Introduction
Few biological processes are as fundamental to individuality or collective identity as memory. Understanding the mechanisms underlying memory formation is dependent on our increased understanding of how gene transcription in the nucleus produces specific proteins underlying synaptic function (Figure 1). The synapse is hypothesized to be the physiological unit of memory, and the singular role of epigenetic modifications in regulating dynamic changes at the synapse during memory and neurodegeneration has been recently review in 1–3. Nuclear reprograming by epigenetic mechanisms is a process indispensable for memory function 4, and the study of these mechanisms has led to the emergence of the field of neuroepigenetics. Encompassing not only the classic stable, Warrington epigenetic marks necessary for cellular differentiation, neuroepigenetics is the study of epigenetic modifications that enable gene transcription programs necessary for cellular function in response to environmental stimuli 5–7. Though a complete understanding of the fundamental mechanisms underlying memory continues to elude us, the study of epigenetic regulation of gene transcription in brain regions such as the hippocampus, has begun to shed light on the underpinning of memory formation and maintenance.
Figure 1.
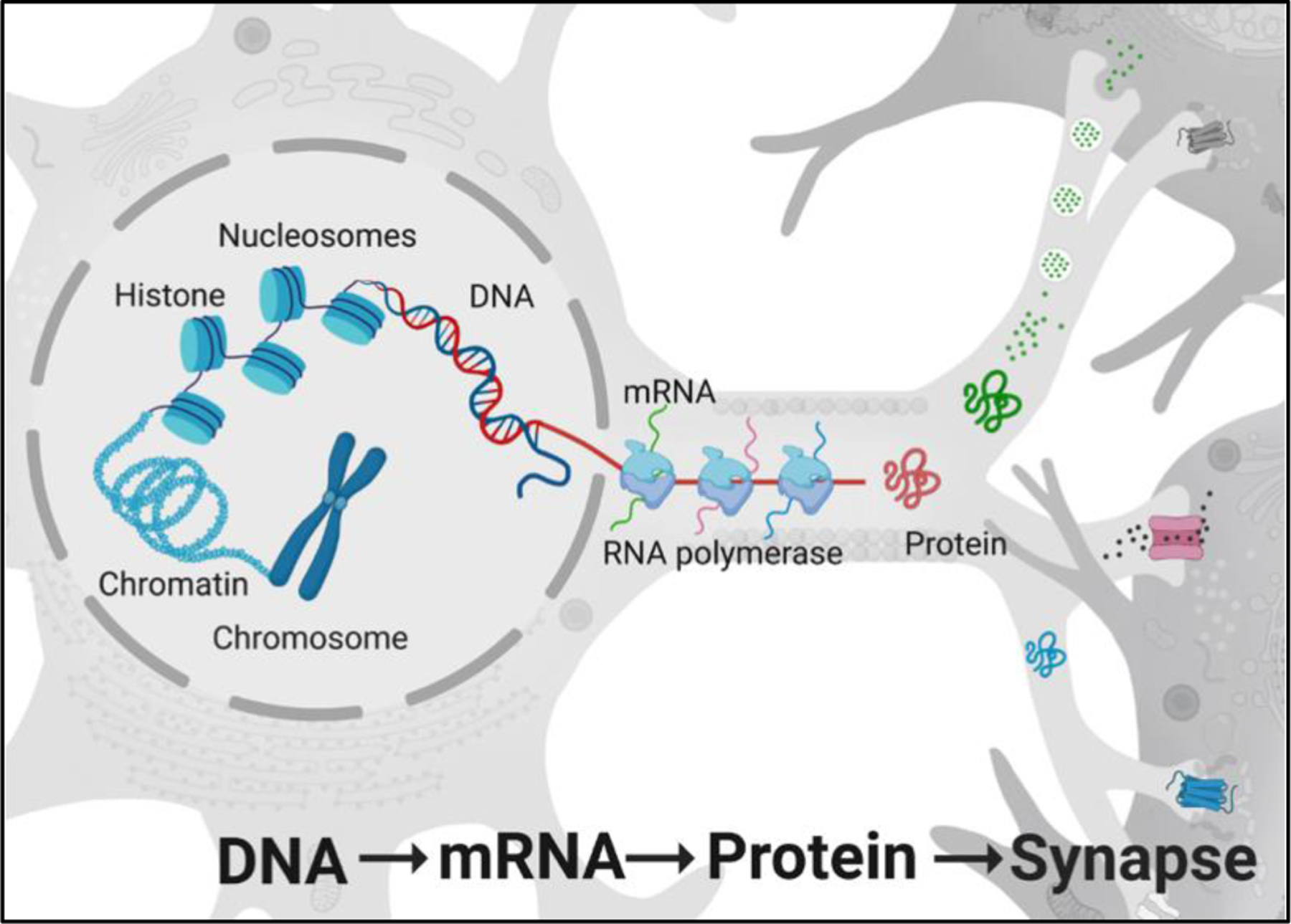
Schematic representation of the central dogma (DNA → mRNA → protein) of molecular biology in the neuron as it relates to synapse function and memory.
Considerable progress has been made in our understanding of how certain epigenetic mechanisms, including DNA methylation and posttranslational modification of histones, contribute to memory formation. Long considered a static mark with the ability to sustain enduring cellular phenotypes, these epigenetic modifications are now known to be dynamically regulated in non-dividing and terminally differentiated neurons, and responsible for established transcriptional regulation of memory associated genes 8–13. For example, inhibition of DNA methyltransferases (DNMTs), which are responsible for the addition of a methyl group to the 5’ position of the cytosine ring, have been shown to attenuate expression of Bdnf in area CA1 of the hippocampus and interfere with contextual fear memory formation 14. Another example involves a diverse group of histone post-translational modifications, impacting chromatin structure around gene regions to contribute to the formation and maintenance of memory. One study found that contextual fear conditioning (CFC) resulted in increases in the transcriptionally repressive dimethylation of histone H3 at lysine 9 (H3K9me2) in CA1 of the hippocampus 15. Interestingly, inhibition of the G9a/GLP methyltransferase complex in the entorhinal cortex enhanced memory in a CFC paradigm via H3K9me2-mediated silencing of the memory-related gene COMT in hippocampal CA1 16. Likewise, methylation of histone H3 at lysine 4 (H3K4me), associated with an open chromatin state, has been found to be necessary for the CFC memory formation process, reviewed extensively in 17.
Over the past 15 years, transcriptional programs necessary for synaptic function and memory formation have been demonstrated to be influenced by epigenetic mechanisms18–20. Despite our progress in the epigenetics research field, much remains to understand about the role of long non-coding RNAs (lncRNAs) in mediating epigenetic regulation of memory. Less than 2% of the genome contains protein coding transcripts 21, the remainder consisting of noncoding transcripts that were originally believed to be “junk” DNA. Though initially overlooked, the biological significance of non-coding RNAs (ncRNAs) appears indisputable as increased appreciation is gained for the profound regulatory capacities of lncRNAs. NcRNA, categorized as lncRNAs or small non-coding RNAs (which include microRNAs, ribosomal RNAs, small nuclear RNAs, piwi-interacting RNAS, transfer RNAs and small interfering RNAs; Figure 2), play a significant role in both normal cellular function and disease 22–25. Very recently a handful of studies have begun to show that lncRNA targeting to the synapse influences synaptic plasticity and likely learning and memory26–28. As it seems is often the case, our understanding of the role of ncRNAs in the brain has lagged behind other fields such as cancer biology, however a growing body of literature now implicates ncRNAs as potent regulators of cognition 29–32. The significance of lncRNAs in memory in particular, is an area ready for further exploration.
Figure 2. Non-coding RNAs.
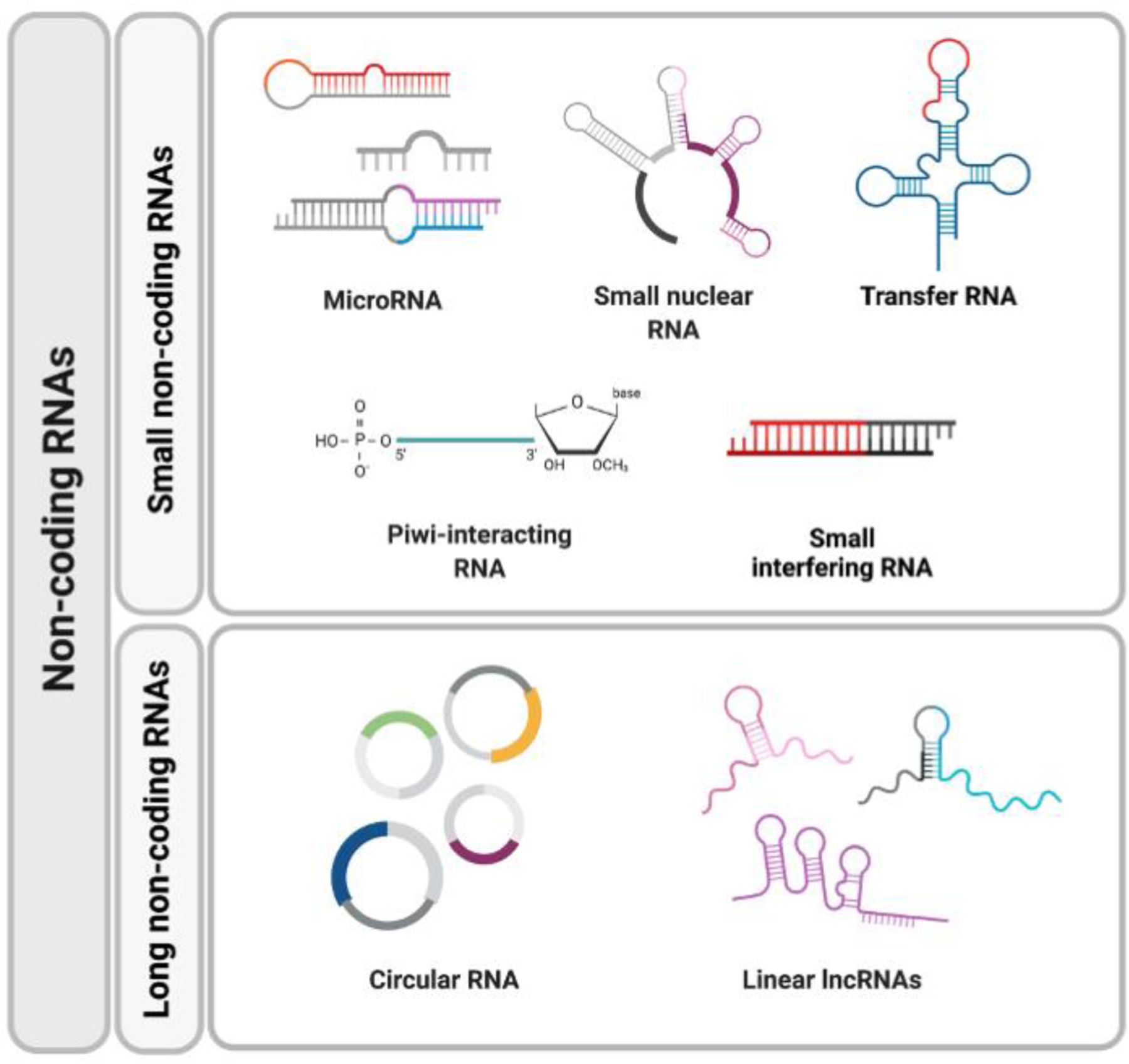
Non-coding RNA (ncRNA) are functional RNA molecules that are not translated into proteins. ncRNA can be classified into long ncRNAs (lncRNAs) and Small ncRNAs. Small ncRNAs include many different RNAs, such as microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), transfer RNA (tRNA), piwi-interacting RNAs (piRNAs) and small interfering RNA (siRNA). lncRNAs are the most ubiquitous and functionally diverse class, they include linear lncRNAs and circular RNAs (circRNAs).
LncRNAs are endogenous regulatory RNA molecules defined somewhat arbitrarily as transcripts greater that 200 base pairs 33. Lacking an open reading frame, and thus protein coding capacity, lncRNAs are involved in numerous biological functions and regulate gene expression through a diverse array of mechanisms 34,35. LncRNAs display temporal, spatial and cell-type specific expression in the brain 36–38, suggesting the potential for unique functional roles. The diversity of mechanisms linked to lncRNA mediated gene transcription has led to the examination of epigenetic crosstalk across the genome. In this review we define epigenetic crosstalk as the ability of one epigenetic mechanism (e.g., lncRNAs) to modify or direct additional epigenetic marks (e.g., histone modifications) with indirect effects on gene expression and subsequently on memory formation. Prior investigations on how modification of chromatin structure by epigenetic enzymes are targeted to gene loci have been unclear. However, the predominantly nuclear localization of lncRNAs, which are heavily enriched in chromatin fractions 21, suggests a role for lncRNAs in chromatin restructuring. In fact, lncRNAs have been shown to bind to numerous chromatin-modifying enzymes, resulting in lncRNA modification or the guiding of regulatory complexes to specific genomic sequences by lncRNAs 39. Moreover, numerous studies have shown a significant role for lncRNAs in behavior 40, cognitive function 41,42, and disease 43–45.
In this review we will place a specific focus on lncRNA crosstalk with other epigenetic mechanisms both in the brain and neurological disease, with the goal of increasing understanding of lncRNA function such that it might be applied to a better understanding of learning and memory.
First, we discuss lncRNA interactions with two epigenetic mechanisms which are critical to normal memory function (see Figure 3): 1) modifications directly to genomic DNA (DNA methylation), 2) mechanisms effecting chromatin availability via histone modification This is followed by an examination of what little is currently known about how lncRNAs are themselves regulated, specifically by epigenetic crosstalk. Next, we consider the role of lncRNA dysregulation in memory disorders, including age-associated memory impairment, Alzheimer’s disease (AD) and epilepsy. Finally, we discuss what, in our view, are critical gaps in the current knowledge in terms of lncRNA regulation of memory, as well as the promise of novel therapeutic options for memory disorders.
Figure 3. Epigenetic mechanisms of gene expression regulation.
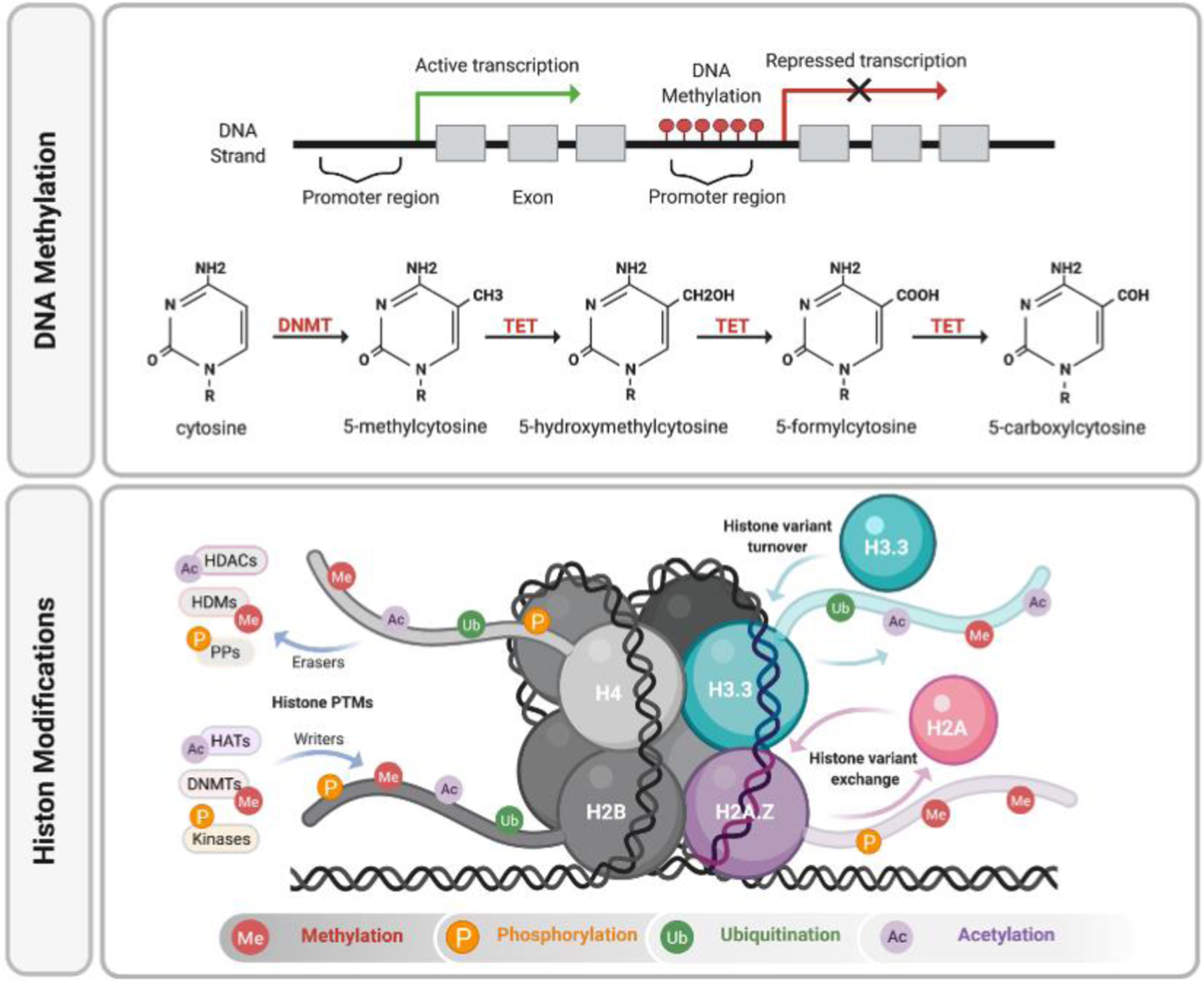
Several types of epigenetic mechanisms play a role in gene regulation, including (1) DNA methylation of gene promoter regions reflect that gene transcriptional activity; if the promoter region is hypermethylated then the gene transcription is repressed, and vice versa, hypomethylated promoter region favors active genes. In this reaction, DNA methyltransferases (DNMTs) modulate gene transcription via the addition of methyl group to the fifth position of cytosines to be converted to 5-methylcytosine (5mC), which then can be demethylated via ten-eleven translocation (TET) dioxygenase to 5-hydroxymethylcytosine (5-hmC) → 5-formylcytosine (5-fC) → 5-carboxycytosine (5-caC). (2) The post-translational modifications (PTMs) of the histone proteins by methylation on lysine or arginine, phosphorylation on serine or threonine residues, ubiquitylation of lysines, acetylation, and deacetylation of lysines. Histone tails can be modified by “writer” enzymes that catalyze the addition of epigenetic marks on histone tails such as histone acetyltransferases (HATs), histone methyltransferases (HMTs), and Kinases, and removed by “eraser” enzymes, such as histone deacetylases (HDACs), histone demethylases (HDMs) and Protein phosphatase (PPs); Histone variant functions is mediated via in histone variant exchange and turnover.
The emerging importance of lncRNA function in the brain has recently been highlighted by a number of quality reviews. Thus far there is significant evidence to demonstrate a role for lncRNA function in neural development46–49 and aging50–53. Even more data is available exploring the association of lncRNAs with psychiatric disorders54–59 and neurological disorders such as CNS/PNS injury and inflammation60–64, ischemic stroke65–70, gliomas71–73, and neurodegenerative disease74–78. In a recent review, Grinman et al., nicely summarizes the conservation, evolution and expression of lncRNAs in the brain, as well as what little is known about lncRNA and the neurobiology of learning and memory, including transcriptional and post-transcriptional regulation. In particular, they emphasize the critical role of cis or trans-acting lncRNA regulation of gene expression via either direct interaction or as part of transcriptional complexes26.
What is missing from the literature is a comprehensive understanding of how lncRNAs influence gene transcription programs necessary for learning and memory, both in the healthy brain and in disease. Thus, in this review we attempt to specifically address a potential role for lncRNAs and epigenetic crosstalk in regulation of gene expression that may in turn be applied to the study of learning and memory.
Epigenetic regulation by lncRNAs
To understand the mechanisms underlying memory, and to develop treatments for disorders of memory, it is necessary to understand how large-scale gene transcription changes are unlocked to allow for memory formation. While our understanding of how lncRNA function to epigenetically control memory-associated gene expression is still in its infancy, much more is known about the function of lncRNA in other fields. Here we will review the known epigenetic cross talk between lncRNA, histone modifications and DNAme in a variety of contexts in the hopes of driving further study and providing insight into how these molecules are directing gene expression changes to enable memory function.
The significant enrichment of many lncRNAs within chromatin identified through the ENCODE transcriptome analysis 21 strongly suggests a role for lncRNA in epigenetic regulation of gene transcription. Indeed, higher order chromatin structure requires RNA-chromatin interactions 79–81. This is particularly true in the brain where lncRNA frequently act to direct chromatin modifying enzymes to specific genomic locations, thus altering chromatin state and inducing changes in gene expression necessary for cellular function 82,83. While the whole of epigenetics includes numerous different mechanisms, this review focuses on the interaction between lncRNA and two significant epigenetic mechanisms responsible for gene expression changes, post-translational histone modification and DNA methylation, as well as how lncRNA themselves are regulated by epigenetic crosstalk.
LncRNA regulation of posttranslational histone modifications
The role of histone modifications in learning and memory is now well-established and has been review extensively17,84–93. However, only relatively recently has the role of ncRNAs in epigenetic control of gene expression been appreciated 94–96. The past few years have seen a rapid advancement of our understanding of how lncRNA interact with a variety of histone modifications 97 including histone methylation 98, acetylation 99,100, and ubiquitination 101. Perhaps the most well-studied role of lncRNA is X-chromosome inactivation (XCI) via the lncRNA Xist. During XCI, a “Xist cloud” coats one X chromosome, recruiting polycomb repressive complex 2 (PRC2) and inducing heterochromatin confirmation via PRC2 as a mechanism of dosage compensation 102. Interestingly, Xist has recently been shown to play a role in maintaining repressive histone marks (H3K27me3 and H2AK119 monoubiquitylation) for purposes of sustained XCI in both neurons and a smaller fraction of astrocytes into adulthood 103. Here we have a prime example of how lncRNA mechanisms associated with development are subsequently coopted for additional purposes across time and in a cell type-specific fashion.
The mechanisms by which lncRNA direct histone modification are diverse and include acting as scaffolds and tethers by binding chromatin modifying enzymes (CME), as well as guiding CME to specific targets 39,97. Polycomb Repressive Complex 2 (PRC2) is responsible for mediating the addition of largely transcriptionally repressive di or tri-methylation of Lys 27 of histone H3 104,105 and several studies have suggested regulation and recruitment of PRC2 by various lncRNA including HOTAIR 106–108, XIST 109,110 and many others 111–113. For example, the long intergenic non-coding RNA (lincRNA) HOTAIR serves as a scaffold for PRC2 (5’) and LSD1/CoREST/REST complex (3’) and the tethering of these complexes results in coupled H3K27methylation and K4 demethylation at target genes 114. These same mechanisms have the potential to play a role in memory formation as this kind of intricate regulation of gene expression by epigenetic mechanisms is critical for memory. Indeed, histone demethylase LSD1 is necessary for synaptic plasticity and hippocampus dependent memory115–120 and has been shown to be dysregulated in memory-related diseases121,122. REST is a significant transcriptional regulator in a variety of neurodegenerative diseases123, while CoREST has recently been shown to mediate memory cosolidation in Drosophila124. Similarly, a component of the PRC2, the histone lysine methyltransferase EZH2, is a critical regulator of gene expression during fear memory125,126. Evidence for the direct interaction of Polycomb repressive complexes and lncRNA is still under debate with many elaborate RNA interactions believed to play a role in PRC2 direction of gene expression 127, however compelling evidence for a direct association with PRC2 by at least some lncRNA was recently reviewed in 128.
Much of what is known about epigenetic regulation of gene expression profiles by lncRNAs has been derived through the study of lncRNAs in cancer. Several lncRNAs are differentially expressed in glia-derived tumors and many studies are examining their capacity to serve as biomarkers. One such example, AGAP2-AS1, interacts with the active component of the polycomb repressive complex, EZH2 to direct them to the promoter region of TFPI2 and inhibiting transcription 129. The lncRNA TUG1 with an EZH2 binding domain has also been shown to recruit PRC2 in glioma cells repressing differentiation relevant genes through increased H3K27me 130. Similarly, the lncRNAs HOTAIRM1 and PXN-AS1 have been found to promote proliferation and migration of glioblastoma cells via sequestration of G9a and EZH2, mediating dimethylation of H3K9 and H327 at the transcription start site of the HOXA1 and DKK1 promoter genes respectively 131,132. While these interactions are described in the context of glioma cells, EZH2 is a key mediator of memory associated gene expression during fear memory125,126.
Extending mechanisms observed in neoplastic tissues to other disease processes or healthy tissues must of course be done with caution. However, recent studies demonstrate that lncRNA interaction with chromatin remodeling mechanisms is not limited to oncogenic processes. With improved sequencing technologies, the ability to probe deeper and more thoroughly into the functions of these transcripts in the brain has advanced considerably. Recent RNA immunoprecipitation (RIP) sequencing studies show extensive binding of various lncRNA to the catalytic subunit of PRC2, EZH2, in numerous tissues including the brain 133, and that many of these interactions may be significant for neurological disease 134. For example, H19 knockdown reverses hypoxic stroke induced upregulation of HDAC1 and downregulation of acetyl-histone H3 and acetyl-histone H4, whereas HDAC1 overexpression negated the beneficial effects of H19 knockdown on infarct volume and brain edema 135. It is well-established that histone acetylation and deacetylation driven regulation of gene expression contributes to memory function, and the use of HDAC inhibitors to treat neurological disorders characterized by memory dysfunction has garnered significant interest88,91,136,137. Given known interactions between HDACs and lncRNAs in other neurological conditions, it appears prudent to explore their likely role in regulating key memory-related epigenetic mechanisms. Indeed, as will be discussed in greater detail below, the lncRNA Neat1 which has been studied extensively in cancer biology, is now known to transcriptionally represses c-fos via H3K9me2, possibly through interaction with the histone methyltransferase G9a in the context of fear memory 37. Collectively, these studies demonstrate a significant role for lncRNA in directing histone post-translational modifications and subsequent gene transcription.
LncRNA regulation of DNA methylation
The dynamic regulation of DNA methylation is often choreographed and influenced by the expression of various lncRNA 138–140. For example, during development, the lncRNA Evf2 both recruits DLX and Methyl CpG binding protein 2 (MeCP2)141, and inhibits DNA methylation, modulating competition between the DLX1/2 activator and MeCP2 repressor, enabling differential control of adjacent genes with shared DNA regulatory elements 142. MeCP2 regulation of transcription has a well-established impact on synaptic function143 and learning and memory144–146.
Beyond development, lncRNA continue to mediate gene expression throughout the normal lifespan, as well as in the case of disease. Regulation of gene expression by DNAme and its associated readers, writers and erasers, is critical for synaptic plasticity and in vivo measures of memory10,147–152. Thus, any potential recruitment or regulation of these mechanisms by lncRNA in the brain is likely to impact memory-associated gene expression.
In one such example, Diabetes Mellitus associated reduction in neurogenesis is followed by cognitive decline that can be linked to upregulation of the lncRNA H19. H19 binds specifically to the IGF2 gene promoter region, resulting in hypermethylation through enrichment of DNA methyltransferase and ultimately silencing IGF2 expression 153. Similarly, decreasing expression of the lncRNA PCAI can protect against neuroinflammation induced cognitive impairment, and does so via negative regulation of SUZ12, which in turn serves as a recruiting platform for DNA methyltransferases 154.
While there are few other examples from the field of learning and memory, the study of cancer has yielded significant insights into the role of lncRNA in epigenetic control of gene transcription profiles. Recruitment of DNA methyltransferases by lncRNA to promoter regions significantly alters proliferation and invasion-permissive genes, as that seen by the lncRNA MCM3AP-AS1 which recruits DNMT1/3 (A/B) to the promoter region of NPY1R resulting in its down regulation and activation of the MAPK pathway in prostate cancer 155. Interestingly, NPY1R expression has recently been shown to mediate spatial learning in adult mice156. It may then be reasonable to ask if, in the context of memory formation, the lncRNA MCM3AP-AS1, which is also expressed in the brain, might contribute to NPY1R transcription regulation through control of DNA methylation at its promoter. Beyond recruitment and direction of DNMTs, lncRNAs have also been shown to modulate the stability of methyltransferases, inhibiting expression of tumor suppressors via increased DNAme 157.
Understanding the role of lncRNA-mediated epigenetic mechanisms in the context of behavior is still in its infancy. However much remains to be learned about how lncRNA mediation of DNA methylation contributes to learning and memory, and how these mechanisms are disrupted in cognitive impairment.
Regulation of lncRNA expression by epigenetic mechanisms
It can be safely surmised based on the studies described above, and the work of many others, that lncRNA are critical players in the control of gene expression. Less however is known about the signaling pathways that facilitate expression of lncRNAs themselves. The tissue, and time specific expression of many lncRNA argues for a tightly controlled regulation of lncRNA transcription. Once again there are few explicit examples of how regulation of lncRNA expression impacts memory formation. Instead, we must explore what has been elucidated from cancer biology and neurological disease to form a starting point from which to investigate the role of lncRNA regulation in memory. For example, in breast cancer tissue, IGF/Insulin signaling arbitrates expression of a subset of lncRNA including SNHG7, which is downregulated by IGF via MAPK-driven post transcriptional mechanisms 158. Interestingly, transcriptional control of SNHG7 also appears to occur through C-myc binding of the promoter region increasing expression and governing glycolysis through the miR-34a-5p/LDHA axis in breast cancer cells 159. These studies demonstrate multiple levels of transcription regulation of a single lncRNA.
Indeed, there are many broad potential mechanisms by which lncRNA expression including can be regulated including second messenger signaling160–162, drugs of abuse163–165, neuronal activation166–169, and many others which have been described elsewhere170,171. Perhaps unsurprisingly then, lncRNAs are subject to regulation themselves by various epigenetic mechanisms. In fact, it is likely that multiple levels of epigenetic regulation will be affected in the case of disease, such as the H3K27me3 facilitation of the lncRNA HOTAIR, leading to altered HOXA1 DNA methylation in chemoresistant small cell lung cancer 172. Complex governance of lncRNAs expression appears to play a role normal healthy development, such as Ezh2-mediated H3K27me of various lncRNAs in embryonic stem cells 173, as well as in disease. Interestingly, in some cases this regulation appears to be bidirectional with differential DNAme at promoter and transcriptional start sites of lncRNAs. For example, decreased DNAme at the promoter region of the lncRNA SNHG12 results in upregulation of its expression and development of TMZ resistance in glioblastoma cells 174,175. In a number of human cancers, loss of MEG2 due to hypermethylation and promoter and intronic regions is associated with tumor growth 176. Similarly, aberrant methylation patterns at multiple lncRNA have been linked to both paranoid and undifferentiated schizophrenia 177. Four lncRNAs (UCA1, ADARB2-AS1, LINC324 and MAP3K14-AS1) were found to be differentially methylated (hypermethylated) in temporal lobe epilepsy, further showing transcriptional control of lncRNA by DNAme 178. In reality, multiple epigenetic mechanisms undoubtedly converge to maintain the delicate homeostasis necessary for cellular function and potentially memory formation.
LncRNAs in Memory disorders
Prior sections of this review attempted to impart the significance of lncRNA in regulating gene expression and the general mechanisms by which this might occur. A growing body of literature implicates aberrant lncRNA expression with cellular dysfunction in memory associate diseases (Figure 4; Table 1). It is imperative to obtain a solid understanding of lncRNA mediated gene expression changes in the healthy brain in order to target these transcripts for therapeutic manipulation under pathological conditions. The following is a discussion of lncRNA involvement in three highly prevalent disorders of memory. Taken together, age-related memory impairment, Alzheimer’s disease and Epilepsy represent a monumental global health burden for which there are currently very limited therapeutic options, and for which exploitation of lncRNAs holds particular promise.
Figure 4. Proposed Molecular Functions of lncRNA in memory disorders.
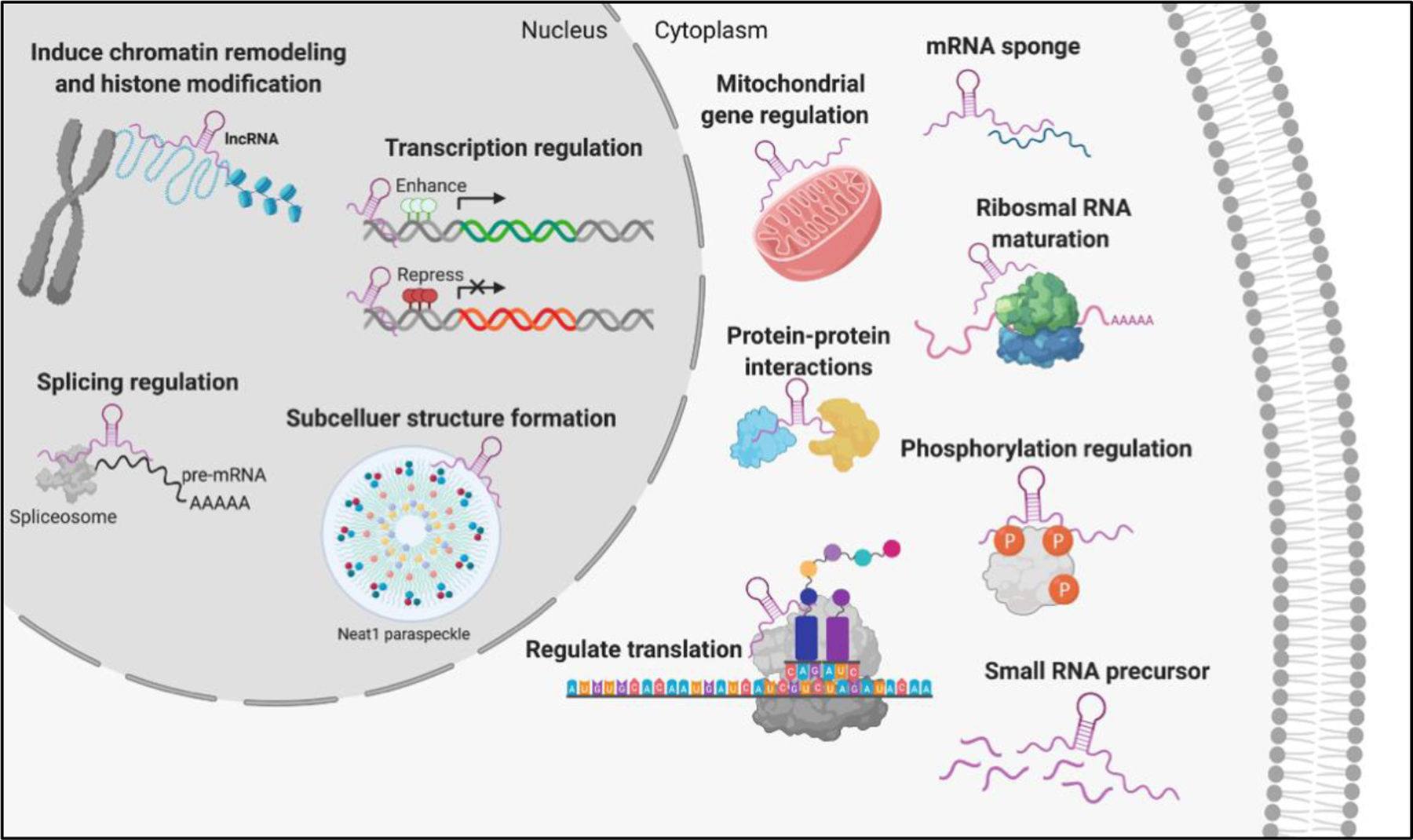
lncRNAs contribute to numerous processes necessary for cellular function and homeostasis. As a result, aberrant expression of lncRNAs seen in disease can significantly alter cellular function resulting in impaired learning and memory. See text for detailed discussion.
Table 1.
Selected lncRNAs and Their Altered Expression in Memory Related Conditions lncRNA Description Regulation
| lncRNA | Description | Regulation | Associated disease | Related Biological Processes | Functions & Implications | References |
|---|---|---|---|---|---|---|
| 17A | LncRNA 17A | Up | AD | Cognitive decline, neurodegeneration | Regulates alternative splicing and signaling. Linked to Aβ secretion and elevation of Aβ42 production. Dysregulation leads to deactivation of GABAB signaling, autophagy and neurodegeneration. | 246–248 |
| ANRIL | Antisense Noncoding RNA in The INK4 Locus (CDKN2BAS1) | Up | AD | Neurodegeneration | Regulates gene transcription repression. Involved in chromatin modifications via PRC2 recruitment. | 249–251 |
| BACE1-AS | Beta-Secretase 1-Antisense RNA | Up | AD | Neurodegeneration, protein aggregation | Involved in post-transcriptional regulation and BACE1 mRNA stability, competes with miR-485–5p for binding to BACE1 and prevents it’s targeting on BACE1 mRNA. Increases Aβ 1–42 accumulation. | 252–255 |
| BC1 | LncRNA BC1 (BC1-FMRP) | Up | AD | Spatial learning and memory impairments, protein aggregation | Involved in mRNA translation and downregulation of BC1. Leads to accumulation of Aβ peptides. | 227,256–259 |
| BC200 | LncRNA BC200 | Soma: Up Dendritic: Down | AD | Cognitive decline | Regulates local translation at the synapse, long-term synaptic plasticity and enhances BACE1 and Aβ1–42 expression. | 256–258,260–263 |
| EBF3-AS | Early B Cell Transcription Factor 3-Antisense RNA | Up | AD | Neurodegeneration | Promotes neuronal apoptosis through Aβ25–35- and okadaic acid. | 264 |
| GDNF-AS1 | Glial Cell Derived Neurotrophic Factor (GDNF Antisense RNA 1) | -- | AD | Neurodegeneration | Involved in mRNA translation. | 206 |
| GDNF-AS | Glial Cell Derived Neurotrophic Factor -Antisense RNA | down | PD | Cognitive decline; neurodegeneration | Involved in mRNA stability. | 263 |
| LRP1‐AS | LDL Receptor Related Protein 1-Antisense RNA | Up | AD | Cognitive decline. neurodegeneration | Transcription repression by sequestration of chromatin-regulatory proteins. Linked to the increasing Aβ formation and decreased clearance. Regulate LRP1 expression. | 265–268 |
| MEG3 | Maternally Expressed 3 | Down | AD | Cognitive decline, Involved with spatial learning and memory ability | Upregulation of MEG3 inhibits the pathological injury and hippocampal neurons apoptosis, decreased Aβ expression, inhibited oxidative stress and inflammatory injury. Involved in induced astrocytes activation through blocking PI3/Akt pathway. | 226 |
| MEG3 | Maternally Expressed 3 | Down | HD | Neurodegeneration | Involved in gene regulation. MEG3 is a direct target of REST and modulate mHTT aggregation. | 251,269–271 |
| NEAT1 | Nuclear Paraspeckle Assembly Transcript 1 | Up | AD | Cognitive decline | Essential for Paraspeckles formation, integrity, gene expression regulation and miRNA sponging. | 28 |
| NEAT1, NEAT1-L, NEAT1-S | Nuclear Paraspeckle Assembly Transcript 1 | Up | HD | Cognitive decline | Decreasing NEAT1 expression lowers mHTT aggregates and TP53 expression in HD. NEAT1 provides neuroprotection against mHtt-induced cytotoxicity NEAT1-L) and oxidative stress-induced injury NEAT1-S). | 251,271–273 |
| NEAT1 | Nuclear Paraspeckle Assembly Transcript 1 | Up | PD | Cognitive decline | Upregulation of NEAT1 supports Bax/BCl ratio, caspase 3 activity, α-synuclein expression, MPTP concentration, LC3-II/LC3-I level and promotes PINK1 protein stability. NEAT1 serves as miR-124 decoy and promotes cell death and apoptosis. | 251,274–276 |
| MIAT | Myocardial Infarction Associated Transcript | Down | AD | Cognitive decline, Neurodegeneration, Protein aggregation | MIAT regulates Aβ clearance through LRP1 expression. Downregulation of MIAT promotes miR-150–5p/VEGF-mediated fibrillogenesis, reduces the number of microvessels and the expression of tight junction proteins. Loss of MIAT increases Aβ40 and Aβ42 levels and promotes neuronal loss. | 251,277 |
| NAT-RAD18 | Natural antisense transcript against RAD18 E3 Ubiquitin Protein Ligase | Up | AD | Neurodegeneration | Promotes neuron loss through the down regulation of RAD18 expression. | 278 |
| NDM29 | Neuroblastoma Differentiation Marker 29 | Up | AD | Neurodegeneration, Protein aggregation | Promotes Alu-induced inflammation and processing of APP and amyloid β secretion. | 247,279 |
| SORL1-AS | Sortilin Related Receptor 1-Antisense RNA | Up | AD | Protein aggregation, Cognitive decline | Decreases SORL1 expression by altering mRNA splicing and impairs APP processing. | 251,280 |
| SOX2-OT | SRY-Box Transcription Factor 2-Overlapping Transcript | Up | AD, PD | Neurodegeneration | Regulates co-transcribed Sox2 gene expression, reduces Frizzled 3/5 FZD3/5)-mediated Wnt signaling and triggers oxidative stress generation that leads to apoptosis and neuronal loss. | 281,282 |
| HAR1A, HAR1F | Highly Accelerated Region 1A, F | Down | HD | Neurodegeneration | Direct targets of REST. Mutated huntingtin gene lead to abnormal nuclear-cytoplasmic REST/NRSF trafficking leading to downregulation of HAR1 expression and subsequently repression of numerous neuronal genes. | 251,283–286 |
| HTT-AS | HTT Antisense RNA | Down | HD | -- | Overexpression of HTT-AS downgrades endogenous HTT transcript levels. | 248,287 |
| LINC003 41 | SYNE3: Spectrin Repeat Containing Nuclear Envelope Family Member 3 | Up | HD | -- | Unknown | 251,269 |
| LINC003 42 | Long Intergenic Non-Protein Coding RNA 342 | Down | HD | -- | Unknown | 251,269 |
| RPS20P 22 | Ribosomal Protein S20 Pseudogene 22 | Up | HD | -- | RPS20P22 regulates RPS20 expression. Reduction of RPS20P22 leads to accumulation of p53. | 251,269 |
| TUG1 | Taurine Up-Regulated 1 | Up | HD, Aging | Cognitive decline, neurodegeneration | Direct downstream target of p53. Binds to the PRC2 epigenetic regulatory complex of genes and sponge/decoy function. | 250,288,289 |
| TUNA (TUNAR) | Tcl1 Upstream Neuron-Associated lincRNA | Down | HD | -- | TUNA expression declines significantly with increased HD disease grade. | 251,290 |
| HOTAIR | HOX Transcript Antisense RNA | Up | PD | Neurodegeneration | HOTAIR upregulation is associated with LRRK2 upregulation and the induction of caspase 3-dependent apoptosis. | 291–293 |
| MALAT1 | Metastasis Associated Lung Adenocarcino ma Transcript 1 | Up | PD | Neurodegeneration | Involved in synapse development by regulating synapse formation and maintenance of genes expression. Modulates the recruitment of SR family pre-mRNA-splicing factors to the transcription site. | 294,295 |
| MALAT1 | Metastasis Associated Lung Adenocarcino ma Transcript 1 | -- | AD | Neurodegeneration | Negatively regulates the CDK5R1/p35 complex and promotes cell death by controlling expression of the miR-15/107 family. | 217,251 |
| NORAD | Non-Coding RNA Activated by DNA Damage | Down | PD | --- | NORAD stabilizes the genome through PUMILIO proteins. Downregulation of NORAD induces cytotoxicity through caspase3/7, ROS and LDH activity. | 251,296 |
| P21 | Long non-coding RNA-p21 | Up | PD | Neurodegeneration | p21 is a miR-1277–5p decoy and regulates α-Synuclein through miR-1277–5p. Upregulation of p21 inhibits cell viability, promotes caspase 3 activation, and increases Bcl family-initiated apoptosis. | 297,298 |
| PINK1-AS | PTEN Induced Kinase 1-Antisense RNA | Up | PD | Neurodegeneration | Regulates the stability of Pink1 transcript, involved in mitochondrial biogenesis and increases the sensitivity to apoptosis. | 299 |
| SNHG1 | Small Nucleolar RNA Host Gene 1 | Up | PD | Neurodegeneration | Upregulation of SNHG1 promotes neuroinflammation. | 300–302 |
| SNHG1 | Small Nucleolar RNA Host Gene 1 | Down | PD | Neurodegeneration | Involved in miR-15 decoy and inhibits miR-15 function. | 268,303 |
| UCHL1-AS1 | Ubiquitin C-Terminal Hydrolase L1-Antisense RNA 1 | Down | PD | Neurodegeneration | Involved in dopaminergic neuron differentiation and maintenance, cellular stress response and miRNA decoy. Promotes Uchl1 expression by upregulating the translation process. | 304–306 |
| RMST | Rhabdomyosar coma 2-associated Transcript | -- | -- | Neurogenesis, Neurodegeneration | Transcriptionally repressed by REST, required for the binding of SOX2 to promoter regions of neurogenic transcription factors and involved in neurogenesis. | 307 |
| GAS5 | Growth-arrest-specific 5 | up | Aging | Cognitive decline, neurodegeneration | Prepares the cell to apoptosis. Upregulation correlates with impaired learning and novelty-induced behavior. | 308–310 |
| DGCR5 | DiGeorge syndrome critical region gene 5 | Down | HD | Neurodegeneration | DGCR5 is downstream target of REST in HD disease. | 269,311,312 |
Age-associated memory impairment
Why some individuals age with cognition relatively intact and others slip precariously into dementia is a question that has intrigued and beleaguered the scientific community and layperson alike. In translating external experience or stimuli into functionally relevant gene expression changes, epigenetics mechanisms are a critical component of the aging process 179–181. Studies investigating the various hallmarks of aging have revealed significant differences in lncRNA expression 182. Differential lncRNA expression is particularly pronounced in the brain, including thousands of novel lncRNA identified as “altered” in the synaptosomes of aging mice 183, as well as age-related expression of two lincRNAs (LINC-RBE and LINC-RSAS) described in the rat brain 184,185. These findings are consistent with trends seen in humans during aging; for example, post-operative cognitive dysfunction is particularly significant in elderly patients, and has been correlated with 868 differentially expressed lncRNAs, as well as 690 differentially expressed mRNAs related to inflammation and apoptotic pathways 41. Similarly, studies of post-cardiac arrest cognitive impairment revealed significant changes in hippocampal expression of the lncRNA RNANONMMUT113601.1 and the mRNA Shc1, also an inflammation and apoptosis coupled gene 186. From these data we have two significant takeaways: first, differentially expressed lncRNA or groups of lncRNA have the potential to serve as biomarkers for age-associated cognitive impairment depending on the timeline with which their expression changes. Second, exosomes or membrane nanovesicles secreted by most cell types including those in the CNS 187, are carriers of a variety of RNAs, including lncRNA 188. This means there is the potential for minimally invasive (e.g., blood draw) means of measuring brain-derived lncRNA in order to identify those with or predisposed to age-associated cognitive decline.
One lncRNA that has been well studied in the context of aging is Neat1. There is an increase in lncRNA Neat1 expression in the brain of both humans and animal modes of normal aging 37. Neat1 mediates age-related impairment in hippocampus dependent memory formation 37. Downregulation of Neat1 (via nimodipine used to treat subarachnoid hemorrhage) resulted in upregulation of miR-27a and subsequent downregulation of MAPT, contributing to improved cognitive function 42. Interestingly, Neat1 knockout mice showed no deficits in memory 189 likely indicating redundant pathways capable of compensatory function in the case of constitutive knockout.
Alzheimer’s disease
The most significant risk factor for developing Alzheimer’s disease is aging. Therefore, with our rapidly aging population, significant funding and research effort has been devoted to the study of the mechanisms underlying AD in hopes of identifying novel therapeutic targets. Clinical trials targeting the accumulation of A β have been largely unsuccessful 190 necessitating a different approach. A number of lncRNAs have been implicated in the pathophysiology of AD and were well reviewed recently by 43,76,77,191–193. For example, 16 age-associated and 12 gender-associated lncRNAs were identified as dysregulated in AD; Specifically, SNHG19 and LNC00672 were significantly correlated with Braak stage, while AS1, LY86-AS1 and LINC00639 were negatively correlated with Braak stage 194. Interestingly, dysregulated lncRNA expression appears to be consistent across various AD models, including Intranasal LPS-mediated AD disease model in mice 195. Likewise, 315 lncRNAs and 311 mRNAs showed significantly altered expression in the hippocampus of a rat model of AD 196. However, understanding the mechanisms that result in differentially expressed lncRNA largely remains a mystery, although at least one study suggests that expression of many lncRNA may be dependent on histone modifications in AD 197.
Extensive research in humans and animal models suggests a role for epigenetic regulation of gene transcription in the development and progression of AD198–204. Altered DNA methylation205,206 and hydroxymethylation207 patterns have been described in humans with AD, including at known susceptibility genes including APOE208, BIN1,209 and TREM2210,211. Likewise, alterations in post-translational histone modification patterns are associated with synaptic dysfunction and memory impairment in AD212–215. Further, studies using a mouse model of AD indicated that a substantial number of differentially expressed lncRNAs and subject to transcriptional regulation by histone modifications197. Based on our previous discussion regarding lncRNA regulation gene expression through via epigenetic crosstalk, it stands to reason that this aberrant lncRNA expression likely contributes to AD pathology. As argued earlier, there is a long way to go towards understanding the governance of lncRNA expression both in the healthy brain and disease.
In the search for a viable biomarker for AD and potential progression, lncRNA are proving a promising target. For example, cyclin-dependent kinase 5 (CDK5) deregulation is highly correlated with progression of AD 216. Two lncRNA NEAT1 and HOTAIR have been shown to negatively regulate CDK5R1 while the lncRNA MALAT1 appears to positively regulate CDK5R1. Together with human data showing positive correlation between CDK5R1 and NEAT1 in brain tissue from AD patients, these lncRNAs may serve as biomarkers and potential neuroprotective agents against AD progression 217. An additional potential biomarker for AD identified recently includes BACE1-AS has been found to be elevated in the exosomes of AD patient 218.
LncRNA appear to also be involved in the pathology of AD. The neuronal RNA-binding protein HuD stabilizes the lncRNA BACE1AS contributing to enhanced BACE1 expression and APP levels in patients with AD and HuD overexpressing mice 219. Perhaps unsurprisingly given the significant role Neat1 appears to play in normal aging, the lncRNA Neat1 is upregulated in the APP/PS1 transgenic model of AD and interacts with NEDD4L to promote PINK1 ubiquitination and degradation, further promoting the pathogenesis of AD 220. Neuron-specific lncRNA neuroLNC interacts with the RNA-binding protein TDP-43 resulting in the stabilization of mRNAs encoding for presynaptic proteins, thus influencing neuronal excitability 221. Alterations in expression of several lncRNAs, either endogenously or artificially is also capable of halting the progression or limiting AD-associated pathology. For example, the apolipoprotein A-I mimetic D4F decreases expression of Aβ through up-regulation of long non-coding RNA SIRT1-AS 222. Silencing of the lncRNA SOX21-AS1 resulted in decreased oxidative stress injury and reduced apoptosis on hippocampal neurons of and AD mouse model 223. In an A β25–35 treated hippocampal mouse neurons, decreasing expression of the lncRNA TUG1 limits apoptosis via elevation of miR-15a and suppression of ROCK1224
Perhaps most importantly, targeting of various lncRNA appears to hold significant promise for future therapeutics. BACE1-AS inhibition via lentiviral siRNA expression improved memory and learning behaviors in SAMP8 mice 225. Up-regulation of the lncRNA Meg3 in the hippocampus of an AD rat model improved spatial learning and memory, inhibited apoptosis of hippocampal neurons and oxidative stress injury via the PI3/Akt pathway. 226. Finally, the lncRNA BC1 induces APP mRNA translation in an AD mouse model, while inhibition of BC1 protects against spatial learning and memory deficits 227.
Epilepsy
Epigenetic control of gene transcription contributes to the aberrant network excitability and recurrent seizures 228,229 however, the functional role of lncRNA in the pathogenesis of epilepsy is still not completely understood, although the state of their role in the disease has been recently reviewed 230–232. Differential expression of 497 lncRNAs have been identified in mesial temporal lobe epilepsy (TLE) patients with hippocampal sclerosis, along with co-dysregulated mRNAs correlated with inflammatory response and neuropeptide receptor activity predicted to play a role in epileptogenesis 233. For example, hippocampal and serum levels of the lncRNA ILF3-AS1 were increased in TLE patients. Ectopic expression of ILF3-AS1 in astrocytes increased expression of several metalloproteinases connected with epilepsy and decreased expression of miR-212 which is consistent with lower levels observed in TLE patients 234.
Nearly a third of epileptic patients develop resistance to available anti-epileptic drug therapeutic options. As such, there is emergent need to identify novel mechanisms and biomarkers for the progression of epilepsy. LncRNAs are emerging as interesting potential biomarker in in epilepsy as well. To date numerous different lncRNAs have been identified as differentially expressed in epilepsy 235,236, with some displaying additional sex-specific differences 237.
Rodent models of epilepsy have been invaluable in identifying the various roles lncRNA might play in the pathogenesis of epilepsy. For example, H19 is significantly upregulated in the hippocampus of a rat model of TLE and aggravates seizure induced neuronal apoptosis via sponging the microRNA let-7b 238. In a rat model of epilepsy downregulation of MALAT1 results in activation of the PI3K/Akt pathways decreasing autophagy and apoptosis in hippocampal neurons 239. Inhibition of the lncRNA PVT1 decreases the loss of neurons and astrocyte activation, as well as increases expression of BDNF in the hippocampus by downregulating the Wnt signaling pathway 240. Once again, the lncRNA Neat1 has been shown to be altered in the disease condition, binding epilepsy associated potassium channel interacting proteins and knockdown induces a neuronal hyper-potentiation phenotypes in iPSCs. Neat1 is also acutely down-regulated in response to neuronal activity, however it becomes unresponsive with chronic stimulation in a rat model of TLE 168.
Cognitive deficits are well-documented in intractable epilepsy 241,242, however mechanisms underlying these cognitive deficits have not been fully elucidated. Expression of the lncRNA UCA1 and NF-□B mRNA are higher in brain tissues of the pilocarpine model of Epilepsy 243. NF-□B is well-known to mediate the gene expression dependent process of synaptic function and memory 244, making its regulation of particular interest in terms of identifying novel therapeutic targets. Indeed, lncRNA interaction with NF- B signaling is a reappearing theme, with downregulation of the lncRNA ANRIL restoring learning and memory via the NF-□B signaling pathway in streptozotocin-induced diabetic rats 245.
Future outlook
The studies reviewed here support a significant role for lncRNAs in epigenetic regulation of transcriptional programs; however, our understanding of how lncRNAs function in the brain is still in relative infancy. Here, we discuss critical questions remaining in the field regarding how lncRNAs function in the context of memory and associated disorders. To better understand how specific lncRNAs contribute to memory formation, lncRNAs must be studied in a region-specific, sex-specific and cell-type-specific manner. Finally, we address the available technologies that can serve to probe important remaining research questions in the field, as well as the advantages and limitations of these molecular genetic approaches.
Brain region and sex specificity
In this section, we consider what is known about brain region specific functions in memory, and subsequently how that knowledge can be applied to the study of lncRNAmediated transcription of memory-permissive genes. Differential expression of several lncRNAs exists between various brain regions 36,38,313, and can be altered in the case of disease 74. It is well established that specific brain regions such as the hippocampus play a critical role in the acquisition and retrieval of memory 314–318. Furthermore, both human studies and rodent models demonstrate that hippocampal subfields show specialization associated with memory 319–324. Thus, it seems likely that lncRNA-mediated regulation of epigenetic mechanisms plays a role in the region-specific transcriptome critical for memory formation.
Given that epigenetic integration of stimuli can confer significant differences in gene expression based on sex 325, expression of specific lncRNAs may vary by sex. Indeed, that appears to be the case in humans and animal models, with differential expression of lncRNAs between the sexes occurring in both the healthy brain and disease states 194,326–328. The examination of sex differences in lncRNAs and influence on memory formation remain to be studied, and further, how functional control of lncRNAs might be leveraged for more precision directed therapeutics.
Cell type specificity
While research evidence has revealed glia specific enrichment of lncRNAs, most studies continue to focus on the role of lncRNAs in neuronal populations. Similarly, numerous studies examining the effects of manipulating lncRNAs in different brain regions did not determine if lncRNAs in specific cell-types is driving behavioral changes.
For example, overexpression of the lncRNA MEG3 via third ventricle infusion of overexpression plasmid led to improved learning and memory in a rodent model of AD 226, a significant finding at a time when novel treatments for AD are desperately needed. However, these broad manipulations did not distinguish if the impacts on memory were due to reduced neurodegeneration or limited astrocyte activation, or some combination thereof. This is an important distinction, as broad overexpression of the lncRNA MEG3 is also reported to play a role in ischemic stroke and may accelerate associated pathological progression 329. Additionally, determining differences in functional lncRNAs in major brain cell-types (neurons, astrocytes, microglia) should be considered in future studies, as cellular subpopulations exist with distinct lncRNA gene signatures 330. Moreover, lncRNAs impact microglia activation and associated inflammatory cascades 331–335. The well-studied lncRNA Xist was recently discovered to have microglia-specific functions, downregulating apoptosis and inflammatory associated with microglia following spinal cord injury 336. The potential functional implications of cell-type specific differences cannot be overstated given the growing body of literature demonstrating the profound impact of altered glia function on synaptic function 337,338, memory, 339–342 and disease 340,343,344. Therefore, distinguishing cell-type specific contributions of lncRNA during memory formation may lead to novel translational approaches for treating neurological disorders while limiting unintended, off-target effects.
Technological advances and limitations
Recent advances in our understanding of lncRNA are greatly indebt to rapidly progressing sequencing technologies. Despite our expanding catalogue of known lncRNAs, the functional roles of these transcripts will depend on techniques designed to study cell-specific function. Increasing use of single nucleus RNA sequencing (snRNA seq) has already provided an abundance of data, particularly in the context of disease states 345,346. For purposes of studying the functionality of lncRNA in animal models, innovative techniques are required in order to isolate of cell-type specific nuclear fractions, as well as manipulating transcripts in a cell-type specific manner. Fluorescent activated cell sorting (FACS) and Magnetically activated cell sorting (MACS) are both widely utilize cellular techniques that enable efficient cell-type enrichment and high viability for subsequent culture 347. In particular, MACS has proven valuable for isolating multiple cell types from the same brain and limiting damage to fragile glia projections which frequently occurs with FACS 348,349. For the purposes of deep sequencing, FACS has been shown to deliver cleaner microglia fractions 347. Difficulties arise when attempting to combine region specific and cell-type specific studies, given the relatively small volumes of tissues involved. However, these studies are critical as we have discussed significant differences in both cell type and regional functions of lncRNAs. In cases such as this, in situ hybridization methods provide spatial information and can be combined with cell-type specific markers for further detail.
There are numerous methods used to manipulate lncRNAs for functional studies 350, however cell-type specific manipulation of lncRNAs is a more challenging task. Most RNAi based methods (siRNA or shRNA) are adequate for cell culture designs 351,352, but in vivo lack the specificity necessary to exclusively target lesser studies cells such as astrocytes or microglia 353. There are multiple technological approaches designed to address this problem. Recently, the use of aptamer-siRNA chimeras has gained considerable interest as a treatment strategy, particularly for the treatment of cancer 354. Aptamers are small single-stranded oligonucleotides which bind with high affinity to their targets which can include lipids, proteins or other small molecules 355. The development of aptamer-siRNA chimeric RNAs, which can subsequently be internalized by the target cell and processed by Dicer, has enabled cell type specific delivery of functional siRNA 356. Despite ongoing challenges to therapeutic application of aptamer-siRNA chimeras, 357,358, initial studies have begun using this technique for treatment of Glioblastoma both in vitro 359 and in vivo mouse models 360.
There is also great potential for Adeno-Associated Viral (AAV) delivery, which is already capable of targeting specific cell types for many over expression studies using cell-type specific promoters 361. A more extensive review of the various techniques which can be used in combination with AAVs for targeting neuronal populations can be found in 362. These cell-type specific promoters are often not however suitable for short siRNA/shRNA sequences necessary for knockdown studies in that they require Pol III recruitment for expression of non-polyadenylated sequences 363,364. Lentiviral vectors are larger with the potential to house shRNA targeting lncRNA of interest, however lentivirus is already known to result in increased expression of the lncRNA Neat1 365, and thus its application used with caution.
Metabolic signaling and lncRNA
An additional research area that deserves further exploration, is investigation of lncRNAs involved in metabolic function, and the reciprocal regulation of lncRNAs by metabolic signaling. Metabolic signaling is mediated at multiple signaling and tissue levels, including the brain 366,367. There is significant interest in the effect of diet on cognitive function 368 and dietary approaches to disorders of memory 369. The so-called ketogenic diet has proven promising as an adjuvant or alternative therapy for pediatric patients with intractable epilepsy and other neurological disorder 370,371. Despite the tentative success of dietary therapeutics, very little is understood about the mechanisms by which these metabolic changes occur, and how they impact memory function. LncRNA have been found to participate in the establishment of metabolic homeostasis 372, representing a promising therapeutic avenue for many diseases. Metabolic reprograming is present with aging 373, cancer 374 and neurodegenerative diseases 375 and we are just beginning to understand the regulatory roles lncRNA may play and the therapeutic applications of targeting these lncRNA 376. Inspiration can be drawn from the cancer research field that aims to understand how lncRNAs contribute to metabolically relevant gene transcription programming. Thus, there is growing appreciation for similar approaches in understanding how lncRNAs control the epigenome and subsequent transcription programs to impact memory formation in health and in memory impairments.
Concluding remarks
The studies discussed here, and likely many others, demonstrate a complex epigenetic regulatory process driving dynamic and or persistent gene transcription necessary for memory. In this review, we have described how lncRNAs provide a valuable window by which we can view the crosstalk of epigenetic marks both in the healthy brain, and disease states. Finally, we discussed several questions that remain to be answered regarding lncRNAs crosstalk with epigenetic mechanisms in specific brain regions or specialized cell types affects memory, and how this crosstalk may be altered in disorders of memory. The contribution of lncRNAs to this epigenetic crosstalk is only now being fully appreciated, and much of what we know about lncRNAs, has yet to be fully investigated in the context of memory. Future work should emphasize studies on lncRNA-epigenetic mediated gene transcription changes and determine if these mechanisms are transcript specific. Overall, these lncRNA-epigenetic mechanisms are engaged in an intricate, multi-leveled crosstalk geared towards homeostatic cellular function, with consequences for dysregulation at specific genes, not necessarily bulk changes in epigenetic marks, driving pathology. Importantly, the therapeutic potential of lncRNA-epigenetic transcriptional processes may be harnessed, and additional studies are crucial to elucidating the consequences of differential lncRNAs and the various epigenetic mechanisms by which they function to control large transcriptional programs in the brain to sub serve the process of memory formation.
Highlights.
Epigenetic mechanisms drive transcriptional programs necessary for memory formation.
LncRNAs interact with key epigenetic mechanisms to regulate gene expression.
Aberrant expression of lncRNAs is associated with cellular dysfunction in cognitive disorders.
Expression of LncRNAs in the brain is region-specific, sex-specific and cell-type specific.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors declare no conflict of interest.
References
- 1.Campbell RR & Wood MA How the epigenome integrates information and reshapes the synapse. Nature reviews. Neuroscience 20, 133–147, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xylaki M, Atzler B & Outeiro TF Epigenetics of the Synapse in Neurodegeneration. Current neurology and neuroscience reports 19, 72, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortés-Mendoza J, Díaz de León-Guerrero S, Pedraza-Alva G & Pérez-Martínez L Shaping synaptic plasticity: the role of activity-mediated epigenetic regulation on gene transcription. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience 31, 359–369, (2013). [DOI] [PubMed] [Google Scholar]
- 4.Lubin FD, Gupta S, Parrish RR, Grissom NM & Davis RL Epigenetic mechanisms: critical contributors to long-term memory formation. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry 17, 616–632, (2011). [DOI] [PubMed] [Google Scholar]
- 5.Jarome TJ & Lubin FD Epigenetic mechanisms of memory formation and reconsolidation. Neurobiology of learning and memory 115, 116–127, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cholewa-Waclaw J et al. The Role of Epigenetic Mechanisms in the Regulation of Gene Expression in the Nervous System. The Journal of neuroscience : the official journal of the Society for Neuroscience 36, 11427–11434, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sweatt JD The emerging field of neuroepigenetics. Neuron 80, 624–632, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halder R et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nature neuroscience 19, 102–110, (2016). [DOI] [PubMed] [Google Scholar]
- 9.Morris MJ & Monteggia LM Role of DNA methylation and the DNA methyltransferases in learning and memory. Dialogues in clinical neuroscience 16, 359–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day JJ & Sweatt JD DNA methylation and memory formation. Nature neuroscience 13, 1319–1323, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyward FD & Sweatt JD DNA Methylation in Memory Formation: Emerging Insights. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry 21, 475–489, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira AM DNA methylation: a permissive mark in memory formation and maintenance. Learning & memory (Cold Spring Harbor, N.Y.) 23, 587–593, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarome TJ, Butler AA, Nichols JN, Pacheco NL & Lubin FD NF-κB mediates Gadd45β expression and DNA demethylation in the hippocampus during fear memory formation. Frontiers in molecular neuroscience 8, 54, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lubin FD, Roth TL & Sweatt JD Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci 28, 10576–10586, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta S et al. Histone methylation regulates memory formation. J Neurosci 30, 3589–3599, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta-Agarwal S et al. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J Neurosci 32, 5440–5453, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins BE, Greer CB, Coleman BC & Sweatt JD Histone H3 lysine K4 methylation and its role in learning and memory. Epigenetics & chromatin 12, 7, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sultan FA & Day JJ Epigenetic mechanisms in memory and synaptic function. Epigenomics 3, 157–181, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puckett RE & Lubin FD Epigenetic mechanisms in experience-driven memory formation and behavior. Epigenomics 3, 649–664, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudenko A & Tsai LH Epigenetic regulation in memory and cognitive disorders. Neuroscience 264, 51–63, (2014). [DOI] [PubMed] [Google Scholar]
- 21.Djebali S et al. Landscape of transcription in human cells. Nature 489, 101–108, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anastasiadou E, Jacob LS & Slack FJ Non-coding RNA networks in cancer. Nature reviews. Cancer 18, 5–18, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL & Barry G Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron 88, 861–877, (2015). [DOI] [PubMed] [Google Scholar]
- 24.Esteller M Non-coding RNAs in human disease. Nature reviews. Genetics 12, 861–874, (2011). [DOI] [PubMed] [Google Scholar]
- 25.Ponting CP, Oliver PL & Reik W Evolution and functions of long noncoding RNAs. Cell 136, 629–641, (2009). [DOI] [PubMed] [Google Scholar]
- 26.Grinman E, Espadas I & Puthanveettil SV Emerging roles for long noncoding RNAs in learning, memory and associated disorders. Neurobiol Learn Mem 163, 107034, (2019). [DOI] [PubMed] [Google Scholar]
- 27.Grinman E et al. Activity-regulated synaptic targeting of lncRNA ADEPTR mediates structural plasticity by localizing Sptn1 and AnkB in dendrites. Sci Adv 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liau WS, Samaddar S, Banerjee S & Bredy TW On the functional relevance of spatiotemporally-specific patterns of experience-dependent long noncoding RNA expression in the brain. RNA Biol 18, 1025–1036, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler AA, Webb WM & Lubin FD Regulatory RNAs and control of epigenetic mechanisms: expectations for cognition and cognitive dysfunction. Epigenomics 8, 135–151, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qureshi IA & Mehler MF Non-coding RNA networks underlying cognitive disorders across the lifespan. Trends Mol Med 17, 337–346, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattick JS The central role of RNA in human development and cognition. FEBS Lett 585, 1600–1616, (2011). [DOI] [PubMed] [Google Scholar]
- 32.Woldemichael BT & Mansuy IM Micro-RNAs in cognition and cognitive disorders: Potential for novel biomarkers and therapeutics. Biochem Pharmacol 104, 1–7, (2016). [DOI] [PubMed] [Google Scholar]
- 33.Mattick JS & Rinn JL Discovery and annotation of long noncoding RNAs. Nature structural & molecular biology 22, 5–7, (2015). [DOI] [PubMed] [Google Scholar]
- 34.Gil N & Ulitsky I Regulation of gene expression by cis-acting long non-coding RNAs. Nature reviews. Genetics 21, 102–117, (2020). [DOI] [PubMed] [Google Scholar]
- 35.Quinodoz S & Guttman M Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends in cell biology 24, 651–663, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadakkuzha BM et al. Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Frontiers in cellular neuroscience 9, 63, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butler AA, Johnston DR, Kaur S & Lubin FD Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Science signaling 12, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goff LA et al. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proceedings of the National Academy of Sciences of the United States of America 112, 6855–6862, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang KC & Chang HY Molecular mechanisms of long noncoding RNAs. Molecular cell 43, 904–914, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labonté B et al. Regulation of impulsive and aggressive behaviours by a novel lncRNA. Molecular psychiatry, (2020). [DOI] [PMC free article] [PubMed]
- 41.Li M et al. Identification of the Potential Key Long Non-coding RNAs in Aged Mice With Postoperative Cognitive Dysfunction. Frontiers in aging neuroscience 11, 181, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li JW et al. Nimodipine Improves Cognitive Impairment After Subarachnoid Hemorrhage in Rats Through IncRNA NEAT1/miR-27a/MAPT Axis. Drug design, development and therapy 14, 2295–2306, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li D et al. Insights into lncRNAs in Alzheimer’s disease mechanisms. RNA biology, 1–11, (2020). [DOI] [PMC free article] [PubMed]
- 44.Pan YB et al. Prognostic and Predictive Value of a Long Non-coding RNA Signature in Glioma: A lncRNA Expression Analysis. Frontiers in oncology 10, 1057, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren D, Chen W, Cao K, Wang Z & Zheng P Expression Profiles of Long Non-coding RNA and Messenger RNA in Human Traumatic Brain Injury. Molecular therapy. Nucleic acids 22, 99–113, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen KW & Chen JA Functional Roles of Long Non-coding RNAs in Motor Neuron Development and Disease. J Biomed Sci 27, 38, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmer-Bensch G Emerging Roles of Long Non-Coding RNAs as Drivers of Brain Evolution. Cells 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark BS & Blackshaw S Understanding the Role of lncRNAs in Nervous System Development. Adv Exp Med Biol 1008, 253–282, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hart RP & Goff LA Long noncoding RNAs: Central to nervous system development. Int J Dev Neurosci 55, 109–116, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dolati S et al. The role of exosomal non-coding RNAs in aging-related diseases. Biofactors, (2021). [DOI] [PubMed]
- 51.He J, Tu C & Liu Y Role of lncRNAs in aging and age-related diseases. Aging Med (Milton) 1, 158–175, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pereira Fernandes D, Bitar M, Jacobs FMJ & Barry G Long Non-Coding RNAs in Neuronal Aging. Noncoding RNA 4, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szafranski K, Abraham KJ & Mekhail K Non-coding RNA in neural function, disease, and aging. Front Genet 6, 87, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mishra P & Kumar S Association of lncRNA with regulatory molecular factors in brain and their role in the pathophysiology of schizophrenia. Metab Brain Dis, (2021). [DOI] [PubMed]
- 55.Rusconi F, Battaglioli E & Venturin M Psychiatric Disorders and lncRNAs: A Synaptic Match. Int J Mol Sci 21, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu N, Wang ZZ, Zhao M, Zhang Y & Chen NH Role of non-coding RNA in the pathogenesis of depression. Gene 735, 144276, (2020). [DOI] [PubMed] [Google Scholar]
- 57.Punzi G, Bharadwaj R & Ursini G Neuroepigenetics of Schizophrenia. Prog Mol Biol Transl Sci 158, 195–226, (2018). [DOI] [PubMed] [Google Scholar]
- 58.Tang J, Yu Y & Yang W Long noncoding RNA and its contribution to autism spectrum disorders. CNS Neurosci Ther 23, 645–656, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang X, Luo YL, Mao YS & Ji JL The link between long noncoding RNAs and depression. Prog Neuropsychopharmacol Biol Psychiatry 73, 73–78, (2017). [DOI] [PubMed] [Google Scholar]
- 60.Tripathi S et al. The Expanding Regulatory Mechanisms and Cellular Functions of Long Non-coding RNAs (lncRNAs) in Neuroinflammation. Mol Neurobiol, (2021). [DOI] [PubMed]
- 61.Lim KH, Yang S, Kim SH, Chun S & Joo JY Discoveries for Long Non-Coding RNA Dynamics in Traumatic Brain Injury. Biology (Basel) 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Z et al. Long non-coding RNAs in the spinal cord injury: Novel spotlight. J Cell Mol Med 23, 4883–4890, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Z et al. The role of long noncoding RNA in traumatic brain injury. Neuropsychiatr Dis Treat 15, 1671–1677, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chandran R, Mehta SL & Vemuganti R Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochem Int 111, 12–22, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolska M et al. Long Non-coding RNAs as Promising Therapeutic Approach in Ischemic Stroke: a Comprehensive Review. Mol Neurobiol 58, 1664–1682, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akella A, Bhattarai S & Dharap A Long Noncoding RNAs in the Pathophysiology of Ischemic Stroke. Neuromolecular Med 21, 474–483, (2019). [DOI] [PubMed] [Google Scholar]
- 67.Alishahi M et al. Long non-coding RNAs and cell death following ischemic stroke. Metab Brain Dis 34, 1243–1251, (2019). [DOI] [PubMed] [Google Scholar]
- 68.Chen R, Xu X, Huang L, Zhong W & Cui L The Regulatory Role of Long Noncoding RNAs in Different Brain Cell Types Involved in Ischemic Stroke. Front Mol Neurosci 12, 61, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Q, Liu X & Zhu R Long Noncoding RNAs as Diagnostic and Therapeutic Targets for Ischemic Stroke. Curr Pharm Des 25, 1115–1121, (2019). [DOI] [PubMed] [Google Scholar]
- 70.Bao MH et al. Long non-coding RNAs in ischemic stroke. Cell Death Dis 9, 281, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janaki Ramaiah M, Divyapriya K, Kartik Kumar S & Rajesh Y Drug-induced modifications and modulations of microRNAs and long non-coding RNAs for future therapy against Glioblastoma Multiforme. Gene 723, 144126, (2020). [DOI] [PubMed] [Google Scholar]
- 72.Zhou Q et al. lncRNAs as potential molecular biomarkers for the clinicopathology and prognosis of glioma: A systematic review and meta-analysis. Gene 668, 77–86, (2018). [DOI] [PubMed] [Google Scholar]
- 73.Wang L et al. Long non-coding RNAs: potential molecular biomarkers for gliomas diagnosis and prognosis. Rev Neurosci 28, 375–380, (2017). [DOI] [PubMed] [Google Scholar]
- 74.Zhou M, Zhao H, Wang X, Sun J & Su J Analysis of long noncoding RNAs highlights region-specific altered expression patterns and diagnostic roles in Alzheimer’s disease. Brief Bioinform 20, 598–608, (2019). [DOI] [PubMed] [Google Scholar]
- 75.Maniati MS, Maniati M, Yousefi T, Ahmadi-Ahangar A & Tehrani SS New insights into the role of microRNAs and long noncoding RNAs in most common neurodegenerative diseases. J Cell Biochem 120, 8908–8918, (2019). [DOI] [PubMed] [Google Scholar]
- 76.Cortini F, Roma F & Villa C Emerging roles of long non-coding RNAs in the pathogenesis of Alzheimer’s disease. Ageing research reviews 50, 19–26, (2019). [DOI] [PubMed] [Google Scholar]
- 77.Shi C, Zhang L & Qin C Long non-coding RNAs in brain development, synaptic biology, and Alzheimer’s disease. Brain research bulletin 132, 160–169, (2017). [DOI] [PubMed] [Google Scholar]
- 78.Wan P, Su W & Zhuo Y The Role of Long Noncoding RNAs in Neurodegenerative Diseases. Mol Neurobiol 54, 2012–2021, (2017). [DOI] [PubMed] [Google Scholar]
- 79.Maison C et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nature genetics 30, 329–334, (2002). [DOI] [PubMed] [Google Scholar]
- 80.Saxena A & Carninci P Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. BioEssays : news and reviews in molecular, cellular and developmental biology 33, 830–839, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han P & Chang CP Long non-coding RNA and chromatin remodeling. RNA biology 12, 1094–1098, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rinn JL lncRNAs: linking RNA to chromatin. Cold Spring Harbor perspectives in biology 6, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rinn JL & Chang HY Genome regulation by long noncoding RNAs. Annual review of biochemistry 81, 145–166, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burns AM & Gräff J Cognitive epigenetic priming: leveraging histone acetylation for memory amelioration. Curr Opin Neurobiol 67, 75–84, (2020). [DOI] [PubMed] [Google Scholar]
- 85.Jarome TJ & Lubin FD Histone lysine methylation: critical regulator of memory and behavior. Rev Neurosci 24, 375–387, (2013). [DOI] [PubMed] [Google Scholar]
- 86.Keiser AA & Wood MA Examining the contribution of histone modification to sex differences in learning and memory. Learn Mem 26, 318–331, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lopez-Atalaya JP & Barco A Can changes in histone acetylation contribute to memory formation? Trends Genet 30, 529–539, (2014). [DOI] [PubMed] [Google Scholar]
- 88.Mahgoub M & Monteggia LM A role for histone deacetylases in the cellular and behavioral mechanisms underlying learning and memory. Learn Mem 21, 564–568, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pang KKL, Sharma M & Sajikumar S Epigenetics and memory: Emerging role of histone lysine methyltransferase G9a/GLP complex as bidirectional regulator of synaptic plasticity. Neurobiol Learn Mem 159, 1–5, (2019). [DOI] [PubMed] [Google Scholar]
- 90.Peixoto L & Abel T The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 38, 62–76, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Penney J & Tsai LH Histone deacetylases in memory and cognition. Sci Signal 7, re12, (2014). [DOI] [PubMed] [Google Scholar]
- 92.Schmauss C The roles of class I histone deacetylases (HDACs) in memory, learning, and executive cognitive functions: A review. Neurosci Biobehav Rev 83, 63–71, (2017). [DOI] [PubMed] [Google Scholar]
- 93.Stilling RM & Fischer A The role of histone acetylation in age-associated memory impairment and Alzheimer’s disease. Neurobiol Learn Mem 96, 19–26, (2011). [DOI] [PubMed] [Google Scholar]
- 94.Schaukowitch K & Kim TK Emerging epigenetic mechanisms of long non-coding RNAs. Neuroscience 264, 25–38, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marchese FP & Huarte M Long non-coding RNAs and chromatin modifiers: their place in the epigenetic code. Epigenetics 9, 21–26, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nakagawa S & Kageyama Y Nuclear lncRNAs as epigenetic regulators-beyond skepticism. Biochimica et biophysica acta 1839, 215–222, (2014). [DOI] [PubMed] [Google Scholar]
- 97.Zhang X et al. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. International journal of molecular sciences 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gaballa JM et al. The Role of Histone Methyltransferases and Long Non-coding RNAs in the Regulation of T Cell Fate Decisions. Frontiers in immunology 9, 2955, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Daneshvar K et al. lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nature cell biology, (2020). [DOI] [PMC free article] [PubMed]
- 100.Ding H et al. LncRNA MALAT1 induces the dysfunction of β cells via reducing the histone acetylation of the PDX-1 promoter in type 1 diabetes. Experimental and molecular pathology 114, 104432, (2020). [DOI] [PubMed] [Google Scholar]
- 101.Żylicz JJ et al. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell 176, 182–197.e123, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao J, Sun BK, Erwin JA, Song JJ & Lee JT Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science (New York, N.Y.) 322, 750–756, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Adrianse RL et al. Perturbed maintenance of transcriptional repression on the inactive X-chromosome in the mouse brain after Xist deletion. Epigenetics & chromatin 11, 50, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Margueron R & Reinberg D The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.O’Meara MM & Simon JA Inner workings and regulatory inputs that control Polycomb repressive complex 2. Chromosoma 121, 221–234, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gupta RA et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Song Y et al. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. International journal of oncology 54, 77–86, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Imai-Sumida M et al. Genistein Represses HOTAIR/Chromatin Remodeling Pathways to Suppress Kidney Cancer. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology 54, 53–70, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Colognori D, Sunwoo H, Wang D, Wang CY & Lee JT Xist Repeats A and B Account for Two Distinct Phases of X Inactivation Establishment. Developmental cell 54, 21–32.e25, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bousard A et al. The role of Xist-mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO reports 20, e48019, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Achour C & Aguilo F Long non-coding RNA and Polycomb: an intricate partnership in cancer biology. Frontiers in bioscience (Landmark edition) 23, 2106–2132 (2018). [DOI] [PubMed] [Google Scholar]
- 112.Li X et al. NCBP3/SNHG6 inhibits GBX2 transcription in a histone modification manner to facilitate the malignant biological behaviour of glioma cells. RNA biology, 1–17, (2020). [DOI] [PMC free article] [PubMed]
- 113.Jin C et al. Long non-coding RNA GAS5, by up-regulating PRC2 and targeting the promoter methylation of miR-424, suppresses multiple malignant phenotypes of glioma. Journal of neuro-oncology 148, 529–543, (2020). [DOI] [PubMed] [Google Scholar]
- 114.Tsai MC et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science (New York, N.Y.) 329, 689–693, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lim CS et al. PKCα-mediated phosphorylation of LSD1 is required for presynaptic plasticity and hippocampal learning and memory. Sci Rep 7, 4912, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Neelamegam R et al. Brain-penetrant LSD1 inhibitors can block memory consolidation. ACS Chem Neurosci 3, 120–128, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Christopher MA et al. LSD1 protects against hippocampal and cortical neurodegeneration. Nat Commun 8, 805, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang L et al. Inhibition of KDM1A activity restores adult neurogenesis and improves hippocampal memory in a mouse model of Kabuki syndrome. Mol Ther Methods Clin Dev 20, 779–791, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Maes T et al. Modulation of KDM1A with vafidemstat rescues memory deficit and behavioral alterations. PLoS One 15, e0233468, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang J et al. LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat Neurosci 18, 1256–1264, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Engstrom AK et al. The inhibition of LSD1 via sequestration contributes to tau-mediated neurodegeneration. Proc Natl Acad Sci U S A 117, 29133–29143, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Maes T et al. KDM1 histone lysine demethylases as targets for treatments of oncological and neurodegenerative disease. Epigenomics 7, 609–626, (2015). [DOI] [PubMed] [Google Scholar]
- 123.Hwang JY & Zukin RS REST, a master transcriptional regulator in neurodegenerative disease. Curr Opin Neurobiol 48, 193–200, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Takakura M et al. Rpd3/CoRest-mediated activity-dependent transcription regulates the flexibility in memory updating in Drosophila. Nat Commun 12, 628, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Butler AA, Sanchez RG, Jarome TJ, Webb WM & Lubin FD O-GlcNAc and EZH2-mediated epigenetic regulation of gene expression during consolidation of fear memories. Learn Mem 26, 373–379, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jarome TJ, Perez GA, Hauser RM, Hatch KM & Lubin FD EZH2 Methyltransferase Activity Controls Pten Expression and mTOR Signaling during Fear Memory Reconsolidation. J Neurosci 38, 7635–7648, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Almeida M, Bowness JS & Brockdorff N The many faces of Polycomb regulation by RNA. Current opinion in genetics & development 61, 53–61, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cerase A & Tartaglia GG Long non-coding RNA-polycomb intimate rendezvous. Open biology 10, 200126, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Luo W, Li X, Song Z, Zhu X & Zhao S Long non-coding RNA AGAP2-AS1 exerts oncogenic properties in glioblastoma by epigenetically silencing TFPI2 through EZH2 and LSD1. Aging 11, 3811–3823, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Katsushima K et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nature communications 7, 13616, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li Q, Dong C, Cui J, Wang Y & Hong X Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. Journal of experimental & clinical cancer research : CR 37, 265, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen H et al. SOX9-activated PXN-AS1 promotes the tumorigenesis of glioblastoma by EZH2-mediated methylation of DKK1. Journal of cellular and molecular medicine 24, 6070–6082, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang Y et al. EZH2 RIP-seq Identifies Tissue-specific Long Non-coding RNAs. Current gene therapy 18, 275–285, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ye M et al. Determination of long non-coding RNAs associated with EZH2 in neuroblastoma by RIP-seq, RNA-seq and ChIP-seq. Oncology letters 20, 1, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang J et al. Long Noncoding RNA H19 Promotes Neuroinflammation in Ischemic Stroke by Driving Histone Deacetylase 1-Dependent M1 Microglial Polarization. Stroke 48, 2211–2221, (2017). [DOI] [PubMed] [Google Scholar]
- 136.Ganai SA, Ramadoss M & Mahadevan V Histone Deacetylase (HDAC) Inhibitors - emerging roles in neuronal memory, learning, synaptic plasticity and neural regeneration. Curr Neuropharmacol 14, 55–71, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fischer A, Sananbenesi F, Mungenast A & Tsai LH Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci 31, 605–617, (2010). [DOI] [PubMed] [Google Scholar]
- 138.O’Leary VB, Ovsepian SV, Smida J & Atkinson MJ PARTICLE - The RNA podium for genomic silencers. Journal of cellular physiology 234, 19464–19470, (2019). [DOI] [PubMed] [Google Scholar]
- 139.Di Ruscio A et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 503, 371–376, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Park J et al. Long non-coding RNA ChRO1 facilitates ATRX/DAXX-dependent H3.3 deposition for transcription-associated heterochromatin reorganization. Nucleic acids research 46, 11759–11775, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bond AM et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci 12, 1020–1027, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Berghoff EG et al. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development (Cambridge, England) 140, 4407–4416, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Na ES, Nelson ED, Kavalali ET & Monteggia LM The impact of MeCP2 loss- or gain-of-function on synaptic plasticity. Neuropsychopharmacology 38, 212–219, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Robinson HA & Pozzo-Miller L The role of MeCP2 in learning and memory. Learn Mem 26, 343–350, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gulmez Karaca K, Brito DVC, Zeuch B & Oliveira AMM Adult hippocampal MeCP2 preserves the genomic responsiveness to learning required for long-term memory formation. Neurobiol Learn Mem 149, 84–97, (2018). [DOI] [PubMed] [Google Scholar]
- 146.Moretti P et al. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci 26, 319–327, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bayraktar G & Kreutz MR Neuronal DNA Methyltransferases: Epigenetic Mediators between Synaptic Activity and Gene Expression? Neuroscientist 24, 171–185, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cui D & Xu X DNA 0. Int J Mol Sci 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Maag JL et al. Widespread promoter methylation of synaptic plasticity genes in long-term potentiation in the adult brain in vivo. BMC Genomics 18, 250,(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Muñoz P et al. Inhibition of DNA Methylation Impairs Synaptic Plasticity during an Early Time Window in Rats. Neural Plast 2016, 4783836, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Day JJ et al. DNA methylation regulates associative reward learning. Nat Neurosci 16, 1445–1452, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Levenson JM et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem 281, 15763–15773, (2006). [DOI] [PubMed] [Google Scholar]
- 153.Yu JL, Li C, Che LH, Zhao YH & Guo YB Downregulation of long noncoding RNA H19 rescues hippocampal neurons from apoptosis and oxidative stress by inhibiting IGF2 methylation in mice with streptozotocin-induced diabetes mellitus. Journal of cellular physiology 234, 10655–10670, (2019). [DOI] [PubMed] [Google Scholar]
- 154.Chen Y et al. Knockdown of lncRNA PCAI protects against cognitive decline induced by hippocampal neuroinflammation via regulating SUZ12. Life sciences 253, 117626, (2020). [DOI] [PubMed] [Google Scholar]
- 155.Li X, Lv J & Liu S MCM3AP-AS1 KD Inhibits Proliferation, Invasion, and Migration of PCa Cells via DNMT1/DNMT3 (A/B) Methylation-Mediated Upregulation of NPY1R. Molecular therapy. Nucleic acids 20, 265–278, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 156.Bertocchi I et al. NPY-Y1 receptor signaling controls spatial learning and perineuronal net expression. Neuropharmacology 184, 108425, (2021). [DOI] [PubMed] [Google Scholar]
- 157.Yoon JH et al. The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer letters 417, 47–57, (2018). [DOI] [PubMed] [Google Scholar]
- 158.Boone DN, Warburton A, Som S & Lee AV SNHG7 is a lncRNA oncogene controlled by Insulin-like Growth Factor signaling through a negative feedback loop to tightly regulate proliferation. Scientific reports 10, 8583, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang L, Fu Y & Guo H c-Myc-Induced Long Non-Coding RNA Small Nucleolar RNA Host Gene 7 Regulates Glycolysis in Breast Cancer. Journal of breast cancer 22, 533–547, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhao J, Zhang X, Zhou Y, Ansell PJ & Klibanski A Cyclic AMP stimulates MEG3 gene expression in cells through a cAMP-response element (CRE) in the MEG3 proximal promoter region. Int J Biochem Cell Biol 38, 1808–1820, (2006). [DOI] [PubMed] [Google Scholar]
- 161.Chen Z et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest 126, 2267–2279, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhou C et al. Hippocampus-specific regulation of long non-coding RNA and mRNA expression in germ-free mice. Funct Integr Genomics 20, 355–365, (2020). [DOI] [PubMed] [Google Scholar]
- 163.Saad MH et al. Differentially expressed gene networks, biomarkers, long noncoding RNAs, and shared responses with cocaine identified in the midbrains of human opioid abusers. Sci Rep 9, 1534, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bannon MJ et al. Identification of long noncoding RNAs dysregulated in the midbrain of human cocaine abusers. J Neurochem 135, 50–59, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhu L et al. Methamphetamine induces alterations in the long non-coding RNAs expression profile in the nucleus accumbens of the mouse. BMC Neurosci 16, 18, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Denkena J et al. Neuronal activity regulates alternative exon usage. Mol Brain 13, 148, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lipovich L et al. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics 192, 1133–1148, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Barry G et al. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Scientific reports 7, 40127, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Barry G et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry 19, 486–494, (2014). [DOI] [PubMed] [Google Scholar]
- 170.Wu Z et al. Regulation of lncRNA expression. Cell Mol Biol Lett 19, 561–575, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Quinn JJ & Chang HY Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17, 47–62, (2016). [DOI] [PubMed] [Google Scholar]
- 172.Fang S et al. H3K27me3 induces multidrug resistance in small cell lung cancer by affecting HOXA1 DNA methylation via regulation of the lncRNA HOTAIR. Annals of translational medicine 6, 440, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wu SC, Kallin EM & Zhang Y Role of H3K27 methylation in the regulation of lncRNA expression. Cell research 20, 1109–1116, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lu C et al. DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Molecular cancer 19, 28, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liao Q et al. DNA methylation patterns of protein-coding genes and long non-coding RNAs in males with schizophrenia. Molecular medicine reports 12, 6568–6576, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhou Y, Zhang X & Klibanski A MEG3 noncoding RNA: a tumor suppressor. Journal of molecular endocrinology 48, R45–53, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liao Q et al. DNA methylation patterns of protein coding genes and long noncoding RNAs in female schizophrenic patients. European journal of medical genetics 58, 95–104, (2015). [DOI] [PubMed] [Google Scholar]
- 178.Miller-Delaney SF et al. Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy. Brain : a journal of neurology 138, 616–631, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sen P, Shah PP, Nativio R & Berger SL Epigenetic Mechanisms of Longevity and Aging. Cell 166, 822–839, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Horvath S & Raj K DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nature reviews. Genetics 19, 371–384, (2018). [DOI] [PubMed] [Google Scholar]
- 181.Barter JD & Foster TC Aging in the Brain: New Roles of Epigenetics in Cognitive Decline. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry 24, 516–525, (2018). [DOI] [PubMed] [Google Scholar]
- 182.Grammatikakis I, Panda AC, Abdelmohsen K & Gorospe M Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging 6, 992–1009, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chen BJ et al. RNA sequencing reveals pronounced changes in the noncoding transcriptome of aging synaptosomes. Neurobiology of aging 56, 67–77, (2017). [DOI] [PubMed] [Google Scholar]
- 184.Kour S & Rath PC Age-dependent differential expression profile of a novel intergenic long noncoding RNA in rat brain. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience 47, 286–297, (2015). [DOI] [PubMed] [Google Scholar]
- 185.Kour S & Rath PC Age-Related Expression of a Repeat-Rich Intergenic Long Noncoding RNA in the Rat Brain. Molecular neurobiology 54, 639–660, (2017). [DOI] [PubMed] [Google Scholar]
- 186.Chen C et al. RNA-seq analysis of the key long noncoding RNAs and mRNAs related to cognitive impairment after cardiac arrest and cardiopulmonary resuscitation. Aging 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hornung S, Dutta S & Bitan G CNS-Derived Blood Exosomes as a Promising Source of Biomarkers: Opportunities and Challenges. Frontiers in molecular neuroscience 13, 38, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gezer U, Özgür E, Cetinkaya M, Isin M & Dalay N Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell biology international 38, 1076–1079, (2014). [DOI] [PubMed] [Google Scholar]
- 189.Kukharsky MS et al. Long non-coding RNA Neat1 regulates adaptive behavioural response to stress in mice. Translational psychiatry 10, 171, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mehta D, Jackson R, Paul G, Shi J & Sabbagh M Why do trials for Alzheimer’s disease drugs keep failing? A discontinued drug perspective for 2010–2015. Expert opinion on investigational drugs 26, 735–739, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chen L, Guo X, Li Z & He Y Relationship between long non-coding RNAs and Alzheimer’s disease: a systematic review. Pathology, research and practice 215, 12–20, (2019). [DOI] [PubMed] [Google Scholar]
- 192.Luo Q & Chen Y Long noncoding RNAs and Alzheimer’s disease. Clinical interventions in aging 11, 867–872, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wu J et al. Co-expression Network Analysis Revealing the Potential Regulatory Roles of lncRNAs in Alzheimer’s Disease. Interdisciplinary sciences, computational life sciences 11, 645–654, (2019). [DOI] [PubMed] [Google Scholar]
- 194.Cao M, Li H, Zhao J, Cui J & Hu G Identification of age- and gender-associated long noncoding RNAs in the human brain with Alzheimer’s disease. Neurobiology of aging 81, 116–126, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tang L et al. Expression Profiles of Long Noncoding RNAs in Intranasal LPS-Mediated Alzheimer’s Disease Model in Mice. BioMed research international 2019, 9642589, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yang B et al. Distinct Hippocampal Expression Profiles of Long Non-coding RNAs in an Alzheimer’s Disease Model. Molecular neurobiology 54, 4833–4846, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wan G et al. Transcriptional Regulation of lncRNA Genes by Histone Modification in Alzheimer’s Disease. Biomed Res Int 2016, 3164238, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nikolac Perkovic M et al. Epigenetics of Alzheimer’s Disease. Biomolecules 11, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Xiao X, Liu X & Jiao B Epigenetics: Recent Advances and Its Role in the Treatment of Alzheimer’s Disease. Front Neurol 11, 538301, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Blanco-Luquin I et al. Early epigenetic changes of Alzheimer’s disease in the human hippocampus. Epigenetics 15, 1083–1092, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Liu X, Jiao B & Shen L The Epigenetics of Alzheimer’s Disease: Factors and Therapeutic Implications. Front Genet 9, 579, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fenoglio C, Scarpini E, Serpente M & Galimberti D Role of Genetics and Epigenetics in the Pathogenesis of Alzheimer’s Disease and Frontotemporal Dementia. J Alzheimers Dis 62, 913–932, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gjoneska E et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518, 365–369, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bennett DA et al. Epigenomics of Alzheimer’s disease. Transl Res 165, 200–220, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhang L et al. Epigenome-wide meta-analysis of DNA methylation differences in prefrontal cortex implicates the immune processes in Alzheimer’s disease. Nat Commun 11, 6114, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Airavaara M et al. Identification of novel GDNF isoforms and cis-antisense GDNFOS gene and their regulation in human middle temporal gyrus of Alzheimer disease. J Biol Chem 286, 45093–45102, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Smith AR et al. Parallel profiling of DNA methylation and hydroxymethylation highlights neuropathology-associated epigenetic variation in Alzheimer’s disease. Clin Epigenetics 11, 52, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Foraker J et al. The APOE Gene is Differentially Methylated in Alzheimer’s Disease. J Alzheimers Dis 48, 745–755, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.De Jager PL et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci 17, 1156–1163, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Smith AR et al. Increased DNA methylation near TREM2 is consistently seen in the superior temporal gyrus in Alzheimer’s disease brain. Neurobiol Aging 47, 35–40, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Celarain N et al. TREM2 upregulation correlates with 5-hydroxymethycytosine enrichment in Alzheimer’s disease hippocampus. Clin Epigenetics 8, 37, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lee MY et al. Epigenome signatures landscaped by histone H3K9me3 are associated with the synaptic dysfunction in Alzheimer’s disease. Aging Cell 19, e13153, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Cuadrado-Tejedor M et al. Concomitant histone deacetylase and phosphodiesterase 5 inhibition synergistically prevents the disruption in synaptic plasticity and it reverses cognitive impairment in a mouse model of Alzheimer’s disease. Clin Epigenetics 7, 108, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Marzi SJ et al. A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat Neurosci 21, 1618–1627, (2018). [DOI] [PubMed] [Google Scholar]
- 215.Narayan P & Dragunow M Alzheimer’s Disease and Histone Code Alterations. Adv Exp Med Biol 978, 321–336, (2017). [DOI] [PubMed] [Google Scholar]
- 216.Liu SL et al. The Role of Cdk5 in Alzheimer’s Disease. Molecular neurobiology 53, 4328–4342, (2016). [DOI] [PubMed] [Google Scholar]
- 217.Spreafico M, Grillo B, Rusconi F, Battaglioli E & Venturin M Multiple Layers of CDK5R1 Regulation in Alzheimer’s Disease Implicate Long Non-Coding RNAs. Int J Mol Sci 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang D et al. Elevated plasma levels of exosomal BACE1‐AS combined with the volume and thickness of the right entorhinal cortex may serve as a biomarker for the detection of Alzheimer’s disease. Molecular medicine reports 22, 227–238, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kang MJ et al. HuD regulates coding and noncoding RNA to induce APP→Aβ processing. Cell reports 7, 1401–1409, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Huang Z, Zhao J, Wang W, Zhou J & Zhang J Depletion of LncRNA NEAT1 Rescues Mitochondrial Dysfunction Through NEDD4L-Dependent PINK1 Degradation in Animal Models of Alzheimer’s Disease. Frontiers in cellular neuroscience 14, 28, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Keihani S et al. The long noncoding RNA neuroLNC regulates presynaptic activity by interacting with the neurodegeneration-associated protein TDP-43. Science advances 5, eaay2670, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ding XH, Han J, Liu Y, Jin Y & Ye P D-4F decreases the expression of Aβ protein through up-regulating long non coding RNA sirt1-as in SAMP8 mice. Saudi pharmaceutical journal : SPJ : the official publication of the Saudi Pharmaceutical Society 25, 517–522, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhang L, Fang Y, Cheng X, Lian YJ & Xu HL Silencing of Long Noncoding RNA SOX21-AS1 Relieves Neuronal Oxidative Stress Injury in Mice with Alzheimer’s Disease by Upregulating FZD3/5 via the Wnt Signaling Pathway. Molecular neurobiology 56, 3522–3537, (2019). [DOI] [PubMed] [Google Scholar]
- 224.Li X, Wang SW, Li XL, Yu FY & Cong HM Knockdown of long non-coding RNA TUG1 depresses apoptosis of hippocampal neurons in Alzheimer’s disease by elevating microRNA-15a and repressing ROCK1 expression. Inflammation research : official journal of the European Histamine Research Society .. [et al. ] 69, 897–910, (2020). [DOI] [PubMed] [Google Scholar]
- 225.Zhang W, Zhao H, Wu Q, Xu W & Xia M Knockdown of BACE1-AS by siRNA improves memory and learning behaviors in Alzheimer’s disease animal model. Experimental and therapeutic medicine 16, 2080–2086, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Yi J et al. Upregulation of the lncRNA MEG3 improves cognitive impairment, alleviates neuronal damage, and inhibits activation of astrocytes in hippocampus tissues in Alzheimer’s disease through inactivating the PI3K/Akt signaling pathway. Journal of cellular biochemistry 120, 18053–18065, (2019). [DOI] [PubMed] [Google Scholar]
- 227.Zhang T et al. Expression of BC1 Impairs Spatial Learning and Memory in Alzheimer’s Disease Via APP Translation. Molecular neurobiology 55, 6007–6020, (2018). [DOI] [PubMed] [Google Scholar]
- 228.Hauser RM, Henshall DC & Lubin FD The Epigenetics of Epilepsy and Its Progression. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry 24, 186–200, (2018). [DOI] [PubMed] [Google Scholar]
- 229.Lubin FD Epileptogenesis: can the science of epigenetics give us answers? Epilepsy currents 12, 105–110, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Villa C, Lavitrano M & Combi R Long Non-Coding RNAs and Related Molecular Pathways in the Pathogenesis of Epilepsy. International journal of molecular sciences 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Jang Y et al. Dysregulated long non-coding RNAs in the temporal lobe epilepsy mouse model. Seizure 58, 110–119, (2018). [DOI] [PubMed] [Google Scholar]
- 232.Henshall DC Epigenetics and noncoding RNA: Recent developments and future therapeutic opportunities. European journal of paediatric neurology : EJPN : official journal of the European Paediatric Neurology Society 24, 30–34, (2020). [DOI] [PubMed] [Google Scholar]
- 233.Cui Z et al. Differential long non-coding RNA (lncRNA) profiles associated with hippocampal sclerosis in human mesial temporal lobe epilepsy. International journal of clinical and experimental pathology 12, 259–266 (2019). [PMC free article] [PubMed] [Google Scholar]
- 234.Cai X et al. LncRNA ILF3-AS1 mediated the occurrence of epilepsy through suppressing hippocampal miR-212 expression. Aging 12, 8413–8422, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hashemian F et al. Epilepsy Is Associated With Dysregulation of Long Non-coding RNAs in the Peripheral Blood. Frontiers in molecular biosciences 6, 113, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mirzajani S et al. Expression Analysis of lncRNAs in Refractory and Non-Refractory Epileptic Patients. Journal of molecular neuroscience : MN 70, 689–698, (2020). [DOI] [PubMed] [Google Scholar]
- 237.Mazdeh M et al. Expression analysis of vitamin D receptor-associated lncRNAs in epileptic patients. Metabolic brain disease 34, 1457–1465, (2019). [DOI] [PubMed] [Google Scholar]
- 238.Han CL et al. Long non-coding RNA H19 contributes to apoptosis of hippocampal neurons by inhibiting let-7b in a rat model of temporal lobe epilepsy. Cell death & disease 9, 617, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wu Q & Yi X Down-regulation of Long Noncoding RNA MALAT1 Protects Hippocampal Neurons Against Excessive Autophagy and Apoptosis via the PI3K/Akt Signaling Pathway in Rats with Epilepsy. Journal of molecular neuroscience : MN 65, 234–245, (2018). [DOI] [PubMed] [Google Scholar]
- 240.Zhao T, Ding Y, Li M, Zhou C & Lin W Silencing lncRNA PVT1 inhibits activation of astrocytes and increases BDNF expression in hippocampus tissues of rats with epilepsy by downregulating the Wnt signaling pathway. Journal of cellular physiology, (2019). [DOI] [PubMed]
- 241.Nickels KC, Zaccariello MJ, Hamiwka LD & Wirrell EC Cognitive and neurodevelopmental comorbidities in paediatric epilepsy. Nature reviews. Neurology 12, 465–476, (2016). [DOI] [PubMed] [Google Scholar]
- 242.Vrinda M, Arun S, Srikumar BN, Kutty BM & Shankaranarayana Rao BS Temporal lobe epilepsy-induced neurodegeneration and cognitive deficits: Implications for aging. Journal of chemical neuroanatomy 95, 146–153, (2019). [DOI] [PubMed] [Google Scholar]
- 243.Wang HK, Yan H, Wang K & Wang J Dynamic regulation effect of long non-coding RNA-UCA1 on NF-kB in hippocampus of epilepsy rats. European review for medical and pharmacological sciences 21, 3113–3119 (2017). [PubMed] [Google Scholar]
- 244.Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS & Baltimore D NF-kappa B functions in synaptic signaling and behavior. Nature neuroscience 6, 1072–1078, (2003). [DOI] [PubMed] [Google Scholar]
- 245.Wen X et al. Down-regulated long non-coding RNA ANRIL restores the learning and memory abilities and rescues hippocampal pyramidal neurons from apoptosis in streptozotocin-induced diabetic rats via the NF-κB signaling pathway. Journal of cellular biochemistry 119, 5821–5833, (2018). [DOI] [PubMed] [Google Scholar]
- 246.Wang X, Zhang M & Liu H LncRNA17A regulates autophagy and apoptosis of SH-SY5Y cell line as an in vitro model for Alzheimer’s disease. Biosci Biotechnol Biochem 83, 609–621, (2019). [DOI] [PubMed] [Google Scholar]
- 247.Massone S et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis 41, 308–317, (2011). [DOI] [PubMed] [Google Scholar]
- 248.Wu P et al. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Research Bulletin 97, 69–80, (2013). [DOI] [PubMed] [Google Scholar]
- 249.Arendt T, Holzer M & Gärtner U Neuronal expression of cycline dependent kinase inhibitors of the INK4 family in Alzheimer’s disease. Journal of Neural Transmission 105, 949–960, (1998). [DOI] [PubMed] [Google Scholar]
- 250.Pereira Fernandes D, Bitar M, Jacobs FMJ & Barry G Long Non-Coding RNAs in Neuronal Aging. Noncoding RNA 4, 12, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wu Y-Y & Kuo H-C Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. Journal of Biomedical Science 27, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Paola R, Antonia R & Marco V The Long Non-Coding RNAs in Neurodegenerative Diseases: Novel Mechanisms of Pathogenesis. Current Alzheimer Research 13, 1219–1231, (2016). [DOI] [PubMed] [Google Scholar]
- 253.Zhou M, Zhao H, Wang X, Sun J & Su J Analysis of long noncoding RNAs highlights region-specific altered expression patterns and diagnostic roles in Alzheimer’s disease. Briefings in Bioinformatics 20, 598–608, (2018). [DOI] [PubMed] [Google Scholar]
- 254.Faghihi MA et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nature Medicine 14, 723–730, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Liu T et al. Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1‐AS expression. Mol Med Rep 10, 1275–1281, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wang H et al. Dendritic BC1 RNA: functional role in regulation of translation initiation. J Neurosci 22, 10232–10241, (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Lin D, Pestova TV, Hellen CU & Tiedge H Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol 28, 3008–3019, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Kondrashov AV et al. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP). J Mol Biol 353, 88–103, (2005). [DOI] [PubMed] [Google Scholar]
- 259.Lewejohann L et al. Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behav Brain Res 154, 273–289, (2004). [DOI] [PubMed] [Google Scholar]
- 260.Li H, Zheng L, Jiang A, Mo Y & Gong Q Identification of the biological affection of long noncoding RNA BC200 in Alzheimer’s disease. Neuroreport 29, 1061–1067, (2018). [DOI] [PubMed] [Google Scholar]
- 261.Feng L et al. Plasma long non-coding RNA BACE1 as a novel biomarker for diagnosis of Alzheimer disease. BMC Neurol 18, 4, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Mus E, Hof PR & Tiedge H Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A 104, 10679–10684, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Modarresi F et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol 30, 453–459, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Gu C et al. Long Noncoding RNA EBF3-AS Promotes Neuron Apoptosis in Alzheimer’s Disease. DNA and Cell Biology 37, 220–226, (2018). [DOI] [PubMed] [Google Scholar]
- 265.Yamanaka Y et al. Antisense RNA controls LRP1 Sense transcript expression through interaction with a chromatin-associated protein, HMGB2. Cell Rep 11, 967–976, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Holtzman DM, Herz J & Bu G Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med 2, a006312, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Tsuiji H et al. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells 16, 479–490, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.He J, Tu C & Liu Y Role of lncRNAs in aging and age-related diseases. Aging Medicine 1, 158–175, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Johnson R Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis 46, 245–254, (2012). [DOI] [PubMed] [Google Scholar]
- 270.Chang KH, Wu YR & Chen CM Down-regulation of miR-9* in the peripheral leukocytes of Huntington’s disease patients. Orphanet J Rare Dis 12, 185, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Chanda K et al. Altered Levels of Long NcRNAs Meg3 and Neat1 in Cell And Animal Models Of Huntington’s Disease. RNA Biol 15, 1348–1363, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Sunwoo J-S et al. Altered Expression of the Long Noncoding RNA NEAT1 in Huntington’s Disease. Molecular Neurobiology 54, 1577–1586, (2017). [DOI] [PubMed] [Google Scholar]
- 273.Cheng C et al. The long non-coding RNA NEAT1 is elevated in polyglutamine repeat expansion diseases and protects from disease gene-dependent toxicities. Hum Mol Genet 27, 4303–4314, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Liu Y & Lu Z Long non-coding RNA NEAT1 mediates the toxic of Parkinson’s disease induced by MPTP/MPP+ via regulation of gene expression. Clinical and Experimental Pharmacology and Physiology 45, 841–848, (2018). [DOI] [PubMed] [Google Scholar]
- 275.Xie SP, Zhou F, Li J & Duan SJ NEAT1 regulates MPP(+)-induced neuronal injury by targeting miR-124 in neuroblastoma cells. Neurosci Lett 708, 134340, (2019). [DOI] [PubMed] [Google Scholar]
- 276.Yan W, Chen Z-Y, Chen J-Q & Chen H-M LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein. Biochemical and Biophysical Research Communications 496, 1019–1024, (2018). [DOI] [PubMed] [Google Scholar]
- 277.Jiang Q et al. Long non-coding RNA-MIAT promotes neurovascular remodeling in the eye and brain. Oncotarget 7, 49688–49698, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Parenti R, Paratore S, Torrisi A & Cavallaro S A natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during beta-amyloid-induced apoptosis. Eur J Neurosci 26, 2444–2457, (2007). [DOI] [PubMed] [Google Scholar]
- 279.Massone S et al. NDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid β secretion. Biochimica et Biophysica Acta - Molecular Cell Research 1823, 1170–1177, (2012). [DOI] [PubMed] [Google Scholar]
- 280.Ciarlo E et al. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer's disease brain samples. Disease Models & Mechanisms 6, 424, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Arisi I et al. Gene expression biomarkers in the brain of a mouse model for Alzheimer’s disease: mining of microarray data by logic classification and feature selection. J Alzheimers Dis 24, 721–738, (2011). [DOI] [PubMed] [Google Scholar]
- 282.Wunderlich G et al. Temporal lobe epilepsy with sensory aura: interictal glucose hypometabolism 38, 139–149, (2000). [DOI] [PubMed] [Google Scholar]
- 283.Johnson R et al. Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol Genomics 41, 269–274, (2010). [DOI] [PubMed] [Google Scholar]
- 284.Zuccato C et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet 35, 76–83, (2003). [DOI] [PubMed] [Google Scholar]
- 285.Shimojo M Huntingtin regulates RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) nuclear trafficking indirectly through a complex with REST/NRSF-interacting LIM domain protein (RILP) and dynactin p150 Glued. The Journal of biological chemistry 283, 34880–34886, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Seong IS et al. Huntingtin facilitates polycomb repressive complex 2. Human molecular genetics 19, 573–583, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Chung DW, Rudnicki DD, Yu L & Margolis RL A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Human molecular genetics 20, 3467–3477, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Khalil AM et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106, 11667–11672, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Barry G, Guennewig B, Fung S, Kaczorowski D & Weickert CS Long Non-Coding RNA Expression during Aging in the Human Subependymal Zone. Frontiers in Neurology 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Lin N et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Molecular cell 53, 1005–1019, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Wang S, Zhang X, Guo Y, Rong H & Liu T The long noncoding RNA HOTAIR promotes Parkinson’s disease by upregulating LRRK2 expression. Oncotarget 8, 24449–24456, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Lin Q, Hou S, Dai Y, Jiang N & Lin Y LncRNA HOTAIR targets miR-126–5p to promote the progression of Parkinson’s disease through RAB3IP 400, 1217, (2019). [DOI] [PubMed] [Google Scholar]
- 293.Liu S et al. Long Non-coding RNA HOTAIR Promotes Parkinson’s Disease Induced by MPTP Through up-regulating the Expression of LRRK2. Curr Neurovasc Res 13, 115–120, (2016). [DOI] [PubMed] [Google Scholar]
- 294.Kraus TFJ et al. Altered Long Noncoding RNA Expression Precedes the Course of Parkinson’s Disease—a Preliminary Report. Molecular Neurobiology 54, 2869–2877, (2017). [DOI] [PubMed] [Google Scholar]
- 295.Bernard D et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. The EMBO Journal 29, 3082–3093, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Sang Q et al. CircSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR-7. Aging (Albany NY) 10, 1281–1293, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Ding XM, Zhao LJ, Qiao HY, Wu SL & Wang XH Long non-coding RNA-p21 regulates MPP(+)-induced neuronal injury by targeting miR-625 and derepressing TRPM2 in SH-SY5Y cells. Chem Biol Interact 307, 73–81, (2019). [DOI] [PubMed] [Google Scholar]
- 298.Xu X et al. LincRNA-p21 Inhibits Cell Viability and Promotes Cell Apoptosis in Parkinson’s Disease through Activating α-Synuclein Expression. BioMed research international 2018, 8181374–8181374, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Scheele C et al. The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics 8, 74, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Qian C et al. Downregulated lncRNA-SNHG1 enhances autophagy and prevents cell death through the miR-221/222 /p27/mTOR pathway in Parkinson’s disease. Exp Cell Res 384, 111614, (2019). [DOI] [PubMed] [Google Scholar]
- 301.Soreq L et al. Long non-coding RNA and alternative splicing modulations in Parkinson’s leukocytes identified by RNA sequencing. PLoS Comput Biol 10, e1003517, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Chen Y et al. LncRNA SNHG1 promotes α-synuclein aggregation and toxicity by targeting miR-15b-5p to activate SIAH1 in human neuroblastoma SH-SY5Y cells. Neurotoxicology 68, 212–221, (2018). [DOI] [PubMed] [Google Scholar]
- 303.Cao B, Wang T, Qu Q, Kang T & Yang Q Long Noncoding RNA SNHG1 Promotes Neuroinflammation in Parkinson’s Disease via Regulating miR-7/NLRP3 Pathway. Neuroscience 388, 118–127, (2018). [DOI] [PubMed] [Google Scholar]
- 304.Carrieri C et al. Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front Cell Neurosci 9, 114, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Carrieri C et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457, (2012). [DOI] [PubMed] [Google Scholar]
- 306.Choi J et al. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem 279, 13256–13264, (2004). [DOI] [PubMed] [Google Scholar]
- 307.Ng S-Y, Gireesh, Boon & Lawrence. The Long Noncoding RNA RMST Interacts with SOX2 to Regulate Neurogenesis. Molecular Cell 51, 349–359, (2013). [DOI] [PubMed] [Google Scholar]
- 308.Wapinski O & Chang HY Long noncoding RNAs and human disease. Trends in Cell Biology 21, 354–361, (2011). [DOI] [PubMed] [Google Scholar]
- 309.Kim J et al. Long noncoding RNAs in diseases of aging. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1859, 209–221, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Meier I, Fellini L, Jakovcevski M, Schachner M & Morellini F Expression of the snoRNA host gene gas5 in the hippocampus is upregulated by age and psychogenic stress and correlates with reduced novelty-induced behavior in C57BL/6 mice. Hippocampus 20, 1027–1036, (2010). [DOI] [PubMed] [Google Scholar]
- 311.Johnson R et al. Regulation of neural macroRNAs by the transcriptional repressor REST. RNA 15, 85–96, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Johnson R & Buckley NJ Gene Dysregulation in Huntington’s Disease: REST, MicroRNAs and Beyond. NeuroMolecular Medicine 11, 183–199, (2009). [DOI] [PubMed] [Google Scholar]
- 313.Derrien T et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research 22, 1775–1789, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Eichenbaum H Memory: Organization and Control. Annual review of psychology 68, 19–45, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Eichenbaum H On the Integration of Space, Time, and Memory. Neuron 95, 1007–1018, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Eichenbaum H Prefrontal-hippocampal interactions in episodic memory. Nature reviews. Neuroscience 18, 547–558, (2017). [DOI] [PubMed] [Google Scholar]
- 317.Lisman J et al. Viewpoints: how the hippocampus contributes to memory, navigation and cognition. Nature neuroscience 20, 1434–1447, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zeidman P & Maguire EA Anterior hippocampus: the anatomy of perception, imagination and episodic memory. Nature reviews. Neuroscience 17, 173–182, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Carey D, Nolan H, Kenny RA & Meaney J Dissociable age and memory relationships with hippocampal subfield volumes in vivo:Data from the Irish Longitudinal Study on Ageing (TILDA). Scientific reports 9, 10981, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Tamnes CK et al. Regional hippocampal volumes and development predict learning and memory. Developmental neuroscience 36, 161–174, (2014). [DOI] [PubMed] [Google Scholar]
- 321.Lee I, Yoganarasimha D, Rao G & Knierim JJ Comparison of population coherence of place cells in hippocampal subfields CA1 and CA3. Nature 430, 456–459, (2004). [DOI] [PubMed] [Google Scholar]
- 322.Leutgeb JK, Leutgeb S, Moser MB & Moser EI Pattern separation in the dentate gyrus and CA3 of the hippocampus. Science (New York, N.Y.) 315, 961–966, (2007). [DOI] [PubMed] [Google Scholar]
- 323.Lee I & Kesner RP Encoding versus retrieval of spatial memory: double dissociation between the dentate gyrus and the perforant path inputs into CA3 in the dorsal hippocampus. Hippocampus 14, 66–76, (2004). [DOI] [PubMed] [Google Scholar]
- 324.Coras R et al. Differential influence of hippocampal subfields to memory formation: insights from patients with temporal lobe epilepsy. Brain : a journal of neurology 137, 1945–1957, (2014). [DOI] [PubMed] [Google Scholar]
- 325.Ratnu VS, Emami MR & Bredy TW Genetic and epigenetic factors underlying sex differences in the regulation of gene expression in the brain. Journal of neuroscience research 95, 301–310, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Liu S et al. Annotation and cluster analysis of spatiotemporal- and sex-related lncRNA expression in rhesus macaque brain. Genome research 27, 1608–1620, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Yuan W et al. Transcriptome profiling analysis of sex-based differentially expressed mRNAs and lncRNAs in the brains of mature zebrafish (Danio rerio). BMC genomics 20, 830, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Fallah H et al. Sex-specific up-regulation of lncRNAs in peripheral blood of patients with schizophrenia. Scientific reports 9, 12737, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Xiang Y et al. LncRNA MEG3 targeting miR-424–5p via MAPK signaling pathway mediates neuronal apoptosis in ischemic stroke. Aging 12, 3156–3174, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Cuevas-Diaz Duran R, Wang CY, Zheng H, Deneen B & Wu JQ Brain Region-Specific Gene Signatures Revealed by Distinct Astrocyte Subpopulations Unveil Links to Glioma and Neurodegenerative Diseases. eNeuro 6, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Cheng S, Zhang Y, Chen S & Zhou Y LncRNA HOTAIR Participates in Microglia Activation and Inflammatory Factor Release by Regulating the Ubiquitination of MYD88 in Traumatic Brain Injury. Journal of molecular neuroscience : MN, (2020). [DOI] [PubMed]
- 332.Gu XH et al. Long non-coding RNA uc.80- overexpression promotes M2 polarization of microglias to ameliorate depression in rats. IUBMB life, (2020). [DOI] [PubMed]
- 333.Xu W, Zhang L, Geng Y, Liu Y & Zhang N Long noncoding RNA GAS5 promotes microglial inflammatory response in Parkinson’s disease by regulating NLRP3 pathway through sponging miR-223–3p. International immunopharmacology 85, 106614, (2020). [DOI] [PubMed] [Google Scholar]
- 334.Li Z et al. Modulating lncRNA SNHG15/CDK6/miR-627 circuit by palbociclib, overcomes temozolomide resistance and reduces M2-polarization of glioma associated microglia in glioblastoma multiforme. Journal of experimental & clinical cancer research : CR 38, 380, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Shao M et al. Exosomes from Long Noncoding RNA-Gm37494-ADSCs Repair Spinal Cord Injury via Shifting Microglial M1/M2 Polarization. Inflammation 43, 1536–1547, (2020). [DOI] [PubMed] [Google Scholar]
- 336.Zhao Q et al. Knockdown of long noncoding RNA XIST mitigates the apoptosis and inflammatory injury of microglia cells after spinal cord injury through miR-27a/Smurf1 axis. Neuroscience letters 715, 134649, (2020). [DOI] [PubMed] [Google Scholar]
- 337.De Pittà M, Brunel N & Volterra A Astrocytes: Orchestrating synaptic plasticity? Neuroscience 323, 43–61, (2016). [DOI] [PubMed] [Google Scholar]
- 338.Volterra A & Steinhäuser C Glial modulation of synaptic transmission in the hippocampus. Glia 47, 249–257, (2004). [DOI] [PubMed] [Google Scholar]
- 339.Adamsky A & Goshen I Astrocytes in Memory Function: Pioneering Findings and Future Directions. Neuroscience 370, 14–26, (2018). [DOI] [PubMed] [Google Scholar]
- 340.Fakhoury M Microglia and Astrocytes in Alzheimer’s Disease: Implications for Therapy. Current neuropharmacology 16, 508–518, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Hassanpoor H, Fallah A & Raza M Mechanisms of hippocampal astrocytes mediation of spatial memory and theta rhythm by gliotransmitters and growth factors. Cell biology international 38, 1355–1366, (2014). [DOI] [PubMed] [Google Scholar]
- 342.Santello M, Toni N & Volterra A Astrocyte function from information processing to cognition and cognitive impairment. Nature neuroscience 22, 154–166, (2019). [DOI] [PubMed] [Google Scholar]
- 343.Verkhratsky A, Parpura V, Rodriguez-Arellano JJ & Zorec R Astroglia in Alzheimer’s Disease. Advances in experimental medicine and biology 1175, 273–324, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Verkhratsky A, Zorec R, Rodriguez JJ & Parpura V Neuroglia: Functional Paralysis and Reactivity in Alzheimer’s Disease and Other Neurodegenerative Pathologies. Advances in neurobiology 15, 427–449, (2017). [DOI] [PubMed] [Google Scholar]
- 345.Grubman A et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nature neuroscience 22, 2087–2097, (2019). [DOI] [PubMed] [Google Scholar]
- 346.Schirmer L et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 573, 75–82, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Pan J & Wan J Methodological comparison of FACS and MACS isolation of enriched microglia and astrocytes from mouse brain. Journal of immunological methods, 112834, (2020). [DOI] [PubMed]
- 348.Holt LM & Olsen ML Novel Applications of Magnetic Cell Sorting to Analyze Cell-Type Specific Gene and Protein Expression in the Central Nervous System. PloS one 11, e0150290, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Holt LM, Stoyanof ST & Olsen ML Magnetic Cell Sorting for In Vivo and In Vitro Astrocyte, Neuron, and Microglia Analysis. Current protocols in neuroscience 88, e71, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Liu SJ & Lim DA Modulating the expression of long non-coding RNAs for functional studies. EMBO reports 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Yu JY, DeRuiter SL & Turner DL RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 99, 6047–6052, (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Ki KH et al. The optimal concentration of siRNA for gene silencing in primary cultured astrocytes and microglial cells of rats. Korean journal of anesthesiology 59, 403–410, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Oliveira S, Storm G & Schiffelers RM Targeted delivery of siRNA. Journal of biomedicine & biotechnology 2006, 63675, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.de Franciscis V Challenging cancer targets for aptamer delivery. Biochimie 145, 45–52, (2018). [DOI] [PubMed] [Google Scholar]
- 355.Ellington AD & Szostak JW In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822, (1990). [DOI] [PubMed] [Google Scholar]
- 356.McNamara JO 2nd et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nature biotechnology 24, 1005–1015, (2006). [DOI] [PubMed] [Google Scholar]
- 357.Kruspe S & Giangrande PH Aptamer-siRNA Chimeras: Discovery, Progress, and Future Prospects. Biomedicines 5, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Esposito CL, Catuogno S, Condorelli G, Ungaro P & de Franciscis V Aptamer Chimeras for Therapeutic Delivery: The Challenging Perspectives. Genes 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Yoon S, Wu X, Armstrong B, Habib N & Rossi JJ An RNA Aptamer Targeting the Receptor Tyrosine Kinase PDGFRα Induces Anti-tumor Effects through STAT3 and p53 in Glioblastoma. Molecular therapy. Nucleic acids 14, 131–141, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Esposito CL et al. STAT3 Gene Silencing by Aptamer-siRNA Chimera as Selective Therapeutic for Glioblastoma. Molecular therapy. Nucleic acids 10, 398–411, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Koh W, Park YM, Lee SE & Lee CJ AAV-Mediated Astrocyte-Specific Gene Expression under Human ALDH1L1 Promoter in Mouse Thalamus. Experimental neurobiology 26, 350–361, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Haery L et al. Adeno-Associated Virus Technologies and Methods for Targeted Neuronal Manipulation. Frontiers in neuroanatomy 13, 93, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Ma H et al. Pol III Promoters to Express Small RNAs: Delineation of Transcription Initiation. Molecular therapy. Nucleic acids 3, e161, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Miyagishi M & Taira K U6 promoter-driven siRNAs with four uridine 3’ overhangs efficiently suppress targeted gene expression in mammalian cells. Nature biotechnology 20, 497–500, (2002). [DOI] [PubMed] [Google Scholar]
- 365.Zhang Q, Chen CY, Yedavalli VS & Jeang KT NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio 4, e00596–00512, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Roh E & Kim MS Brain Regulation of Energy Metabolism. Endocrinology and metabolism (Seoul, Korea) 31, 519–524, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Lam TK Neuronal regulation of homeostasis by nutrient sensing. Nature medicine 16, 392–395, (2010). [DOI] [PubMed] [Google Scholar]
- 368.Cordner ZA & Tamashiro KL Effects of high-fat diet exposure on learning & memory. Physiology & behavior 152, 363–371, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Valls-Pedret C et al. Mediterranean Diet and Age-Related Cognitive Decline: A Randomized Clinical Trial. JAMA internal medicine 175, 1094–1103, (2015). [DOI] [PubMed] [Google Scholar]
- 370.Verrotti A, Iapadre G, Pisano S & Coppola G Ketogenic diet and childhood neurological disorders other than epilepsy: an overview. Expert review of neurotherapeutics 17, 461–473, (2017). [DOI] [PubMed] [Google Scholar]
- 371.Jagadish S et al. The Ketogenic and Modified Atkins Diet Therapy for Children With Refractory Epilepsy of Genetic Etiology. Pediatric neurology 94, 32–37, (2019). [DOI] [PubMed] [Google Scholar]
- 372.Kornfeld JW & Brüning JC Regulation of metabolism by long, non-coding RNAs. Frontiers in genetics 5, 57, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Takeuchi Y et al. Intravenous Bone Marrow Mononuclear Cells Transplantation in Aged Mice Increases Transcription of Glucose Transporter 1 and Na(+)/K(+)-ATPase at Hippocampus Followed by Restored Neurological Functions. Frontiers in aging neuroscience 12, 170, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Faubert B, Solmonson A & DeBerardinis RJ Metabolic reprogramming and cancer progression. Science (New York, N.Y.) 368, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Baik SH et al. A Breakdown in Metabolic Reprogramming Causes Microglia Dysfunction in Alzheimer’s Disease. Cell metabolism 30, 493–507.e496, (2019). [DOI] [PubMed] [Google Scholar]
- 376.Lin W et al. LncRNAs regulate metabolism in cancer. International journal of biological sciences 16, 1194–1206, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]


