Abstract
Objectives
The global spread of SARS-CoV-2 is a serious public health issue. Large-scale surveillance screenings are crucial but can exceed test capacities. We (A) optimized test conditions and (B) implemented pool testing of respiratory swabs into SARS-CoV-2 diagnostics.
Study design
(A) We determined the optimal pooling strategy and pool size. In addition, we measured the impact of vortexing prior to sample processing, compared a pipette-pooling method (by combining transport medium of several specimens) and a swab-pooling method (by combining several swabs into a test tube filled with PBS) as well as determined the sensitivities of three PCR assays. (B) Finally, we applied high-throughput pool testing for diagnostics.
Results
(A) In a low prevalence setting, we defined a preferable pool size of ten in a two-stage hierarchical pool testing strategy. Vortexing of swabs (n = 33) increased cellular yield by a factor of 2.34. By comparing Ct-values of 16 pools generated with two different pooling strategies, pipette-pooling was more efficient compared to swab-pooling. Measuring dilution series of 20 SARS-CoV-2 positive samples in three PCR assays simultaneously revealed detection rates of 85% (assay I), 50% (assay II), and 95% (assay III) at a 1:100 dilution. (B) We systematically pooled 55,690 samples in a period of 44 weeks resulting in a reduction of 47,369 PCR reactions.
Conclusions
For implementing pooling strategies into high-throughput diagnostics, we recommend utilizing a pipette-pooling method, performing sensitivity validation of the PCR assays used, and vortexing swabs prior to analyses. Pool testing for SARS-CoV-2 detection is feasible and effective in a low prevalence setting.
Keywords: SARS-CoV-2, Pool testing, Surveillance
1. Introduction
The SARS-CoV-2 pandemic is a serious public health problem of unprecedented magnitude in recent times. In particular, individuals at older age or with comorbidities are at a high risk to require hospitalization and intensive care [1]. Therefore, it is essential to control person-to-person transmission in order to protect vulnerable individuals and limit the number of severe cases. Until immunity is achieved by vaccination, nonpharmaceutical interventions need to be applied. Many countries could successfully contain the spread of COVID-19 through social distancing or lock-down measures, contact tracing, quarantine, and large-scale testing in the ongoing pandemic [2, 3]. In order to control viral transmission when lifting lock-down strategies, large-scale testing and surveillance are critical interventions. These approaches are based on frequent tests of individuals, e.g. by rapid antigen-based tests or reverse transcription-real-time PCR, to detect SARS-CoV-2 in swab specimens. However, large-scale surveillance screenings can exceed test capacities of diagnostic laboratories.
Pooling swab specimens for PCR-testing can increase test capacities and limit the consumption of reagents [4]. To this end, swab samples are combined and tested in a single PCR reaction. If the pool test is positive, the remaining sampling material of the included specimens can be retested separately to detect the infected individual(s). If the pool test is negative, all individuals are declared as not infected [5], [6], [7] (two-stage hierarchical pool testing).
Pool testing is highly efficient in a setting of low disease prevalence and availability of highly sensitive test methods [6]. It can be applied to enable surveillance screenings of asymptomatic individuals in public institutions e.g. hospitals, schools or retirement homes, which carry a high risk for superspreading events and severe disease courses. When pool testing is established, test conditions need to be optimized including (a) the pooling strategy and pool sizes, (b) sample preparation and pooling method, and (c) the quality of SARS-CoV-2 detection by PCR. In this study, we determined and implemented the optimized pool testing procedure into the diagnostic routine for SARS-CoV-2 detection.
2. Materials and methods
2.1. Pool testing algorithm
Pooling efficiency was computed using a web tool published by Bilder and colleagues [6, 8]. Calculations were performed assuming a PCR-test sensitivity of 99% or 95% and a test specificity of 99%. The expected number of tests was computed for different pool sizes as described [6].
2.2. Swab specimens
Oropharyngeal or combined nasal/oropharyngeal swabs were collected and transferred into MSwab™ Medium, UTM-RT/mini (COPAN Diagnostics, Murrieta, CA), BD ESwab™ (Becton & Dickinson, Sparks, MD, USA) or Sigma Transwab®Purflock® (Medical Wire & Equipment, Corsham, Wiltshire, England). All specimens were processed at the Institute of Virology, University Hospital Cologne within 12 h after collection. Samples were stored for validation procedures at −80 °C.
2.3. Clinical data
All samples and clinical data were collected at the University Hospital Cologne. No identifying data were used for the patients' characterization. According to §15 subparagraph 3 (Professional Code for Physicians in Germany) ethical principles of WMA Declaration of Helsinki were respected. The retrospective data analysis was approved by the Institutional Review Committee of the Medical Faculty, University Hospital Cologne, Germany (ethical vote no. 20–1638).
2.4. Sample preparation and evaluation of the pooling strategy
To determine the cellular content of the same n = 33 specimens before and after vortexing, human β-globin-gene quantification was performed as published [9].
For the pipette pooling, an aliquot of the transport medium from each of ten storage tubes were combined into one test tube. For the swab pooling, transport medium was removed, PBS added to a first tube containing the swab, vortexed, and transferred into a second swab tube followed by vortexing. After the PBS had traveled through all ten swab tubes, it was transferred into a test tube (Fig 2C,E). 16 different pools were conducted using each of the two pooling methods. Preparation time was measured.
To simulate various pool sizes, 25 positive specimens with various Ct-values were diluted 1:5, 1:10, 1:20 and 1:50 in negative samples, respectively.
To compare detection rates of three PCR systems, ten-fold dilution series of n = 20 SARS-CoV-2-positive samples were simultaneously tested in three assays.
2.5. Integration of pool testing into the diagnostic routine
After arrival at our laboratory, samples were vortexed 5 s, and preselected to be tested individually or in pools. Samples of symptomatic individuals and persons recently tested positive were excluded from pool testing. Within 44 weeks, 55,690 samples were tested in pools using the pipette pooling method.
2.6. Reverse transcription, amplification and detection
(Assay I) Nucleic acid extraction was performed using the MagNAPure® 96 DNA and Viral NA Kit, followed by amplification on LightCycler® 480II (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. Detection of SARS-CoV-2 was conducted using the RealStar® SARS-CoV-2 RT-PCR Kit1.0 (altona Diagnostics, Hamburg, Germany) or LightMix®SarbecoV E-gene plus equine arteritis virus (EAV) control (TibMolBiol, Berlin, Germany). (Assay II) Processing of swabs was implemented with Panther Fusion® Hologic® and SARS-CoV-2 was detected using 5 μl of total RNA in 20 μl of LightMix®SarbecoV E-gene plus β-globin as internal control (TibMolBiol, Berlin, Germany). As second target the N-gene was amplified (inhouse primer sets in multiplex PCR, data unpublished). (Assay III) Detection of SARS-CoV-2 was performed on a Roche cobas®6800 using the cobas® SARS-CoV-2 kit targeting the E-gene/ORF-1a/b regions according to manufacturer's protocol.
SARS-CoV-2 was quantified using dilution series of cell culture supernatant extrapolated to approved standards (INSTAND e.V., Düsseldorf, Germany), measured in all assays, and Ct-values adjusted to Assay III (Cta).
2.7. Data analysis
For correlation analysis, a spearman's rank correlation was used. For comparing β-globin-gene concentrations, a Mann-Whitney test was performed. To assess statistical differences in Ct-values comparing pooling methods or PCR assays, a multiple comparison one-way ANOVA was used. For comparing preparation times and for matched-pair analysis, a paired t-test was used. The amplification factor was calculated as published [10], Kruskal-Wallis test was performed. GraphPadPrism 7.0 (GraphPad Software, Inc.) was used for analysis. Figures were created using Adobe Illustrator 18.1 (Adobe Inc.).
3. Results
3.1. Hierarchical pool testing
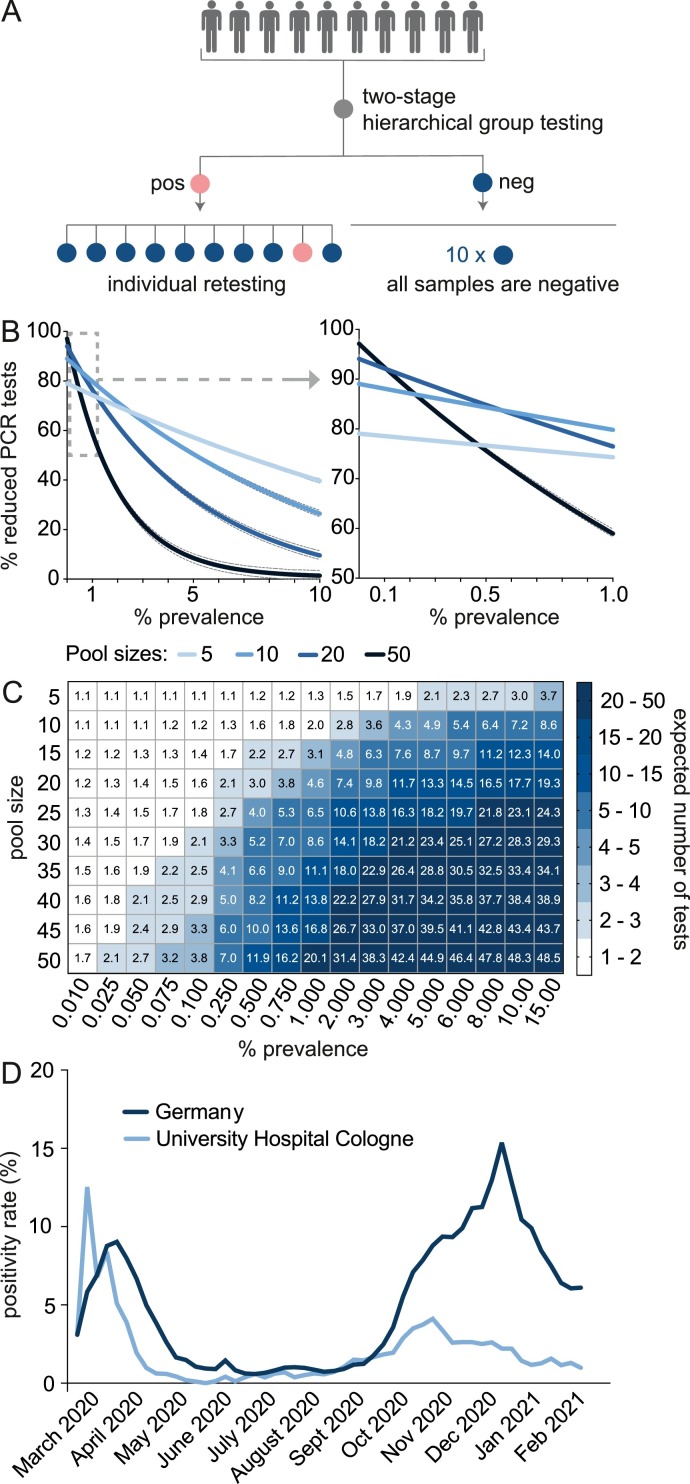
Pool testing can be performed using different strategies. In this study we conducted two-stage hierarchical pooling procedures (Fig. 1 A).
Fig. 1.
Hierarchical pool testing for SARS-CoV-2 detection. A: Illustration of the two-stage hierarchical pool testing strategy. B: The reduction of PCR-tests compared to individual testing (continuous lines are nonlinear regression curves, outer dotted lines are 95% or 99% test sensitivity, respectively) and C: The expected number of tests for different pool sizes are shown. Data visualized in Figures B and C are generated using the shiny app of Christopher Bilder, which is based on an algorithm to compute the expected number of tests when performing two-stage hierarchical pool testing [5,8]. D: The mean positivity rate per week of tests performed at the University Hospital of Cologne and in Germany (as published [11]) are shown.
Pool testing efficiency depends on the disease prevalence. Bilder and colleagues [6, 8] proposed an algorithm to compute the expected number of tests when performing two-stage hierarchical pool testing (Fig. 1B, C). As the disease prevalence increases, the reduction of PCR-tests declines due to the retesting of individual samples of positive pools. However, the pooling efficiency of smaller pool sizes declines more slowly compared to pooling of 20 or more samples.
At the time pool testing was initiated, the positivity rate at the University Hospital Cologne was 3.88% (Fig. 1D). However, by excluding samples of symptomatic individuals and recently infected persons, the positivity rate of pooled samples was below 0.1%.
3.2. Pre-analytics
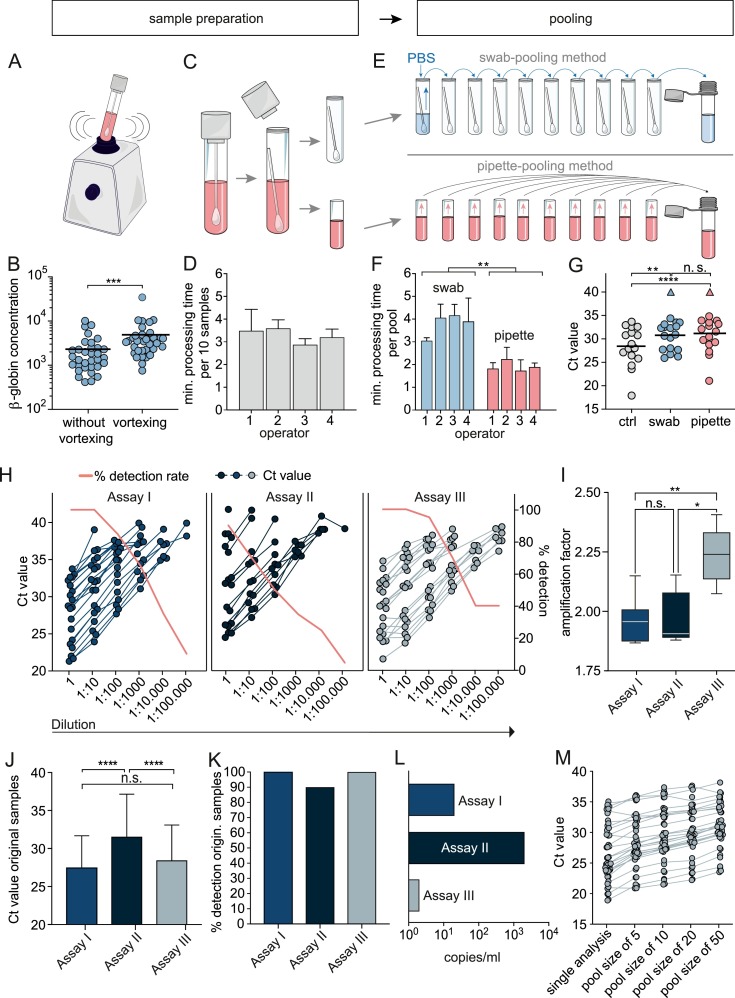
Pool testing requires optimal sample conditions in order to minimize false negative results. Pre-analytic factors can influence the test results. We could not detect a significant difference in viral loads indicated by Ct-values comparing oropharyngeal swabs (n = 39) with combined nasal/oropharyngeal swabs (n = 124). However, samples taken in the late phase of infection had significantly lower viral loads compared to earlier timepoints (n = 79) (Supplementary Figure 2A, B). Therefore, pool testing needs to be sensitive to detect even low virus concentrations. We investigated whether vortexing increases the number of cells released from the swabs (Fig. 2 A,B). We measured the β-globin-gene concentration in the medium of n = 33 swabs without vortexing and of the same specimens after 5 s of vortexing. The average increase of β-globin concentration after vortexing was 2.34-fold (95% CI 1.622–3.057; p = 0.0001). In addition, we could detect SARS-CoV-2 in three specimens in which vortexing reduced the Ct-value from 33.39 to 32.79, from 28.50 to 28.19 and from 35.58 to 32.87, respectively.
Fig. 2.
Validation of the pooling method and determining PCR sensitivity. A and B: β-globin concentration in individual specimens before and after vortexing (n = 33). A Mann-Whitney test was performed. C and D: Sample preparation time was measured for four different operators preparing n = 10 samples in 6 replicates. E and F: Swab-pooling and pipette-pooling are illustrated, and processing time was measured for four operators preparing n = 6 pools with a size of 10 each (paired t-test was performed). G: Ct-values are displayed for n = 16 single positive specimens (ctrl) as well as for each positive specimen in a pool prepared either by the pipette or swab pooling method, respectively, and tested in assay I. Negative test results are highlighted by the triangle shape. H: Ten-fold dilution series of n = 20 SARS-CoV-2-positive samples, tested with three PCR assays. I: The amplification factor was calculated for dilution series containing five Ct-values. J: The mean and standard deviation of Ct-values and K: Detection rate of n = 20 undiluted samples are shown. L: Lowest detectable SARS-CoV-2 copy number as determined using dilution series of cell culture supernatant extrapolated to approved standards measured in all assays. M: Ct-values of n = 25 positive samples combined with a stock of negative specimens in a 1:5, 1:10, 1:20 and 1:50 dilution, respectively, tested in assay III. Ctrl: control, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
3.3. Validation of two different pooling methods
To test feasibility of the pipette- and swab-pooling method, four operators conducted sample preparation (Fig 2C,D) and processed n = 6 pools applying both methods, respectively (Fig. 2E,F). The mean processing time was 3 min, 47 s for a swab-based pool (95% CI: (2 min,59 s)–(4 min,36 s)) and 1 min, 55 s for a pipette-based pool (95% CI: (1 min,33 s)–(2 min,16 s)).
To investigate the sensitivity of the different pooling methods, we generated 16 different pools with each of the two pooling methods, by spiking one SARS-CoV-2-positive sample into nine negative samples, respectively (Fig. 2G). The mean Ct of individually tested samples was 28.41 (95% CI 26.17–30.66), 30.77 for the swab pooling method (95% CI 28.79–32.75), and 31.18 for the pipette pooling method (95% CI 28.92–33.44). There was no significant difference between Ct-values of the two pooling methods. With both methods there was a single pool, which yielded a negative test result (triangle shape in Fig. 2G).
3.4. Sensitivity of three PCR assays
To compare the detection rates of three PCR systems , ten-fold dilution series of n = 20 SARS-CoV-2-positive samples were simultaneously tested in three assays (I, II, and III, referring to the Roche LightCycler® 480II, the Hologic Panther Fusion®, and Roche Cobas®6800 System). Ct-values for e-gene amplification were analyzed as they yielded similar Ct-values compared to the second viral target, respectively (Supplementary Figure 1C). As shown in Fig. 2H and K, assay I and III could detect all undiluted samples whereas assay II only detected 18 out of 20 samples. The mean Ct-values of the undiluted samples were 27.48 (95% CI 25.4–29.57) for assay I, 31.55 (95% CI 28.77–34.33) for assay II and 28.44 (95% CI 26.13–30.75) for assay III (Fig. 2J). When diluting the samples 10-fold, assays I and III could still detect all samples, whereas the detection rate of assay II was 70%. Assay III could still detect 95% of samples at a 1:100 dilution and showed a lower decline of the detection rate compared to assays I and II. The amplification factors for the three assays were 1.957 (CI 1.867–2.149), 1.906 (CI 1.879–2.153) and 2.240 (CI 2.074–2.407), respectively (Fig. 2I). The lowest detectable copy number was 200 copies per reaction for assay I, 2000 for assay II, and 20 copies for assay III as determined using two approved standards (Fig. 2L).
To determine the detection-rate for different pool sizes, 25 positive samples with Ct-values ranging from 18.96 to 34.99 were each diluted in a stock of negative specimens and tested in duplicates in assay III. All pools were SARS-CoV-2-positive (Fig. 2M), however, for two pools the 1:20 and 1:50 dilution resulted in one negative and one positive replicate, respectively.
3.5. Integration of pool testing into the diagnostic routine of SARS-CoV-2 detection
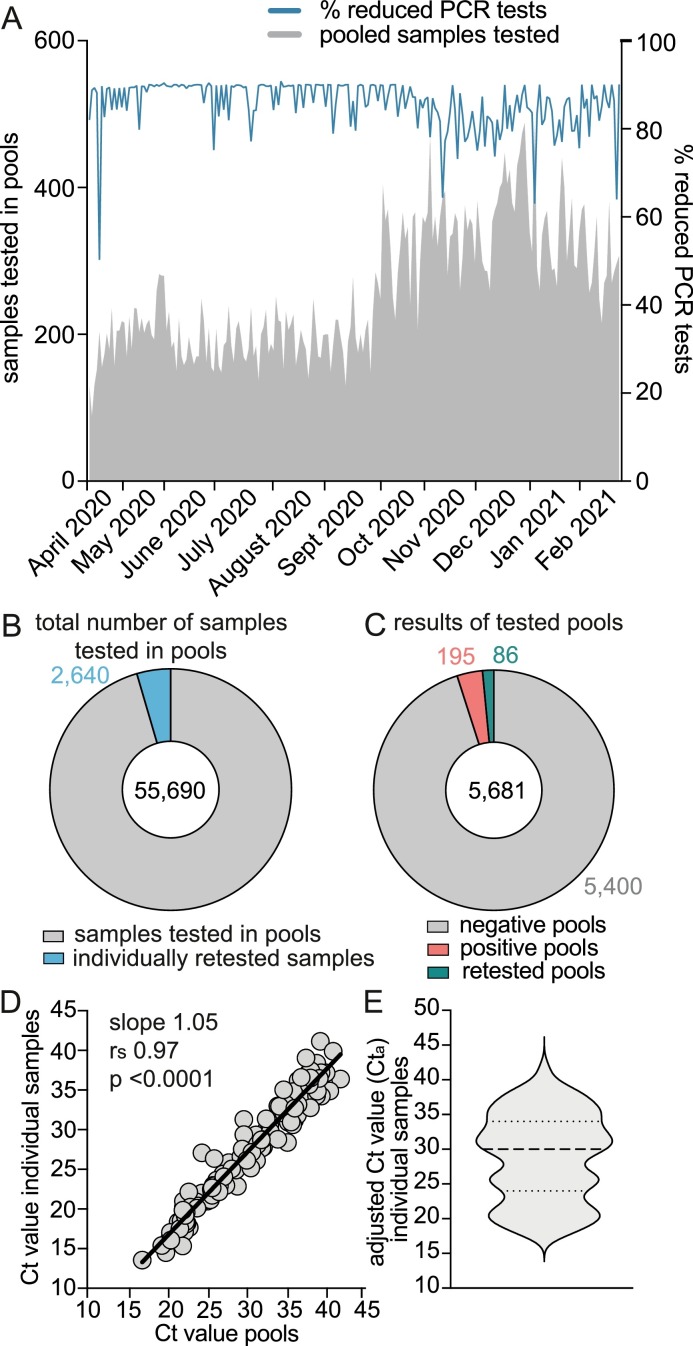
The above experiments suggested the following as the optimal pool test conditions for SARS-CoV-2 detection: (a) pooling of 10 samples using the two-stage hierarchical strategy; (b) vortexing the swab specimens before pooling; (c) applying the pipette-pooling method, and (d) utilizing assay III for PCR-testing. We set up a pool testing facility, implemented features for pool testing into the laboratory software, and systematically pooled up to 488 samples per day (Fig. 3 A). In order to limit the number of positive samples run in pools, we preselected samples supported by algorithms of the laboratory software. Patients that had been tested positive for SARS-CoV-2 before or showed COVID-19-like symptoms were tested individually. Pool testing was performed for internal surveillance screenings of patients and staff at the University Hospital Cologne.
Fig. 3.
Performance of high-throughput pool testing for SARS-CoV-2 detection. A: Pool testing started on April 9, 2020. The number of pooled samples per day and the percentage of reduced PCR-tests compared to individual testing (blue line) are displayed. B: The number of samples tested in pools and C: The number of pools tested during a period of 44 weeks are shown. D: Correlation of Ct-values of n = 128 positive pools and the respective individual positive sample. Correlation was performed only if a pool and the respective positive individual sample was analyzed with the same assay. E: Violin plot of adjusted Ct-values (Cta) of n = 175 individual positive samples detected in pools. The reduced number of data points is due to a software problem, so that some data could not be retrieved. Dotted lines represent quartiles and the dashed line the median.
The mean percentage of reduced PCR-tests was 85.77%. Decreased savings of PCR reactions were due to retesting caused by technical issues or positive tested pools. Within 44 weeks, 55,690 samples were tested in pools and only 4.7% (n = 2640 samples) had to be retested individually (Fig. 3B). As Fig. 3C shows, 5,681 pools were analyzed from which 195 were positive. We mostly detected one, rarely two positive samples in the same positive pool. Another 86 pools were retested due to technical issues. In total, 47,369 PCR reactions were saved by pool testing. The Ct-values of 128 positive pools and the respective individual positive sample strongly correlated (rs = 0.97, CI 0.96–0.98, p < 0.0001) with a mean Ct-value of 30.39 for pools and 27.38 for individual samples (Figs. 3D). Ct-values of individual positive samples were adjusted to assay III (Figs. 3E). 82.86% of the samples displayed Ct-values ≤35 and 17.14% Ct-values>35.
4. Discussion
Large-scale testing and surveillance screenings enable the rapid detection of clusters of infections and help preventing superspreading events and uncontrolled transmission of the virus until immunity by vaccination is reached. However, test capacities are limited and PCR-tests are cost-intensive. Pool testing is a feasible option to enable high-throughput screenings without overwhelming capacities of diagnostic laboratories. To our knowledge, this is the first systematic investigation addressing various aspects of pool testing for SARS-CoV-2 detection. However, reports and a review on this topic have recently been published [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27].
Eberhardt and colleagues suggest forming subgroups if a pool yields a positive result [15]. The samples of the positive subgroup are then tested individually. This strategy can further increase efficiency, as fewer tests need to be performed, but it also requires additional hands-on-time and a sensitive, rapid detection method to ensure that the test result for the individual specimen is not delayed.
Pooling strategies need to be time efficient to rapidly detect SARS-CoV-2-infected individuals as well as suitable for high-throughput screenings. Following these considerations, we decided to use two-stage hierarchical pool testing. In a low prevalence setting with restricted test resources and a neglectable time aspect, increasing pool sizes and forming subgroups is a reasonable option. Another approach is the combinatorial pool testing strategy [19, 20, 28]. Here, samples are assigned into multiple pools which enables the detection of infected individuals in a single round of testing.
Pre-analytical handling can substantially impact test sensitivity, however, limited data on this topic are available. Test results are influenced by improper transport conditions, variations of the sampling device (flocked vs. cotton swabs), the transport media [29, 30] as well as various specimen collection procedures. We could not observe differences in Ct-values comparing oropharyngeal and combined nasal/oropharyngeal specimens. This is in line with findings by Wölfel et al., describing no differences in viral loads or detection rates when comparing nasopharyngeal and oropharyngeal specimens [31]. In addition, SARS-CoV-2 can be detected in saliva, which has also been used for pool testing [24, 32] [33].
Pre-analytics can potentially influence the inhibition rate which in our setting seems to be a rather rare multifactorial event, possibly due to high quality sampling material, trained staff etc. Data on inhibition rates are of great interest but are exceeding the focus of this study.
Compared to individual testing, pool testing requires additional processing as well as documentation steps. By integrating a special feature for pool testing into our laboratory software, we were able to reduce additional hands-on-time. The already labelled tubes are scanned and assigned to a pool. If a pool tests positive, a request for re-testing of the individual samples is automatically generated by the lab software. The individual samples are stored according to the pool numbering, so that samples for re-testing can easily be identified. However, in order to save personnel capacities, it might be advisable to use a pipette robot, performing both pooling and documentation.
A critical point in the context of pool testing is the time aspect. To ensure, that the test result of the individual sample is available and communicated on the same day, we have set a time limit up to which we perform pool testing. Samples arriving later that day are tested individually.
Pooling of individual samples using the swab-method is more time-consuming and has additional limitations regarding handling and risk of contamination compared to the pipette-method. However, recent developments in PCR diagnostics allow a rapid detection of SARS-CoV-2 in swab specimens [34], and swab-pooling is valuable if pools are prepared directly after collection in schools, old people's homes, hospitals, etc.
We developed a feasible pooling procedure that can readily be implemented in diagnostic routines. The data communicated here will contribute to the process of finding a consensus pool testing strategy enabling larger test capacities to effectively combat the SARS-CoV-2 pandemic.
5. Funding
This work was supported by the German Federal Ministry of Education and Research „NaFoUniMedCovid19“—COVIM [01KX2021].
6. Authors’ contributions
Planned and conducted experiments (M.W., D.A., E.H., D.E., J.K., G.H.R., I.F., M.S., S.S., M.H., A.S.S.); conceptualised the laboratory work (M.W., E.H., S.S., F.K., E.K., R.K.); performed/helped with data analysis and interpretation (M.W., H.G., M.A., S.S., F.K.); wrote the manuscript draft (M.W.); provided clinical data, conceptualised sample collection (F.D., M.A., C.L.); made substantial revisions to the article drafts for important intellectual content (E.H., H.G., S.S., F.K.); gave final approval for publication (F.K.); conceived and designed the overall study (F.K.); contributed equally to this article (F.K., S.S.). All authors read and approved the final manuscript.
Declaration of Competing Interest
We declare no competing interests.
Acknowledgments
We would like to thank all members of the Institute of Virology and our colleagues of the Department I of Internal Medicine, University Hospital Cologne and the Institute of Medical Statistics and Computational Biology, University of Cologne for supporting our work.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2021.105018.
Appendix. Supplementary materials
References
- 1.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert M., Dewatripont M., Muraille E., Platteau J.-.P., Goldman M. Preparing for a responsible lockdown exit strategy. Nat. Med. 2020;26:643–644. doi: 10.1038/s41591-020-0871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dehning J., Zierenberg J., Spitzner F.P., Wibral M., Neto J.P., Wilczek M., et al. Inferring change points in the spread of COVID-19 reveals the effectiveness of interventions. Science. 2020 doi: 10.1126/science.abb9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van T.T., Miller J., Warshauer D.M., Reisdorf E., Jernigan D., Humes R., et al. Pooling nasopharyngeal/throat swab specimens to increase testing capacity for influenza viruses by PCR. J. Clin. Microbiol. 2012;50:891–896. doi: 10.1128/JCM.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilder C.R., Iwen P.C., Abdalhamid B. Pool size selection when testing for SARS-CoV-2. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilder C.R., Iwen P.C., Abdalhamid B., Tebbs J.M., McMahan C.S. Tests in short supply? Try group testing. Significance (Oxford, England) 2020;17:15. doi: 10.1111/1740-9713.01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black M.S., Bilder C.R., Tebbs J.M. Optimal retesting configurations for hierarchical group testing. J. R. Stat. Soc. Ser. C Appl. Stat. 2015;64:693. doi: 10.1111/rssc.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilder C.R., A Shiny app for pooled testing. (https://bilder.shinyapps.io/PooledTesting/), accessed 26 June 2020.
- 9.van Duin M., Snijders P.J., Schrijnemakers H.F., Voorhorst F.J., Rozendaal L., Nobbenhuis M.A., et al. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int. J. Cancer. 2002;98:590–595. doi: 10.1002/ijc.10232. [DOI] [PubMed] [Google Scholar]
- 10.Ruijter J., Ramakers C., Hoogaars W., Karlen Y., Bakker O., Van den Hoff M., et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic. Acids. Res. 2009;37 doi: 10.1093/nar/gkp045. e45-e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robert-Koch-Institute, Coronavirus disease 2019 (COVID-19). Daily situation report of the Robert Koch Institute, updated status for Germany 24 June 2020. (https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Situationsberichte/2020-06-24-de.pdf?__blob=publicationFile), accessed 15 July 2020.
- 12.Lohse S., Pfuhl T., Berkó-Göttel B., Rissland J., Geißler T., Gärtner B., et al. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogan C.A., Sahoo M.K., Pinsky B.A. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA. 2020;323:1967–1969. doi: 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra B., Behera B., Mohanty M., Ravindra A., Ranjan J. Challenges and issues of SARS-CoV-2 pool testing. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberhardt J.N., Breuckmann N.P., Eberhardt C.S. Challenges and issues of SARS-CoV-2 pool testing. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30467-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J., Kim S.Y., Sung H., Lee S.W., Lee H., Roh K.H., et al. Challenges and issues of SARS-CoV-2 pool testing. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30516-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Ami R., Klochendler A., Seidel M., Sido T., Gurel-Gurevich O., Yassour M., et al. Large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yelin I., Aharony N., Shaer Tamar E., Argoetti A., Messer E., Berenbaum D., et al. Evaluation of COVID-19 RT-qPCR test in multi-sample pools. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleary B., Hay J.A., Blumenstiel B., Gabriel S., Regev A., Mina M.J. Efficient prevalence estimation and infected sample identification with group testing for SARS-CoV-2. medRxiv. 2020 [Google Scholar]
- 20.Shental N., Levy S., Wuvshet V., Skorniakov S., Shalem B., Ottolenghi A., et al. Efficient high-throughput SARS-CoV-2 testing to detect asymptomatic carriers. Sci. Adv. 2020:eabc5961. doi: 10.1126/sciadv.abc5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garg J., Singh V., Pandey P., Verma A., Sen M., Das A., et al. Evaluation of sample pooling for diagnosis of COVID-19 by Real time PCR- A resource saving combat strategy. J. Med. Virol. 2020 doi: 10.1002/jmv.26475. [DOI] [PubMed] [Google Scholar]
- 22.Clark A.E., Lee F.M. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) screening with specimen pools: time to swim, or too deep for comfort? Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulu A., Alemayehu D.H., Alemu F., Tefera D.A., Wolde S., Aseffa G., et al. Evaluation of sample pooling for screening of SARS CoV-2. PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0247767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambrosis N., Martin Aispuro P., Belhart K., Bottero D., Crisp R.L., Dansey M.V., et al. Active surveillance of asymptomatic, presymptomatic, and oligosymptomatic SARS-CoV-2-infected individuals in communities inhabiting closed or semi-closed institutions. Front. Med. (Lausanne) 2021;8 doi: 10.3389/fmed.2021.640688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bish D.R., Bish E.K., El-Hajj H., Aprahamian H. A robust pooled testing approach to expand COVID-19 screening capacity. PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0246285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawicki R., Korona-Glowniak I., Boguszewska A., Stec A., Polz-Dacewicz M. Sample pooling as a strategy for community monitoring for SARS-CoV-2. Sci. Rep. 2021;11:3122. doi: 10.1038/s41598-021-82765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deka S., Kalita D. Effectiveness of sample pooling strategies for SARS-CoV-2 mass screening by RT-PCR: a scoping review. J. Lab. Physicians. 2020;12:212–218. doi: 10.1055/s-0040-1721159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleary B., Hay J.A., Blumenstiel B., Harden M., Cipicchio M., Bezney J., et al. Using viral load and epidemic dynamics to optimize pooled testing in resource-constrained settings. Sci. Transl. Med. 2021 doi: 10.1126/scitranslmed.abf1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daley P., Castriciano S., Chernesky M., Smieja M. Comparison of flocked and rayon swabs for collection of respiratory epithelial cells from uninfected volunteers and symptomatic patients. J. Clin. Microbiol. 2006;44:2265–2267. doi: 10.1128/JCM.02055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore C., Corden S., Sinha J., Jones R. Dry cotton or flocked respiratory swabs as a simple collection technique for the molecular detection of respiratory viruses using real-time NASBA. J. Virol. Methods. 2008;153:84–89. doi: 10.1016/j.jviromet.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 32.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watkins A.E., Fenichel E.P., Weinberger D.M., Vogels C.B., Brackney D.E., Casanovas-Massana A., et al. Pooling saliva to increase SARS-CoV-2 testing capacity. MedRxiv. 2020 doi: 10.3201/eid2704.204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandrasekaran A.R., Zhou L., Halvorsen K. Rapid one-step detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020;4:1123–1124. doi: 10.1038/s41551-020-00663-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.