Abstract
The cell cycle inhibitor p15INK4b is frequently inactivated by homozygous deletion together with p16INK4a and p19ARF in some types of tumors. Although the tumor suppressor capability of p15INK4b is still questioned, it has been found to be specifically inactivated by hypermethylation in hematopoietic malignancies in the absence of p16INK4a alterations. Here we show that, in vitro, p15INK4b is a strong inhibitor of cellular transformation by Ras. Surprisingly, p15INK4b is induced in cultured cells by oncogenic Ras to an extent similar to that of p16INK4a, and their expression is associated with premature G1 arrest and senescence. Ras-dependent induction of these two INK4 genes is mediated mainly by the Raf-Mek-Erk pathway. Studies with activated and dominant negative forms of Ras effectors indicate that the Raf-Mek-Erk pathway is essential for induction of both the p15INK4b and p16INK4a promoters, although other Ras effector pathways can collaborate, giving rise to a stronger response. Our results indicate that p15INK4b, by itself, is able to stop cell transformation by Ras and other oncogenes such as Rgr (a new oncogene member of the Ral-GDS family, whose action is mediated through Ras). In fact, embryonic fibroblasts isolated from p15INK4b knockout mice are susceptible to transformation by the Ras or Rgr oncogene whereas wild-type embryonic fibroblasts are not. Similarly, p15INK4b-deficient mouse embryo fibroblasts are more sensitive than wild-type cells to transformation by a combination of the Rgr and E1A oncogenes. The cell cycle inhibitor p15INK4b is therefore involved, at least in some cell types, in the tumor suppressor activity triggered after inappropriate oncogenic Ras activation in the cell.
The transforming activity of oncogenes has been extensively studied in the last 2 decades. Early on, transformation of primary cells was observed to require two cooperating oncogenes to convert normal cells to a tumorigenic state (27, 59). The resistance of primary cells to transformation by single oncogenes can now be explained as the effect of the induction of tumor suppressor genes by inappropriate oncogenic signals (reviewed in reference 73). Thus, prolonged oncogenic Ras activity produces in primary cells an increase in the levels of p16INK4a, p21Cip1, and p53, which, in turn, is dependent on the transcriptional induction of its positive regulator p19ARF (32, 48, 64). Induction of the expression of these genes by Ras is accompanied by growth arrest in the G1 phase of the cell cycle and a phenotype indistinguishable from premature senescence (33, 64). Both the p16INK4a and p19ARF proteins are expressed from a complex gene structure, the INK4a locus (for reviews, see references 13 and 67). Each of the two proteins uses a different exon 1, and both use the same exon 2, but each protein is translated in a different reading frame (53). Although their amino acid sequences are completely different, both proteins are cell cycle inhibitors. p16INK4a is a potent inhibitor of cyclin-dependent kinases 4 and 6 (Cdk4/6) (61), whereas p19ARF stabilizes the p53 tumor suppressor gene (for reviews, see references 7, 66, and 67).
In both humans and mice, the INK4a locus is located close to a second gene of the INK4 family, p15INK4b, which also functions as a Cdk4/6 inhibitor and is strongly induced by transforming growth factor β (TGF-β) (14, 22, 54). Both loci, INK4a and INK4b, are frequently deleted in a variety of tumors and cell lines (22, 58). In addition, these proteins can also be inactivated by point mutations or methylation (reviewed in references 50 and 58). The expression of proteins p16INK4a, p15INK4b, and p19ARF can be decreased by hypermethylation of the CpG island upstream of corresponding exon 1 in both humans (17, 41, 56) and rodents (36, 69). No clear tumor suppressor role has been assigned to the other two members of the INK4 family, p18INK4c and p19INK4d.
Whereas the evidence for a tumor suppressor role of p16INK4a is abundant, the role of p15INK4b in tumor suppression is more controversial. In most tumors, homozygous deletions affect both the INK4a and INK4b loci or the INK4a locus alone. In only a few cases have specific deletions of p15INK4b sequences been reported, i.e., leukemias and lymphomas, which are among the tumors with higher involvement of p15INK4b deletions (58). Point mutations, which are relatively frequent in INK4a, only rarely occur in p15INK4b (36, 50). In contrast, inactivation of p15INK4b by hypermethylation seems to be selectively frequent in leukemias and lymphomas and does occur independently of p16INK4a status (4, 17, 18, 36, 38), suggesting a tissue-specific tumor suppressor role for p15INK4b in hematopoietic malignancies. In concordance with these data, Lois et al. (34) demonstrated an inverse relationship between p15INK4b expression and proliferation of lymphocytes after mitogenic stimuli, suggesting a specific role for this gene in maintaining cell quiescence in lymphocytes.
Early studies on Ras mitogenic potential demonstrated that Ras induces and is required for DNA synthesis in serum-stimulated cells (44). Only recently have the pathways linking Ras activity with cell cycle control begun to be dissected. Ras acts on the cell cycle machinery by inactivating Cdk inhibitors such as p27Kip1 and inducing cyclins, giving rise to an increase in Cdk4/6 and Cdk2 kinase activities (for reviews, see references 11 and 35). Thus, Ras activity is linked directly to the G1/S transition of the cell cycle and, in fact, G1 is the only phase in which inhibition of Ras affects cell cycle progression. Ras is required for activation of both Cdk2 and Cdk4/6 complexes until 2 h before the G1/S transition, a time corresponding to the so-called restriction point. Once cells have entered S phase, Ras becomes dispensable until the next cell cycle (19, 44). Although Ras signals through a growing number of different effector pathways, effects on both cyclin D induction and p27Kip1 degradation seem to be dependent on the Raf1-Erk pathway. The specific activation of the Erk pathway, however, is not sufficient to trigger p27Kip1 degradation, and it seems to be involved in a RhoA-associated pathway that could require a phosphatidylinositol 3′-kinase (PI3K)-dependent but protein kinase B-independent pathway (for a review, see reference 35).
Whereas different experiments have clearly shown that p16INK4a is able to suppress cellular transformation by Ras and can contribute to cellular senescence (2, 20, 47, 62), the ability of p15INK4b to inhibit cellular transformation has not been studied. In this article, we show that the cell cycle inhibitor p15INK4b is able to produce cell cycle arrest and stop cellular transformation by Ras. Interestingly, this Cdk4/6 inhibitor is strongly induced in cultured cells by oncogenic Ras and, thus, can cooperate in causing the premature cellular senescence resulting from oncogenic signals. Using luciferase constructs carrying the p15INK4b and p16INK4a promoters, we show that the Raf-Mek-Erk pathway is the main effector pathway in the induction of the INK4 promoters although other Ras effectors cooperate in the transcriptional induction of both p15INK4b and p16INK4a in NIH 3T3 cells. Finally, using mouse embryonic fibroblasts (MEFs) from p15INK4b knockout mice, we show that the lack of p15INK4b protein is sufficient to render these fibroblasts susceptible to transformation by the Ras or Rgr oncogene.
MATERIALS AND METHODS
Plasmids and DNA manipulation.
Mouse genomic N-ras containing a codon 61 point mutation (pMZNT-17) or the wild-type sequence (pMZNN-1) was subcloned into the Zeocin-resistant vector pcDNA3.1/Zeo(+) (Invitrogen). Mouse p15INK4b cDNA was isolated by reverse transcription-PCR and subcloned into the pCR3.1 (Invitrogen) or pMAMneo (Clontech) vector, giving rise to plasmids pHM414 (cytomegalovirus [CMV] promoter orientation), pHM411 (opposite orientation), and pMAMneo-p15. The same fragment was subcloned into the pBabe-puro vector for retroviral transduction of primary cells. A 6-kb genomic fragment containing the p15INK4b and 5′ upstream sequences was amplified by long-template PCR (Expand System; Boehringer Mannheim) using primers Mp15-P-1F (5′-GGC CAA AAC AGG ATC CCT TGG GAT GTG TTA-3′) and Mp15-3′-1R (5′-TAA CCA TGG AGA TCT CTC CAG GCT CCA-3′) as described previously (37). This fragment includes about 700 bp upstream of the p15INK4b coding region and was subcloned into the bacterial plasmid pCR2.1 (Invitrogen) to generate pMM134. A different 8-kb genomic fragment containing the mouse p15INK4b gene was obtained from pmp15 (36) and subcloned into pCR3.1 in the orientation opposite to that of the CMV promoter (pCRpmp15). The integrity and orientation of the inserts were confirmed by sequencing with an automatic 373 DNA Sequencer (Applied Biosystems). p16INK4a, p27Kip1 and retinoblastoma expression plasmids were a gift from M. Pagano. Plasmids expressing wild-type, activated, or dominant negative forms of Raf (25), Mek1 (8), Erk1 and Erk2 (74), RalA (71, 76), p53 (70), or the PI3K p110 and p85 subunits (15, 55) were described previously. Ras effector domain mutants containing an activating G12V mutation (57) were used to specifically study the activity of the Raf, RalGDS, and PI3K effector pathways. The p16-luc5 plasmid contained a 1.1-kb promoter fragment of p16INK4a upstream of the luciferase reporter gene (29). The plasmids pGal4-ElkC and p5xGal-luc were for assaying transactivation of Elk-1 (74). RhoA, Rac1, Cdc42, JNK1, PKCζ, and MEKK1 expression plasmids were graciously provided by P. Crespo. The mouse RalGDS gene (1) was subcloned into the pCR3.1 (Invitrogen) vector for expression in mammalian cells (pCR-RalGDS).
For hybridizations, digested DNA or RNA was separated on agarose gels and transferred to nitrocellulose membranes (Schleicher & Schuell). DNA probes were labeled with [α-32P]dCTP (3,000 Ci/mmol; Dupont-NEN, Boston, Mass.) using a random primed labeling kit (Boehringer Mannheim, Indianapolis, Ind.) in accordance with the manufacturer's protocol. Hybridizations were visualized and quantified by use of a PhosphorImager (Molecular Dynamics) or by exposure of X-ray films (Kodak), digital scanning, and analysis with the NIH Image software.
Cell culture, transfection, and retroviral infection assays.
All cultures were maintained in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 10% calf serum and 1% penicillin G–streptomycin sulfate (Gibco). Stable transfection of mammalian cells was performed using the calcium phosphate technique, and G418 (Gibco) at 400 μg/ml or Zeocin (Invitrogen) at 500 μg/ml was used for selection of clones. Expression of recombinant proteins in pooled transfected cells or in individual clones was analyzed by Western blotting. Inducible expression of p15INK4b in NIH 3T3 clones containing pMAMneo-p15 was achieved after addition of 1 μM dexamethasone dissolved in ethanol. Uninduced cells were treated with an equivalent amount of ethanol. For the focus assay, cells were cotransfected with 1 μg of pMZNT-17 (Ras) or pNM11 (a pMEXneo-derivative plasmid expressing the rabbit Rgr oncogene [9]) and 3 μg of other expression plasmids or empty vectors. After transfection, cells were split 1:5 and maintained in DMEM with 5% calf serum for 2 weeks. Foci were scored after staining with cresyl violet.
For soft-agar assays, cells were resuspended in 0.33% agar in DMEM supplemented with 10% calf serum at a density of 5,000/65-mm-diameter plate and seeded onto solidified 0.5% agar-containing culture medium. Cultures were fed weekly, and photomicrographs of colonies were taken 2 weeks postplating.
MEFs were isolated from C57BL/6J × DBA2 mice using E14.5 embryos. Embryonic tissues were disaggregated with trypsin, and primary fibroblasts were cultured at intermediate densities in DMEM with 10% calf serum. p15INK4b-deficient MEFs and those of wild-type littermates were kindly provided by E. Latres and M. Barbacid (unpublished data). For the transformation assays, 106 cells were plated and transfected with 10 μg of a Ras (pMZNT-17), Rgr (pNM11), or E1A (a gift of M. Serrano) expression plasmid. When needed, transfection mixtures were supplemented with up to 20 μg of mouse genomic DNA as carrier DNA. After transfection, MEFs were maintained in DMEM plus 10% fetal bovine serum and visible foci (>2 mm in diameter) were scored after 2 weeks of culture and cresyl violet staining. Infection of three-passage MEFs with a pZIPneoSV(X)-derivative retrovirus carrying the oncogenic N-ras cDNA (pZip4) was performed essentially as previously described (39) using clone Ψ2A2. Mouse p15INK4b and N-ras N61 cDNAs were cloned in the pBabe-puro retroviral vector. One day before the infection, MEFs were plated at 8 × 105/10-cm dish. Infections were performed by replacing the medium with the virus-containing supernatant which had previously been filtered (0.45-μm-pore-size filter; Millipore) and supplemented with Polybrene (Sigma) at 4 μg/ml. Infected cell populations were selected for 4 days using puromycin at 2 μg/ml. At 1 and 3 days after selection, the cells were incubated for 5 h with 10 μM bromodeoxyuridine (BrdU) (Boehringer Mannheim) and BrdU incorporation was analyzed by flow cytometry (see below).
The activity of senescence-associated β-galactosidase (SA-βGal) was detected by following the original protocol described by Dimri et al. (10). About 100,000 MEFs were infected with the retrovirus carrying p15INK4b, oncogenic Ras, or the empty vector, and SA-βGal activity was assayed 7 days postinfection.
Protein expression.
Western blotting was used to assess the level of p15INK4b in cultured cells. Samples were homogenized with a Tissumizer in ice-cold, freshly prepared lysis buffer (1% NP-40, 20 mM HEPES [pH 7.5], 5 mM MgCl2, aprotinin at 10 μg/ml, leupeptin at 2 μg/ml, pepstatin A at 1 μg/ml, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol) and spun at 100,000 × g for 45 min at 4°C. The amount of protein in the supernatants was quantified by the Bradford method using bovine serum albumin as the standard. Aliquots containing 50 μg of protein per sample were subjected to sodium dodecyl sulfate–10 to 18% polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes, which were blocked with 5% nonfat dried milk in TBST buffer (0.1% Tween 20, 132 mM NaCl, 20 mM Tris [pH 7.5]) for 12 h. The polyclonal antibody that recognizes the murine p15INK4b protein was kindly provided by E. Latres and M. Barbacid. A commercial antibody was used to detect the Erk proteins (C-16; Santa Cruz Biotechnologies) as a control for protein loading. Peroxidase signal was detected by the enhanced-chemiluminescence method (Amersham). Protein band intensity after different exposure times was quantified using a Umax PowerLook scanner and the NIH Image software.
Flow cytometry.
The cell cycle status of mammalian cells was analyzed by propidium iodide staining of DNA. For double staining of DNA content and BrdU, cells were pulsed with 10 μM BrdU, washed, and fixed in 70% ethanol. After a 30-min treatment with 2 N HCl and pepsin at 0.2 mg/ml, the pH was stabilized with 0.1 M sodium tetraborate (pH 8.5) and the cells were preincubated in 0.5% Tween 20–2% normal mouse serum in phosphate-buffered saline. Incorporated BrdU was detected with a fluorescein isothiocyanate-conjugated anti-BrdU antibody (Boehringer Mannheim) and resuspended in a solution containing propidium iodide at 50 μg/ml and DNase-free RNase at 0.5 mg/ml in phosphate-buffered saline. Fluorescence was analyzed with a FACScan cytometer, and data were interpreted using the CellQuest and ModFitLT applications (Becton Dickinson).
Luciferase assays.
The pLUC+ luciferase reporter plasmid used in this work and its derivatives were described previously (3). Mouse p15INK4b genomic sequences upstream of the coding region were obtained from plasmid pmp15 (36). For deletion mapping of the murine p15INK4b promoter, different fragments carrying p15INK4b upstream sequences were amplified by PCR and subcloned into pLUC+ digested with XhoI and HindIII. The following oligonucleotides that incorporate a HindIII (forward primers) or XhoI (reverse primers) recognition site were used for priming: 1F (5′-AGG GGA AGC TTG TAA AGA CAG GCC-3′), 2F (5′-TGC GCA AGC TTC TAA GAT CTT CCG AC-3′), 3F (5′-AAC AAG CTT GGG GGA GGG GTT AG-3′), 6F (5′-GCT AAG CTT CTG CGG GCT CCC C-3′), 1R (5′-CAG AAC TCG AGG TTT CCT AGT CTG GAA C-3′), and 2R (5′-CCC CTC TCG AGA CCC AGT AGC TTC GG-3′). DNA fragments were digested with HindIII and XhoI and subcloned into the pLUC+ vector to generate p15-1F1R-luc, p15-2F1R-luc, and so on. The identities and orientations of the new constructs were confirmed by DNA sequencing. About 500 ng of the pLUC+ constructs, 100 ng of β-galactosidase plasmid pCH110 (Pharmacia) or pQP-CH110 (a pCH110 derivative in which the CMV promoter has been replaced with the Q fragment of the mouse N-ras promoter [21]), and 3 μg of the expression plasmids described above were used to cotransfect different cell lines. When several expression plasmids were used in one transfection, the total amount of the mixture was 3 μg. Transient transfections were performed by the calcium phosphate method (NIH 3T3 and HaCaT cells and MEFs) or Lipofectin (A431 cells). Cells were plated onto six-well plates at a density of 100,000 per well, grown for 24 h, and transfected. Forty hours after transfection, cells were collected and lysed using the buffer provided in the Luciferase Assay System (Promega). In some cases, TGF-β1 (Boehringer Mannheim) was added to transfected HaCaT cells to a concentration of 100 pM and luciferase activity was measured 20 h later, following the timing described by Li et al. (28). Luciferase was assayed in accordance with the manufacturer's recommendations (Luciferase Assay System; Promega Corp.). The same protein extracts were used then to measure the chemiluminescence produced by β-galactosidase using the Galacto-Light Plus system (Tropix). All of the luciferase experiments were performed at least in triplicate.
RESULTS
p15INK4b inhibits cellular transformation by Ras.
NIH 3T3 fibroblasts, similar to many other immortal cell lines, lack the INK4a and INK4b loci due to homozygous deletion of their chromosomal region (31, 54). In order to analyze the effect of the reintroduction of p15INK4b, NIH 3T3 cells were stably transfected with pMAMneo-p15, where p15INK4b expression is controlled by the dexamethasone-responsive mouse mammary tumor virus promoter. Addition of 1 μM dexamethasone produces enforced expression of p15INK4b compared to cells grown in the presence of the solvent (ethanol). The effect of p15INK4b induction was investigated by growth curve analysis of transfected clones and double staining for BrdU incorporation and DNA content. NIH 3T3 cells expressing p15INK4b grew more slowly than noninduced cells or nontransfected NIH 3T3 cells (Fig. 1).
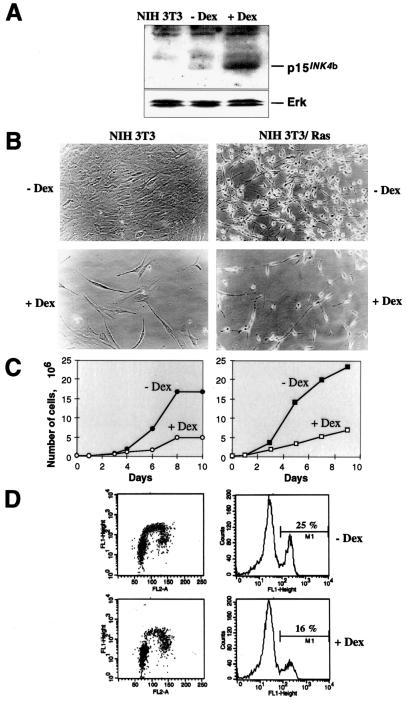
FIG. 1.
Cell cycle-inhibitory activity of p15INK4b in Ras-transformed or untransformed NIH 3T3 cells. (A) Western blot of NIH 3T3 cell lysates. Control NIH 3T3 cells have no p15INK4b protein, whereas fibroblasts containing pMAMneo-p15 express the protein at low levels without dexamethasone treatment (− Dex). Twenty hours after the addition of dexamethasone (+ Dex), the levels of p15INK4b are strongly increased. Erk proteins were detected as loading controls. (B) Photomicrographs of untransformed or Ras-transformed NIH 3T3/pMAMneop-15 fibroblasts. About 100,000 cells were seeded and grown in DMEM–10% calf serum without or with dexamethasone. Pictures were taken after 7 days of culture. (C) Growth curve of the same cultures. (D) Double staining for BrdU incorporation and DNA content using a flow cytometer. Ras-transformed NIH 3T3 cells containing plasmid pMAMneo-p15 were grown in the absence or presence of dexamethasone for 24 h, pulsed with BrdU for 2 h, and fixed in 70% ethanol. Incorporation of BrdU was measured in a FACScan cytometer (FL1-Height) and plotted against propidium iodide staining (FL2-A) (left panels). BrdU-positive cells (M1 marker in the right panels) decreased from 25 to 16% after p15INK4b induction.
The growth-inhibitory effect of p15INK4b expression was maintained even in Ras-transformed cells. NIH 3T3 cells were transfected with pMZNT-17, containing the activated N-ras gene in a Zeocin-resistant vector. Transformed clones that expressed the Ras oncogene were selected with Zeocin. Clones expressing the N-ras oncogene and showing transformed morphology were chosen and transfected with pMAMneo-p15. Zeocin- and G418-resistant pooled cells were treated with dexamethasone or left untreated. Induction of p15INK4b by dexamethasone was able to provoke cell cycle arrest in the presence of activated Ras, as in untransformed NIH 3T3 cells. Ras-transformed NIH 3T3 cells grew more slowly, and the percentage of BrdU-incorporating cells decreased from 25 to 16% when p15INK4b was induced. However, the morphology of Ras-transformed NIH 3T3 cells remained unchanged in the presence of p15INK4b. These cells maintained the typical morphology of Ras-transformed cells, being small and highly refractile and presenting long, thin cellular prolongations (Fig. 1).
In the focus formation assay, p15INK4b was also able to inhibit the formation of foci by Ras to an extent similar to that of the p16INK4a or p27Kip1 protein (Fig. 2A and B). Only about 30% of the foci produced by oncogenic Ras were scored when Ras was cotransfected with pHM414, a G418-resistance plasmid in which p15INK4b is driven by the CMV promoter. The reduction in the number of foci was similar to that found when Ras was cotransfected with the p16INK4a or p27Kip1 expression plasmid but not when p15INK4b was placed in the orientation opposite to that of the CMV promoter (pHM411). Individual clones resistant to Zeocin and G418 were selected, and Ras and p15INK4b expression was determined by Western blotting. Some of these clones presented a typical Ras-transformed morphology even in the presence of the p15INK4b protein. However, clones expressing both oncogenic Ras and p15INK4b grew more slowly than the Ras-positive, p15INK4b-deficient clones (data not shown). In addition, the presence of p15INK4b decreased the number and size of colonies formed in soft agar by Ras-transformed NIH 3T3 cells (Fig. 2C).
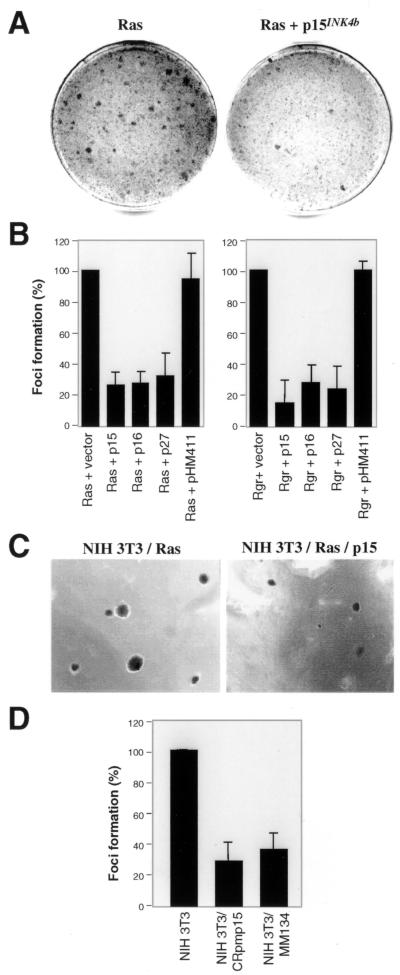
FIG. 2.
Effects of several cell cycle inhibitors on NIH 3T3 cellular transformation by Ras. (A) Photograph of foci obtained after cotransfection of NIH 3T3 cells with pMZNT-17 (expressing oncogenic N-ras N61) and the empty vector pCR3.1 (left) or pMZNT-17 and pHM414 (expressing p15INK4b) (right). (B) Suppression activity of p15INK4b and other cell cycle inhibitors (p16INK4a and p27Kip1) on focus formation by Ras (left panel) or Rgr (right panel) in NIH 3T3 cells. Cells were cotransfected with pMZNT-17 (Ras) or pNM11 (Rgr) and the expression plasmid for p15INK4b, p16INK4a, or p27Kip1. The empty vector or the vector containing p15INK4b in the opposite orientation (pHM411) was used as a control. Results are represented as percentages of the number of foci obtained with Ras or Rgr and the empty vector (taken as 100%). The mean and standard error were calculated from three independent experiments. (C) Effect of p15INK4b expression on anchorage-independent growth of Ras-transformed NIH 3T3 cells. Transformed cells expressing or not expressing p15INK4b were seeded in agar-containing medium and grown for 2 weeks. The presence of this cell cycle inhibitor was accompanied by a decrease in the number and size of colonies in two separate experiments. A sample of the colonies obtained in cells expressing Ras or Ras plus p15INK4b is shown. (D) Focus formation by oncogenic Ras in NIH 3T3 cells containing a p15INK4b genomic fragment. The same number of NIH 3T3, NIH 3T3/CRpmp15, or NIH 3T3/MM134 cells was transfected with pMZNT-17, and the number of foci was scored after 15 days. Results are presented as percentages of the number of foci obtained with Ras in NIH 3T3 cells (taken as 100%). The mean and standard error were calculated from three independent experiments.
p15INK4b and the other Cdk inhibitors (p16INK4a and p27Kip1) are also able to inhibit transformation by Rgr (Fig. 2B), an oncogene belonging to the Ral family of guanine nucleotide exchange factors (9). This oncogene has recently been shown to activate the Erk kinases and to induce Fos expression and cyclin D1 transcription by using pathways similar to those used by Ras (M. I. Hernández and A. Pellicer, unpublished data).
Oncogenic Ras induces p15INK4b expression and growth arrest.
Since oncogenic Ras produces transformation in NIH 3T3 cells but G1 arrest in primary fibroblasts, we analyzed the effect of oncogenic Ras on NIH 3T3 cells containing a p15INK4b genomic fragment. NIH 3T3 cells were transfected with pCRpmp15 or pMM134, containing an 8- or 6-kb genomic p15INK4b fragment, respectively, and selected with G418. Both NIH 3T3/CRpmp15 and NIH 3T3/MM134 cells are highly resistant to focus formation when transfected with the oncogenic Ras plasmid pMZNT-17, compared to parental NIH 3T3 cells in the focus formation assay (Fig. 2D). The expression of p15INK4b in NIH 3T3 cells decreased the tumorigenic potential of Ras in these cells similarly to what happens in primary cultures. Expression of the oncogenic N-ras N61 mutant in MEFs produced the same morphological changes and cell cycle arrest found previously by Serrano et al. (64) using H-ras V12. Indeed, similar results were observed when MEFs were infected with a p15INK4b-expressing retrovirus (Fig. 3A). p15INK4b-transduced cells became flat and enlarged, stained positive for SA-βGal, and grew more slowly than cells infected with the Ras retrovirus or an empty vector. BrdU incorporation and cell cycle analysis by flow cytometry revealed that this phenomenon was produced through arrest in the G1 phase of the cell cycle and a decrease in the number of cells that progressed to S phase (Fig. 3B). Thus, similar to that of Ras, retroviral expression of p15INK4b in primary fibroblasts produces a senescence morphology accompanied by an arrest in cell proliferation. These results indicate that Ras-dependent induction of p15INK4b is physiologically relevant, since only overexpression of p15INK4b is able to produce G1 arrest and a senescence phenotype in primary cells similar to that described for oncogenic Ras.
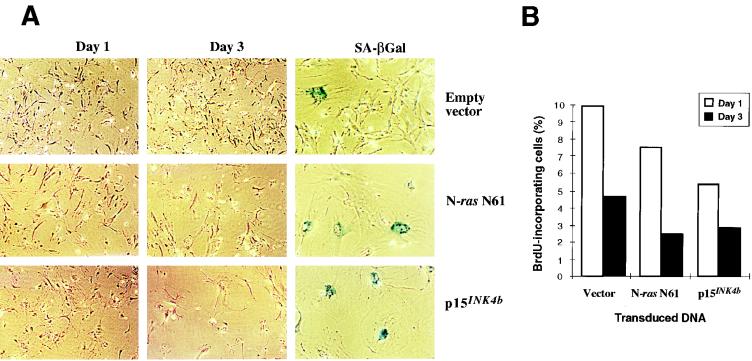
FIG. 3.
Retrovirus-mediated ectopic expression of N-ras N61 and p15INK4b in mouse primary fibroblasts. Early-passage MEFs were transduced with a retrovirus expressing p15INK4b or N-ras N61 (as a positive control for premature cell senescence) or with an empty pBabe-puro vector. Infected cells were selected with puromycin for 4 days. (A) Cell morphology at days 1 and 3 after selection with puromycin and staining for SA-βGal. SA-βGal panels were photographed at an original magnification of ×1,000 to show in detail the morphology of the β-Gal-positive cells. All of the other panels were photographed at an original magnification of ×400. (B) Levels of BrdU incorporation by these MEFs on days 1 and 3. The decrease in BrdU-incorporating cells is correlated with G1 arrest, as shown previously by others (72). These results are representative of three separate experiments.
We next measured the expression of the p15INK4b and Ras proteins in transfected NIH 3T3 cells or infected primary cultures. Surprisingly, when analyzing NIH 3T3/MM134 cells, carrying the genomic p15INK4b region, we observed strong induction of p15INK4b expression in Ras-transfected cells (Fig. 4A). A similar induction was not found when a p15INK4b cDNA driven by the CMV promoter (pHM414) was used instead of the genomic fragment, suggesting the presence of Ras-responsive sequences in the p15INK4b genomic region.
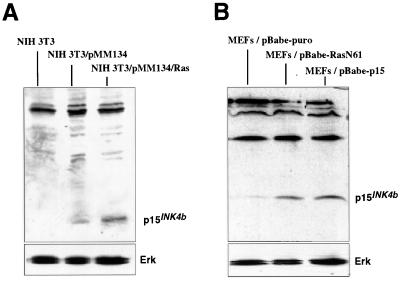
FIG. 4.
Oncogenic Ras induces the expression of p15INK4b in MEFs. (A) NIH 3T3 cells were cotransfected with pMM134, carrying p15INK4b genomic DNA with 5′-upstream sequences, and pMZNT-17, containing oncogenic N-ras. Forty-eight hours after the transfection, cell lysates were prepared and p15INK4b expression was analyzed by Western blotting. The levels of p15INK4b protein were greatly increased in cells expressing activated Ras (NIH 3T3/pMM134/Ras cells) compared to those in fibroblasts transfected with pMM134 and an empty pcDNA3.1 vector (NIH 3T3/pMM134 cells). (B) Expression of p15INK4b in MEFs infected with an oncogenic Ras retrovirus (pBabe-RasN61), a p15INK4b retrovirus (pBabe-p15), or the empty vector (pBabe-puro). Ras activity induced endogenous expression of p15INK4b at levels similar to those obtained with the p15INK4b retrovirus. As in Fig. 1, detection of the Erk proteins was used as a loading control.
We then analyzed the induction of p15INK4b in early-passage MEFs, where both p15INK4b and p16INK4a are expressed. MEFs were infected with pZip4, a retroviral vector containing the N-ras oncogene, and after 48 h, cell lysates were analyzed for protein expression. As shown in Fig. 4B, p15INK4b was overexpressed after infection of MEFs with the Ras-expressing retrovirus. This overexpression is similar to that obtained when p15INK4b was retrovirally transduced into the same cells.
Ras-dependent induction of p15INK4b is mediated by sequences located in the promoter region.
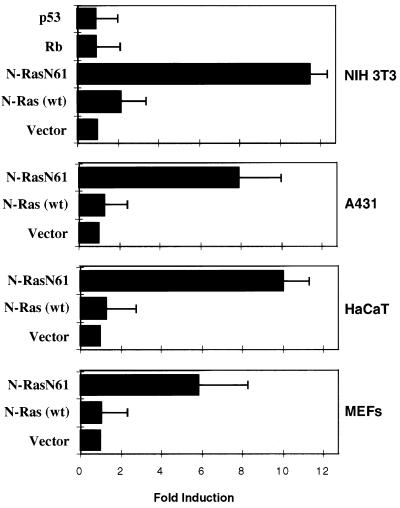
The effect of Ras activation on the p15INK4b promoter was analyzed next using luciferase reporter plasmids. Since the sequences needed for the Ras-dependent induction of p15INK4b were present in pMM134, we subcloned the 720-bp DNA fragment upstream of the p15INK4b coding region (fragment 1F2R) present in pMM134 into the pLUC+ vector. Plasmid p15-1F2R-luc was then cotransfected into NIH 3T3 cells with an expression plasmid for the N-ras proto-oncogene (pMZNN-1) or N-ras oncogene N61 (pMZNT-17), and luciferase activity was measured after 40 h. Whereas the Ras proto-oncogene had a negligible effect on the p15INK4b promoter in the assay, luciferase activity was increased 12-fold in cotransfections with the Ras oncogene (Fig. 5). This effect was independent of the Ras isoform used, and similar results were obtained with oncogenic N-RasN61, H-RasV12, and K-RasV12 constructs (data not shown). Similar induction was also found when the Rgr oncogene was used instead of Ras. However, an expression plasmid carrying the retinoblastoma (pCMV-Rb) or p53 (p11-4) gene had no effect on the p15INK4b promoter under these conditions. To analyze the effect of Ras on different cell lines, luciferase constructs were cotransfected with the Ras expression plasmids in the A431 (human squamous carcinoma cells) and HaCaT (human normal keratinocytes) cell lines and in primary MEFs (Fig. 5). In all cases, the p15INK4b promoter was induced by oncogenic Ras and this induction was similar to that obtained with p16-luc5, a reporter plasmid carrying the p16INK4a promoter (see below).
FIG. 5.
Induction of the murine p15INK4b promoter by oncogenic Ras in cultured cells. Plasmid p15-1F2R-luc, containing 720 bp of p15INK4b 5′ sequences driving a luciferase reporter gene, was cotransfected with different expression plasmids in NIH 3T3, A431, or HaCaT cells or in primary MEFs. Luciferase activities from three separate experiments with each cell line were measured. The promoter activity of p15-1F2R-luc cotransfected with an empty vector was arbitrarily chosen as 1. Cotransfection with an N-ras proto-oncogene, p53, or retinoblastoma expression plasmid had little or no effect on promoter induction. However, there was a 12-fold increase in luciferase activity when an oncogenic N-ras plasmid (pMZNT-17) was used in NIH 3T3 cells and similar results were obtained with the other cells. wt, wild type.
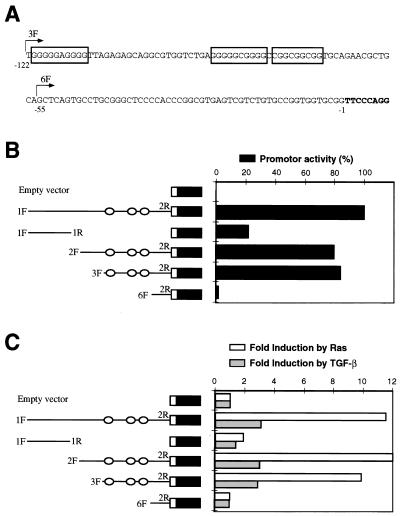
In order to identify the sequence elements involved in the response to Ras, progressive deletions of the murine p15INK4b promoter were amplified by PCR and subcloned into the pLUC+ reporter plasmid. These constructs were cotransfected with pMZNT-17 or the empty vector (pcDNA3.1), and luciferase activity was assayed (Fig. 6). Induction by Ras was obtained with all constructions carrying at least 120 bp proximal to exon 1. However, no response was found with p15-1F1R-luc, carrying a distal fragment, or p15-6F2R-luc, containing only 55 bp upstream of exon 1. Thus, some Ras response elements must be located at positions −122 to −65. Computer searches revealed the presence of three Sp1 consensus sites between these positions (Fig. 6A), in an organization similar to that of the human sequences (28). The Sp1 and Sp3 sites present in the human gene seem to be responsible for its induction by TGF-β (28). To determine whether these homologous sequences are involved in the induction by TGF-β in the murine promoter, the deletion constructs described above were transfected into HaCaT keratinocytes. Twenty hours after transfection, cells were treated with 100 pM TGF-β1 or left untreated and luciferase activity was assayed 20 h later. Only construct p6F2R-luc had lost inducibility by TGF-β, showing that the same region responsible for Ras induction seems to be involved, at least partially, in the TGF-β response. These results agree with those of Li et al. (28) showing that the promoter response to TGF-β resides in Sp1 sites proximal (positions −79 to −55) to the p15INK4b gene. However, when we used a 720-bp fragment upstream of the mouse p15INK4b coding sequence (similar to the human fragment used as described in reference 28), only a threefold induction of p15INK4b could be observed after TGF-β treatment. Since a 30-fold induction of p15INK4b was observed in keratinocytes after TGF-β treatment (14), other elements needed for a complete TGF-β response could be located outside this promoter region (our results and A. Iavarone and J. Massagué, personal communication).
FIG. 6.
Deletion analysis of the mouse p15INK4b promoter. (A) Proximal sequence upstream p15INK4b exon 1. Nucleotide positions are numbered backward from the first base of the cDNA sequence (nucleotides in boldface). Different fragments upstream of the p15INK4b gene were amplified by PCR and subcloned into the luciferase reporter vector. The 5′-end position of some primers used for amplification (3F and 6F) is shown by arrows. Primer 2R (not shown) is a reverse oligonucleotide inside the cDNA sequence. Boxes indicate putative recognition sites for the Sp1 family of transcription factors. (B) Scheme of the deletion fragments and promoter activity (black bars). Circles indicate the positions of putative Sp1-binding sites. The data shown are as percentages of the activity of p15-1F2R-luc (100%) carrying a 720-bp insert. (C) Induction of promoter activity by Ras and TGF-β. The constructs were cotransfected with an activated Ras plasmid (pMZNT-17) or the vector pcDNA3.1. Fold induction by oncogenic Ras (white bars) was compared to the luciferase activity of the same fragment cotransfected with the empty vector. Fold induction by TGF-β (gray bars) corresponded to the increase in luciferase activity after TGF-β treatment of HaCaT cells transfected with the constructs. All of the data shown here were normalized to β-galactosidase activity as described in Materials and Methods and are representative of three separate experiments.
The Raf-Mek-Erk effector pathway is essential for Ras-dependent INK4 induction.
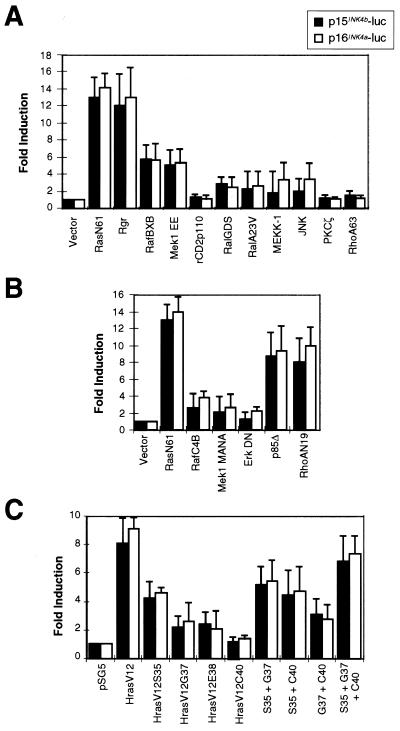
Cellular effects of Ras activation are diverse and are mediated by several different effector pathways (5, 24, 35). In order to investigate which of the signaling cascades downstream of Ras are involved in the induction of the p15INK4b and p16INK4a cell cycle inhibitors, several expression plasmids containing activated versions of signaling proteins were cotransfected with the p15-1F2R-luc (p15INK4b promoter) and p16-luc5 (p16INK4a promoter) plasmids. Both oncogenic Ras and Rgr induce the INK4 promoters to similar extents (about 12-fold). Activated versions of Raf1 (RafBXB) and Mek1 (Mek1 EE) showed partial induction of p15INK4b and p16INK4a compared with Ras (Fig. 7A). Other activated proteins, such as RalA (RalA23V), RhoA (RhoA63), and the catalytic subunit of PI3K (rCD2p110), or overexpression of the wild-type RalGDS, JNK, or PKCζ protein had a reduced or no effect on the p15INK4b or p16INK4a promoter. To ensure the functional integrity of the Raf1 and Mek1 constructs, these proteins were used in Elk-1 transactivation assays using luciferase reporters (74). Although the three proteins strongly induced Elk-1 transactivation (data not shown), only a fivefold increase on p15INK4b or p16INK4a promoter activity was observed (Fig. 7A).
FIG. 7.
Analysis of the Ras effector pathways involved in p15INK4b and p16INK4a promoter induction. The p15INK4b (p15-1F2R-luc; black columns) and p16INK4a (p16-luc5, white columns) reporter plasmids were cotransfected into NIH 3T3 cells with empty plasmids (Vector) or with expression plasmids carrying wild-type, activated, or dominant negative forms of molecules involved in the pathways downstream of Ras. Luciferase activity was then compared to the effect of oncogenic Ras (NrasN61 in pMZNT-17 or HrasV12 in pSG5), and the data are shown as fold induction with respect to the effect of the empty vector. Averages and standard errors from at least three different experiments are shown. Luciferase activity was normalized to β-galactosidase activity as a control for transfection efficiency. (A) Effect of overexpression of wild-type or activated proteins in p15INK4b and p16INK4a promoters. Whereas the Ras or Rgr oncogene produced a 12-fold increase in luciferase activity, only partial induction was found with single molecules downstream of Ras. (B) NIH 3T3 cells were cotransfected with the p15INK4b and p16INK4a reporter plasmids, oncogenic Ras (pMZNT-17), and the dominant negative forms of Raf1, Mek1, Erk1, PI3K (p85Δ), and RhoA. All of the Raf1-Mek1-Erk inhibitory forms suppressed Ras-dependent induction, whereas the other proteins had little or no effect. (C) p15INK4b and p16INK4a promoter induction by effector domain mutant forms of Ras. Only partial induction was achieved with HasV12S35 (Raf1-interacting mutant), and a very modest effect was obtained with the other forms. However, the three different forms cooperated to produce a stronger response.
Using a similar experimental approach, dominant negative variants of several proteins involved in the signaling cascades downstream of Ras were cotransfected with the luciferase reporter plasmids and the oncogenic Ras construct (pMZNT-17). Only the dominant negative forms of Raf1 (RafC4B), Mek1 (Mek1 MANA), and Erks (KR-Erk1 and K52R-Erk2) were able to suppress the luciferase activity induced by Ras (Fig. 7B). No significant effect was observed with a RalA (RalAN28), Rac1 (Rac1N17), Cdc42 (Cdc42N17), or PKCζ (PKCζW281) inhibitory mutant protein (data not shown), and only a slight decrease in luciferase activity was detected when the PI3K (p85Δ) and RhoA (RhoAN19) dominant negative proteins were used. We also took advantage of several effector domain mutant forms of Ras which are impaired in specific downstream pathways. Thus, S35 Ras mutant proteins signal through the Raf1 pathway but not through the RalGDS or PI3K effector. G37 and E38 mutant proteins are specific for the RalGDS pathway, and C40 interacts with PI3K but not with the other two effectors (57, 75). We cotransfected activated forms (V12) of these mutant proteins with a p15-1F2R-luc or p16-luc5 reporter using the HrasV12 mutant protein (with full effector response) or the empty vector as a control. As shown in Fig. 7C, HrasV12 produced 8- to 10-fold induction of the promoters whereas Raf1-interacting mutant HrasV12S35 mutant showed partial (3-fold) induction. A slight increase in luciferase activity was found with HrasV12G37 and HrasV12E38 (less than twofold induction in the RalGDS-interacting forms). No significant increase in luciferase activity was found with PI3K-interacting Ras mutant HrasV12C40. Interestingly, higher induction of the INK4 promoters was observed when combinations of these mutants were used, especially in the case of Raf1- and RalGDS-interacting proteins or when a mixture of the three Ras mutant proteins was used, indicating that, at least in NIH 3T3 cells, several Ras effector pathways can cooperate in p15INK4b and p16INK4a induction.
p15INK4b-deficient MEFs are susceptible to transformation by a single oncogene.
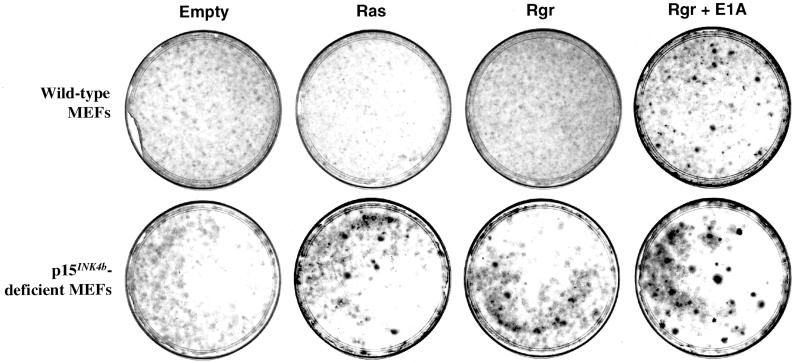
Since the in vitro results described above suggested a role for p15INK4b in suppressing the oncogenic activity of Ras in cultured fibroblasts, we decided to analyze the effect of Ras activation in MEFs obtained from p15INK4b knockout mice. These MEFs and those from wild-type littermates were transfected at early passages with plasmids expressing oncogenic Ras, Rgr, or the E1A protein. After transfection, cells were cultured for 2 weeks and visible foci were scored. Whereas no foci were observed in wild-type MEFs transfected with either the Ras or the Rgr oncogene, a few foci were scored in p15INK4b-deficient MEFs transfected with either of these proteins (Fig. 8). The efficiency of transformation of p15INK4b-deficient MEFs (averages of five foci per assay with Ras and seven foci per assay with Rgr) was, however, lower than that of INK4aΔ2,3 knockout MEFs lacking both the p16INK4a and p19ARF proteins (an average of 36 foci per assay; data not shown). These results are similar to those obtained by Latres and Barbacid (unpublished) showing that p15INK4b knockout MEFs are more susceptible to transformation by Ras or a combination of the Ras and Myc oncogenes. The numbers of foci obtained with Ras and Rgr were very similar, supporting an oncogenic mechanism common to both proteins (Hernández and Pellicer, unpublished). When Rgr was cotransfected with the E1A oncoprotein, transformed foci were observed in wild-type MEFs (an average of 10 foci per assay). However, the lack of p15INK4b in the fibroblasts resulted in higher susceptibility to transformation, since the number of foci increased (about 27 per assay) and they were larger than those obtained with wild-type MEFs (Fig. 8).
FIG. 8.
Transformation assays with MEFs derived from p15INK4b knockout mice. p15INK4b-deficient MEFs or those from wild-type littermates were transfected with a plasmid expressing the Ras or Rgr oncogene or a combination of Rgr plus E1A. Whereas no foci were observed in wild-type MEFs after transfection with Ras or Rgr, foci developed in cells lacking p15INK4b. Similarly, p15INK4b-deficient MEFs were more sensitive than wild-type cells to cotransfection with both the Rgr and E1A oncogenes.
DISCUSSION
Members of the two families of Cdk inhibitors, such as p16INK4a (62), p21Cip1 (42), and p57Kip2 (72), have been previously shown to suppress cell transformation by oncogenic Ras or other oncogenes. Our results obtained with NIH 3T3 cells show that p15INK4b is as potent an inhibitor of Ras transformation as p16INK4a or p27Kip1. p15INK4b produced a similar G1 arrest in Ras-transformed cells and decreased the tumorigenic potential of Ras or other oncogenes such as Rgr in the focus formation assay (Fig. 1 and 2). BrdU incorporation assays show that p15INK4b is able to suppress the entry into S phase of the cell cycle induced by oncogenic Ras, as previously reported for p16INK4a (62). In addition, cells expressing both Ras and p15INK4b formed fewer and smaller colonies in soft agar, indicating that ectopic expression of p15INK4b inhibits Ras-induced anchorage-independent growth of NIH 3T3 cells (Fig. 2). A similar effect has also been described for p21Cip1 (42). Interestingly, the morphology of Ras-transformed NIH 3T3 cells did not change after the ectopic introduction of p15INK4b. Cells expressing both Ras and p15INK4b maintained the characteristic phenotype of Ras-transformed cells (small and refractile) even when they entered p15INK4b-induced G1 arrest (Fig. 1B). These results suggest that the effect of p15INK4b on cell transformation by Ras is exclusively suppression of mitogenic activity. A dissociation between Ras-dependent growth arrest and cell morphology has been recently described in the premature-senescence phenotype of primary fibroblasts. Whereas Ras-arrested cells became large and flat (64), cells arrested by an activated Mek protein were small and refractile (30). In this system, the Raf-Mek-Erk pathway provides the antiproliferative signals to arrest growth without modifying cell morphology. Thus, our data support a specific role for p15INK4b in the mitogenic arrest of Ras-transformed NIH 3T3 cells without affecting other Ras-dependent pathways involved in producing the transformed morphology.
p15INK4b induction by Ras.
Transfection of oncogenic Ras into NIH 3T3 cells carrying a p15INK4b genomic fragment resulted in inhibition of focus formation compared to control NIH 3T3 cells (Fig. 2D). We realized that p15INK4b is overexpressed in Ras-transfected cells (Fig. 4), and since this effect is not observed with a p15INK4b cDNA, it is therefore induced by Ras through noncoding genomic sequences. As we have shown in this work, overexpression of p15INK4b is able to suppress Ras transformation and induction of p15INK4b is likely to antagonize the oncogenic effects of Ras.
Although p15INK4b is not expressed in mouse embryos, this gene is strongly induced in the first 4 days after embryos are disrupted and cultured as MEFs (47, 81). This behavior is similar to that described for p16INK4a, although its expression is delayed compared to that of p15INK4b. Two days after MEF plating, p15INK4b is highly expressed while the expression of p16INK4a is only barely detectable. After 4 days, p16INK4a continues accumulating as the MEFs divide in culture and approach crisis whereas p15INK4b expression only slightly increases (47, 81). We have shown here that the retroviral overexpression of p15INK4b is sufficient to cause G1 arrest and senescence morphology (Fig. 3). In fact, it has been recently reported that p15INK4b can induce G1 arrest and senescence even more effectively than p16INK4a or the Cip and Kip proteins (40). The Ras-dependent induction of p15INK4b described above is also similar to that described for p16INK4a in primary fibroblasts (64). Ras oncogenic activity in human and murine primary fibroblasts provokes the induction of p16INK4a and p53, which is associated with premature senescence of these cells. As shown in Fig. 4, p15INK4b is also induced in mouse primary fibroblasts after infection with an oncogenic Ras retrovirus.
This effect on cell cycle arrest and senescence could be even more dramatic in lymphoid tissues, where p15INK4b has been shown to play an important role in lymphocyte activation (34) or tumorigenesis (4, 17, 36). T lymphocytes accumulate p15INK4b protein during successive population doublings and display high levels of this molecule as they enter replicative senescence (12). Accumulation of INK4 proteins is accompanied by their increased binding to Cdk6 and decreased Cdk6 and Cdk2 activity. Thus, absence of the p15INK4b protein could make it possible for tumoral lymphocytes to proliferate, avoiding replicative senescence.
Transfection experiments using pMM134 (p15INK4b genomic DNA containing about 720 bp of the promoter sequence) and luciferase reporter assays show that Ras-dependent induction of p15INK4b and p16INK4a occurs through sequences located in the promoter region. In the case of p15INK4b, induction by Ras is blocked when 60 bp located between positions −122 and −55 are deleted from the promoter. Similar sequences also seem to be necessary for response to TGF-β in the human p15INK4b promoter (28). Sp1-binding sites are also important for the regulation of the p19ARF promoter (56), and since p19ARF is also induced by Ras activation (48), we are currently analyzing whether these sequences are also involved in Ras-dependent expression of the p19ARF protein. Ras, therefore, could use the same sequence motifs that TGF-β uses for induction of p15INK4b. In fact, a role for Ras in TGF-β transcriptional activation has been suggested (16, 45, 78). In epithelial cells, growth inhibition by TGF-β1 and TGF-β2 is associated with rapid activation of both Ras and Erk1 (45). Moreover, expression of the Ras dominant negative form (RasN17) partially abrogates Erk1 activation and inhibition of DNA synthesis by TGF-β (16) and Ras activation is also needed for TGF-β upregulation of p21Cip1 and p27Kip1 (78). Since p27Kip1 upregulation by TGF-β has been described as an indirect effect of p15INK4b induction and displacement of p27Kip1 from cyclin D-Cdk4/6 complexes (49, 51, 60), Ras could be physiologically involved in p15INK4b induction. If this is the case, the ability of Ras to induce the expression of some Cdk inhibitors could not only be a protective or stress response of the cell but have a function in normal physiological stages.
The constant levels of p15INK4b expression during the cell cycle and its induction by TGF-β have suggested a role for p15INK4b as an important effector in TGF-β-induced growth arrest, rather than in regulation of the timing of events in the cell cycle itself (14, 68). Our results suggest that p15INK4b could also provoke G1 arrest in response to cellular signals mediated by not only TGF-β but also other stimuli depending on Ras signaling. When Ras is required to send a mitogenic stimulus, other signals must cooperate to suppress the induction of p15INK4b and other inhibitors. For instance, in Ras-dependent induction of p21Cip1, once Ras has been activated, Rho signaling is required for suppression of p21Cip1 induction (46). When signaling through Rho is inhibited, constitutively active Ras induces p21Cip1 and entry into the DNA synthesis phase of the cell cycle is blocked.
Ras effector pathways and cell cycle control.
Since p15INK4b is also induced by Ras in NIH 3T3 cells containing the genomic INK4b gene, the downstream effector pathways used by Ras seem to be intact in these cells. Among the Ras effectors, the Raf-Mek-Erk pathway has been shown to function not only in the stimulation of cellular proliferation but also in growth arrest. There is growing evidence that a sustained increase in Erk activity can lead to inhibition of Cdk activity and cell cycle arrest (30, 33, 52, 77). In some systems, these inhibitory effects have been attributed to induction of the Cdk inhibitor p21Cip1 or a decrease in cyclin A levels (32, 65). This induction has been proposed to occur in a p53-dependent manner in primary rat Schwann cells (33) but independently of p53 in mouse primary fibroblasts (77). In other cases, the increase in p16INK4a expression seems more relevant and activated Raf or Mek protein is sufficient to provoke its induction and premature senescence in primary fibroblasts (30, 79).
In our study, analysis of the pathways involved in the Ras-dependent transcriptional activation of the p15INK4b and p16INK4a promoters showed that in NIH 3T3 cells, different cooperating pathways can be required to emulate the effect of oncogenic Ras. Activated forms of the Raf1 and Mek (MEK EE) kinases only partially increased luciferase activity from the p15INK4b and p16INK4a promoter, whereas the effect was almost null in other pathways (Fig. 7). Interestingly, a recently described oncogene named Rgr (9) is able to induce the INK4 proteins to an extent similar to that of oncogenic Ras. Rgr is a strong oncogene belonging to the RalGDS family of exchange factors. Rgr is able to activate RalA, similar to the other members of the family, but is also able to activate some Rho-mediated pathways and the mitogen-activated protein kinases, and this activation is at least partially dependent on Ras (Hernández and Pellicer, unpublished). All of these pathways are strongly related to transcriptional activation and could therefore account for the induction of the INK4 inhibitors.
The use of dominant negative proteins shows that the Raf-Mek-Erk pathway is essential for Ras-dependent activation of p15INK4b and p16INK4a. These results agree with the recent reports by Lin et al. (30) and Zhu et al. (79) showing that activation of the mitogen-activated protein kinase cascade is essential for induction of Cdk inhibitors and promotion of premature senescence. However, while they described comparable data on growth arrest and senescence after Ras, Raf1, or Mek activation, we repeatedly found only partial induction of the INK4 promoters when using activated Raf or Mek protein. These results can be explained by differences between the experimental approaches. While we analyzed the transcriptional activation of individual promoters, the work of Lin et al. (30) was based mainly on measurement of growth arrest and cellular senescence. Thus, small increases in the individual levels of p16INK4a, p15INK4b, and perhaps p21Cip1 induced by H-RasV12S35 could still cooperate and produce cell cycle arrest. On the other hand, strong mutant proteins used in their work, such as MekQ56P, could produce a stronger signal than the activated forms used in the present work. In fact, activated MekQ56P causes cell cycle arrest even more rapidly than oncogenic Ras (30) and differences in the strength of the signal have been previously shown to be essential in the stimulation of the Raf-Mek-Erk cascade outcome (26, 77). It has been suggested that the ultimate cellular response (cell cycle arrest or cellular proliferation) to activation of the Mek-Erk pathway can depend on the strength and duration of the Erk signal, which is the transient or cyclical activation responsible for the proliferative output, while sustained high levels of Erk activity could result in cell cycle arrest (52, 77). Thus, low levels of Raf activity lead to activation of cyclin D1-Cdk4 and cyclin E-Cdk2 complexes and cell cycle progression, whereas higher Raf activity elicited cell cycle arrest correlating with p21Cip1 induction and inhibition of cyclin-Cdk activity (77). Finally, the signaling pathways from Ras to p16INK4a and p15INK4b induction could be modified in NIH 3T3 cells, so that the Raf and Mek proteins might not be able to produce as high levels of INK4 induction as Ras does. We therefore do not exclude the possibility that different pathways could cooperate with the Raf-Mek-Erk cascade in NIH 3T3 and other cell types to produce growth arrest mediated by the Cdk inhibitors. In fact, in NIH 3T3 cells expressing human TrkA, PD98059 (a Mek inhibitor) blocked the ability of nerve growth factor (NGF) to inhibit Cdk2 and Cdk4 activities but only partially prevented the NGF induction of p21Cip1. Thus, NGF-dependent up-regulation of the p21Cip1 protein appears to be only partially mediated through the Mek-Erk pathway (52), suggesting that other pathways can also be involved.
Susceptibility of p15INK4b-deficient MEFs to cell transformation by single oncogenes.
The need for oncogene cooperation in the transformation of primary cells was reported almost 20 years ago (27, 59). In the last few years, it has been demonstrated both in vitro and in vivo that oncogenic activation and loss of inhibitory effects might cooperate in cell transformation. In fact, primary cells from INK4aΔ2,3-, p19ARF-, p21Cip1-, or p53-deficient mice can be transformed by Ras alone (23, 43, 63). On the other hand, Ras transgenics in an INK4aΔ2,3 background have a significantly higher tumor incidence (6). We show here that p15INK4b-deficient MEFs are susceptible to transformation by the Ras or Rgr oncogene, although with a lower efficiency than INK4aΔ2,3 MEFs. These MEFs lack both the p16INK4a and p19ARF proteins, and therefore, the contribution of each protein to this phenotype has not been assessed yet. Moreover, MEFs lacking only p19ARF are also susceptible to transformation by Ras (23) and the effect of p16INK4a inactivation in primary MEFs remains to be determined. The absence of the other INK4 proteins (p18INK4c and p19INK4d) in the corresponding knockout MEFs does not result in increased sensitivity to transformation by Ras (82; Latres and Barbacid, unpublished). Our results and those obtained by Latres and Barbacid (unpublished) indicate that p15INK4b has a suppressor effect on MEF transformation by either the Ras or the Rgr oncogene or by the combination of Ras and different oncogenes such as Myc or E1A.
The role of p15INK4b as a tumor suppressor in lymphocytes has also been recently highlighted as a result of methylation studies with both human and mouse cells (4, 17, 18, 36, 38, 80). Thus, p15INK4b is frequently (up to 88%) inactivated by methylation of the promoter region in both humans (4, 17, 18) and experimental systems (36). In agreement with these observations, p15INK4b knockout mice frequently develop lymphoproliferative disorders (Latres and Barbacid, unpublished). The fact that oncogenic Ras can activate p16INK4a and p15INK4b therefore provides a new insight into their role as tumor suppressor genes and suggests that Ras oncogenic signals might cooperate with p15INK4b inactivation in tumorigenesis in vivo. These INK4 proteins have similar activities in vitro and are likely to participate in the growth arrest response to oncogenic signals. Their different role in vivo could be dependent on their specific pattern of expression or in their ability to be induced by different stimuli. Among them, Ras seems to be a strong INK4 inducer and an important one due to its ability to participate in physiological and tumorigenic processes.
ACKNOWLEDGMENTS
We thank J. Altschmied, M. Barbacid, J. L. Bos, D. A. Brenner, P. Crespo, C. J. Der, L. A. Feig, M. Kasuga, D. Levy, J. Massagué, M. Pagano, U. R. Rapp, K. Reif, P. Rodríguez-Viciana, M. Serrano, R. A. Weinberg, and Y. Xiong for kindly providing some of the plasmids and reagents used in this work. We are specially indebted to E. Latres and M. Barbacid for providing us with the p15INK4b-deficient MEFs.
M.M., M.I.H., and I.P.C. received fellowships from the Ministerio de Educación (Madrid, Spain). This work was supported by grants CA 36327 and CA 50434 from NIH to A.P.
REFERENCES
- 1.Albright C F, Giddings B W, Liu J, Vito M, Weinberg R A. Characterization of a guanine nucleotide dissociation stimulator for a ras-related GTPase. EMBO J. 1993;12:339–347. doi: 10.1002/j.1460-2075.1993.tb05662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcorta D A, Xiong Y, Phelps D, Hannon G, Beach D, Barret J C. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci USA. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschmied J, Duschl J. Set of optimized luciferase reporter gene plasmids compatible with widely used CAT vectors. BioTechniques. 1997;23:436–438. doi: 10.2144/97233bm19. [DOI] [PubMed] [Google Scholar]
- 4.Batova A, Diccianni M B, Yu J C, Nobori T, Link M P, Pullen J, Yu A L. Frequent and selective methylation of p15 and deletion of both p15 and p16 in T-cell acute lymphoblastic leukemia. Cancer Res. 1997;57:832–836. [PubMed] [Google Scholar]
- 5.Campbell S L, Khosravi-Far R, Rossman K L, Clark G J, Der C J. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- 6.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner J W, 2nd, DePinho R A. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin L, Pomerantz J, DePinho R A. The INK4a/ARF tumor suppressor: one gene—two products—two pathways. Trends Biochem Sci. 1998;23:291–296. doi: 10.1016/s0968-0004(98)01236-5. [DOI] [PubMed] [Google Scholar]
- 8.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 9.D'Adamo D R, Novick S, Kahn J M, Leonardi P, Pellicer A. rsc: a novel oncogene with structural and functional homology with the gene family of exchange factors for Ral. Oncogene. 1997;14:1295–1305. doi: 10.1038/sj.onc.1200950. [DOI] [PubMed] [Google Scholar]
- 10.Dimri G P, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano E E, Linskens M, Rubelj I, Pereira-Smith O, Peacocke M, Campisi J. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downward J. Routine role for Ras. Curr Biol. 1997;7:R258–R260. doi: 10.1016/s0960-9822(06)00116-3. [DOI] [PubMed] [Google Scholar]
- 12.Erickson S, Sangfelt O, Heyman M, Castro J, Einhorn S, Grandér D. Involvement of the Ink4 proteins p16 and p15 in T-lymphocyte senescence. Oncogene. 1998;17:595–602. doi: 10.1038/sj.onc.1201965. [DOI] [PubMed] [Google Scholar]
- 13.Haber D A. Splicing into senescence: the curious case of p16 and p19ARF. Cell. 1997;91:555–558. doi: 10.1016/s0092-8674(00)80441-9. [DOI] [PubMed] [Google Scholar]
- 14.Hannon G J, Beach D. p15INK4b is a potential effector of TGF-β-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 15.Hara K, Yonezawa K, Sakaue H, Ando A, Kotani K, Kitamura T, Kitamura Y, Ueda H, Stephens L, Jackson T R, et al. 1-Phosphatidylinositol 3-kinase activity is required for insulin-stimulated glucose transport but not for RAS activation in CHO cells. Proc Natl Acad Sci USA. 1994;91:7415–7419. doi: 10.1073/pnas.91.16.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartsough M T, Frey R S, Zipfel P A, Buard A, Cook S J, McCormick F, Mulder K M. Altered transforming growth factor β signaling in epithelial cells when Ras activation is blocked. J Biol Chem. 1996;271:22368–22375. doi: 10.1074/jbc.271.37.22368. [DOI] [PubMed] [Google Scholar]
- 17.Herman J G, Jen J, Merlo A, Baylin S B. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- 18.Herman J G, Civin C I, Issa J P, Collector M I, Sharkis S J, Baylin S B. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res. 1997;57:837–841. [PubMed] [Google Scholar]
- 19.Hitomi M, Stacey D W. Cellular Ras and cyclin D1 are required during different cell cycle periods in cycling NIH 3T3 cells. Mol Cell Biol. 1999;19:4623–4632. doi: 10.1128/mcb.19.7.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huschtscha L I, Reddel R R. p16INK4a and the control of cellular proliferative life span. Carcinogenesis. 1999;20:921–926. doi: 10.1093/carcin/20.6.921. [DOI] [PubMed] [Google Scholar]
- 21.Jeffers M, Pellicer A. Identification of multiple promoters within the N-ras proto-oncogene. Biochim Biophys Acta. 1994;1219:623–635. doi: 10.1016/0167-4781(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 22.Kamb A, Gruis N A, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian S V, Stockert E, Day R S, 3rd, Johnson B E, Skolnick M H. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 23.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 24.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 25.Kerkhoff E, Rapp U R. Induction of cell proliferation in quiescent NIH 3T3 cells by oncogenic c-Raf-1. Mol Cell Biol. 1997;17:2576–2586. doi: 10.1128/mcb.17.5.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerkhoff E, Rapp U R. High-intensity Raf signals convert mitotic cell cycling into cellular growth. Cancer Res. 1998;58:1636–1640. [PubMed] [Google Scholar]
- 27.Land J, Parada L F, Weinberg R A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 28.Li J M, Nichols M A, Chandrasekharan S, Xiong Y, Wang X F. Transforming growth factor activates the promoter of cyclin-dependent kinase inhibitor p15INK4B through an Sp1 consensus site. J Biol Chem. 1995;270:26750–26753. doi: 10.1074/jbc.270.45.26750. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Nichols M A, Shay J W, Xiong Y. Transcriptional repression of the D-type cyclin-dependent kinase inhibitor p16 by the retinoblastoma susceptibility gene product pRb. Cancer Res. 1994;54:6078–6082. [PubMed] [Google Scholar]
- 30.Lin A W, Barradas M, Stone J C, van Aelst L, Serrano M, Lowe S W. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linardopoulos S, Street A J, Quelle D E, Parry D, Peters G, Sherr C J, Balmain A. Deletion and altered regulation of p16INK4a and p15INK4b in undifferentiated mouse skin tumors. Cancer Res. 1995;55:5168–5172. [PubMed] [Google Scholar]
- 32.Lloyd A C. Ras versus cyclin-dependent kinase inhibitors. Curr Opin Genet Dev. 1998;8:43–48. doi: 10.1016/s0959-437x(98)80060-9. [DOI] [PubMed] [Google Scholar]
- 33.Lloyd A C, Obermuller F, Staddon S, Barth C F, McMahon M, Land H. Cooperating oncogenes converge to regulate cyclin/cdk complexes. Genes Dev. 1997;11:663–677. doi: 10.1101/gad.11.5.663. [DOI] [PubMed] [Google Scholar]
- 34.Lois A F, Cooper L T, Geng Y, Nobori T, Carson D. Expression of the p16 and p15 cyclin-dependent kinase inhibitors in lymphocyte activation and neuronal differentiation. Cancer Res. 1995;55:4010–4013. [PubMed] [Google Scholar]
- 35.Malumbres M, Pellicer A. Ras pathways to cell cycle control and cell transformation. Front Biosci. 1998;3:887–912. doi: 10.2741/a331. [DOI] [PubMed] [Google Scholar]
- 36.Malumbres M, Pérez de Castro I, Santos J, Meléndez B, Mangues R, Serrano M, Pellicer A, Fernández-Piqueras J. Inactivation of the cyclin-dependent kinase inhibitor p15INK4b by deletion and de novo methylation with independence of p16INK4a alterations in murine primary T-cell lymphomas. Oncogene. 1997;14:1361–1370. doi: 10.1038/sj.onc.1200969. [DOI] [PubMed] [Google Scholar]
- 37.Malumbres M, Mangues R, Ferrer N, Lu S, Pellicer A. Isolation of high molecular weight DNA for reliable genotyping of transgenic mice. BioTechniques. 1997;22:1114–1119. doi: 10.2144/97226st03. [DOI] [PubMed] [Google Scholar]
- 38.Malumbres M, Pérez de Castro I, Santos J, Fernández-Piqueras J, Pellicer A. Hypermethylation of the cell cycle inhibitor p15INK4b 3′-untranslated region interferes with its transcriptional regulation in primary lymphomas. Oncogene. 1999;18:385–396. doi: 10.1038/sj.onc.1202299. [DOI] [PubMed] [Google Scholar]
- 39.Matesanz F, Pellicer A. In vivo and in vitro analysis of retroviral vectors carrying the N-ras oncogene. Int J Oncol. 1995;7:443–451. doi: 10.3892/ijo.7.3.443. [DOI] [PubMed] [Google Scholar]
- 40.McConnell B B, Starborg M, Brookes S, Peters G. Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr Biol. 1998;8:351–354. doi: 10.1016/s0960-9822(98)70137-x. [DOI] [PubMed] [Google Scholar]
- 41.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 42.Michieli P, Li W, Lorenzi M V, Miki T, Zakut R, Givol D, Pierce J H. Inhibition of oncogene-mediated transformation by ectopic expression p21Waf1 in NIH3T3 cells. Oncogene. 1996;12:775–784. [PubMed] [Google Scholar]
- 43.Missero C, Di Cunto F, Kiyokawa H, Koff A, Dotto G P. The absence of p21Cip1/WAF1 alters keratinocyte growth and differentiation and promotes ras-tumor progression. Genes Dev. 1996;10:3065–3075. doi: 10.1101/gad.10.23.3065. [DOI] [PubMed] [Google Scholar]
- 44.Mulcahy L S, Smith M R, Stacey D W. Requirements for ras proto-oncogene function during serum-stimulated growth of NIH 3T3 cells. Nature. 1985;313:241–243. doi: 10.1038/313241a0. [DOI] [PubMed] [Google Scholar]
- 45.Mulder K M, Morris S L. Activation of p21ras by transforming growth factor beta in epithelial cells. J Biol Chem. 1992;267:5029–5031. [PubMed] [Google Scholar]
- 46.Olson M F, Paterson H F, Marshall C J. Signals from Ras and Rho GTPases interact to regulate expression of p21Waf1/Cip1. Nature. 1998;394:295–299. doi: 10.1038/28425. [DOI] [PubMed] [Google Scholar]
- 47.Palmero I, McConnell B, Parry D, Brookes S, Hara E, Bates S, Jat P, Peters G. Accumulation of p16INK4a in mouse fibroblasts as a function of replicative senescence and not of retinoblastoma gene status. Oncogene. 1997;15:495–503. doi: 10.1038/sj.onc.1201212. [DOI] [PubMed] [Google Scholar]
- 48.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 49.Peters G. Stifled by inhibitors. Nature. 1994;371:204–205. doi: 10.1038/371204a0. [DOI] [PubMed] [Google Scholar]
- 50.Pollock P M, Pearson J V, Hayward N K. Compilation of somatic mutations of the CDKN2 gene in human cancers: non-random distribution of base substitutions. Genes Chromosomes Cancer. 1996;15:77–88. doi: 10.1002/(SICI)1098-2264(199602)15:2<77::AID-GCC1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 51.Polyak K, Kato J Y, Solomon M J, Sherr C J, Massague J, Roberts J M, Koff A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-β and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 52.Pumiglia K M, Decker S J. Cell cycle arrest mediated by the MEK/mitogen-activated protein kinase pathway. Proc Natl Acad Sci USA. 1997;94:448–452. doi: 10.1073/pnas.94.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 54.Quelle D E, Ashmun R A, Hannon G J, Rehberger P A, Trono D, Richter K H, Walker C, Beach D, Sherr C J, Serrano M. Cloning and characterization of murine p16INK4a and p15INK4b genes. Oncogene. 1995;11:635–645. [PubMed] [Google Scholar]
- 55.Reif K, Nobles C D, Thomas G, Hall A, Cantrell D A. Phosphatidylinositol 3-kinase signals activate a selective subset of Rac/Rho-dependent effector pathways. Curr Biol. 1996;6:1445–1455. doi: 10.1016/s0960-9822(96)00749-x. [DOI] [PubMed] [Google Scholar]
- 56.Robertson K D, Jones P A. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez-Viciana P, Warne P H, Khwaja A, Marte B M, Pappin D, Das P, Waterfield M D, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 58.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 59.Ruley H E. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- 60.Sandhu C, Garbe J, Bhattacharya N, Daksis J, Pan C H, Yaswen P, Koh J, Slingerland J M, Stampfer M R. Transforming growth factor β stabilizes p15INK4B protein, increases p15INK4B-cdk4 complexes, and inhibits cyclin D1-cdk4 association in human mammary epithelial cells. Mol Cell Biol. 1997;17:2458–2467. doi: 10.1128/mcb.17.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serrano M, Hannon G J, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 62.Serrano M, Gómez-Lahoz E, DePinho R A, Beach D, Bar-Sagi D. Inhibition of Ras-induced proliferation and cellular transformation by p16INK4. Science. 1995;267:249–252. doi: 10.1126/science.7809631. [DOI] [PubMed] [Google Scholar]
- 63.Serrano M, Lee H W, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 64.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 65.Sewing A, Wiseman B, Lloyd A C, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharpless N E, DePinho R A. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 67.Sherr C J. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 68.Stone S, Dayananth P, Jiang P, Weaver-Feldhaus J M, Tavtigian S V, Cannon-Albright L, Kamb A. Genomic structure, expression and mutational analysis of the P15 (MTS2) gene. Oncogene. 1995;11:987–991. [PubMed] [Google Scholar]
- 69.Swafford D S, Middleton S K, Palmisano W A, Nikula K J, Tesfaigzi J, Baylin S B, Herman J G, Belinsky S A. Frequent aberrant methylation of p16INK4a in primary rat lung tumors. Mol Cell Biol. 1997;17:1366–1374. doi: 10.1128/mcb.17.3.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan T-H, Wallis J, Levine A J. Identification of the p53 protein domain involved in formation of the simian virus 40 large T-antigen–p53 protein complex. J Virol. 1986;59:574–583. doi: 10.1128/jvi.59.3.574-583.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Urano T, Emkey R, Feig L A. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- 72.Watanabe H, Pan Z Q, Schreiber-Agus N, DePinho R A, Hurwitz J, Xiong Y. Suppression of cell transformation by the cyclin-dependent kinase inhibitor p57KIP2 requires binding to proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 1998;95:1392–1397. doi: 10.1073/pnas.95.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinberg R A. The cat and mouse games that genes, viruses, and cells play. Cell. 1997;88:573–575. doi: 10.1016/s0092-8674(00)81897-8. [DOI] [PubMed] [Google Scholar]
- 74.Westwick J K, Cox A D, Der C J, Cobb M H, Hibi M, Karin M, Brenner D A. Oncogenic Ras activates c-Jun via a separate pathway from the activation of extracellular signal-regulated kinases. Proc Natl Acad Sci USA. 1994;91:6030–6034. doi: 10.1073/pnas.91.13.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.White M A, Nocolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler M H. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 76.Wolthuis R M, de Ruiter N D, Cool R H, Bos J L. Stimulation of gene induction and cell growth by the Ras effector Rlf. EMBO J. 1997;16:6748–6761. doi: 10.1093/emboj/16.22.6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woods D, Parry D, Cherwinski H, Bosch E, Lees E, McMahon M. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yue J, Buard A, Mulder K M. Blockade of TGFbeta3 up-regulation of p27Kip1 and p21Cip1 by expression of RasN17 in epithelial cells. Oncogene. 1998;17:47–55. doi: 10.1038/sj.onc.1201903. [DOI] [PubMed] [Google Scholar]
- 79.Zhu J, Woods D, McMahon M, Bishop J M. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhuang S M, Schippert A, Haugen-Strano A, Wiseman R W, Söderkvist P. Inactivations of p16INK4a-α, p16INK4a-β and p15INK4b genes in 2′,3′-dideoxycytidine- and 1,3-butadiene-induced murine lymphomas. Oncogene. 1998;16:803–808. doi: 10.1038/sj.onc.1201600. [DOI] [PubMed] [Google Scholar]
- 81.Zindy F, Quelle D E, Roussel M F, Sherr C J. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 82.Zindy F, van Deursen J, Grosveld G, Sherr C J, Roussel M. INK4d-deficient mice are fertile despite testicular atrophy. Mol Cell Biol. 2000;20:372–378. doi: 10.1128/mcb.20.1.372-378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]