Abstract
The structure of the highly efficient simian virus 40 late polyadenylation signal (LPA signal) is more complex than those of most known mammalian polyadenylation signals. It contains efficiency elements both upstream and downstream of the AAUAAA region, and the downstream region contains three defined elements (two U-rich elements and one G-rich element) instead of the single U- or GU-rich element found in most polyadenylation signals. Since many reports have indicated that the secondary structure in RNA may play a significant role in RNA processing, we have used nuclease structure analysis techniques to determine the secondary structure of the LPA signal. We find that the LPA signal has a functionally significant secondary structure. Much of the region upstream of AAUAAA is sensitive to single-strand-specific nucleases. The region downstream of AAUAAA has both double- and single-stranded characteristics. Both U-rich elements are predominately sensitive to the double-strand-specific nuclease RNase V1, while the G-rich element is primarily single stranded. The U-rich element closest to AAUAAA contains four distinct RNase V1-sensitive regions, which we have designated structural region 1 (SR1), SR2, SR3, and SR4. Linker scanning mutants in the downstream region were analyzed both for structure and for function by in vitro cleavage analyses. These data show that the ability of the downstream region, particularly SR3, to form double-stranded structures correlates with efficient in vitro cleavage. We discuss the possibility that secondary structure downstream of the AAUAAA may be important for the functions of polyadenylation signals in general.
Polyadenylation is the process by which the 3′ ends of most mammalian mRNAs are formed. In a tightly coupled set of reactions the precursor RNA is endonucleolytically cleaved at a specific site. Next, approximately 250 adenosine residues are polymerized to the cleaved end, forming the poly(A) tail of the final mRNA. It has been clearly established that a poly(A) tail is essential for the survival, transport, stability, and translation of most mRNAs (reviewed in references 8, 32, and 42 to 44).
The elements of the polyadenylation signal in the precursor RNA define the site of polyadenylation through specific binding with a complex of proteins which orchestrate cleavage and polyadenylation. The central, nearly invariant, element of mammalian polyadenylation signals is the AAUAAA sequence, which is the binding site for the cleavage and polyadenylation specificity factor (CPSF) (19). AAUAAA is located 11 to 25 nucleotides upstream of the actual cleavage and polyadenylation site. In addition, it is now well established that sequence elements located 14 to 70 nucleotides downstream of AAUAAA greatly increase the efficiency of utilization of the polyadenylation signal (4, 9, 14, 15, 23, 24, 25, 31, 33, 34, 45, 46, 49). Comparison of the downstream elements (DSEs) of many polyadenylation signals has not provided a clear consensus sequence other than various lengths (6 to 20 nucleotides) of GU- or U-rich sequences. However, the position of the DSE relative to the AAUAAA sequence appears to be important, since the DSE is the binding site for the cleavage-stimulatory factor (CStF) (38), which interacts with CPSF bound at AAUAAA to form a stable polyadenylation complex. Interestingly, the site of polyadenylation and DSE of the human T-cell leukemia virus type 1 polyadenylation signal, which is relatively efficient, are located more than 200 nucleotides downstream of AAUAAA. However, a large stem-loop secondary structure brings the DSE into an optimal position (i.e., 14 to 70 nucleotides away) relative to AAUAAA (1, 3A).
Many mammalian polyadenylation signals appear to conform to the simple structure of an AAUAAA and a GU- or U-rich DSE. However, a growing number of polyadenylation signals have been shown to contain additional efficiency elements located upstream of AAUAAA. These upstream elements (USEs) were originally studied in viral polyadenylation signals, such as the polyadenylation signals of the simian virus 40 (SV40) late genes (7, 36), human immunodeficiency virus (HIV) (6, 13, 40, 41), the adenovirus major late region (12), cauliflower mosaic virus (35), and ground squirrel hepatitis virus (29, 30). More recently, USEs have been found in some cellular genes as well (5, 27). Our studies of the SV40 late polyadenylation signal (LPA signal) suggest that the USEs impart characteristics which provide efficiency, or special levels of control, to the polyadenylation signal. For example, mutation of the LPA signal USEs reduces polyadenylation efficiency by 75 to 85% both in vivo and in vitro (7, 36).
Few generalizations can be made about USEs. Among the USEs which have been studied, no definitive characteristics have been noted. However, functional upstream motifs in the LPA signal and the HIV polyadenylation signal have been characterized (36, 40, 41). In HIV these motifs appear to be involved in the formation of a secondary structure which aids CPSF recruitment to the HIV type 1 polyadenylation signal (16).
The LPA signal is a very efficient polyadenylation signal which contains a more complex structure than most. As shown in Fig. 1, it contains both USEs and DSEs. In addition, the downstream region is more unusual in that it contains three defined elements, two U-rich elements (DSE-U and DSE-U′) (Fig. 1) (9, 33, 34) and a G-rich region (DSE-G) (Fig. 1) (3, 28) in between. The DSE closest to AAUAAA (DSE-U) contains the site for CStF binding. The G-rich region has been shown to interact with a protein of the hnRNP H family, which increases polyadenylation efficiency (2, 3, 28). The other downstream U-rich DSE (DSE-U′) can bind hnRNP C proteins (45, 46) and functions as a DSE under certain conditions. Specifically, in an in vitro polyadenylation reaction with a substrate representing the entire LPA signal, deletion or mutation of DSE-U′ had little effect on polyadenylation efficiency (our unpublished observations). However, if the LPA signal is combined with a splicing cassette, such that a coupled splicing and polyadenylation substrate is formed, mutation of DSE-U′ causes diminished polyadenylation and splicing, suggesting that the element may function under conditions of coupled polyadenylation and splicing (10).
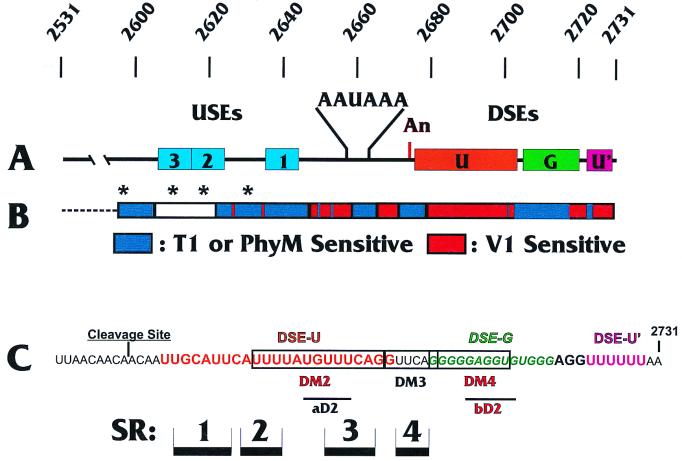
FIG. 1.
SV40 LPA signal. (A) Features of the SV40 LPA signal. AAUAAA and the cleavage site (An) are shown along with the three USEs (USE1 to -3; blue boxes) and the three DSEs (DSE-U [orange box], a predominantly U-rich element just downstream of the cleavage site; DSE-G [green box], a G-rich region downstream of DSE-U; and DSE-U′ [purple box], a second U-rich element further downstream). The numbering above the diagram indicates the SV40 nucleotide numbering. (B) Summary of nuclease structure analyses of the LPA signal. Regions sensitive to the double-strand-specific RNase V1 are indicated in red, and regions sensitive to the single-strand-specific RNase T1 or PhyM are indicated in blue. The white box and asterisks indicate regions which appeared to be inaccessible to the nucleases in the structural analyses (see the text). Note that the diagram does not continue as far upstream as does the structural analysis shown in Fig. 2. (C) Sequence of the region downstream of the cleavage site. The approximate regions of the three DSEs are shown; DSE-U is in orange letters, DSE-G is in green italic letters, and DSE-U′ is in purple letters. Regions of the double-stranded structures SR1 to SR4 identified in this study are shown as black bars. The locations of the various linker substitution mutations used in these studies are indicated (see the text).
The USEs of the LPA signal consist of three regions with sequences similar to the sequence AUUUGURA (36). We have previously shown that the U1A protein interacts with the LPA signal and that the USEs are necessary for efficient U1A binding (21). Our data suggest that the interaction of U1A protein may facilitate polyadenylation complex formation or be involved in the coupling of polyadenylation and splicing (22).
The existence of secondary structures in human T-cell leukemia virus type 1 (1, 3A) and HIV polyadenylation signals (11, 16) has been mentioned above. In addition, secondary structures have been suggested to be significant in the functions of several other polyadenylation signals, including those of the equine infectious anemia virus (18), the bovine growth hormone gene (17), the adenovirus L4 gene (37), the human CD59 gene (39), and the murine immunoglobulin M secretory gene (27a). Given these findings and the complexity of the LPA signal, we have used nuclease structure analysis techniques to analyze the secondary structure in the LPA signal. We find that the LPA signal forms a functionally significant secondary structure. Much of the region upstream of AAUAAA is sensitive to single-strand-specific nucleases. The downstream region has both double- and single-stranded characteristics. Both U-rich DSEs are predominately sensitive to the double-strand-specific nuclease RNase V1, while the G-rich element is primarily single stranded. We find that the U-rich element closest to AAUAAA contains four distinct RNase V1-sensitive regions. Mutational analyses indicate that the ability to form downstream double-stranded regions correlates with efficient in vitro cleavage. We discuss the possibility that a secondary structure downstream of AAUAAA may be important for the function of polyadenylation signals in general.
MATERIALS AND METHODS
Plasmids and substrate RNAs.
Substrate RNAs containing the LPA signal were produced by in vitro transcription using T7 RNA polymerase and the plasmid templates described below. RNAs were synthesized either unlabeled or labeled internally with [32P]UTP. Unlabeled RNAs were transcribed either with (for 3′-end labeling) or without (for 5′-end labeling) a 5′ cap structure. RNAs were then dephosphorylated using calf intestinal alkaline phosphatase. After calf intestinal alkaline phosphatase inactivation, the RNAs were extracted with phenol-chloroform and precipitated with ethanol. 5′-end labeling was performed using [γ-32P]ATP and polynucleotide kinase, while 3′-end labeling was accomplished using 32P-pCp and T4 RNA ligase (20). All RNAs were gel isolated by separating the full-length RNAs from smaller transcripts on 5% polyacrylamide–8 M urea gels. The full-length transcripts were eluted from gel slices by overnight incubation in 20 mM Tris HCl (pH 7.5)–400 mM NaCl–0.1% sodium dodecyl sulfate (SDS). The eluted RNAs were phenol extracted and ethanol precipitated.
The plasmid templates for the substrate RNAs included the following. (i) pUPAS (7, 36) encodes the wild-type LPA signal between nucleotides 2533 and 2770 inserted into the polylinker of pGEM2. All of the plasmids used for in vitro transcription were linearized at a DraI site at the end of the downstream region of the LPA signal (SV40 nucleotide 2731). (ii) pPAS (7, 36) is similar to pUPAS except that an XbaI linker has been inserted 5′ of AAUAAA and an XhoII linker has been inserted 3′ of AAUAAA. These sites were used along with the restriction sites found in the polylinker to create the following constructions: pUM123, which was constructed by inserting linkers to substitute for the sequences containing the three USEs (36), and pdlUSEs, which lacks the sequence upstream of AAUAAA and was created by removing the XbaI-to-XbaI fragment from pPAS and inserting it into the pGEM2 vector.
A set of linker substitution mutants with mutations through the downstream region, mutants DM2 to DM4 (10) and mutants aD2, bD2, and abD2, was constructed with pUPAS by using previously described PCR-based linker scanning mutagenesis techniques (36, 47, 48). The positions of their mutations are shown in Fig. 1C. The wild-type nucleotides were replaced with the following sequences: 5′…CGCGGGAGGTACC…3′ (DM2), 5…GGTACC…3′ (DM3), 5′…ATAGGTACC…3′ (DM4), 5′…GGTACC…3′ (aD2), 5′…GGTACC…3′ (bD2), and a construction that combines the aD2 and bD2 mutations (abD2). In all cases, the wild-type sequences were exactly replaced with either a KpnI linker or a KpnI linker plus additional sequences.
In vitro polyadenylation cleavage assays.
Substrate RNAs were synthesized by in vitro transcription and internally labeled with [32P]UTP as described above. Labeled RNAs (105 cpm) were incubated in a final volume of 25 μl containing 64 μg of HeLa nuclear extract (26), 250 μM ATP, 1 mM cordycepin, 250 mM phosphocreatine, and 6.5 μl of 10% polyvinyl alcohol. The cleavage reaction mixtures were incubated at 30°C for 30 min. Reactions were stopped by the addition of a high-concentration salt–SDS buffer (20 mM Tris HCl [pH 7.5], 400 mM NaCl, 0.1% SDS), and RNAs were purified by phenol-chloroform extraction and then ethanol precipitation. RNA products were fractionated on a 5% acrylamide–8 M urea gel and visualized by autoradiography. Products were quantitated using a Molecular Dynamics Storm PhosphorImager.
RNA sequencing and structure analysis.
RNAs, either 3′ or 5′ end labeled, were analyzed by both sequencing and structure-probing reactions which were modified from previously established procedures (20). Prior to all reactions, the RNAs were thawed slowly and then incubated at 37°C for 15 min.
RNAs were sequenced by incubating 104 cpm of RNA in RNA sequencing buffer (7 M urea, 0.025% xylene cyanol, 0.025% bromphenol blue, 20 mM sodium citrate [pH 5], 1 mM EDTA) in the presence of 7.5 × 10−2 U of RNase T1 or 3 U of RNase PhyM for 15 min at 50°C in a final volume of 4 μl. Nonspecific RNA degradation was determined by incubating the RNAs with RNA sequencing buffer for 15 min at 50°C. RNA ladders were formed by partial hydrolysis of RNAs using 1 M NaHCO3, pH 9.2, for 5 min at 90°C.
Structural analyses of RNAs were performed by incubating RNA substrates in standard structure-probing buffer (10 mM Tris HCl [pH 7], 10 mM MgCl2, 100 mM KCl) with various RNases (7.5 × 10−3 U of RNase T1, 1.5 U of RNase PhyM, or 1.5 × 10−3 U of RNase V1) for 20 min at 37°C in a final volume of 4 μl.
Immediately following all analyses, the reactions were stopped by adding an equal volume of 2× sequencing gel loading solution (9 M urea, 0.05% xylene cyanol, 0.05% bromphenol blue, 10% glycerol) and quick freezing in dry ice-ethanol. Prior to electrophoretic analysis, samples were heated at 80°C for 1 min. The samples were separated on prerun 10% polyacrylamide–7 M urea gels.
RESULTS
Secondary-structure analysis of the wild-type SV40 LPA signal.
We made use of RNase sequencing and structure analysis techniques to determine the RNA secondary structure of the LPA signal. The RNases used included RNase T1, which cleaves at single-stranded G residues; RNase PhyM, which cleaves at single-stranded A and U residues; and RNase V1, which cleaves double-stranded RNA with no nucleotide specificity.
The wild-type LPA signal RNA substrate analyzed consists of SV40 nucleotides 2531 to 2731 (Fig. 1A), which contain all the characterized elements (see the introduction) known to be needed for efficient cleavage and polyadenylation both in vitro and in vivo (7, 10, 36). To obtain both sequence and structure data from the entire LPA signal, the RNA substrates were 32P labeled at either the 5′ or the 3′ end. The substrates were then treated with RNases using (i) high-temperature, denaturing conditions for sequence analysis or (ii) lower-temperature, native conditions for structure analysis (see Materials and Methods). Figures 2 (region upstream of AAUAAA) and 3 (region downstream of AAUAAA) show examples of data upon which we based our structural conclusions. The overall structural conclusions are based on the consensus results of numerous structure analysis experiments, the data from which were examined by several individuals in the laboratory in order to obtain unbiased interpretations.
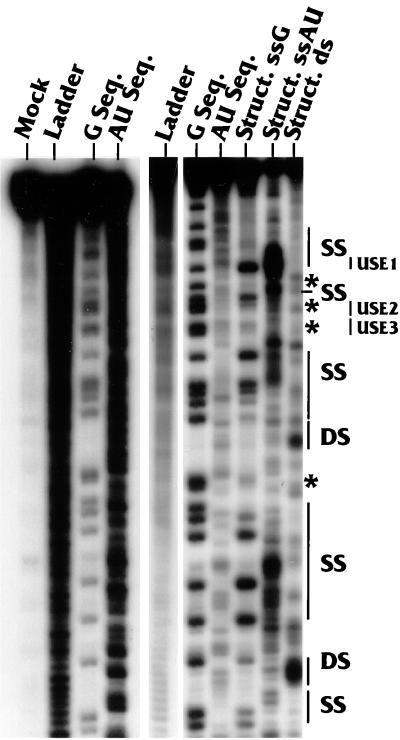
FIG. 2.
RNase sequencing and structure analyses of the region upstream of AAUAAA. Sequencing and structure analysis reactions were carried out with a substrate in which the wild-type LPA signal was labeled at its 5′ end with 32P as described in Materials and Methods. The labeling of the lanes indicates the nucleotide(s) (G or AU) being analyzed and whether the analysis is for sequence (Seq.) or structure (Struct.). The lane marked Struct. ds reflects results of the structural analysis using RNase V1, which is specific for double-stranded RNA with no nucleotide preference. The lane marked Ladder contains the oligonucleotide ladder generated by alkaline hydrolysis of the substrate RNA. The panel on the left shows the mock-digested sample (lane Mock), an additional hydrolysis ladder, and the results of repeated sequencing analyses, which provided for better analysis of the structural data. The positions of single-stranded (SS) and double-stranded (DS) regions are indicated on the right as well as the positions of the three USEs, USE1, USE2, and USE3. The asterisks indicate G nucleotides which were not well cleaved by either RNase T1 or V1 in the structure analyses (see the text).
As indicated in Figures 2 and 3, our data suggested that regions both upstream and downstream of AAUAAA contained single- and double-stranded characteristics as indicated by specific nuclease sensitivity. Figure 1B shows a graphic summary of the data indicating the RNase T1- and PhyM-sensitive regions (single strand specific) in blue and the RNase V1-sensitive regions (double strand specific) in red. The data suggest that the upstream region (Fig. 1B and 2) is predominantly sensitive to single-strand-specific nucleases but that the region downstream of AAUAAA is significantly more sensitive to the double-strand-specific nuclease (Fig. 1B and 3).
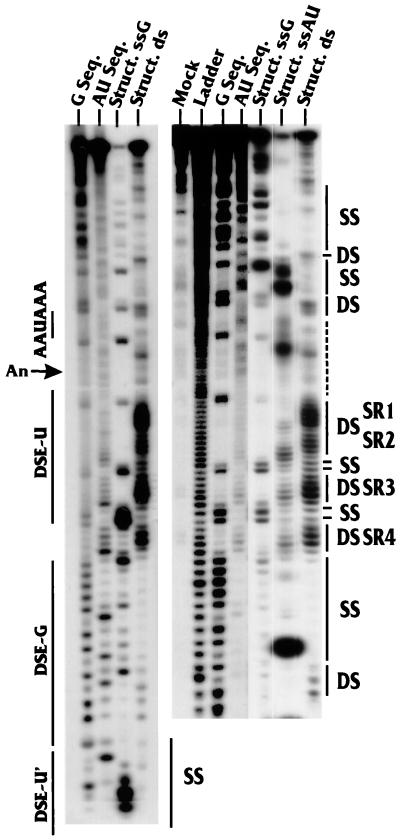
FIG. 3.
RNase sequencing and structure analyses of the region downstream of AAUAAA. Sequencing and structure analysis reactions were carried out with an RNA substrate in which the wild-type LPA signal was labeled at its 3′ end with 32P as described in Materials and Methods. The lanes indicate the nucleotide(s) (G or AU) being analyzed and whether the analysis is for sequence (Seq.) or structure (Struct.). The lane marked Struct. ds shows the results of the structural analysis using RNase V1, which is specific for double-stranded RNA with no nucleotide preference. The lane marked Ladder contains the oligonucleotide ladder generated by alkaline hydrolysis of the substrate RNA. The lane marked Mock contains the mock-digested sample. The panel on the left provides additional sequence and structural analyses to show reproducibility. The locations of AAUAAA, the cleavage site (An), DSE-U, DSE-G, and part of DSE-U′ are indicated at the left of the panels. The positions of single-stranded (SS) and double-stranded (DS) regions as well as the positions of the four prominent double-stranded regions, SR1 to SR4 (see the text), are indicated at the right of the panels. It should be noted that the sample used in the Struct. ssAU lane was overly digested with RNase PhyM, which resulted in low levels of cleavage at A's and U's in the double-stranded regions SR1 to SR4.
Structure of the regions upstream of AAUAAA.
A more detailed examination of the data for the upstream region (Fig. 1B and 2) shows that USE1, the upstream element closest to the AAUAAA (Fig. 1), was consistently sensitive to the single-strand-specific nucleases and resistant to RNase V1. This element is very AU rich, and its single-stranded nature can be seen by the prominent RNase PhyM cleavages. Previous linker substitution mutational analysis (36) to examine the individual effects of the three USEs indicated that USE1 has the most significant effect on polyadenylation efficiency. Thus, the marked single-stranded characteristic of USE1 may be functionally significant. Interestingly, it was difficult to determine the structures of USE2 and USE3. The asterisks in Fig. 1B and 2 indicate specific examples of G's which were cleaved by RNase T1 in the sequencing reactions but failed to be cleaved by either RNase T1 or RNase V1 in the structure analysis. This result indicates that there are structural features in the RNA which protect these nucleotides from access by the relatively large nucleases. Please note that the results of our sequencing and structure analyses in Fig. 2 extend further upstream than is diagrammed in Fig. 1B.
Structure of the regions downstream of AAUAAA.
As mentioned above, a significant portion of the downstream region is sensitive to the double-strand-specific nuclease RNase V1. A distinct pattern of four double-stranded regions can be seen. These are denoted structural regions 1, 2, 3, and 4 (SR1 to -4) (Fig. 3). The positions of these regions with respect to those of the three DSEs (DSE-U, DSE-G, and DSE-U′) and the nucleotide sequence of the downstream region are shown in Fig. 1C. The first U-rich DSE (DSE-U) (Fig. 1C), nearest AAUAAA, is predominantly double stranded, whereas the G-rich DSE (DSE-G) is single stranded. The final U-rich DSE (DSE-U′) is again within a double-stranded region as indicated by RNase V1 sensitivity (note that the RNase V1 sensitivity of DSE-U′ is not shown in Fig. 3). Figure 3 and results of additional structural analyses (not shown) suggest that the structure of AAUAAA is predominantly single stranded; however, the first two A's of AAUAAA consistently show sensitivity to RNase V1.
Integrity of the downstream structure during mutation of the upstream region.
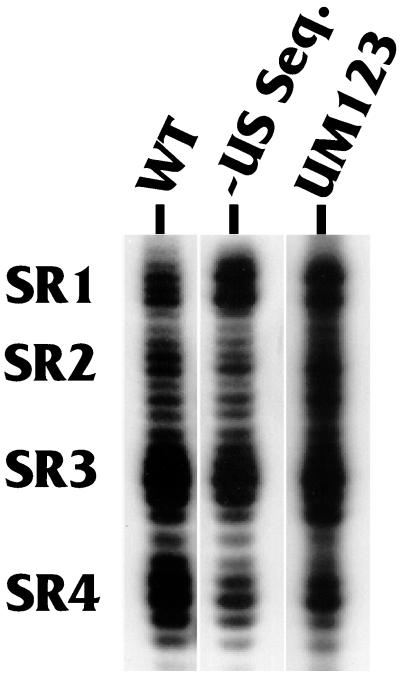
We next asked whether the formation of the four downstream double-stranded regions, SR1 to SR4, was dependent on the presence of the upstream region. Figure 4 shows the structures of SR1 to SR4 as analyzed with RNase V1. It is clear that the wild-type structure of the region (lane WT) changed very little either by the deletion of all sequences upstream of AAUAAA (lane −US Seq.), or by specific linker substitution mutagenesis of the three USEs (lane UM123). Thus, the formation of the downstream SRs, SR1 to SR4, is independent of upstream sequences.
FIG. 4.
Integrity of SR1 to SR4 during mutation of the upstream region. The RNase V1 sensitivity of the prominent downstream double-stranded regions SR1 to SR4 was analyzed using substrates in which the wild-type LPA signal RNA was labeled at its 3′ end with 32P (lane WT) and two mutants in which (i) the entire region upstream of AAUAAA was deleted (lane −US Seq.) or (ii) the three USEs were mutated by specific linker substitution mutagenesis (lane UM123).
Correlation of function with the formation of the downstream double-stranded structure.
To determine the functional significance of the downstream double-stranded regions (SR1 to -4), we examined several linker substitution mutations in the downstream region. Each mutation was examined for its effect on structure, which was correlated with its effect on in vitro polyadenylation efficiency. The mutations tested are shown in Fig. 1C, where the exact bases substituted are indicated by boxes (mutants DM2 to -4) or lines (mutants aD2 and bD2). The mutant sequences substituted in each case are described in Materials and Methods.
The wild-type and mutant LPA signal substrates were assayed in in vitro polyadenylation cleavage reactions. The efficiency of cleavage for each substrate was determined and compared to that of the wild type, which was set at 100%. These experiments were repeated at least three times, resulting in a standard error of less than 5%. The results are shown in the boxes at the bottom of Fig. 5, below the RNase V1 structural analysis of the downstream region of each substrate.
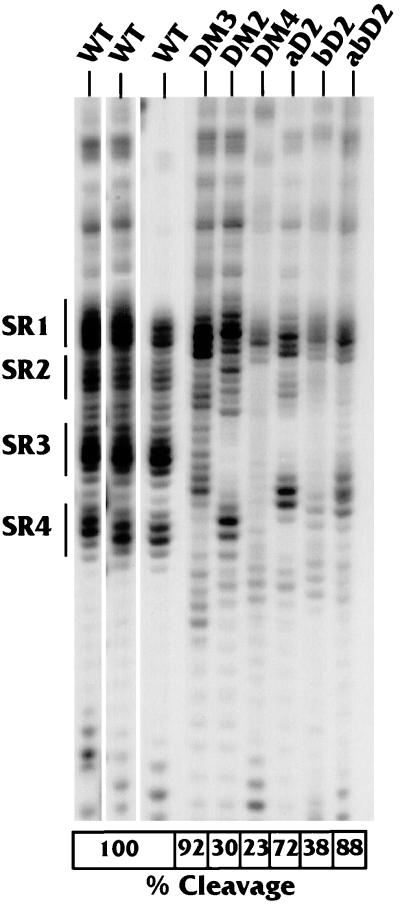
FIG. 5.
Correlation of downstream structure with in vitro cleavage efficiency. The RNase V1 sensitivity of the downstream region, particularly SR1 to SR4, was analyzed using 32P-3′-end-labeled substrates representing the wild-type LPA signal (WT) and the LPA signal containing linker substitution mutants in the downstream region (DM2, DM3, DM4, aD2, bD2, and abD2). The locations of these mutations in the LPA signal are shown in Fig. 1C. The results of three WT structure analyses are provided to demonstrate reproducibility. The boxes at the bottom show the percentage of cleavage by each of the substrates as measured in an in vitro cleavage reaction by using a HeLa cell extract (see Materials and Methods). The wild-type cleavage activity is set at 100%, and the standard error of the analyses is ±5%.
We show three different analyses of the wild-type LPA signal to indicate the consistency of detection of the RNase V1-sensitive regions SR1 to SR4. Mutant DM3, which had very little effect on in vitro cleavage (92% of the wild-type level), primarily had mutated nucleotides in the RNase V1-sensitive region SR4 (Fig. 1C). These mutations affected the structure in SR4 (Fig. 5), but the RNase V1-sensitive structure remained in SR1, -2, and -3, although it appeared reduced in intensity compared to that of the wild type, suggesting that it may not be as stable. In addition, the position of the V1 sensitivity of SR3 is shifted 2 to 4 nucleotides downstream. The fact that this mutation had little effect on function suggests that (i) the residual RNase V1-sensitive regions are sufficient for cleavage and (ii) the SR4 region is not essential for efficient cleavage of the LPA signal.
Mutant DM2 markedly lowered cleavage efficiency to 30% of that of the wild type. Its mutations also altered a significant number of nucleotides in DSE-U, affecting nucleotides in SR2 and SR3 (Fig. 1C). The mutations completely eliminated RNase V1 sensitivity in SR3 and altered that of SR2; however, RNase V1 sensitivity remained in SR1 and -4. The deleterious effect of the mutations in DM2 on function, coupled with the disruption of SR3 and possibly SR2, suggests that these double-stranded regions may be significant for cleavage.
Mutants DM4 and bD2 can be considered together since they overlap, altering bases in DSE-G. Importantly, these mutations affect sequences which are predominately single stranded by nuclease analysis. No nucleotides within SR1 to SR4 were mutated. Both mutations were functionally quite deleterious (producing levels 23 and 38% of that of the wild type, respectively), and despite their location each eliminated RNase V1 sensitivity throughout SR2, SR3, and SR4. These data again show that disruption of the secondary structure in SR3, and possibly SR2, correlates with loss of cleavage efficiency.
The mutations in aD2 affected fewer bases in DSE-U than did those in DM2, and it had only a moderate effect on function (72% of wild-type cleavage). The nucleotides mutated included some in SR3 (Fig. 1C), and the double-stranded nature of SR3 was disrupted. However, a new region with a double-stranded structure appeared 2 to 4 nucleotides further downstream of the wild-type position of SR3 (Fig. 5). Thus, structure at or near the position of SR3 (relative to AAUAAA) again correlates with function. Further, the observation that this mutation had little effect on cleavage suggests that the ability to form a secondary structure at or near the position of SR3 may be more significant than the actual nucleotide sequence.
This idea is supported by mutant abD2, which combines the mutations in aD2 and bD2 in the same RNA substrate. This mutant substrate was efficiently processed (88% of wild-type cleavage), indicating that the deleterious effect of the mutations in bD2 on cleavage efficiency is compensated for by aD2's mutations. In addition, the double-stranded region introduced near SR3 by mutant aD2 was again present. Further, the double-stranded region near SR3 was the only significant RNase V1-sensitive region in the abD2 substrate other than SR1 (which was present to some extent in all of the mutants). Hence, the ability of the mutations in aD2 to form this SR3-like structure appears to have compensated for the deleterious effects of the mutations in bD2. These results argue that double-stranded structure at or near the position of SR3 (relative to AAUAAA) is important for function.
DISCUSSION
In this report we have described RNA secondary structure as a functional feature of the LPA signal. In the introduction we noted a number of examples where the secondary structure of a polyadenylation signal appears to play a role in function. Many of these examples were based on a computer-predicted structure with minimal experimental conformation. We have provided experimental characterization of the secondary structure, concluding that the upstream region is predominantly single stranded and that the downstream region contains functionally significant double-stranded regions. We have not presented a diagram of a base-paired secondary structure, since attempts to generate such structures using available programs have resulted in either structures which do not completely correspond to the nuclease sensitivity data or structures which are thermodynamically unfavorable when the programs are forced to consider the nuclease sensitivity data. Our nuclease sensitivity analysis provides a first approximation of the solution structure of the LPA signal in the absence of proteins. Although our data suggest that this structure correlates with function, there are clearly many additional aspects of the structure which are yet to be determined as discussed below.
The upstream region.
Our data indicate that the region upstream of AAUAAA is predominantly sensitive to single-strand-specific nucleases, suggesting a relatively large region of linear, nonstructured RNA. USE1, the USE closest to AAUAAA, is clearly single stranded; this is the USE with the greatest effect on polyadenylation efficiency. However, the structures of USE2 and -3 were difficult to determine because these regions were relatively resistant to both single- and double-strand-specific nucleases. This result is interesting, since it suggests a higher-order structure in which the nucleotides in the region of USE2 and -3 are folded such that they are not accessible to the comparatively large nucleases.
The downstream region.
The downstream region's predominant characteristics are the double-stranded SRs, SR1 to SR4, within the U-rich DSE closest to AAUAAA (DSE-U), followed by the starkly single-stranded region encompassing the majority of the G-rich DSE (DSE-G). Our data show that the structures formed in SR1 to SR4 are solely a property of the downstream region, since mutation or deletion of sequences upstream of AAUAAA had no effect on the formation of SR1 to SR4.
Our data strongly suggest that the double-stranded nature of SR3 is functionally significant. The most compelling evidence for this came from mutant aD2, which altered nucleotides within SR3 but reconstituted an alternate RNase V1-sensitive region within 2 to 4 nucleotides of the wild-type position of SR3 relative to that of AAUAAA. It should be reiterated that this alternate structure contains none of the wild-type bases normally found in this region but that the mutant functions at more than 70% of the wild-type level of cleavage. This observation suggests that structure at this position in the polyadenylation signal is more significant than exact sequence. The significance of SR1 in the function of the LPA signal cannot be predicted from our data since none of the mutants tested disrupted its RNase V1 sensitivity.
Mutations in DM4 and bD2, within DSE-G, provided surprising results. Each dramatically eliminated the double-stranded character of SR2, SR3, and SR4. In addition, these mutations had the greatest negative effect on in vitro cleavage. One could argue that the loss of in vitro cleavage is simply due to mutation of the G-rich element, which is a binding site for an hnRNP H family member (known also as DSEF-1) previously shown to be significant for LPA signal function (2, 3, 28). However, using mutant abD2 (the combination of mutants aD2 and bD2), we have shown that the loss of in vitro cleavage mediated by mutant bD2 can be overcome by the formation of an alternate double-stranded structure near the position of SR3 by mutant aD2 (discussed above). These data suggest that the functional defect of mutant bD2 is not simply the mutation of the hnRNP H site but that it is, at least in part, the result of the mutations' disruption of the double-stranded structure in the SR3 region. The wild-type region SR3 begins at a position 35 nucleotides downstream of AAUAAA. Our data suggest that such a structure, at a position between 35 and 39 nucleotides downstream of AAUAAA, is important for the functioning of the polyadenylation signal. Further, our data suggest that the structure in this region is more functionally significant than the sequence.
Downstream secondary structure as a general characteristic of polyadenylation signals.
Our data indicate the functional significance of the secondary structure in the downstream region of the LPA signal. However, the specific mechanism requiring this secondary structure for function is yet to be identified. The sequences encompassing SR1 to SR4 contain the site for CStF binding. It is possible that the secondary structure aids the CStF interaction. Indeed, the downstream secondary structure in the murine immunoglobulin M gene polyadenylation signal may affect polyadenylation complex formation (27a).
It is intriguing to consider that RNA structure in the downstream region may perform a catalytic role in the cleavage process. In this regard it should be noted that, although the cleavage factors of the polyadenylation complex have been examined, no nucleolytic activity has yet been noted. Hence, it is possible that the combination of protein and structure in the substrate RNA provides the cleavage activity.
How general is the feature of secondary structure in the downstream regions of polyadenylation signals? Clearly not enough polyadenylation signals have been examined to answer this question. However, it is well established that the DSEs of most mammalian polyadenylation signals consist of a region (8 to 12 or more nucleotides) which is GU or U rich. Due to the multiple ways G's and U's can base pair in RNA, a GU- or U-rich region would provide the most versatility in forming base-paired regions with surrounding sequences. In the LPA signal, a very efficient polyadenylation signal, the downstream structure was stable enough to be examined in solution. However, in a simpler polyadenylation signal, the needed secondary structure in the DSE may form transiently or coordinately with the binding of polyadenylation factors. Indeed the initial structure, or the structure determined in solution, may be altered by the binding of proteins. Thus, we suggest that stable or transient formation of secondary structure involving the DSE may be an important feature of mammalian polyadenylation signals.
ACKNOWLEDGMENTS
We thank the members of the Alwine laboratory for aid in and discussion of the experiments.
H.H. was supported by NIH training grant 5-T32-AI 07325. This work was supported by NIH grant GM45773 provided to J.C.A. by the Public Health Service.
REFERENCES
- 1.Ahmed Y F, Gilmartin G M, Hanly S M, Nevins J R, Greene W C. The HTLV-I rex response element mediates a novel form of mRNA polyadenylation. Cell. 1991;64:727–737. doi: 10.1016/0092-8674(91)90502-p. [DOI] [PubMed] [Google Scholar]
- 2.Bagga P S, Arhin G K, Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagga P S, Ford L P, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Bar-Shira A, Panet A, Honigman A. An RNA secondary structure juxtaposes two remote genetic signals for human T-cell leukemia virus type I RNA 3′-end processing. J Virol. 1991;65:5165–5173. doi: 10.1128/jvi.65.10.5165-5173.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhat B M, Wold W S M. ATTAAA as well as downstream sequences are required for RNA 3′-end formation in the E3 complex transcription unit of adenovirus. Mol Cell Biol. 1985;5:3183–3193. doi: 10.1128/mcb.5.11.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brackenridge S, Ashe H L, Giacca M, Proudfoot N J. Transcription and polyadenylation in a short human intergenic region. Nucleic Acids Res. 1997;25:2326–2335. doi: 10.1093/nar/25.12.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown P H, Tiley L S, Cullen B R. Efficient polyadenylation with the human immunodeficiency virus type I long terminal repeat requires flanking U3-specific sequences. J Virol. 1991;65:3340–3343. doi: 10.1128/jvi.65.6.3340-3343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carswell S, Alwine J C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989;9:4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 9.Conway L, Wickens M. A sequence downstream of AAUAAA is required for formation of simian virus 40 late mRNA 3′ termini in frog oocytes. Proc Natl Acad Sci USA. 1985;82:3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooke C, Hans H, Alwine J C. Utilization of splicing elements and polyadenylation elements in the coupling of polyadenylation and last intron removal. Mol Cell Biol. 1999;19:4971–4979. doi: 10.1128/mcb.19.7.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A T, Klaver B, Berkhout B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeZazzo J D, Imperiale M J. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol Cell Biol. 1989;9:4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeZazzo J D, Kilpatrick J E, Imperiale M J. Involvement of long terminal repeat U3 sequences overlapping the transcription control region in human immunodeficiency virus type I mRNA 3′ end formation. Mol Cell Biol. 1991;11:1624–1630. doi: 10.1128/mcb.11.3.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gil A, Proudfoot N J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit β-globin mRNA formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 15.Gil A, Proudfoot N J. A sequence downstream of AAUAAA is required for rabbit β-globin mTNS 3′-end formation. Nature. 1984;312:473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- 16.Gilmartin G M, Fleming E S, Oetjen J. Activation of HIV-1 pre-mRNA 3′ processing in vitro requires both an upstream element and TAR. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gimmi E R, Reff M E, Deckman I C. Alterations in the pre-mRNA topology of the bovine growth hormone polyadenylation region decrease poly(A) site efficiency. Nucleic Acids Res. 1989;17:6983–6998. doi: 10.1093/nar/17.17.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graveley B R, Gilmartin G M. A common mechanism for the enhancement of mRNA 3′ processing by U3 sequences in two distantly related lentiviruses. J Virol. 1996;70:1612–1617. doi: 10.1128/jvi.70.3.1612-1617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller W, Bienroth S, Lang K M, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing AAUAAA. EMBO J. 1991;10:4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knapp G. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- 21.Lutz C, Alwine J C. Direct interaction of the U1snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 22.Lutz C S, Murthy K G, Schek N, Manley J L, Alwine J C. Interaction between the U1snRNP-A protein and the 160 kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 23.McDevitt M A, Hart R P, Wong W W, Nevins J R. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 1986;5:2907–2913. doi: 10.1002/j.1460-2075.1986.tb04586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDevitt M A, Imperiale M J, Ali H, Nevins J R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984;37:992–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- 25.McLauchlan J, Gaffney D, Whitton J L, Clements J B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res. 1985;13:1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore C L. Preparation of mammalian extracts active in polyadenylation. Methods Enzymol. 1990;181:49–74. doi: 10.1016/0076-6879(90)81112-8. [DOI] [PubMed] [Google Scholar]
- 27.Moreira A, Wollerton M, Monks J, Proudfoot N J. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogentically conserved. EMBO J. 1995;14:3809–3819. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Phillips C, Kyriakopoulou C B, Virtanen A. Identification of a stem-loop structure important for polyadenylation at the murine IgM secertory poly(A) site. Nucleic Acids Res. 1999;27:429–438. doi: 10.1093/nar/27.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian Z, Wilusz J. An RNA-binding protein specifically interacts with a functionally important domain of the downstream element of the simian virus 40 late polyadenylation signal. Mol Cell Biol. 1991;11:5312–5320. doi: 10.1128/mcb.11.10.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russnak R. Regulation of polyadenylation in hepatitis B viruses: stimulation by the upstream activating signal PS1 is orientation-dependent, distance-dependent, and additive. Nucleic Acids Res. 1991;19:6449–6456. doi: 10.1093/nar/19.23.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russnak R, Ganem D. Sequences 5′ to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 1990;4:764–776. doi: 10.1101/gad.4.5.764. [DOI] [PubMed] [Google Scholar]
- 31.Ryner L C, Takagaki Y, Manley J L. Sequences downstream of AAUAAA signals affect pre-mRNA cleavage and polyadenylation in vitro both directly and indirectly. Mol Cell Biol. 1989;9:1759–1771. doi: 10.1128/mcb.9.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachs A, Wahle E. Poly(A) tail metabolism and function in eucaryotes. J Biol Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- 33.Sadofsky M, Alwine J C. Sequences on the 3′-side of hexanucleotide AAUAAA affect efficiency of cleavage at the polydenylation site. Mol Cell Biol. 1984;4:1460–1468. doi: 10.1128/mcb.4.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadofsky M, Connelly S, Manley J L, Alwine J C. Identification of a sequence element on the 3′ side of AAUAAA which is necessary for simian virus 40 late mRNA 3′-end processing. Mol Cell Biol. 1985;5:2713–2719. doi: 10.1128/mcb.5.10.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanfacon H, Brodmann P, Hohn T. A dissection of the cauliflower mosaic virus polyadenylation signal. Genes Dev. 1991;5:141–149. doi: 10.1101/gad.5.1.141. [DOI] [PubMed] [Google Scholar]
- 36.Schek N, Cooke C, Alwine J C. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal using in vitro cleavage and polyadenylation analyses. Mol Cell Biol. 1992;12:5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sittler A, Gallinaro H, Jacob M. The secondary structure of the adenovirus-2 L4 polyadenylation domain: evidence for a hairpin structure exposing the AAUAAA signal in the loop. J Mol Biol. 1995;248:525–540. doi: 10.1006/jmbi.1995.0240. [DOI] [PubMed] [Google Scholar]
- 38.Takagaki Y, Manley J L. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tone M, Walsh L A, Waldmann H. Gene structure of human CD59 and demonstration that discrete mRNAs are generated by alternative polyadenylation. J Mol Biol. 1992;227:971–976. doi: 10.1016/0022-2836(92)90239-g. [DOI] [PubMed] [Google Scholar]
- 40.Valsamakis A, Schek N, Alwine J C. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol Cell Biol. 1992;12:3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valsamakis A, Zeichner S, Carswell S, Alwine J C. The human immunodeficiency virus type 1 polyadenylation signal: a 3′-LTR element upstream of the AAUAAA necessary for efficient polyadenylation. Proc Natl Acad Sci USA. 1991;88:2108–2112. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahle E, Keller W. The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 43.Wahle E, Keller W. The biochemistry of polyadenylation. Trends Biochem Sci. 1996;27:247–250. [PubMed] [Google Scholar]
- 44.Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990;15:277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- 45.Wilusz J, Feig D I, Shenk T. The C proteins of heterogeneous nuclear ribonucleoprotein complexes interact with RNA sequences downstream of polyadenylation cleavage sites. Mol Cell Biol. 1988;8:4477–4483. doi: 10.1128/mcb.8.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilusz J, Shenk T. A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein-RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol Cell Biol. 1990;10:6397–6407. doi: 10.1128/mcb.10.12.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaret K S, Lin J, DePersio C M. Site-directed mutagenesis reveals a liver transcription factor essential for the albumin transcriptional enhancer. Proc Natl Acad Sci USA. 1990;87:5469–5473. doi: 10.1073/pnas.87.14.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeichner S L, Kim J Y H, Alwine J C. Linker scanning mutational analysis of the transcriptional activity of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1991;65:2436–2444. doi: 10.1128/jvi.65.5.2436-2444.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F, Cole C N. Identification of a complex associated with processing and polyadenylation in vitro of herpes simplex virus type 1 thymidine kinase precursor RNA. Mol Cell Biol. 1987;7:3277–3286. doi: 10.1128/mcb.7.9.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]