Abstract
Coronavirus disease 19 (COVID-19) is a highly contagious respiratory viral infection. Dysregulated immune response is an important feature of disease, and cytokines are among the most important mediators of dysregulated immunity. Interleukin-37 (IL-37) is one such cytokine and studies have indicated its role in pathogenesis of COVID-19. However, IL37 gene polymorphisms have not been identified in patients with COVID-19. Therefore, this case-control study (100 patients and 100 controls) was performed to understand the role six single nucleotide polymorphisms of IL37 gene (SNPs: rs3811042, rs3811043, rs2466449, rs3811045, rs3811046 and rs3811047) in susceptibility to COVID-19 among cases with severe disease. These polymorphisms were identified by Sanger DNA sequencing. Results revealed that TG genotype of rs3811046 showed a significantly increased frequency in patients compared to controls (61.0 vs. 38.0%; odds ratio [OR] = 2.55; 95% confidence interval [CI] = 1.45–4.50; probability [p] = 0.002; corrected p [pc] = 0.01). GA genotype of rs3811047 also showed an increased frequency in patients but the pc-value was not significant (39.0 vs. 24.0%; OR = 2.02; 95% CI = 1.10–3.71; p = 0.033; pc = 0.165). Haplotype analysis revealed a significantly increased frequency of the haplotype G-C-A-T-T-A (in the order: rs3811042, rs3811043, rs2466449, rs3811045, rs3811046 and rs3811047) in COVID-19 patients compared to controls (0.055 vs. 0.006; OR = 10.23; 95% CI = 1.53–68.14; p = 0.003; pc = 0.03). In conclusion, the study indicated that two variants of IL37 gene (rs3811046 and rs3811047) may be associated with susceptibility to COVID-19 among Iraqi population.
Keywords: COVID-19, Interleukin-37, Single nucleotide polymorphism, Haplotype
1. Introduction
Coronavirus disease 19 (COVID-19) is a highly contagious respiratory viral infection caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The first cases were reported in December 2019 in the Chinese city of Wuhan, and since then the disease has spread rapidly to more than 200 countries with millions of infected cases and deaths, forcing the World Health Organization (WHO) to declare COVID-19 a global pandemic (Vannabouathong et al., 2020). Several viral, environmental and host factors have been described as being associated with an increased risk of disease. Advanced age, male gender, diabetes, cardiovascular disease, and malignancy are among the most important proposed host-related risk factors (Rashedi et al., 2020). Further, humoral and cellular components of the innate and adaptive immune systems can also be considered as additional risk factors and dysregulated immune responses have been indicated in COVID-19 patients (Mohamed Khosroshahi and Rezaei, 2021). These include, but are not limited to, cytokines, the ratio of neutrophils to lymphocytes and lymphopenia (Darif et al., 2021; Imran et al., 2021).
Cytokines are perhaps the most important factors of immunity because of their involvement in mediating and controlling immune responses in various infectious and inflammatory diseases (Lin and Leonard, 2019). In COVID-19 patients, especially those with severe disease, there has been accumulating evidence indicating a robust and uncontrolled systemic inflammatory response known as cytokine release syndrome. High levels of pro-inflammatory cytokines and chemokines have been reported in the serum of these patients and are associated with respiratory inflammation and severe lung damage (Darif et al., 2021). In addition, SARS-CoV-2 infection has been shown to promote coagulopathy through the effects of pro-inflammatory cytokines and other immune components. Accordingly, the term cytokine storm has been introduced to describe the uncontrolled excessive production of inflammatory markers in COVID-19 patients (Coperchini et al., 2020).
Interleukin (IL)-37 is one of the cytokines recently recognized to play a role in the pathogenesis of COVID-19 (Conti et al., 2020; Li et al., 2021). It is a new member of the IL-1 cytokine family that is expressed by many immune and non-immune cells including monocytes, natural killer cells, stimulated B lymphocytes and epithelial cells (Quirk and Agrawal, 2014). Functionally, IL-37 is an anti-inflammatory cytokine that has inhibitory effects on inflammatory responses by affecting the production of pro-inflammatory cytokines (Jia et al., 2018). Because of this immune function, IL-37 has been indicated to play a key role in the pathogenesis of a variety of inflammatory and autoimmune diseases, and in fact, dysregulated expression of IL-37 has been reported under these conditions (Ding et al., 2018; Wang et al., 2018). Besides, the anti-viral, anti-bacterial and anti-fungal properties of IL-37 have also been recognized (Allam et al., 2020). With respect to COVID-19, IL-37 plasma levels have been significantly associated with the clinical prognosis of disease, and it has been predicted that severe illness can occur due to the lack of an IL-37-mediated response (Li et al., 2021). In addition, the therapeutic potential of IL-37 in COVID-19 has recently been suggested (Conti et al., 2020).
IL37 gene (Gene ID: 27178) is mapped to the long arm of human chromosome 2 (2q14.1), spans 3.617 kb and consists of seven exons (www.ncbi.nlm.nih.gov/gene/27178). The gene harbors many single nucleotide polymorphisms (SNPs), and genetic association studies have linked some of these SNPs with susceptibility to several autoimmune, inflammatory and infectious diseases (for instance, Behçet disease, systemic lupus erythematosus, periodontal inflammation, hepatitis viral infection and tuberculosis)(Al-Anazi et al., 2019; Allam et al., 2016; Lin et al., 2018; Offenbacher et al., 2018; Özgüçlü et al., 2019). Of particular interest is the exon SNP rs3811047, and some of these studies have suggested a role for this variant in the risk of various human diseases. Therefore, a pilot study was designed to sequence a DNA region of IL37 gene comprising SNP rs3811047 in patients with severe COVID-19, in order to reveal the significance of this SNP and other SNPs in the region in susceptibility to disease.
2. Materials and methods
2.1. Populations studied
A case-control study was conducted on 100 patients with severe COVID-19 (mean ± standard deviation: 56.2 ± 13.9 years; 78.0% male) and 100 healthy controls (35.0 ± 9.8 years, 76% male) during October 1–November 15, 2020. Patients presenting with signs and symptoms of COVID-19 were admitted to the COVID-19 care units in Baghdad, and upon admission, nasopharyngeal swabs were obtained and tested for SARS-CoV-2 RNA. Virus RNA was isolated using the ExtractNow Virus RNA Swab kit (Minerva Biolabs GmbH), while the RealLine SARS-CoV-2 kit (Bioron Diagnostics GmbH) was used to detect SARS-CoV-2 RNA, following the manufacturer's instructions. The diagnosis was confirmed by computed tomography (CT) of the chest. Included patients were those who gave written consent to participate in the study, and showed a positive molecular test along with a CT profile indicating COVID-19. Most of the admitted patients were in severe condition because mild cases tended to leave the hospital; therefore, only severe COVID-19 patients were included. The severity of COVID-19 was determined according to the WHO Interim Guidance (severe respiratory distress, respiratory rate ≥ 30 breaths/min or pulse oxygen saturation (SpO2) ≤ 93% on resting state) (World Health Organization, 2020). Control subjects were blood donors and health service personnel. They were healthy and had no respiratory infections in the past 12 months or chronic diseases (diabetes and cardiovascular). Besides, the serum C-reactive protein (CRP) test was negative and the erythrocyte sedimentation rate (ESR) was <20 mm/h. The protocol of study was approved by the Ethics Committee at the Iraqi Ministry of Health and Environment.
2.2. IL37 gene polymorphism
The complete DNA sequence and SNP data for IL37 gene were downloaded (http://asia.ensembl.org). Forward (5’-GGGGGAGAACTCAGGAGTGA-3′) and reverse (5′-GAAAGACTTCAGCCCCATCCA-3′) primers were designed to amplify a DNA region of IL37 gene comprising SNP rs3811047 using Geneious software version 11.1.2 (Kearse et al., 2012). In-silico PCR analysis was applied to test the specificity of primers (https://genome.ucsc.edu/cgi-bin/hgPcr). The analysis revealed that the target region had a molecular size of 971 bp (Chromosome 2:112913024 + 112913994). This region was explored for SNPs with a minor allele frequency ≥ 10% (selection criterion). By applying this criterion, six SNPs were found (rs3811042 G/A, rs3811043 C/A/G/T, rs2466449 G/A/C/T, rs3811045 T/A/C, rs3811046 G/A/C/T and rs3811047 A/G/T) and analyzed.
A PCR reaction mix of 25 μL was prepared to include 12.5 μL GoTaq Green Master Mix (2×: Promega, U.S.A), 1 μL of each primer (10 pmol); 8.5 μL nuclease free water and 2 μL of template DNA. The PCR machine (Thermal Cycler, BioRad, USA) was programmed for the following optimized conditions: initial denaturation at 95 °C for 5 min (one cycle), followed by 30 cycles of denaturation (95 °C for 30 s), annealing (60 °C for 30 s) and extension (72 °C for 60 s). A final extension cycle was accomplished (72 °C for 7 min), followed by 10 min hold at 4 °C. The PCR products were sent for DNA sequencing (Sanger sequencing, Macrogen Corporation, South Korea). The received DNA sequences were subjected to alignments with reference DNA sequences of IL37 gene SNPs (http://asia.ensembl.org). Geneious software version 11.1.2 was used to reveal the genotypes of IL37 gene SNPs (Kearse et al., 2012). Fig. 1 shows a representative chromatogram of a DNA sequence of SNP rs3811046.
Fig. 1.
DNA sequence chromatogram of IL37 gene SNP (rs3811046) showing three genotypes: GG (sample 2), GT (sample 4) and TT (sample 3). In addition, the reference sequence (rs3811046) is also given.
2.3. Statistical analysis
Allele and genotype frequencies were presented in number and percentage. Genotype frequencies of SNPs in controls were tested for Hardy-Weinberg equilibrium (HWE) using the Pearson's Chi-square goodness-of-fit test. Significant differences between patients and controls with regard to allele and genotype frequencies were evaluated using Fisher's exact test with two-tailed probability (p). Logistic regression analysis was employed to calculate odds ratio (OR) and 95% confidence interval (CI). Bonferroni correction was applied to correct for statistical significance due multiple comparisons (corrected probability; pc). A p-value ≤ 0.05 was taken significant. The statistical package IBM SPSS Statistics 25.0 (Armonk, NY: IBM Corp.) was used to perform these analyses. Haplotype frequencies between SNPs of IL37 gene were estimated using SHEsis software, which was also used to assess linkage disequilibrium (LD) between SNPs. The LD coefficient (D′) was used to define LD. The D′ value has a range between 0 (no LD) and 1.0 (complete LD) (Shi and He, 2005).
3. Results
Genotype frequencies of rs3811042, rs3811043, rs2466449, rs3811045, rs3811046 and rs3811047 were in good agreement with HWE in controls, as there were no significant differences between observed and expected genotype frequencies. When COVID-19 patients were compared to controls, only two SNPs showed significant variations (rs3811046 and rs3811047). TG genotype of SNP rs3811046 showed a significantly increased frequency in patients compared to controls (61.0 vs. 38.0%; OR = 2.55; 95% CI = 1.45–4.50; p = 0.002; pc = 0.01). GA genotype of SNP rs3811047 also showed an increased frequency in patients compared to controls, but the pc-value was not significant (39.0 vs. 24.0%; OR = 2.02; 95% CI = 1.10–3.71; p = 0.033; pc = 0.165) (Table 1 ).
Table 1.
Allele and genotype distribution of six SNPs in IL37 gene among COVID-19 patients and controls.
| SNP | Allele/Genotype | Patients (N = 100) |
Controls (N = 100) |
OR | 95% CI | p-value (pc) | ||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | |||||
| rs3811042 | A | 72 | 36.0 | 79 | 39.5 | Reference | ||
| G | 128 | 64.0 | 121 | 60.5 | 1.16 | 0.78–1.74 | 0.536 (1.0) | |
| AA | 20 | 20.0 | 19 | 19.0 | Reference | |||
| AG | 32 | 32.0 | 41 | 41.0 | 0.68 | 0.38–1.20 | 0.240 (1.0) | |
| GG | 48 | 48.0 | 40 | 40.0 | 1.38 | 0.79–2.42 | 0.319 (1.0) | |
| HWE-p | 0.155 | |||||||
| rs3811043 | C | 80 | 40.0 | 64 | 32.0 | Reference | ||
| G | 120 | 60.0 | 136 | 68.0 | 0.71 | 0.47–1.06 | 0.118 (0.59) | |
| CC | 13 | 13.0 | 9 | 9.0 | Reference | |||
| CG | 54 | 54.0 | 46 | 46.0 | 1.38 | 0.79–2.40 | 0.322 (1.0) | |
| GG | 33 | 33.0 | 45 | 45.0 | 0.60 | 0.34–1.07 | 0.111 (0.555) | |
| HWE-p | 0.568 | |||||||
| rs2466449 | A | 186 | 93.0 | 183 | 91.5 | Reference | ||
| G | 14 | 7.0 | 17 | 8.5 | 0.81 | 0.39–1.69 | 0.709 (1.0) | |
| AA | 86 | 86.0 | 83 | 83.0 | Reference | |||
| AG | 14 | 14.0 | 17 | 17.0 | 0.79 | 0.37–1.71 | 0.696 (1.0) | |
| HWE-p | 0.352 | |||||||
| rs3811045 | C | 132 | 66.0 | 143 | 71.5 | Reference | ||
| T | 68 | 34.0 | 57 | 28.5 | 1.29 | 0.85–1.97 | 0.281 (1.0) | |
| CC | 45 | 45.0 | 53 | 53.0 | Reference | |||
| CT | 42 | 42.0 | 37 | 37.0 | 1.23 | 0.70–2.17 | 0.563 (1.0) | |
| TT | 13 | 130 | 10 | 10.0 | 1.34 | 0.56–3.21 | 0.658 (1.0) | |
| HWE-p | 0.356 | |||||||
| rs3811046 | T | 125 | 62.5 | 138 | 69.0 | Reference | ||
| G | 75 | 37.5 | 62 | 31.0 | 1.34 | 0.88–2.02 | 0.206 (1.0) | |
| TT | 32 | 32.0 | 50 | 50.0 | Reference | |||
| TG | 61 | 61.0 | 38 | 38.0 | 2.55 | 1.45–4.50 | 0.002 (0.01) | |
| GG | 7 | 7.0 | 12 | 12.0 | 0.55 | 0.21–1.46 | 0.335 (1.0) | |
| HWE-p | 0.263 | |||||||
| rs3811047 | G | 155 | 77.5 | 170 | 85.0 | Reference | ||
| A | 45 | 22.5 | 30 | 15.0 | 1.65 | 0.99–2.74 | 0.072 (0.36) | |
| GG | 58 | 58.0 | 73 | 73.0 | Reference | |||
| GA | 39 | 39.0 | 24 | 24.0 | 2.02 | 1.10–3.71 | 0.033 (0.165) | |
| AA | 3 | 3.0 | 3 | 3.0 | 1.00 | 0.20–5.04 | 1.0 (1.0) | |
| HWE-p | 0.556 | |||||||
SNP: Single nucleotide polymorphism; HWE-p: Hardy-Weinberg probability; N: Absolute number; OR: Odds ratio; CI: Confidence interval; p: Two-tailed Fisher's exact probability; pc: Bonferroni correction probability. Significant p-value is bold-marked.
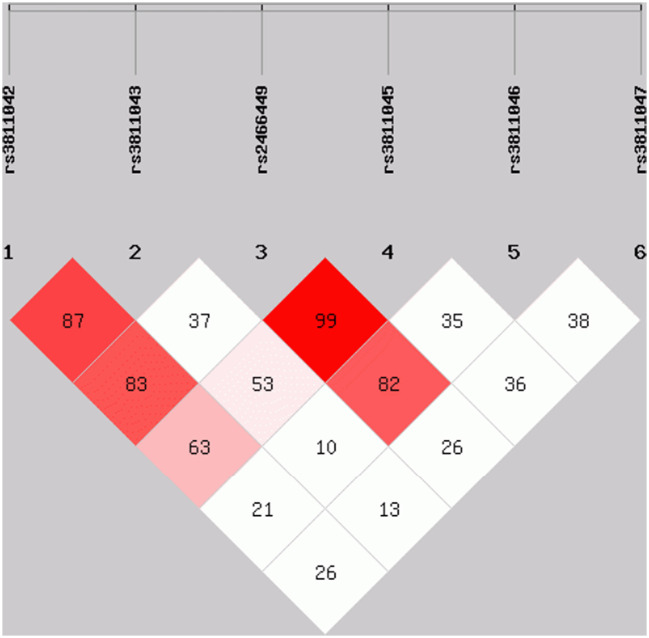
Pairwise linkage analysis revealed that some of the tagged SNPs were in strong LD; rs3811042 with rs3811043 (D′ = 0.87), rs3811042 with rs2466449 (D′ = 0.83), rs2466449 with rs3811045 (D′ = 0.99), and rs2466449 with rs3811046 (D′ = 0.82) (Fig. 2 ). Haplotype analysis revealed a significantly increased frequency of the haplotype G-C-A-T-T-A (in the order: rs3811042, rs3811043, rs2466449, rs3811045, rs3811046 and rs3811047) in COVID-19 patients compared to controls (0.055 vs. 0.006; OR = 10.23; 95% CI = 1.53–68.14; p = 0.003; pc = 0.03). Conversely, the G-A-C-T-G haplotype showed a lower frequency in patients than in controls, but the pc-value was not significant (0.126 vs. 0.204; OR = 0.57; 95% CI = 0.33–1.01; p = 0.05; pc = 0.5) (Table 2 ).
Fig. 2.
Pairwise linkage disequilibrium (LD) map of six IL37 gene SNPs (rs3811042, rs3811043, rs2466449, rs3811045, rs3811046 and rs3811047) genotyped using SHEsis software. The LD is expressed between any pair of SNPs with a value of D′ multiplied by 100. Values approaching zero indicate no LD, and those approaching 100 indicate complete LD. The squares colored red represent varying degrees of LD and darker shades indicate stronger LD.
Table 2.
Haplotype analysis of IL37 gene SNPs (in the order: rs3811042, rs3811043, rs2466449, rs3811045, rs3811046 and rs3811047) in COVID-19 patients and controls.
| Haplotype | N (Frequency) |
OR | 95% CI | p-value (pc) | |
|---|---|---|---|---|---|
| Patients (N = 100) | Controls (N = 100) | ||||
| A-G-A-C-G-G | 13.05 (0.065) | 11.49 (0.057) | 1.21 | 0.53–2.76 | 0.650 (1.0) |
| A-G-A-C-T-G | 44.09 (0.220) | 51.38 (0.257) | 0.86 | 0.53–1.39 | 0.544 (1.0) |
| G-C-A-C-G-G | 8.77 (0.044) | 5.94 (0.030) | 1.58 | 0.54–4.59 | 0.393 (1.0) |
| G-C-A-C-T-G | 17.61 (0.088) | 14.13 (0.071) | 1.34 | 0.64–2.81 | 0.426 (1.0) |
| G-C-A-T-G-A | 7.17 (0.036) | 3.54 (0.018) | 2.18 | 0.59–7.95 | 0.226 (1.0) |
| G-C-A-T-G-G | 7.70 (0.039) | 9.67 (0.048) | 0.82 | 0.31–2.19 | 0.703 (1.0) |
| G-C-A-T-T-A | 10.98 (0.055) | 1.20 (0.006) | 10.23 | 1.53–68.14 | 0.003 (0.03) |
| G-C-A-T-T-G | 15.55 (0.078) | 15.87 (0.079) | 1.03 | 0.49–2.15 | 0.932 (1.0) |
| G-G-A-C-T-G | 25.16 (0.126) | 40.89 (0.204) | 0.57 | 0.33–1.01 | 0.05 (0.5) |
| G-G-G-T-G-G | 6.06 (0.030) | 9.86 (0.049) | 0.63 | 0.22–1.78 | 0.381 (1.0) |
N: Absolute number; OR: Odds ratio; CI: Confidence interval; p: Two-tailed Fisher's exact probability; pc: Bonferroni correction probability. Significant p-value is bold-marked.
4. Discussion
Although cytokines have been implicated in initiating anti-viral immune responses and involved in virus-mediated pathogenesis, host genetic variations may influence their responses to respiratory viral infections (Forbester and Humphreys, 2020). Among these cytokines is IL-37 and its immunological role in viral, bacterial and fungal infections has received increasing attention (Allam et al., 2020). Besides, accumulating data indicated that SNPs of IL37 gene are conserved among humans by selective force, and their potential involvement in the regulation of immune responses and susceptibility to human diseases has been proposed (Kang et al., 2015). Therefore, it is reasonable to postulate that IL37 gene SNPs may be linked with susceptibility to COVID-19.
In this initial study, six SNPs of IL37 gene were analyzed in COVID-19 patients, but only two were indicated as markers of susceptibility to disease (rs3811046 and rs3811047). Both SNPs are located in exon 2, 31 nucleotides apart, and are missense mutations leading to different amino acid codons (Gly/Val at rs3811046 and Thr/Ala at rs3811047) (Yan et al., 2015). The current study showed that TG genotype of rs3811046 may increase the risk of developing COVID-19 by 2.55-fold. As presented by the 1000 Genomes Project Phase 3, rs3811046 is characterized by two alleles in South Asian populations, G (29%) and T (71%) (Auton et al., 2015). The present study matched these figures and the corresponding allele frequencies in control subjects were 31 and 69%, respectively. Regarding disease association studies, SNP rs3811046 has not been well investigated and limited observations have been reported. In primary open-angle glaucoma (Mookherjee et al., 2016), tuberculosis (Allam et al., 2016), rheumatoid arthritis (Zhang et al., 2018) and Hashimoto's thyroiditis (Yan et al., 2015), no association with rs3811046 was found. However, two significant associations were reported with periodontal inflammation. In the first, rs3811046 was linked with severe chronic periodontitis (OR = 1.50), 10-year incident tooth loss (relative risk = 1.33) and aggressive periodontitis (OR = 1.12) in German/Dutch adults (Offenbacher et al., 2018). In a more recent study, it has been demonstrated that rs3811046 was associated with an increased risk of developing moderate periodontitis (OR = 2.58) in a Brazilian population (Cirelli et al., 2021). In Graves' disease, the rs3811046 variant was significantly associated with a decreased risk in female patients (OR = 0.777) (Yan et al., 2015). In the case of viral infections, rs3811046 has not been investigated and the present study is probably the first to reveal the significance of this polymorphism in susceptibility to COVID-19. Taken together, these data may propose a role for rs3811046 in inflammation-mediated diseases, but the evidence has not been overwhelmed and further studies are warranted.
The second SNP that showed an association with the risk of COVID-19 infection was rs3811047, and although the pc value was not significant, individuals with the GA genotype may have a 2.02-fold susceptibility to disease. Studies have highlighted this variant more than rs3811046. This SNP has been associated with susceptibility to or protection against several infectious, inflammatory and autoimmune diseases (Al-Anazi et al., 2019; Allam et al., 2016; Lin et al., 2018; Offenbacher et al., 2018; Özgüçlü et al., 2019). Tuberculosis is the only infectious disease of the respiratory tract studied and susceptibility to disease was not associated with rs3811047, but C allele and CC genotype of another IL37 gene SNP (rs2723176) occurred significantly more frequently in patients with active tuberculosis (Allam et al., 2016). Recently, rs3811047 SNP was investigated in Helicobacter pylori-infected patients with peptic ulcer, and G allele and GG genotype were expressed more frequently in patients than in controls and were associated with increased susceptibility to peptic ulcer (Davarpanah et al., 2020). With respect to inflammatory and autoimmune diseases, G allele and GG genotype of rs3811047 SNP showed significantly increased frequencies in Chinese patients with Behcet's disease in comparison with controls (Tan et al., 2016), but these findings were not confirmed in Turkish patients, and Behcet's disease did not show association with rs3811047 (Özgüçlü et al., 2019). A meta-analysis of multiple autoimmune diseases in Chinese patients (ankylosing spondylitis, rheumatoid arthritis, autoimmune thyroid disease, Behcet's disease and Vogt-Koyanagi-Harada disease) revealed a significant association between rs3811047 variant and susceptibility to autoimmune diseases in four genetic models (allele, recessive, dominant and heterozygous) (Lin et al., 2018). In ankylosing spondylitis patients, it was interesting to note that expressions of C-reactive protein and erythrocyte sedimentation rate (inflammatory markers) were influenced by GG genotype of rs3811047 (Chen et al., 2011). These data indicate that rs3811047 may influence susceptibility to infectious and inflammatory diseases.
Regarding the remaining SNPs of IL37 gene (rs3811042, rs3811043, rs2466449 and rs3811045), allele and genotype frequencies did not show significant differences between COVID-19 patients and controls. To the best of our knowledge, these SNPs have not been investigated in any human disease. However, when the six SNPs of IL37 gene were included in a haplotype analysis, interesting results were found. Some of the SNPs were in strong LD, and the haplotype G-C-A-T-T-A (in the order: rs3811042, rs3811043, rs2466449, rs3811045, rs3811046 and rs3811047) was significantly associated with a 10.23-fold increased risk of COVID-19. Haplotypes can be considered as the principal unit of heredity and may provide the opportunity to better understand the role of polymorphic traits (i.e., SNPs) in disease susceptibility rather than genotypes at a single locus (Glusman et al., 2014). This issue was highlighted in a study of three IL37 gene SNPs (rs3811047, rs2723176 and rs2723816) in Hashimoto's thyroiditis. The three SNPs showed no association with susceptibility to disease, but haplotype analysis showed that ACG haplotype was associated with a 1.57-fold risk of Hashimoto's thyroiditis (Yan et al., 2015). Therefore, haplotype analysis is a useful approach to understand the genetic association between IL37 gene SNPs and diseases, but due to the limited data available, further studies are needed.
It is worth noting that the study has some limitations. First, only severe cases of COVID-19 were included, and asymptomatic and moderate cases should have been considered in the analysis. Second, data for viral load were not obtained, and it would be important to correlate these data with genotypes of IL37 gene SNPs. Third, sample size may limit study results and a larger number of patients and controls may provide more insight on the role of IL37 gene SNPs in susceptibility to COVID-19.
In conclusion, the study indicated that two variants of IL37 gene (rs3811046 and rs3811047) may be associated with susceptibility to COVID-19 among Iraqi population.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that there were no conflicts of interest.
Acknowledgments
The authors sincerely thank the medical staff at the COVID-19 care units in Baghdad for their kind cooperation.
References
- Al-Anazi M.R., Matou-Nasri S., Al-Qahtani Arwa A., Alghamdi J., Abdo A.A., Sanai F.M., Al-Hamoudi W.K., Alswat K.A., Al-Ashgar H.I., Khan M.Q., Albenmousa A., Shamsi M.B., Alanazi S.K., Dela Cruz D., Bohol M.F.F., Al-Ahdal M.N., Al-Qahtani Ahmed A. Association between IL-37 gene polymorphisms and risk of HBV-related liver disease in a Saudi Arabian population. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-42808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allam G., Mohamed I.A.A., Alswat K.A., Abbadi S.H., Nassif R., Alharthi B.J., Nasr A. Association of IL-37 gene polymorphisms with susceptibility to tuberculosis in Saudi subjects. Microbiol. Immunol. 2016;60:778–786. doi: 10.1111/1348-0421.12444. [DOI] [PubMed] [Google Scholar]
- Allam G., Gaber A.M., Othman S.I., Abdel-Moneim A. The potential role of interleukin-37 in infectious diseases. Int. Rev. Immunol. 2020;39:3–10. doi: 10.1080/08830185.2019.1677644. [DOI] [PubMed] [Google Scholar]
- Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E., Flicek P., Gabriel S.B., Gibbs R.A., Green E.D., Hurles M.E., Knoppers B.M., Korbel J.O., Lander E.S., Lee C., Lehrach H., Mardis E.R., Marth G.T., McVean G.A., Nickerson D.A., Schmidt J.P., Sherry S.T., Wang J., Wilson R.K., Boerwinkle E., Doddapaneni H., Han Y., Korchina V., Kovar C., Lee S., Muzny D., Reid J.G., Zhu Y., Chang Y., Feng Q., Fang X., Guo X., Jian M., Jiang H., Jin X., Lan T., Li G., Li J., Li Yingrui, Liu S., Liu Xiao, Lu Y., Ma X., Tang M., Wang B., Wang G., Wu H., Wu R., Xu X., Yin Y., Zhang D., Zhang W., Zhao J., Zhao M., Zheng X., Gupta N., Gharani N., Toji L.H., Gerry N.P., Resch A.M., Barker J., Clarke L., Gil L., Hunt S.E., Kelman G., Kulesha E., Leinonen R., McLaren W.M., Radhakrishnan R., Roa A., Smirnov D., Smith R.E., Streeter I., Thormann A., Toneva I., Vaughan B., Zheng-Bradley X., Grocock R., Humphray S., James T., Kingsbury Z., Sudbrak R., Albrecht M.W., Amstislavskiy V.S., Borodina T.A., Lienhard M., Mertes F., Sultan M., Timmermann B., Yaspo M.L., Fulton L., Ananiev V., Belaia Z., Beloslyudtsev D., Bouk N., Chen C., Church D., Cohen R., Cook C., Garner J., Hefferon T., Kimelman M., Liu C., Lopez J., Meric P., O’Sullivan C., Ostapchuk Y., Phan L., Ponomarov S., Schneider V., Shekhtman E., Sirotkin K., Slotta D., Zhang H., Balasubramaniam S., Burton J., Danecek P., Keane T.M., Kolb-Kokocinski A., McCarthy S., Stalker J., Quail M., Davies C.J., Gollub J., Webster T., Wong B., Zhan Y., Campbell C.L., Kong Y., Marcketta A., Yu F., Antunes L., Bainbridge M., Sabo A., Huang Z., Coin L.J.M., Fang L., Li Q., Li Z., Lin H., Liu B., Luo R., Shao H., Xie Y., Ye C., Yu C., Zhang F., Zheng H., Zhu H., Alkan C., Dal E., Kahveci F., Garrison E.P., Kural D., Lee W.P., Leong W.F., Stromberg M., Ward A.N., Wu J., Zhang M., Daly M.J., DePristo M.A., Handsaker R.E., Banks E., Bhatia G., Del Angel G., Genovese G., Li H., Kashin S., McCarroll S.A., Nemesh J.C., Poplin R.E., Yoon S.C., Lihm J., Makarov V., Gottipati S., Keinan A., Rodriguez-Flores J.L., Rausch T., Fritz M.H., Stütz A.M., Beal K., Datta A., Herrero J., Ritchie G.R.S., Zerbino D., Sabeti P.C., Shlyakhter I., Schaffner S.F., Vitti J., Cooper D.N., Ball E.V., Stenson P.D., Barnes B., Bauer M., Cheetham R.K., Cox A., Eberle M., Kahn S., Murray L., Peden J., Shaw R., Kenny E.E., Batzer M.A., Konkel M.K., Walker J.A., MacArthur D.G., Lek M., Herwig R., Ding L., Koboldt D.C., Larson D., Ye Kai, Gravel S., Swaroop A., Chew E., Lappalainen T., Erlich Y., Gymrek M., Willems T.F., Simpson J.T., Shriver M.D., Rosenfeld J.A., Bustamante C.D., Montgomery S.B., De La Vega F.M., Byrnes J.K., Carroll A.W., DeGorter M.K., Lacroute P., Maples B.K., Martin A.R., Moreno-Estrada A., Shringarpure S.S., Zakharia F., Halperin E., Baran Y., Cerveira E., Hwang J., Malhotra A., Plewczynski D., Radew K., Romanovitch M., Zhang C., Hyland F.C.L., Craig D.W., Christoforides A., Homer N., Izatt T., Kurdoglu A.A., Sinari S.A., Squire K., Xiao C., Sebat J., Antaki D., Gujral M., Noor A., Ye Kenny, Burchard E.G., Hernandez R.D., Gignoux C.R., Haussler D., Katzman S.J., Kent W.J., Howie B., Ruiz-Linares A., Dermitzakis E.T., Devine S.E., Kang H.M., Kidd J.M., Blackwell T., Caron S., Chen W., Emery S., Fritsche L., Fuchsberger C., Jun G., Li B., Lyons R., Scheller C., Sidore C., Song S., Sliwerska E., Taliun D., Tan A., Welch R., Wing M.K., Zhan X., Awadalla P., Hodgkinson A., Li Yun, Shi X., Quitadamo A., Lunter G., Marchini J.L., Myers S., Churchhouse C., Delaneau O., Gupta-Hinch A., Kretzschmar W., Iqbal Z., Mathieson I., Menelaou A., Rimmer A., Xifara D.K., Oleksyk T.K., Fu Yunxin, Liu Xiaoming, Xiong M., Jorde L., Witherspoon D., Xing J., Browning B.L., Browning S.R., Hormozdiari F., Sudmant P.H., Khurana E., Tyler-Smith C., Albers C.A., Ayub Q., Chen Y., Colonna V., Jostins L., Walter K., Xue Y., Gerstein M.B., Abyzov A., Balasubramanian S., Chen J., Clarke D., Fu Yao, Harmanci A.O., Jin M., Lee D., Liu J., Mu X.J., Zhang J., Zhang Yan, Hartl C., Shakir K., Degenhardt J., Meiers S., Raeder B., Casale F.P., Stegle O., Lameijer E.W., Hall I., Bafna V., Michaelson J., Gardner E.J., Mills R.E., Dayama G., Chen K., Fan X., Chong Z., Chen T., Chaisson M.J., Huddleston J., Malig M., Nelson B.J., Parrish N.F., Blackburne B., Lindsay S.J., Ning Z., Zhang Yujun, Lam H., Sisu C., Challis D., Evani U.S., Lu J., Nagaswamy U., Yu J., Li W., Habegger L., Yu H., Cunningham F., Dunham I., Lage K., Jespersen J.B., Horn H., Kim D., Desalle R., Narechania A., Sayres M.A.W., Mendez F.L., Poznik G.D., Underhill P.A., Mittelman D., Banerjee R., Cerezo M., Fitzgerald T.W., Louzada S., Massaia A., Yang F., Kalra D., Hale W., Dan X., Barnes K.C., Beiswanger C., Cai H., Cao H., Henn B., Jones D., Kaye J.S., Kent A., Kerasidou A., Mathias R., Ossorio P.N., Parker M., Rotimi C.N., Royal C.D., Sandoval K., Su Y., Tian Z., Tishkoff S., Via M., Wang Y., Yang H., Yang L., Zhu J., Bodmer W., Bedoya G., Cai Z., Gao Y., Chu J., Peltonen L., Garcia-Montero A., Orfao A., Dutil J., Martinez-Cruzado J.C., Mathias R.A., Hennis A., Watson H., McKenzie C., Qadri F., LaRocque R., Deng X., Asogun D., Folarin O., Happi C., Omoniwa O., Stremlau M., Tariyal R., Jallow M., Joof F.S., Corrah T., Rockett K., Kwiatkowski D., Kooner J., Hien T.T., Dunstan S.J., ThuyHang N., Fonnie R., Garry R., Kanneh L., Moses L., Schieffelin J., Grant D.S., Gallo C., Poletti G., Saleheen D., Rasheed A., Brooks L.D., Felsenfeld A.L., McEwen J.E., Vaydylevich Y., Duncanson A., Dunn M., Schloss J.A. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Xu S., Pan F., Xu J. Study of the interleukin-1F7 gene single nucleotide polymorphism in patients with ankylosing spondylitis. Chin. J. Rheumatol. 2011;15:546–549. doi: 10.3760/cma.j.issn.1007-7480.2011.08.008. [DOI] [Google Scholar]
- Cirelli T., Nepomuceno R., Orrico S.R.P., Rossa C., Cirelli J.A., North K.E., Graff M., Barros S.P., Scarel-Caminaga R.M. Validation in a Brazilian population of gene markers of periodontitis previously investigated by GWAS and bioinformatic studies. J. Periodontol. 2021;92:689–703. doi: 10.1002/JPER.20-0126. [DOI] [PubMed] [Google Scholar]
- Conti P., Caraffa A., Gallenga C.E., Ross R., Kritas S.K., Frydas I., Younes A., Ronconi G. Coronavirus-19 (Sars-cov-2) induces acute severe lung inflammation via il-1 causing cytokine storm in covid-19: a promising inhibitory strategy. J. Biol. Regul. Homeost. Agents. 2020;34:1971–1975. doi: 10.23812/20-1-E. [DOI] [PubMed] [Google Scholar]
- Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darif D., Hammi I., Kihel A., El Idrissi Saik I., Guessous F., Akarid K. The pro-inflammatory cytokines in COVID-19 pathogenesis: what goes wrong? Microb. Pathog. 2021;153 doi: 10.1016/j.micpath.2021.104799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davarpanah E., Jafarzadeh A., Nemati M., Bassagh A., Abasi M.H., Khosravimashizi A., Kazemipoor N., Ghazizadeh M., Mirzaee M. Circulating concentration of interleukin-37 in Helicobacter pylori-infected patients with peptic ulcer: its association with IL-37 related gene polymorphisms and bacterial virulence factor CagA. Cytokine. 2020;126 doi: 10.1016/j.cyto.2019.154928. [DOI] [PubMed] [Google Scholar]
- Ding L., Hong X., Liu D. The role of Interleukin-37 in inflammation: suppression or pro-motion? J. Rheum. Dis. Treat. 2018;4:058. doi: 10.23937/2469-5726/1510058. [DOI] [Google Scholar]
- Forbester J.L., Humphreys I.R. Genetic influences on viral-induced cytokine responses in the lung. Mucosal Immunol. 2020;141(14):14–25. doi: 10.1038/s41385-020-00355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G., Cox H.C., Roach J.C. Whole-genome haplotyping approaches and genomic medicine. Genome Med. 2014;69(6):1–16. doi: 10.1186/S13073-014-0073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imran M.M., Ahmad U., Usman U., Ali M., Shaukat A., Gul N. Neutrophil/lymphocyte ratio—a marker of COVID-19 pneumonia severity. Int. J. Clin. Pract. 2021;75 doi: 10.1111/ijcp.13698. [DOI] [PubMed] [Google Scholar]
- Jia H., Liu J., Han B. Reviews of interleukin-37: functions, receptors, and roles in diseases. Biomed. Res. Int. 2018;2018 doi: 10.1155/2018/3058640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B., Cheng S., Peng J., Yan J., Zhang S. Interleukin-37 gene variants segregated anciently coexist during hominid evolution. Eur. J. Hum. Genet. 2015;2310(23):1392–1398. doi: 10.1038/ejhg.2014.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Ling Y., Song Z., Cheng X., Ding L., Jiang R., Fu W., Liu Y., Hu H., Yuan S., Chen J., Zhu C., Fan J., Wang J., Jin Y., Zhang M., Zhu L., Sun P., Zhang Linxia, Qin R., Zhang W., Qiu C., Shen Y., Zhang Lin, Shi Z., Zhao C., Zhu T., Lu H., Zhang X., Xu J. Correlation between early plasma interleukin 37 responses with low inflammatory cytokine levels and benign clinical outcomes in severe acute respiratory syndrome coronavirus 2 infection. J. Infect. Dis. 2021;223:568–580. doi: 10.1093/infdis/jiaa713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.X., Leonard W.J. Fine-tuning cytokine signals. Annu. Rev. Immunol. 2019;37:295–324. doi: 10.1146/annurev-immunol-042718-041447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X.Y., Guo X.J., He Y.Z., Hou S.F., Zhu H. Bin, Cheng Y., Nan Z. Association between interleukin 37 (rs3811047) polymorphism and multiple autoimmune diseases in a Chinese population. Med. (United States) 2018;97 doi: 10.1097/MD.0000000000010386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed Khosroshahi L., Rezaei N. Dysregulation of the immune response in coronavirus disease 2019. Cell Biol. Int. 2021;45:702–707. doi: 10.1002/cbin.11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mookherjee S., Banerjee D., Chakraborty S., Mukhopadhyay I., Sen A., Ray K. Evaluation of the IL1 gene cluster single nucleotide polymorphisms in primary open-angle glaucoma pathogenesis. Genet. Test. Mol. Biomark. 2016;20:633–636. doi: 10.1089/gtmb.2015.0344. [DOI] [PubMed] [Google Scholar]
- Offenbacher S., Jiao Y., Kim S.J., Marchesan J., Moss K.L., Jing L., Divaris K., Bencharit S., Agler C.S., Morelli T., Zhang S., Sun L., Seaman W.T., Cowley D., Barros S.P., Beck J.D., Munz M., Schaefer A.S., North K.E. GWAS for Interleukin-1β levels in gingival crevicular fluid identifies IL37 variants in periodontal inflammation. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-05940-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özgüçlü S., Duman T., Ateş F.S.Ö., Küçükşahin O., Çolak S., Ölmez Ü. Serum interleukin-37 level and interleukin-37 gene polymorphism in patients with Behçet disease. Clin. Rheumatol. 2019;38:495–502. doi: 10.1007/s10067-018-4288-7. [DOI] [PubMed] [Google Scholar]
- Quirk S., Agrawal D.K. Immunobiology of IL-37: mechanism of action and clinical perspectives. Expert. Rev. Clin. Immunol. 2014;10:1703–1709. doi: 10.1586/1744666X.2014.971014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashedi J., Poor B.M., Asgharzadeh V., Pourostadi M., Kafil H.S., Vegari A., Tayebi-Khosroshahi H., Asgharzadeh M. Risk factors for covid-19. Le Infez. Med. 2020;28:469–474. [PubMed] [Google Scholar]
- Shi Y.Y., He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- Tan H., Deng B., Yu H., Yang Y., Ding L., Zhang Q., Qin J., Kijlstra A., Chen R., Yang P. Genetic analysis of innate immunity in Behcet’s disease identifies an association with IL-37 and IL-18RAP. Sci. Rep. 2016;6:35802. doi: 10.1038/srep35802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannabouathong C., Devji T., Ekhtiari S., Chang Y., Phillips S.A., Zhu M., Chagla Z., Main C., Bhandari M. Novel coronavirus COVID-19 current evidence and evolving strategies. J. Bone Jt. Surg. Am. 2020;102:734–744. doi: 10.2106/JBJS.20.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xu K., Chen S., Li Y., Li M. Role of interleukin-37 in inflammatory and autoimmune diseases. Iran. J. Immunol. 2018;15:165–174. doi: 10.22034/IJI.2018.39386. [DOI] [PubMed] [Google Scholar]
- World Health Organization . WHO; 2020. Clinical Management of Severe Acute Respiratory Infection When Novel Coronavirus (2019-nCoV) Infection is Suspected: Interim Guidance 28 January 2020. [Google Scholar]
- Yan N., Meng S., Song R.H., Qin Q., Wang X., Yao Q., Jiang Y., Jiang W., Shi L., Xu J., Zhang J. Polymorphism of IL37 gene as a protective factor for autoimmune thyroid disease. J. Mol. Endocrinol. 2015;55:209–218. doi: 10.1530/JME-15-0144. [DOI] [PubMed] [Google Scholar]
- Zhang X.Y., Zuo Y., Li C., Tu X., Xu H.J., Guo J.P., Li Z.G., Mu R. IL1F7 gene polymorphism is not associated with rheumatoid arthritis susceptibility in the northern Chinese Han population: a case-control study. Chin. Med. J. 2018;131:171–179. doi: 10.4103/0366-6999.222340. [DOI] [PMC free article] [PubMed] [Google Scholar]