SUMMARY
Elevated levels of fetal globin protect against β-hemoglobinopathies, such as sickle cell disease and β-thalassemia. Two zinc-finger (ZF) repressors, BCL11A and ZBTB7A/LRF, bind directly to the fetal globin promoter elements positioned at −115 and −200, respectively. Here, we describe X-ray structures of the ZBTB7A DNA-binding domain, consisting of four adjacent ZFs, in complex with the −200 sequence element, which contains two copies of four consecutive C:G base pairs. ZF1 and ZF2 recognize the 5′ C:G quadruple, and ZF4 contacts the 3′ C:G quadruple. Natural non-coding DNA mutations associated with hereditary persistence of fetal hemoglobin (HPFH) impair ZBTB7A DNA binding, with the most severe disruptions resulting from mutations in the base pairs recognized by ZF1 and ZF2. Our results firmly establish ZBTB7A/LRF as a key molecular regulator of fetal globin expression and inform genome-editing strategies that inhibit repressor binding and boost fetal globin expression to treat hemoglobinopathies.
Graphical abstract
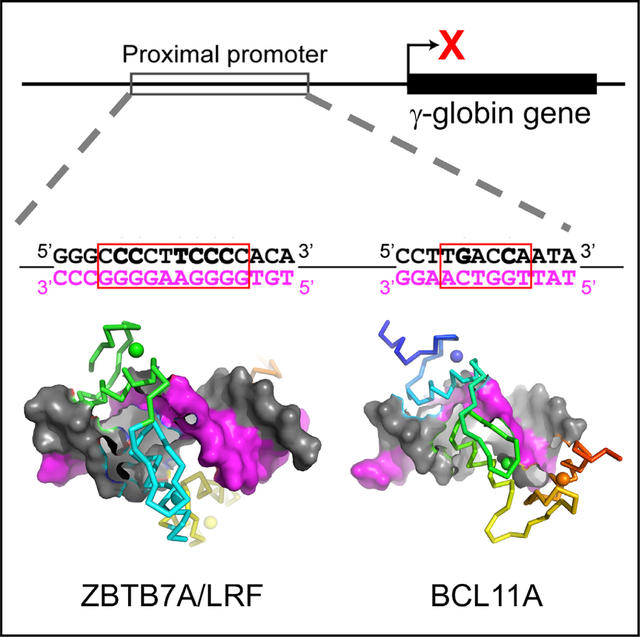
In brief
Yang et al. show that the transcription factor ZBTB7A has features that deviate from conventional one finger-three bases recognition. Among the four fingers, ZF1 and ZF2 each contact two DNA bases. ZF3 does not make base-specific contacts but serves as a spacer to position ZF4 into the right location.
INTRODUCTION
De-repression of fetal globin gene expression is a promising strategy for treating diseases caused by mutations in the adult globin genes, such as sickle cell disease and β-thalassemia. Accordingly, the mechanisms of fetal globin repression have been recently investigated (Masuda et al., 2016; Orkin and Bauer, 2019; Wienert et al., 2018). Two C2H2 zinc-finger (ZF) DNA-binding proteins, BCL11A and ZBTB7A (also known as leukemia/lymphoma-related factor [LRF]; Kukita et al., 1999; Liu et al., 2004), directly bind the fetal globin promoter at the −115 and −200 sites, respectively (Liu et al., 2018; Martyn et al., 2018). BCL11A binds a 5′-TGACCA-3′ element (spanning nucleotides −118 to −113) using its three C-terminal ZFs (Yang et al., 2019). ZBTB7A has an array of four ZFs and binds a 10-base-pair (bp) pyrimidine-rich site 5′-CCCCTTCCCC-3′ (or 3′-GGGGAAGGGG-5′ on the complementary strand), spanning nucleotides −203 to −194 (Martyn et al., 2018). At least seven naturally occurring mutations at six positions within the −200 element are associated with fetal globin derepression and the benign genetic condition of hereditary persistence of fetal hemoglobin (HPFH) (Wienert et al., 2017; Martyn et al., 2018, 2019).
Given the importance of understanding how naturally occurring HPFH mutations impair ZBTB7A binding, we used X-ray crystallography to investigate the detailed interactions of the ZBTB7A DNA-binding domain with the −200 element as well as biophysical methods to determine precisely how the known human mutations interfered with repressor binding. Together, our study provides a mechanism for ZBTB7A recognition of eight C:G base pairs within the −200 element and a framework for future work focused on, for instance, precise gene editing that can interfere with this repressive mechanism to alleviate fetal globin silencing and treat β-hemoglobinopathies (Weber et al., 2020).
RESULTS
ZF1 and ZF2 of ZBTB7A are important for binding DNA
The ZBTB7A gene encodes a protein with four ZFs toward its C terminus (Kukita et al., 1999). It is known to regulate gene expression with multiple cellular processes (Constantinou et al., 2019), but of relevance here is the consequence of it binding to the −200 element in the fetal globin promoter and repressing the fetal globin expression in adult erythroid cells (Figure 1A). In order to investigate which ZFs were required for binding to the −200 element, we first carried out an electrophoretic mobility shift assay (EMSA). Nuclear extracts were prepared from COS-7 cells transiently overexpressing a number of deletion constructs containing different ZFs and tested their binding to a probe encompassing the −200 element (Figures 1B and 1C). A construct containing ZF1–ZF4 bound strongly to the probe (lane 2 in Figure 1C). Removing ZF4 or both ZF3 and ZF4 did not eradicate binding (lanes 3 or 6 in Figure 1C). However, fragments without ZF1, ZF2–ZF4 (lanes 4 and 5 in Figure 1C) and ZF2 and ZF3 (lanes 7–9 in Figure 1C), strongly reduced binding, indicating that ZF1 is important for binding of the −200 element in vitro. Quantitative conclusions should not be made from these data because the expressed levels of all constructs are not identical (Figure 1B).
Figure 1. ZBTB7A DNA-binding domain.
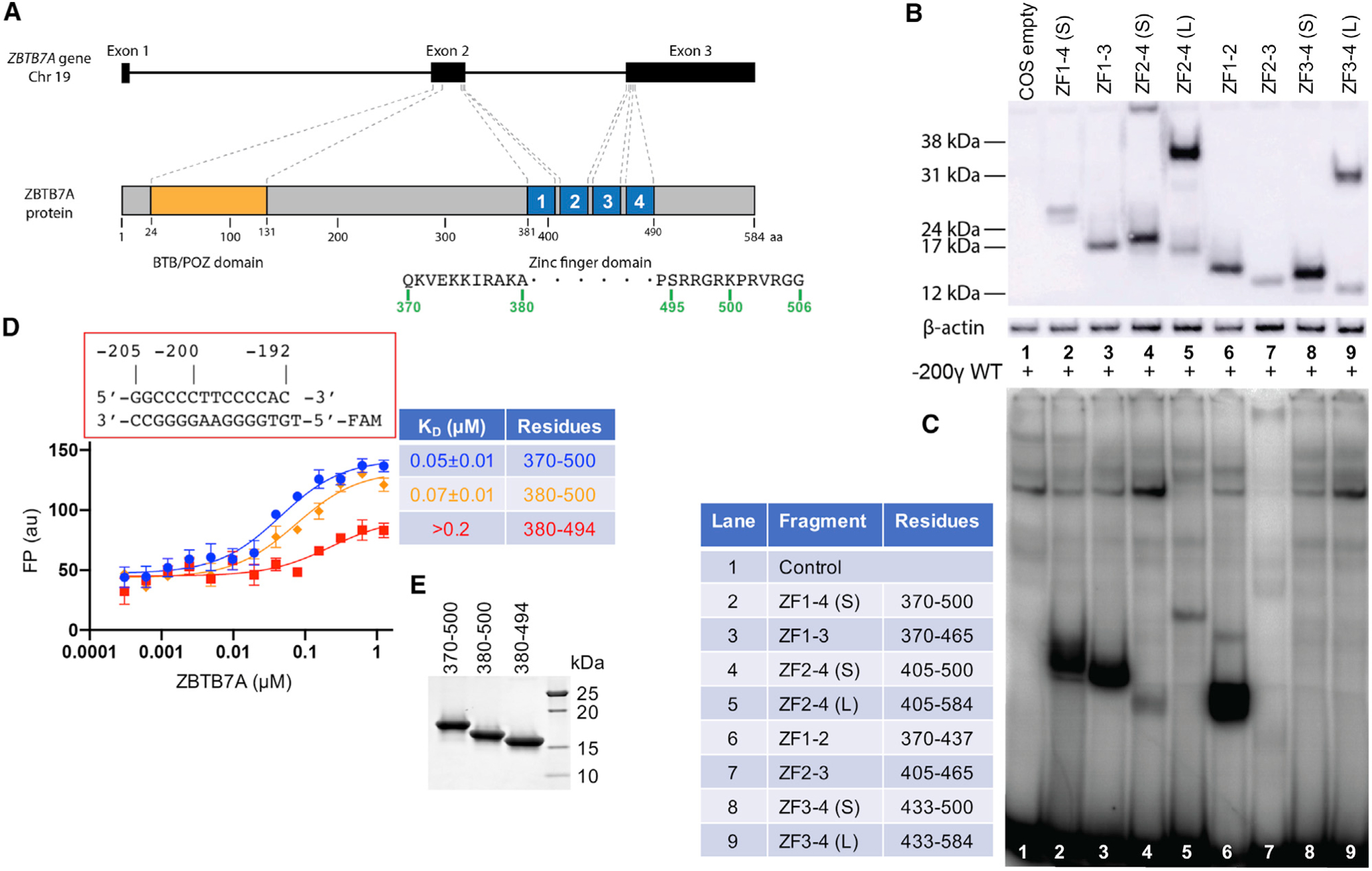
(A) Schematic diagram of ZBTB7A gene and protein.
(B) A western blot of the nuclear extracts prepared from COS-7 cells transiently expressing constructs containing different numbers of ZFs of ZBTB7A using an antibody against the FLAG tag.
(C) An EMSA assay of the nuclear extracts assessing binding to a radiolabeled DNA probe covering the −200 element (−200γ wild type [WT]).
(D) FP assay of ZBTB7A ZF fragments expressed in E. coli with and without N- and C-terminal additions. Data represent the mean ± SD of N number of independent determinations (N = 3). For the percentage of saturation, see Figure S2.
(E) A 4%–20% gradient gel with Coomassie blue staining showing the examples of purified recombinant proteins used in the FP assays.
In order to identify a suitable construct for X-ray crystallography, we generated three different human ZBTB7A constructs expressed in Escherichia coli: the first encompassed residues 380–494 and thus included the ZF DNA-binding domain, the next had six additional C-terminal residues (380–500), and the last had both N- and C-terminal additions (370–500). We compared the binding of the three constructs to a double-stranded oligonucleotide spanning positions −205 to −192 of the fetal globin gene promoter. Fluorescence polarization (FP) was used to measure the dissociation constants (KDs) of the three fragments with the oligo (Figure 1D–1E). The N-terminal ten residues contributed little to the binding affinities; the KD values (~50–70 nM) were approximately the same for the constructs with or without the N-terminal additional 10 residues (370–379). However, including the C-terminal six residues imparted ~3× stronger binding to DNA (from 0.2 μM to 70 nM). These binding data suggest that ZF4 and its associated extended C-terminal residues, although largely dispensable in EMSA assays, do contribute to the DNA binding. Thus, we used the constructs with the C terminus extending to residue 500 in the subsequent structural analyses.
General features of ZBTB7A DNA-binding domain
We crystallized the four-finger DNA-binding domain with two different 14-base-pair oligos derived from the −200 site (Figure 2A), the first contains base pairs −205 to −192 and the second −206 to −193. In both cases, we used a 5′ overhang at each end (either A and T or G and C) (Figures 2B and 2C). These complexes crystallized in space group P2221 (PDB: 7N5T) and P2 (PDB: 7N5S), and both structures were determined to a resolution of ~2.9 Å (Table S1). In the crystallographic asymmetric unit of both space groups, there was one ZBTB7A-DNA complex. The two structures were highly similar with root-mean-square deviations (RMSDs) of 0.7 Å between 108 pairs of Cα atoms. The largest difference between the two structures lay in the crystallographic packing interactions. In the P2221 space group, the two neighboring complexes were rotated 180° in respect to each other, forming tail to tail (or head to head) at the joint (Figure 2B). In the P2 space group, the two neighboring complexes were stacked head to tail at the joint (Figure 2C).
Figure 2. Structures of ZBTB7A DNA-binding domain in complex with DNA.
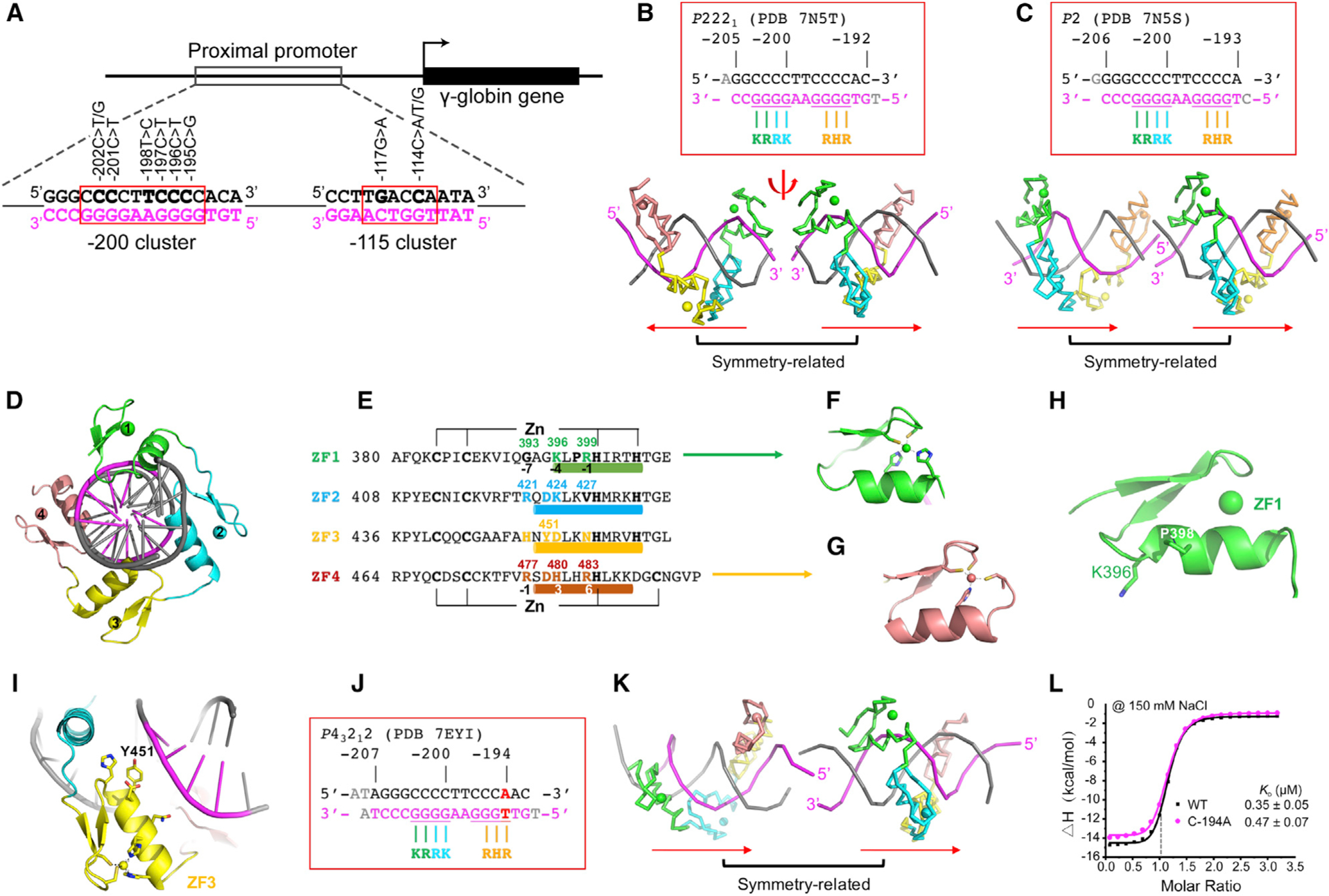
(A) Schematic diagram of the fetal globin promoter, with the known HPFH-associated mutations clustered at elements of −115 and −200.
(B and C) ZBTB7A-DNA complexes in space group P2221 (PDB: 7N5T) and space group P2 (PDB: 7N5S) (N = 2 of independent determined structures). Two symmetry-related complexes are shown.
(D) A view of ZBTB7A-DNA complex along the DNA axis.
(E) Sequence alignment of the four ZFs of ZBTB7A with DNA base-interacting positions −1, −4, and −7 shown below ZF1. The structure-based numbering scheme shown below ZF4 refers to the position immediately prior to the helix (−1) and positions within the helix (3 and 6).
(F) The Zn-coordinating residues C2H2 of ZF1.
(G) The Zn-coordinating residues C3H1 of ZF4.
(H) ZF1 has a shorter helix due to Pro398.
(I) ZF3 has a bulky Tyr451 blocking closer contact with DNA major groove.
(J and K) ZBTB7A in complex with a base pair (bp) substitution (C:G to A:T) at −194 position (complex H in PDB: 7EYI).
(L) ITC measurement of ZBTB7A against oligos with C:G (gray) or A:T (magenta) at −194 position (see Table S2 for derived ITC binding parameters). Each KD value is presented as fitted value ± SD against 19 data points for WT and the mutant.
There were four major differences between ZBTB7A and conventional C2H2 ZF proteins. In conventional C2H2 ZF proteins (Choo and Klug, 1997; Klug, 2010; Wolfe et al., 2000), each finger comprises two β strands and an α helix, with two His residues in the helix together with one Cys in each strand coordinating a Zn ion, forming a characteristic tetrahedral C2-Zn-H2 structural unit that confers rigidity to the finger (Figure 2D). This is true for the first three fingers of ZBTB7A (Figures 2E and 2F), but the fourth finger has an atypical CCHC (or C3H1) coordination (Figures 2E and 2G). What is usually the last His is here replaced by a Cys, and the inter-residue distance increases from three residues between the two His to five residues between the His and the Cys coordination. We note that the C3H1 Zn coordination has also been used in ZF11 of CTCF (see Figure S1 of Hashimoto et al. [2017]).
When bound to DNA, ZF1–ZF4 occupies the DNA major groove, with their α helices toward DNA and the strands facing outward (Figures 2B–2D). In conventional C2H2 ZF proteins, the amino acids occupying three key “canonical” positions of the helix, namely −1, 3, and 6, specify a DNA target sequence of three adjacent DNA base pairs, which is called the “triplet element.” This structure-based numbering scheme refers to the position immediately prior to the helix (−1) and positions within the helix (3 and 6). Interestingly, the conventional rule of one finger-three bases recognition was no longer valid for ZF1 of ZBTB7A, which had a shorter helix due to Pro398 (Figure 2H). We note a Pro also exists in the first finger of ZNF410, a transcription factor important in indirectly silencing the fetal-type β-globin genes in adult erythroid cells (Lan et al., 2021). Interestingly, substituting Glu446 with Pro (E446P) in KLF4 (Hashimoto et al., 2016) or Q369P and E427P in Wilms tumor protein WT1 (Hashimoto et al., 2014) increased their affinities for 5-methylcytosine DNA, respectively.
To reduce possible ambiguity, we used the first Zn-coordination His in each finger (which is almost always separated from the preceding Zn-binding Cys by 12 residues) as reference position 0, with residues prior to this, at sequence positions −1, −4, and −7, corresponding to the 6, 3, and −1 of the structure-based numbering (Liu et al., 2013). The new numbering scheme (residues at positions −1, −4, and −7) corresponds to Arg399, Lys396, and Gly393 of ZF1 (Figure 2E). Among them, Gly393—the smallest amino acid—was not involved in base-specific contact and ZF1 only recognized two bases. The one finger-two base recognition was also observed in ZF2, which has Arg421 at −7 position, Lys424 at −4 position, and Val427 at −1 position (Figure 2E). We note that variation of individual ZFs contacting 2, 3, or 4 bases has been observed previously in ZFP568 (Patel et al., 2018).
ZF3 has His, Asp, and Asn at base-interacting positions (−7, −4, and −1), and each of these residues could form base-specific interactions, e.g., His for recognition of guanine (Patel et al., 2016b), Asp for cytosine (Ren et al., 2018), and Asn for adenine (Luscombe et al., 2001). Interestingly, in the current structures, ZF3 was not involved in base-specific interactions, probably due to a bulky side chain of Tyr451 at the −5 position, which blocked the base-interacting residues from getting close to the major groove (Figure 2I), but served as a spacer to position the next finger ZF4 into the right location (see discussion section). In functional C2H2 ZF proteins, position −5 usually contains a small amino acid, e.g., Ala, Ser, or Thr (Patel et al., 2018; Patel et al., 2017). The manner in which the position −5 residue interacted with DNA differed from triplet to triplet. Nevertheless, most interacted with the non-recognition strand (here, the pyrimidine-rich top strand) and with the nucleotide immediately before the first base pair of the cognate triplet (for example, see below). In addition, the appearance of Asp at −5 position was often together with Arg at −7 position, forming an Arg-Asp pair as observed in ZF2 and ZF4 of ZBTB7A as well as in other ZFs, EGR1 (Pavletich and Pabo, 1991), KLF4 (Liu et al., 2014), and WT1 (Hashimoto et al., 2014).
The identities of the amino acids at the base-interacting positions (−7, −4, and −1) are the principle determinants of the DNA sequence recognized (Choo and Klug, 1997; Klug, 2010; Wolfe et al., 2000). Here, in ZF1, ZF2, and ZF4, four Arg, two Lys, and one His were found at these positions, and all were involved in DNA guanine recognition (Luscombe et al., 2001; Patel et al., 2016b; Vanamee et al., 2005). Indeed, ZF1 and ZF2 were responsible for the recognition of four guanines at positions −203 to −200. However, there are four guanines at positions −197 to −194 of the same (bottom) strand. Thus, the binding of ZBTB7A will involve the protein identifying the optimal register for its key fingers along the DNA. We did note that moderate-to-severe anisotropy along the DNA axis and high Wilson B-factors (77 Å2 for PDB: 7N5T and 96 Å2 for PDB: 5N5S) for the two data-sets of X-ray diffraction collected from the wild-type sequence covering the −200 element. The high Wilson B-factors indicate there is a significant disorder in the crystal (Evans, 2011; Wilson, 1942).
To help the protein register correctly, we designed another oligo with a C:G base pair at −194 position changed to A:T (Figures 2J and 2K). Our rationale was that the −194 position is just outside of known human mutations occurring at the −200 site, with all of them resulting in reduced DNA binding (see below). Indeed, isothermal titration calorimetry (ITC) measurement revealed nearly identical dissociation constants between wild type (KD = 0.35 μM for a C:G pair at −194) and the mutant (KD = 0.47 μM for an A:T pair at −194) (Figure 2L). The DNA-binding domain of ZBTB7A in complex with the mutant oligo was crystallized in space group P43212 with a higher resolution at 2.4 Å and a reduced Wilson B-factor (39 Å2) (PDB: 7EYI; Table S1). There were two protein-DNA complexes (complexes H and G) per crystallographic asymmetric unit (Figures S1A–S1C), and one of them (complex H) was stacked head to tail (Figure 2K). We discuss the differences between the two.
Base-specific interactions
The numbering used for nucleotides is shown in Figure 3A. Base pairs of the oligo are numbered based on the positions of the fetal globin promoter, with the purine-rich sequence as the “recognition” strand and the pyrimidine-rich sequence as the “top” strand. The four C:G base pairs at positions −203 to −200 were recognized primarily by hydrogen bonds (H-bonds) between the guanines and Lys396 and Arg399 of ZF1 and Arg421 and Lys424 of ZF2, respectively (Figures 3B–3E). Two terminal N atoms of Arg donate H-bonds to the guanine O6 and N7 atoms, a pattern specific to guanine. Many sequence-specific proteins recognize guanine in this same way as, for example, the dimeric SfiI endonuclease (recognition sequence: GGCCN5GGCC), where three of the four guanines in each half site form identical H-bonds with Arg and the fourth guanine H bonds with Lys in almost the same manner (Vanamee et al., 2005). Similar observations were made for BCL11A binding the −115 element where the two guanines at −115 and −114 positions are recognized by Lys784 and Arg787, respectively, of BCL11A (Yang et al., 2019). As noted above, Asp423 of ZF2 formed an intra-ZF charge-charge interaction with Arg421 (Figure 3D) and accepted a H-bond from the N4 group of cytosine at −202 on the top strand in the preceding base pair (Figure 3C).
Figure 3. Details of ZBTB7A-base interactions.
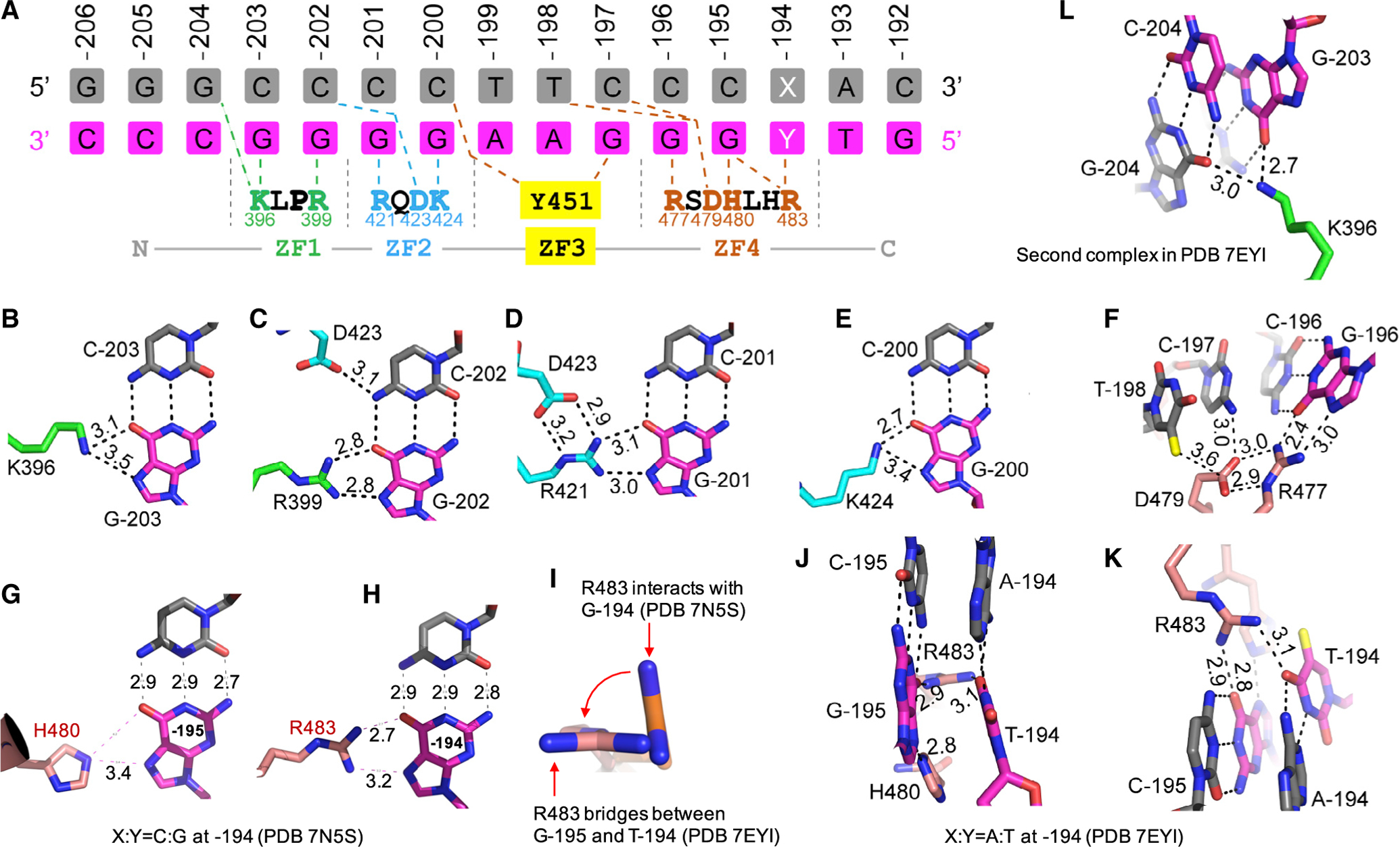
(A) General scheme of interactions between ZF1–ZF4 and DNA. The top three lines indicate base pairs of the fetal globin promoter positions from −206 to −192. The amino acids from N-to-C terminus (shown in the bottom) run antiparallel with the DNA recognition strand (magenta) from 3′ to 5′. X:Y = C:G or A:T
(B–K) Examples of base-specific contacts, taken from PDB: 7EYI or otherwise as indicated. Interatomic distances are measured in angstroms.
(B) Lys396 of ZF1 interacts with G at −203.
(C) Arg399 of ZF1 and Asp423 of ZF2 interact with C:G base pair at −202.
(D) Arg421 of ZF2 interacts with G at −201.
(E) Lys424 of ZF2 interacts with G at −200.
(F) Arg477 of ZF4 interacts with G at −196. Asp479 is in contact with preceding T (at −198) and C (at −197).
(G) His480 of ZF4 interacts with G at −195.
(H) Arg483 of ZF4 interacts with G at −194.
(I) Arg483 undergoes a rotation from C:G to A:T at −194.
(J and K) When C:G at −194 is substituted by A:T, Arg483 of ZF4 bridges between the two neighboring bases T (at −194) and G (at −195). For the corresponding electron density, see Figure S1H.
(L) In the second complex of PDB: 7EYI, Lys396 of ZF1 has a strand-cross bridge between two guanines, G at −203 and G at −204. For the corresponding electron density, see Figure S1I.
Among the 3′ end four C:G base pairs, only three of them at positions −196 to −194 are recognized, by Arg477, His480, and Arg483 of ZF4, respectively (Figures 3F–3H). Like Asp423 of ZF2 (Figures 3C and 3D), Asp479 formed an interaction with Arg477 and accepted a similar H-bond from cytosine at −197 position (Figure 3F). In addition, Asp479 formed a van der Waals interaction with the methyl group of thymine at −198 (Figure 3F). While the Asp479-methyl group interaction contributed to enhanced binding affinity, this interaction was non-specific, which made a variable interaction (e.g., comparing with the equivalent Asp423 of ZF2).
When the C:G base pair was substituted by an A:T at −194 position, Arg483 rotated its side-chain guanidine group ~90° (Figure 3I) to bridge between two oxygen atoms of thymine (at −194) and guanine (at −195), effectively occupying two base pairs (Figures 3J and 3K). Although Arg483 underwent a conformational change, the number of H-bonds that Arg483 engages remained the same, with two H-bonds with guanine at −194 (Figure 3H) or one each with guanine at −195 and thymine at −194 (Figures 3J and 3K), resulting in nearly identical binding strength (Figure 2L). This property of ZBTB7A can be traced to the ability of specific residues in each ZF unit to adopt alternative conformations, allowing it to establish versatile H-bonds with some bases. Examples of the versatile binding by C2H2 ZF include PRDM9 (Patel et al., 2016b) and CTCF (Hashimoto et al., 2017), where side-chain conformational switching allows identical ZF modules to recognize different sequences. In this regard, we also note Lys396 of ZF1 adopts altered different conformations between the two complexes (Figures 3B and 3L). In addition, the C-terminal residues beyond 495, which contributes to DNA binding (Figure 1D) possibly via non-specific DNA interactions, were apparently disordered in the current structures.
Effects of HPFH mutations on binding of ZBTB7A
We next considered the naturally occurring mutations at six promoter positions −202, −201, −198, −197, −196, and −195 (Figure 4). The mutation at position −198 has not been previously examined with respect to its effect on ZBTB7A binding, whereas some mutations at the other five positions had been evaluated in our previous study (Martyn et al., 2018). To be complete, we substituted each base pair at the six positions to the other three possible base pairs. We used two biophysical methods, EMSA and ITC, to assay the protein-DNA binding strengths. As one moves across the element, the first base pair mutation that had an impact was at position −203 and the last was at −195 (Figure 4A). The negative effect of naturally occurring human mutations at −202, −201, −197, −196, and −195 on ZBTB7A binding is evident (Figure 4A).
Figure 4. The effects of HPFH-associated mutations on ZBTB7A binding.
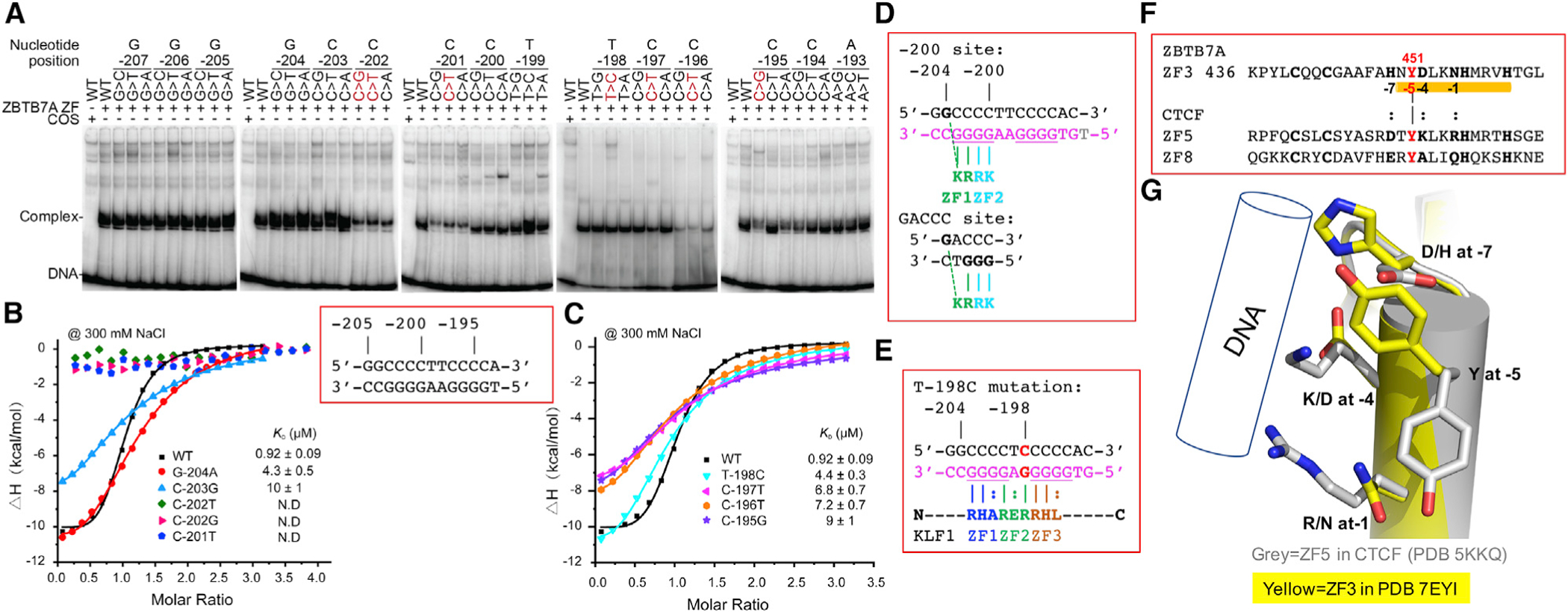
(A) An EMSA showing the effect on ZBTB7A (using a FLAG-tagged residues 370–500) binding to DNA of all possible single base pair changes across the −200 element (from −207 to −193) with naturally occurring HPFH mutations (shown in red). For quantification of shifted bands, see Figure S3.
(B and C) ITC evaluation of base pair substitution at (B) positions of −204 to −201 and (C) positions of −198 to −195 (see Table S2 for derived ITC binding parameters). Each KD value is presented as fitted value ± SD against 19 data points for every DNA variant.
(D) Alignment of −200 site of the fetal globin promoter and the consensus GACCC sequence.
(E) T:A-to-C:G substitution at −198 creates a KLF1 binding site. The base-interacting residues are indicated, and each finger of KLF1 accommodates a variable site (indicted by a colon).
(F) Sequence alignment of three fingers containing a Tyr at position −5, ZF3 of ZBTB7A and ZF5 and ZF8 of CTCF.
(G) Superimpositions of ZF5 of CTCF with ZF3 of ZBTB7A.
To assess the quantitative binding affinities, we used ITC and measured the KD values of ZBTB7A with mutated oligonucleotides. ZBTB7A displayed the weakest binding to mutations at positions −202 (C>T/G) and −201 (C>T); all three mutations abolished the interactions (Figure 4B). For comparison, under constant conditions, ZBTB7A demonstrated reduced binding to mutations at positions −203 and −195 (~10–11× reduced binding), followed by mutations at −196 and −197 (~7–8×) and mutations at −198 and −204 (~5×) (Figures 4B and 4C). The reductions in binding were the greatest with respect to interactions medicated by ZF1 (interacting with C:G at −202 and −203) and ZF2 (interacting with C:G at −201), followed by ZF4 (interacting with C:G at −195 and −196), and are largely consistent with the EMSA data (Figure 1C).
The two mutations that had the least impact (~5×) were G:C-to-A:T substitution at −204 and T:A-to-C:G substitution at −198, the latter being a naturally occurring mutation associated with HPFH. We note that in the second complex of the P43212 space group (PDB: 7EYI), the tip of side chain of Lys396 adopted a slightly different conformation with the Nε atom bridging between two guanines at −204 and −203 of the opposite strands (Figure 3L). The missing interaction with O6 atom of guanine at −204 could lower the binding due to G-to-A transition. Similarly, the loss of the van der Waals contact with the methyl group of thymine at −198 (Figure 3F) could be responsible for the reduced binding affinity of T-to-C substitution.
DISCUSSION
Here, we describe X-ray structures of ZBTB7A bound to a critical DNA element involved in repression of human fetal globin expression and detail how naturally occurring HPFH mutations disrupt molecular contacts. The extended 11-base pair interactions (ranging from −204 to −194), as expected for a tandem array of four fingers (based on estimation of the one finger-three bases rule), does not match the previously reported consensus sequences for ZBTB7A/LRF (Maeda et al., 2005; Masuda et al., 2016; Pessler and Hernandez, 2003), which is essentially GACCC. This discrepancy could be understood from two observations made in our study. First, as shown in EMSA (Figure 1C), the first two fingers, ZF1 and ZF2, are sufficient to achieve binding. Second, Lys396 of ZF1 is able to make a cross-strand interaction with guanine at −204 position (Figure 3L). Thus, GACCC could be viewed as a variation of GCCCC (from −204 to −200) (Figure 4D). The cross-strand Lys-guanine interaction has been previously observed in Lys328 of ZNF410 and Lys413 of KLF4 (Lan et al., 2021; Liu et al., 2014).
Here, we show how the key DNA bases implicated in HPFH disrupt specific molecular contacts and also investigate the effect of T-to-C HPFH mutation at −198 on ZBTB7A binding. This mutation caused a small but detectable reduction in the DNA-binding affinity, with an ~5× reduction observed in the calorimetry, although this small effect was not evident in the less quantitative EMSA assay (Figure 4A). Importantly, this mutation is known to create a de novo site for the strong activator KLF1 (Wienert et al., 2017). As shown in Figure 4E, the T:A-to-C:G mutation created the binding site for ZF2 Arg in KLF1. Like Arg-guanine interactions observed in ZF1 and ZF2 of ZBTB7A, the corresponding Arg-guanine interaction in KLF1 is a sequence-discriminatory contact (as illustrated in the case of KLF4, which shares the invariant DNA-binding domain with KLF1 [Liu et al., 2014]). This is in close analogy to the two highly recurrent, cancer-specific C-to-T mutations in the promoter of TERT (telomerase reverse transcriptase), creating an identical 11-base pair sequence (5′-CCCCT(C>T) CCGGG-3′) that contains an ETS binding site (Horn et al., 2013; Huang et al., 2013). Thus, our analysis presented here together with the X-ray data and calorimetry suggest that the −198 T-to-C mutation works through two cooperative mechanisms: it both weakens binding of the repressor ZBTB7A and creates a de novo site for the activator KLF1. In summary, all of the mutations that disrupt ZBTB7A binding are associated with significant derepression in fetal globin in HPFH individuals, but correlating the disruption of binding with total fetal globin readouts is not possible since the latter depends on multiple other factors, such as the status of the other globin allele, broader genetic background effects, and the method of measuring fetal globin levels.
As mentioned above, ZF3 makes little contact with the −200 site, but we observed a difference between the two complexes in the space group P43212 (PDB: 7EYI). Whereas complex H shares high similarity to the individual complex observed in PDB: 7N5S and PDB: 7N5T (RMSD < 1 Å in Figure S1D), ZF3 of complex G deviates from the other three complexes particularly in the conformation of Tyr451 (Figure S1E). Interestingly, a boric acid (used in the crystallization) was observed sitting in the DNA major groove between the protein and DNA interface in complex G (Figure S1F). The boric acid made two H-bonds with guanine at −197 position, mimicking the Arg guanidinium moiety in the interactions with guanine. In addition, the boric acid formed a weak H-bond with Asp452 of ZF3 and a van der Waals stacking contact with Arg477 of ZF4 (Figure S1G). We suggest that the boric-acid-binding site may provide a target location for designing inhibitors of small molecules that disrupt ZBTB7A-DNA binding—an interface historically viewed as “undruggable” (Bushweller, 2019).
We previously observed two individual ZFs, ZF5 and ZF8 in CTCF, possessing a Tyr at the corresponding position of Tyr451 of ZBTB7A (Figure 4F) (Hashimoto et al., 2017). Like ZF3 of ZBTB7A, ZF8 of CTCF functions as a spacer spanning the DNA minor groove (Hashimoto et al., 2017). However, ZF5 of CTCF is capable of interacting with a triplet of GGN (where N is a variable base). We superimposed the two Tyr-containing fingers ZF3 of ZBTB7A and ZF5 of CTCF (Figure 4G). It is evident that the Tyr takes an alternative conformation and base-interacting residues in CTCF (R, K, and D), Arg at −1 position and Lys at −4 position, are the longest side chains that can still reach to the guanine bases, while the shorter Asp at −7 position for the interaction with the variable base. The corresponding residues of ZBTB7A (N, D, and H), Asn at −1 and Asp at −4 positions, have shorter side chains to contact the base in the presence of Tyr451. The same reason might also apply to the lacking of specific interactions of ZF8 of CTCF, which has shorter and smaller side chains (E, A, and Q) at the base-interacting positions, but does increase non-specific DNA binding along the DNA phosphate backbone (Hashimoto et al., 2017). We note that ZBTB7A is also involved in tumorigenesis and metastasis (Singh et al., 2021, and references therein). It is likely ZBTB7A has other important but uncharacterized target genes at which ZF3 may make base-specific interactions.
Taken together with BCL11A-DNA interactions (Martyn et al., 2018; Yang et al., 2019), our results support the view that ZBTB7A and BCL11A are two essential and direct repressors of fetal globin expression (Masuda et al., 2016). The two repressors may use distinct mechanisms of interaction with the nucleosome-remodeling and deacetylase NuRD complex for local chromatin compaction. ZBTB7A uses its N-terminal BTB domain (also known as POZ domain) for interaction with NuRD components (Masuda et al., 2016), whereas BCL11A uses its histone H3 mimicry N-terminal tail for binding RBBP4, a common component of NuRD and PRC2 complexes (Moody et al., 2018). The detailed information of ZBTB7A binding across the −200 element explains the mechanism of action of specific naturally occurring mutations and also provides a framework for future work focused on the design of inhibitors disrupting protein-DNA binding (Bushweller, 2019) or gene-editing strategies that may specifically interfere with this repressive mechanism to alleviate fetal globin silencing and treat β-hemoglobinopathies (Weber et al., 2020).
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Requests for further information, resources, and reagents should be directed to and will be fulfilled by the Lead Contact, Xiaodong Cheng (xcheng5@mdanderson.org).
Materials availability
All plasmids and reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The X-ray structures (coordinates and structure factor files) of the ZBTB7A ZF domain with bound DNA have been deposited to PDB and are publicly available as of the date of publications. Accession numbers and DOI are listed in the key resource table as well as in Table S1.
The paper does not report original code.
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Human ZBTB7A residues 370–500 expressed in E. coil | This paper | pXC2222 |
| Human ZBTB7A residues 380–494 expressed in E. coli | This paper | pXC2238 |
| Human ZBTB7A residues 380–500 expressed in E. coli | This paper | pXC2243 |
| Human ZBTB7A residues 382–506 expressed in E. coli | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| ZF1-4 (residues 370–500) | This paper | 370–500 |
| ZF1-4 (residues 380–500) | This paper | 380–500 |
| ZF1-4 (residues 380–494) | This paper | 380–494 |
| ZF1-4 (residues 382–506) | This paper | 382–506 |
| Deposited data | ||
| ZF1-4 (residues 370–500)-DNA complex structure | This paper | PDB: 7N5S |
| https://doi.org/10.2210/pdb7N5T/pdb | ||
| ZF1-4 (residues 370–500)-DNA complex structure | This paper | PDB: 7N5T |
| https://doi.org/10.2210/pdb7N5T/pdb | ||
| ZF1-4 (residues 382–506)-DNA complex structure | This paper | PDB: 7EYI |
| https://doi.org/10.2210/pdb7EYI/pdb | ||
| Experimental models: Cell lines | ||
| COS-7 | Stuart H. Orkin Lab, Harvard | N/A |
| Oligonucleotides | ||
| Table S3 | This paper | N/A |
| Recombinant DNA | ||
| pcDNA3.1 | Thermo Fisher | Catalog # V79020 |
| pcDNA3-FLAG-ZBTB7A residues 370–437 | This paper | 370–437 = ZF1–2 |
| pcDNA3-FLAG-ZBTB7A residues 370–465 | This paper | 370–465 = ZF1–3 |
| pcDNA3-FLAG-ZBTB7A residues 370–500 | This paper | 370–500 = ZF1–4 (S) |
| pcDNA3-FLAG-ZBTB7A residues 405–465 | This paper | 405–465 = ZF2–3 |
| pcDNA3-FLAG-ZBTB7A residues 405–500 | This paper | 405–500 = ZF2–4 (S) |
| pcDNA3-FLAG-ZBTB7A residues 405–584 | This paper | 405–584 = ZF2–4 (L) |
| pcDNA3-FLAG-ZBTB7A residues 433–500 | This paper | 433–500 = ZF3–4 (S) |
| pcDNA3-FLAG-ZBTB7A residues 433–584 | This paper | 433–584 = ZF3–4 (L) |
| Software and algorithms | ||
| HKL2000 | Otwinowski et al., 2003 | https://hkl-xray.com |
| PHENIX | Terwilliger et al., 2009 | https://phenix-online.org |
| SHELX | Fu, Z.-Q, 2007 | http://shelx.uni-goettingen.de/ |
| CCP4 | Collaborative Computational Project, Number 4, 1994 | https://www.ccp4.ac.uk |
| COOT | Emsley and Cowtan, 2004 | https://strucbio.biologie.uni-konstanz.de/ccp4wiki/index.php/Coot |
| PHENIX.Refine | Headd et al., 2012 | https://phenix-online.org |
| PyMOL | DeLano Scientific LLC | https://www.schrodinger.com/products/pymol |
| Graph-pad prim (version 8.0) | GraphPad Software | https://www.graphpad.com |
| MicroCal PEAQ-ITC | Malvern Panalytical | https://www.malvernpanalytical.com/en/products/product-range/microcal-range/microcal-itc-range/microcal-peaq-itc |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Nuclear extracts were prepared from COS-7 cells transiently overexpressing the various ZBTB7A constructs that encoded the various ZF regions and an N-terminal FLAG tag expressed via pcDNA3. Recombinant ZBTB7A fragments were purified from expression in Escherichia coli strain BL21(DE3).
METHOD DETAILS
Protein expression and Purification
The C2H2 zinc finger coding region of human ZBTB7A (NCBI Reference Sequence: NP_001304919.1; residues 382–506) with an additional tryptophan at the end of C terminus was amplified by PCR from the bone marrow cDNA library, then cloned into a modified pGEX-4T1 vector (GE Healthcare) with a TEV protease cleavage site after the GST tag (pGEX-4T1-TEV), further verified by DNA sequencing.
The protein was expressed in Escherichia coli BL21 (DE3) cells (Novagen) that were cultured at 37°C in LeMaster and Richards minimal medium (LR medium), supplemented with 0.1 mM ZnSO4. When OD600 reached to 1.0, the cultured medium was shifted to 16°C and induced with 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 20 h. The cells were resuspended in the 1M NaCl Tris buffer (20 mM Tris-HCl, pH 7.5, and 1 M NaCl) supplemented with 1 tablet/1 mL protease inhibitor cocktail (Roche) and DNAaseI (20U; Thermo Scientific), and lysed by high-pressure cell disrupter. The supernatant was incubated with glutathione Sepharose (GE Healthcare) for 6 h and eluted with 30 mM reduced glutathione in the 1M NaCl buffer, then cleaved in the 0.4M NaCl buffer by TEV protease overnight at 4°C to remove GST tag. The tag cleaved proteins were then purified using size-exclusion chromatography on a HiLoad 16/60 Superdex 75 column (GE Healthcare) in the 1M NaCl buffer. The purified proteins were dialyzed and concentrated in 0.15M NaCl or 0.3M NaCl buffer for respectively subsequent crystallization or ITC experiments.
In parallel, the fragments of human ZBTB7A (residues 370–500, pXC2222; residues 380–494, pXC2238; and residues 380–500, pXC2243) were cloned into pGEX-6P-1 vector with a GST fusion tag. All three plasmids were transformed into Escherichia coli strain BL21-Codon-plus (DE3)-RIL. Bacteria was grown in LB broth at 37°C until reaching the log phase (OD600nm between 0.4 and 0.5), the shake temperature was set to 16°C and 25 μM ZnCl2 was added to the cell culture. When the shake temperature reached 16°C and OD600nm reached ~0.8, addition of 0.2 mM IPTG induced the expression for 20 h at 16°C. Cell harvesting and protein purification were carried out at 4°C through a three-column chromatography protocol, conducted in a BIO-RAD NGC™ system. Cells were collected by centrifugation and pellet was suspended in the lysis buffer consisting of 20 mM Tris-HCl pH 7.5, 5% glycerol, 0.5 mM tris(2-carboxyethl)phosphine (TCEP), supplemented with 0.5 M NaCl and 25 μM ZnCl2. Cells were lysed by sonication and 0.3% (w/v) polyethylenimine was slowly titrated into the cell lysate before centrifugation (Patel et al., 2016a). The debris was removed by centrifugation for 30 min at 47,000 g and the supernatant was loaded onto a 5 mL GSTrap column (GE Healthcare). The resin was washed by the lysis buffer and bound protein was eluted in 100 mM Tris-HCl pH 8.0, 0.5 M NaCl, 5% glycerol, 0.5 mM TCEP and 20 mM glutathione (reduced form). The proteins were digested with PreScission protease (produced in-house) to remove the GST fusion tag. The cleaved proteins were loaded onto a 5 mL Heparin column (GE Healthcare). The proteins were eluted by NaCl gradient from 0.25 to 1M in the lysis buffer. The peak fractions were pooled, concentrated and loaded onto a HiLoad 16/60 Superdex S200 column (GE Healthcare) equilibrated with 0.25M NaCl supplemented lysis buffer. The proteins were frozen and stored at −80°C.
Binding assays of protein-nucleic acids interaction
We used three binding assays to characterize the binding affinity of ZBTB7A to oligodeoxynucleotides: (i) EMSA, (ii) fluorescence polarization (FP) (Moerke, 2009; Patel et al., 2016a) and (iii) isothermal titration calorimetry (ITC) (Gontier et al., 2021).
The EMSA experiments were carried out as previously described (Crossley et al., 1996). Briefly, the sense oligonucleotide 5′-TGG GGG CCC CTT CCC CAC ACT AT-3′ was labeled with [γ−32P] triphosphate using T4 polynucleotide kinase by incubating at 37°C for 30 min then boiling for 1 min at 100°C. The unlabeled antisense strand was annealed via slow cooling from 100°C to room temperature. The radiolabeled probes were purified using the Quick Spin Columns for radiolabeled DNA Purification Kit. Nuclear extracts were prepared from COS-7 cells transiently overexpressing constructs that encoded the various ZF regions of ZBTB7A and an N-terminal FLAG tag expressed from pcDNA3. A pcDNA3 “empty” COS-7 lane was used as a negative control to show binding of any endogenous background proteins to the probe. Each lane contained 5 μL of nuclear extract (approximately 5 μg of protein) and 1 μL of radiolabeled probe. Samples were incubated for 10 min at 4°C before being loaded on a 6% native polyacrylamide gel in 0.5X TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA). Gels were run at 250 V for 1 h 45 min at 4°C and dried using a Bio-Rad vacuum heated gel dryer (80°C for 30–45 min). Gels were visualized using the Typhoon FLA 9500 Laser Scanner following overnight exposure to a Fujifilm BAS Cassette 2025 phosphor screen.
The FP experiments were used to measure the binding affinity with Synergy 4 Microplate Reader (BioTek). Aliquots (5 nM) of 6-carboxy-fluorescein (FAM)-labeled DNA duplex (FAM-5′-TGT GGG GAA GGG GCC-3′ and 3′-CAC CCC TTC CCC GG-5′) was incubated in 20 μL with varied amount of proteins (0 to 2.5 μM) in 20 mM Tris-HCl pH 7.5, 225 mM NaCl, 5% glycerol and 0.5 mM TCEP for 10 min at room temperature. The mixture was transferred to a black opaque 384-well plate before measurement. The data were processed using Graph-pad prim (version 8.0) with equation [mP] = [maximum mP] × [C] / (KD +[C]) + [baseline mP], in which mP is milli-polarization and [C] is protein concentration. The KD value for each protein-DNA interaction was derived from three replicated experiments.
The ITC experiments were performed on a Microcal PEAQ-ITC instrument (Malvern) at 20°C. HPLC grade single-stranded oligonucleotides in powder form was dissolved to 100 μM with 0.15 M NaCl or 0.3 M NaCl in 20 mM Tris-HCl, mixed with equal molar and incubated at 95°C for 5 min in a metal bath and slowly cooling to room temperature. The concentration of proteins (300–400 μM) and DNA oligos (to 20 μM) in the same buffer were determined spectrophotometrically. The double-stranded oligonucleotides were diluted to the target concentration and were loaded with 200 μL into sample cell, whereas the proteins were loaded with 40 μL into syringe. The titration protocol was the same for all the measurements, which were composed of a single initial injection of 1 μL protein, followed by 19 injections of 2 μL protein into DNA samples, the intervals between injections was set to 150 s and a reference power is 5 μcal s−1. Curves fitting to a single-site binding model were performed by MicroCal PEAQ-ITC Analysis Software provided by the manufacturer and summarized in Table S2.
Crystallography
The protein-DNA complex was prepared by mixing the purified fragment (residues 382–506) with the double-stranded oligonucleotides (5′-ATA GGG CCC CTT CCC AAC-3′ and 3′-ATC CCG GGG AAG GGT TGT-5′ with underlined substitution of C:G to A:T) in a 1:1.2 molar ratio, followed by dialysis in 20 mM Tris-HCl pH 7.5, 150 mM NaCl overnight. The complex was further purified via size-exclusion chromatography on a HiLoad 16/60 Superdex 75 column and concentrated to ~1.2 mM and 0.5 mM TCEP was added prior to crystallization. The crystals were grown at 293 K via the hanging-drop vapor diffusion method by mixing 1 μL protein-DNA complex and 1 μL reservoir buffer composed of 0.1 M malonate, imidazole, and boric acid (produced by mixing sodium malonate, imidazole, and boric acid in the molar ratio of 2:3:3), pH 7.0, 25% (w/v) polyethylene glycol (PEG) 1500. The crystals were soaked in cryo-protector made of mother liquor supplemented with 25% (v/v) glycerol before being flash-frozen in liquid nitrogen.
X-ray diffraction data of the crystal (PDB 7EYI) was collected on beamline 19U1 at the Shanghai Synchrotron Radiation Facility (SSRF) at a wavelength of 0.979 Å. The diffraction dataset was processed and scaled using the HKL-2000 (Otwinowski et al., 2003). The crystallographic phasing was performed by using the AutoSolve module in the PHENIX (Terwilliger et al., 2009), with the zinc atom sites being found by the SHELX C/D program (Fu et al., 2007). The initial protein model was built using the Buccaneer program in the CCP4 suite (Collaborative Computational Project, Number 4, 1994). Next, the DNA model was further built using Coot (Emsley and Cowtan, 2004), and refinement was performed using the PHENIX.Refine program (Headd et al., 2012). The Phenix.composite_omit_map program (Headd et al., 2012) was used to generate 2Fo-Fc composite omit electron density maps. All structures in the figures were generated with PyMOL (DeLano Scientific LLC).
In parallel, the ZBTB7A fragment (residues 370–500; 0.9 mM) was mixed with oligonucleotides (annealed in 10 mM Tris-HCl pH 7.5, 50 mM NaCl) with molar ratio 1:1.2 of protein to DNA on ice for 30 min incubation. An Art Robbins Gryphon Crystallization Robot was used to set up screens of the sitting drop at ~19°C via vapor diffusion. The complex crystals (PDB 7N5T) with oligonucleotides (5′-AGG CCC CTT CCC CAC-3′ and 3′-CCG GGG AAG GGG TGT-5′) were obtained under the condition of 0.2 M sodium bromide, 0.1 M Tris pH 7.5, 25% PEG 3350. The complex crystals (PDB 7N5S) with oligonucleotides (5′-GGG GCC CCT TCC CCA-3′ and 3′-CCC GGG GAA GGG GTC-5′) were grown under the condition of 0.2 M trimethylamine N-oxide dihydrate, 0.1 M Tris pH 8.5, 20% (w/v) PEG monomethyl ether 2,000.
Crystals were flash frozen using 20% (v/v) ethylene glycol as the cryo-protector. The X-ray diffraction data were collected at SER-CAT 22-ID beamline of the Advanced Photon Source at Argonne National Laboratory utilizing X-ray beams at 1.0 Å wavelength and processed by HKL2000. Molecular replacement was performed with the PHENIX PHASER program utilizing the two-finger module (ZF1 and ZF2) of PDB 7N5U as the search model and further model building commenced with COOT. The initial electron density showed recognizable molecular features of the β sheets and α helices of the zinc fingers and helical DNA. Manual building proceeded in COOT to build protein and the DNA duplex. COOT was also utilized for corrections between PHENIX refinement rounds. In all three structures, the C-terminal residues beyond amino acid 491 were disordered.
QUANTIFICATION AND STATISTICAL ANALYSIS
X-ray crystallographic data were measured quantitatively and processed with HKL2000. Structure refinements were performed by PHENIX Refine with 5% randomly chosen reflections for validation by R-free values. The data collection and refinement statistics are shown in Table S1. Structure quality was analyzed during rounds of PHENIX refinements and validated by the PDB server. Statistics details on FP and ITC experiments can be found in legends of Figures 1D, 2L, 4B and 4C, and Table S2.
Supplementary Material
Highlights.
ZBTB7A DNA-binding domain recognizes two stretches of G-rich sequence
HPFH-associated regulatory DNA mutations disrupt ZBTB7A-DNA interactions
ZF1 contains a Pro resulting in a shorter recognition helix
ZF4 has C3H1, instead of C2H2, Zn coordination
ACKNOWLEDGMENTS
We thank Ms. Yu Cao of MDACC for technical assistance and Dr. Xing Zhang of MDACC for discussion throughout. We thank the beamline scientists of the BL19U1 at the National Center for Protein Science Shanghai and Shanghai Synchrotron Radiation Facility for assistance during data collection. We thank the beamline scientists of Southeast Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source (APS), Argonne National Laboratory. SER-CAT is supported by its member institutions and equipment grants (S10_RR25528, S10_RR028976, and S10_OD027000) from the National Institutes of Health. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31-109-Eng-38. The work presented here were independently performed and supported in three laboratories. The work in the Shi laboratory was supported by the Ministry of Science and Technology of the China (2016YFA0500700 and 2019YFA0508403), National Natural Science Foundation of China (32090040, 31870760, and U1932122), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB39000000). Y.Y. was supported by China Postdoctoral Science Foundation (2021M690148). The work in the Crossley laboratory was supported by Australian National Health and Medical Research Council (APP1164920). K.G.R.Q. was a UNSW Sydney Scientia Associate Professor. L.C.L. was supported by an Australian Government Research Training Program (RTP) scholarship. The work in the Cheng laboratory was supported by National Institutes of Health (R35GM134744) and Cancer Prevention and Research Institute of Texas (RR160029). X.C. is a CPRIT Scholar in Cancer Research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109759.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Bushweller JH (2019). Targeting transcription factors in cancer - from undruggable to reality. Nat. Rev. Cancer 19, 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Y, and Klug A (1997). Physical basis of a protein-DNA recognition code. Curr. Opin. Struct. Biol 7, 117–125. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Constantinou C, Spella M, Chondrou V, Patrinos GP, Papachatzopoulou A, and Sgourou A (2019). The multi-faceted functioning portrait of LRF/ZBTB7A. Hum. Genomics 13, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, and Orkin SH (1996). Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol 16, 1695–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Evans PR (2011). An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr 67, 282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z-Q, Chrzes J, Sheldrick GM, Rose J, and Wang B-C (2007). A parallel program using SHELXD for quick heavy-atom partial structureal solution on high-performance computers. J. Appl. Crystallogr 40, 387–390. [Google Scholar]
- Gontier A, Varela PF, Nemoz C, Ropars V, Aumont-Nicaise M, Desmadril M, and Charbonnier JB (2021). Measurements of Protein-DNA Complexes Interactions by Isothermal Titration Calorimetry (ITC) and Microscale Thermophoresis (MST). Methods Mol. Biol 2247, 125–143. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Olanrewaju YO, Zheng Y, Wilson GG, Zhang X, and Cheng X (2014). Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev 28, 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Wang D, Steves AN, Jin P, Blumenthal RM, Zhang X, and Cheng X (2016). Distinctive Klf4 mutants determine preference for DNA methylation status. Nucleic Acids Res 44, 10177–10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Wang D, Horton JR, Zhang X, Corces VG, and Cheng X (2017). Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA. Mol. Cell 66, 711–720.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headd JJ, Echols N, Afonine PV, Grosse-Kunstleve RW, Chen VB, Moriarty NW, Richardson DC, Richardson JS, and Adams PD (2012). Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Crystallogr. D Biol. Crystallogr 68, 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. (2013). TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961. [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, and Garraway LA (2013). Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A (2010). The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem 79, 213–231. [DOI] [PubMed] [Google Scholar]
- Kukita A, Kukita T, Ouchida M, Maeda H, Yatsuki H, and Kohashi O (1999). Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood 94, 1987–1997. [PubMed] [Google Scholar]
- Lan X, Ren R, Feng R, Ly LC, Lan Y, Zhang Z, Aboreden N, Qin K, Horton JR, Grevet JD, et al. (2021). ZNF410 Uniquely Activates the NuRD Component CHD4 to Silence Fetal Hemoglobin Expression. Mol. Cell 81, 239–254.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CJ, Prazak L, Fajardo M, Yu S, Tyagi N, and Di Cesare PE (2004). Leukemia/lymphoma-related factor, a POZ domain-containing transcriptional repressor, interacts with histone deacetylase-1 and inhibits cartilage oligomeric matrix protein gene expression and chondrogenesis. J. Biol. Chem 279, 47081–47091. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang X, Blumenthal RM, and Cheng X (2013). A common mode of recognition for methylated CpG. Trends Biochem. Sci 38, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Olanrewaju YO, Zheng Y, Hashimoto H, Blumenthal RM, Zhang X, and Cheng X (2014). Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res 42, 4859–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Hargreaves VV, Zhu Q, Kurland JV, Hong J, Kim W, Sher F, Macias-Trevino C, Rogers JM, Kurita R, et al. (2018). Direct Promoter Repression by BCL11A Controls the Fetal to Adult Hemoglobin Switch. Cell 173, 430–442.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, and Thornton JM (2001). Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res 29, 2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, Teruya-Feldstein J, and Pandolfi PP (2005). Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature 433, 278–285. [DOI] [PubMed] [Google Scholar]
- Martyn GE, Wienert B, Yang L, Shah M, Norton LJ, Burdach J, Kurita R, Nakamura Y, Pearson RCM, Funnell APW, et al. (2018). Natural regulatory mutations elevate the fetal globin gene via disruption of BCL11A or ZBTB7A binding. Nat. Genet 50, 498–503. [DOI] [PubMed] [Google Scholar]
- Martyn GE, Wienert B, Kurita R, Nakamura Y, Quinlan KGR, and Crossley M (2019). A natural regulatory mutation in the proximal promoter elevates fetal globin expression by creating a de novo GATA1 site. Blood 133, 852–856. [DOI] [PubMed] [Google Scholar]
- Masuda T, Wang X, Maeda M, Canver MC, Sher F, Funnell AP, Fisher C, Suciu M, Martyn GE, Norton LJ, et al. (2016). Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science 351, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ (2009). Fluorescence Polarization (FP) Assays for Monitoring Peptide-Protein or Nucleic Acid-Protein Binding. Curr. Protoc. Chem. Biol 1, 1–15. [DOI] [PubMed] [Google Scholar]
- Moody RR, Lo MC, Meagher JL, Lin CC, Stevers NO, Tinsley SL, Jung I, Matvekas A, Stuckey JA, and Sun D (2018). Probing the interaction between the histone methyltransferase/deacetylase subunit RBBP4/7 and the transcription factor BCL11A in epigenetic complexes. J. Biol. Chem 293, 2125–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, and Bauer DE (2019). Emerging Genetic Therapy for Sickle Cell Disease. Annu. Rev. Med 70, 257–271. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Borek D, Majewski W, and Minor W (2003). Multiparametric scaling of diffraction intensities. Acta Crystallogr. A 59, 228–234. [DOI] [PubMed] [Google Scholar]
- Patel A, Hashimoto H, Zhang X, and Cheng X (2016a). Characterization of How DNA Modifications Affect DNA Binding by C2H2 Zinc Finger Proteins. Methods Enzymol 573, 387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Horton JR, Wilson GG, Zhang X, and Cheng X (2016b). Structural basis for human PRDM9 action at recombination hot spots. Genes Dev 30, 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Zhang X, Blumenthal RM, and Cheng X (2017). Structural basis of human PR/SET domain 9 (PRDM9) allele C-specific recognition of its cognate DNA sequence. J. Biol. Chem 292, 15994–16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Yang P, Tinkham M, Pradhan M, Sun MA, Wang Y, Hoang D, Wolf G, Horton JR, Zhang X, et al. (2018). DNA conformation induces adaptable binding by tandem zinc finger proteins. Cell 173, 221–233.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, and Pabo CO (1991). Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science 252, 809–817. [DOI] [PubMed] [Google Scholar]
- Pessler F, and Hernandez N (2003). Flexible DNA binding of the BTB/POZ-domain protein FBI-1. J. Biol. Chem 278, 29327–29335. [DOI] [PubMed] [Google Scholar]
- Ren R, Horton JR, Zhang X, Blumenthal RM, and Cheng X (2018). Detecting and interpreting DNA methylation marks. Curr. Opin. Struct. Biol 53, 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Verma S, Kushwaha PP, Prajapati KS, Shuaib M, Kumar S, and Gupta S (2021). Role of ZBTB7A zinc finger in tumorigenesis and metastasis. Mol. Biol. Rep 48, 4703–4719. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, and Hung LW (2009). Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr 65, 582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanamee ES, Viadiu H, Kucera R, Dorner L, Picone S, Schildkraut I, and Aggarwal AK (2005). A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA. EMBO J 24, 4198–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber L, Frati G, Felix T, Hardouin G, Casini A, Wollenschlaeger C, Meneghini V, Masson C, De Cian A, Chalumeau A, et al. (2020). Editing a γ-globin repressor binding site restores fetal hemoglobin synthesis and corrects the sickle cell disease phenotype. Sci. Adv 6, eaay9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienert B, Martyn GE, Kurita R, Nakamura Y, Quinlan KGR, and Crossley M (2017). KLF1 drives the expression of fetal hemoglobin in British HPFH. Blood 130, 803–807. [DOI] [PubMed] [Google Scholar]
- Wienert B, Martyn GE, Funnell APW, Quinlan KGR, and Crossley M (2018). Wake-up Sleepy Gene: Reactivating Fetal Globin for β-Hemoglobinopathies. Trends Genet 34, 927–940. [DOI] [PubMed] [Google Scholar]
- Wilson AJC (1942). Determination of Absolute from Relative X-Ray Intensity Data. Nature 150, 151–152. [Google Scholar]
- Wolfe SA, Nekludova L, and Pabo CO (2000). DNA recognition by Cy-s2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct 29, 183–212. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xu Z, He C, Zhang B, Shi Y, and Li F (2019). Structural insights into the recognition of γ-globin gene promoter by BCL11A. Cell Res 29, 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The X-ray structures (coordinates and structure factor files) of the ZBTB7A ZF domain with bound DNA have been deposited to PDB and are publicly available as of the date of publications. Accession numbers and DOI are listed in the key resource table as well as in Table S1.
The paper does not report original code.
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Human ZBTB7A residues 370–500 expressed in E. coil | This paper | pXC2222 |
| Human ZBTB7A residues 380–494 expressed in E. coli | This paper | pXC2238 |
| Human ZBTB7A residues 380–500 expressed in E. coli | This paper | pXC2243 |
| Human ZBTB7A residues 382–506 expressed in E. coli | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| ZF1-4 (residues 370–500) | This paper | 370–500 |
| ZF1-4 (residues 380–500) | This paper | 380–500 |
| ZF1-4 (residues 380–494) | This paper | 380–494 |
| ZF1-4 (residues 382–506) | This paper | 382–506 |
| Deposited data | ||
| ZF1-4 (residues 370–500)-DNA complex structure | This paper | PDB: 7N5S |
| https://doi.org/10.2210/pdb7N5T/pdb | ||
| ZF1-4 (residues 370–500)-DNA complex structure | This paper | PDB: 7N5T |
| https://doi.org/10.2210/pdb7N5T/pdb | ||
| ZF1-4 (residues 382–506)-DNA complex structure | This paper | PDB: 7EYI |
| https://doi.org/10.2210/pdb7EYI/pdb | ||
| Experimental models: Cell lines | ||
| COS-7 | Stuart H. Orkin Lab, Harvard | N/A |
| Oligonucleotides | ||
| Table S3 | This paper | N/A |
| Recombinant DNA | ||
| pcDNA3.1 | Thermo Fisher | Catalog # V79020 |
| pcDNA3-FLAG-ZBTB7A residues 370–437 | This paper | 370–437 = ZF1–2 |
| pcDNA3-FLAG-ZBTB7A residues 370–465 | This paper | 370–465 = ZF1–3 |
| pcDNA3-FLAG-ZBTB7A residues 370–500 | This paper | 370–500 = ZF1–4 (S) |
| pcDNA3-FLAG-ZBTB7A residues 405–465 | This paper | 405–465 = ZF2–3 |
| pcDNA3-FLAG-ZBTB7A residues 405–500 | This paper | 405–500 = ZF2–4 (S) |
| pcDNA3-FLAG-ZBTB7A residues 405–584 | This paper | 405–584 = ZF2–4 (L) |
| pcDNA3-FLAG-ZBTB7A residues 433–500 | This paper | 433–500 = ZF3–4 (S) |
| pcDNA3-FLAG-ZBTB7A residues 433–584 | This paper | 433–584 = ZF3–4 (L) |
| Software and algorithms | ||
| HKL2000 | Otwinowski et al., 2003 | https://hkl-xray.com |
| PHENIX | Terwilliger et al., 2009 | https://phenix-online.org |
| SHELX | Fu, Z.-Q, 2007 | http://shelx.uni-goettingen.de/ |
| CCP4 | Collaborative Computational Project, Number 4, 1994 | https://www.ccp4.ac.uk |
| COOT | Emsley and Cowtan, 2004 | https://strucbio.biologie.uni-konstanz.de/ccp4wiki/index.php/Coot |
| PHENIX.Refine | Headd et al., 2012 | https://phenix-online.org |
| PyMOL | DeLano Scientific LLC | https://www.schrodinger.com/products/pymol |
| Graph-pad prim (version 8.0) | GraphPad Software | https://www.graphpad.com |
| MicroCal PEAQ-ITC | Malvern Panalytical | https://www.malvernpanalytical.com/en/products/product-range/microcal-range/microcal-itc-range/microcal-peaq-itc |


