profiles were obtained for the 17 strains tested. Two Japanese (J5 and J4) and two African (AF2 and AF1) isolates yielded identical hybridization profiles with this probe. Partial clustering of the Japanese (four of five) and African (three of five) isolates was obtained, but no specific profile could be associated with a given region. Therefore, we tested another molecular technique to determine it could further differentiate isolates based on their geographical origins.
FIG. 1.
Schematic representation of and dendogram generated from the Dice coefficient complement computed from the CNRE-1 patterns of environmental isolates from Japan (J), France (F), the United States (US), and Africa (AF).
Previous studies using RAPD techniques demonstrated geographical clustering among isolates of C. neoformans var. gattii (2, 27). RAPD analysis has been applied in several epidemiological studies of C. neoformans; however, some questions have been raised concerning the interpretation of the profiles generated. The advantages RAPD offers over other methods, particularly speed and ease of execution, made it suitable for investigation of the ability to discriminate between C. neoformans var. grubii isolates according to their geographical origins.
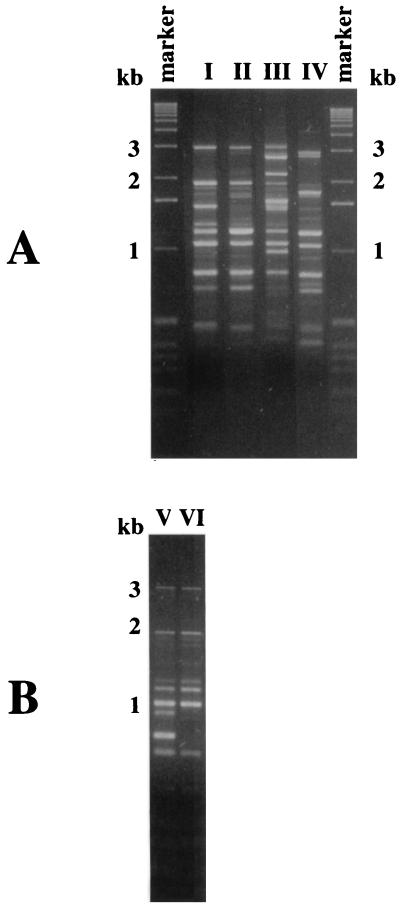
Six different primers previously used in some epidemiological studies of C. neoformans were chosen. Using different amplification temperatures, they were tested on the control group of 17 environmental isolates to which 3 more environmental isolates from France were added. Only primers ERIC1 and (GACA)4 revealed roughly the same patterns for strains coming from the same geographical region and different patterns for strains coming from different continents. Representative RAPD profiles obtained with the primer (GACA)4 are shown in Fig. 2. Using this primer, all five Japanese isolates had profile II, five of the six French isolates exhibited profile I, and four of the five African isolates gave profile V or VI. Of the four North American isolates, two exhibited profile I and two showed profile II.
FIG. 2.
Representative RAPD profiles of C. neoformans isolates generated with the (GACA)4 primer. Profile identifications are indicated as roman numerals (I through VI). Profiles V and VI were obtained only with environmental isolates. Molecular size standards were obtained with the 1-kb ladder (Gibco BRL, Gaithersburg, Md.).
Evaluation of clinical isolates with the selected techniques.
One hundred and three C. neoformans var. grubii clinical isolates were then analyzed with the two primers described above. Four different profiles were obtained with the primer ERIC1. However, no association between a given profile and the corresponding patient’s continent of origin could be established (data not shown). Typing of the clinical isolates with primer (GACA)4 revealed four different RAPD profiles, whose distributions are reported in Table 1. All of the strains exhibiting profile III were isolated from European patients. Of the 15 patients with profile I strains, 11 were born in Europe. None of the strains generating profile II or IV were isolated from patients born in Europe. It is important to note that all of the isolates recovered from a particular patient gave the same profile with both primers. Thus, the analysis of clinical isolates showed that the distribution of profiles according to the geographical origin of the patients was not random. This bias of distribution was statistically significant (P < 0.0005) when the European and African patients were being compared (Table 2). None of the clinical isolates tested yielded profile V or VI, both of which were characteristic of environmental African isolates. On the other hand, profiles III and IV were specific to clinical isolates in this study. Finally, profile II, which was characteristic of the Japanese environmental isolates, was generated mostly by strains isolated from African patients. These results might be explained by the facts that environmental and clinical African strains did not come from the same country and that no Japanese patients were included in our study. Moreover, the discriminatory power of this RAPD method was low.
TABLE 2.
Distribution of RAPD profiles obtained with the (GACA)4 primer
| Geographical region | Total no. of patients | No. of patients with RAPD profilea:
|
|||
|---|---|---|---|---|---|
| I | II | III | IV | ||
| Europe | 17 | 11 (65) | 0 | 6 (35) | 0 |
| Africa | 9 | 3 (33) | 5 (56) | 0 | 1 (11) |
The numbers in parentheses are the percentaes of patients whose isolates showed the corresponding profile.
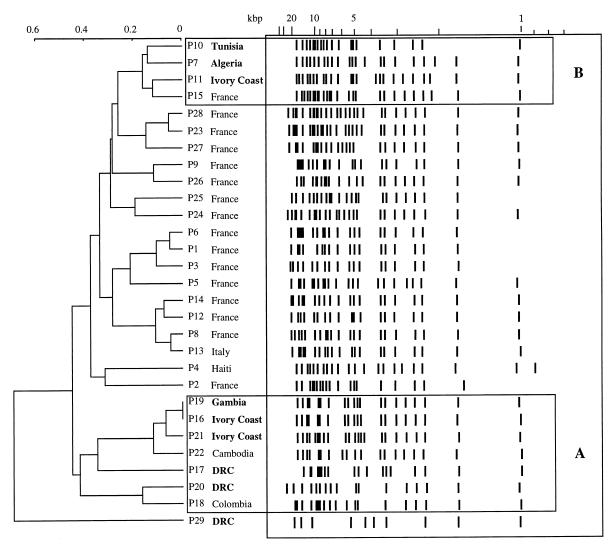
We then tested the 103 strains with the CNRE-1 probe. As previously described in other reports (29, 32), we found that isolates from the same patient showed identical hybridization patterns, although there were a few examples of microevolution (15, 30). Thus, for the remainder of the analysis, one representative isolate from each patient was chosen, and their hybridization patterns are presented in Fig. 3; 28 different profiles were obtained from 29 isolates studied.
FIG. 3.
Schematic representation of and dendogram generated from the Dice coefficient complement computed from the CNRE-1 patterns of 29 clinical isolates. African strains (in boldface) are regrouped in clusters A and B (boxed in this figure). DRC, Democratic Republic of Congo.
The Taxotron software analysis generated a dendogram in which two boxed clusters, named A and B, can be seen (Fig. 3). Cluster A contained the seven strains with the profile II subtype as determined by the (GACA)4 RAPD technique. Five of these strains were isolated from patients born in Africa, one was from a patient born in Colombia, and one was from a patient born in Cambodia. None of the strains from this cluster was isolated from a European patient. Cluster B contained four strains which were all the profile I subtype, as determined by the (GACA)4 RAPD technique. Three of them were isolated from patients born in Africa, and one was from a patient born in Europe. The strain from the African patient P29 seemed to be completely different from the others. It was also the only strain for which the profile IV subtype was generated by the RAPD technique.
DISCUSSION
After analyzing the epidemiology of cryptococcosis in France (10), we were especially interested in learning more about the pathophysiology of this infection. An important issue was the time of acquisition of the infecting isolate compared to the time of diagnosis: were the infectious particles inhaled daily and killed as long as host defense mechanisms were efficient, or, on the contrary, would the immune system normally achieve local control without eradication? Since this latter mechanism has already been evoked or demonstrated for other microorganisms, such as Leishmania spp. (1) and Histoplasma capsulatum (34), and since several lines of evidence suggest that it could also occur with C. neoformans, we looked for a way to verify this hypothesis. To do so required control samples composed of environmental isolates from remote areas (to ascertain their geographical origin), clinical isolates recovered from patients whose travels and clinical histories were known, and the assessment of various typing methods since none has yet been able to correlate geographical origin with a specific pattern.
Indeed, several groups have studied the molecular epidemiology of C. neoformans infections and attempted to regroup isolates based on their geographical origins. However, the genetic differentiation generated by CNRE-1 RFLP analysis showed no geographical correlation among strains isolated from Brazil and the United States (13). Using UT-4p, Varma and colleagues reported no obvious clustering of the C. neoformans var. grubii isolates according to geographical origin (33), although Garcia-Hermoso and coworkers evoked the possibility of geographical clustering (16) when they compared the patterns obtained for French isolates to those observed by Varma et al. (33) and Kohno (20). Finally, using the multilocus enzyme electrophoresis technique, Brandt and colleagues found that some subtypes were more common in some areas of the United States than in others; however, that finding was not confirmed when another typing method (RAPD) was used (3). In light of our results, we think that the lack of clear-cut regional differences in the previously published studies may be due to the population sample from which the clinical isolates were recovered (with its lack of patients coming from remote places and with known travel histories), the lack of environmental isolates from remote areas, and/or the technique selected. Indeed, our data show that depending on the sample chosen (environmental or clinical isolates) and the technique tested (six primers selected for RAPD or CNRE-1 RFLP), we could have concluded that either there was or was not a geographical clustering of isolates and that the isolates tested exhibited or did not exhibit genetic variability. These discrepancies clearly demonstrate the importance of the sample choice and the technique selected to answer a specific epidemiological question. To address the question of geographical clustering, we needed an epidemiological tool with adequate discriminatory power: not too high (to prevent further strain delineation among isolates from the same geographical origin) and not too low (to enable the differentiation of the isolates based on their geographical origin). The RAPD method using the primer (GACA)4 fulfilled this requirement.
Based on the RAPD profiles obtained, we showed that the distribution of clinical isolates from nine African patients diagnosed with cryptococcosis in France was significantly different from that of clinical isolates recovered from the 17 European patients (P < 0.0005). Furthermore, a second, independent typing method (CNRE-1 RFLP) confirmed the results, showing two clusters that contained the isolates from eight of the nine African patients. This finding suggests that the infecting organism can be acquired long before the infection develops, since these patients had been living in France a median of 110 months and had not been in contact with an African environment for as long as 13 years. That African patients were infected with African isolates strongly suggests that these isolates had been sequestered and contained somewhere in the body, most likely the alveolar macrophages. Then, as soon as some kind of immune system defect occurred, which in most of the patients was AIDS, the fungus could multiply, disseminate, and cause infection. The clinical histories of these patients and the demonstration of a geographical clustering of isolates based on the generated profiles are consistent with a dormant phase of C. neoformans within all individuals. Why infection would be caused by a dormant strain of C. neoformans rather than a newly acquired one, what form the dormant form of C. neoformans would assume, and why some isolates would be more virulent than others remain to be determined.
The observation that the infecting organism had been (or at least could have been) acquired long before the infection was diagnosed should be taken into account in the prospective development of prophylactic programs, such as vaccination or antifungal therapy, for populations at particularly high risk of developing cryptococcosis, such as AIDS patients in central or southern Africa, South Africa, or Southeast Asia (24).
ACKNOWLEDGMENTS
We thank A. Casadevall (Albert Einstein College of Medicine, Bronx, N.Y.), J. M. Clauson (Western Kentucky University, Bowling Green, Ky.), S. Kohno (Nagasaki University School of Medicine, Nagasaki, Japan), and D. Swinne (Institute of Tropical Medicine, Antwerp, Belgium) for supplying the environmental isolates used in this study. We are grateful to Eric Spitzer and Silvia Spitzer for the generous gift of the CNRE-1 probe. We are grateful for the collaboration of the French Cryptococcosis Study Group, which includes clinicians and microbiologists from various hospitals in France. Those who participated in collection of the data used for this specific study are listed here (in alphabetical order by city): M. E. Bougnoux, S. Morelon, and E. Rouveix (Boulogne); C. Passa Gaudouen, B. Michel, G. Otterbein, and J. Roucoules (Bry-sur-Marne); J. M. Korach (Châlon en Champagne); B. Salles and C. Sire (Chalon/Saône); M. Gauthier and O. Salmon (Evry); F. Botterel and J. F. Delfraissy (Le Kremlin Bicêtre); M. A. Desailly and H. Maisonneuve (La Roche/Yon); D. Bouhour, E. Dannaoui, D. Peyramond, and M. A. Piens (Lyon); O. Morin and P. Poirier (Nantes); M. Gari Toussaint and P. Dellamonica (Nice); and in Paris C. Chochillon, X. Duval, and J. L. Vildé (Hôpital Bichat); M. Kazatchkine, V. Lavarde, and C. Piketty (Hôpital Broussais); B. Dupont (Institut Pasteur); L. Baril, F. Bricaire, J. Carrière, A. Datry, S. Herson, and C. Trivalle (Hôpital Pitié Salpétrière); G. Delzant and G. Kac (Hôpital Tenon); J. Gilquin (Hôpital St. Joseph); D. Toubas (Reims); B. Hery and J. Y. Leberre (St. Nazaire); M. F. Biava and C. Rabaud (Vandoeuvre les Nancy); and C. Fontier and E. Mazards (Valenciennes). We also thank Olivier Ronin for serotyping the cryptococcal isolates, K. Sitbon and A. de Gouvello for help in collecting the clinical data, and J. Jacobson for editorial assistance.
Financial support for this work was provided by SIDACTION (a postdoctoral fellowship for G.J. and a grant for F.D.), Pfizer Laboratory (a scholarship for D.G.-H.), and the Pasteur Institute (Contrat de Recherche Clinique for F.D.).
REFERENCES
- 1.Alvar J, Cañavate C, Gutiérrez-Solar B, Jiménez M, Laguna F, López-Vélez R, Molina R, Moreno J. Leishmania and human immunodeficiency virus coinfection: the first 10 years. Clin Microbiol Rev. 1997;10:298–319. doi: 10.1128/cmr.10.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boekhout T, van Belkum A, Leenders A C A P, Verbrugh H A, Mukamurangwa P, Swinne D, Scheffers W A. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int J Syst Bacteriol. 1997;47:432–442. doi: 10.1099/00207713-47-2-432. [DOI] [PubMed] [Google Scholar]
- 3.Brandt M E, Hutwagner L C, Klug L A, Baughman W S, Rimland D, Graviss E A, Hamill R J, Thomas C, Pappas P G, Reingold A L, Pinner R W the Cryptococcal Disease Active Surveillance Group. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. J Clin Microbiol. 1996;34:912–917. doi: 10.1128/jcm.34.4.912-917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, D.C: ASM Press; 1998. pp. 41–70. [Google Scholar]
- 5.Chen S C A, Brownlee A G, Sorrell T C, Ruma P, Ellis D H, Pfeiffer T J, Speed B R, Nimmo G. Identification by random amplification of polymorphic DNA of a common molecular type of Cryptococcus neoformans var. neoformans in patients with AIDS or other immunosuppressive conditions. J Infect Dis. 1996;173:754–758. doi: 10.1093/infdis/173.3.754. [DOI] [PubMed] [Google Scholar]
- 6.Currie B P, Freundlich L F, Casadevall A. Restriction fragment length polymorphism analysis of Cryptococcus neoformans isolates from environmental (pigeon excreta) and clinical sources in New York City. J Clin Microbiol. 1994;32:1188–1192. doi: 10.1128/jcm.32.5.1188-1192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dromer F, Aucouturier P, Clauvel J P, Saimot G, Yeni P. Cryptococcus neoformans antibody levels in patients with AIDS. Scand J Infect Dis. 1988;20:283–285. doi: 10.3109/00365548809032452. [DOI] [PubMed] [Google Scholar]
- 8.Dromer F, Ronin O, Dupont B. Isolation of Cryptococcus neoformans var. gattii from an Asian patient in France: evidence for dormant infection in healthy subjects. J Med Vet Mycol. 1992;30:395–397. [PubMed] [Google Scholar]
- 9.Dromer F, Gueho E, Ronin O, Dupont B. Serotyping of Cryptococcus neoformans by using a monoclonal antibody specific for capsular polysaccharide. J Clin Microbiol. 1993;31:359–363. doi: 10.1128/jcm.31.2.359-363.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dromer F, Mathoulin S the French Cryptococcosis Study Group. Epidemiology of cryptococcosis in France: a 9-year survey (1985–1993) Clin Infect Dis. 1996;23:82–90. doi: 10.1093/clinids/23.1.82. [DOI] [PubMed] [Google Scholar]
- 11.Emmons C W. Isolation of Cryptococcus neoformans from soil. J Bacteriol. 1951;62:685–690. doi: 10.1128/jb.62.6.685-690.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emmons C W. Saprophytic sources of Cryptococcus neoformans associated with the pigeon (Columba livia) Am J Hyg. 1955;62:227–232. doi: 10.1093/oxfordjournals.aje.a119775. [DOI] [PubMed] [Google Scholar]
- 13.Franzot S P, Hamdan J S, Currie B P, Casadevall A. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J Clin Microbiol. 1997;35:2243–2251. doi: 10.1128/jcm.35.9.2243-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franzot S P, Salkin I F, Casadevall A. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J Clin Microbiol. 1999;37:838–840. doi: 10.1128/jcm.37.3.838-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Hermoso, D. Unpublished data.
- 16.Garcia-Hermoso D, Mathoulin-Pélissier S, Couprie B, Ronin O, Dupont B, Dromer F. DNA typing suggests pigeon droppings as a source of pathogenic Cryptococcus neoformans serotype D. J Clin Microbiol. 1997;35:2683–2685. doi: 10.1128/jcm.35.10.2683-2685.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimont P A D. Taxotron’s user’s manual. Paris, France: Institut Pasteur; 1998. [Google Scholar]
- 18.Henderson D K, Bennett J E, Huber M A. Long-lasting, specific immunologic unresponsiveness associated with cryptococcal meningitis. J Clin Investig. 1982;69:1185–1190. doi: 10.1172/JCI110555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard D H. The commensalism of Cryptococcus neoformans. Sabouraudia. 1973;11:171–174. [PubMed] [Google Scholar]
- 20.Kohno S. Abstracts of the Third International Conference on Cryptococcus and Cryptococcosis. Paris, France: Institut Pasteur; 1996. Epidemiology of cryptococcosis in Japan, abstr. II.3; pp. 41–42. [Google Scholar]
- 21.Kwon-Chung K J, Bennett J E. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am J Epidemiol. 1984;120:123–130. doi: 10.1093/oxfordjournals.aje.a113861. [DOI] [PubMed] [Google Scholar]
- 22.Kwon-Chung K J, Bennett J E, editors. Medical mycology. Philadelphia, Pa: Lea & Febiger; 1992. pp. 397–446. [Google Scholar]
- 23.Meyer W, Mitchell T G, Freedman E Z, Vilgalys R. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J Clin Microbiol. 1993;31:2274–2280. doi: 10.1128/jcm.31.9.2274-2280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira R P, Macedo A M, Chiari E, Pena S D J. An alternative approach to evaluating the intraspecific genetic variability of parasites. Parasitol Today. 1997;13:196–200. doi: 10.1016/s0169-4758(97)01044-2. [DOI] [PubMed] [Google Scholar]
- 26.Randhawa H S, Paliwal D K. Survey of Cryptococcus neoformans in the respiratory tract of patients with bronchopulmonary disorders and in the air. Sabouraudia. 1979;17:399–404. doi: 10.1080/00362177985380591. [DOI] [PubMed] [Google Scholar]
- 27.Sorrell T C, Chen S C A, Ruma P, Meyer W, Pfeiffer T J, Ellis D H, Brownlee A G. Concordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting. J Clin Microbiol. 1996;34:1253–1260. doi: 10.1128/jcm.34.5.1253-1260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spitzer E D, Spitzer S G. Use of a dispersed repetitive DNA element to distinguish clinical isolates of Cryptococcus neoformans. J Clin Microbiol. 1992;30:1094–1097. doi: 10.1128/jcm.30.5.1094-1097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spitzer E D, Spitzer S G, Freundlich L F, Casadevall A. Persistence of initial infection in recurrent Cryptococcus neoformans meningitis. Lancet. 1993;341:595–596. doi: 10.1016/0140-6736(93)90354-j. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan D, Haynes K, Moran G, Shanley D, Coleman D. Persistence, replacement, and microevolution of Cryptococcus neoformans strains in recurrent meningitis in AIDS patients. J Clin Microbiol. 1996;34:1739–1744. doi: 10.1128/jcm.34.7.1739-1744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varma A, Kwon-Chung K J. Rapid method to extract DNA from Cryptococcus neoformans. J Clin Microbiol. 1991;29:810–812. doi: 10.1128/jcm.29.4.810-812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varma A, Kwon-Chung K J. DNA probe for typing of Cryptococcus neoformans. J Clin Microbiol. 1992;30:2960–2967. doi: 10.1128/jcm.30.11.2960-2967.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varma A, Swinne D, Staib F, Bennett J E, Kwon-Chung K J. Diversity of DNA fingerprints in Cryptococcus neoformans. J Clin Microbiol. 1995;33:1807–1814. doi: 10.1128/jcm.33.7.1807-1814.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warnock D W, Dupont B, Kauffman C A, Sirisanthana T. Imported mycoses in Europe. Med Mycol. 1998;36:87–94. [PubMed] [Google Scholar]
- 35.Wickes B L, Mayorga M E, Edman U, Edman J C. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc Natl Acad Sci USA. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto Y, Kohno S, Koga H, Kakeya H, Tomono K, Kaku M, Yamazaki T, Arisawa M, Hara K. Random amplified polymorphic DNA analysis of clinically and environmentally isolated Cryptococcus neoformans in Nagasaki. J Clin Microbiol. 1995;33:3328–3332. doi: 10.1128/jcm.33.12.3328-3332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]