Abstract
To explore how the immune system controls clearance of SARS-CoV-2, we used a single-cell, mass cytometry–based proteomics platform to profile the immune systems of 21 patients who had recovered from SARS-CoV-2 infection without need for admission to an intensive care unit or for mechanical ventilation. We focused on receptors involved in interactions between immune cells and virus-infected cells. We found that the diversity of receptor repertoires on natural killer (NK) cells was negatively correlated with the viral clearance rate. In addition, NK subsets expressing the receptor DNAM1 were increased in patients who more rapidly recovered from infection. Ex vivo functional studies revealed that NK subpopulations with high DNAM1 expression had cytolytic activities in response to target cell stimulation. We also found that SARS-CoV-2 infection induced the expression of CD155 and nectin-4, ligands of DNAM1 and its paired coinhibitory receptor TIGIT, which counterbalanced the cytolytic activities of NK cells. Collectively, our results link the cytolytic immune responses of NK cells to the clearance of SARS-CoV-2 and show that the DNAM1 pathway modulates host-pathogen interactions during SARS-CoV-2 infection.
Keywords: COVID-19
Keywords: NK cells
Introduction
The COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread globally since it first appeared in Wuhan, China. As of September 8, 2021, there were over 221 million reported cases of COVID-19, and the death toll is over 4.5 million worldwide according to the WHO Coronavirus (COVID-19) Dashboard (https://covid19.who.int/; accessed September 8, 2021). Although most of those infected with SARS-CoV-2 are either asymptomatic or have mild symptoms, a significant number of cases progress into acute respiratory distress syndrome with diffuse alveolar damage (1–3). The immune system is critical in the progression of the disease. The innate immune response is activated in the early phase of the response to infections with SARS-CoV-2, Middle East respiratory syndrome virus, and influenza virus. This response is characterized by the secretion of proinflammatory cytokines, especially type I interferon, and the recruitment of innate immune cells such as neutrophils and natural killer (NK) cells (4–6). After this initial response from the innate immune system, virus-specific cytolytic T cells and antibodies are generated, and viral antigens are recognized by these adaptive immune system components (7, 8). Without coordinated responses from the immune system, effective control of the infection is not achieved, and immunopathogenesis, such as cytokine storms, can occur, contributing to multi-organ failure and eventually fatality in severe cases of COVID-19 (8). Although distinct immune signatures have been identified that correlate with the severity of COVID-19, little is known about the immune features relevant to the viral clearance, especially in mild to moderate cases of COVID-19 (9). A better understanding of the interactions between the virus and the host immune system will guide the identification of therapeutic targets and the design of effective viral control strategies.
Several studies have profiled the composition of immune cells in peripheral blood from patients at different stages and with different severities of COVID-19 (8, 10–16). It has been consistently found that lower proportions of lymphocytes and higher neutrophil-to-lymphocyte ratios are correlated with disease severity (10, 15, 17). In addition, systematic immune perturbations have been observed in patients with severe COVID-19 (10–13, 15, 18). Analyses of bronchoalveolar lavage samples from COVID-19 patients revealed that NK cells are enriched at the sites of infection, suggesting that these cells interact with virus-infected host cells (19–21). NK cells are innate immune cells that play critical roles in antiviral and antitumor responses. Unlike cytolytic T cells, which utilize recombined antigen-specific receptors to detect infected or transformed cells, NK cells employ an array of germline-encoded activating and inhibitory surface receptors to recognize virus-infected cells and tumor cells (22). The balance between signaling from activating and inhibitory receptors determines the responses of NK cells (22). Interestingly, the existence of paired activating and inhibitory receptors, such as DNAM1 and TIGIT, which share the same ligands but transduce opposite signals upon binding, suggests that the delicate tuning of NK activities might be achieved by combinations of receptors (23, 24). Owing to recent advances in single-cell technologies, including mass cytometry, the unprecedented heterogeneity of the NK receptor repertoire has been revealed (25). The diversity of the NK receptor repertoire is correlated with susceptibility to viral infection and is affected by previous viral infections (26, 27).
In this study, we used a single-cell, mass cytometry–based proteomics platform to profile the immunophenotypes of NK cells from convalescent COVID-19 patients in depth, with a focus on the relationship between NK-related immune features and viral load. Our results showed that there is a negative correlation between the diversity of the NK receptor repertoire and the viral clearance rate. In addition, patients with a higher abundance of NK subsets expressing DNAM1 and its paired coinhibitory receptor TIGIT had faster viral clearance than did patients with low levels of these receptors. We also demonstrated the cytolytically skewed character of the NK subset that expresses higher levels of DNAM1 and TIGIT. Finally, our data indicate that CD155 and nectin-4, ligands that preferentially bind to DNAM1 paired receptors, are upregulated after SARS-CoV-2 infection and could modulate NK cell interactions with virally infected cells.
Results
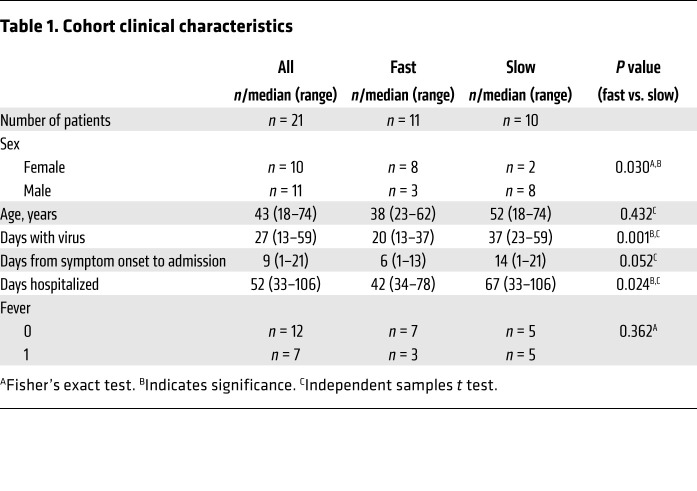
Plasma and peripheral blood mononuclear cells (PBMCs) were collected from healthy donors (n = 12) and convalescent patients (n = 21) who had recovered from SARS-CoV-2 infection without admission to an intensive care unit or mechanical ventilation; all had been confirmed positive for SARS-CoV-2 by RT-PCR. The demographics and clinical characteristics, killer cell immunoglobulin-like receptors, and HLA genotypes of the subjects separated into groups with fast and slow viral clearance are listed in Table 1 and Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/JCI146408DS1). Interestingly, there were significantly more females than males in the fast clearance group, which is consistent with the previous observation that men with COVID-19 are more at risk for worse outcomes (28).
Table 1. Cohort clinical characteristics.
To identify immune features associated with viral clearance, we designed an in-depth phenotypic analysis using mass cytometry (CyTOF) with a staining panel composed of antibodies against lineage markers that define distinct immune cells and receptors known to mediate cell-cell interactions between immune cells and virus-infected cells, particularly focusing on receptors known to be expressed on NK cells (Supplemental Table 2). The rate of viral clearance was defined by the slope of the linear regression curve of the viral loads from serial nasal swabs from each patient (Supplemental Figure 1A). It is important to emphasize that the persistence of low levels of viral RNA might not be associated with the presence of infectious virus but instead may indicate that the immune response is unable to promote viral RNA clearance (29, 30). In total, we measured 2.9 million live CD45+ cells with an average of 88,692 cells per sample.
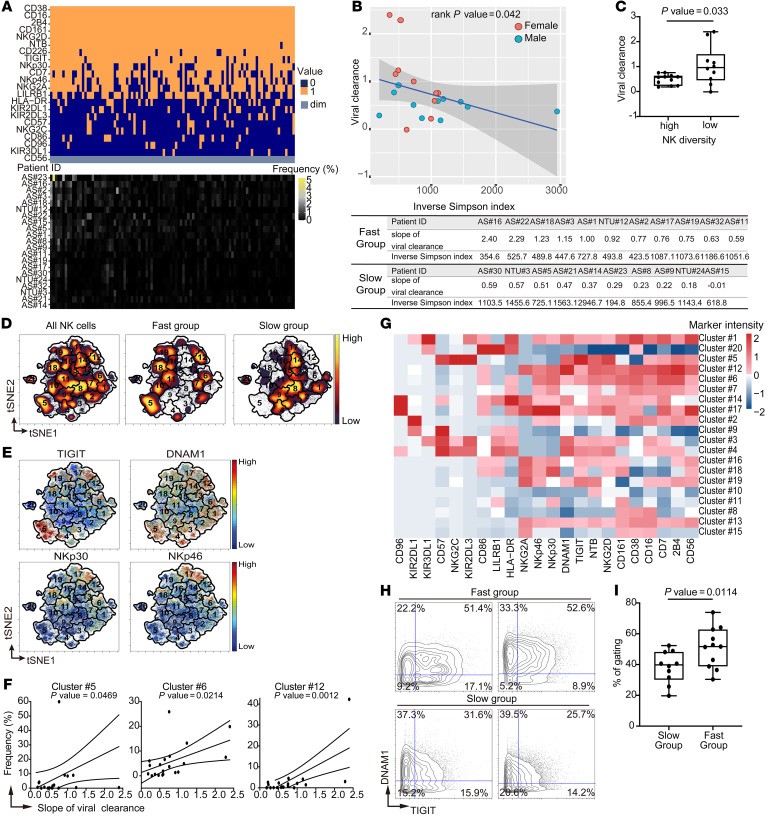
To investigate the relationship between the NK cell compartment and SARS-CoV-2 clearance, we first analyzed the NK receptor repertoire using a Boolean gating strategy described previously (25, 27). All NK cells were classified as positive or negative for the expression of each of 22 NK cell receptors. This allowed us to define 222 (or 4,194,304) NK subsets based on the unique single-cell combinations for individual patients (Figure 1A). In line with previous reports, there was a high degree of both inter- and intra-individual diversity in the NK receptor repertoire (25, 27). We detected 39,435 unique NK immunophenotypic combinations from all patients in the cohort, and no single immunophenotype accounted for more than 5% of the total NK cells in any individual. To quantify the heterogeneity of NK cells, we utilized a previously described measurement, the inverse Simpson index, as the indicator of the NK repertoire diversity (27). We observed a negative coefficient between the inverse Simpson index and the clearance rate (Figure 1B; rank P = 0.042), which indicates that more diversity in NK repertoire is correlated with a slower SARS-CoV-2 clearance rate. After adjusting for age and sex, there was a significantly higher viral clearance rate in the low-NK-diversity patients compared with high-NK-diversity patients (Figure 1C; P = 0.033).
Figure 1. SARS-CoV-2 clearance rate is associated with features of NK cells.
(A) Upper: Frequencies of top 100 marker combinations from Boolean gating over the 21 patient samples. Each column represents a specific marker combination. Orange, dark blue, and light gray indicate positive (1: marker intensity higher than the threshold [CD56dim]) and negative (0: marker intensity lower than the threshold [CD56dim]) for the indicated markers and CD56dim, respectively. Lower: Frequencies of the indicated marker combinations for each patient. (B) Upper: Viral clearance rate plotted versus the inverse Simpson index of receptor repertoire diversity. Circles, individual patients. Blue, male. Red, female. Blue line, linear regression; gray shading, 95% confidence interval (rank P = 0.042). Lower: Table of slopes and inverse Simpson indices for each patient. (C) Box-and-whisker plot of slopes for each patient in high (n = 11) and low (n = 10) NK diversity groups. Maximums, 75th, 50th, 25th percentiles, and minimums are 0.757, 0.633, 0.573, 0.229, and 0.184 for the high group and 2.40, 1.50, 0.96, 0.451, and –0.012 for the low group. P = 0.033 by linear regression of the binary NK diversity coefficient. (D) Abundances of NK cells with indicated characteristics visualized in tSNE maps color coded by cell density (yellow, high; purple, low). The number indicates the cluster ID of each FlowSOM metacluster. (E) Expression levels of indicated markers visualized in tSNE maps. (F) Frequencies of indicated clusters in individual patients versus the viral clearance (P = 0.0469, 0.0214, and 0.0012, for clusters 5, 6, and 12, respectively). (G) Heatmap of median arcsinh-transformed intensities of indicated markers in each metacluster. (H) Biaxial contour plots of TIGIT and DNAM1 from patients in fast (n = 11) and slow (n = 10) clearance groups. The numbers indicate the percentage of cells in each quadrant. (I) Box-and-whisker plot of frequencies of DNAM1hiTIGIThi cells in patients from slow and fast groups. Maximums, 75th, 50th, 25th percentiles, and minimums are 52.3, 48.28, 39.80, 30.13, and 19.70 for the slow group and 73.90, 62.80, 51.70, 38.80, and 30.40 for the fast group. P = 0.0114 by 1-tailed, unpaired Student’s t test.
To further investigate whether specific NK subsets correlate with more rapid SARS-CoV-2 clearance, we applied t-distributed stochastic neighbor embedding (tSNE) analysis to compare the immunophenotypic differences between patients with faster and slower viral clearance rates. There is a clear separation of NK cell subsets between the 2 groups of patients in the tSNE map (Figure 1D and Supplemental Figure 1B). Specifically, NK subsets expressing higher levels of activating receptors NKp46, NKp30, and DNAM1 and the paired coinhibitory receptor of DNAM1, TIGIT, were enriched in patients with faster viral clearance (Figure 1, D and E, and Supplemental Figure 2). We next performed cluster analysis on all NK cells using FlowSOM (31) and identified the clusters with abundance correlated with the viral clearance rate. Three clusters, 5, 6, and 12, were significantly positively correlated with viral clearance rate (Figure 1F and Supplemental Figure 3A). All 3 of these clusters are composed of cells that have high levels of expression of DNAM1 and TIGIT, and 2 (clusters 6 and 12) are characterized by high levels of NKp46 and NKp30 (Figure 1G). Interestingly, cluster 12 is also characterized by high levels of NKG2A (Figure 1G). This cluster might contain educated NK cells that have enhanced effector functions (32). Furthermore, these specific NK subsets were recapitulated using conventional manual gating on only DNAM1 and TIGIT (slow vs. fast: 39.80% vs. 51.70%, P = 0.0114; Figure 1, H and I) or NKp30 and NKp46 (slow vs. fast: 31.70% vs. 41.80%, P = 0.0461; Supplemental Figure 3, B and C), indicating the robustness of the analysis.
To rule out the possibility that the observed differential expression is due to technical batch effects, we barcoded samples before pooling and staining simultaneously. Immunophenotypic features specific to the fast (DNAM1hiTIGIThi) and slow (DNAM1loTIGITlo) groups were observed in the same batches of staining, demonstrating that the group-specific immune features were not due to batch effects (Supplemental Figure 3D). Also, the observed immunophenotypic features were validated with fluorescent flow cytometry (Supplemental Figure 3E). Thus, the results of this comprehensive high-dimensional immune profiling of the NK cell compartment revealed unique NK immune features that correlate with the SARS-CoV-2 clearance rate.
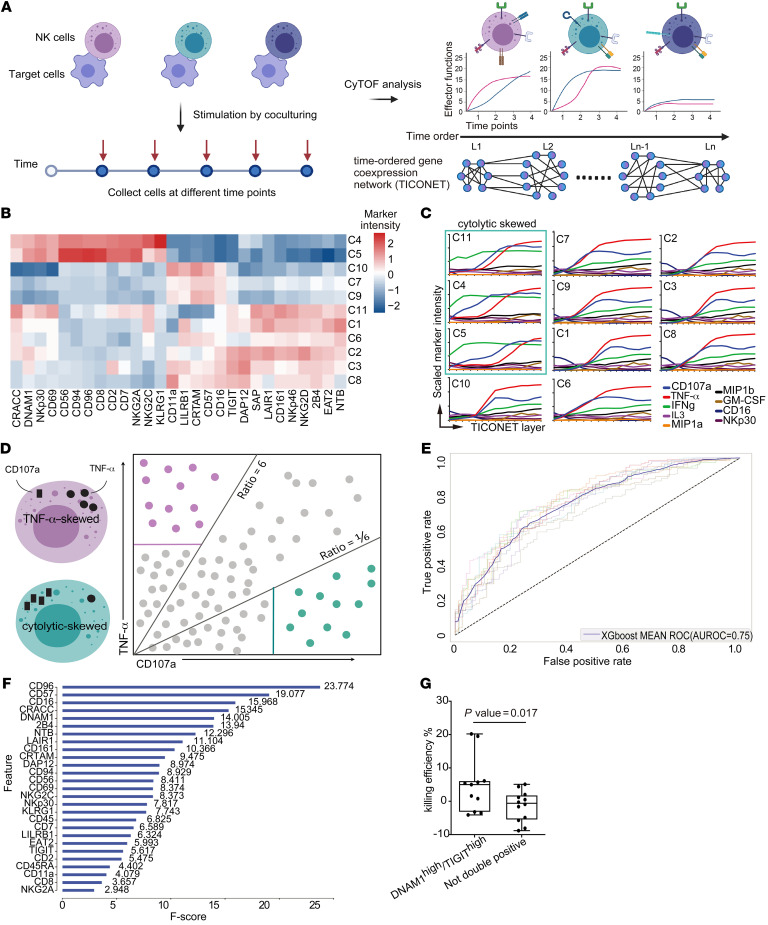
Given the association of distinct NK subsets with viral clearance rate, we evaluated the functional characteristics of different NK subsets in response to the target cell stimulation. We hypothesized that NK cells in the faster viral clearance group not only have a unique cell-surface receptor repertoire but also have distinct functional capabilities. We reasoned that time-resolved progression analysis would detect transitions resulting from activation due to the dynamic changes of NK receptors when target cells are encountered. We utilized the EBV-transformed cell line 721.221 as the model system (33). The 721.221 cells were cocultured with IL-2–primed or unprimed primary NK cells from healthy donors, samples were collected every hour over 9 hours, and surface immunophenotypes and intracellular cytokine and cytotoxic responses were quantified in the samples using mass cytometry (Figure 2A and Supplemental Table 3). First, Pearson’s correlations were calculated between individual markers from the single-cell mass cytometry data sets to determine the coexpression patterns of NK cell immunophenotypes and effector functions (Supplemental Figure 4A). As expected, the expression of NKG2A was strongly correlated with expression of its coreceptor CD94 but negatively correlated with the expression of the maturation marker CD57. In line with previous reports, strong positive correlations were detected between MIP1a and MIP1b, and among IFN-γ, TNF-α, and CD107a (34). In contrast, activating receptors, such as CD16 and NKG2D, which are known to become downregulated after activation, showed a negative association with the functional effectors, including IFN-γ, TNF-α, and CD107a (35, 36).
Figure 2. NK cell subsets have distinct kinetics and effector functions in response to target cell stimulation.
(A) Schematic diagram of the experiment. Primary NK cells were cocultured with 721.221 cells, and collected at different time points for immunophenotyping and intracellular functional effector staining followed by mass cytometry (CyTOF) analysis. The resultant data set was utilized to build the TICONET. The levels (L1 to Ln) in the TICONET represent the time order of coexpressed gene sets. Each TICONET starts from a cluster of 0-hour time point cells, labeled L1. Cells in the next level are the most similar to the cells in the previous level. (B) Heatmap of arcsinh-transformed marker intensities of protein expression from the 0-hour time point NK cells in clusters C1 to C11. (C) Line graphs of arcsinh-transformed intensities of the indicated markers from NK cells in each TICONET. The cytolytically skewed clusters are designated by the green box. (D) Schematic illustration of the strategy of machine learning. Purple dots represent the expression profiles of TNF-α–skewed NK cells, and green dots represent the cytolytically skewed NK cells. Purple and solid green lines indicate TNF-α– and CD107a-positive thresholds, respectively. (E) AUROC curves of XGBoost models during 10-fold cross validation. The solid curve represents the average curve of 10 ROC curves. (F) Markers in order of feature importance in the final XGBoost model trained on the entirety of selected data. F scores are indicated. (G) Box-and-whisker plot of quantification of the killing efficiency of SARS-CoV-2–infected cells by DNAM1hiTIGIThi cells (n = 11) and as not-double-positive cells (n = 12) in 2 independent experiments. Maximums, 75th, 50th, 25th percentiles, and minimums are 20.20, 6.10, 5.00, –3.20, and –4.10 for DNAM1hiTIGIThi NK cells and 5.10, 1.80, –0.60, –5.50, and –8.80 for the not-double-positive NK cells. P = 0.017 by 1-tailed, unpaired Student’s t test.
To further dissect the kinetics of NK cell responses to target cell stimulation, the time-course single-cell data sets were analyzed using TICONET, a time-ordered coexpression network analysis (37), to trace the dynamic trajectories of individual NK subsets from the beginning to the end of coculturing (Figure 2A). Briefly, cells in each NK subpopulation from the 0-hour time point were defined as the original state of the NK subsets. A cell coexpression network was built by connecting cells with highly similar expression profiles. The breadth-first search algorithm was then applied to assign the time-ordered layer for all cells in the coexpression network. By tracing the trajectories from each original state to its fully activated state, the entire path composed of multiple transitional states was obtained for each NK subset; these paths cannot be revealed by analysis of cross-sectional data sets. To demonstrate the validity of TICONET in time-course mass cytometry data analysis, we analyzed a published data set that includes serial time point single-cell mass cytometry profiles from mouse embryonic stem cells differentiating into the 3 germ layers: endoderm, mesoderm, and ectoderm (38). The expression levels of individual markers from the dynamic trajectories ordered by TICONET showed strong correlations with their expression at the actual time points (r = 0.8, 0.69, 0.94, and 0.73 for combined 3 cell types, ectoderm, endoderm, and mesoderm, respectively; Supplemental Figure 4B).
We then applied TICONET to examine the coexpression networks of the kinetics of NK cell responses to target cell stimulation. Although factors such as CD107a, IFN-γ, and TNF-α were often coexpressed in fully activated NK cells, how coexpression was reached differed. For example, some NK subsets (clusters C4, C5, and C11) showed early and high expression of cytolytic degranulation marker CD107a and then expressed TNF-α, whereas CD57+ NK subsets first secreted TNF-α and subsequently CD107a (Figure 2, B and C). Interestingly, all cytolytically skewed clusters expressed higher levels of DNAM1 and NKp30 than other clusters, which is similar to the immunophenotypes of the clusters positively correlated with viral clearance rate. In addition, without IL-2 priming, DNAM1hi and NKp30hi clusters were identified and showed early and high cytolytic activities upon target cell stimulation (Supplemental Figure 4, C and D).
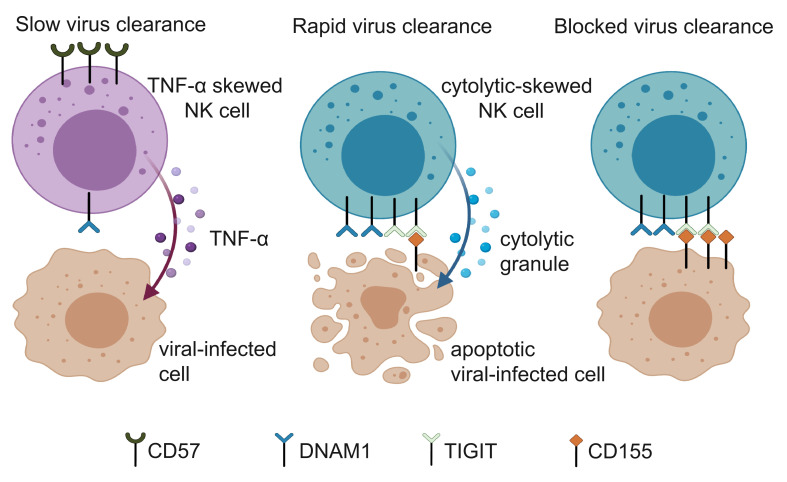
Consistent with the TICONET analysis, when we applied a machine learning algorithm to predict different NK functional phenotypes using surface receptors only (Figure 2D), we were able to identify the NK subsets that predominantly expressed CD107a with good accuracy (average area under the receiver operating characteristic curve of 75%) during cross validation (Figure 2E). Notably, DNAM1 and its paired coreceptor, CD96, are enriched in the CD107a-skewed subset (Figure 2B), and ranked as 2 of the top 5 most informative markers for the prediction (Figure 2F and Supplemental Figure 4E). In line with these findings, when DNAM1hiTIGIThi NK cells and not-double-positive NK cells were isolated and cocultured with SARS-CoV-2–infected Calu-1 cells, which are of lung epithelial cell origin, we observed stronger killing efficiencies for the NK cells that expressed DNAM1 and TIGIT than for NK cells that did not express these proteins (P = 0.017; Figure 2G). These results implied that even upon stimulation with the same target cells, NK subsets with different receptor repertoires have differential effector functions. In addition, NK subsets with similar immunophenotypes to those enriched in the fast viral clearance group tended to respond to target cell stimulation with cytolytic activities first and then with TNF-α secretion. Interestingly, previous studies reported that patients with severe COVID-19 had higher frequencies of CD57+NKG2C+ adaptive NK cells than patients with mild symptoms (21, 39). Our finding that CD57+ NK subsets initially respond to target cells with TNF-α secretion, whereas DNAM1hiTIGIThi NK cells respond with cytolytic activities, suggests a potential mechanistic link that should be further explored.
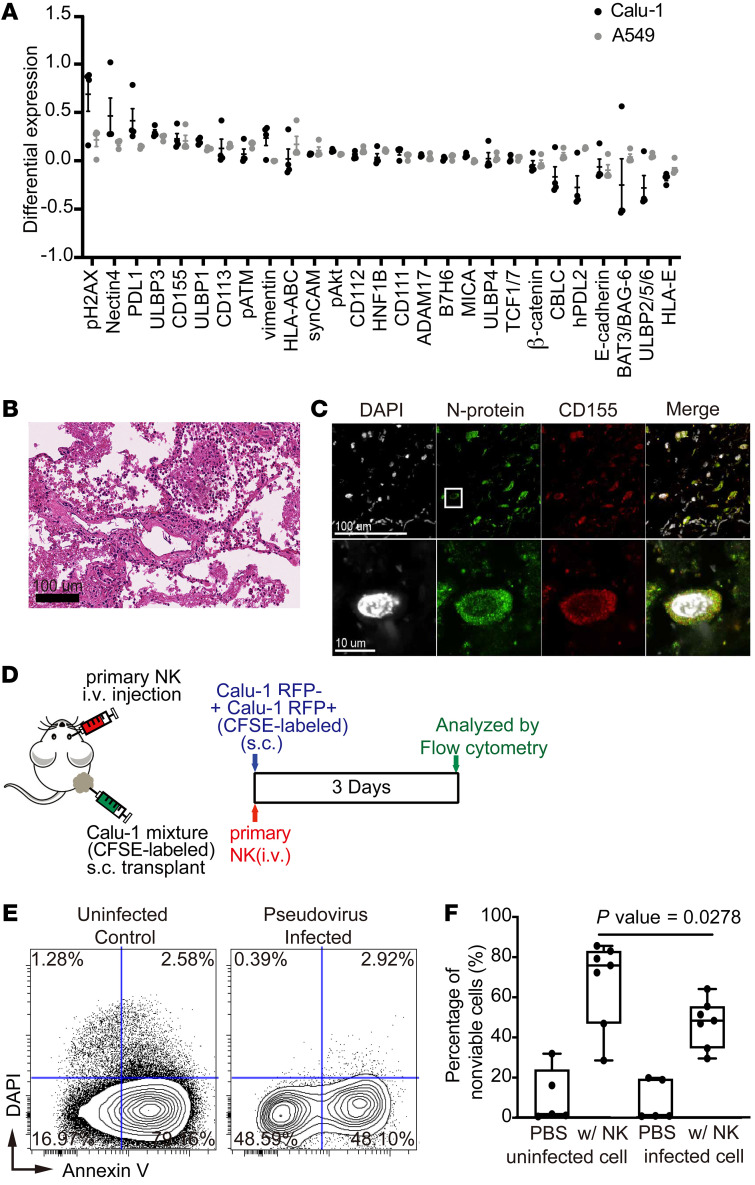
Finally, we infected 2 different cell lines of lung epithelial cell origin, A549 and Calu-1 cells, with SARS-CoV-2 or with a pseudovirus that displays the SARS-CoV-2 glycoprotein on its surface and investigated the phenotypic changes induced by the virus using mass cytometry with a panel of antibodies against ligands known to bind to receptors on immune cells (Supplemental Table 4). Interestingly, CD155 was among the ligands that were highly upregulated in both cell lines after SARS-CoV-2 or pseudovirus infection (Figure 3A and Supplemental Figure 5A). The upregulation of CD155 in SARS-CoV-2–infected lung epithelial cells was further confirmed by the colocalization of CD155 with virus N protein by immunostaining of lung sections from a COVID-19 patient (Figure 3, B and C). Interestingly, nectin-4, another ligand known to bind TIGIT (40), was also upregulated in SARS-CoV-2–infected cells (Figure 3A and Supplemental Figure 5B).
Figure 3. SARS-CoV-2 induces the expression of factors in infected cells that allow evasion of NK-mediated cytotoxicity.
(A) Dot plots of differential expression of indicated ligands induced upon infection of A549 cells (gray) and Calu-1 cells (black) with SARS-CoV-2. Differential expression was calculated by subtracting the arcsinh-transformed intensities of ligands in cells that do not express N protein from N protein–positive cells. Results were validated in 4 independent experiments (n = 4). Data represent mean ± SEM. (B) Representative image of H&E-stained lung tissue from a COVID-19 patient demonstrating organizing diffuse alveolar damage. (C) Representative immunofluorescence images of a lung tissue section from a SARS-CoV-2–infected patient stained for CD155 (red), N protein (green), and DAPI (white). The rectangle in the upper image indicates the region magnified in the lower image. (D) Schematic of the experiment used to evaluate cytotoxicity of primary NK cells against SARS-CoV-2 pseudovirus–infected cells. A mixture of infected (RFP+) and uninfected (RFP–) Calu-1 cells were labeled with CFSE and injected subcutaneously into mice on day 0. Primary NK cells were expanded and injected intravenously on the same day. Control mice were injected with PBS rather than NK cells. Calu-1 cells were analyzed by flow cytometry on day 3. (E) Representative biaxial plot of dissociated Calu-1 cells from mice treated as described in panel D stained with annexin V and DAPI. (F) Quantification of dead Calu-1 cells from 7 mice from 2 independent experiments described in panel D. Maximums, 75th, 50th, 25th percentiles, and minimums for uninfected cells in mice treated with PBS were 31.89, 23.99, 1.83, 0.90, and 0.63, respectively; for uninfected cells in mice treated with NK cells were 85.64, 83.02, 75.88, 46.94, and 28.54, respectively; for infected cells in mice treated with PBS were 19.82, 19.37, 0.93, 0.83, and 0.76, respectively; for infected cells in mice treated with NK cells were 64.12, 55,45, 48.35, 34.65, and 29.54, respectively. P = 0.0278 by Tukey’s multiple comparison test.
Given that CD155 and nectin-4 are known to interact with DNAM1 paired receptors, including TIGIT and CD96, on NK cells, we hypothesized that SARS-CoV-2 evades NK cell–mediated cytolytic activities (23, 24). To test this hypothesis, we established a Calu-1 cell line that stably expresses pseudovirus and RFP and coinjected it with uninfected Calu-1 cells into immunocompromised mice (Figure 3D). After mice were injected with expanded primary NK cells, preferential killing of uninfected Calu-1 cells was observed (uninfected vs. infected: 75.88% vs. 48.35%, P = 0.0248; Figure 3, E and F), suggesting that the virally infected cells can escape from the immunosurveillance mediated by NK cells. Interestingly, the ligands HLA-ABC and PDL-1 are also induced by SARS-CoV-2 infection (Figure 3A) and could potentially contribute to the immune escape mediated by SARS-CoV-2 (41).
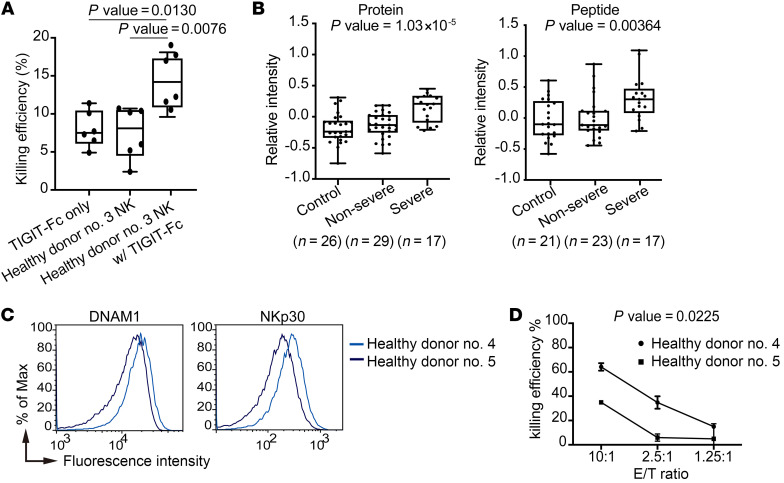
To further confirm that SARS-CoV-2 immune escape occurs through the DNAM1 paired receptor pathways, we cocultured SARS-CoV-2–infected target cells with primary NK cells in the absence or presence of recombinant TIGIT-Fc protein. We found that the killing efficiency of virus N protein–positive cells was enhanced by the recombinant TIGIT-Fc protein (with TIGIT-Fc vs. without TIGIT-Fc: 14.2% vs. 8.1%, P = 0.005; Figure 4A). By analyzing published proteomic data on serum samples from COVID-19 patients, we found that the level of CD155 is highly associated with the disease severity (P = 1.03 × 10–5 and 0.00364 for protein and peptide, respectively; Figure 4B). Moreover, healthy donor cells with DNAM1hi and NKp30hi immunophenotypes (Figure 4C) had stronger killing efficiency against pseudovirus-infected A549 target cells in vitro than did donor cells with lower levels of these proteins (Figure 4D). These results corroborate the essential role of the DNAM1 paired receptor pathways in the immune surveillance against SARS-CoV-2.
Figure 4. SARS-CoV-2 escapes immune surveillance through the DNAM1 paired receptor pathway.
(A) Killing efficiency of SARS-CoV-2–infected Calu-1 cells by primary NK cells with or without TIGIT-Fc from 2 independent experiments (n = 6). Maximums, 75th, 50th, 25th percentiles and minimums for TIGIT-Fc alone were 11.40, 10.43, 7.50, 6.10, 4.90; for NK cells alone were 10.70, 10.48, 8.10, 4.50, 2.40; and for NK cells with TIGIT-Fc were 18.10, 17.28, 14.20, 10.88, 9.60, respectively. P = 0.0130 between TIGIT-Fc alone and NK cells with TIGIT-Fc and P = 0.0076 between NK cells alone and NK cells with TIGIT-Fc; Tukey’s multiple comparison test. (B) Box-and-whisker plots of relative CD155 protein (left) and peptide (right) levels in control sera, sera from patients with mild COVID-19, and sera from patients with severe COVID-19. Maximums, 75th, 50th, 25th percentiles and minimums for control, mild COVID-19, and severe COVID-19 in the protein data set were 0.31, –0.07, –0.23, –0.33, and –0.75; 0.18, 0.02, –0.13, –0.25, and –0.59; and 0.45, 0.33, 0.21, –0.09, and –0.21, respectively (n = 72). Values in the peptide data set were 0.61, 0.27, –0.10, –0.27, and –0.58; 0.87, 0.11, –0.11, –0.20, and –0.44; and 1.09, 0.47, 0.30, 0.08, and –0.21, respectively (n = 61). P = 1.03 × 10–5 for protein data set and P = 0.00364 for peptide data set; linear regression. (C) Histogram overlays of percentage of the maximum events for DNAM1 and NKp30 in NK cells from 2 healthy donors versus fluorescence intensity. (D) The efficiencies of killing of pseudovirus-infected A549 cells (target, T) by NK cells from 2 healthy donors (effector, E) at 3 effector/target (E-T) ratios. The means ± SEM for NK cells from healthy donor no. 4 (DNAM1hi and NKp30hi) and healthy donor no. 5 (DNAM1lo and NKp30lo), respectively, were 64.0% ± 1.79% and 34.90% ± 0.40% in 10:1 conditions, 34.78% ± 2.99% and 5.92% ± 1.73% in 2.5:1 conditions, and 14.93% ± 0.93% and 4.91% ± 7.16% in 0.25:1 conditions. P = 0.0225 by linear regression.
Discussion
There are several limitations to our study. First, our cohort size was small, and predominantly peripheral blood was evaluated. Second, although we estimated the rate of viral clearance by fitting the curve of the viral loads from serial nasal swabs from each patient after admission, samples between exposure of patients to the virus and admission were not collected; therefore, these estimates may be flawed. Third, the NK profiles of uninfected family members in the same household would be of interest but were not available. Finally, profiling NK cells in the peripheral blood during active infection might be biased due to the recruitment of NK cells to the primary sites of infection; therefore, we focused on samples collected from convalescent patients. Additional studies that include analyses of preexposure samples will be required for validation of the correlations.
In sum, our mass cytometry–based analysis of 21 patients who had been infected with SARS-CoV-2 demonstrated a link between NK cell receptor repertoires and SARS-CoV-2 clearance rate. In addition, we also demonstrated that the extent of immune evasion after SARS-CoV-2 infection could play a role in determining the viral clearance or disease severity. Our data indicate that NK subsets expressing the receptor DNAM1 are critical to rapid recovery from SARS-CoV-2 infection. Our data suggest that therapeutic agents that block DNAM1 paired receptors should facilitate the clearance of SARS-CoV-2.
Methods
Viral load quantification by real-time PCR.
Specimens, including oropharyngeal swab and sputum, for each patient were collected and analyzed for SARS-CoV-2 RNA using quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) in accordance with the Taiwan Centers for Disease Control (TCDC) guidance (42, 43). The cycle threshold (Ct) values of qRT-PCR were used to quantify the viral loads for each patient.
Sample preparation for mass cytometry.
Samples were processed as previously described, with some modifications (44, 45). Briefly, human PBMCs were isolated by density gradient centrifugation using Ficoll-Paque PLUS (GE Healthcare Bio-Sciences AB) and stored frozen. The PBMCs were thawed for experiments and rested in RPMI with 10% fetal bovine serum (FBS) for 15 minutes. Then, cells were incubated with a cell-surface antibody cocktail prepared in cell staining media (CSM: PBS with 0.5% protease-free bovine serum albumin and 0.02% NaN3) in a final volume of 100 μL for 30 minutes to 1 hour at room temperature. After washing the cells once with CSM and once with serum-free RPMI medium, cells were stained with cisplatin (Sigma-Aldrich) at a final concentration of 25 μM for 1 minute at room temperature to label dead cells and then quenched by adding an equal volume of RPMI with 10% FBS for 3 minutes for viability staining. Cells were fixed in 1.6% paraformaldehyde (Electron Microscopy Sciences) in PBS at room temperature for 10 minutes, and 100% ice-cold methanol was used to permeabilize cells on ice for 10 minutes. For intracellular marker staining, cells were washed twice with CSM and stained with an intracellular antibody cocktail prepared in CSM in a final volume 100 μL for 30 minutes to 1 hour at room temperature. The antibodies used are listed in Supplemental Tables 2–4. Cells were then washed twice with CSM and stained with Cell-ID Intercalator-Ir (191Ir and 193Ir; Fluidigm) at a final concentration of 125 nM in 500 μL 4% fresh paraformaldehyde (diluted in PBS) overnight at 4°C to stain DNA. Finally, cells were resuspended in MilliQ water containing EQ Four Element Calibration Beads (Fluidigm) for normalization, and data were acquired on a CyTOF2 mass cytometer (Fluidigm). For the live-cell barcoding, PBMCs from 1 patient and 1 healthy donor were incubated with individual platinum-labeled CD45 antibody conjugates separately. Cells were then washed and pooled together for subsequent incubation with cell-surface antibody cocktail, as described previously (46). For the coculture experiments with 721.221 cells and for epithelial cell ligand profiling experiments, cells were stained with cisplatin, fixed, barcoded using palladium and permeabilized with saponin as previously described (44, 45, 50), and stained with indicated antibody panels.
Data acquisition, analysis, and diversity calculation for mass cytometry.
Data in raw FCS files were normalized using the Fluidigm Helios software or the Premessa R package and gated in Cytobank (Supplemental Figure 5C). The marker intensities were arcsinh transformed with a cofactor of 5, analyzed, and visualized using viSNE and FlowSOM (Cytobank). Diversity scores for each patient were calculated based on the Boolean expression of 22 NK surface markers (CD38, NTB, CD16, 2B4, CD161, NKG2D, DNAM1, TIGIT, NKp46, NKp30, NKG2A, CD7, LILRB1, CD86, CD57, NKG2C, CD96, HLA-DR, KIR2DL1, KIR3DL1, KIR2DL3, and CD56) using the inverse Simpson index (27). The mouse A20 cell line was stained as the negative control, and the 99th percentile intensity of each marker was defined as the threshold for Boolean gating.
Machine learning.
The eXtreme Gradient Boosting (XGBoost), a robust algorithm in classifying tasks (47), was applied to classify subsets of NK cells using 30 surface markers. Because of the difference in intensity value ranges between TNF-α and CD107a, we first normalized their intensities by subtraction of their positive threshold values for each cell (99th percentile intensity at time point 0 was defined as the positive threshold). Then, we divided the NK cells based on the normalized intensity ratios of TNF-α to CD107a into TNF-αhi (intensity ratio ≥ 6) and CD107ahi (intensity ratio ≤ 1/6) groups. The XGBoost library was implemented in the Scikit-Learn for Machine Learning analysis (48). To avoid overfitting, we applied 10-fold cross validation in which 80% of the data were used for training models and the remaining 20% of data were withheld for evaluation of the prediction performance. In the training process, accuracy and area under the receiver operating characteristic (AUROC) curve were required to improve at least once every 2 rounds. In addition to trainable parameters, hyperparameters of XGBoost were tuned and optimized. The model that achieved the highest AUROC score had parameters ETA = 0.1, MAX_DEPTH = 8, SUBSAMPLE = 0.8, MIN_CHILD_WEIGHT = 0.05, and L1_ALPHA = 1; other parameters were set to default values. These hyperparameters were then used to train the final model on the entirety of the data sets with 2,262 cells. Additionally, we implemented the built-in function of XGBoost library to obtain an importance score for each feature from the final model.
Cells and culture conditions.
HEK293T cells were obtained from American Type Culture Collection. The 721.221 cells were a gift from Garry Nolan’s lab at Stanford University. ACE2-expressing A549 and Calu-1 cells (49) were cultured in DMEM/F12 with 10% FBS and 1% penicillin-streptomycin. A549-ACE2-luciferase cells were generated by infecting A549-ACE2 cells with pLAS5w.FLuc.PeGFP lentivirus (RNAi core, Academia Sinica). HEK293T and 721.221 cells were cultured in DMEM with 10% FBS and 1% penicillin-streptomycin or RPMI 1640 with 10% FBS and 1% penicillin-streptomycin, respectively. Cells were incubated at 37°C in 5% CO2.
Coculture of 721.221 cells and NK cells.
Primary NK cells were processed as previously described, with some modifications (50, 51). Primary NK cells were purified from PBMCs using an NK cell isolation kit (Miltenyi Biotec), which results in at least 90% purity, and were incubated overnight with or without 10 IU/mL IL-2 (R&D Systems). For coculturing, 721.221 cells were incubated with primary NK cells at a 1:1 ratio in U-bottomed 96-well plates containing RPMI 1640 with 10% FBS and 1× Brefeldin plus Monensin (BD Biosciences) and anti-CD107a antibody. The cells were maintained at 37°C in 5% CO2 atmosphere. Samples were collected at hourly intervals over a 9-hour time course and were analyzed by mass cytometry. The expression level of DNAM1 and TIGIT was measured for IL-2–primed or unprimed NK cells using flow cytometry (Supplemental Figure 5D). The DNAM1hi and DNAM1lo NK cells expressed similar levels of IL-2 receptors (Supplemental Figure 5E).
TICONET construction on the 721.221 cells and NK cell coculture and the FLOW-MAP data set.
The pipeline to build the TICONET based on the 37 marker intensities from NK cells (37) and 32 marker intensities from the mouse embryonic stem cells (38) consisted of 3 steps. First, all cells in the 0-hour time point were clustered into 11 clusters using FlowSOM (52), and the mouse embryonic stem cells collected on day 0 were used as the initial nodes in the network. Second, the network was built by calculating Pearson’s correlation coefficients of all cell-cell pairs with the cutoff set at 0.99. Third, the medians of the marker intensities in each time-ordered level in the TICONET were calculated and visualized using R language.
KIR genotyping.
Genomic DNA from each patient was extracted from peripheral blood leukocytes using the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions, and samples were stored at −20°C until analysis. KIR ligand genotyping was performed using the KIR HLA ligand kit (CareDx) as per the manufacturer’s instructions.
Immunofluorescence and immunohistochemistry.
Lung tissue samples from patients with COVID-19 were embedded into paraffin following standard protocols and sectioned at a thickness of 4 μm using a microtome. For immunohistochemistry and immunofluorescence staining, procedures described previously (44) were followed, with some modifications. The sections were deparaffinized, rehydrated, and immersed in epitope retrieval buffer (10 mM sodium citrate, pH 6) in a pressure cooker. The sections were then rinsed twice with water and once with wash buffer (Tris-buffered saline plus 0.1% Tween, pH 7.2). After blocking with normal human serum in Tris-buffered saline for 1 hour, sections were incubated with primary antibodies (anti–N protein antibody, clone 36, a gift from An-Suei Yang, Genomics Research Center, Academia Sinica; anti-CD155 antibody, clone D8A5G, Cell Signaling Technology; anti–nectin-4 antibody, clone EPR15613-68) overnight at 4°C in a humidified chamber. Sections were then washed twice with wash buffer and incubated for 1 hour at 25°C with secondary antibodies and counterstained with DAPI. Images were obtained on a Leica Eclipse E400 microscope equipped with a SPOT RT color digital camera model 2.1.1 (Diagnostic Instruments).
Ex vivo NK cell activation, expansion, and isolation.
To expand and promote NK cytolytic activity, PBMCs were cocultured with K562-41BBL-mbIL-15 cells (a gift from Chang Yu-Hsiang, National Taiwan University) in a G-Rex 24-well culture plate (Wilson Wolf) in X-VIVO 15 medium with 5% human serum and 10 IU/mL IL-2 (R&D Systems). The medium was refreshed on days 3 and 5 with medium containing IL-2. After expansion for 7 days, NK cells were enriched with an NK cell isolation kit (Miltenyi Biotec) to at least 90% purity. The immunophenotypes of unexpanded or expanded NK cells were profiled using mass cytometry (Supplemental Figure 5F).
Murine xenograft model.
An aliquot of 5 × 106 Calu-1 RFP– and Calu-1 RFP+ (1:1) was injected subcutaneously into the right flank of NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(IL15)1Sz/SzJ mice (NSG-TG-HuIL15, Jackson Laboratory, 32–40 weeks old, male), followed by intravenous injection of PBS (n = 7) or 1 × 106 primary NK cells (n = 7) per mouse. After 3 days, mice were sacrificed and the tumor tissues were collected. The tumor tissues were minced in RPMI containing 10% FBS, collagenase IV (1 mg/mL), and DNase I (0.1 mg/mL) to dissociate the tumor cells. The cells were then stained with Annexin V (BD Biosciences) and DAPI in Annexin V assay buffer (BD Biosciences) for 15 minutes. The percentage of dead cells was determined by flow cytometry, and the background death rate (determined using dissociated cells from PBS-treated control animals) was subtracted. Mice were maintained as small breeding colonies in a specific pathogen–free environment of the animal facilities of the Institute of Biomedical Sciences, Academia Sinica. Protocols for the mouse experiments were approved by the Institutional Animal Care and Use Committee, Academia Sinica.
SARS-CoV-2 isolation and infection.
Target cells were infected with patient-derived SARS-CoV-2 at MOI 0.1 (2,000 PFU/well). Virus was isolated by the TCDC and amplified in Vero E6 cells in MEM supplemented with 2% FBS at 37°C and 5% CO2. The ACE2-expressing A549 and Calu-1 cells were plated at a density of 5 × 105 cells per well in a 6-well plate and infected with SARS-CoV-2 for 24 hours. All procedures were conducted following the laboratory biosafety guidelines of the TCDC and were conducted in a biosafety level 3 facility in the Institute of Biomedical Sciences, Academia Sinica.
SARS-CoV-2 spike pseudovirus production and infection.
pCMV-SARS-CoV-2G was constructed by replacing the open reading frame of VSV-G envelope protein in pCMV-VSV-G vector (Addgene, 8454) with the human codon-optimized spike gene of SARS-CoV-2. SARS-CoV-2–based pseudoviruses were produced by transient transfection of HEK293T cells using X-tremeGENE HP DNA Transfection Reagent (Roche) with 30 μg of vector plasmid (pLAS2w.RFP-C.Ppuro, National RNAi Core Facility Platform at the Institute of Molecular Biology/Genomics Research Center, Academia Sinica), 30 μg of the lentiviral helper plasmid pCMV-dR8.2 dvpr (Addgene, 8455), and 15 μg pCMV-SARS-CoV-2G. Transfection was carried out in 15-cm dishes according to the manufacturer’s instructions. SARS-CoV-2 spike pseudovirions encoding RFP were efficiently released into the supernatant. The A549 cells and the Calu-1 cells, both expressing ACE2, were infected with SARS-CoV-2–based pseudovirus for 24 hours.
In vitro killing assay.
Calu-1 cells that overexpress ACE2 were seeded at 10,000 cells per well in 96-well plates after 24 hours of SARS-CoV-2 infection or mock treatment and then incubated with or without 10 μg/mL human TIGIT protein, human IgG1 Fc tag (GeneTex) at 37°C for 30 minutes and then cocultured with primary NK cells that were isolated from PBMCs of healthy donors using an NK cell isolation kit (Miltenyi Biotec) or by sorting using a FACSAria (BD Biosciences) for the DNAM1+TIGIT+ population. After coculture at 37°C for 2 hours, the cells were fixed with 4% paraformaldehyde at room temperature for 30 minutes, permeabilized using cold 100% methanol at –80°C overnight, and washed in FACS buffer (PBS, 0.5%–1% bovine serum albumin, 0.1% NaN3). Samples were then stained with BV510 anti–human CD45 antibody (BioLegend, clone HI30) at 4°C for 30 minutes, and then incubated with anti–N protein antibody (a gift from An-Suei Yang’s Lab), PE anti–cleaved PARP (BioLegend, clone QA17A17), and Alexa Fluor 594 goat anti–human IgG secondary antibody (Jackson ImmunoResearch). The killing efficiency was determined by flow cytometry and calculated based on the percentage of cleaved PARP in N protein–positive cells relative to signal from wells containing only the target cells.
A549 cells that express ACE2, SARS-CoV-2 spike pseudovirus, and luciferase were seeded at 5,000 cells per well in 96-well plates. For the killing assay, enriched primary NK cells were incubated with these target cells at ratios of 10:1, 2.5:1, and 1.25:1 at 37°C for 4 hours. The killing efficiency was determined by quantification of the decrease in cell-associated luciferase activity, as previously described (53).
Quantification of IL-2 receptor in PBMCs.
Human PBMCs were stained with PE anti–human CD56 (BioLegend, clone MEM-188), PE-Cy7 anti–human CD3 (BD Biosciences, clone UCHT1), APC anti–human CD226 (DNAM-1; BioLegend, clone 11A8), PE/Cy7 anti–human CD122 IL-2Rβ (BioLegend, clone TU27), PerCP/Cy5.5 anti–human CD25 (BioLegend, clone BC96), and BUV563 rat anti–human CD132 (BD Biosciences, clone TUGh4) antibodies at 4°C for 30 minutes, and then cells were washed in FACS buffer. Data were acquired on an LSRII cytometer (BD Biosciences) and were analyzed based on fluorescence minus one or biological comparison controls using FlowJo software (BD Biosciences) (Supplemental Figure 5E).
Statistics.
GraphPad Prism was used to prepare box-and-whisker plots and for unpaired comparisons and linear regression. Fitting of generalized linear models and visualization were performed using stats, ggplot2, and pheatmap from the R package. A P value of less than 0.05 was considered statistically significant.
Study approval.
Peripheral blood samples were collected from 21 donors who had COVID-19 as confirmed by RT-PCR and who provided written informed consent (IRB protocol: AS-IRB-BM-20006 v.2.). Peripheral blood samples from healthy donors were obtained from the Stanford Blood Center or donors in Taiwan who provided written informed consent. Mouse experimental procedures were reviewed and approved by the Animal Care and Use Committee of Academia Sinica, Taipei, Taiwan.
Author contributions
WCH, YFW, YST, YT Lu, and SY Chen designed and performed the experiments, analyzed and interpreted the data, and wrote the manuscript. EYL, YT Liu, HCN, YMC, JHH, HKT, and YTH analyzed and interpreted the data and wrote the manuscript. DYT, JJL, CCL, LC, GW, YJL, HCH, CWL, JTW, SY Chang, CYC, and HCT designed and performed experiments. LJ wrote the manuscript. KIL and YLL assisted in experimental design and interpreted data. MTL coordinated patient sample collection.
Supplementary Material
Acknowledgments
We thank all the medical staff who participated in the treatment of patients, the doctors who collected the specimens, and the patients who provided the specimens. We thank the Taiwan Central Epidemic Command Center for coordinating the collection of specimens and providing SARS-CoV-2 TCDC 4 (hCoV-19/Taiwan/4/2020). The Institute of Biomedical Sciences P3 facility was supported by funding from Academia Sinica (AS-CFII-108-102), Taipei, Taiwan, and the Ministry of Science and Technology (MOST), Taiwan (grant 109-3114-Y-001-001). We thank Ya-Ping Lin and Jung-Lee Lin for the technical contributions. We thank Garry Nolan’s lab for reagents and technical support. We thank An-Suei Yang for reagents. We acknowledge the service provided by the core facility of the Institute of Biomedical Sciences, Academia Sinica and Translational Medicine of BioTReC (National Biotechnology Research Park, Academic Sinica) and Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII108-113) for flow cytometric analysis and cell sorting. Mass cytometry analyses were performed by Common Mass Spectrometry Facilities for Proteomics and Protein Modification Analysis located at the Genomics Research Center, Academia Sinica (AS-CFII-108107) and Stanford University, Palo Alto, CA, USA. BioRender (https://biorender.com/) was used to generate the schematic summary of our proposed model. LJ holds an Undergraduate Research Award at MOST. SY Chen holds a Career Development Award at Academia Sinica. This work was supported by MOST grants 108-2320-B-001-034-MY2, AS-CDA-110-L09, AS-GC-110-05, AS-KPQ-110-EIMD, NTUH 110-T20, and NTUH-109-P13.
Version 1. 11/01/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(21):e146408.https://doi.org/10.1172/JCI146408.
Contributor Information
Wan-Chen Hsieh, Email: judyjoy918@gmail.com.
En-Yu Lai, Email: junelai@stat.sinica.edu.tw.
Yu-Ting Liu, Email: meow23571379@gmail.com.
Yi-Fu Wang, Email: yfwang@ibms.sinica.edu.tw.
Yi-Shiuan Tzeng, Email: yishiuantzeng@gmail.com.
Lu Cui, Email: lucui@stanford.edu.
Yun-Ju Lai, Email: yunju_lai@uml.edu.
Hsiang-Chi Huang, Email: hsiangchi2009@gmail.com.
Jia-Hsin Huang, Email: jiahsin.huang@gmail.com.
Hung-Chih Ni, Email: jimmy101062341@gmail.com.
Dong-Yan Tsai, Email: johninstinct87@gmail.com.
Jian-Jong Liang, Email: jjliang@ibms.sinica.edu.tw.
Chun-Che Liao, Email: jfliao@gmail.com.
Ya-Ting Lu, Email: yatinglu@ibms.sinica.edu.tw.
Laurence Jiang, Email: jianglaurent@gmail.com.
Ming-Tsan Liu, Email: mtliu@cdc.gov.tw.
Jann-Tay Wang, Email: wang.jt1968@gmail.com.
Sui-Yuan Chang, Email: sychang@ntu.edu.tw.
Chung-Yu Chen, Email: peterx1peterx4@gmail.com.
Hsing-Chen Tsai, Email: htsai@ntu.edu.tw.
Yao-Ming Chang, Email: petitming@gmail.com.
Gerlinde Wernig, Email: gwernig@stanford.edu.
Chia-Wei Li, Email: cwli@ibms.sinica.edu.tw.
Kuo-I Lin, Email: kuoilin@gate.sinica.edu.tw.
Yi-Ling Lin, Email: yll@ibms.sinica.edu.tw.
Huai-Kuang Tsai, Email: hktsai@iis.sinica.edu.tw.
Yen-Tsung Huang, Email: ythuang@stat.sinica.edu.tw.
Shih-Yu Chen, Email: sychen@ibms.sinica.edu.tw.
References
- 1.Grasselli G, et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020;323(16):1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson S, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323(20):2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arons MM, et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382(22):2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKechnie JL, Blish CA. The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe. 2020;27(6):863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trouillet-Assant S, et al. Type I IFN immunoprofiling in COVID-19 patients. J Allergy Clin Immunol. 2020;146(1):206–208. doi: 10.1016/j.jaci.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taefehshokr N, et al. Covid-19: perspectives on innate immune evasion. Front Immunol. 2020;11:2549. doi: 10.3389/fimmu.2020.580641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vabret N, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rydyznski Moderbacher C, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183(4):996–1012. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maggi E, et al. COVID-19: Unanswered questions on immune response and pathogenesis. J Allergy Clin Immunol. 2020;146(1):18–22. doi: 10.1016/j.jaci.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuri-Cervantes L, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5(49):eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arunachalam PS, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369(6508):1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulte-Schrepping J, et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182(6):1419–1440. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silvin A, et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182(6):1401–1418. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernardes JP, et al. Longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19. Immunity. 2020;53(6):1296–1314. doi: 10.1016/j.immuni.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giamarellos-Bourboulis EJ, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolouri H, et al. The COVID-19 immune landscape is dynamically and reversibly correlated with disease severity. J Clin Invest. 2021;131(3):e143648. doi: 10.1172/JCI143648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varchetta S, et al. Unique immunological profile in patients with COVID-19. Cell Mol Immunol. 2021;18(3):604–612. doi: 10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 20.Bost P, et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181(7):1475–1488. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maucourant C, et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol. 2020;5(50):eabd6832. doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol. 2015;15(4):243–254. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- 24.Stanietsky N, Mandelboim O. Paired NK cell receptors controlling NK cytotoxicity. FEBS Lett. 2010;584(24):4895–4900. doi: 10.1016/j.febslet.2010.08.047. [DOI] [PubMed] [Google Scholar]
- 25.Horowitz A, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. 2013;5(208):208ra145. doi: 10.1126/scitranslmed.3006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strunz B, et al. Chronic hepatitis C virus infection irreversibly impacts human natural killer cell repertoire diversity. Nat Commun. 2018;9(1):2275. doi: 10.1038/s41467-018-04685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strauss-Albee DM, et al. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci Transl Med. 2015;7(297):297ra115–297ra115. doi: 10.1126/scitranslmed.aac5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin JM, et al. Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim M-C, et al. Duration of culturable SARS-CoV-2 in hospitalized patients with Covid-19. N Engl J Med. 2021;384(7):671–673. doi: 10.1056/NEJMc2027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singanayagam A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Gassen S, et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry Part A. 2015;87(7):636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 32.Enqvist M, et al. Coordinated expression of DNAM-1 and LFA-1 in educated NK cells. J Immunol. 2015;194(9):4518–4527. doi: 10.4049/jimmunol.1401972. [DOI] [PubMed] [Google Scholar]
- 33.Litwin V, et al. Specificity of HLA class I antigen recognition by human NK clones: evidence for clonal heterogeneity, protection by self and non-self alleles, and influence of the target cell type. J Exp Med. 1993;178(4):1321–1336. doi: 10.1084/jem.178.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fauriat C, et al. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115(11):2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogasawara K, et al. Impairment of NK cell function by NKG2D modulation in NOD mice. Immunity. 2003;18(1):41–51. doi: 10.1016/S1074-7613(02)00505-8. [DOI] [PubMed] [Google Scholar]
- 36.Romee R, et al. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17) Blood. 2013;121(18):3599–3608. doi: 10.1182/blood-2012-04-425397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang YM, et al. Comparative transcriptomics method to infer gene coexpression networks and its applications to maize and rice leaf transcriptomes. Proc Natl Acad Sci U S A. 2019;116(8):3091–3099. doi: 10.1073/pnas.1817621116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko ME, et al. FLOW-MAP: a graph-based, force-directed layout algorithm for trajectory mapping in single-cell time course datasets. Nat Protoc. 2020;15(2):398–420. doi: 10.1038/s41596-019-0246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varchetta S, et al. Unique immunological profile in patients with COVID-19. Cell Mol Immunol. 2021;18(3):604–612. doi: 10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reches A, et al. Nectin4 is a novel TIGIT ligand which combines checkpoint inhibition and tumor specificity. J Immunother Cancer. 2020;8(1):e000266. doi: 10.1136/jitc-2019-000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabbatino F, et al. PD-L1 dysregulation in COVID-19 patients. Front Immunol. 2021;12:695242. doi: 10.3389/fimmu.2021.695242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corman VM, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang JR, et al. Building the national SARS-CoV-2 laboratory diagnostic capacity in Taiwan. Health Secur. 2020;18(5):383–391. doi: 10.1089/hs.2020.0056. [DOI] [PubMed] [Google Scholar]
- 44.Cui L, et al. Activation of JUN in fibroblasts promotes pro-fibrotic programme and modulates protective immunity. Nat Commun. 2020;11(1):2795. doi: 10.1038/s41467-020-16466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wernig G, et al. Unifying mechanism for different fibrotic diseases. Proc Natl Acad Sci U S A. 2017;114(18):4757–4762. doi: 10.1073/pnas.1621375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mei HE, et al. Platinum-conjugated antibodies for application in mass cytometry. Cytometry A. 2016;89(3):292–300. doi: 10.1002/cyto.a.22778. [DOI] [PubMed] [Google Scholar]
- 47. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. Paper presented at: 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; August 13–17, 2016; San Francisco, California, USA. Accessed September 20, 2021. [DOI]
- 48.Pedregosa F, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12(1):2825–2830. [Google Scholar]
- 49.Lai Y-J, et al. Epithelial-mesenchymal transition induced by SARS-CoV-2 required transcriptional upregulation of Snail. Am J Cancer Res. 2021;11(5):2278–2290. [PMC free article] [PubMed] [Google Scholar]
- 50.Jensen H, et al. EBI3 regulates the NK cell response to mouse cytomegalovirus infection. Proc Natl Acad Sci U S A. 2017;114(7):1625–1630. doi: 10.1073/pnas.1700231114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukherjee S, et al. In silico modeling identifies CD45 as a regulator of IL-2 synergy in the NKG2D-mediated activation of immature human NK cells. Sci Signal. 2017;10(485):eaai9062. doi: 10.1126/scisignal.aai9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Gassen S, et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87(7):636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 53.Fu X, et al. A simple and sensitive method for measuring tumor-specific T cell cytotoxicity. PLoS One. 2010;5(7):e11867. doi: 10.1371/journal.pone.0011867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.