Abstract
Central obesity with cardiometabolic syndrome (CMS) is a major global contributor to human disease, and effective therapies are needed. Here, we show that cyclic GMP–selective phosphodiesterase 9A inhibition (PDE9-I) in both male and ovariectomized female mice suppresses preestablished severe diet-induced obesity/CMS with or without superimposed mild cardiac pressure load. PDE9-I reduces total body, inguinal, hepatic, and myocardial fat; stimulates mitochondrial activity in brown and white fat; and improves CMS, without significantly altering activity or food intake. PDE9 localized at mitochondria, and its inhibition in vitro stimulated lipolysis in a PPARα-dependent manner and increased mitochondrial respiration in both adipocytes and myocytes. PPARα upregulation was required to achieve the lipolytic, antiobesity, and metabolic effects of PDE9-I. All these PDE9-I–induced changes were not observed in obese/CMS nonovariectomized females, indicating a strong sexual dimorphism. We found that PPARα chromatin binding was reoriented away from fat metabolism–regulating genes when stimulated in the presence of coactivated estrogen receptor-α, and this may underlie the dimorphism. These findings have translational relevance given that PDE9-I is already being studied in humans for indications including heart failure, and efficacy against obesity/CMS would enhance its therapeutic utility.
Keywords: Metabolism
Keywords: Fatty acid oxidation, Obesity, Phosphodiesterases
Introduction
Nearly one-fifth of all humans are currently obese (1), and in the United States this proportion exceeds 40% (2). The obesity pandemic is impacting global health, increasing risk for diabetes, dyslipidemia, nonalcoholic fatty liver disease, and cardiovascular disease such as hypertension, all major components of cardiometabolic syndrome (CMS) (1, 3, 4). Studies in humans show that abdominal (visceral) obesity is most pathogenic (5), whereas metabolically active adipose tissue known as brown fat appears protective (6). Obesity-related disorders also exhibit a sexual dimorphism, as women are relatively protected prior to menopause (7, 8), but visceral fat and CMS risks rise after menopause (9, 10). Obesity has also become a major comorbidity in patients with heart failure and preserved ejection fraction (HFpEF) (9, 11), a syndrome conferring high morbidity, with little effective therapy (10, 12). Treatments to reduce obesity are being sought. Glucagon-like peptide 1 agonists are promising (13) but largely suppress appetite with dose-limiting gastrointestinal side effects. Methods that stimulate fat catabolism remain lacking.
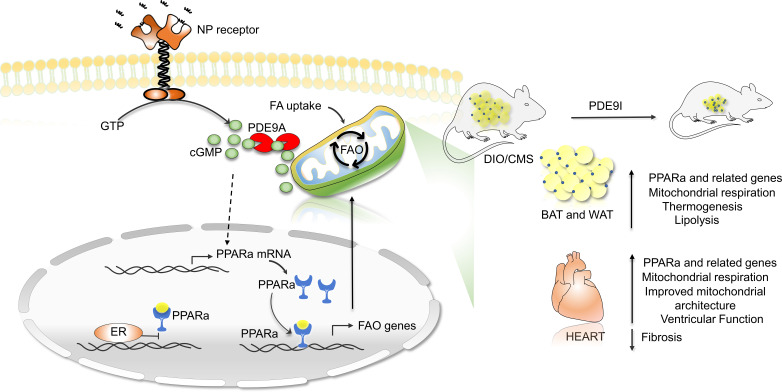
One intrinsic lipolytic pathway is coupled to cyclic GMP (cGMP)/protein kinase G (PKG) signaling stimulated by natriuretic peptides (NPs) or nitric oxide. PKG is the primary kinase effector of cGMP and it phosphorylates hormone-sensitive lipase (HSL) and perilipin, stimulates mitochondrial biogenesis and oxidative activity, and improves insulin signaling, suppressing diet-induced obesity (DIO) (14–16). PKG activation is therapeutically achieved by increasing cGMP synthesis or blocking its hydrolysis by specific phosphodiesterases (particularly PDE5 and PDE9). Stimulation with synthetic NPs (17), nitrates/nitrites (18), or soluble guanylate cyclase stimulators (19) to augment cGMP has limitations, as these molecules act quickly with a short half-life, can potently lower blood pressure, and can require parental administration. PDE5 inhibition is clinically used as a vasodilator to treat erectile dysfunction and pulmonary hypertension, and while it also ameliorates cardiac pressure-overload pathobiology (20), there is scant evidence for potent effects countering obesity. PDE5 also primarily regulates NO-dependent cGMP, and its activation of PKG and consequently its cardioprotective effects against pressure-overload stress are lost in ovariectomized (OVX) females (21, 22). This poses another potential limitation, as most women with obesity/CMS are postmenopausal.
PDE9 is the other cGMP-selective member of the superfamily, and its pharmacological or genetic inhibition improves hearts subjected to pathological pressure overload (20, 23) and renal function in dilated heart failure (24). PDE9 regulates NO-independent cGMP, i.e., from NP signaling (20, 25), suggesting that unlike PDE5, its inhibition could remain effective even in OVX females. PDE9 is expressed in heart, but is also found in adipose tissue and liver (20, 26), yet expressed at low levels in systemic vessels, so its inhibition has minimal effects on arterial blood pressure (20, 27).
Here, we tested the impact of chronic oral PDE9 inhibition (PDE9-I) in a model of preestablished severe DIO/CMS with or without superimposed mild cardiac pressure overload in males and intact or OVX females. In both OVX females and males, we found that PDE9-I prevents weight accumulation, reducing total body fat, fat depots, and lipid accumulation in multiple tissues, including the heart and liver. It also stimulates mitochondrial respiration and fatty acid oxidation in conjunction with upregulating peroxisome proliferator–activated receptor-α (PPARα) signaling. None of these changes were observed in non-OVX females, and this sexual dimorphism may result from interference with PPARα binding at promoter regions of genes coupled to fat metabolism by the coactivation of estrogen receptor-α.
Results
Model of combined severe DIO and CMS.
To test the impact of PDE9-I on obesity/CMS, we subjected C57BL/6N mice to DIO for approximately 6 months (Figure 1A). For the primary cohort, the last 2 months of DIO was combined with mild cardiac pressure overload (transaortic constriction, mTAC) to mimic pressure stress commonly observed with CMS. This also served to stimulate NP synthesis to provide more cGMP substrate for PDE9 regulation. A second cohort was also studied with DIO alone. In the final 6 to 7 weeks (1 week after mTAC if used), mice were randomized to receive either oral PDE9-I (PF-04447943) or placebo (vehicle). In OVX females, DIO increased body weight 2- to 3-fold, similar to changes in males (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI148798DS1). After 4 months of HFD but before mTAC and/or drug randomization, OVX mice developed profound visceral adiposity (Supplemental Figure 1B), elevated blood sugar and glucose intolerance (Supplemental Figure 1C), and marked hepatic steatosis (Supplemental Figure 1D). The results that follow are from the combined DIO/mTAC model.
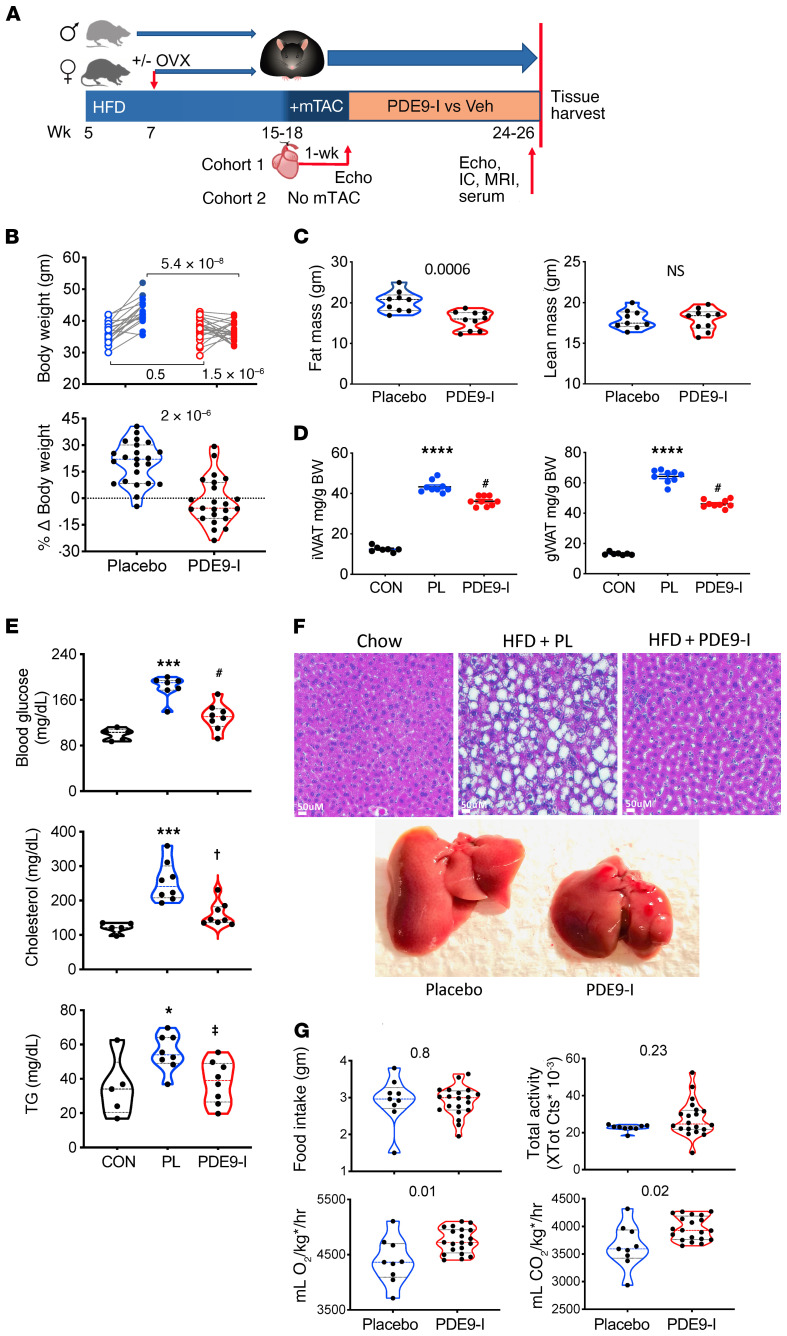
Figure 1. PDE9 inhibition suppresses diet-induced obesity and metabolic defects in ovariectomized females with cardiometabolic syndrome.
(A) Protocol for generating DIO/CMS with and without ventricular pressure stress. Mice were fed a high-fat diet (HFD) for approximately 4 months (with and without ovariectomy, OVX) in females), mild aortic constriction (mTAC) was applied in the main cohort (withheld in Cohort 2), and 1 week later, randomized to receive 6 to 8 weeks of placebo versus PF-7943. IC, indirect calorimetry; MRI, magnetic resonance imaging for body fat vs. lean tissue; EC, echocardiogram; Veh, vehicle. (B) Left: Body weight of OVX mice before/after 8 weeks of placebo (n = 19) or PDE9-I (n = 23) treatment (2-way ANOVA; P value lower right, drug × time effect interaction; others, Sidak’s multiple comparison test). Right: Same data plotted as percentage weight change for each group. Mann-Whitney test. (C) MRI-derived total body fat and lean mass for placebo (n = 9) or PDE9-I (n = 10) in OVX mice (P value by Mann-Whitney test). (D) Inguinal (iWAT) and gonadal (gWAT) white adipose tissue weight in OVX females treated with placebo or PDE9-I (n = 7–9/group). ****P = 0.00001 vs. normal diet control (CON); #P = 0.028 vs. placebo; by Kruskal-Wallis with Dunn’s multiple comparison test. (E) Serum fasting blood glucose and lipids in OVX mice on control versus HFD with and without PDE9-I. ***P = 0.0004, *P = 0.03 vs. CON; #P = 0.03, †P = 0.037, ‡P = 0.047 vs. placebo [PL]) by 1-way ANOVA with Holm-Sidak multiple comparison test. (F) Upper: Representative liver histology in OVX with standard diet (Chow) or HFD with placebo vs. PDE9-I. Marked steatosis is seen with placebo versus near normal histology with PDE9-I (replicated n = 5/group). Lower: Examples of whole liver showing a marked reduction of liver mass by PDE9-I. Group results in Supplemental Figure 2A. (G) Food intake, activity, and whole-body indirect calorimetry in OVX mice with and without PDE9-I (P value by Mann-Whitney test). kg* = (lean mass + fat mass × 0.2), as described previously (60).
PDE9-I lowers obesity and improves CMS in OVX-female and male mice.
OVX mice receiving placebo continued to gain weight (median 22%) over the 8-week drug-trial period, whereas those treated with PDE9-I exhibited a –5.6% weight decline (P = 2 × 10–6; Figure 1B). MRI body composition analysis found PDE9-I treatment reduced total fat mass but left lean mass unchanged (Figure 1C). There was less inguinal white adipose tissue (subcutaneous, iWAT) and gonadal (visceral, gWAT) fat mass (Figure 1D) with PDE9-I. CMS in the placebo arm was reflected by elevated fasting blood glucose, triglycerides, and cholesterol, and all were reduced by PDE9-I treatment (Figure 1E). Liver mass and steatosis increased in OVX placebo-treated mice but were at near-normal levels with PDE9-I (Figure 1F and Supplemental Figure 2, A and B). Importantly, PDE9-I had no statistically significant impact on food intake or activity when compared to placebo control. However, total body O2 consumption and CO2 production were higher in PDE9-I–treated mice (Figure 1G) without a change in core temperature (Supplemental Figure 2C). These indirect calorimetry and activity results were similarly observed during the light and dark circadian phases (Supplemental Figure 3A). Age-matched males were subjected to the same DIO/mTAC obesity/CMS protocol and treatment randomization and also displayed lower body weight with PDE9-I, associated with reduced total fat, iWAT, and gWAT mass (Supplemental Figure 4, A and B), less dyslipidemia (Supplemental Figure 4C), and reduced hepatic steatosis (Supplemental Figure 2D). PDE9-I also had no significant impact in males on their food intake or activity but increased total body O2 consumption and CO2 production (Supplemental Figures 3B and 4D). Despite fat and weight loss, PDE9-I did not significantly alter insulin resistance as assessed by the glucose tolerance test in OVX or males (Supplemental Figure 5).
PDE9-I improves left ventricular function, blunts hypertrophy/fibrosis, and augments myocardial PPARα signaling over placebo in OVX and male mice.
Prior studies found that PDE9-I reduces cardiac hypertrophy and profibrotic signaling cascades and improves left ventricular (LV) function in nonobese males subjected to pressure overload and that this efficacy was maintained despite inhibiting NO synthase with NG-nitro-L-arginine methyl ester (L-NAME) (20). This led us to predict that PDE9-I would retain its efficacy even in mice with OVX that have estrogen-dependent declines in NO stimulation (21, 22). This was the case, as PDE9-I augmented myocardial cGMP in OVX mice (Supplemental Figure 6A). OVX also resulted in mild cardiac hypertrophy and a slight decline in LV ejection fraction compared with non-OVX mice (Supplemental Figure 6B). PDE9-I improved ejection fraction (2-way ANOVA, interaction for drug × time P = 0.001) in OVX while attenuating LV hypertrophy (P = 0.005 for interaction) compared with placebo (Figure 2A). Similar results were obtained in males (Supplemental Figure 7A). LV diastolic dysfunction in the OVX-placebo group also improved toward lean control values with PDE9-I (Figure 2B). PDE9-I reduced prohypertrophic and profibrotic gene expression compared with placebo in both OVX females and males (Figure 2C and Supplemental Figure 7B), and this correlated with histological analysis of LV interstitial fibrosis that was from 4% in the placebo group versus 1.5% with PDE9-I (P = 0.008; Supplemental Figure 2E).
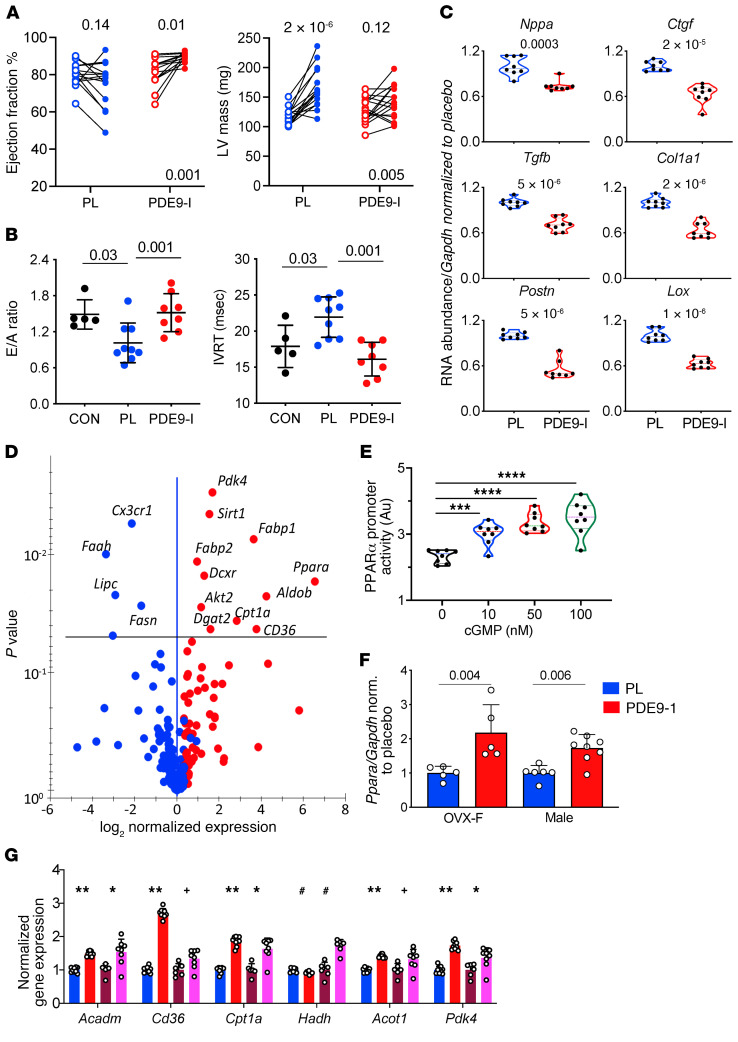
Figure 2. PDE9-I improves cardiac function, suppresses pathological hypertrophic gene expression, and stimulates PPARα signaling in OVX myocardium.
(A) Paired data from OVX mice at start and end of 8-week treatment period for left ventricular (LV) ejection fraction and mass (n = 15 and 16 for placebo and PDE9-I, respectively; repeated measures ANOVA, P value lower right for treatment × time interaction; upper values Tukey’s multiple comparison test). (B) Diastolic function assessed by mitral filling ratio (early/atrial, E/A) and isovolumic relaxation time (IVRT) in normal-diet controls (CON), and DIO/mTAC OVX mice treated either placebo (PL) or PDE9-I; n = 5–9/group, Kruskal-Wallis with Dunn’s multiple comparison test. (C) mRNA abundance normalized to Gapdh for A-type natriuretic peptide (Nppa), transforming growth factor β (Tgfb), collagen type 1α1 (Col1a1), lysyl oxidase (Lox), connective tissue growth factor (Ctgf), and periostin (Postn) in OVX myocardium (8/group). P values by Mann-Whitney test. (D) Volcano plot of differential gene expression in OVX-DIO/mTAC myocardium for PDE9-I vs. placebo. P values from Benjamini-Hochberg–adjusted P value versus log2(fold change) (n = 10/group). (E) Activation of PPARα promoter by cGMP in HepG2 cells (n = 8/group). ***P = 0.007, ****P ≤ 2 × 10–6 by 1-way ANOVA with Holm-Sidak multiple comparison test. (F) Ppara/Gapdh expression in brown adipose tissue (BAT) from OVX (n = 5) and males (n = 6, 8) treated with placebo versus PDE9-I (P values by Mann-Whitney test). G) PPARα-regulated fatty acid metabolism genes in myocardium from OVX or male mice treated with placebo (P; n = 9, 6) or PDE9-I (n = 9, 8). Data normalized to Gapdh and then to placebo for each gene; analysis by 2-step Benjamini-Krieger-Yekutieli multiple comparison Kruskal-Wallis test; q values shown. **P = 0.00005; #P = 0.008; +P = 0.03; *P = 0.02 vs. placebo.
Dietary nitrate intake has been reported to enhance fatty acid (FA) oxidation coupled to an upregulation of the transcription factor PPARα (28). Given the impact of PDE9-I on fat stores, we tested if such upregulation occurs and found greater myocardial mRNA abundance for PPARα and many of its downstream regulated genes with PDE9-I treatment (Figure 2D). cGMP stimulation increased PPARα promotor activity in a dose-dependent manner in HepG2 (liver) cells (Figure 2E), supporting prior results (28), and PPARα mRNA increased in myocardium of OVX and male mice treated with PDE9-I (Figure 2F). Evidence of enhanced PPARα activity was provided by an increased mRNA abundance of multiple regulated genes in the PDE9-I treatment groups (Figure 2G). Metabolite profiling of myocardial acylcarnitines found significant increases most notably in long-chain FAs in OVX and males receiving placebo as compared with controls, and these were lowered by PDE9-I (Supplemental Figure 7C). Collectively, these data show that PDE9-I ameliorates cardiac dysfunction and remodeling, reduces profibrotic signaling and interstitial fibrosis, and enhances FA metabolism in concert with PPARα activation.
PDE9-I induces adipose tissue browning and enhances FA catabolism.
The most substantial effects of PDE9-I in our model were in fat, so we explored their impact and mechanisms in more detail. Increased FA oxidation by PPARα manifests in brown (BAT) and white (WAT) adipose tissue by increasing mitochondrial biogenesis and FA oxidation. Figure 3A shows example histology of BAT from a nonobese control and obese OVX or male mice treated with placebo or PDE9-I. Control BAT was dark, with small lipid droplets and higher mitochondrial density. BAT from placebo-treated DIO/mTAC mice was pale with larger fat vacuoles and fewer mitochondria, and this appearance was closer to normal after PDE9-I treatment. PDE9-I also resulted in increased mRNA abundance of multiple genes related to FA metabolism and thermogenesis (Figure 3B) and was accompanied by a broad decline in acylcarnitine levels (Figure 3C). Expression of genes regulating mitochondrial biogenesis in BAT (browning) (Figure 3D) and FA metabolism (Figure 3E) and thermogenesis (Figure 3F) in WAT were also upregulated by PDE9-I relative to placebo. Together, these results show that PDE9-I activates adipocyte catabolic and thermogenic programs associated with PPARα activation.
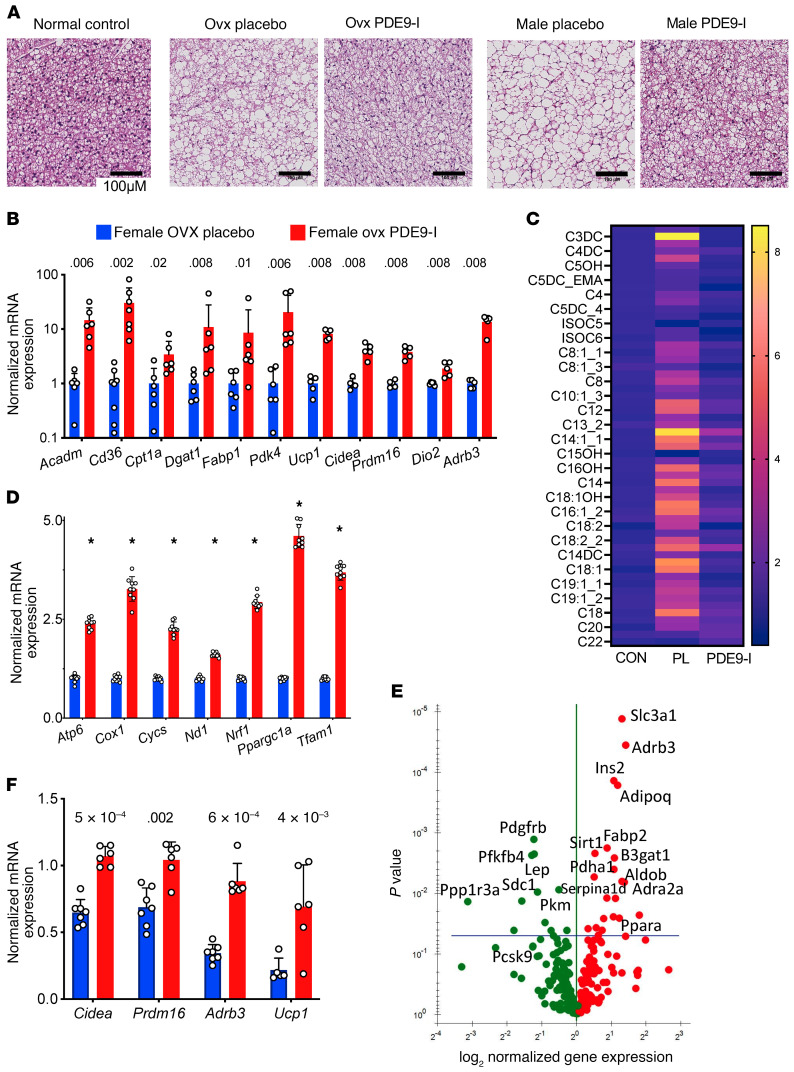
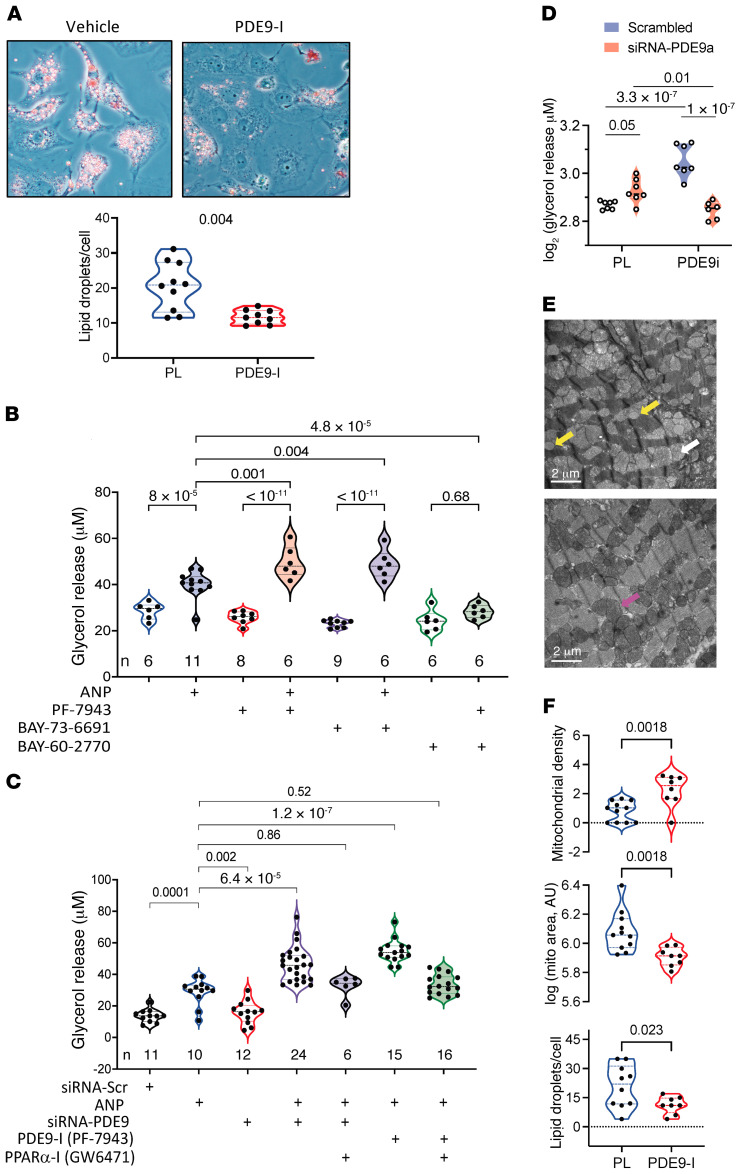
Figure 3. PDE9-I reduces multi-organ lipid accumulation by enhancing lipolysis and mitochondrial respiration.
(A) Representative histology of brown fat in normal control (left) and obese/CMS OVX mice after placebo or PDE9-I treatment (each replicated n = 7/group). Scale bar: 100 μm. See text for details. (B) BAT mRNA abundance for fat metabolism and thermogenic genes in OVX mice with placebo or PDE9-I. Results normalized as in Figure 2G, mean ± SD and individual data shown; q values from 2-step Benjamini-Krieger-Yekutieli multiple comparison Kruskal-Wallis test. (C) Metabolomic analysis of acylcarnitines in BAT from OVX obese/CMS mice with placebo (PL) or PDE9-I treatment (n = 5 per group). Analysis by 2-step Benjamini-Krieger-Yekutieli multiple comparison Mann-Whitney test; all metabolites with q values < 0.008. (D) mRNA abundance of mitochondrial biogenesis genes in BAT from OVX with PL or PDE9-I (n = 9/group). *P < 1 × 10–11. (E) Volcano plot of differential gene expression for lipid metabolism PCR array in WAT tissues from OVX model treated with PDE9-I versus placebo. Benjamini-Hochberg–adjusted P value versus log2(fold change), n = 8/group. (F) mRNA abundance of mitochondrial oxidative genes in WAT from OVX treated with placebo or PDE9-I (n = 5–7/group); 2-step Benjamini-Krieger-Yekutieli multiple comparison Kruskal-Wallis q values shown.
PDE9-I directly stimulates fat lipolysis and increases mitochondrial respiration.
The preceding findings suggested PDE9-I may stimulate lipolysis in multiple cell types, including adipocytes. In cardiomyocytes fed free FA (FFA) with vehicle or PDE9-I for 48 hours, we found that PDE9-I reduced intracellular lipid droplets (Figure 4A and Supplemental Figure 8A). Lipolysis was then assessed in matured adipocytes by glycerol release. Lipolysis increased as expected after 24-hour exposure to atrial NP (ANP). Exposure to either of 2 selective small molecule PDE9 inhibitors (PF-7943 as used in vivo, or BAY-73-6691) alone had no impact on glycerol release, but each significantly augmented the ANP response (Figure 4B). By contrast, PDE9-I did not augment lipolysis in cells cotreated with a soluble guanylate cyclase activator (BAY-60-2770), supporting a selective interaction of PDE9 with NP-stimulated cGMP, as previously reported in myocytes (20). Knockdown of the PDE9 gene in adipocytes with siRNA (Supplemental Figure 8B) also augmented ANP-stimulated lipolysis to levels similar to those achieved with PDE9-I (PF-7943; Figure 4C). In this experiment, we further tested the impact of coinhibiting PPARα (GW6471) and found it blocked lipolysis augmentation by PDE9-I, reducing levels to that with ANP alone. Consistent with requiring PPARα transcriptional regulation, subacute ANP exposure (4 hours) did not stimulate lipolysis, whereas longer exposure (e.g., 24 hours) did (Supplemental Figure 8C). Similar augmentation of ANP-stimulated lipolysis from PDE9-I in a PPARα-dependent manner was confirmed in neonatal rat ventricular myocytes (NRVMs) and hepatic HepG2 cells (Supplemental Figure 8D). Lastly, PDE9-I amplification of ANP-stimulated lipolysis was lacking in adipocytes preincubated with siRNA-Pde9a, supporting selectivity of the inhibitor response (Figure 4D).
Figure 4. PDE9-I increases lipolysis in fat, liver, and myocardium.
(A) Phase-contrast microscopy with Oil Red O staining of cardiomyocytes fed a lipid mixture for 48 hours and cotreated with vehicle or PDE9-I (replicated 4 times); summary of lipid droplets/myocyte on right (n = 10/group). P value by Mann-Whitney test. Original magnification, 400×. (B) Lipolysis measured by glycerol released in human subcutaneous preadipocytes (SP-2096, Zen-Bio) incubated for 24 hours with combinations of ANP, PDE9-I (PF-7493 or BAY-73-6691), or soluble guanylate cyclase activator (BAY 60-2770). The n values for each are provided in the figure; 1-way ANOVA with Sidak’s multiple comparison test P values shown. (C) Glycerol release in adipocytes exposed to chronic ANP with and without siRNA against Pde9a or scrambled control (Scr), PDE9-I, or PPARα-I. The n for each group is provided in the figure. Brown-Forsythe-Welch ANOVA with Dunnet’s multiple comparison test P values shown. (D) Glycerol release in adipocytes pretreated with siRNA against Pde9a or scrambled control, and then exposed for 24 hours to PDE9-I (PF-7943) or vehicle. Two-way ANOVA, lower left P value for interaction term, upper P values for Sidak’s multiple comparison test between pairs shown. (E) Example transmission EM images of LV myocardium from DIO/mTAC OVX with placebo vs. PDE9-I treatment. With vehicle, mitochondria were less dense and swollen (white arrow) and there was deposition of macro-lipid droplets (yellow arrows). Both are quantified from multiple images in each group in Supplemental Figure 7A. PDE9-I restored the mitochondrial architecture to a more normal dense appearance (e.g., pink arrow) and there was minimal lipid deposition observed. (F) Quantitation of mitochondrial density (pixels/area), cross-sectional area (arbitrary units, log plot), and lipid droplet counts based on these EM images (n = 9–20 individual mitochondria analyzed within each EM image quantified and averaged, n = 11 for placebo [PL], n = 9 for PDE9-I). Analysis by Kruskal-Wallis test, with P values displayed.
DIO/mTAC induces fat accumulation in multiple tissues, including the heart. In myocardium, this is associated with characteristic swelling of mitochondrial cristae and presence of macro-lipid droplets (29, 30). We observed similar abnormalities in OVX and male mouse myocardium in the present model and that PDE9-I substantially reversed them (Figure 4, E and F). The mitochondrial changes with DIO/mTAC resulted in mitochondria that were less electron dense than myofibrils in the vehicle-treated group, but denser than myofibrils as occurs normally in the PDE9-I cohort. Similar findings were obtained in male myocardium (Supplemental Figure 9). These results show that PDE9-I stimulates lipolysis when added to background NP stimulation, and reduces mitochondrial swelling, lipid accumulation, and morphological disruption in DIO/mTAC.
PDE9 localizes to mitochondria and stimulates fat oxidation.
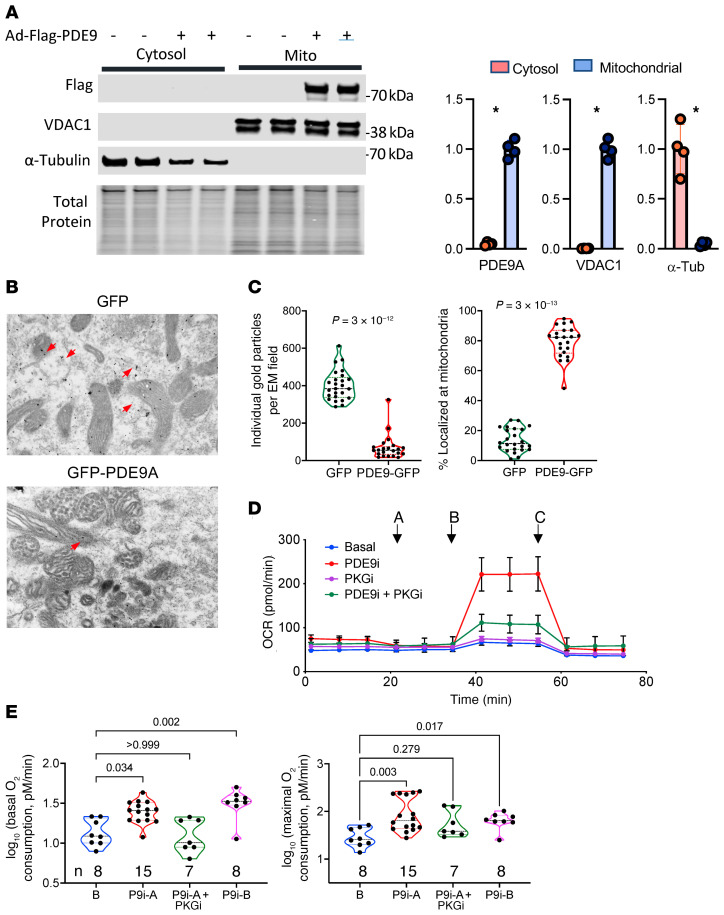
We previously reported that PDE9 colocalizes with the sarcoplasmic reticular ATPase at T-tubules in cardiac myocytes (20). The proximity of T-tubular membranes with mitochondria in the dyadic cleft raised the possibility that PDE9 also localized with these organelles. To test this, Pde9a-Flag was expressed in cardiomyocytes, the cells then fractionated into mitochondrial and cytosolic components, and Flag-Ab used to detect PDE9 protein (PDE9 antibodies are not sufficiently selective for this analysis). PDE9-Flag was found in the mitochondrial and not cytosolic fraction (Figure 5A). This localization was further supported by immunogold labeling of cardiomyocytes expressing either GPF-PDE9 or GFP alone. GFP particles were diffusely distributed, whereas GFP-PDE9 was more selectively localized at mitochondria (Figure 5B). Quantitation of 24 separate images found many more GFP-positive particles, with approximately 80% of GFP-PDE9 at mitochondria versus approximately 10% for GFP (P = 3 × 10–13; Figure 5C). Together, these results support PDE9 localization to mitochondria.
Figure 5. PDE9 localizes to mitochondria and its inhibition augments fat oxidation.
(A) PDE9-Flag expressed in cardiomyocytes was identified by Flag immunoblot primarily in the mitochondrial subfraction and not cytosol. VDAC1 is mitochondrial control, and α-tubulin (α-tub) cytosolic control. TP, total protein. Summary results n = 4, mean ± SD. *P = 0.028 by Mann-Whitney test. (B) Representative EM images for immunogold-labeled PDE9-GFP and GFP. Dark black dots identify each protein. Gold-stained GFP (red arrows) is diffusely distributed, whereas PDE9-GFP (yellow arrows) primarily colocalizes to mitochondria (n = 12/group). Each rectangular panel is 2.4 × 3.6 µM. (C) Summary analysis of 24 such images (12 per group) showing total number of particles identified in each field for each protein, and their percentage localization at mitochondria. P values from Mann-Whitney test. (D) Oxygen consumption rate (OCR) in adipocytes pretreated with vehicle (Veh, n = 8) or PDE9-I (5 μM, n = 8, mean ± SD), with or without PKG inhibitor (DT3 1 μM). The protocol includes administering: A, oligomycin; B, FCCP; or C, rotenone + antimycin. Mean ± SD. (E) Summary results from this experiment for basal and maximal oxygen consumption shown on log scale (n/group provided in figure). Kruskal-Wallis test with Dunn’s multiple comparison test.
To test the impact of PDE9-I on mitochondrial FA oxidation, adipocytes in the presence of ANP were treated with either PDE9-I or vehicle for 24 hours and the Seahorse mitochondrial stress test was performed. Figure 5D shows an example time tracing, and Figure 5E shows summary data. Basal and maximal oxygen consumption increased with either PDE9-I (5 μM PF-7943 [P9i-A] or 10 μM BAY-73-6691 [P9i-B]), and this increase was suppressed by coinhibition of PKG. When a similar protocol was performed using acute (1 hour) PDE9-I incubation, we found only a modest PKG-dependent increase in basal but no change in maximal O2 consumption (Supplemental Figure 10A). Similar studies performed in isolated cardiomyocytes found that chronic, but not acute, PDE9-I increases basal and maximal respiration (Supplemental Figure 10, B and C). In myocytes, we further tested the impact of PDE9 gain of function by using adenovirus to express Gfp and Pde9a (Adv-Gfp-Pde9a) in the cells; this lowered both basal and maximal respiration (Supplemental Figure 10D). Together, these results show that PDE9 localizes to mitochondria where it can suppress mitochondrial FA oxidation and respiration, and that its inhibition augments both.
PPARα activation is required for PDE9-I to reduce obesity and CMS.
To test whether PPARα activity is required for the antiobesity effects of PDE9-I, OVX females with preestablished DIO/mTAC were randomized to PDE9-I alone or PDE9-I plus PPARα inhibition (GW6471, 3 mg/kg i.p. every other day) for an additional 6 weeks. We chose this approach over using PPARα KO because we could first establish severe obesity/CMS prior to suppressing PPARα. GW6471 cotreatment reduced mRNA abundance of PPARα-regulated genes by 50%, supporting on-target efficacy (Figure 6A). Mice receiving only PDE9-I again displayed reduced total body weight and fat mass, no significant change in lean body mass, and increased VO2 and VCO2 over placebo. Coinhibition of PPARα prevented these changes (Figure 6B). Total food intake and daily activity were not significantly different between the groups (Supplemental Figure 11A). Compared with placebo, PDE9-I reduced iWAT and gWAT mass (Figure 6C) and modestly improved cardiac fractional shortening (Figure 6D), and these effects were also largely abrogated by concomitant PPARα blockade. As a further control, we randomized a cohort of DIO OVX mice to receive PPARα-I or vehicle but not PDE9-I over a similar duration. Changes in body weight and fat pad mass over time were similar in both groups (Supplemental Figure 11, B and C). Fractional shortening declined slightly with placebo but not PPARα-I, and there was no significant drug-time interaction by 2-way ANOVA (Supplemental Figure 11D). Thus, PPARα-I itself did not stimulate weight gain or LV dysfunction in this time frame. Together, these results show that in vivo efficacy of PDE9-I in DIO/mTAC requires PPARα activation.
Figure 6. Inhibition of PPARα blocks beneficial effects of PDE9-I in OVX model.
(A) Quantitative PCR of PPARα-associated genes in myocardial tissue from OVX obese/CMS mice treated with PDE9-I with and without PPARα-I. Mean ± SD, 2-step Benjamini-Krieger-Yekutieli multiple comparison Mann-Whitney, q values shown. *P ≤ 5 × 10–5; #P = 0.007. (B) Effect of PPARα coinhibition on PDE9-I–induced reductions in total body, fat, and lean mass, and increases in VO2 and CO2 in OVX obese/CMS mice. Results for placebo-treated OVX mice (derived from data in Figure 1) are plotted with median (dark line) and 75% to 25% confidence intervals (shaded). The addition of PPARα-I reversed PDE9-I–mediated responses in these parameters, returning their values to those with placebo for all by lean mass that rose by 5% (n = 10/group). P values displayed from Kruskal-Wallis (includes all 3 groups) with Dunn’s multiple comparison test. (C) iWAT and gWAT weight in OVX mice PDE9-I with and without PPARα-I versus placebo (n = 9/group). One-way ANOVA with Tukey’s multiple comparison test. (D) Percentage fractional shortening (FS) from the same experiment (n = 7, 5); 2-way ANOVA with Sidak’s multiple comparison test.
PDE9-I is effective in countering DIO in the absence of concomitant mTAC.
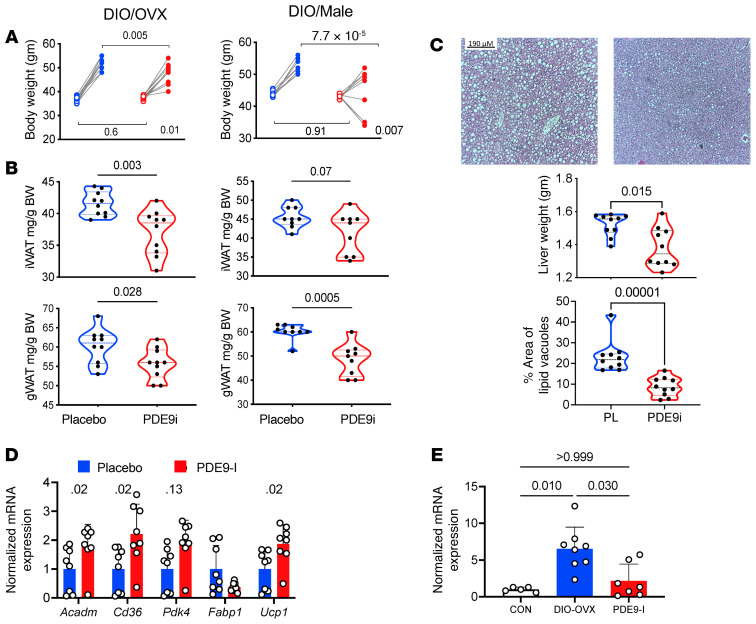
Our primary DIO/mTAC model was used to mimic key components of CMS and modestly enhance cGMP coupled to NP stimulation that is in turn regulated by PDE9. This leaves open the question of whether the therapy is also effective with DIO alone. To test this, we performed a second series of studies using the same duration of DIO and drug randomization, but without mTAC. PDE9-I also suppressed weight gain with DIO alone in both OVX female and male mice (Figure 7A), accompanied by reduced iWAT and gWAT mass (Figure 7B). Hepatic steatosis also declined substantially with active treatment (Figure 7C). PDE9-I also stimulated expression of PPARα-related FA metabolic genes in BAT in both OVX female (Figure 7D) and male (Supplemental Figure 12A) mice, though the magnitude of changes was smaller than that in the primary DIO/mTAC model (c.f. Figure 3B). Myocardial Nppa expression also rose with DIO-OVX alone and declined with PDE9-I (Figure 7E) as observed with DIO/mTAC (c.f. Figure 2C). These results establish that mTAC is not required to observe improvement in obesity and associated FA signaling from PDE9-I.
Figure 7. PDE9-I reduces obesity and fat mass and improves hepatic steatosis in OVX and male mice with DIO without superimposed pressure overload (mTAC).
(A) Body weight before and after 6 weeks of treatment with vehicle or PF-7493 (PDE9-I) in DIO OVX (n = 10) and male (n = 9) mice. P value in lower right is for interaction of drug treatment and time-change, by 2-way ANOVA. P values above are from Sidak’s multiple comparison test. Baseline values for each group were essentially identical. (B) Fat mass for iWAT and gWAT in both OVX and male DIO mice from same study. P values from Kruskal-Wallis test. (C) Example liver histology and summary data (n = 10/group). There was substantial steatosis reflected by increased hepatic mass and increased percentage area occupied by large lipid vacuoles. P values by Kruskal-Wallis test. (D) Expression of PPARα-associated FA regulatory genes normalized to Gapdh in BAT from DIO OVX mice (n = 7 or 8). Two-step Benjamini-Krieger-Yekutieli multiple comparison Kruskal-Wallis q values shown. (E) Expression of Nppa/Gapdh in normal diet control versus DIO with vehicle or PDE9-I. P values from Kruskal-Wallis with Dunn’s multiple comparison test.
PDE9-I is ineffective in non-OVX females with ERα shifting PPARα DNA binding away from FA oxidation–regulating genes.
Endogenous estrogen is suggested to blunt PPARα-stimulated gene expression and its corresponding regulation of FA oxidation (31). While the mechanisms remain unclear, this, along with the current findings that PPARα inhibition prevents PDE9-I from reducing obesity, led us to hypothesize that sexual dimorphism may also apply to effects from PDE9-I. We tested this in a separate group of non-OVX females subjected to the identical DIO/mTAC protocol. Interestingly, in non-OVX females, PDE9-I had negligible impact in these mice on total body, fat, or lean mass (Figure 8A), BAT adipocyte size or browning (Figure 8B), serum lipids (Supplemental Figure 13A), iWAT or gWAT mass (Supplemental Figure 13B), food intake, activity, indirect colorimetry (Supplemental Figure 13C), and hepatic steatosis (Supplemental Figure 13D). We also examined responses to PDE9-I in non-OVX females with DIO only and also found no impact on weight or fat mass increase (Supplemental Figure 12, B and C).
Figure 8. PDE9-I does not impact body weight, fat, or PPARα downstream signaling in OVX DIO/mTAC mice.
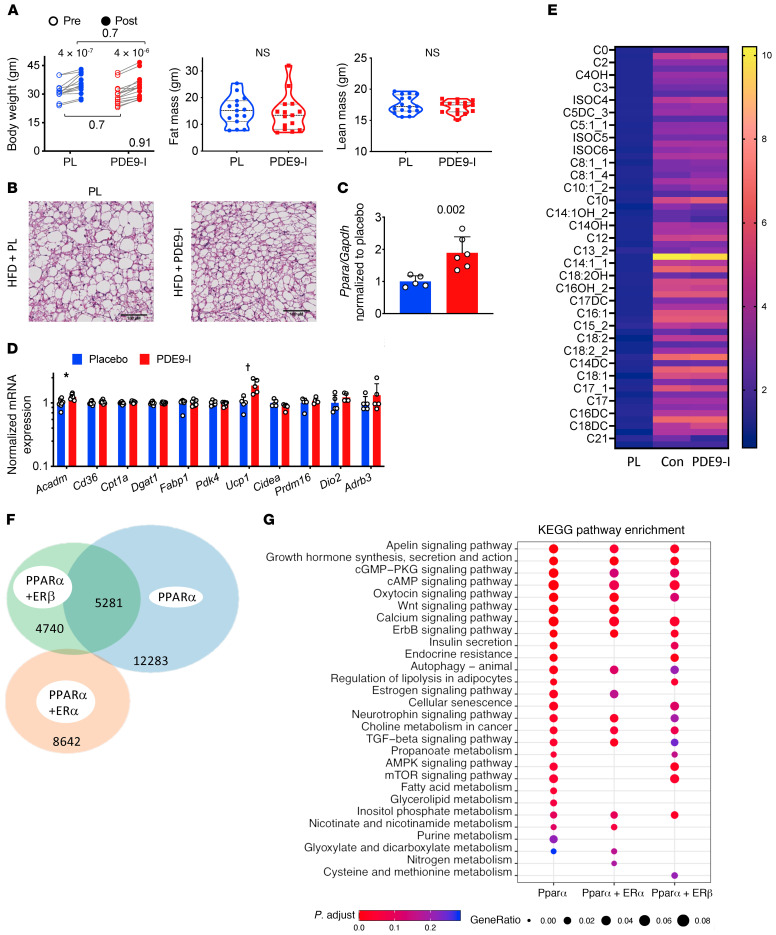
(A) Total body weight, fat, and lean mass in non-OVX obese/CMS female mice before and after 8 weeks of treatment with either placebo or PDE9-I. n = 15/group; 2-way ANOVA, P value for interaction of treatment × time shown at lower right; paired Sidak’s multiple comparison test also displayed. (B) BAT histology shows enlarged adipocytes and reduced mitochondria density in the placebo group, which was not changed with PDE9-I (repeated 3 times). Scale bars: 100 μm. (C) Ppara mRNA expression increases in non-OVX to levels similar to those for male and OVX mice (n = 5 or 6/group). P value by Mann-Whitney test. (D) qPCR results for fatty acid metabolism and mitochondrial respiration genes in BAT from non-OVX model (log-transformed, 2-step Benjamini-Krieger-Yekutieli multiple comparison Mann-Whitney test; *q = 0.03, †q = 0.04, all others q > 0.5). (E) Metabolomics of acylcarnitines in BAT from normal controls and obese/CMS non-OVX with placebo vs. PDE9-I (n = 5/group, 2-step Benjamini-Krieger-Yekutieli multiple comparison Mann-Whitney test; for placebo vs. PDE9-I all q values > 0.5; for control chow (nonobese) versus placebo, all but one P ≤ 0.001 (C22, P = 0.006). (F) Venn diagram of ChIP-Seq–identified PPARα binding sites in HepG2 cells with PPARα stimulation alone or combined with ERα or ERβ costimulation. (G) KEGG pathway analysis from ChIP-Seq–identified genes with PPARα binding shows loss of fat metabolism–related pathways by coactivation with ERα or ERβ.
The failure of PDE9-I to alter DIO/mTAC obesity/CMS in non-OVX females was not due to a lack of increased Ppara mRNA abundance, which rose similarly in these mice and in OVX females and males (Figure 8C, c.f. Figure 2F). However, mRNA abundance of genes regulated by PPARα was minimally altered by PDE9-I in these mice (Figure 8D) as well as non-OVX females with DIO only (Supplemental Figure 12D). Acylcarnitines in BAT were also minimally altered by active treatment (Figure 8E).
These results suggested that sex hormones in intact females impeded the capacity of PDE9-I to increase PPARα-regulated FA catabolism. Since Ppara expression still increased while mRNA abundance of its downstream regulated genes did not, we speculated that coincident estrogen receptor (ER) signaling may impede PPARα DNA binding to alter the latter’s transcriptional regulation. To test this, chromatin immunoprecipitation and sequencing (ChIP-Seq) for PPARα binding was performed using HepG2 cells transfected with either PPARα (+PPARα agonist) alone or in combination with ERα or ERβ transfection and their respective agonists. This cell model was used because it is well defined for studies of both fat metabolism and ER and PPARα signaling. We identified approximately 17,500 PPARα DNA binding sites when it was stimulated alone, but this number fell by nearly half and engaged mostly different sites if ERα was coactivated (Figure 8F). With ERβ coactivation, approximately one-third of binding sites with PPARα alone were maintained and one-third were altered, and there was also an overall reduction. Gene lists based on PPARα chromatin binding for the 3 conditions were subjected to KEGG pathway enrichment analysis, and we found that ERα coactivation particularly reduced PPARα binding to genes involved with FA metabolism (Figure 8G). This is consistent with the mRNA analysis (Figure 8, C and D) and the lack of obesity/CMS effects by PDE9-I in non-OVX mice.
Discussion
This study reveals that PDE9 localizes to mitochondria, and its suppression increases lipolysis and mitochondrial respiration, reducing total body fat, and fat accumulation in BAT, liver, and myocardium in mice with DIO/CMS. Reduced hepatic and visceral fat are important, as both are proinflammatory and worsen CMS morbidity (32). Cardiac effects include reduction of hypertrophy and fibrosis and corresponding molecular signaling, and improved heart function. PDE9-I did not appear to alter insulin resistance. While our main model combined DIO and mild cardiac pressure overload to better mimic CMS and augment NP signaling, we also found PDE9-I to be effective even without pressure overload, expanding its potential therapeutic use. At a molecular level, PDE9-I upregulates Ppara expression and activity, increasing the abundance of many genes that control fat uptake and catabolism and mitochondrial respiration and biogenesis. Inhibiting PPARα abrogates lipolytic and antiobesity effects of PDE9-I. Lastly, we find a striking sexual dimorphism, as PDE9-I induces these changes in OVX females and males but not non-OVX females. Redirection of PPARα chromatin binding away from genes regulating FA metabolism by ERα coactivation may underlie this dimorphism. PDE9 inhibitors have been tested in humans for other disorders and appear safe and well tolerated. Our findings support expansion into men and postmenopausal women with obesity/CMS, with or without concomitant pressure overload.
Several mechanisms linking PKG stimulation to lipolytic and/or antiobesity effects have been previously proposed. They include phosphorylation and activation of HSL, p38 MAP kinase (by an unknown intermediate; ref. 33) similar to the pathway used by PKA (34), and AMP-activated kinase (35). Their activation has been suggested to converge on transcriptional regulators PPARγ coactivator 1α (PGC1α) and PPARα (14, 15) to control metabolism. PKG-dependent PPARα activation has also been linked to mitochondrial protection against hypoxia-induced cardiac injury (18) and increased FA oxidation and catabolism in skeletal muscle (28), but not previously to reducing obesity. Furthermore, to our knowledge, none of these proposed effectors of PKG/obesity modulation have been reported to display sexual dimorphism. Yet crosstalk between estrogen signaling and PPARα activation has been reported (36). The similar dimorphism in PDE9-I responses coupled to differential downstream PPARα signaling supports the notion that the latter is central to the mechanism for PDE9-I metabolic and antiobesity effects. Estrogen itself stimulates fat metabolism and mitochondrial oxidative respiration (8), and its decrease after menopause or with OVX stimulates visceral adiposity, CMS, and reduced PPARα- and PGC1α-regulated signaling (7). ERα and PPARα share DNA binding motifs and protein partners, including RXR and coactivator PGC1α (36), and interference between ERα and PPARα has been reported in liver cancer (37) and apolipoprotein regulation (38). To our knowledge, the present data are the first to examine this interaction by ChIP-Seq and identify its specific targeting of FA metabolic genes. It seems likely that before menopause, ER signaling can substitute for PPARα regulation, whereas after menopause, PPARα becomes more prominent so its activation (e.g., from PDE9-I) counters obesity/CMS. In males, the situation is always similar to OVX females. The mechanisms for the transcriptional competition and their extension to other cell types remain to be explored.
While our results support a connection between PDE9 efficacy against DIO/CMS and PPARα activation, they do not explain how PDE9 localization to mitochondria (at least in myocytes) is key to this efficacy. Importantly, sustained but not acute PDE9-I increased mitochondrial respiration, suggesting PDE9-I is unlikely to do so by enhancing PKG phosphorylation of mitochondrial proteins directly controlling FA metabolism. Still, we find PKG activity to be important, so there is likely some phosphorylation target, and we suspect this is a yet-to-be identified protein that serves as a messenger to the nucleus to modulate PPARα and related gene expression. That PPARα promoter activity is augmented by cGMP does not mean this is a direct PKG effect but could involve other intermediates. Future studies are needed to dissect how such communication occurs. Another potential mechanism to couple PKG with PPARα may be mediated by mTORC1 activity. We have shown that PKG phosphorylates tuberous sclerosis complex protein 2 (39) at 1 of 2 tandem serines, resulting in a Rheb-dependent suppression of mTORC1. Prior studies found that mTORC1 is also a negative regulator of PPARα (40), so PKG suppression of mTORC1 could in turn enhance PPARα expression and activity. This mechanism is also being explored.
The metabolic effects of PDE9-I as shown here are compatible with its modulation of NP-derived cGMP (20, 25) and the demonstrated efficacy of NP to stimulate fat catabolism (14, 41–44) by enhancing FA oxidation. NP stimulation is also associated with enhanced adipocyte glucose uptake (45) and insulin sensitization (46, 47). These effects as well as diminution of DIO appear largely due to NP action in adipocytes as opposed to skeletal muscle (48), identifying fat as a major signaling nexus. The lack of change in a glucose-challenge response from PDE9-I despite increased lipolysis and fat/weight loss appears to differ from NP stimulation, but this could relate to different tissue-specific targeting. While Pde9a mRNA is detectable in skeletal muscle, its functionality remains unknown but may not be as relevant as in adipose tissue, heart, and liver. Skeletal muscle plays a key role in regulating postprandial insulin-mediated glucose uptake, so a disparity could contribute to the present observations. The data suggest that added benefit may come from combining PDE9-I with insulin sensitizers.
Compared with exogenous NP, PDE9-I has some therapeutic advantages. For one, it is a small molecule as opposed to a peptide and so can be administered orally. Furthermore, obesity blunts NP signaling by increasing expression of the NP-clearance receptor (NPRC, NPR3) in adipose tissue (48), greater NP glycosylation that suppresses cleavage of the pro-peptide form to the active peptide (49, 50), and reducing NP signaling receptors (51). However, PDE9 inhibition can still augment cGMP so long as there is some upstream stimulation. Our finding that PDE9-I is effective even without coactivation of NP by cardiac hemodynamic stress supports this. Further, while prior studies required gene deletion of NPRC to achieve weight reduction from exogenous NP (33), this was not needed for PDE9-I to be effective. Lastly, PDE9-I augments cGMP in cells in which it is expressed, whereas cGMP stimulation methods activate this pathway broadly. Its inhibition induces little to no change in systemic artery pressure in preclinical (23, 24) and clinical trials (52), whereas hypotension is common with cGMP-stimulation approaches.
Several antidiabetic therapies are being tested for weight loss, with the most recent success appearing from glucagon-like peptide 1 (13). This increases insulin secretion, enhancing glucose uptake and storage as glycogen, and its antiobesity effects are thought primarily to occur from appetite suppression with increased satiety (53) that can also be a dose-limiting side effect. Sodium-glucose cotransporter 2 (SGLT2) antagonists have also been studied for weight loss (54), although their impact is modest. Lastly, it is worth noting that the combination of sacubitril and valsartan (neprilysin inhibitor and AT-1 receptor blocker) for patients with HFpEF appears to have some efficacy (55) and was approved by the FDA for patients with an ejection fraction of at least 40%. The mechanism of benefit over AT-1 blockade alone is generally attributed to suppression of NP proteolysis by neprilysin (56). While no evidence to date has found concomitant changes in PPARα signaling or weight reduction (57), our findings suggest a potential synergy between these drugs and PDE9-I that could augment lipolysis, and this reduces obesity. This would be impactful given the high prevalence of obesity in patients with HFpEF.
We used PDE9 pharmacological inhibition rather than gene deletion to test its role, as we wanted to generate substantial obesity and CMS before impeding PDE9 activity. Results from a global Pde9-KO mouse subjected to DIO are presented in a separate study and also reveal less weight gain and metabolic improvement over time (58). While our model of obesity/CMS provides insights into HFpEF, it did not recapitulate true HFpEF, as the mice did not develop frank heart failure — with fluid retention and increased mortality. That could require greater pressure stress and/or a longer time for the combined stimuli. With DIO alone, we found essentially normal basal heart function, while dysfunction is well recognized with chronic obesity. Nonetheless, it is a reasonable model of severe obesity/CMS, with mTAC providing a second hemodynamic hit, as often occurs in patients with CMS. Notably, our model generated substantial obesity prior to testing any therapy, an approach that is less often taken but translationally important. We did not show direct evidence for PDE9-I–increased PKG activity, as this remains difficult to detect despite cGMP increases by this regulatory pathway. This likely relates to a membrane-localized nano-compartment whereby PDE9 cGMP signaling occurs but is difficult to find in assays of cell/tissue lysates. Still, the molecular signatures we report in the myocardium are consistent with PKG activation. Lastly, how PPARα and ERα interfere with each other at the level of chromatin binding remains unknown, as is the universality of the finding. However, our results identify a striking functional interaction consistent with the sexual dimorphism observed in the PDE9-I–mediated increase in Ppara-regulated genes and their physiological effects.
Gene and protein expression of PDE9 is most readily detected in the brain, so not surprisingly, initial pharmacological development of selective inhibitors focused on neurocognitive disorders, including schizophrenia and Alzheimer’s. While effective human translation remains elusive for these indications, preclinical data in the heart have since spawned clinical trials testing its impact on heart failure. Clinical studies are now ongoing, but data in humans support safety and tolerability of multiple PDE9 inhibitors as well as negligible impact on arterial blood pressure or heart rate. Given this, the present results suggest translation to humans with obesity and CMS is feasible and worth pursuing.
Methods
Obesity/CMS model.
C57BL/6N male and female mice (Jackson Laboratory) were fed standard chow or 60% HFD (D12492, Research Diets) starting at age 5 weeks. At 7 to 8 weeks, female mice were randomized to bilateral oophorectomy (OVX) or sham surgery, and all mice were then continued on their respective diet for approximately 4 months. For the majority of studies, a baseline echocardiogram was then obtained, and mice subsequently underwent an mTAC surgical procedure (stenosis sized by 26-G needle). The goal was to subject the heart to modest pressure load and stimulate some NP release. One week after mTAC, mice were further randomized to placebo (vehicle) or the selective PDE9 inhibitor PF-04447943 (PF-7493; 40 mg/kg/day) delivered in their oral chow (yields 10 nM free plasma concentration; see ref. 59) for 2 additional months. Another cohort followed the same basic protocol but without mTAC. In a third set of studies, DIO/mTAC OVX mice were randomized to PDE9-I alone or PDE9-I plus PPARα-I (GW6471, catalog 4618, Tocris Biosciences) dissolved in 1% DMSO, 95% saline, 4% polyethylene glycol 400, 3 mg/kg i.p. every other day) or vehicle. This treatment was then continued for 6 weeks. A control group of DIO OVX was treated with vehicle or PPARα-I only for 6 weeks. At the end of the treatment protocol, animals were assessed by echocardiography, indirect calorimetry, total body fat/lean mass by MRI, fat pad (inguinal and gonadal fat) mass, and serum glucose and lipids. Tissues were then harvested for histology and molecular assays, and frozen in liquid nitrogen and stored at –80°C until analyzed.
Mouse echocardiography.
Mouse echocardiography was performed in conscious mice using a Vevo 2100 (VisualSonics) with an 18 to 38 MHz transducer (SanoSite Inc.), and images analyzed with VisualSonics software. The sonographer was blinded to genotype and treatment. Ejection fraction and estimated wall mass were derived by using standard methods (20).
Blood collection and analysis.
Blood samples were collected by tail vein bleeding, and serum isolated using Microvette CB 300 (Sarstedt). Serum triglyceride and cholesterol were measured using an Infinity kit (Thermo Fisher Scientific).
Glucose tolerance test.
Mice were subjected to overnight food deprivation (approximately 16 hours), after which glucose (1 mg/kg) was injected intraperitoneally and baseline blood glucose measurement was taken from a clean tail blood droplet using a glucometer (BD Logic, NovaMax strips, catalog 8548043523). Repeat measurements were made at 15, 30, 60, and 120 minutes.
MRI for body composition analysis.
Total body fat and lean mass composition was determined using an EchoMRI 100 (Echo Medical Systems) by the Johns Hopkins University School of Medicine Mouse Phenotyping Core using standard protocols.
Indirect calorimetry, caloric intake, and physical activity measurements.
Indirect calorimetry and additional measures of food intake and physical activity were performed in open-circuit indirect calorimetry cages (Comprehensive Lab Animal Monitoring System, Columbus Instruments). Mice were monitored individually and continuously for 4 days: 2 to 3 days to monitor adaptation to the novel environment, and the third or fourth day for reporting data. The system periodically measured rates of O2 consumption (VO2) and CO2 production (VCO2) for each cage, as well as food intake and physical activity (infrared beam array). Data were processed in time segments (daily 24-hour, and 12-hour dark/12-hour light), and averages calculated for each as well as for the 24-hour period. VO2 and VCO2 were normalized to (lean mass + 0.2 × fat mass), as described previously (60), using EchoMRI 100 data acquired proximate to indirect calorimetry.
Lipid loading and impact of PDE9-I in cardiomyocytes.
Rat neonatal cardiomyocytes were isolated from newborn pups and cultured for 2 days prior to study. Cells were then incubated with 2.5 μM PF-7943 or vehicle for 24 hours prior to exposure to an FA mixture: oleic acid (catalog O1008, Sigma-Aldrich), linoleic acid (catalog L1376, Sigma-Aldrich), and palmitate acid (catalog P0500, Sigma-Aldrich) complexed (3:1 ratio) with 3% FA-free bovine serum albumin (BSA) at a concentration of 0.4 mmol/L. After an additional 24-hour incubation, cells were fixed in 4% formaldehyde for 30 minutes, washed with PBS 3 times, and stained with Oil Red O. After gentle washing with PBS 3 times, cells were imaged using phase-contrast microscopy (Zeiss AxioObserver A1 inverted microscope and Olympus DP80 dual RGB and monochrome camera) to identify lipid-laden vesicles. Images were processed with ImageJ (NIH) to determine vesicles/cell. In another assay, lipid-fed myocytes were washed with PBS, loaded with 5 μM BODIPY 493/503 (D3922, Invitrogen) for 20 minutes at 37°C, fixed in 4% paraformaldehyde, washed, permeabilized, and counterstained with 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) (D1306, Invitrogen). Images were taken with a Revolve microscope (Echo Laboratories) and analyzed (ImageJ, v1.52) after background subtraction and threshold detection.
Lipolysis assays.
Human subcutaneous preadipocytes (SP-2096, Zen-Bio), were grown and differentiated per the manufacturer’s instructions. Differentiated adipocytes were maintained in adipocyte maintenance medium and incubated with combinations of the following compounds for 24 hours: vehicle (0.1% DMSO), ANP (10 μM, catalog S-20647, Anaspec), PDE9-I (5 μM PF-7943, 5 μM BAY 73-6691, catalog B3561, MilliporeSigma), PPARα inhibitor GW6471 (10 μM, catalog 4618, Tocris), and soluble guanylate cyclase activator BAY 60-2770 (1 nmol/L, catalog SML2281, MilliporeSigma). Cells were first incubated with the respective comodulators for 3 hours prior to the addition of ANP. In another study, adipocytes were transfected with either scrambled control siRNA (25 nM, catalog D-001810-10-20) or human PDE9–targeted siRNA (25 nM, catalog L-003820-00-0005, Horizon Discovery) on day 10 of differentiation and 48 hours thereafter, and treated with ANP and PDE9-I for an additional 24 hours. Lipolysis was assessed by glycerol release using an adipocyte lipolysis assay kit (LIP-1-NC, Zen-Bio) per the manufacturer’s protocol.
Lipolysis was also assessed in HepG2 cells (human, ATCC cell line HB-8065) and in NRVMs. For NRVMs, day 1 to 2 newborn Sprague-Dawley rats (Envigo) were placed in an anesthesia chamber with a paper towel that had been saturated with isoflurane and then sterilized in ethanol. The chest was opened via midline incision, the heart removed, placed in Krebs-Henseleit buffer, and mechanistically dissociated. The disaggregated myocytes were then placed in cell culture dishes and incubated in DMEM at 37°C for 1 to 2 days prior to use. HepG2 cells were seeded at a density of 5 × 103 cells/well and NRVMs plated at a density of 40,000/well in 96-well plates. Cells were maintained in medium containing FA mixture and treated with ANP, PDE9-I (PF-7943), and PPARα-I (all at 10 μM), for 24 hours. In one set of NRVM studies, myocytes were transfected with either scrambled control siRNA (25 nM, catalog D-001810-10) or rat PDE9–targeted siRNA (25 nM, catalog L-098890-02) from Horizon Discovery.
Effect of PDE9 on mitochondrial oxygen consumption and ATP generation.
Mitochondrial respiration was determined from oxygen consumption rate (OCR) measured by a Seahorse XF96 Extracellular Flux Analyzer (Seahorse Biosciences), per the manufacturer’s protocol. Both adipocytes and NRCMs were studied. For NRCMs, cells were seeded onto fibronectin-coated wells of an Agilent Seahorse XF96 cell culture plate in 10% FBS containing DMEM. Twenty-four hours later, a confluent monolayer of spontaneously beating NRCMs formed. Media were replaced with DMEM plus FA mixture as noted above plus 0.5 mM carnitine and cultured for an additional 24 hours. For acute studies, NRCMs were treated with vehicle or PDE9-I (PF-7943 5 μM) for 1 hour, and for chronic, for 24 hours prior to the exposure to FA-containing medium. In another study, NRCMs were infected with adenovirus expressing GFP or PDE9-GFP (C-terminus, MOI 10, incubated for 48 hours prior to OCR measurements). To assess OCR, cells were tested in DMEM without phenol red but supplemented with sodium pyruvate (pH 7.4). After calibration of the sensor cartridge, the XF96 plate was placed into the Seahorse instrument and OCR measured in pmol/min/cell. Analysis of mitochondrial function was made using changes in OCR in the presence of 1.0 μM oligomycin (inhibits complex V – basal OCR determined), 1.0 μM FCCP (uncoupling agent revealing maximal OCR through complex IV), 0.5 μM rotenone and antimycin A (inhibits complex 1, shutting down mitochondrial respiration to calculate nonmitochondrial respiration). After each injection, OCR was assessed 4 times at 5 different time points. The results were normalized to cell protein in each well and averaged.
Similar analysis was performed in differentiated human subcutaneous preadipocytes. Cells were maintained in adipocyte maintenance medium for 24 hours and then incubated with a combination of the following for 1 hour (acute) or 24 hours (chronic): vehicle (0.1% DMSO), PDE9-I (5 μM PF-7943, 5 μM BAY 73-6691, catalog B3561, MilliporeSigma), with and without the PKG inhibitor DT3 (1 μM) added 1 hour prior to treatment with PDE9-I.
PPARα promoter activity assay.
HepG2 (human, ATCC cell line HB-8065) cells were cultured in DMEM (10% FBS, 1% penicillin/streptomycin, Thermo Fisher Scientific) to 70% confluence. Cells were transfected using Xfect transfection reagent (catalog 631317, Takara Bio) with recombinant plasmid containing the 1206-bp upstream TSS of human PPARα promoter-luciferase reporter (0.3 μg) (provided by Bert Staels, University of Lille, Lille, France). Renilla-luciferase plasmid (0.001 μg; Promega) was transfected as an internal control. After 24 hours, cells were incubated with 8-pCPT-cGMP (C5438, Sigma-Aldrich) at varying concentrations for 6 hours. Cell lysates were then extracted using passive lysis buffer (E1910, Promega), and luciferase activity measured by Dual-Luciferase Reporter Assay (E1910, Promega) using a GloMax96 plate reader (BioTek).
Mitochondrial PDE9-localization analysis.
NRCMs were infected with adenovirus expressing PDE9-Flag (10 MOI) and maintained in DMEM with 10% FBS. After 48 hours, cells were scraped into ice-cold PBS and spun at 800g for 2 minutes, and mitochondria isolated using a kit per the manufacturer’s instructions (Thermo Fisher Scientific, catalog 89874, lot TJ272027). Mitochondria were reconstituted in SDS sample loading buffer (Licor, catalog 928-40004), run in 4% to 20% Mini-Protean TGX gels (Bio-Rad), and blotted onto a nitrocellulose membrane (Bio-Rad). Antibodies against the following proteins were used: α-tubulin (Cell Signaling Technology, 3873S, lot 12), GFP (Invitrogen, A6455, lot 2185052), Flag (Sigma-Aldrich, F1804, lot SLBW5142), VDAC1/porin (Abcam, ab15895, lot GR3237200-1), and Total Protein Stain (Licor, 926-11016). Antibody binding was visualized with an infrared imaging system (Odyssey, Licor) and quantification by Odyssey Application Software 3.1. See complete unedited blots in the supplemental material.
Light microscopy histology.
Immediately after removal, liver, iWAT, gWAT, and BAT were fixed overnight by immersion at 4°C in 4% paraformaldehyde. Paraffin-embedded, dewaxed, 4-μm sections were stained using hematoxylin and eosin.
Transmission electron microscopy and immunogold staining.
Freshly extracted heart tissues were fixed in 2.5% glutaraldehyde, 3 mM MgCl2, 0.1 M sodium phosphate (Sorenson’s) buffer, pH 7.2, for 1 hour at room temperature. After a buffer rinse, samples were postfixed in 1% osmium tetroxide and 0.8% potassium ferrocyanide in 0.1 M Sorenson’s buffer (1 hour) on ice in the dark. Following a 0.1 M maleate buffer rinse, samples were stained with 2% uranyl acetate (0.22 μm filtered, 1 hour in the dark) in maleate buffer, dehydrated in a graded series of ethanol, followed by propylene oxide and embedded in Eponate 12 (Ted Pella, Inc.) resin. Samples were polymerized at 60°C overnight. Thin sections, 60 to 90 nm, were cut with a diamond knife on the Reichert-Jung Ultracut E ultramicrotome and picked up with 2 × 1 mm formvar-coated copper slot grids. Grids were stained with 2% uranyl acetate in 50% methanol and 0.4% lead citrate before imaging on a Hitachi 7600 TEM at 80 kV. Images were captured with an AMT XR50 CCD (5 megapixel) camera at the Johns Hopkins University School of Medicine Microscopy Core. Mitochondrial density and area measurements were performed using ImageJ (v1.52). Scales were set based on the 500-nm legend for each image. The adjust threshold function was used to highlight mitochondria and the measure function to determine pixel intensity and area of the highlighted region.
For immunogold staining, NRVMs were plated in a 6-well plate (1 × 105 cells per well). Twenty-four hours later, they were infected with adenovirus expressing either GFP alone or GFP-tagged PDE9 (MOI 10). Forty-eight hours after transfection, cells were fixed in EM grade 4% paraformaldehyde and 0.1% glutaraldehyde in 80 mM phosphate buffer (Sorenson’s buffer) with 3 mM magnesium chloride, pH 7.2. The subsequent preparation steps to obtain the stained EM grids are provided in Supplemental Methods. Images were then captured at 120 kV on a Ceta Camera (8 megapixel CCD, 16 bit, 40 fps) on a Talos L120C G2 Transmission Electron Microscope at the Johns Hopkins University School of Medicine Microscopy Core.
RNA isolation and gene expression analysis.
Total RNA from heart, WAT, and BAT was extracted using TRIzol reagent (catalog 15596026, Invitrogen) per the manufacturer’s instructions. A High-Capacity RNA-to-cDNA Kit (catalog 4388950, Applied Biosystems) was used to reverse transcribe the RNA into cDNA. The PCR-array gene expression profiling was performed using the Bio-Rad Prime PCR plates dedicated to Carbohydrate (10029626, M384) and Lipid metabolism (10040365, M384) gene panels according to the manufacturer’s instruction.
Quantitative real-time PCR analysis was carried out using TaqMan specific primers that are listed in Supplemental Methods. The threshold cycle (Ct) values were determined by the crossing-point method and normalized to GAPDH (Applied Biosystems) values for each run.
cGMP assay.
Tissue cGMP levels were measured by EIA assay (Cyclex), as described previously (20).
Metabolic profiling of BAT and myocardial tissue for acylcarnitines.
Upon sacrifice, 5 replicates of each experimental series of heart and BAT tissues were washed for 5 seconds in ice-cold PBS to remove excess blood and then snap-frozen in liquid nitrogen. Tissues were ground in liquid nitrogen. Frozen ground tissue powder (15 mg) was further homogenized using ice -cold extraction solution (40% acetonitrile/40% methanol/20% water, spiked with internal standards). Homogenized samples were then incubated at –20°C for 1 hour, after which all the samples were centrifuged at 16.2g (Sorvall Legend Micro 21R Microcentrifuge) for 15 minutes at 4°C. Supernatants were transferred to new glass tubes and dried using a speed vac. Pellets were used for protein estimation using the BCA method.
Extracted myocardial tissue and BAT samples were assayed using stable-isotope dilution (2H3-carnitine (d3C0), 2H3-acetyl- (d3C2), 2H3-propionyl- (d3C3), 2H3-butyryl- (d3C4), 2H9-isovaleryl- (d9C5), 2H3-octanoyl- (d3C8), 2H9-lauroyl- (d9C12), 2H9-miristoyl- (d9C14), 2H3-palmitoyl- (d3C16) and 2H3-stearoylcarnitine (d3C18). Cambridge Isotope Laboratories (CIL) NSK-B-US and NSK-B-US-G1 standards were reconstituted in 0.5 mL 10% methanol in water plus 0.1% formic acid by means of 10 minutes of orbital shaking and thorough vortexing.
One hundred twelve species comprising aliphatic acylcarnitines ranging from C0 (free) to docosanoyl- (C22), and including odd-chain, branched-chain, unsaturated, hydroxylated, and dicarboxylic moieties, were analyzed using a Waters Acquity UHPLC in a ACE C18-PFP column (2.1 × 100 mm, 3 μm particle size) at 0.3 mL/min and 30°C with water partitioning in 3:1 acetonitrile/methanol (by volume), both with 0.1% formic in gradient elution (5 minutes, 0% B; 8 minutes to 23% B and then to 100% B in 18 minutes) and monitored in a Sciex 4500 triple quadrupole mass spectrometer using optimized specific transitions to class-specific product ion m/z 85 at defined retention times. Neutral loss of trimethylamine from the parent ion fragment was also monitored as a confirming transition. Injection volume was 5 μL.
Species were quantified using 6 non-zero-level calibration curves using the authentic standard (CIL, NSK-B-US and NSK-B-US-G1) where possible, spiked in analyte stripped serum (Biocell Laboratories); if not available, the closest in the homolog series was used. Unsaturated species were assayed using the same response factor as the corresponding aliphatic species of the same chain length. Hydroxylated are referred to as hydroxyisovaleryl- (C5OH) or hydroxylpalmitoylcarnitine (C16OH) and dicarboxylic to glutarylcarnitine (C5DC). Replicates showed acceptable coefficients of variation (20%).
ChIP-Seq analysis of PPARα binding and influence of ERα and ERβ coactivation.
ChIP was performed in HepG2 cells with 2 independent full biological replicates. Approximately 5 × 106 HepG2 cells (human, ATCC cell line, HB-8065) were cultured in three 15-cm-diameter plates in DMEM with 10% FBS. After 24 hours, cells were transfected with expression vectors for either PPARα alone (human-tagged ORF, RC216176, Origene), or combined with ERα (pCMV ERalpha, 101141, Addgene) or ERβ (pcDNA Flag ERbeta, 35562 Addgene). Forty-eight hours after transfection, cells were treated with a PPARα agonist alone for 2 hours (10 μM, Wy14643, 1312, Sigma-Aldrich) or first with an ERα agonist (10 nM, PPT, 1426/50, Tocris Biosciences) or ERβ agonist (10 nM, DPN, 1494/50, Tocris Biosciences) for 45 minutes, followed by a 2-hour incubation with the PPARα agonist. A SimpleChIP Enzymatic Chromatin IP kit (catalog 9003, Cell Signaling Technology) was used for the immunoprecipitation per the manufacturer’s instructions. Cells were washed twice with ice-cold PBS and then crosslinked by incubating with 1% formaldehyde for 10 minutes. The reaction was stopped by the addition of 2.5 M glycine and the plates washed twice with ice-cold PBS. Chromatin was treated with nuclease and sonicated. Immunoprecipitation was performed with PPARα-specific antibody (ab227074 ChIP-grade, Abcam) and Protein G magnetic beads. Chromatin was eluted from the beads, decrosslinked, and purified. ChIP and input DNA libraries were prepared using a Clontech DNA SMART kit (catalog 634866, Takara Bio). ChIP-Seq was performed according to the ENCODE (The Encyclopedia of DNA Elements) consortium’s standards and guidelines. Sequencing was carried out by the JHU Genomics Core Facility using an Illumina NextSeq 500 sequencing system. Irreproducible discovery rate was used to evaluate reproducibility of the experiments by measuring consistency between our biological replicates within an experiment. The raw ChIP-Seq data have been deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE156956). Data quality was validated using FastQC (v0.11.9). Sequenced reads were trimmed for adaptor sequence, and for low-quality sequence, using Trim Galore v0.6.4_dev. Gene Alignment was done to hg38 whole genome using bowtie (v1.0.1) and peak locations were determined using the MACS2 algorithm v2.2.7.1 (0.01 FDR cutoff, fold enrichment 2, 3, and 5). Peaks were annotated using Homer v4.11 and functional analysis was performed using R (version 3.3.6), ChIPSeeker (v1.18.0, TSS region –2500, 2500) and DiffBind (v2.16) packages. We identified clearly distinct grouping of PPARα alone versus combined with either ERα or ERβ by non-negative matrix factorization and principal component analysis (Supplemental Figure 14).
Statistics.
All statistical analysis was performed using GraphPad Prism 9.2. For data involving 2 grouping factors, e.g., effect of time and drug intervention, analysis was by 2-way ANOVA, with Sidak’s multiple comparison test. For multiple group comparison, 1-way ANOVA, Welch’s ANOVA (does not assume equal variances among groups), or a Kruskal-Wallis nonparametric test was employed, with corresponding multiple comparison tests. The majority of unpaired comparisons between 2 groups was performed using a nonparametric Mann-Whitney U test. Much of the data are presented as violin plots, showing the distribution, median, 25% and 75% IQRs, and all individual data. Others are provided as bar graphs with all individual data displayed and mean ± SD shown. Sample size is provided for each figure legend. Data that did not have sufficient sample size (e.g., <6/group) to assess for normality were analyzed using nonparametric statistics. A P value of less than 0.05 was considered significant. All absolute P values are displayed in the figures themselves or provided in the corresponding legends.
Study approval.
The animal protocol was approved by the Johns Hopkins University Animal and Care Use Committee.
Author contributions
SM performed the majority of experiments and performed data analysis. NS maintained and performed the studies with the in vivo DIO/mTAC model. BDE performed mitochondrial/cytosol fraction assays. SR and DCS performed metabolic assays. RPC contributed BAT histology. SAM and HK assisted with ChIP-Seq informatics. VPJ assisted with lipolysis assays. DA helped with Seahorse analysis. CUO performed echo imaging. JAG and DDS performed and helped analyze metabolic profiling. BOR, GWW, and SC assisted with data interpretation and analysis and in editing the manuscript. DAK wrote and organized the manuscript and figures, performed data analysis, and was responsible for the study overall.
Supplementary Material
Acknowledgments
The authors thank the Johns Hopkins University School of Medicine (JHUSOM) Rodent Metabolism Core at the Center for Metabolism and Obesity Research for indirect calorimetry and glucose tolerance tests, the Phenotyping Core for MRI body composition analysis, the Small Animal CV Phenotyping and Models Core for echocardiography imaging and analysis and pressure-overload surgery, the JHU Imaging Core for electron microscopy slide preparation, and the JHU Deep Sequencing and Microarray Core Facility for sequencing. The study was supported by NIH grants R35-HL135827, RO1-HL-119012, P01HL10715, and AHA 16SFRN28620000 (to DAK); DK084171 (to GWW); RO1-HL134821 (to BOR); and DK116625 (to SC) and by AHA grants 16SFRN28620000 (to SC) and 16SFRN28420002 (to DDS) NIH grant F32 DK-116520 (to RPC), and OE 688/1-1 (Deutsche Forschungsgemeinschaft, to CUO). PF-04447943 was provided by Pfizer Inc. under a material transfer agreement.
Version 1. 10/07/2021
In-Press Preview
Version 2. 11/01/2021
Electronic publication
Footnotes
Conflict of interest: DAK is a consultant and advisory board member for Cytokinetics, Inc., and has received research support from Boehringer Ingelheim, Intra-Cellular Therapies, and Cytokinetics, Inc. Both SM and DAK are coinventors on a patent (PCT WO1010076752A1, US patent pending) filed by Johns Hopkins University titled “Phosphodiesterase 9 inhibition as treatment for obesity-related conditions.”
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(21):e148798.https://doi.org/10.1172/JCI148798.
Contributor Information
Sumita Mishra, Email: smishr12@jhmi.edu.
Nandhini Sadagopan, Email: nandhini.sadagopan@gladstone.ucsf.edu.
Susana Rodriguez, Email: srodri12@jhmi.edu.
Dylan C. Sarver, Email: dsarver1@jhmi.edu.
Ryan P. Ceddia, Email: ryan.p.ceddia@vanderbilt.edu.
Sean A. Murphy, Email: smurph50@jhu.edu.
Hildur Knutsdottir, Email: hknutsd1@jhu.edu.
Vivek P. Jani, Email: vjani1@jhmi.edu.
Deepthi Ashok, Email: dashok1@jhmi.edu.
Christian U. Oeing, Email: Christian.Oeing@gmx.de.
Brian O’Rourke, Email: bor@jhmi.edu.
Jon A. Gangoiti, Email: jgangoiti@ucsd.edu.
Dorothy D. Sears, Email: Dorothy.Sears@asu.edu.
G. William Wong, Email: gwwong@jhmi.edu.
Sheila Collins, Email: sheila.collins@vanderbilt.edu.
References
- 1.Koliaki C, et al. Obesity and cardiovascular disease: revisiting an old relationship. Metabolism. 2019;92:98–107. doi: 10.1016/j.metabol.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Blüher M. Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol. 2019;15(5):288–298. doi: 10.1038/s41574-019-0176-8. [DOI] [PubMed] [Google Scholar]
- 3.Piché ME, et al. Obesity phenotypes, diabetes, and cardiovascular diseases. Circ Res. 2020;126(11):1477–1500. doi: 10.1161/CIRCRESAHA.120.316101. [DOI] [PubMed] [Google Scholar]
- 4.Kahn CR, et al. Altered adipose tissue and adipocyte function in the pathogenesis of metabolic syndrome. J Clin Invest. 2019;129(10):3990–4000. doi: 10.1172/JCI129187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao VN, et al. Regional adiposity and heart failure with preserved ejection fraction. Eur J Heart Fail. 2020;22(9):1540–1550. doi: 10.1002/ejhf.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becher T, et al. Brown adipose tissue is associated with cardiometabolic health. Nat Med. 2021;27(1):58–65. doi: 10.1038/s41591-020-1126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morselli E, et al. The effects of oestrogens and their receptors on cardiometabolic health. Nat Rev Endocrinol. 2017;13(6):352–364. doi: 10.1038/nrendo.2017.12. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Z, et al. Estrogen receptor α controls metabolism in white and brown adipocytes by regulating Polg1 and mitochondrial remodeling. Sci Transl Med. 2020;12(555):eaax8096. doi: 10.1126/scitranslmed.aax8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obokata M, et al. Evidence supporting the existence of a distinct obese phenotype of heart failure with preserved ejection fraction. Circulation. 2017;136(1):6–19. doi: 10.1161/CIRCULATIONAHA.116.026807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitzman DW, Nicklas BJ. Pivotal role of excess intra-abdominal adipose in the pathogenesis of metabolic/obese HFpEF. JACC Heart Fail. 2018;6(12):1008–1010. doi: 10.1016/j.jchf.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra S, Kass DA. Cellular and molecular pathobiology of heart failure with preserved ejection fraction. Nat Rev Cardiol. 2021;18(6):400–423. doi: 10.1038/s41569-020-00480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitzman DW, Lam CSP. Obese heart failure with preserved ejection fraction phenotype: from pariah to central player. Circulation. 2017;136(1):20–23. doi: 10.1161/CIRCULATIONAHA.117.028365. [DOI] [PubMed] [Google Scholar]
- 13.Wilding JPH, et al. Once-weekly semaglutide in adults with overweight or obesity. N Eng J Med. 2021;384(11):989. doi: 10.1056/NEJMoa2032183. [DOI] [PubMed] [Google Scholar]
- 14.Collins S. A heart-adipose tissue connection in the regulation of energy metabolism. Nat Rev Endocrinol. 2014;10(3):157–163. doi: 10.1038/nrendo.2013.234. [DOI] [PubMed] [Google Scholar]
- 15.Vinnakota S, Chen HH. The importance of natriuretic peptides in cardiometabolic diseases. J Endocr Soc. 2020;4(6):bvaa052. doi: 10.1210/jendso/bvaa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moro C. Targeting cardiac natriuretic peptides in the therapy of diabetes and obesity. Expert Opin Ther Targets. 2016;20(12):1445–1452. doi: 10.1080/14728222.2016.1254198. [DOI] [PubMed] [Google Scholar]
- 17.Cannone V, et al. Atrial natriuretic peptide: a molecular target of novel therapeutic approaches to cardio-metabolic disease. Int J Mol Sci. 2019;20(13):3265. doi: 10.3390/ijms20133265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horscroft JA, et al. Inorganic nitrate, hypoxia, and the regulation of cardiac mitochondrial respiration-probing the role of PPARα. FASEB J. 2019;33(6):7563–7577. doi: 10.1096/fj.201900067R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann LS, et al. Stimulation of soluble guanylyl cyclase protects against obesity by recruiting brown adipose tissue. Nat Commun. 2015;6:7235. doi: 10.1038/ncomms8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee DI, et al. Phosphodiesterase 9A controls nitric-oxide-independent cGMP and hypertrophic heart disease. Nature. 2015;519(7544):472–476. doi: 10.1038/nature14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasaki H, et al. PDE5 inhibitor efficacy is estrogen dependent in female heart disease. J Clin Invest. 2014;124(6):2464–2471. doi: 10.1172/JCI70731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukuma N, et al. Estrogen receptor-α non-nuclear signaling confers cardioprotection and is essential to cGMP-PDE5 inhibition Efficacy. JACC Basic Transl Sci. 2020;5(3):282–295. doi: 10.1016/j.jacbts.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards DA, et al. CRD-733, a novel PDE9 (phosphodiesterase 9) inhibitor, reverses pressure overload-induced heart failure. Circ Heart Fail. 2021;14(1):e007300. doi: 10.1161/CIRCHEARTFAILURE.120.007300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott NJA, et al. Hemodynamic, hormonal, and renal actions of phosphodiesterase-9 inhibition in experimental heart failure. J Am Coll Cardiol. 2019;74(7):889–901. doi: 10.1016/j.jacc.2019.05.067. [DOI] [PubMed] [Google Scholar]
- 25.Harms JF, et al. Phosphodiesterase 9A in brain regulates cGMP signaling independent of nitric-oxide. Front Neurosci. 2019;13:837. doi: 10.3389/fnins.2019.00837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Omar B, et al. Alterations in cyclic nucleotide phosphodiesterase activities in omental and subcutaneous adipose tissues in human obesity. Nutr Diabetes. 2011;1:e13. doi: 10.1038/nutd.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Methawasin M, et al. Phosphodiesterase 9a inhibition in mouse models of diastolic dysfunction. Circ Heart Fail. 2020;13(5):e006609. doi: 10.1161/CIRCHEARTFAILURE.119.006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashmore T, et al. Nitrate enhances skeletal muscle fatty acid oxidation via a nitric oxide-cGMP-PPAR-mediated mechanism. BMC Biol. 2015;13:110. doi: 10.1186/s12915-015-0221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shao D, et al. Increasing fatty acid oxidation prevents high-fat diet-induced cardiomyopathy through regulating parkin-mediated mitophagy. Circulation. 2020;142(10):983–997. doi: 10.1161/CIRCULATIONAHA.119.043319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsushima K, et al. Mitochondrial reactive oxygen species in lipotoxic hearts induce post-translational modifications of AKAP121, DRP1, and OPA1 that promote mitochondrial fission. Circ Res. 2018;122(1):58–73. doi: 10.1161/CIRCRESAHA.117.311307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeong S, Yoon M. Inhibition of the actions of peroxisome proliferator-activated receptor alpha on obesity by estrogen. Obesity (Silver Spring) 2007;15(6):1430–1440. doi: 10.1038/oby.2007.171. [DOI] [PubMed] [Google Scholar]
- 32.Ferrara D, et al. Impact of different ectopic fat depots on cardiovascular and metabolic diseases. J Cell Physiol. 2019;234(12):21630–21641. doi: 10.1002/jcp.28821. [DOI] [PubMed] [Google Scholar]
- 33.Bordicchia M, et al. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J Clin Invest. 2012;122(3):1022–1036. doi: 10.1172/JCI59701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robidoux J, et al. Selective activation of mitogen-activated protein (MAP) kinase kinase 3 and p38alpha MAP kinase is essential for cyclic AMP-dependent UCP1 expression in adipocytes. Mol Cell Biol. 2005;25(13):5466–5479. doi: 10.1128/MCB.25.13.5466-5479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Souza SC, et al. Atrial natriuretic peptide regulates lipid mobilization and oxygen consumption in human adipocytes by activating AMPK. Biochem Biophys Res Commun. 2011;410(3):398–403. doi: 10.1016/j.bbrc.2011.05.143. [DOI] [PubMed] [Google Scholar]
- 36.Keller H, et al. Signaling cross-talk between peroxisome proliferator-activated receptor/retinoid X receptor and estrogen receptor through estrogen response elements. Mol Endocrinol. 1995;9(7):794–804. doi: 10.1210/mend.9.7.7476963. [DOI] [PubMed] [Google Scholar]
- 37.Jeng LB, et al. Fenofibrate induced PPAR alpha expression was attenuated by oestrogen receptor alpha overexpression in Hep3B cells. Environ Toxicol. 2018;33(2):234–247. doi: 10.1002/tox.22511. [DOI] [PubMed] [Google Scholar]
- 38.Puckey LH, Knight BL. Interaction of oestrogen and peroxisome proliferator-activated receptors with apolipoprotein(a) gene enhancers. Biochem J. 2002;366(pt 1):157–163. doi: 10.1042/BJ20020293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranek MJ, et al. PKG1-modified TSC2 regulates mTORC1 activity to counter adverse cardiac stress. Nature. 2019;566(7743):264–269. doi: 10.1038/s41586-019-0895-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sengupta S, et al. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468(7327):1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 41.Moro C, Lafontan M. Natriuretic peptides and cGMP signaling control of energy homeostasis. Am J Physiol Heart Circ Physiol. 2013;304(3):H358–H368. doi: 10.1152/ajpheart.00704.2012. [DOI] [PubMed] [Google Scholar]
- 42.Schlueter N, et al. Metabolic actions of natriuretic peptides and therapeutic potential in the metabolic syndrome. Pharmacol Ther. 2014;144(1):12–27. doi: 10.1016/j.pharmthera.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 43.Sanyal A, et al. Interplay between obesity-induced inflammation and cGMP signaling in white adipose tissue. Cell Rep. 2017;18(1):225–236. doi: 10.1016/j.celrep.2016.12.028. [DOI] [PubMed] [Google Scholar]
- 44.Neeland IJ, et al. Higher natriuretic peptide levels associate with a favorable adipose tissue distribution profile. J Am Coll Cardiol. 2013;62(8):752–760. doi: 10.1016/j.jacc.2013.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coue M, et al. Natriuretic peptides promote glucose uptake in a cGMP-dependent manner in human adipocytes. Sci Rep. 2018;8(1):1097. doi: 10.1038/s41598-018-19619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimura H, et al. Treatment with atrial natriuretic peptide induces adipose tissue browning and exerts thermogenic actions in vivo. Sci Rep. 2021;11(1):17466. doi: 10.1038/s41598-021-96970-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verboven K, et al. Natriuretic peptides in the control of lipid metabolism and insulin sensitivity. Obes Rev. 2017;18(11):1243–1259. doi: 10.1111/obr.12598. [DOI] [PubMed] [Google Scholar]
- 48.Wu W, et al. Enhancing natriuretic peptide signaling in adipose tissue, but not in muscle, protects against diet-induced obesity and insulin resistance. Sci Signal. 2017;10(489):eaam6870. doi: 10.1126/scisignal.aam6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Semenov AG, et al. Processing of pro-brain natriuretic peptide is suppressed by O-glycosylation in the region close to the cleavage site. Clin Chem. 2009;55(3):489–498. doi: 10.1373/clinchem.2008.113373. [DOI] [PubMed] [Google Scholar]
- 50.Goetze JP, et al. Cardiac natriuretic peptides. Nat Rev Cardiol. 2020;17(11):698–717. doi: 10.1038/s41569-020-0381-0. [DOI] [PubMed] [Google Scholar]
- 51.Verboven K, et al. Attenuated atrial natriuretic peptide-mediated lipolysis in subcutaneous adipocytes of obese type 2 diabetic men. Clin Sci (Lond) 2016;130(13):1105–1114. doi: 10.1042/CS20160220. [DOI] [PubMed] [Google Scholar]
- 52.Schwam EM, et al. A multicenter, double-blind, placebo-controlled trial of the PDE9A inhibitor, PF-04447943, in Alzheimer’s disease. Curr Alzheimer Res. 2014;11(5):413–421. doi: 10.2174/1567205011666140505100858. [DOI] [PubMed] [Google Scholar]
- 53.Brown E, et al. Weight loss variability with SGLT2 inhibitors and GLP-1 receptor agonists in type 2 diabetes mellitus and obesity: Mechanistic possibilities. Obes Rev. 2019;20(6):816–828. doi: 10.1111/obr.12841. [DOI] [PubMed] [Google Scholar]
- 54.Pereira MJ, Eriksson JW. Emerging role of SGLT-2 inhibitors for the treatment of obesity. Drugs. 2019;79(3):219–230. doi: 10.1007/s40265-019-1057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Solomon SD, et al. Angiotensin-neprilysin inhibition in heart failure with preserved ejection fraction. N Engl J Med. 2019;381(17):1609–1620. doi: 10.1056/NEJMoa1908655. [DOI] [PubMed] [Google Scholar]
- 56.Nougué H, et al. Effects of sacubitril/valsartan on neprilysin targets and the metabolism of natriuretic peptides in chronic heart failure: a mechanistic clinical study. Eur J Heart Fail. 2019;21(5):598–605. doi: 10.1002/ejhf.1342. [DOI] [PubMed] [Google Scholar]
- 57.Croteau D, et al. Differential effects of sacubitril/valsartan on diastolic function in mice with obesity-related metabolic heart disease. JACC Basic Transl Sci. 2020;5(9):916–927. doi: 10.1016/j.jacbts.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ceddia RP, Liu D, Shi F, Crowder MK, Mishra S, Kass DA, Collins S. Increased energy expenditure and protection from diet-induced obesity in mice lacking the cGMP-specific phosphodiesterase PDE9. Diabetes. 2021 doi: 10.2337/db21-0100. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verhoest PR, et al. Design and discovery of 6-[(3S,4S)-4-methyl-1-(pyrimidin-2-ylmethyl)pyrrolidin-3-yl]-1-(tetrahydro-2H-pyr an-4-yl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one (PF-04447943), a selective brain penetrant PDE9A inhibitor for the treatment of cognitive disorders. J Med Chem. 2012;55(21):9045–9054. doi: 10.1021/jm3007799. [DOI] [PubMed] [Google Scholar]
- 60.Even PC, Nadkarni NA. Indirect calorimetry in laboratory mice and rats: principles, practical considerations, interpretation and perspectives. Am J Physiol Regul Integr Comp Physiol. 2012;303(5):R459–R476. doi: 10.1152/ajpregu.00137.2012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.