Abstract
Real-time polymerase chain reaction (RT-PCR) remains the gold standard for detection of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). This study tested the performance of a pooled testing strategy for RT-PCR and its cost-effectiveness. In total, 1280 leftover respiratory samples collected between 19 April and 6 May 2021 were tested in 128 pools of 10 samples each, out of which 16 pools were positive. The positivity rate of the unpooled samples was 1.9% (24/1280). After parallel testing using the individual and pooled testing strategies, positive agreement was 100% and negative agreement was 99.8%. The overall median cycle threshold (Ct) value of the unpooled samples was 29.8 (interquartile range 22.3–34.3). Pools that remained positive when compared with the results of individual samples had lower median Ct values compared with those that turned out to be negative (28.8 versus 34.8; P=0.0.035). Pooled testing reduced the cost >4-fold. Pooled testing may be a more cost-effective approach to diagnose SARS-CoV-2 in resource-limited settings without compromising diagnostic performance.
Keywords: COVID-19, Pooled testing, RT-PCR, SARS-CoV-2, LMIC
Introduction
Coronavirus disease 2019 (COVID-19) continues to be an important global health problem with a significant impact on individual and global public health (Fauci et al., 2020). In the face of a rapidly spreading disease with a shortage of vaccines and/or effective treatment, rapid mass testing has been suggested as one of the measures to map, contain and mitigate the COVID-19 pandemic (Denny et al., 2020).
In Uganda, COVID-19 testing is largely performed using real-time polymerase chain reaction (RT-PCR) for detection of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in nasopharyngeal swabs. However, the current diagnostic platform falls short of the clinical needs as: (a) the population that needs testing far exceeds the available capacity due to the limited number of available RT-PCR testing centres in Uganda; (b) the positivity rate of SARS-CoV-2 in Uganda fluctuated between 1% and 5% in the first wave and between 12% and 20% in the second wave (Woldometer, 2021), so individual testing is not cost-effective as the majority of samples are negative; and (c) RT-PCR testing kits are expensive and there is a limited supply due to a high global demand.
Pooled sample testing for SARS-CoV-2 has been shown to be feasible to detect incident cases of COVID-19 and determine transmission dynamics in communities and schools (Joachim et al., 2021). The strategy has been found to be cost-effective compared with individual testing (Abdalhamid et al., 2020). The present study assessed the performance of a pooled testing strategy for RT-PCR, and determined its effect on diagnostic accuracy while mitigating the cost of testing for COVID-19 compared with individual testing in Uganda.
Methods
Study design and setting
This study conducted cross-sectional, laboratory-based verification of the performance of pooled sample testing using leftover nasopharyngeal samples obtained for routine SARS-CoV-2 testing between 19 April and 6 May 2021, and biobanked at the Genomics and Molecular Biology Laboratory of the College of Health Sciences at Makerere University in Kampala, Uganda.
Study procedure
Leftover samples were selected at random from the pool of samples at the Genomics and Molecular Biology Laboratory using the Random Number Generator function in Microsoft Excel (Microsoft Corp., Redmond, WA, USA). A 100-µL aliquot was obtained from each leftover sample and mixed in a 1.5-mL microcentrifuge in batches of 10 samples to form the different pools, as described previously (Deka and Kalita, 2020; Barak et al., 2021). After mixing to obtain a homogenous mixture, 100 µL of the pooled samples was transferred to a clean 1.5-mL microcentrifuge tube, and RNA was extracted using a viral RNA mini kit (Qiagen, Hilden, Germany). RT-PCR was performed using an Applied Biosystems PCR platform.
If a pool tested positive on RT-PCR analysis, the samples were deconvoluted and the process was repeated on the individual samples that comprised that pool to identify the positive sample. If a pool tested negative on RT-PCR analysis, all samples in that pool were declared negative and underwent no further testing. The results were compared in parallel with real-time, routine RT-PCR testing.
Data analysis and cost-effectiveness analysis
The results were compared using a two-by-two table, and positive agreement (percentage of cases that matched when the reference method was positive), negative agreement (percentage of cases that matched when the reference method was negative) and Cohen's Kappa statistic were calculated. The average cost-effectiveness ratio and incremental cost-effective ratio taking a provider's perspective and ingredient costing approach were calculated using the current market cost of 56 US$ for each RT-PCR test in Uganda. Data analysis was performed using Microsoft Excel and GraphPad Prism 8.4.
Results
In total, 1280 samples were tested using both individual and pooled sample testing methods. The positivity rate of the unpooled samples was 1.9% (24/1280). In total, 128 pools of 10 samples each were made, out of which 16 pools were positive. These were deconvoluted and the samples were run individually to identify those that were truly positive. Deconvolution identified 21 samples that were positive out of the 24 obtained using individual testing. Three of the pools which tested negative contained one positive sample each, while three of the 16 pools that tested positive contained more than one positive sample. Positive and negative agreements between pooled and individual testing were 100% and 99.8%, respectively (Table 1 ). The probability of agreement was 99.8% and Cohen's kappa statistic was 0.932. The overall median cycle threshold (Ct) value of the unpooled samples was 29.8 (interquartile range 22.3–34.3). Pools that remained positive when compared with the results of individual samples had lower median Ct values compared with those that turned out to be negative (28.8 versus 34.8; P=0.0.035).
Table 1.
Pooled testing results compared with individual real-time polymerase chain reaction (RT-PCR) reference assays
| Individual RT-PCR testing |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Pooled testing | Positive | 21 | 0 | 21 |
| Negative | 3 | 1256 | 1259 | |
| Total | 24 | 1256 | 1280 | |
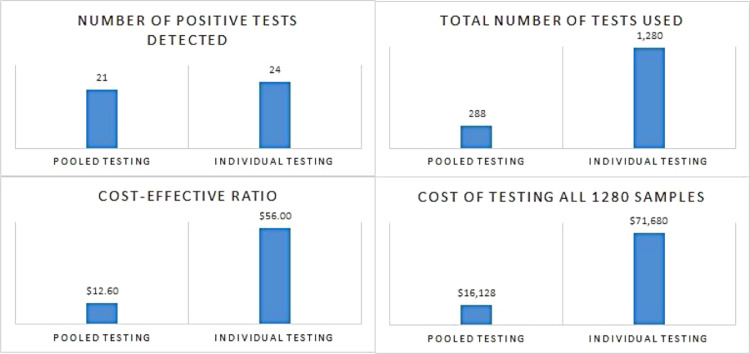
The average cost-effectiveness ratio was four-fold lower using pooled sample testing compared with individual sample testing (12.6 US$ per test versus 56 US$ per test, respectively) (Figure 1 ). The total cost for pooled testing was 77.5% less than individual testing (16,128 US$ versus 71,680 US$, respectively). The incremental cost-effectiveness ratio was 55,552 US$ for further identification of three positive tests using the individual testing method.
Figure 1.
Comparison of the number of positive tests detected, total cost of testing 1280 samples, cost-effectiveness ratio and number of tests performed between pooled testing and individual testing.
Discussion
This study to assess the diagnostic performance and cost-effectiveness of a pooled testing strategy for RT-PCR had two main findings. Firstly, pooled testing using a pool size of 10 had almost-perfect agreement with individual testing. Secondly, the pooled testing strategy reduced the cost of the test >4-fold and reduced the cost of testing by 77% while maintaining a high level of agreement with individual testing. The findings are similar to those in other studies which showed that pooled testing increases cost-effectiveness without much influence on the accuracy of PCR testing (Borillo et al., 2020; Alcoba-Florez et al., 2021; Barak et al., 2021; Deckert et al., 2021; Kagan et al., 2021; Lim et al., 2020). Previous studies have indicated that a pool size ≤10 is required to maintain the sensitivity and cost-effectiveness of pooled testing (Barak et al., 2021).
As the vast majority of people with COVID-19 remain asymptomatic (Dhama et al., 2020), large-scale screening using a pooled sample strategy may be employed in settings such as schools, the community and other high-volume test settings for rapid identification of cases and limitation of SARS-CoV-2 transmission (Joachim et al., 2021). Moreover, batched testing of pooled specimens to detect SARS-CoV-2 has been shown to be an effective strategy to conserve resources and to substantially increase testing capacity, especially in resource-limited settings (Abdalhamid et al., 2020). The present study found that the pooled strategy reduced the costs significantly. This finding is consistent with previous studies which showed that combining samples and testing them in groups significantly reduces the number of tests required, substantially lowering costs without reducing the diagnostic performance of this highly sensitive RT-PCR assay (Singh et al., 2021).
The results of this study have far-reaching implications for health ministries in resource-limited settings which have to plan to increase testing services in their countries at the same time as providing adequate medical services to individuals with severe COVID-19. A sensitive testing strategy that cuts costs would be very helpful in resource-limited settings, as the money saved could be used to purchase other medical supplies while increasing the number of tests performed. Larger population-level trials utilizing pooled testing are underway in Rwanda and South Africa (Mutesa et al., 2021). Importantly, pooled testing may be influenced by the population positivity rate for SARS-CoV-2. Previous studies have shown that pooled testing is appropriate in settings with positivity rates <10% (Deka and Kalita, 2020).
This study has some limitations. It was conducted as a single laboratory experiment, and only one pool size (10 samples) was tested. More studies are needed to optimize sample pooling strategies and define clearly the clinical significance of missing a few positive samples on the spread of COVID-19. The three positive samples that were falsely detected as negative using pooled testing in this study highlight the pitfalls of pooled testing, and caution should be taken, especially when adopting pooled testing as a routine method of testing. Other algorithms to increase the sensitivity of pooled testing, such as testing close contacts together, reducing the pool size and testing multiple times, may be needed. These should be studied in future research.
Conclusion
This study found that a pooled testing strategy for RT-PCR is a feasible and cost-effective option compared with individual testing in Uganda. These results are crucial in planning for strategies to be used in mass testing campaigns in the face of the current high cost of RT-PCR testing of individual samples.
Acknowledgements
The authors wish to thank the staff at the Genomics and Molecular Biology Laboratory, Department of Immunology and Molecular Biology, College of Health Sciences, Makerere University. The laboratory analyses, sample retrieval and pooling were carried out at these facilities.
Conflict of interest statement
None declared.
Funding
Financial support was provided by the Government of Uganda through the Makerere University Research and Innovation Fund for COVID-19 research. The funders had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Ethical approval
This study was approved by Makerere University School of Medicine Research and Ethics Committee (Ref. No. 2020-175), which also provided a waiver of consent. All samples were anonymized.
References
- Abdalhamid B, Bilder CR, Garrett JL, Iwen PC. Cost effectiveness of sample pooling to test for SARS-CoV-2. J Infect Dev Ctries. 2020;14:1136–1137. doi: 10.3855/jidc.13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcoba-Florez J, Gil-Campesino H, García-Martínez de Artola D, Díez-Gil O, Valenzuela-Fernández A, González-Montelongo R, et al. Increasing SARS-CoV-2 RT-qPCR testing capacity by sample pooling. Int J Infect Dis. 2021;103:19–22. doi: 10.1016/j.ijid.2020.11.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak N, Ben Ami R, Sido T, Perri A, Shtoyer A, Rivkin M, et al. Lessons from applied large-scale pooling of 133,816 SARS-CoV-2 RT-PCR tests. Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abf2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borillo GA, Kagan RM, Baumann RE, Fainstein BM, Umaru L, Li HR, et al. Pooling of upper respiratory specimens using a SARS-CoV-2 real-time RT-PCR assay authorized for emergency use in low-prevalence populations for high-throughput testing. Open Forum Infect Dis. 2020;7 doi: 10.1093/ofid/ofaa466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert A, Anders S, de Allegri M, Nguyen HT, Souares A, McMahon S, et al. Effectiveness and cost-effectiveness of four different strategies for SARS-CoV-2 surveillance in the general population (CoV-Surv Study): a structured summary of a study protocol for a cluster-randomised, two-factorial controlled trial. Trials. 2021;22:39. doi: 10.1186/s13063-020-04982-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deka S, Kalita D. Effectiveness of sample pooling strategies for SARS-CoV-2 mass screening by RT-PCR: a scoping review. J Lab Physicians. 2020;12:212–218. doi: 10.1055/s-0040-1721159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny TN, Andrews L, Bonsignori M, Cavanaugh K, Datto MB, Deckard A, et al. Implementation of a pooled surveillance testing program for asymptomatic SARS-CoV-2 infections on a college campus — Duke University, Durham, North Carolina, August 2–October 11, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1743–1747. doi: 10.15585/mmwr.mm6946e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama K, Khan S, Tiwari R, Sircar S, Bhat S, Malik YS, et al. Coronavirus disease 2019–COVID-19. Clin Microbiol Rev. 2020;33 doi: 10.1128/CMR.00028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci AS, Lane HC, Redfield RR. Covid-19 – navigating the uncharted. N Engl J Med. 2020 doi: 10.1056/nejme2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachim A, Dewald F, Suárez I, Zemlin M, Lang I, Stutz R, et al. Pooled RT-qPCR testing for SARS-CoV-2 surveillance in schools – a cluster randomised trial. EClinicalMedicine. 2021;39 doi: 10.1016/j.eclinm.2021.101082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan RM, Rogers AA, Borillo GA, Clarke NJ, Marlowe EM. Performance of unobserved self-collected nasal swabs for detection of SARS-CoV-2 by RT-PCR utilizing a remote specimen collection strategy. Open Forum Infect Dis. 2021;8 doi: 10.1093/ofid/ofab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KL, Johari NA, Wong ST, Khaw LT, Tan BK, Chan KK, et al. A novel strategy for community screening of SARS-CoV-2 (COVID-19): sample pooling method. PLoS One. 2020;15 doi: 10.1371/journal.pone.0238417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutesa L, Ndishimye P, Butera Y, Souopgui J, Uwineza A, Rutayisire R, et al. A pooled testing strategy for identifying SARS-CoV-2 at low prevalence. Nature. 2021;589:276–280. doi: 10.1038/s41586-020-2885-5. [DOI] [PubMed] [Google Scholar]
- Singh L, Anyaneji UJ, Ndifon W, Turok N, Mattison SA, Lessells R, et al. Implementation of an efficient SARS-CoV-2 specimen pooling strategy for high throughput diagnostic testing. Sci Rep. 2021;11:17793. doi: 10.1038/s41598-021-96934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldometer. COVID-19 coronavirus pandemic. 2021. https://www.worldometers.info/coronavirus/.