Abstract
The Nsp9 replicase is a conserved coronaviral protein that acts as an essential accessory component of the multi-subunit viral replication/transcription complex. Nsp9 is the predominant substrate for the essential nucleotidylation activity of Nsp12. Compounds specifically interfering with this viral activity would facilitate its study. Using a native mass-spectrometry-based approach to screen a natural product library for Nsp9 binders, we identified an ent-kaurane natural product, oridonin, capable of binding to purified SARS-CoV-2 Nsp9 with micromolar affinities. By determining the crystal structure of the Nsp9-oridonin complex, we showed that oridonin binds through a conserved site near Nsp9’s C-terminal GxxxG-helix. In enzymatic assays, oridonin’s binding to Nsp9 reduces its potential to act as substrate for Nsp12’s Nidovirus RdRp-Associated Nucleotidyl transferase (NiRAN) domain. We also showed using in vitro cellular assays oridonin, while cytotoxic at higher doses has broad antiviral activity, reducing viral titer following infection with either SARS-CoV-2 or, to a lesser extent, MERS-CoV. Accordingly, these preliminary findings suggest that the oridonin molecular scaffold may have the potential to be developed into an antiviral compound to inhibit the function of Nsp9 during coronaviral replication.
Keywords: SARS-CoV-2, Nsp9, kaurane, oridonin, replication transcription complex
Abbreviations: ESI-FT-ICR-MS, electrospray ionization–Fourier transform–ion cyclotron resonance–mass spectrometry; DMSO, dimethylsulfoxide; NiRAN, nidovirus RdRp-associated nucleotidyl transferase; Nsp, nonstructural protein; RdRp, RNA-dependent RNA polymerase; RTC, replication transcription complex; TEV, tobacco etch virus
The global spread of SARS-CoV-2 throughout the human population has inundated healthcare systems worldwide (1, 2). The newly developed vaccines now being distributed are expected to drastically reduce morbidity and mortality (3, 4, 5). However, interest remains in identifying new antiviral compounds targeting coronaviral proteins. The utility in such antivirals would reside in their ability to act as a bulwark against new viral strains arising with reduced sensitivity to deployed spike-focused vaccines and in their likely activity against any new zoonotic CoV that might emerge in the future. Due to its obvious successes (6) the current global stratagem for combating COVID19 focuses intensely upon inducing immunity to the spike glycoprotein responsible for binding host-cell receptors. One potential drawback to this approach is it requires continued surveillance of mutations within this specific viral protein (7, 8) and rapid adaption in the event of large-scale changes.
As a consequence of the SARS and MERS epidemics, a number of alternate therapeutic targets within coronaviruses came under investigation. The lead candidates were mainly enzymatic nonstructural proteins (Nsp) that facilitate replication inside host cells, with much interest focusing upon Nsp12’s RNA-dependent RNA polymerase (RdRp). Nsp12, along with Nsp7 and Nsp8, is central component of the viral replication transcription complex (RTC) targeted by remdesivir (9) and is responsible for duplicating genomic mRNA alongside production of subgenomic transcripts. Other therapeutic targets considered include the catalytic site of Nsp5 (10, 11) a glutamate-specific protease and the Nsp3 deubiquitinase (12, 13, 14, 15, 16). These three viral proteins all have well-defined enzymatic activities with active sites akin to molecular targets already under investigation in mature drug discovery pipelines.
Other coronaviral Nsps are highly conserved and known to be essential for viral replication yet their physiological roles were unclear from sequence homology resulting in their therapeutic potential being unclear. One such protein is the replicase Nsp9, a small 12 kDa component of the PP1ab polyprotein directly transcribed from viral genomic mRNA. Nsp9 is well conserved among coronaviruses and adopts a unique architecture that had previously been ascribed as having affinity for single-stranded oligonucleotides (17, 18, 19, 20). The Nsp9 homologue from SARS-CoV (Nsp9SARS) shares 98% sequence identity with that of Nsp9COV19 (21) and is essential for viral replication (22). While its precise function remains unclear (23, 24), regions of the Nsp9 protein necessary for replication have been identified. Namely, the loss of residues within the GxxxG protein–protein interaction motif reduces viral titers in SARS-CoV (22) and SARS-CoV-2 (24) infection assays.
A homodimeric form of Nsp9 predominates in solution in vitro with self-association occurring via the conserved GxxxG motif (17, 22). But recent structural insights into the extended SARS-CoV-2 replication transcription complex also identified monomeric Nsp9 utilizing the same interface as a means to associate with the N-terminal NiRAN (Nidovirus RdRp-associated nucleotidyl transferase) domain of Nsp12 (23). When bound in this manner, the N-terminus of Nsp9 is able to enter the NiRAN enzymatic site, abutting the phosphates of a bound nucleotide within. The nucleotidylation activity of RdRp NiRAN domains is essential for coronaviral replication (22) but the physiological reason for this remains under study. NiRAN pseudokinases catalyze a novel NMPylation reaction, transferring an entire nucleoside monophosphate group onto a protein (and/or RNA) substrate (25). The SARS-CoV-2 NiRAN domain retains an intact kinase N-lobe structure including catalytic DFG-motif, whereas only some components of the C-lobe are retained (23).
The Nsp9:Nsp12 interaction was first described as an inhibitory mechanism regulating mRNA-cap formation (23). But nucleotidylation assays also suggest that the α-amide of Nsp9’s Asn-1 residue is a major substrate for the reaction (24). Substitution of the first two residues of Nsp9 with alanine curtailed NiRAN activity and also viral replication (24). Together these data imply that posttranscriptional nucleotide modification of viral RTC components may be required for their proper function (24). The role of Nsp9 and its NiRAN domain interaction within the RTC is essential for viral replication (24) potentially recruiting the Nsp10/Nsp14 co-complex to facilitate genome 5′-capping and/or proofreading (26). We thus sought to identify Nsp9-binding small molecules targeting essential features of this protein.
Here we describe a subset of ent-Kaurane compounds capable of binding to recombinant SARS-CoV-2 Nsp9 in vitro. Ent-kauranes are natural phytochemical diterpenoids consisting of a perhydrophenantrene ring system fused to cyclopentane, a chemical skeleton found within a number of plants from the Lamiaceae family (27). Indeed, several hundred ent-kauranes have been catalogued within the Isodon Genus alone, and their biological activities have been particularly well studied due to their use in Chinese medicine (28). The polycyclic bridged ring molecule identified, oridonin, displayed micromolar affinity for Nsp9COV19, with its binding site characterized alongside the fold’s GxxxG α-helix. Binding is mediated via contact with three of the four hydroxyl groups of the compound; the compound’s lone enone group also forms an adduct with Cys-73, a residue conserved throughout α/β-coronaviridae. This binding site partially overlaps the region of Nsp9 responsible for engaging Nsp12 and within in vitro infection assays, oridonin reduced viral titer following infection with SARS-CoV-2. The ent-kaurane binding-site of Nsp9 is conserved among diverse coronaviruses, and accordingly we also observed reduced viral titers upon infection with MERS-CoV. The ent-kaurane polycyclic ring-structure presented herein may thus represent a framework for the further development of more potent, and less cytotoxic small molecules with the potential to specifically target the function of coronaviral Nsp9 replicases.
Results
Identification of Nsp9-binding compounds by native mass spectrometry
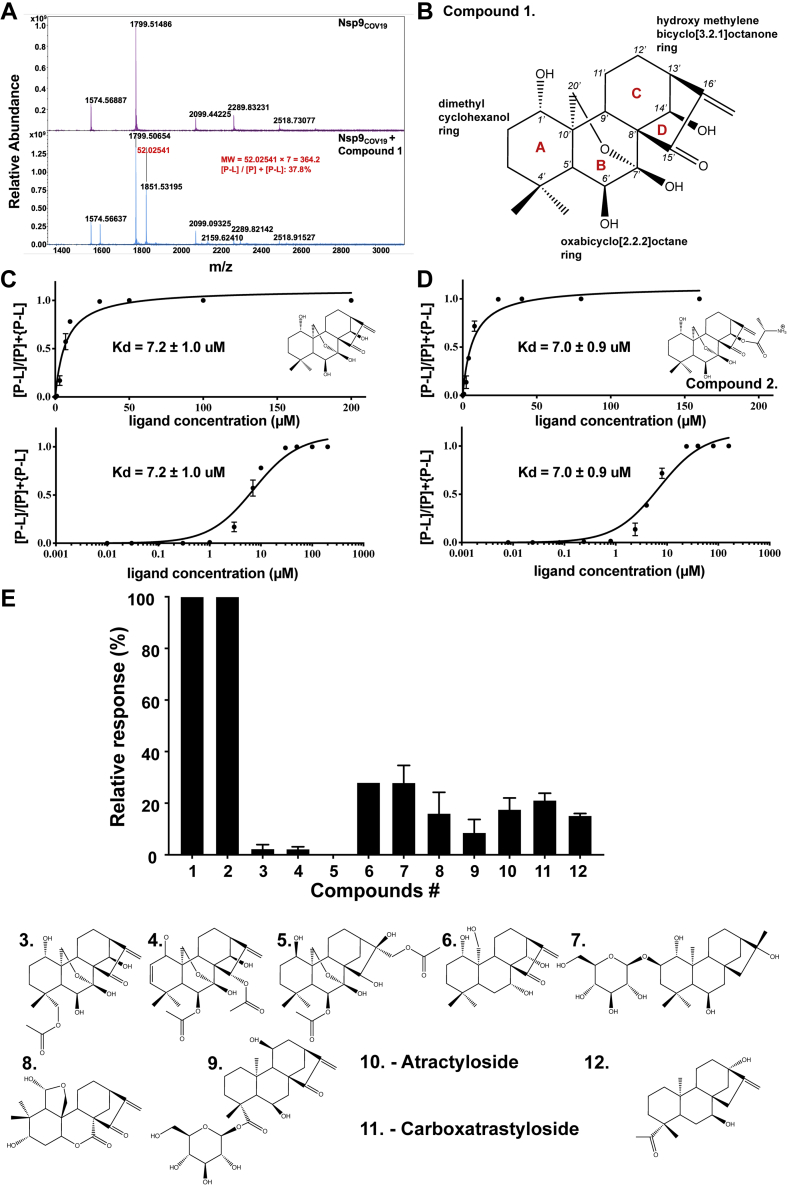
To identify small molecules that bind to Nsp9COV19, recombinant protein was produced as described (21) and screened by native mass spectrometry against an in-house natural product library of 1614 molecules with drug-like physicochemical properties. Native MS screening using high-resolution electrospray ionization–Fourier transform–ion cyclotron resonance–mass spectrometry (ESI-FT-ICR-MS) is a fast, label-free, accurate method that allows direct detection of covalent and noncovalent protein−ligand complexes (29). Nsp9COV19 recombinant protein was subjected to mass spectrometry under optimized conditions, yielding protein signals at low charged states between +5 and +8 with high protein signal (109 counts) (Fig. 1A, top panel). Nsp9COV19 was subsequently screened with the natural product library that identified the strongest binder as compound 1 (Fig. 1A, bottom panel). The difference between the mass-to-charge ratio (Δm/z) for the unbound protein and the protein–ligand complex ions was multiplied by the charge state (z) to directly afford the molecular weight of the bound ligand, 364.2 Da, which is consistent with compound 1. This compound, oridonin, is an ent-kaurane with two bicyclic rings: a central di-hydroxy 2-oxabicyclo[2.2.2]octane ring (B in Fig. 1B) ortho-fused on one side to hydroxy methylene bicyclo[3.2.1]octanone ring (C/D in Fig. 1B) and on its other side to 4,4-dimethyl cyclohexanol. The identified compound has previously been shown to inhibit SARS-CoV-2 replication in in vitro cellular assays (30) but its mechanism of action was unknown. Oridonin is also known to act on NLRP3 inducing an anti-inflammasome response (31). Related ent-kauranes display antiviral activity in vitro against HIV (32, 33, 34). Accordingly, a native mass spectrometry screen identified oridonin that bound to Nsp9COV19.
Figure 1.
Native mass spectrometry of SARS-CoV-2 Nsp9.A, top, FTMS confirmation of the recombinant Nsp9 with 19 μl Nsp9 incubated with 1 μl methanol; bottom, 19 μl Nsp9 incubated with 1 μl Compound 1. B, chemical structure of the identified ent-Kaurane compound. The ring systems are labeled with red lettering and the carbon atoms numbered. C, dose-responsive binding of Compound 1 to Nsp9 calculated at 11 concentrations from the 7+ m/z peak and (D) an equivalent dose–response curve for the modified compound 2. Errors represent the S.D. of three independent recordings. E, relative Nsp9COV19 binding ratios of similar kaurene-type ligands for which binding ratios were calculated following incubation with 50 μM of each compound.
Affinity measurement
To determine the affinity between Nsp9COV19 and oridonin (compound 1), 12 concentrations ranging from 0.02 μM to 1 mM were incubated with the protein before repeating the mass spectrometry analysis. The percentage of ligand-bound complex within the sample was derived from the ratio of unliganded to liganded +6, +7, +8 m/z peaks (materials and methods), which were used to plot a dissociation curve from which a proxy Kd could be obtained of ∼7.2 ± 1.0 μM (Fig. 1C). Oridonin is thus able to bind to Nsp9COV19 with micromolar affinity.
Ent-kaurane compounds
Oridonin has a number of functional groups that could interact with residues within Nsp9COV19, including 4-hydroxyl moieties, a dimethyl group, and an enone within the bridging D-ring (Fig. 1B). The angular fusion of the three major nonplanar ring systems presents these functional groups on what could be described as a spherical wedge-shaped molecule with concave and convex faces, with most functional groups occurring on the latter. To determine which groups contribute to the compound’s affinity for Nsp9COV19 we repeated the mass-spectrometry-based binding assays after cocomplexation with a series of related ent-kauranes. Several such compounds had relatively minor chemical differences and yielded comparable affinities: Compound 2 had a measured Kd of ∼7.0 ± 0.9 μM and carries only an L-alanine ester modification to the 14′-hydroxyl of compound 1 (Fig. 1D). Compounds 3 to 5 include bulky acetyl additions to convex facing functional groups, and all display markedly reduced affinities (Fig. 1E). However, if a convex-facing functional group is merely lacking, such as the 6′-hydroxyl within compound 6 (Kd of ∼574.0 ± 87.4 μM), some ability to cocomplex with Nsp9COV19 is retained (Fig. 1E). Larger-scale modifications to the chemical scaffold occur within compounds 7 to 12, and these mostly reduce or ablate each molecule’s ability to cocomplex with Nsp9COV19 (Fig. 1E). It is noteworthy that compound 8 carries a bulky addition on the concave surface of the kaurene scaffold yet still retains some Nsp9COV19-binding capability. Compounds 1 and 2 retained micromolar affinities for Nsp9, but more drastic chemical modifications to the scaffold had reduced binding.
Oridonin binds to Nsp9 at the base of the GxxxG helix
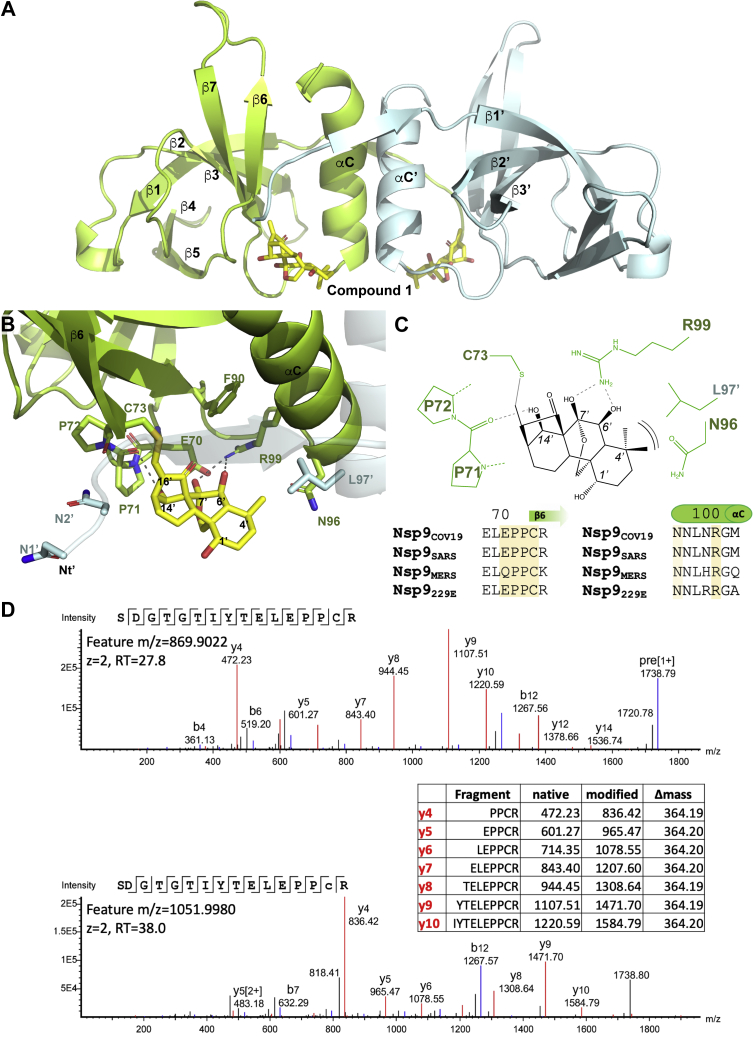
To characterize the binding mode of oridonin with Nsp9COV19, we crystallized the cocomplex and determined its structure (35) (see Experimental procedures and Table S1 for data collection and refinement statistics). The crystal’s asymmetric unit contained eight molecules of Nsp9COV19 within four homodimers; these were seen to be largely identical except at their extreme C-termini. After initial rounds of refinement, a large section of unmodeled density was observed between the base of GxxxG helix and strand β6 within an identical position of each protomer (Fig. S1). Oridonin could be modeled within this density and refined to yield a good fit (Fig. 2A). The geometry of the final refined oridonin molecule was constrained to that determined to high resolution by small-molecule crystallography where appropriate (KUYHIV, 1201993).
Figure 2.
Cocomplex structure of Nsp9COV19with oridonin.A, an overall view using cartoon representation for the 2.95 Å crystal structure of Nsp9COV19 with oridonin. The two bound compounds are displayed in stick representation and nearby secondary structural elements are labeled. B, a zoomed in view of the compound-binding site in which the side chains of nearby residues labeled and the potential hydrogen bonds made with the protein highlighted. C, conservation of coronaviral Nsp9 residues interacting with oridonin. In the schematic the ligand is in black and protein side-chain residues in green. Potential hydrogen bonds are indicated with dashed lines and the A-ring van der Waals interaction with brackets. D, example spectrum of native tryptic peptide SDGTGTIYTELEPPCR (top) and its modified counterpart containing the oridonin adduct (bottom). The modified peptide is characterized by a mass shift of +364.2 Da as well as prominent y ions differing by m/z +364.2 from their counterparts in the native peptide (inset table). Annotated spectra were extracted from PEAKS X Pro, lower case c denotes the Cys residue modified with oridonin. RT denotes retention time.
Binding to Nsp9COV19 occurs with the compound’s convex face nestled between the αC-helix and β6-strand (Fig. 2B). Arg-99 from within the helix forms two hydrogen bonds to the central ring’s 6′ and 7′-hydroxyls. A similar interaction occurs between the backbone carbonyl group of a conserved dipropyl motif immediately preceding β6 and the lone hydroxyl group from the bicyclo[3.2.1]octanone ring; this interaction positions the bridging enone group near Cys-73, the first residue of the strand. All protein residues interacting with the ligand appear highly conserved throughout different viral Nsp9 homologues (Fig. 2C). The unbridged dimethyl hydroxy hexanol ring of compound 1 is less involved in binding to Nsp9COV19 and makes fewer obvious contacts with the protein, accordingly this section of the ligand also has less well-defined electron density in the structure. Oridonin’s lone concave facing group is within this ring (the 1′-hydroxyl) and is directed toward solvent making no contact with the protein; in contrast, the dimethyl moiety of this ring is sufficiently proximal to form van der Waals interactions with Asn-96 at the base of the αC-helix and potentially Leu-97 from the homodimer subunit (Fig. 2A). The binding site of oridonin was identified at the base of the fold’s lone α-helix.
Enone adduct formation with Nsp9
During refinement the enone group of oridonin closely approached the sulfhydryl group of Cys-73 potentially indicating a covalent adduct was being formed with the residue. The crystals take 2 to 3 days to grow so we sought to assess whether such an adduct might form with Nsp9COV19 in solution in shorter timeframes (10 min incubation with a twofold molar excess). We thus treated recombinant Nsp9COV19 with oridonin before gel filtration to remove soluble drug and then digested the eluted protein with trypsin without reduction or alkylation of Cys residues. There are three Cys residues within the sequence of Nsp9. Evidence for modification of all Cys residues was observed although only Cys-73 was modified within the crystal structure and in the folded state is the only one expected to be solvent exposed (Fig. 2D). Modified peptides were characterized by a +364.2 Da mass shift compared with the native peptide, as well as low mass ions consistent with fragmentation of oridonin (Fig. S2). This is consistent with the D-ring enone group acting as a Michael acceptor to form a protein thioether. A similar adduct has been reported for oridonin and NLRP3 (31) indicating that oridonin can selectively form thioether adducts with cysteine residues of some proteins.
In vitro coronaviral replication assays
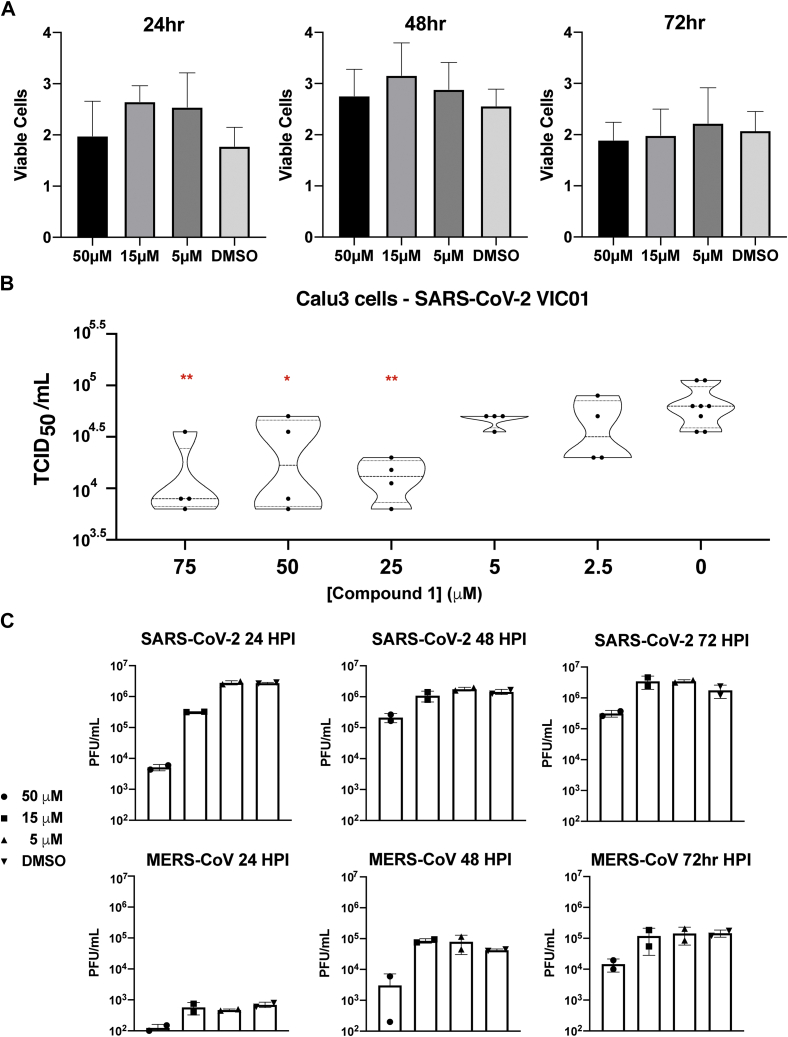
We then assessed whether coronaviral replication was disrupted through the action of oridonin on binding Nsp9COV19. To minimize any influence of compound-induced cellular toxicity upon such measurements, we treated different cell types with varying micromolar concentrations of oridonin and performed an MTT cell proliferation assay. The relative cell survival of Vero 81, Calu-3, and Vero e6 cells was assessed and compared with controls (Fig. S3). Calu-3 cells were seen to be highly tolerant with no effect on cell proliferation detected in this concentration range, an effect maintained up to 72 h post infection (Fig. 3A); in contrast, a degree of reduced proliferation was observed in other cell types at the highest 50 μM concentration point. In light of these results, viral replication assays were performed by taking confluent Calu-3 monolayers preincubated with different concentrations of oridonin then infected with the original cultured patient SARS-CoV-2 isolate: Australia/VIC01/2020 (36). After 24-h, a TCID50 assay was performed to assess the amount of virus present within the supernatant and a dose–response curve plotted (Fig. 3B).
Figure 3.
Oridonin inhibits coronaviral replication.A, MMT proliferation assay of different cell types treated with three different concentrations of compound 1. Error bars represent S.D. of three independent replicates. B, TCID50 assay of Calu-3 cells pretreated with compound 1 prior to infection with SARS-CoV-2. Viral titer was assessed for 14 concentration points after 24 h of growth and compared with untreated controls. Statistical significance is indicated where p-values are less than 0.05 (∗), 0.01 (∗∗), or 0.001 (∗∗∗). C, viral growth kinetics of SARS-CoV-2 and MERS-CoV infection of Calu-3 cells pre- and posttreated with compound 1. Representative of two independent experiments.
The sequence of Nsp9COV19 is relatively conserved between SARS-CoV and SARS-CoV-2, so we sought to assess whether oridonin inhibited replication of more distantly related coronaviruses. Coronaviral replication assays were repeated using Calu-3 cells in which viral titer was monitored at 24, 48, and 72 h post infection. Cells were treated with 5, 15, or 50 μM concentrations of oridonin then infected with either SARS-CoV-2 or MERS-CoV. In total, 50 μM of oridonin was seen to reduce viral tite at all time points following SARS-CoV-2 infection (Fig. 3C); at the 15 μM concentration, viral titer was still approximately fivefold reduced after 24 h, but this was not maintained at later time points (Fig. 3C). Oridonin inhibited MERS-CoV replication by 0.5 to 1 log at 50 μM concentration (Fig. 3C). The reduced sensitivity to oridonin displayed by MERS-CoV may be attributed to genetic dissimilarity as Nsp9SARS and Nsp9MERS share only 52% sequence identity, although the majority of compound-interacting residues remain conserved (Fig. 2C). These data indicate that oridonin reduced both SARS-CoV-2 and MERS-CoV replication in vitro, thereby demonstrating conserved inhibition of these viruses and suggest that specific inhibitors toward conserved Nsps, such as Nsp9, can potentially be effective pan-coronavirus inhibitors.
UMPylation of Nsp9COV19
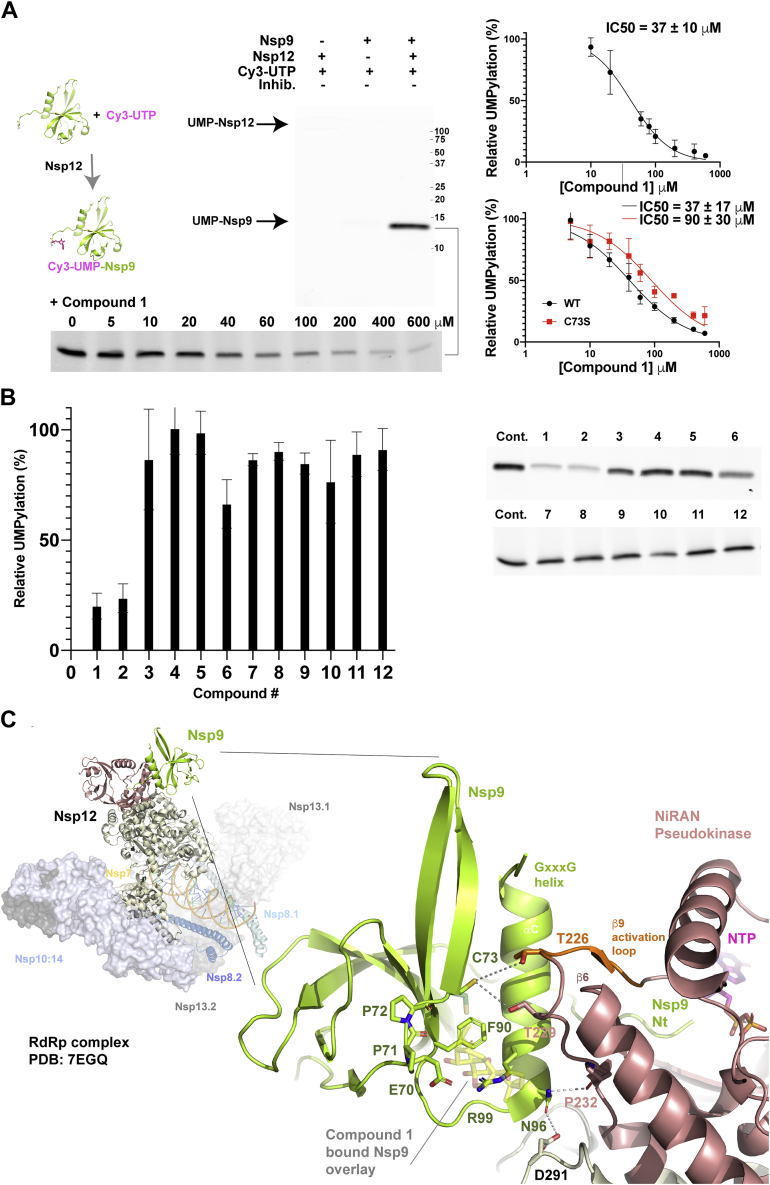
Nsp9 is the major substrate of the essential, and virally unique, coronaviral nucleotidylation reaction catalyzed by the NiRAN domain of Nsp12, with uridine triphosphate (UTP), the preferred substrate for transfer (24). Following the nucleotidylation reaction, uridine monophosphate (UMP) is transferred from UTP to a substrate protein’s N-terminal residue, the α-amide acting as acceptor in the case of Nsp9 (24). To assess whether binding of ent-kaurane compounds to Nsp9 reduced its potential to act as substrate, we established an assay using Cy3-UTP, which was incubated with 0.5 μM Nsp12COV19 and 1.5 μM Nsp9COV19. Reaction products were subsequently separated on standard SDS-PAGE gels and imaged, all reactions were performed in triplicate. In this assay, if UTP is provided in excess, we observed some UMP transfer to Nsp12 (faint band left lane of Fig. 4A) as reported by others (24). However, the N-terminus of Nsp9 is clearly the preferred protein substrate (Fig. 4A). If the reaction is repeated in the presence of a concentration series from 0 to 600 μM of oridonin, we observed clear inhibition of the UMPylation reaction with IC50 measured at 37 ± 10 μM (bottom Fig. 4A). As the highest concentration points essentially eliminated UMPylation, we repeated the reaction in the presence of 500 μM of each compound (1–12) and compared the amount of modified Nsp9 with control reactions (Fig. 4B). In this reaction the ent-kaurene compound 1 reduced Nsp9COV19 Cy3-modification to 20 ± 6% and compound 2 to 24 ± 7% of the signal of dimethylsulfoxide (DMSO)-only control reactions (Fig. 4B) while the other compounds had more nuanced effect, with compound 6 being the next most dramatic reduction of UMPylation to ∼66% of control reactions (Fig. 4B). Finally, to assess whether Cys-73 is required for the inhibitory action of compound 1 within the UMPylation reaction, we repeated the inhibitory concentration-series with wild-type Nsp9COV19 and a Cys-73 to Ser mutant; the level of inhibition was approximately threefold lower when the mutant Nsp9 was the substrate (Fig. 4A). We have not yet been able to produce an R99A mutant, to assess the involvement of this additional key contact residue demonstrated within our crystal structure. Compound binding thus appears to reduce the potential for Nsp9COV19 to act as NiRAN substrate, accordingly if we overlay oridonin-bound Nsp9 upon its Nsp-12 associated form (23) steric clashes would appear to perturb the binding mode (Fig. 4C).
Figure 4.
Oridonin binding mapped onto the SARS-CoV-2 extended replication transcription complex.A, UMPylation assay of Nsp9 by the NiRAN domain of Nsp12. Cy3-labeled UTP was incubated with Nsp9 and Nsp12 with the reaction products, then separated by SDS-PAGE, and imaged. IC50 calculations were obtained from a control-normalized dose–response fit to the data. Errors represent the S.D. of triplicate reaction series. B, the UMPylation reaction was repeated in triplicate, the presence of each of the 12 compounds described previously and compared with in-gel control reactions. Quantification of the Cy3-modified Nsp9 is plotted with errors representing standard deviation of three independent reactions. Note: the upper gel in panel A and cropped compound screen 1 to 6 of panel B are part of the same experiment. C, a cartoon representation of the extended RTC (26) is shown with the Nsp9: NiRAN-domain interaction highlighted. Nsp12 is colored with salmon for the pseudokinase domain and sand for the RdRp. The pseudokinase activation loop is in orange. The engaged Nsp9 is colored green with N-terminal residues labeled. The oridonin-binding site is overlaid in partial transparency onto the RNA complex indicating potential clashes with residues immediately C-terminal to the activation loop and near the Asn-96 engagement point. Potential Nsp9:Nsp12 interface interactions are highlighted with gray lines, the local resolution of cryo-EM map (26) identifies their proximity but precludes specific categorization into, e.g., van der Waals contacts or hydrogen bonds.
Discussion
The coronaviral Nsp9 protein is essential for viral reproduction (22, 24) but its function within the replication/translation complex is unclear (23, 24) as are any secondary roles manipulating host cell metabolism (19). We have identified a subset of ent-kaurane compounds with low micromolar affinity for Nsp9COV19 that bind between the αC-helix and β6-strand, a region of the protein mediating homodimer formation (37) and also Nsp12 NiRAN-domain association (23). While cytotoxic at high concentrations, the presence of these compounds inhibits SARS-CoV-2 and MERS-CoV replication in cell culture. Within the kaurane scaffold, the hydroxy methylene bicyclo[3.2.1]octanone ring is a major point of contact between compound 1 (oridonin) and Nsp9COV19; this ring system facilitates binding between the conserved residues 70EPPCR74 and the lone helix within Nsp9COV19. Three hydroxyl moieties engage the protein orienting the bound ligand within the site such that the bridging enone group is proximal to Cys-73, facilitating thiol-Michael addition.
The addition reaction is clearly sensitive to features outside of the methylene bicyclo[3.2.1]octanone skeleton as this ring system is present within other kauranes we examined (2, 3, 6, 8 and 9), which nonetheless displayed significantly reduced binding response (3 or 9). The dihydroxyl moieties within the central 2-oxabicyclo[2.2.2]octane ring are likely candidates for the increased affinity of compounds 1 and 2 due to their engagement of Arg-99 within our structure. Compound 6 lacks the 6′-hydroxyl and accordingly has a fourfold reduced dose response, while other compounds tested had bulky additions to these groups, which abrogated binding completely (3). Overall our structure suggests that bulky additions to the functional groups on the convex face of the compound are likely to hinder engagement or require an altered binding mode, within this site. The Nsp9COV19 structure in complex with oridonin does not immediately suggest a role for the unbridged ring system, the 1′-hydroxyl upon its concave molecular face is solvent facing and unlikely to contribute to Nsp9 binding. In contrast, this ring’s dimethyl group may provide some van der Waals contacts with the side chains of residues emanating from the N-terminus of the α-helix. The potential interaction with Asn-96 is of note as this residue appears to play a crucial role in Nsp9’s NiRAN-association within the RTC (23).
In the extended-RTC complex, the α-helix of Nsp9 is repurposed such that its coiled-coil homodimerization interface instead contacts a mixed β-sheet within the pseudokinase. In this conformation, the α-helix lies perpendicularly across the β-sheet, facilitating the position of the N-terminus within the NiRAN active site. N-terminal residues of Nsp9 become sandwiched between an elongated NiRAN pseudokinase β1-strand on one side and the catalytic Asp-221 residue from the DFG-motif on the other. The novel nucleotidylation activity of NiRAN pseudokinases will necessitate some enzymatic differences as compared with canonical kinases (38). Despite this, a vast array of kinases have been studied, and key componentry is presumably repurposed to an extent. This is relevant as the NiRAN β-sheet engaged by the Nsp9 α-helix is constructed from remnant pseudokinase C-lobe elements whose kinase counterparts have known catalytic roles. This includes the “β9” activation loop following the DFG-motif and the pseudokinase “β6” catalytic loop, albeit one without an active HRD sequence (39); the third and final β-strand could be considered the returning leg of the activation loop, but the pseudokinase boundary lies along its length so may mark a point of functional divergence. When the oridonin-bound Nsp9COV19 structure is overlaid onto the extended RTC, or Nsp10/Nsp14 engaged Cap(0)-RTC (26), elements of the A-ring clash with the turning point of the activation loop particularly near the kinase-equivalent αE helix as well as perturbing any potential interface role for Cys-73. Binding may also perturb Nsp10 recruitment. This suggests that the A-ring induced steric hindrance may well be an important means by which oridonin inhibits coronaviral replication.
Our UMPylation assays support the hypothesis that some ent-kauranes can inhibit the reaction with Nsp12 as the two parental-class compounds (1 and 2) displayed significant levels of inhibition. If oridonin is to be ultimately developed into a specific-inhibitor targeting coronaviruses, a degree of closer scrutiny will be needed to ensure its action is sufficiently specific to Nsp9. Moreover, a full Ki/Kinact analysis would ideally delineate between the stages of oridonin’s Nsp9 binding, its adduct formation, and consequent reduced NiRAN activity. The overall ent-kaurene framework within compound 2 has been assessed in clinical trials (CTR20150246) at intravenous dosages of up to 320 mg/d (40). We observed some cell-line specific cytotoxicity for oridonin, and this necessitates a degree of caution when drawing conclusions from cellular viral replication assays. Of the cell lines tested, Calu-3 is the most robust cell line for viral replication in the presence of ent-kaurane class compounds; in these cells we saw reduced viral titer upon treatment with oridonin. Given the oridonin binding site of Nsp9 is predominantly constructed from residues conserved among a diverse set of coronaviruses, it is encouraging that we also measured viral inhibition among distinct coronaviruses. Ideally modifications to the framework of oridonin may be identified that reduce cytotoxicity, increase specificity for Nsp9COV19, and improve upon its antiviral response. Our viral experiments indicate a degree of viral recovery 72 HPI; this may be due to reduced concentrations of free compound 1 as more Nsp9 is made and covalently modified.
We note the possibility that modification of Nsp12 within our in vitro UMPylation assays could also contribute to the observed inhibitory effect. While most of Nsp12’s cysteine residues are buried, three are relatively solvent-exposed and present within the NiRAN domain; modification of Cys-53 in particular would clearly influence nucleotide binding (41) albeit it does not appear easily accessible to the bulky oridonin. Cys-22 is more obviously accessible, and while not as obviously a part of the NiRAN enzymatic machinery, it may be involved in RNA substrate binding, which could influence our cellular assays.
In parallel with chemical scaffold modifications, Nsp9 viral mutants should also be made to better define how residues within the oridonin-binding site might influence viral replication. This would help ascertain to what extent our observed antiviral activity is solely dependent upon disruption of the Nsp9:Nsp12 interface; at μM concentrations, oridonin has the potential to act on other proteins such as the NLRP3 inflammasome (31) perturbing alternate cellular pathways upon which viral replication may depend (42).
Experimental procedures
Protein expression and purification
Recombinant protein was produced as described previously (21) for mass spectrometry binding assays. For the UMPylation assays, Nsp9COV19 must have Asn-1 without artificial residues N-terminal to this. The pET-28-LIC-Nsp9 construct was therefore recloned by PCR amplifying of the synthetic gene using a primer that encoded an enterokinase protease site prior to Asn-1. The protein was then purified in an identical manner except with enterokinase replacing 3C-protease for hexahistidine tag removal. When sample shipment was required, protein was frozen in liquid nitrogen and transferred on dry ice. The Nsp9COV19(C73S) mutant was ordered as a synthetic gene and cloned with an enterokinase cleavage site and otherwise purified as per the wild-type protein.
The SARS-CoV-2 Nsp12 expression construct was provided by the Crick institute, Nsp12 is fused to a C-terminal decahistidine tag followed by a Tobacco Etch Virus (TEV) protease cleavable protein A tag cloned into a baculoviral expression system. Following baculoviral infection of 2L of Sf9 insect cells were washed in 50 mM HEPES, pH 7.5, 300 mM NaCl, 10% v/v glycerol, 0.05% w/v octylthioglucoside, and 1 mM dithiothreitol, then lysed by sonication. Following clarification by centrifugation, cell lysate was incubated with 1 ml IgG sepharose, then washed with three volumes of wash buffer. Nsp12 was subsequently eluted through cleavage with TEV protease, concentrated, and loaded on a Superdex 200 10/300GL column equilibrated in 25 mM HEPES pH 7.5, 150 mM NaCl, 5% v/v glycerol, 2 mM MgCl2, and 1 mM DTT. Protein was frozen in liquid nitrogen until required.
Compound sourcing and quality control
Compounds used within the study were the following: 1 Oridonin (28957-04-02); 2: L-alanine ester oridonin; 3: Xerophilusin G (304642-94-2); 4: Odonicin (51419-51-3); 5: Maoyerabdosin (90468-72-7); 6: Kamebakaurin (73981-34-7); 7: Creticoside C (53452-34-9); 8: Enmein (3776-39-4); 9: ent-6,11-Dihydroxy-15-oxo-16-kauren-19-oic acid beta-D-glucopyranosyl ester (81263-97-0); 10: atractyloside (102130-43-8); 11: carboxyatractyloside (77228-71-8); 12: steviol (471-80-7). Compounds were sourced from ChemFaces Wuhan, with 1D NMR and mass spectrometry quality controls (1, 2–11), Nanjing NutriHerb BioTech (1), for Kd determination studies, Merck (12), or synthesized by SYNthesis Melbourne Australia (2).
Native MS: Instrument control and acquisition
Experiments were performed on a Bruker SolariX XR 12 T Fourier transform ion cyclotron resonance mass spectrometry (ESI-FT-ICR-MS) (Bruker Daltonics Inc) equipped with an automated chip-based nano-electrospray system (TriVersa NanoMate, Advion Biosciences). Mass spectra were recorded in positive ion and profile modes with a mass range from 50 to 6000 m/z. Each spectrum was a sum of 16 transients (scans) composed of 1 M data points. All aspects of pulse sequence control and data acquisition were controlled by Solarix control software in a Windows operating system. For initial natural product library screening, pools of ten compounds (1 μl of each compound at 500 μM in DMSO) were dried, resuspended in 1 μl MeOH, and incubated with proteins for 1 h at room temperature and analyzed by ESI-FT-ICR-MS. Final compound/protein molar ratio was 5.6:1. When a protein–ligand complex was found, the molecular weight of the binding compound was calculated from the spectrum using the following equation: MWligand = Δm/z × z. For hit confirmation, the binding of the individual compounds was confirmed in a separate experiment.
Dose-responsive binding of compounds
The percentage of ligand-bound protein was determined using the following equation.
% ligand-bound protein = [P-L]/([P] + [P-L]) Where [P-L] is the total intensity of the protein–ligand complex and [P] is the total intensity of the apo-protein for a single charge state. A binding curve was generated (ligand concentration against percentage of ligand-bound protein), and nonlinear regression using the equation below was fit in GraphPad Prism. Y = Bmax∗X/(Kd + X)
Protein crystallization, data collection, and structure determination
Nsp9COV19 with the His-tag retained was concentrated to ∼15 mg/ml and crystallized via hanging drop vapor diffusion over a reservoir consisting of 20% v/v polyethylene glycol 4000, 0.17 M ammonium sulfate, and 0.1 M Citrate pH 4.0. All diffraction data were collected at the Australian synchrotrons MX2 beamlines (35). Data was processed using the program XDS (43), scaled, and merged with programs from the CCP4 suite (44). Diffraction data were processed P21 with β ∼ 90° but can also be processed in P212121 with some loops having alternate configurations. Initial phases were obtained using the molecular replacement program Phaser (45) with trimmed starting model 6W9Q (21). Subsequent rounds of manual building and refinement were performed in the programs COOT (46) and Phenix (47). Due to the data resolution, during refinement the conformation of the covalent oridonin adduct was restrained against previously determined small-molecule experimental data (Cambridge crystallographic data centre: KUYHIV 1201993) adjusted for sp2 to sp3 orbital changes on C16 due to thioether formation (48).
Protein digestion and LC-MS/MS analysis
Protein was diluted in 100 mM Tris, pH 8, and digested in solution overnight with proteomics grade trypsin at a 1:40 trypsin:Nsp9 ratio. Samples were desalted using OMIX C18 pipette tips, vacuum concentrated, and reconstituted in 2% acetonitrile, 0.1% formic acid. Samples were acquired by liquid chromatography–tandem mass spectrometry (LC-MS/MS) using a SCIEX 5600+ TripleTOF mass spectrometer with a Nanospray III ion source, coupled to a NanoUltra cHiPLC system (Eksigent). Samples were loaded at 5 μl/min onto a cHiPLC trap column (3 μm, ChromXP C18CL, 120 Å, 0.5 mm × 200 μm) in 0.1% formic acid, 2% acetonitrile, prior to separation at 300 nl/min over a cHiPLC column (3 μm, ChromXP C18CL, 120 Å, 15 cm × 75 μm) using increasing acetonitrile in 0.1% formic acid. Data were collected in positive ion mode using a data-dependent acquisition strategy. The MS1 and MS2 mass ranges were 200 to 1800 m/z and 60 to 1800 m/z, respectively, with MS/MS to occur for the top 20 ions per cycle matching the following criteria: >200 m/z, charge state +2 to +5, intensity >40 counts per second, and exclusion of ions after two fragmentations for 30 s. Spectra were annotated using PEAKS X Pro (Bioinformatics Solutions Inc) via database search against the recombinant Nsp9 sequence, considering specific trypsin digestion and variable modifications: Oxidation (M) +15.99, Deamidation (N/Q) +0.98, and several possible Cys modifications +364.19 (Oridonin), +346.18 (Oridonin - 18), +31.99 (dioxidation), and +47.98 (trioxidation).
MTT cell proliferation/viability assay
Calu-3, Vero E6, and Vero 81 cells were seeded on 96-well plates and treated with 100 μM, 50 μM, 15 μM, and 5 μM of compound 1 or DMSO control for 27 h. Media containing the compound were removed and serum-free DMEM and MTT reagent (Abcam MTT Cell Proliferation Assay Kit) were added to cells and incubated for 3 h. After removal of MTT reagent, MTT solvent was added and cells were gently rocked for 15 min. Absorbance was read at OD590nm and percentage of viable cells was normalized to control cells treated with equal amounts of DMSO.
SARS-CoV-2 and MERS-CoV replication assays
All viral work was performed within accredited PC3 laboratories. For the TCID50 assays compound 1 in DMSO was diluted from a concentrated stock solution to 1.5 mM, then further diluted to 150 μM in serum-free media and filter-sterilized. A 14-point dilution series of the compound in media spanning 100 μM to 2.5 μM was made and added to Calu-3 cell monolayers growing in a 24-well plate at >95% confluence, which had previously been washed with infection media. Following addition of treatment, Calu-3 cells were incubated at 37 °C, 5% CO2 for 7 h 100 μl/well of SARS-CoV-2 patient isolate (Australia/VIC01/2020) (36) containing 1000 TCID50/ml (50% Tissue Culture Infectious Dose/ml) was then added. Infected cells were incubated for 1 h prior to addition of 800 μl of infection media with 1.2 μg/ml TPCK Trypsin—plates were then incubated for a further 23 h. At the 24 h timepoint, the viral supernatant was removed and a TCID50 assay performed.
Calu-3 2B4 cells were treated with 50 μM, 15 μM, and 5 μM of compound 1 or DMSO control (0.1% v/v) for 3 h prior to infection with 0.01 MOI of MERS-CoV (EMC/2012) or SARS-CoV-2 (2019-nCoV/USA-WA1/2020). Virus inocula were removed and replaced with media containing compound 1. Virus was collected at 24, 48, and 72 hpi and freeze thawed. MERS-CoV and SARS-CoV-2 supernatants were serially diluted and used to infect Vero81 cells or VeroE6 cells, respectively at 37 °C in 5% CO2 for 1 h. After removing virus inocula, plates were overlaid with 0.6% agarose containing 2% FBS and DMEM supplemented with 10% FBS. After 3 days, agar overlays were removed, fixed with 4% PFA, and stained with 0.1% crystal violet. Viral titers were calculated as pfu/ml.
UMPylation assay
Nucleotidylation reactions were performed using a 1:100 ratio of Cy3-UTP (Jena biosciences) to UTP (Merck) pH adjusted to ∼7.4. Reactions were set up for 20 min at 30 °C with final concentrations of 20 mM HEPES pH 7.4, 150 mM NaCl, 50 μM MnCl2, 25 μM UTP mix, 1.5 μM of enterokinase-treated Nsp9COV19, and 0.5 μM Nsp12COV19. Following dilution of recombinant protein stocks, the final reducing agent concentration within the assay is ∼10 μM DTT and 4% v/v DMSO. Within the 1 to 12 compound series reactions, Figure 4B, 15 μM Nsp9COV19 and 1 μM Nsp12COV19 were instead used. The reaction was terminated through addition of 2× loading dye containing 2% w/v sodium dodecyl sulfate and immediately separated via SDS-PAGE on an 18% w/v polyacrylamide gel. After washing once in 50 mM Tris, 150 mM NaCl, the Cy3 modified bands were imaged on an Amersham Typhoon 5 using Cy3 settings. Band quantification was performed using ImageQuant TL (Cytiva) with fitting done in Prism (GraphPad). The intensity of Cy3 staining is plotted as a percentage of the signal obtained compared with in-gel DMSO controls after background subtraction. The raw data from wild-type Nsp9 and the C73S mutant were not obviously dissimilar.
Data availability
The accession number for the atomic coordinates of the 3C-Nsp9COV19:Oridonin and associated structure factors have been deposited at the protein databank (www.rcsb.org) with accession code 7N3K.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This research was undertaken in part using the MX beamlines at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector. Additionally, we thank Dr Geoffrey Kong of the Monash Molecular Crystallisation Facility for his assistance with crystallographic screening and optimization. We thank A. Riboldi-Tunnicliffe and R. Williamson for assistance with data collection. We thank the Prof. E. Fodor from the William Dunn School of Pathology for supplying the SARS-CoV-2 Nsp12 pFastBac expression vector; Prof. D. Thal for help with baculoviral expression and Prof. J Baell for proofreading the manuscript. The University of Melbourne acknowledges the support of Melbourne Health, through its Victorian Infectious Diseases Reference Laboratory at the Doherty Institute, in providing our laboratory with isolated SARS-CoV-2 material. Computational resources were supported by the R@CMon/Monash Node of the NeCTAR Research Cloud, an initiative of the Australian Government’s Super Science Scheme and the Education Investment Fund.
Author contributions
D. R. L., R. J. Q., and J. R. conceptualization; D. R. L. data curation; D. R. L., M. L., J. L. M., S. A. L., and P. T. I. formal analysis; A. W. P., S. P., D. F. J. P., and J. R. funding acquisition; D. R. L., M. L., J. L. M., S. A. L., P. T. I., and I. R. C. investigation; M. L. methodology; S. P., D. F. J. P., R. J. Q., and J. R. project administration; I. R. C., and J. R. resources; B. S. G., A. W. P., S. P., D. F. J. P., R. J. Q., and J. R. supervision; D. R. L. and R. J. Q. writing–original draft; D. R. L., M. L., J. L. M., S. A. L., P. T. I., B. S. G., A. W. P., S. P., D. F. J. P., and J. R. writing–review and editing.
Funding and additional information
Funding for the work originated from the Australian Research Council Centre of Excellence for Advanced Molecular Imaging and the National Institutes of Health (USA) (PO1 AI060699, RO1 AI129269) (S. P.). A. W. P. is supported by a NHMRC PRF (APP1137739). P. T. I. was supported by a Monash University Faculty of Medicine, Nursing and Health Sciences Senior Postdoctoral Fellowship. J. R. is supported by an Australian Research Council Laureate Fellowship. D. F. J. P. and J. L. M. received antiviral program support from the DHHS COVID-19 Victorian Consortium. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Craig Cameron
Contributor Information
Dene R. Littler, Email: dene.littler@monash.edu.
Ronald J. Quinn, Email: r.quinn@griffith.edu.au.
Jamie Rossjohn, Email: jamie.rossjohn@monash.edu.
Supporting information
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Bibi S., Briner C., Cicconi P., Collins A.M., Colin-Jones R., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Zeng G., Pan H., Li C., Hu Y., Chu K., Han W., Chen Z., Tang R., Yin W., Chen X., Hu Y., Liu X., Jiang C., Li J., et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18-59 years: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021;21:181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., Wang B., Paesen G.C., Lopez-Camacho C., Slon-Campos J., Hallis B., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 11.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Aimon A., Abranyi-Balogh P., Brandao-Neto J., Carbery A., Davison G., Dias A., Downes T.D., et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11:5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunther S., Reinke P.Y.A., Fernandez-Garcia Y., Lieske J., Lane T.J., Ginn H.M., Koua F.H.M., Ehrt C., Ewert W., Oberthuer D., Yefanov O., Meier S., Lorenzen K., Krichel B., Kopicki J.D., et al. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372:642–646. doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daczkowski C.M., Dzimianski J.V., Clasman J.R., Goodwin O., Mesecar A.D., Pegan S.D. Structural insights into the interaction of coronavirus papain-like proteases and interferon-stimulated gene product 15 from different species. J. Mol. Biol. 2017;429:1661–1683. doi: 10.1016/j.jmb.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G., Kuchel N.W., Grohmann C., Shibata Y., Gan Z.Y., Cooney J.P., Doerflinger M., Au A.E., Blackmore T.R., et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39 doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osipiuk J., Azizi S.A., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A., Kang S., Kathayat R.S., Kim Y., Lisnyak V.G., Maki S.L., Nicolaescu V., Taylor C.A., Tesar C., Zhang Y.A., et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021;12:743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., El Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti-COVID-19 drug design. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egloff M.P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L., Diprose J., Alderton D., Walsh M., et al. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., Thai J., Miller Z.D., Lin A.E., Schmidt M.M., Stewart D.G., et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339.e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Littler D.R., Mohanty B., Lowery S.A., Colson R.N., Gully B.S., Perlman S., Scanlon M.J., Rossjohn J. Binding of a pyrimidine RNA base-mimic to SARS-CoV-2 non-structural protein 9. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Littler D.R., Gully B.S., Colson R.N., Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. iScience. 2020;23 doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miknis Z.J., Donaldson E.F., Umland T.C., Rimmer R.A., Baric R.S., Schultz L.W. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J. Virol. 2009;83:3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M., Liu Z., Wang H., Li Y., Chen Y., Guddat L.W., et al. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184:184–193.e10. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slanina H., Madhugiri R., Bylapudi G., Schultheiss K., Karl N., Gulyaeva A., Gorbalenya A.E., Linne U., Ziebuhr J. Coronavirus replication-transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2022310118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sreelatha A., Yee S.S., Lopez V.A., Park B.C., Kinch L.N., Pilch S., Servage K.A., Zhang J., Jiou J., Karasiewicz-Urbanska M., Lobocka M., Grishin N.V., Orth K., Kucharczyk R., Pawlowski K., et al. Protein AMPylation by an evolutionarily conserved pseudokinase. Cell. 2018;175:809–821.e19. doi: 10.1016/j.cell.2018.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan L., Yang Y., Li M., Zhang Y., Zheng L., Ge J., Huang Y.C., Liu Z., Wang T., Gao S., Zhang R., Huang Y.Y., Guddat L.W., Gao Y., Rao Z., et al. Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell. 2021;184:3474–3485.e11. doi: 10.1016/j.cell.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia P.A., de Oliveira A.B., Batista R. Occurrence, biological activities and synthesis of kaurane diterpenes and their glycosides. Molecules. 2007;12:455–483. doi: 10.3390/12030455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H.D., Huang S.X., Han Q.B. Diterpenoids from Isodon species and their biological activities. Nat. Prod. Rep. 2006;23:673–698. doi: 10.1039/b604174d. [DOI] [PubMed] [Google Scholar]
- 29.Elnaas A.R., Grice D., Han J., Feng Y., Capua A.D., Mak T., Laureanti J.A., Buchko G.W., Myler P.J., Cook G., Quinn R.J., Liu M. Discovery of a natural product that binds to the Mycobacterium tuberculosis protein Rv1466 using native mass spectrometry. Molecules. 2020;25:2384. doi: 10.3390/molecules25102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z.R., Zhang Y.N., Li X.D., Zhang H.Q., Xiao S.Q., Deng F., Yuan Z.M., Ye H.Q., Zhang B. A cell-based large-scale screening of natural compounds for inhibitors of SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5:218. doi: 10.1038/s41392-020-00343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kjer-Nielsen L., Corbett A.J., Chen Z., Liu L., Mak J.Y., Godfrey D.I., Rossjohn J., Fairlie D.P., McCluskey J., Eckle S.B. An overview on the identification of MAIT cell antigens. Immunol. Cell Biol. 2018;96:573–587. doi: 10.1111/imcb.12057. [DOI] [PubMed] [Google Scholar]
- 32.Bruno M., Rosselli S., Pibiri I., Kilgore N., Lee K.H. Anti-HIV agents derived from the ent-kaurane diterpenoid linearol. J. Nat. Prod. 2002;65:1594–1597. doi: 10.1021/np020029b. [DOI] [PubMed] [Google Scholar]
- 33.Li W.F., Wang J., Zhang J.J., Song X., Ku C.F., Zou J., Li J.X., Rong L.J., Pan L.T., Zhang H.J. Henrin A: A new anti-HIV ent-kaurane diterpene from pteris henryi. Int. J. Mol. Sci. 2015;16:27978–27987. doi: 10.3390/ijms161126071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y.C., Hung Y.C., Chang F.R., Cosentino M., Wang H.K., Lee K.H. Identification of ent-16 beta, 17-dihydroxykauran-19-oic acid as an anti-HIV principle and isolation of the new diterpenoids annosquamosins A and B from Annona squamosa. J. Nat. Prod. 1996;59:635–637. doi: 10.1021/np960416j. [DOI] [PubMed] [Google Scholar]
- 35.Aragao D., Aishima J., Cherukuvada H., Clarken R., Clift M., Cowieson N.P., Ericsson D.J., Gee C.L., Macedo S., Mudie N., Panjikar S., Price J.R., Riboldi-Tunnicliffe A., Rostan R., Williamson R., et al. MX2: A high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian synchrotron. J. Synchrotron Radiat. 2018;25:885–891. doi: 10.1107/S1600577518003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caly L., Druce J., Roberts J., Bond K., Tran T., Kostecki R., Yoga Y., Naughton W., Taiaroa G., Seemann T., Schultz M.B., Howden B.P., Korman T.M., Lewin S.R., Williamson D.A., et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med. J. Aust. 2020;212:459–462. doi: 10.5694/mja2.50569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponnusamy R., Moll R., Weimar T., Mesters J.R., Hilgenfeld R. Variable oligomerization modes in coronavirus non-structural protein 9. J. Mol. Biol. 2008;383:1081–1096. doi: 10.1016/j.jmb.2008.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang B., Svetlov D., Artsimovitch I. NMPylation and de-NMPylation of SARS-CoV-2 nsp9 by the NiRAN domain. Nucleic Acids Res. 2021;49:8822–8835. doi: 10.1093/nar/gkab677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modi V., Dunbrack R.L., Jr. A structurally-validated multiple sequence alignment of 497 human protein kinase domains. Sci. Rep. 2019;9:19790. doi: 10.1038/s41598-019-56499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu X., Wang Y., Gao X., Xu S., Zang L., Xiao Y., Li Z., Hua H., Xu J., Li D. Recent progress of oridonin and its derivatives for the treatment of acute Myelogenous leukemia. Mini Rev. Med. Chem. 2020;20:483–497. doi: 10.2174/1389557519666191029121809. [DOI] [PubMed] [Google Scholar]
- 41.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreira A.C., Soares V.C., de Azevedo-Quintanilha I.G., Dias S., Fintelman-Rodrigues N., Sacramento C.Q., Mattos M., de Freitas C.S., Temerozo J.R., Teixeira L., Damaceno Hottz E., Barreto E.A., Pao C.R.R., Palhinha L., Miranda M., et al. SARS-CoV-2 engages inflammasome and pyroptosis in human primary monocytes. Cell Death Discov. 2021;7:43. doi: 10.1038/s41420-021-00428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casanal A., Lohkamp B., Emsley P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 2019;29:1069–1078. doi: 10.1002/pro.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., Moriarty N.W., Oeffner R.D., Poon B.K., Prisant M.G., Read R.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei Y., Li Y., Tan Y., Qian Z., Zhou Q., Jia D., Sun Q. Novel mechanistic observations and NES-binding groove features revealed by the CRM1 inhibitors plumbagin and oridonin. J. Nat. Prod. 2021;84:1478–1488. doi: 10.1021/acs.jnatprod.0c01231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the atomic coordinates of the 3C-Nsp9COV19:Oridonin and associated structure factors have been deposited at the protein databank (www.rcsb.org) with accession code 7N3K.