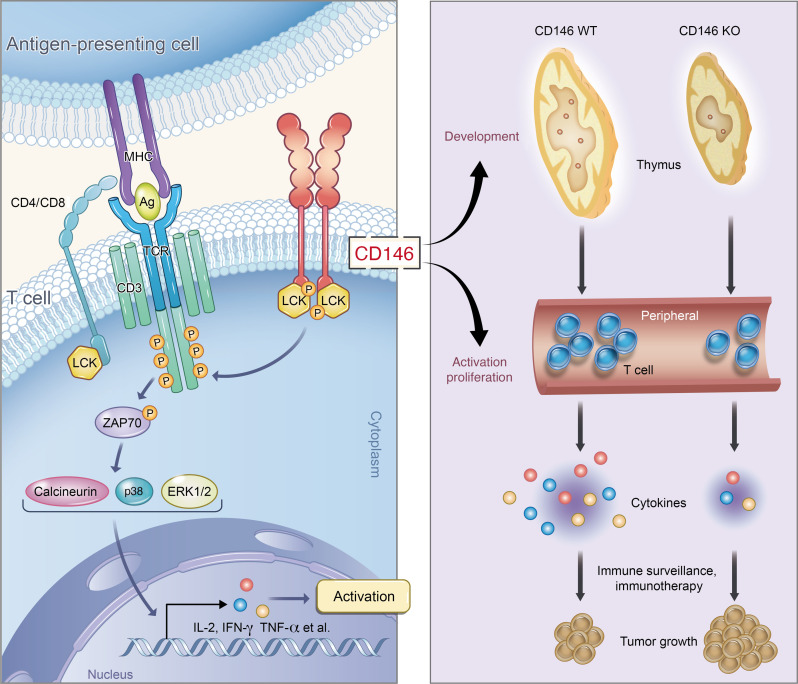
Abstract
Initiation of T cell receptor (TCR) signaling involves the activation of the tyrosine kinase LCK; however, it is currently unclear how LCK is recruited and activated. Here, we have identified the membrane protein CD146 as an essential member of the TCR network for LCK activation. CD146 deficiency in T cells substantially impaired thymocyte development and peripheral activation, both of which depend on TCR signaling. CD146 was found to directly interact with the SH3 domain of coreceptor-free LCK via its cytoplasmic domain. Interestingly, we found CD146 to be present in both monomeric and dimeric forms in T cells, with the dimerized form increasing after TCR ligation. Increased dimerized CD146 recruited LCK and promoted LCK autophosphorylation. In tumor models, CD146 deficiency dramatically impaired the antitumor response of T cells. Together, our data reveal an LCK activation mechanism for TCR initiation. We also underscore a rational intervention based on CD146 for tumor immunotherapy.
Keywords: Cell Biology, Immunology
Keywords: Adaptive immunity, Cancer immunotherapy
Introduction
T cell activation undergoes an orderly and complex process that critically relies on T cell receptor (TCR) signaling, which requires an orchestrated interplay of receptors, receptor-associated factors, kinases, and transcription factors, together termed the “TCR signalosome” (1). In the clinic, harnessing TCR signaling has been a popular area of study and has achieved exciting breakthroughs in immunotherapy for tumors and other immune disorders. Although the intracellular signaling cascade has been extensively studied, exactly how membrane TCR activation leads to intracellular signaling has remained elusive.
Lymphocyte-specific protein tyrosine kinase (LCK), an Src kinase family member, is critical in transducing cell membrane signals that activate intracellular signaling cascades. Deficiency of LCK abolishes proximal TCR signaling and blocks T cell development and activation (2–4). Mounting evidence suggests that the phosphorylation of LCK at Y394 is essential for its activation (5). Crystallization studies reveal that LCK forms an asymmetrical head-to-tail dimer, with the Y394 activation loop of one monomer in the other monomer’s active site. Trans-autophosphorylation is believed to be crucial for the activation of LCK (6, 7); however, little is known about how this process is regulated.
LCK has previously been reported to associate with the coreceptors CD4 and CD8 in thymocytes (8, 9). There is evidence that CD4 forms a dimer that associates with two LCK proteins, resulting in LCK trans-autophosphorylation (10). However, coreceptor-deleted T cells have recently been shown to maintain enough signal to undergo clonal deletion in vivo (8, 11). These results suggest that coreceptors contribute to TCR signaling sensitivity, rather than the activation of T cells (12). Thus, the underlying molecular mechanism for LCK trans-autophosphorylation and activation remains elusive.
Several earlier studies suggest that the adhesion molecule CD146 (also known as MCAM and MUC18) is associated with the activation of T cells during immune cell responses. CD146 is a type I transmembrane glycoprotein consisting of 646 amino acids in humans (13). The cytoplasmic domain is composed of 63 amino acid residues and exhibits 2 putative PKC phosphorylation sites and a dileucine motif (14). Our previous studies and the work of other investigators have shown that CD146 plays multiple roles in a wide variety of cells (15–18). We identified a dimeric form of CD146 on the surface of endothelial cells that regulates endothelial cell activation and migration (19). These studies suggest that dimeric CD146 plays important roles in outside-in signaling transduction.
CD146 expression was first reported on activated T cells in 1997 (20). Subsequent clinical studies have shown that in healthy individuals, CD146 is expressed on subpopulations of CD4+ and CD8+ T cells with effector/memory phenotype (21). In patients with autoimmune diseases, such as multiple sclerosis (22, 23), rheumatoid arthritis, and contact dermatitis (24), the percentage of CD146+ T cells is significantly increased, and, importantly, CD146+ T cells show a proinflammatory phenotype with increased cytokine secretion. In addition, CD146 was found to be expressed on immature cortical thymocytes in humans (25). These studies suggest a potential role of CD146 in T cell development and activation.
In the present study, we addressed this possibility using various CD146-deficient mouse models. We found that CD146 is required for both T cell development and peripheral activation associated with disrupted proximal TCR signaling. Interestingly, the cytoplasmic tail of CD146 directly interacts with LCK, and dimeric CD146 appears to act as a platform mediating LCK dimerization, autophosphorylation, and activation. Furthermore, CD146 deficiency in T cells compromises their function in tumor models. Therefore, our data suggest an important mechanism of dimeric CD146-mediated LCK activation for TCR initiation, which may have potential as an application in tumor immunotherapy.
Results
Deletion of CD146 on thymocytes inhibits T cell development.
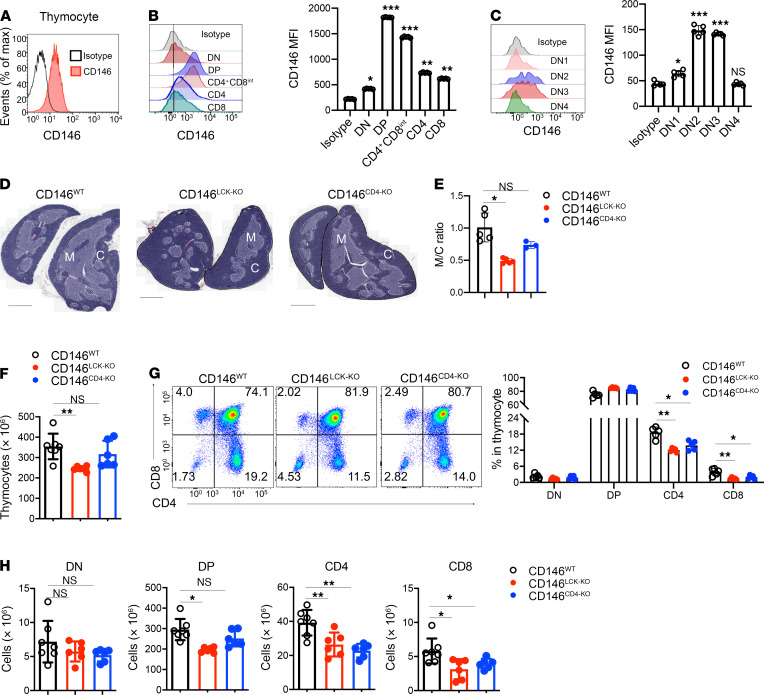
To investigate the role of CD146 in T cell development, we first isolated mouse thymocytes and tested CD146 expression on thymocytes of different developmental stages, including double-negative (DN; CD4–CD8–), double-positive (DP; CD4+CD8+), and single-positive (CD4+ or CD8+ SP) stages (26). As shown in Figure 1, A and B, CD146 expression was detected on thymocytes, especially on DP and CD4+CD8int subsets. During the DN stage, DN2 (CD4–CD8–CD44+CD25+) and DN3 (CD4–CD8–CD44–CD25+) cells had higher levels of CD146 expression compared with that on DN1 (CD4–CD8–CD44+CD25–) and DN4 (CD4–CD8–CD44–CD25–) cells (Figure 1C). CD146 expression decreased as DP thymocytes became SP (Figure 1B). Together, these expression patterns suggest that CD146 may play a regulatory role during thymocyte development, in both DN and DP cells.
Figure 1. Deletion of CD146 on thymocytes inhibits T cell development.
(A) Surface staining of CD146 (antibody clone AA1) on total thymocytes. (B) Left: Surface staining of CD146 on DN, DP, CD4+CD8int, and SP cells. Right: MFI of CD146 on DN, DP, CD4+, and CD8+ subpopulations of thymocytes from CD146WT mice (n = 5). (C) Left: Surface staining of CD146 on DN1–DN4 cells. Right: MFI of CD146 on DN1–DN4 cells (n = 5). (D) Thymus sections from CD146LCK-WT (n = 5), CD146LCK-KO (n = 5), and CD146CD4-KO mice (n = 3), stained with H&E; the darker area constitutes the cortex (C), and the lighter area constitutes the medulla (M). Original magnification, ×4. Scale bars: 1 mm. (E) Ratio of areas of M and C. (F) Total cell numbers in thymuses from WT (n = 7), CD146LCK-KO (n = 6), and CD146CD4-KO (n = 6) mice. (G) Left: Surface staining of CD4 and CD8 on thymocytes. Right: Percentages of DN, DP, and SP subpopulations (n = 5). (H) Cell numbers of DN, DP, CD4+, and CD8+ subpopulations (n = 7 for WT or 6 for KO). Each symbol represents an individual mouse; the short horizontal lines indicate the mean ± SEM. One-way ANOVA followed by Bonferroni’s correction was performed (B, C, and E–H). *P < 0.05, **P < 0.01, ***P < 0.001.
To further clarify the role of T cell–derived CD146 during thymocyte development, we generated 2 conditional Cd146-knockout (KO) mouse strains by crossing Cd146fl/fl mice with mice expressing Cre downstream of the proximal Lck (CD146LCK-KO) or Cd4 (CD146CD4-KO) promoters, which are first activated in DN2 and DP thymocytes, respectively. The genotypes were confirmed by PCR analysis (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI148568DS1). Both CD146LCK-KO and CD146CD4-KO mice exhibited Cd146 mRNA levels in T cells below the detection limit. Furthermore, CD146LCK-KO mice showed lower CD146 protein levels in DN, DP, and SP thymocytes, while CD146CD4-KO mice showed lower CD146 protein expression in DP and SP thymocytes (Supplemental Figure 1, B and C). Compared with WT mice, CD146LCK-KO mice had much smaller thymuses than their control littermates (Supplemental Figure 1D). In addition, the thymic architecture was also altered in CD146LCK-KO mice, with a remarkably reduced medulla/cortex ratio (Figure 1, D and E). The thymic size and medulla/cortex ratio were largely normal in CD146CD4-KO mice. In agreement with these results, the total number of thymocytes was reduced by approximately 35% in CD146LCK-KO mice compared with WT controls, but less affected in CD146CD4-KO mice (Figure 1F). More specifically, while the percentages of DN and DP remained constant, those of CD4+ SP and CD8+ SP cells were reduced in both CD146LCK-KO and CD146CD4-KO mice (Figure 1G). The numbers of CD4+ SP and CD8+ SP cells were also remarkably reduced in both CD146LCK-KO and CD146CD4-KO mice (Figure 1H). In addition, the number of DP cells was lower in CD146LCK-KO mice than in CD146CD4-KO mice (Figure 1H), consistent with the earlier deletion of Cd146 in the former mouse strain. Together, these experiments suggest that T cell–derived CD146 plays important roles during both DN-DP and DP-SP transitions.
CD146 is required for β selection.
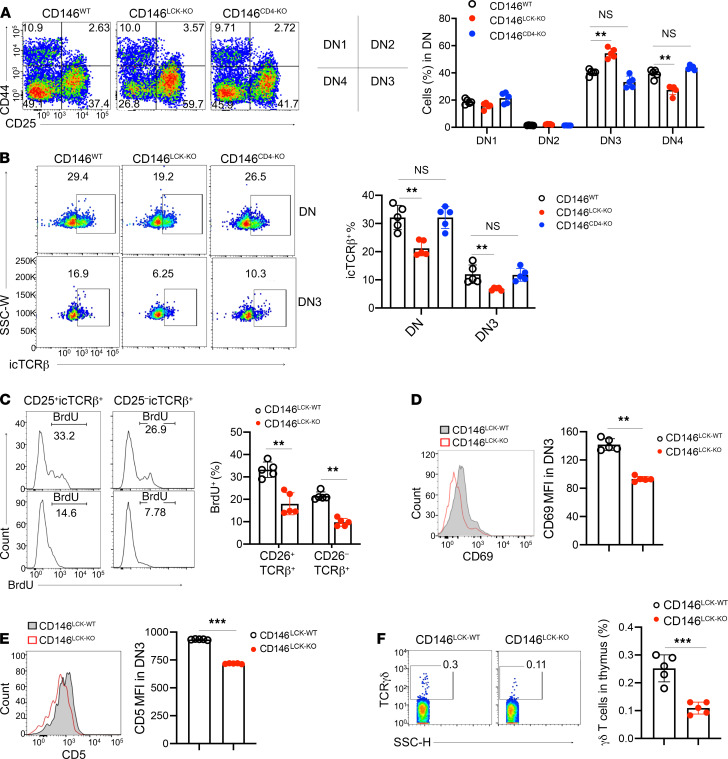
β Selection and positive selection are important steps for T cell development. To examine the role of CD146 in T cell development in more detail, we first analyzed the ratios of DN1–DN4 cells using flow cytometry. As shown in Figure 2A, the percentages of DN1 and DN2 subpopulations were essentially comparable among WT, CD146LCK-KO, and CD146CD4-KO mice. CD146LCK-KO mice exhibited an increased ratio of DN3 cells and a reduced ratio of DN4 cells compared with WT or CD146CD4-KO mice, suggesting that the CD146 deficiency interferes with DN3-to-DN4 transition (β selection) at an early stage. To confirm this, we stained intracellular TCRβ (icTCRβ), a marker for β selection. As shown in Figure 2B, the percentage of icTCRβ+ cells among DN3 cells was indeed significantly lower in CD146LCK-KO mice, while the expression level of membrane TCRβ was not disturbed (Supplemental Figure 2A). In addition, CD146 deficiency resulted in reduced BrdU incorporation in icTCRβ+ cells during an in vivo 2-hour BrdU pulse labeling assay (Figure 2C), suggesting its effect on icTCRβ+ proliferation. The expression levels of CD69 and pan–T antigen CD5 at the DN3 stage were also reduced (Figure 2, D and E). The cell survival issue is unlikely to be involved in the defect of DN3-DN4 transition, given that the 2 groups of DN3 cells had comparable apoptosis rates (Supplemental Figure 2B). Given that icTCRβ expression is essential for pre-TCR signaling (26), these data suggest that CD146-deficient DN3 cells are defective in pre-TCR signaling and fail to proliferate during the DN3-DN4 transition; thus, deletion of CD146 blocks T cell development at the β selection stage. Interestingly, however, this block failed to initiate a shift in lineage commitment toward γδ T cells, as their absolute number was also markedly lower in CD146LCK-KO compared with that in WT mice (Figure 2F). This result indicates that CD146 may also be required for γδ T cell development. Therefore, CD146 appears to be required for pre-TCR signaling, which controls both αβ and γδ T cell development.
Figure 2. CD146 is required for β selection.
(A) Left: Surface staining of CD44 and CD25 on DN thymocytes from WT, CD146LCK-KO, or CD146CD4-KO mice. Middle: Schematic diagram of the left graph. Right: Percentages of DN1–DN4 subpopulations (n = 5). Numbers adjacent to outlined areas indicate percent cells in each gate. (B) Left: Intracellular staining of TCRβ in DN or DN3 cells. Right: Percentages of intracellular TCRβ+ (icTCRβ+) cells among DN or DN3 cells (n = 5). (C) Left: Intracellular staining of BrdU in CD25–icTCRβ+ and CD25+icTCRβ+ cells. Right: Percentages of BrdU+ cells in each subpopulation. Numbers adjacent to graph lines indicate the percentage of cells in each gate (n = 5). (D and E) MFI of CD69 (D) and CD5 (E) in DN3 (n = 5). (F) Left: Surface staining of TCRγδ in thymocytes from CD146LCK-WT and CD146LCK-KO mice. Right: Percentages of γδ T cells. Numbers in the outlined areas indicate percent cells in each gate (n = 5). Each symbol represents an individual mouse; the short horizontal lines indicate the mean ± SEM. One-way ANOVA followed by Bonferroni’s correction (A–C) or 2-tailed t test (D–F) was performed. **P < 0.01, ***P < 0.001.
CD146 is required for positive selection.
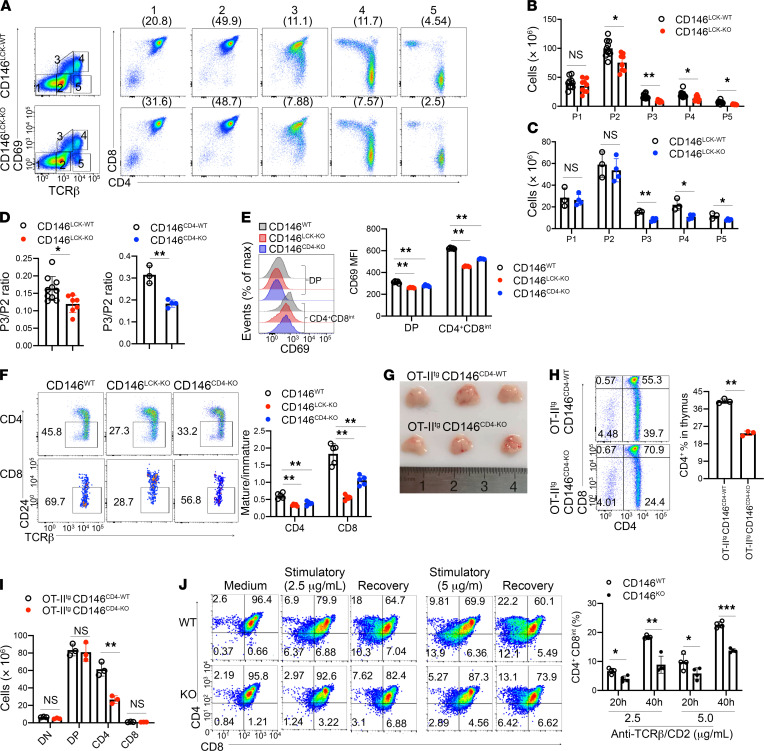
Mature TCR complexes are formed post-DN4, and DP cells require mature TCR signaling to proceed through positive selection and develop into mature SP cells. To evaluate whether CD146 is involved in thymocyte positive selection and SP cell maturation, we next examined thymocyte expression of both TCRβ and CD69 (markers for successful TCR activation and positive selection; ref. 27) in 5 distinct developmental subpopulations (Figure 3A). The number of immature CD69–TCRβ– cells (subpopulation 1, P1) was comparable between CD146LCK-KO mice and their WT littermates. However, all of the subsequent populations were significantly reduced in CD146LCK-KO mice, including subpopulation 2 (CD69–TCRβint, pre-selection DP cells; P2), subpopulation 3 (CD69intTCRβint, cells undergoing positive selection; P3), subpopulation 4 (CD69hiTCRβhi, post–positive selection; P4), and subpopulation 5 (CD69loTCRβhi, mature SP cells; P5) (Figure 3B). By using CD146CD4-KO mice, we also found that cells in P3–P5 were reduced in the absence of CD146 (Figure 3C). A remarkably reduced ratio of P3 to P2 in CD146LCK-KO and CD146CD4-KO mice further verified that positive selection was blocked in the absence of CD146 (Figure 3D). The blocked positive selection and maturation were also confirmed by CD69 and CD5 expression analysis (Figure 3E and Supplemental Figure 3, A and B). Furthermore, the maturation of CD4+ and CD8+ SP thymocytes was substantially impaired, as shown by the lower ratios of mature CD4+ (CD24neg–loTCRβ+) to immature CD4+ (CD24hiTCRβ+) and mature CD8+ (CD24neg–loTCRβ+) to immature CD8+ (CD24hiTCRβ+) thymocytes (Figure 3F). The defect in positive selection was unlikely to be due to apoptosis given that DP cells from WT and KO mice showed similar rates of apoptosis in vivo (Supplemental Figure 3C) and in vitro (Supplemental Figure 3D). Furthermore, the absence of CD146 did not affect the TCR-induced expression of Nur77, a critical regulator of TCR-induced cell death (ref. 28 and Supplemental Figure 3E). However, BrdU incorporation in DP cells was reduced in the absence of CD146 (Supplemental Figure 3F). These data suggest that the CD146 deficiency damaged the TCR-induced positive selection but did not affect the apoptosis of thymocytes.
Figure 3. CD146 is required for positive selection.
(A) Surface staining of CD69 and TCRβ on thymocytes from CD146LCK-WT and CD146LCK-KO mice. Numbers in outlined areas on the left indicate subpopulations gated, which are further analyzed and shown individually on the right, and numbers in parentheses above plots on the right indicate percent thymocytes in each subpopulation. (B and C) The numbers in subpopulations 1–5 of CD146LCK-WT (n = 10) and CD146LCK-KO (n = 7) (B) or CD146CD4-WT (n = 3) and CD146CD4-KO (n = 4) mice (C). (D) Ratio of subpopulation 3 to 2 from CD146WT, CD146LCK-KO, or CD146CD4-KO mice. (E) Left: Surface staining of CD69 on gated CD4+CD8int and DP thymocytes from WT, CD146LCK-KO, or CD146CD4-KO mice. Right: Quantification of CD69 MFI (n = 5). (F) Left: Surface staining of CD24 and TCRβ in CD4+ (top) or CD8+ (bottom) thymocytes from CD146WT, CD146LCK-KO, or CD146CD4-KO mice. The outlined areas indicate mature cells (CD24neg–loTCRβ+). Right: The ratio of mature (CD24neg–lo) to immature (CD24hi) cells among CD4+ or CD8+ cells from WT, CD146LCK-KO, or CD146CD4-KO mice (n = 5). (G) Thymuses from OT-IItgCD146CD4-WT and OT-IItgCD146CD4-KO mice (representative of n = 3). (H) Left: Surface staining of CD4 and CD8 on thymocytes from OT-IItgCD146CD4-WT and OT-IItgCD146CD4-KO mice. Right: Percentage of CD4+ thymocytes. (I) Cell numbers of DN, DP, CD4+, and CD8+ subpopulations (n = 3). (J) Left: Surface staining of CD4 and CD8 on sorted and developmental DP cells from WT and KO mice. Right: Percentages of CD4+CD8int cells among DP cells stimulated as described (n = 4). Each symbol represents an individual mouse; the short horizontal lines indicate the mean ± SEM. One-way ANOVA followed by Bonferroni’s correction (B, C, E, F, I, and J) or 2-tailed t test (D and H) was performed. *P < 0.05, **P < 0.01, ***P < 0.001.
The compensatory changes in the TCR repertoire would mask the developmental defects associated with thymocyte selection. To limit such compensation, we introduced OT-II β TCR transgenes into CD146CD4-KO mice (Supplemental Figure 3G). As shown in Figure 3, G–I, OT-II thymocytes showed a substantial developmental defect in CD146-deficient mice compared with WT control mice.
DP thymocyte commitment to CD4 or CD8 lineage requires downregulation of CD8 expression and transition through the CD4+CD8int stage. To determine whether CD146 deficiency leads to defects during this process, we performed an in vitro 2-stage differentiation assay (29). DP cells isolated from WT or KO mice were stimulated for 20 hours with the indicated concentrations of anti-TCRβ and anti-CD2 antibodies. As shown in Figure 3J, downregulation of CD8 was impaired, and the generation of CD4+CD8int cells was blocked in the recovery culture of KO DP cells. Together, these results strongly suggest that CD146 is required explicitly during thymocyte development at the positive selection stage.
Cell-intrinsic role of CD146 in thymocyte development.
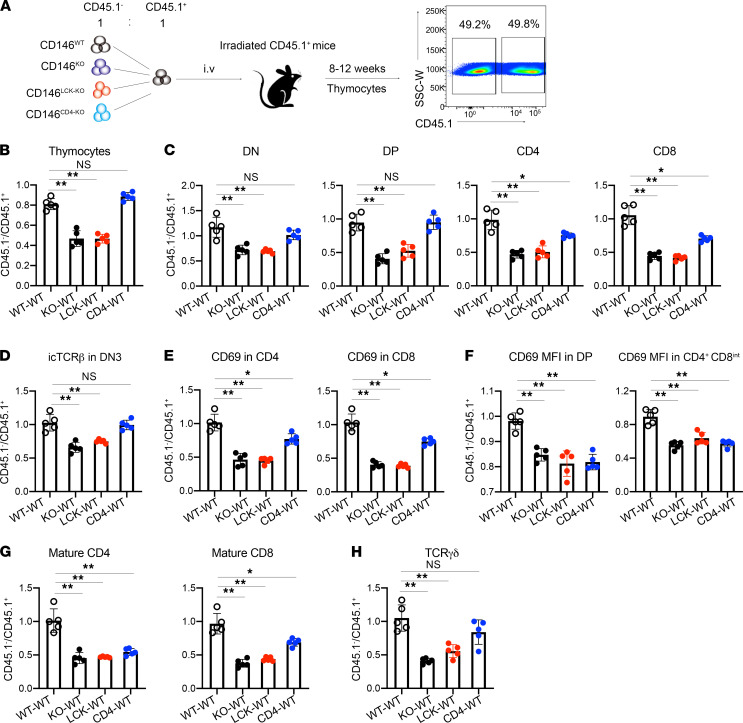
To determine the cell-intrinsic role of CD146 in T cell development, we next used a bone marrow chimeric mouse model. Briefly, bone marrow at a 1:1 ratio from CD45.1+ (WT) donors and CD45.2+CD45.1– donors that were CD146WT (mixed WT-WT), CD146KO (mixed KO-WT), CD146LCK-KO (mixed LCK-WT), or CD146CD4-KO (mixed CD4-WT) was transferred to lethally irradiated CD45.1+ host mice. After 8–12 weeks, thymocytes were isolated, and both the CD45.1+ and the CD45.2+ cells were analyzed by flow cytometry (Figure 4A). Among these 4 groups, mixed KO-WT and mixed LCK-WT mice exhibited similar abnormalities in T cell development, including reduced thymocyte numbers (Figure 4, B and C), inhibited β selection (Figure 4D), positive selection of DP cells (Figure 4, E and F), and SP maturation (Figure 4G). The thymus γδ T cells were also reduced in mixed KO-WT and mixed LCK-WT mice (Figure 4H). Mixed CD4-WT mice showed a defect in DP positive selection and SP maturation (Figure 4, C and E–G). Together, these findings suggest that CD146 plays a thymocyte-intrinsic role in developmental regulation.
Figure 4. Cell-intrinsic role of CD146 in thymocyte development.
(A) Schematic representation of the bone marrow chimeric model. (B and C) Ratio of CD45.1– to CD45.1+ cells among thymocytes (B) and DN, DP, CD4+, and CD8+ subpopulations (C) (n = 5). (D) Ratio of CD45.1– to CD45.1+ cells among icTCRβ+ cells in the DN3 subpopulation (n = 5). (E) Ratio of CD45.1– to CD45.1+ cells among CD69+ cells (n = 5). (F) MFI of CD69 in DP and CD4+CD8int subpopulations (n = 5). (G) Ratio of CD45.1– to CD45.1+ cells in mature CD4+ or CD8+ subpopulations (n = 5). (H) Ratio of CD45.1– to CD45.1+ cells in γδ T cell populations (n = 5). Each symbol represents an individual mouse. One-way ANOVA followed by Bonferroni’s correction was performed. Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01.
CD146 deletion impairs TCR-mediated response in T cells.
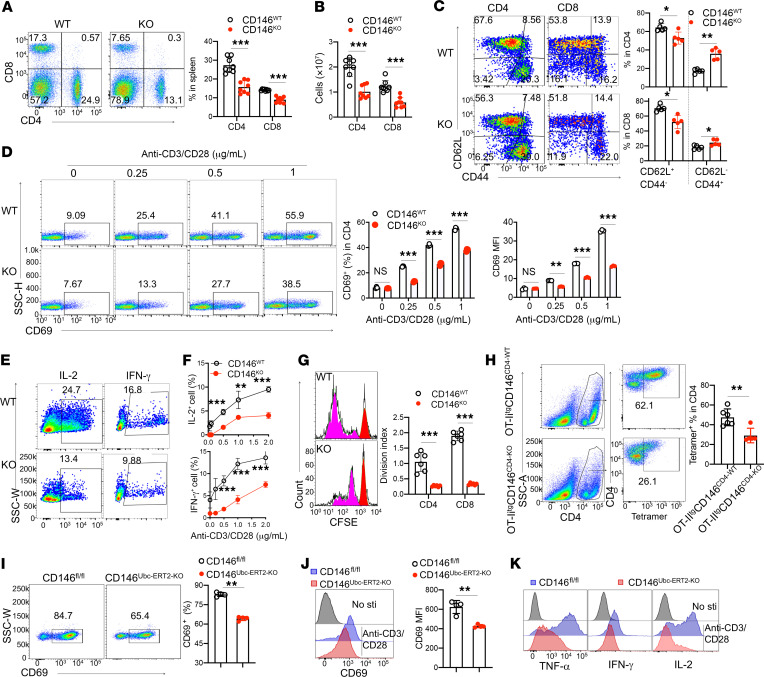
After thymic maturation, T cells migrate to the periphery for immune surveillance and defense. CD146 deficiency reduced the peripheral T cell population in peripheral blood and spleen (Supplemental Figure 4, A and B; and Figure 5, A and B). Moreover, in CD146-KO mice, the proportion of naive T cells (CD62hiCD44lo) in the spleen was lower, while the percentage of accumulated effector cells (CD62–CD44hi) was higher, compared with that in WT mice (Figure 5C), probably because of T cell lymphopenia–induced T cell proliferation. To understand whether CD146 controls T cell activation at the early stage, we measured CD69+ cells in lymph nodes and peripheral blood from WT and KO mice. As shown in Supplemental Figure 4, C and D, the percentages of CD69+CD4+ cells and CD69+CD8+ cells were remarkably lower in KO mice, indicating that the early activation of T cells was defective in CD146-KO mice. To confirm this finding, we directly assessed naive T cell activation using an in vitro anti–CD3/CD28 stimulation assay. As shown in Figure 5D and Supplemental Figure 4E, the percentages of CD69+ cells and the CD69 protein levels in stimulated naive CD4+ and CD8+ cells increased with increasing concentration of anti–CD3/CD28. However, a substantial defect was found in CD146-KO T cells. These results were further confirmed using T cells from CD146LCK-KO or CD146CD4-KO mice (Supplemental Figure 4, F and G). In addition, after stimulation with anti–CD3/CD28, the production of both IL-2 and IFN-γ was greatly reduced in CD146-deficient T cells (Figure 5, E and F, and Supplemental Figure 4H). Granzyme B expression in CD8+ cells was also reduced under anti–CD3/CD28 stimulation (Supplemental Figure 4I). Moreover, in an in vitro proliferation assay using the cytosolic dye CFSE, T cells from KO mice exhibited a much lower proliferation capacity in response to anti–CD3/CD28 stimulation (Figure 5G), suggesting that CD146 is essential for TCR-driven activation of T cells; this was further confirmed in an OT-II antigen–specific scenario (Figure 5H).
Figure 5. CD146 deletion impairs the TCR-mediated response of T cells.
(A) Left: CD4 and CD8 expression on splenic cells. Right: Percentage of cell populations (n = 8). (B) Cell numbers of splenic CD4+ and CD8+ populations (n = 8). (C) Left: CD44 and CD62L expression on splenic CD4+ and CD8+ cells. Right: Percentage of naive T (CD62L+CD44–) and memory T cells (CD62L–CD44+) in the CD4+ or CD8+ population (n = 5). (D) Left: CD69 expression on naive CD4+ T cells left stimulated for 5 hours. Right: Quantification of CD69+ cells and quantification of CD69 MFI (n = 3). (E and F) IL-2+ or IFN-γ+ cells of splenic naive T cells after stimulation for 3 days in vitro (n = 5). (G) Left: CFSE dilution in splenic naive T cells stimulated for 4 days. Red, nonproliferated cells; pink, proliferating cells. Right: Division index (n = 6). (H) Left: Staining of CD4 and OVA-MHC-II tetramer on splenic CD4+ T cells left stimulated with OVA323–339 peptide for 72 hours. Right: Percentage of tetramer+ populations (n = 6). (I) Left: CD69 expression on naive CD4+ T cells from tamoxifen-treated mice left stimulated for 5 hours. Right: Percentages of CD69+ cells (n = 4). (J) Histogram and MFI of CD69 (n = 4). (K) Analysis of TNF-α, IFN-γ, and IL-2 in tamoxifen-treated splenic naive T cells after stimulation for 72 hours (representative of n = 3). Each symbol represents an individual mouse. One-way ANOVA (A–D and G), 2-tailed t test (H–J), or 2-way ANOVA with multiple-comparison test (F) was performed. Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
To exclude the potential thymic developmental effect on peripheral T cell activation and to investigate the direct impact of CD146 deletion on peripheral T cell activation, we generated inducible-KO mice as described in Methods. Tamoxifen treatment was found to inhibit CD146 expression on T cells (Supplemental Figure 4, J and K), and naive T cells with inducible KO of CD146 showed lower CD69 expression in response to anti–CD3/CD28 stimulation (Figure 5, I and J), as well as reduced expression of cytokines such as TNF-α, IFN-γ, and IL-2 (Figure 5K and Supplemental Figure 4L). These data further confirmed that the peripheral activation defect in CD146-KO mice is mainly primary, rather than being secondary to a developmental defect.
CD146 deletion damages proximal TCR signaling.
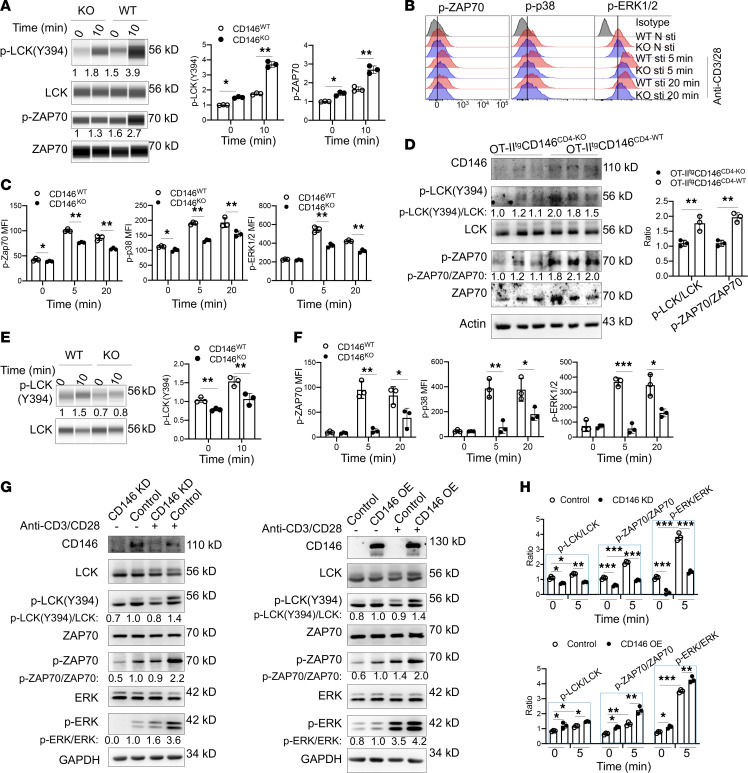
We next examined the mechanisms responsible for CD146 control of T cell activation. Splenic naive T cells from WT and CD146-KO mice were stimulated with anti-CD3 and anti-CD28 for 10 minutes or left unstimulated, and the activation status of various TCR signaling molecules was determined. After stimulation, the phosphorylation levels of LCK and ZAP70, the proximal events associated with TCR activation, were remarkably lower in CD146-deficient T cells compared with WT controls (Figure 6A). Interestingly, the levels of phosphorylated LCK(Y394) [p-LCK(Y394)] and p-ZAP70 were also significantly lower in CD146-deficient T cells, even without antibody stimulation, suggesting a role of CD146 in the resting T cell stage (Figure 6, A–C). Consistent with the impaired LCK and ZAP70 activation, the activation of p38 and ERK1/2 and the downstream calcium flux were also lower in the absence of CD146 (Figure 6, B and C, and Supplemental Figure 5A). These data suggest that CD146 functions at the proximal signaling of TCR. This was also confirmed by in vitro OT-IItg T cell activation assay using OVA329–337 peptide. As shown in Figure 6D, OVA-induced T cells showed defective LCK and ZAP70 activation in the absence of CD146. In addition, we performed a T cell activation assay using phorbol-12-myristate-13-acetate (PMA) and ionomycin, a PKC activator, to bypass proximal TCR signaling. The results showed that upon PMA/ionomycin stimulation, WT and KO T cells had similar levels of p-p38, ERK1/2, calcium flux, and CD69 expression (Supplemental Figure 5, B–D), confirming that CD146 mainly mediated proximal TCR signaling. The impaired proximal TCR signaling defect and downstream effect were also found in CD146-KO thymocytes, as shown by reduced p-LCK(Y394), p-ZAP70, p-p38, p-ERK1/2, and downstream calcium flux (Figure 6, E and F, and Supplemental Figure 5E), suggesting a general function of CD146 in TCR activation (See complete unedited blots in the supplemental material.).
Figure 6. CD146 deletion damages proximal TCR signaling.
(A) Western immunoblot (WB) analysis of p-LCK(Y394), LCK, p-ZAP70(Y319), and ZAP70 in splenic naive T cells stimulated with or without anti-CD3 and -CD28 antibodies (1 μg/mL) for 10 minutes. Total LCK and ZAP70 proteins were included as loading controls. Right: Quantification of p-LCK(Y394)/LCK and quantification of p-ZAP70/ZAP70 (n = 3). (B) Staining of p-ZAP70, p-p38, and p-ERK1/2 in splenic naive T cells stimulated with or without antibodies (1 μg/mL) for the indicated times. (C) MFI of p-ZAP70, p-ERK1/2, and p-p38 in the cells described above (n = 3). (D) Left: WB showing the expression of CD146, p-LCK(Y394), and p-ZAP70(Y319) in isolated CD3+ T cells from OT-IItgCD146CD4-WT or OT-IItgCD146CD4-KO mice; total LCK, ZAP70, and actin proteins were used as loading controls. Right: Quantification of p-LCK(Y394)/LCK or p-ZAP70/ZAP70 (n = 3). (E) Left: WB analysis of p-LCK(Y394) and LCK in thymocytes stimulated with or without anti–CD3/CD28 antibodies (1 μg/mL) for the indicated times. Total LCK protein was included as the loading control. Right: Quantification of p-LCK(Y394)/LCK (n = 3). (F) MFI of p-ZAP70, p-p38, and p-ERK1/2 in thymocytes stimulated as above (n = 3). (G) WB showing p-LCK, p-ZAP70, and p-ERK1/2 protein levels in Jurkat cells with CD146 knockdown (CD146 KD), WT (Control), or CD146 overexpression (CD146 OE) left stimulated with or without anti–CD3/CD28 antibodies (1 μg/mL) for 5 minutes. Total LCK, ZAP70, GAPDH, and ERK1/2 proteins were used as loading controls. (H) Quantifications of p-LCK(Y394)/LCK, p-ZAP70/ZAP70, and p-ERK1/2/ERK1/2 (n = 3). Each symbol represents an individual mouse (C, D, and F) or 1 experiment (A, B, E, and H). Data are representative of 3 independent experiments. One-way ANOVA (A and C–F) or 2-way ANOVA with multiple-comparison test (H) was performed. Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
To avoid the potential developmental influence on T cell activation, we manipulated CD146 expression levels in Jurkat cells. CD146 protein was knocked down using CD146 shRNA or overexpressed using CD146-encoding plasmids (Supplemental Figure 5F). As shown in Figure 6, G and H, significantly reduced p-LCK, p-ZAP70, and p-ERK levels were observed in CD146-knockdown cells, irrespective of whether they were at rest or following TCR stimulation. By contrast, CD146 overexpression enhanced the activation of the aforementioned proteins. Together, these data suggest that CD146 is involved in proximal TCR signaling, independent of T cell development, and at both resting and activating status.
CD146 interacts with LCK and acts as a platform to promote LCK activation.
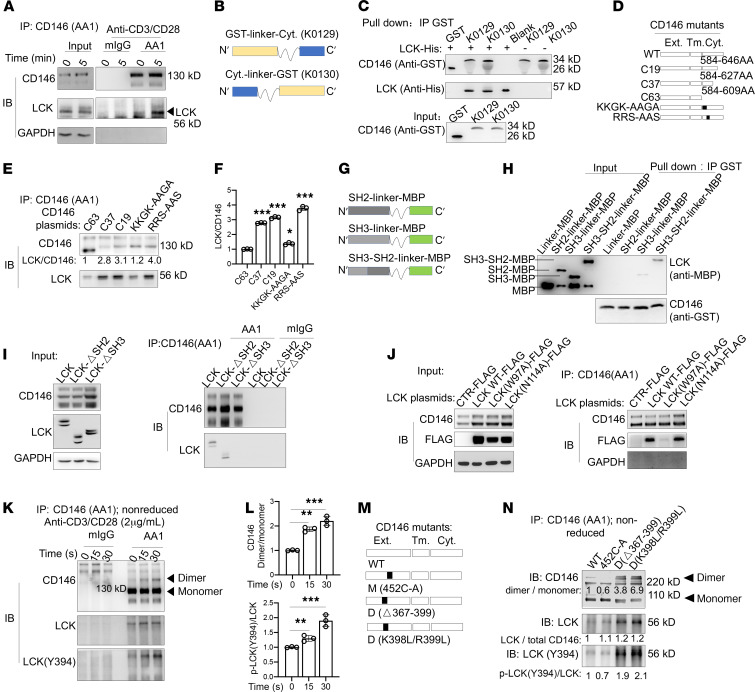
We next proceeded to determine how CD146 regulates the proximal TCR signaling. We first tested whether CD146 may directly or indirectly interact with molecules associated with TCR signaling, such as CD3, CD4, ZAP70, LAT, PLCγ, FYN, and LCK. Using immunoprecipitation (IP) assays, we observed no interactions between CD146 and CD3, CD4, ZAP70, LAT, PLCγ, or FYN; however, we found an interaction between CD146 and LCK (Supplemental Figure 6, A and B). Interestingly, the interaction between CD146 and LCK was already present in resting T cells and was further increased with prolonged stimulation of the TCR using IP and 3-dimensional structured illumination microscopy imaging (3D-SIM) (Figure 7A and Supplemental Figure 6C). Surprisingly, the interaction between CD146 and LCK was independent of coreceptors, such as CD4, suggesting that it is direct (Supplemental Figure 6, B and D). To confirm these findings, we constructed 2 recombinants of the cytoplasmic segment of CD146 with the GST protein linked at either the C- or the N-terminal (CD146-linker-GST or GST-linker-CD146, respectively; Figure 7B), which were used for pull-down experiments. As shown in Figure 7C, both cytoplasmic CD146 recombinants directly interacted with LCK. Thus, these data provide further evidence for the direct interaction between CD146 and LCK.
Figure 7. CD146 interacts with LCK, and its dimerization promotes LCK activation.
(A) Immunoblot (IB) analysis of CD146 and LCK in Jurkat cells immunoprecipitated (IP) with anti-CD146 (AA1) or isotype IgG stimulated for the times indicated. (B) Schematic representations of recombinant CD146 intracellular proteins. (C) Pull-down assay of recombinant CD146 intracellular proteins and LCK-His protein. (D) Schematic representations of CD146 intracellular mutants. (E) IP with AA1 to detect the interaction of CD146 intracellular mutants and LCK in 293T cells. (F) Quantification of the LCK to CD146 ratio (n = 3). (G) Schematic representation of the SH2, SH3, and SH3-SH2 domains of LCK. (H) Pull-down assay of recombinant CD146 intracellular proteins and LCK domain–MBP protein. (I) IB analysis of CD146 and LCK in 293T cells transfected with LCK or LCK-deletion SH2 or SH3 domain plasmid and then immunoprecipitated with anti-CD146 (AA1) or isotype IgG. (J) IB analysis of CD146 and FLAG in 293T cells transfected with FLAG-LCK mutant plasmid and then immunoprecipitated with anti-CD146 (AA1). (K) IB analysis of CD146 dimer and LCK in Jurkat cells immunoprecipitated with anti-CD146 (AA1) or isotype IgG upon anti–CD3/CD28 stimulation for the times indicated. Arrowheads indicate the corresponding proteins. (L) Relative ratio of either CD146 dimer to monomer or LCK(Y394) to total LCK (n = 3). (M) Schematic representation of the CD146 monomer, WT, and dimer mutants. (N) IB of LCK and LCK(Y394) in 293T cells transfected with CD146 monomer, WT, or dimer and LCK plasmids and immunoprecipitated with anti-CD146 (AA1). The number below each band represents the relative ratio to WT. Each symbol represents 1 experiment. One-way ANOVA followed by Bonferroni’s correction (F and L) was performed. Data are representative of 3 independent experiments. Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. ext., extracellular; tm., transmembrane; cyt., cytoplasmic.
To determine which residues of CD146 contribute to its interaction with LCK, we first generated 3 truncations of CD146, namely C19 (627 aa), C37 (609 aa), and C63 (583 aa) (Figure 7D). We transfected these CD146 truncation–encoding plasmids into 293T cells with stably transfected LCK-encoding plasmids. IP experiments using the anti-CD146 antibody AA1 showed that C63 truncation but not C19 and C37 impaired the interaction (Figure 7, E and F), suggesting that the interaction site is located in the cytoplasmic domain 584–609 aa. Our previous study showed that a conserved positively charged amino acid cluster KKGK motif in the juxtamembrane region of the CD146 cytoplasmic tail served as the binding site for ERM proteins (30). Therefore, we hypothesized that this KKGK motif might also be used for the interaction with LCK. To test this, we generated the mutation plasmid KKGK-AAGA (Figure 7D) and transfected this mutant plasmid into 293T cells with stably transfected LCK-encoding plasmids. IP experiments showed that the KKGK mutation comparably reduced LCK interaction, suggesting that KKGK is indeed essential for LCK interaction. The mutation of another positively charged amino acid cluster RRS motif, which is similar to the motif RRR in CD4 or CD44 for their interaction with LCK (31), showed unimpaired interaction with LCK (Figure 7, E and F). Thus, CD146 depends mainly on the KKGK motif for its interaction with LCK.
To determine which LCK region interacts with the cytoplasmic CD146, we constructed 3 plasmids encoding domains SH2, SH3, and SH2-SH3 reported as LCK regulatory domains (ref. 6 and Figure 7G). Pull-down assays showed that the SH3 domain interacted with CD146 (Figure 7H). This was further confirmed by IP using CD146-stably-transfected 293T cells, which were transiently transfected with a plasmid encoding SH2- or SH3-deleted LCK (Figure 7I). In addition, the CD146-LCK SH3 interaction may depend on the cation-π interaction of a cationic side chain (such as lysine, K) and an aromatic side chain (such as tryptophan, W), which is prevalent in protein-protein complexes (32). To confirm this, we generated a plasmid encoding a form of LCK with a mutation in the SH3 domain, namely LCK(W97A). LCK(N114A) served as a control. IP experiments showed that the LCK(W97) site was critical for the interaction of SH3 and CD146 (Figure 7J).
The active form of LCK [p-LCK(Y394)] is constitutively expressed in T cells at the resting stage, and TCR stimulation further enhances de novo phosphorylation of LCK(Y394) (5). The activation of LCK is believed to occur via trans-autophosphorylation of tyrosine in the activation loop (Y394). We previously reported that CD146 dimerizes in endothelial cells and showed that this form facilitates NF-κB signaling (19). Therefore, we investigated whether CD146 dimerizes in T cells and whether dimeric CD146 contributes to LCK activation. Indeed, the dimerized form of CD146 was detected in Jurkat cells at resting status, as shown in Figure 7K. Upon anti–CD3/CD28 stimulation, increased CD146 clusters were observed (Supplemental Figure 6E), and the ratio of the dimer to monomer was also increased (Figure 7L). Concomitantly, activated LCK was increased with prolonged stimulation (Figure 7, K and L), suggesting that CD146 dimerization may promote LCK activation. The CD146 C452 residue is reported to be essential for the dimerization of CD146 (19). To directly test this hypothesis, we constructed a mutant CD146 monomer, M(452C-A) (Figure 7M). WT or mutant monomer CD146 plasmids were transfected into LCK-stably-transfected 293T cells. As shown in Figure 7N, while the interacting LCK levels were similar in cells transfected with plasmids of WT dimeric CD146 or mutant monomeric CD146, activated LCK was markedly reduced in cells transfected with monomeric CD146 compared with cells transfected with dimeric CD146. These results were further confirmed using dimerized CD146 plasmids, D(Δ367–399) and D(K398L/R399L) (Figure 7, M and N), both of which were determined as important residues for CD146 dimerization by structural analysis (33). In addition, CD146 cross-linking by anti-CD146 antibody AA1 or AA98 (34) led to the activation of LCK, ZAP70, and ERK1/2 (Supplemental Figure 6, F and G). These data suggest that dimerized CD146 may serve as a platform that brings LCK together and promotes its trans-autophosphorylation (proposed model, Supplemental Figure 6H).
CD146 on T cells enhances the antitumor activity.
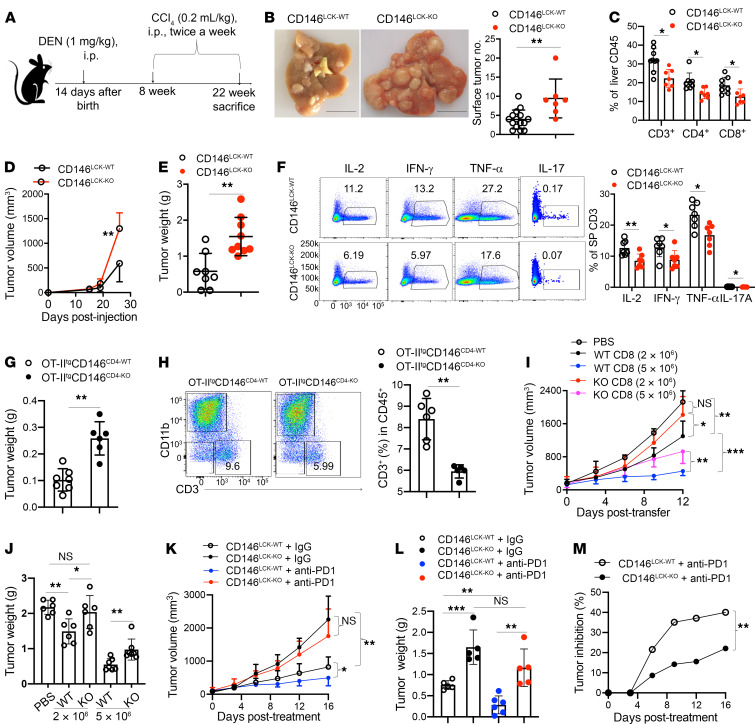
T cells play important roles in the implementation of immune surveillance and immune defense. To elucidate the function of CD146 on T cells in tumor immunosurveillance, we established a diethylnitrosamine-induced hepatocellular carcinoma (HCC) mouse model in CD146LCK-KO mice (Figure 8A). Twenty-two weeks after induction, CD146LCK-KO mice developed more severe HCC compared with WT mice as demonstrated by the increased number and size of tumor nodules (Figure 8B). In addition, the percentages of CD4+ and CD8+ T cells infiltrated into the liver were markedly reduced in CD146LCK-KO HCC mice (Figure 8C), while other cells were slightly affected (Supplemental Figure 7A). These data suggest that T cell–derived CD146 is important in the antitumor response.
Figure 8. CD146 on T cells enhances antitumor activity.
(A) Schematic representation of diethylnitrosamine-induced (DEN-induced) HCC mouse model. (B) Left: DEN-induced liver tumors in CD146LCK-WT (n = 15) and CD146LCK-KO (n = 7) mice. Right: Quantification of surface tumors. Scale bars: 1 cm. (C) Percentage of infiltrated T cells among liver CD45+ cells from DEN-induced HCC mice (n = 7). (D and E) Volume (D) and weight (E) of B16 tumor growth in CD146LCK-WT (n = 8) and CD146LCK-KO mice (n = 9). (F) Left: Intracellular staining of IL-2, IFN-γ, TNF-α, and IL-17 in CD3+ T cells from tumor-bearing mice. Right: Percentages of IL-2+, IFN-γ+, TNF-α+, and IL-17+ cells among CD3+ T cells (n = 7). (G) Weight of B16-OVA tumors (n = 7 for WT or 6 for KO). (H) Percentage of infiltrated CD3+ T cells among tumor CD45+ cells from B16-OVA tumor-bearing mice (n = 6). (I and J) Tumor growth (I) and tumor weights (J) for B16-OVA tumor–bearing mice adoptively transferred with CD8+ T cells (n = 6). (K and L) Tumor growth (K) and tumor weight (L) in CD146LCK-WT or CD146LCK-KO mice treated with anti–PD-l antibody or isotype IgG (200 μg/mouse) (n = 5 or 6). (M) Tumor inhibition analysis between CD146LCK-WT and CD146LCK-KO mice with or without anti–PD-1 treatment (n = 5 or 6). Each symbol represents an individual mouse. Two-tailed t test (B, E, G, and H), 1-way ANOVA followed by Bonferroni’s correction (C, F, J, and L), 2-way ANOVA with multiple-comparison test (D and M), or 2-way repeated-measures ANOVA with Tukey’s test (I and K) was performed. Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
To further determine the function of CD146 in T cell antitumor capacity, we established a xenograft melanoma model using CD146LCK-KO mice. In the transplanted melanoma model induced by s.c. injection of 1 × 106 B16F10 cells, we observed accelerated tumor growth in CD146LCK-KO mice (Figure 8, D and E). In addition, less IL-2, IFN-γ, IL-17A, and TNF-α were produced in CD146LCK-KO splenic T cells compared with control T cells from tumor-bearing mice (Figure 8F), suggesting that the absence of CD146 impairs the antitumor immunity of T cells. These data were further confirmed using OVA-overexpression B16F10 (B16-OVA) tumor-bearing OT-IItgCD146CD4-KO mice (Figure 8G and Supplemental Figure 7B). In addition, the tumor-infiltrating T cells were also dramatically reduced in OT-IItgCD146CD4-KO mice (Figure 8H). To exclude the effect of a development defect, we performed T cell adoptive transfer experiments in a tumor immunotherapy model. As shown in Figure 8, I and J, adoptively transferred CD8+ T cells had a strong capacity to kill tumors in a dose-dependent manner, which was reduced by CD146 deficiency. These data suggest that CD146 is required for antitumor T cell response.
Because CD146 on T cells has been reported to facilitate the transmigration of T cells (23, 35), to exclude this effect, we coinjected B16-OVA cells and preactivated T cells s.c. at a 1:10 ratio into nude mice and examined the tumor development. The result showed that mice coinjected with CD146-deficient T cells developed larger tumors than those injected with WT T cells (Supplemental Figure 7C). To further confirm the antitumor activity of CD146+ T cells, we isolated splenic T cells at day 7 after inoculation, and cocultured T cells with B16-OVA cells in vitro (20:1 or 50:1). We found that fewer tumor cells were killed on day 3 by CD146-deficient T cells than by WT T cells (Supplemental Figure 7D). Moreover, we found that CD146-deficient T cells had lower activation of LCK and ZAP70 than WT T cells (Supplemental Figure 7E). These data suggest that CD146 expression on T cells enhances antitumor immunity.
Tumor immunotherapy with anti–PD-1 has shown great success in some solid tumors. To determine whether CD146 expression on T cells is required for anti–PD-1 immunotherapy, we established an MC38 tumor model with CD146LCK-KO or WT mice and treated the mice with or without anti–PD-1 at day 8 after tumor inoculation. As shown in Figure 8, K–M, CD146LCK-KO mice developed larger tumors than WT mice, which was consistent with the aforementioned results. Moreover, while anti–PD-1 treatment inhibited the tumor growth in WT mice, it had less effect in CD146LCK-KO mice. Together, these data suggest that manipulating the T cell activity by CD146 may be helpful for tumor immunotherapy.
Discussion
Here, we identified an essential role for CD146 in T cell development and activation. Our analysis of mouse models revealed that CD146 is expressed on thymocytes in a finely regulated dynamic pattern throughout T cell development. We found that CD146 is involved in the transition from the DN to the DP stage, a process that requires pre-TCR signaling. We also observed that CD146 regulates activation of peripheral T cells, which requires signaling through the mature TCR complex. Mechanistically, CD146 is directly involved in TCR signaling by interacting with LCK. Interestingly, CD146 was dimerized in T cells and appears to act as a platform for LCK autophosphorylation. Deficiency in CD146 disrupted the proximal events of TCR signaling both in vivo and in vitro. Importantly, mice with T cell–specific CD146 deletion showed markedly defective antitumor immunity compared with those with WT T cells. Thus, CD146 was discovered as an important membrane protein controlling TCR signaling during T cell development and function.
Our study uncovered a previously unrecognized direct interaction between CD146 and LCK. Cytoplasmic membrane LCK is usually considered to be associated with CD4 and CD8 coreceptors (36–38). However, a recent imaging study suggests that CD4 and LCK are localized in their separate clusters with limited interactions in the interfaces between them (9), indicating the presence of a significant amount of coreceptor-free LCK on the cell membrane. In our current study, we found an interaction between LCK and CD146 using IP assay in T cells at both resting and activating status. Moreover, the results of the in vitro pull-down experiment further showed that the intracellular component of CD146 has a direct interaction with LCK, and revealed the interaction face residing mainly on the KKGK motif of the cytoplasmic domain 584–609 aa in CD146 and W97 aa of the SH3 domain in LCK. These data also suggest that the interaction of LCK and CD146 is independent of coreceptors.
A significant amount of LCK is present in its active form in T cells at resting status and is further increased upon TCR stimulation (5, 39, 40). How this is maintained and regulated remains unknown. A previous study has suggested that CD4 dimerization mediates LCK trans-autophosphorylation (10). However, this study does not explain the presence of activated LCK in coreceptor-negative thymocytes (39). In addition, coreceptors are only partially dispensable for T cell development and activation (8). Furthermore, a previous study has demonstrated a significant amount of coreceptor-free LCK on the cell membrane (9). These findings indicate that additional factors controlling LCK activation may exist. In our study, significant reduction of active LCK was found in CD146-deficient resting T cells, suggesting an important role of CD146 in maintaining constitutive LCK activation. This is likely mediated by CD146 dimerization (19). Indeed, dimerized CD146 was clearly detected in resting primary T cells and Jurkat T cells, and dimerized CD146, but not monomeric CD146, promoted LCK activation in 293T cells. These data suggest that CD146 functions as a platform for LCK dimerization and autophosphorylation. In addition, increased CD146 clustering and dimerization were observed upon TCR stimulation, which was accompanied by increased LCK activation. Thus, CD146 dimerization appears to mediate LCK autophosphorylation in both resting and activated T cells and is required for TCR signaling for T cell development and activation.
How CD146 dimerization is regulated is still unknown. However, our data suggest that this might be related to the association between CD146 and lipid rafts. In resting T cells, although a fraction of CD146 is already located in the lipid rafts, protein palmitoylation modification has been reported to be able to steer proteins toward lipid rafts (41). In particular, a functional palmitoylation site at C590 has been reported in CD146, which might promote constitutive CD146 recruitment to lipid rafts (42). In activating T cells, the fraction of CD146 in lipid rafts is further increased, which may be due to the increased content of raft-associated lipids (43), which would further recruit CD146, leading to enhanced CD146 dimerization. Alternatively, CD146 ligands/interacting partners on antigen-presenting cells (APCs) may promote CD146 dimerization during APC–T cell interaction and immunological synapse formation. However, this does not explain the effect of CD146 during anti–CD3/CD28 stimulation–induced T cell activation, when no synapse is formed. Therefore, the exact mechanism underlying CD146 dimerization remains to be studied in the future.
We have provided evidence that CD146 is required for LCK dimerization and activation and is also required for downstream TCR signaling, such as ZAP70 phosphorylation and ERK phosphorylation, and further downstream effector cytokine production and cell proliferation. The impaired LCK activation occurs at the point of early T cell development and remains throughout their peripheral homeostasis and activation. Therefore, it appears that CD146 plays a universal role in LCK activation throughout the entire process of T cell development, homeostasis, and response. Previous studies have suggested that CD146 associates with FYN, another member of the Src family of kinases, in endothelial cells (44). However, in T cells, CD146 does not appear to function via FYN. Indeed, we observed no association between CD146 and FYN in this study; this is probably due to cell type–specific differences.
Although remaining at low expression level in peripheral naive T cells, CD146 indeed plays an important role in the activation of LCK in naive T cells. In addition, CD146 expression is upregulated upon TCR activation, and may further potentiate T cell activation. Moreover, as CD146 is highly expressed on effector/memory T cells, their reactivation during effector/restimulation phases may be also regulated by CD146. In fact, CD146hi T cells were shown to be elevated in various autoimmune diseases and capable of producing several proinflammatory cytokines, including IFN-γ, IL-17, and TNF-α (45). Numerous studies have shown that CD146hi T cells, especially CD146hi Th17 cells, are encephalitogenic (23, 46–49). Whether CD146 plays the same important role for effector/memory T cells during the T cell effector phase as during T cell development and priming activation remains an interesting question for further investigation.
T cell–mediated immunotherapy using chimeric antigen receptors (CARs) has achieved great success in tumor therapy, and enhancing LCK phosphorylation is crucial for the activity of CARs. A recent paper showed that the binding of the LCK SH3 domain to the CD3ε cytoplasmic tail resulted in local augmentation of LCK activity, providing the rational improvement of CAR design for the treatment of cancer (50). Our findings suggest the important role of CD146+ T cells in tumor defense. In addition, the CD146-LCK interaction enhanced the T cell response both in vivo and in vitro. Therefore, manipulating LCK activation by CD146 in T cells is a potential strategy for tumor therapy. It should be noted that CD146 expression has been found upregulated and positively correlated with poor tumor prognosis. In addition, CD146 is a biomarker for tumor angiogenesis (51) and is considered a potential target for blockade for therapy in most tumors (52). Thus, cell type–specific manipulation of CD146 signaling, instead of systemic targeting, is required. For instance, bispecific antibodies may achieve tumor cell–specific CD146 blockade or T cell–specific CD146 activation for enhanced tumor therapy. In addition, increasing CD146 expression on CAR-T cells by engineering may further enhance their efficacy.
Taken together, this study provides the detailed evidence to show how the TCR is initiated by membrane CD146. A small fraction of dimerized CD146 is located in resting T cells and interacts with coreceptor-free LCK to maintain basic LCK activation. TCR engagement further promotes CD146 dimerization, resulting in recruitment of more LCK to the local TCR synapse to activate LCK itself, and coreceptor-bound LCK might further enhance and/or stabilize TCR signaling. Our results demonstrate that CD146 is an integral membrane component mediating LCK activation, important for regulating proximal TCR signaling for both T cell development and activation.
Methods
Mice.
All animal experiments were performed in compliance with the guidelines for the care and use of laboratory animals of the Institute of Biophysics, Chinese Academy of Sciences, and were approved by the institutional biomedical research ethics committee of the Institute. All mice were housed in a pathogen-free facility.
Cd146fl/fl mice were generated at the Nanjing Biomedical Research Institute of Nanjing University (Nanjing, China). All knockout mice were generated using a Cre/loxP recombinase system and backcrossed onto a C57BL/6J background for a minimum of 9 generations. Five-week-old female mice were used for phenotype analysis. EIIA-Cre, Lck-Cre, and Cd4-Cre were purchased from the Nanjing Biomedical Research Institute of Nanjing University. Ly5.1(CD45.1+) mice and transgenic mice expressing OT-II (OT-IItg) were purchased from The Jackson Laboratory. The 5- to 6-week-old female C57BL/6J mice and BALB/c nude mice were obtained from the Department of Laboratory Animal Science, Peking University Health Science Center, Beijing, China.
For generating inducible CD146-knockdown mice, Cd146fl/fl mice were crossed with Ubc-Cre/Ert2 mice (a gift from Shuyang Yu, China Agricultural University, Beijing, China). The F1 Ubccre/+ Cd146fl/+ genotype was backcrossed with Cd146fl/fl mice to generate Ubccre/+ Cd146fl/fl mice. Cd146fl/fl mice were used as control. Five-week-old mice were used for tamoxifen treatment. To prepare a tamoxifen working solution (10 mg/mL), tamoxifen was dissolved in 1 mL ethanol, and the mix was shaken vigorously for 1 minute; then 9 mL of corn oil was added, and the mixture was again shaken vigorously at room temperature for about 30 minutes while light was avoided. Tamoxifen was injected i.p. for 4 consecutive days (1 mg/kg). One week later, the treated mice were used for the experiment.
Antibodies and reagents.
The following anti-CD146 antibodies were used: AA1 and AA98 were generated in our laboratory (34). AA1 was used for immunoprecipitation and immunoblotting; AA1 and AA98 were used for cross-linking. PE/Cy7-conjugated rat anti–mouse CD146 (clone ME-9F1, catalog 134714, BioLegend) was used for flow cytometry; anti-CD146 (catalog ab75769, Abcam) was used for immunoblotting.
Other antibodies and reagents used were as follows: PerCP/Cy5.5–anti–mouse CD3ε (catalog 100327), Brilliant Violet 421TM–anti–mouse NK-1.1 (catalog 108731), APC/Cy7–anti–mouse CD25 (catalog 102025), PE/Cy7–anti–mouse CD5 (catalog 100621), APC–anti–mouse TCRγ/δ (catalog 118115), RBC Lysis Buffer (10×; catalog 420301), Alexa Fluor 647–anti–mouse TCR β chain (catalog 109217), Alexa Fluor 700–anti–mouse CD45.2 (catalog 109822), PerCP/Cy5.5–anti–mouse IL-17A (catalog 506919), Brilliant Violet 421–anti–human/mouse granzyme B (catalog 396413), MojoSort Mouse CD3 T Cell Isolation Kit (catalog 480023), MojoSort Mouse CD4 Naive T Cell Isolation Kit (catalog 480040), MojoSort Mouse CD8 Naive T Cell Isolation Kit (catalog 480043), and MojoSort Mouse CD8 T Cell Isolation Kit (catalog 480008) were obtained from BioLegend. Anti–mouse TCR β–eFluor 450 (catalog 48-5961-82), anti–human/mouse CD44–APC (17-0441-81), anti–mouse CD45.1–FITC (11-0453-82), anti–mouse F4/80 antigen–eFluor 450 (48-4801-82), anti–mouse Ly-6G (Gr-1)–FITC (11-5931-82), anti–mouse CD11b–PerCP-Cyanine5.5 (45-0112-80), anti–human/mouse phospho–p38 MAPK (T180/Y182)–APC (17-9078-41), anti–human/mouse phospho–ZAP70/SYK (Y319/Y352)–PerCP-eFluor 710 (46-9006-41), anti–human/mouse phospho-ERK1/2 (T202/Y204)–PE-eFluor 610 (61-9109-41), anti–mouse TNF-α–PE-Cyanine7 (25-7321-82), anti–mouse IFN-γ–PE (12-7311-82), anti–mouse IL-2–APC (17-7021-82), anti–mouse CD11c–APC (17-0114-81), Anti–Mouse CD2 Functional Grade Purified (16-0031-82), Anti–Mouse TCR β Functional Grade Purified (16-5961-82), Anti–Mouse CD28 Functional Grade Purified (16-0281-82), Anti–Mouse CD3ε Functional Grade Purified (16-0281-82), Cell Stimulation Cocktail (plus protein transport inhibitors) (500×; 00-4975-03), Cell Stimulation Cocktail (500×; 00-4970-93), BrdU Staining Buffer Set for Flow Cytometry Kit (00-5525-00), Mouse IFN-γ ELISA Ready-SET-Go! (88-7314-22), Mouse IL-2 ELISA Ready-SET-Go! (88-7024-77), Permeabilization Buffer (10×; 00-8333-56), and IC Fixation Buffer (00-8222-49) were purchased from eBioscience. Fluo-4 (catalog F14201) was purchased from Invitrogen. FcR Blocking Reagent, mouse (catalog 130-092-575), was purchased from Miltenyi Biotec. Anti-CD24–APC (catalog M30241-11A), anti-CD4–FITC (M10041-02B), anti-CD8–PE (M10083-09B), anti-CD69–FITC (M30691-02B), anti-CD4–APC (M10043-11A), anti-CD62L–PE (M10621-09B), anti-CD19–PE (M10191-09D), anti-GST (KM8005), and the Annexin V–FITC/7-AAD Cell Apoptosis Analysis Kit (AO2001-02A-H) were purchased from Tianjin SUNGENE BIOTECH Ltd. Anti-LCK (catalog ab32149), anti-ZAP70 (phospho Y319) (ab194792), anti-CD4 (ab133616), anti-CD3δ (ab103573), anti-CD3ξ (ab190728), anti-FYN (ab184276), and anti-CD3γ (ab134096) were purchased from Abcam. Anti-ZAP70 (catalog A2195), anti-LCK (phospho Y394) (AP0182), anti-actin (AC028), anti-GAPDH (AC033), anti-His tag (AE003), and anti-MBP (AE016) were purchased from ABclonal. Antibodies against phospho–p44/42 MAPK (ERK1/2) (Thr202/Tyr204) (catalog 4370S), p44/42 MAPK (ERK1/2) (4695S), phospho–p38 MAPK (Thr180/Tyr182) (4511S), p38α MAPK (L53F8) (9228S), PLCγ (2822S), and LAT (9166S) were purchased from Cell Signaling Technology. Mouse IgG2a (catalog M5409), IgG2b (M1395), anti-FLAG (Fl804), BrdU (B5002), and diethylnitrosamine (73861) were obtained from MilliporeSigma. The OVA-I-Ab tetramer (TS-M710-1) was purchased from MBL International. Sangon Biotech synthesized OVA323–339 peptide. InVivoMAb anti–mouse PD-1 (CD279) (BE0146-25MG) and rat IgG2a control (BE0089) were purchased from Bio X Cell.
Cells and plasmids.
Cell lines, including Jurkat, B16F10, and MC38, were purchased from ATCC. The B16-OVA cell line was a gift from Yang-Xin Fu (University of Texas Southwestern Medical Center, Dallas, Texas, USA). Knockdown of CD146 was performed using CD146 shRNA (Supplemental Table 1) constructed in the lentivirus vector GV248. Overexpression of CD146 was performed using pEGFP-N1-hCD146 plasmid or pHS-AVC (for mouse CD146). All CD146 mutants were constructed in a pECMV-3×FLAG-N vector. All human LCK cytoplasmic truncations were constructed in a pIRES2-DsRed-Express vector. Transfection of plasmids in Jurkat cells was performed using the AMAXA 4D-Nucleofector (Lonza).
Generation of bone marrow chimeric models.
Bone marrow cells collected from CD45.1+ mice were mixed at a ratio of 1:1 with bone marrow cells from CD146WT or different versions of CD146KO (CD45.2+) mice, and the mixture (5 × 106) was injected into irradiated CD45.1+ mice (7.5 Gy). Mice were analyzed 8–12 weeks after bone marrow transfer was completed.
BrdU labeling.
Labeling was performed as previously described (53). Briefly, for detection of thymocytes, mice were injected i.p. with 1 mg BrdU for 2 hours before being sacrificed. BrdU incorporation was analyzed by flow cytometry using a BrdU Flow Kit (BioLegend).
DP-to-CD4+CD8int development in vitro.
The DP-to-CD4+CD8int development assay was performed as described previously (29). Briefly, DP cells were isolated from WT and KO thymocytes by FACS and were then resuspended in RPMI 1640 medium to adjust the concentration to 2.5 × 107/mL. One hundred microliters of cells were incubated overnight with 2.5 μg/mL or 5 μg/mL plate-bound anti-TCRβ and anti-CD2 antibodies. Incubated cells were then washed and stained against CD4 and CD8 and finally analyzed for CD4+CD8int cells using flow cytometry (Stimulatory) or cultured for another 24 hours in the same medium before analysis (Recovery).
Ca2+ flux.
Cell suspensions of thymocytes or splenocytes were first labeled for 1 hour at 37°C with 4 μg/mL Fluo-4 (Invitrogen) before being washed with ice-cold PBS and resuspended in PBS. Cells (3 × 106) were surface-labeled for 30 minutes on ice with APC-conjugated anti-CD4 and PE-conjugated anti-CD8, before being warmed for 20 minutes to room temperature and stimulated with 3 μg/mL anti-CD3 antibody (for thymocytes) or anti–CD3/CD28 antibodies (for splenocytes) or PMA/ionomycin immediately before flow cytometry. Mean fluorescence ratios were plotted after analysis with FlowJo software (Tree Star).
Splenocyte CD69 upregulation assay.
Splenic naive T cells isolated from CD146WT, CD146KO, or CD146LCK-KO mice were stimulated with the indicated concentrations of plate-bound anti-CD3 and anti-CD28 antibodies or PMA/ionomycin for 5 hours at 37°C. Cells were collected and stained with FITC-CD69 and analyzed by flow cytometry.
OVA329–337-specific T cell induction.
Splenocytes isolated from OT-IItgCD146 CD4-WT and -KO mice were stimulated with OVA329–337 peptide (1 μg/mL) for 72 hours. OVA-I-Ab tetramer was used to detect the OVA329–337-specific T cells.
Real-time fluorescence quantitative PCR.
The cells were washed twice with 1× PBS, and the total RNA was isolated using TRIzol reagent (15596026, Invitrogen). The mRNA served as a template, and reverse transcriptase PCR was performed according to standard protocols. Quantitative real-time PCR was performed using SYBR Premix Ex TaqII (RR820A, Takara) and the ViiA 7 Real-Time PCR system (Thermo Fisher Scientific). The primers used are listed in Supplemental Table 2.
Pull-down assay for CD146 and LCK proteins.
CD146–intracellular linker–GST protein (K0130) was produced by a prokaryotic expression system (Generay Biotech). Recombinant LCK-His protein (ab82185) was purchased from Abcam. Recombinant CD146 protein was ligated on GST beads. After washing of the conjugates 3 times, recombinant LCK-His protein was added and incubated at 4°C for 1 hour. After being washed 3 times with PBS/lysis buffer (9:1), samples were resolved in 1× SDS reduced sample buffer for SDS-PAGE gels (10%), and then blotted using anti-GST and anti-LCK antibodies.
Recombinant LCK SH2-linker-MBP, SH3-linker-MBP, and SH3-SH2-linker-MBP were produced by prokaryotic expression system. All the recombinant proteins were used for the pull-down assay.
Immunoprecipitation and immunoblot analysis.
Cells were lysed in Cell Lysis Reagent (MilliporeSigma). Cell lysates were precleared with Protein A/G Sepharose beads, and then the supernatants were immunoprecipitated overnight at 4°C with the appropriate antibodies. Protein A/G Sepharose beads were added, and the samples were incubated for an additional 2 hours. After being washed 3 times with PBS/lysis buffer (9:1), samples were resolved in 1× SDS reduced sample buffer for SDS-PAGE gels (10%) and blotted, or resolved in 1× SDS nonreduced sample buffer (no 2-mercaptoethanol) for dimer detection. For immunoblot analysis, cells were lysed in SDS sample buffer by the addition of 1:4 volume of 5× SDS sample buffer directly into cell suspensions. Samples were then boiled for 5 minutes and separated by (10%) SDS-PAGE.
Simple Western immunoblots were performed with Peggy Sue (ProteinSimple) using the Size Separation Master Kit with Split Buffer (12–230 kDa) according to the manufacturer’s instructions, using the following antibodies: anti–p-ZAP70, anti-ZAP70, anti–p-LCK(Y394), and anti-LCK. Compass software (version 2.7.1, ProteinSimple) was used to program the Peggy Sue robot and to quantify the peak values of the Western immunoblots. Output data are displayed from the software-calculated average of 7 exposures (5–480 seconds).
Adoptively transferred T cells in B16-OVA tumor model.
Splenic CD8+ T cells were isolated from OT-IItgCD146CD4-WT and OT-IItgCD146CD4-KO mice bearing B16-OVA tumors for 8 days. BALB/c nude mice were injected s.c. with 2 × 106 B16-OVA cells and received 2 × 106 to 5 × 106 isolated CD8+ T cells by adoptive transfer on day 7. The tumor volumes were measured every 2 days.
Statistics.
GraphPad Prism software was used for statistical analysis. All bar graphs show the mean ± SEM. Differences between groups were examined by Student’s t test or ANOVA for comparison of means, as indicated in each legend. P values less than 0.05 were considered statistically significant.
Study approval.
This study conformed to approvals granted by the institutional biomedical research ethics committee of the Institute of Biophysics, Chinese Academy of Sciences (permit SYXK2018-45). All experimental procedures involving vertebrate animals were conducted in accordance with the approval SYXK2018-45.
Author contributions
HD designed and performed the experiments, analyzed the data, and wrote the manuscript. LJ designed and performed the experiments and analyzed the data. YM, XJ, DW, JX, ZW, HY, JJ, and ZL assisted with the experiments. XC and JF analyzed the data. MZ and XY designed the research and wrote the manuscript.
Supplementary Material
Acknowledgments
We are indebted to Yu Zhang (Peking University), Wei He (Peking Union Medical College), Wanli Liu (Tsinghua University), and Chenqi Xu (Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences) for their encouragement and guidance throughout this study. We thank Torsten Juelich (Peking University) for linguistic assistance during the preparation of the manuscript. This work was supported in part by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (grants XDB29040101 and XDA12020207), the Beijing Natural Science Foundation of China (grant 7192123), the National Natural Science Foundation of China (grants 82025015 and 31770793), and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (grant 2018122).
Version 1. 09/07/2021
In-Press Preview
Version 2. 11/01/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(21):e148568.https://doi.org/10.1172/JCI148568.
Contributor Information
Hongxia Duan, Email: cherryshoen@aliyun.com.
Lin Jing, Email: 18844195017@163.com.
Xiaoqing Jiang, Email: jiangxiaoqing9104@163.com.
Yanbin Ma, Email: mayanbin888@163.com.
Daji Wang, Email: 986976987@qq.com.
Jianquan Xiang, Email: xuexixiang@163.com.
Xuehui Chen, Email: 13621223059@163.com.
Zhenzhen Wu, Email: wuzhenzhen@moon.ibp.ac.cn.
Huiwen Yan, Email: wencaihuicui@163.com.
Junying Jia, Email: jiajy@ibp.ac.cn.
Zheng Liu, Email: 13811165210@163.com.
Jing Feng, Email: fengjing@moon.ibp.ac.cn.
Mingzhao Zhu, Email: zhumz@ibp.ac.cn.
Xiyun Yan, Email: yanxy@ibp.ac.cn.
References
- 1.Lopez-Rodriguez C, et al. Transcription factors and target genes of pre-TCR signaling. Cell Mol Life Sci. 2015;72(12):2305–2321. doi: 10.1007/s00018-015-1864-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin K, et al. Lck domains differentially contribute to pre-T cell receptor (TCR)- and TCR-alpha/beta-regulated developmental transitions. J Exp Med. 2000;191(4):703–716. doi: 10.1084/jem.191.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trobridge PA, Levin SD. Lck plays a critical role in Ca(2+) mobilization and CD28 costimulation in mature primary T cells. Eur J Immunol. 2001;31(12):3567–3579. doi: 10.1002/1521-4141(200112)31:12<3567::AID-IMMU3567>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 4.van Oers NS, et al. Lck regulates the tyrosine phosphorylation of the T cell receptor subunits and ZAP-70 in murine thymocytes. J Exp Med. 1996;183(3):1053–1062. doi: 10.1084/jem.183.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philipsen L, et al. De novo phosphorylation and conformational opening of the tyrosine kinase Lck act in concert to initiate T cell receptor signaling. Sci Signal. 2017;10(462):eaaf4736. doi: 10.1126/scisignal.aaf4736. [DOI] [PubMed] [Google Scholar]
- 6.Eck MJ, et al. Structure of the regulatory domains of the Src-family tyrosine kinase Lck. Nature. 1994;368(6473):764–769. doi: 10.1038/368764a0. [DOI] [PubMed] [Google Scholar]
- 7.Xu Q, et al. Identifying three-dimensional structures of autophosphorylation complexes in crystals of protein kinases. Sci Signal. 2015;8(405):rs13. doi: 10.1126/scisignal.aaa6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Laethem F, et al. Deletion of CD4 and CD8 coreceptors permits generation of αβT cells that recognize antigens independently of the MHC. Immunity. 2007;27(5):735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Roh KH, et al. The coreceptor CD4 is expressed in distinct nanoclusters and does not colocalize with T-cell receptor and active protein tyrosine kinase p56lck. Proc Natl Acad Sci U S A. 2015;112(13):E1604–E1613. doi: 10.1073/pnas.1503532112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moldovan MC, et al. CD4 dimers constitute the functional component required for T cell activation. J Immunol. 2002;169(11):6261–6268. doi: 10.4049/jimmunol.169.11.6261. [DOI] [PubMed] [Google Scholar]
- 11.Van Laethem F, et al. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell. 2013;154(6):1326–1341. doi: 10.1016/j.cell.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irvine DJ, et al. Direct observation of ligand recognition by T cells. Nature. 2002;419(6909):845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann JM, et al. Discrimination between benign and malignant cells of melanocytic lineage by two novel antigens, a glycoprotein with a molecular weight of 113,000 and a protein with a molecular weight of 76,000. Cancer Res. 1987;47(3):841–845. [PubMed] [Google Scholar]
- 14.Guezguez B, et al. A dileucine motif targets MCAM-l cell adhesion molecule to the basolateral membrane in MDCK cells. FEBS Lett. 2006;580(15):3649–3656. doi: 10.1016/j.febslet.2006.05.048. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, et al. CD146 coordinates brain endothelial cell-pericyte communication for blood-brain barrier development. Proc Natl Acad Sci U S A. 2017;114(36):E7622–E7631. doi: 10.1073/pnas.1710848114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Y, et al. Macrophagic CD146 promotes foam cell formation and retention during atherosclerosis. Cell Res. 2017;27(3):352–372. doi: 10.1038/cr.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan H, et al. CD146 is required for VEGF-C-induced lymphatic sprouting during lymphangiogenesis. Sci Rep. 2017;7(1):7442. doi: 10.1038/s41598-017-06637-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Yan X. CD146, a multi-functional molecule beyond adhesion. Cancer Lett. 2013;330(2):150–162. doi: 10.1016/j.canlet.2012.11.049. [DOI] [PubMed] [Google Scholar]
- 19.Bu P, et al. Visualization of CD146 dimerization and its regulation in living cells. Biochim Biophys Acta. 2007;1773(4):513–520. doi: 10.1016/j.bbamcr.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Pickl WF, et al. MUC18/MCAM (CD146), an activation antigen of human T lymphocytes. J Immunol. 1997;158(5):2107–2115. [PubMed] [Google Scholar]
- 21.Elshal MF, et al. CD146 (Mel-CAM), an adhesion marker of endothelial cells, is a novel marker of lymphocyte subset activation in normal peripheral blood. Blood. 2005;106(8):2923–2924. doi: 10.1182/blood-2005-06-2307. [DOI] [PubMed] [Google Scholar]
- 22.Brucklacher-Waldert V, et al. Phenotypical and functional characterization of T helper 17 cells in multiple sclerosis. Brain. 2009;132(pt 12):3329–3341. doi: 10.1093/brain/awp289. [DOI] [PubMed] [Google Scholar]
- 23.Duan H, et al. Targeting endothelial CD146 attenuates neuroinflammation by limiting lymphocyte extravasation to the CNS. Sci Rep. 2013;3:1687. doi: 10.1038/srep01687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dagur PK, et al. MCAM-expressing CD4(+) T cells in peripheral blood secrete IL-17A and are significantly elevated in inflammatory autoimmune diseases. J Autoimmun. 2011;37(4):319–327. doi: 10.1016/j.jaut.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seftalioglu A, Karakoc L. Expression of CD146 adhesion molecules (MUC18 or MCAM) in the thymic microenvironment. Acta Histochem. 2000;102(1):69–83. doi: 10.1078/0065-1281-00544. [DOI] [PubMed] [Google Scholar]
- 26.Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat Immunol. 2011;12(3):666–673. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D, et al. Tespa1 is involved in late thymocyte development through the regulation of TCR-mediated signaling. Nat Immunol. 2012;13(6):560–568. doi: 10.1038/ni.2301. [DOI] [PubMed] [Google Scholar]
- 28.Calnan BJ, et al. A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity. 1995;3(3):273–282. doi: 10.1016/1074-7613(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 29.Cibotti R, et al. Surface molecules that drive T cell development in vitro in the absence of thymic epithelium and in the absence of lineage-specific signals. Immunity. 1997;6(3):245–255. doi: 10.1016/S1074-7613(00)80327-1. [DOI] [PubMed] [Google Scholar]
- 30.Luo Y, et al. Recognition of CD146 as an ERM-binding protein offers novel mechanisms for melanoma cell migration. Oncogene. 2012;31(3):306–321. doi: 10.1038/onc.2011.244. [DOI] [PubMed] [Google Scholar]
- 31.Lefebvre DC, et al. CD44 interacts directly with Lck in a zinc-dependent manner. Mol Immunol. 2010;47(10):1882–1889. doi: 10.1016/j.molimm.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 32.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci U S A. 1999;96(17):9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X, et al. Structure basis for AA98 inhibition on the activation of endothelial cells mediated by CD146. iScience. 2021;24(5):102417. doi: 10.1016/j.isci.2021.102417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, et al. Generation and characterization of a panel of monoclonal antibodies against distinct epitopes of human CD146. Hybridoma (Larchmt) 2008;27(5):345–352. doi: 10.1089/hyb.2008.0034. [DOI] [PubMed] [Google Scholar]
- 35.Zondler L, et al. MCAM/CD146 signaling via PLCγ1 leads to activation of β1-integrins in memory T-cells resulting in increased brain infiltration. Front Immunol. 2020;11:599936. doi: 10.3389/fimmu.2020.599936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veillette A, et al. Engagement of CD4 and CD8 expressed on immature thymocytes induces activation of intracellular tyrosine phosphorylation pathways. J Exp Med. 1989;170(5):1671–1680. doi: 10.1084/jem.170.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiest DL, et al. TCR activation of ZAP70 is impaired in CD4+CD8+ thymocytes as a consequence of intrathymic interactions that diminish available p56lck. Immunity. 1996;4(5):495–504. doi: 10.1016/S1074-7613(00)80415-X. [DOI] [PubMed] [Google Scholar]
- 38.Wiest DL, et al. Regulation of T cell receptor expression in immature CD4+CD8+ thymocytes by p56lck tyrosine kinase: basis for differential signaling by CD4 and CD8 in immature thymocytes expressing both coreceptor molecules. J Exp Med. 1993;178(5):1701–1712. doi: 10.1084/jem.178.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nika K, et al. Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity. 2010;32(6):766–777. doi: 10.1016/j.immuni.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wan R, et al. Biophysical basis underlying dynamic Lck activation visualized by ZapLck FRET biosensor. Sci Adv. 2019;5(6):eaau2001. doi: 10.1126/sciadv.aau2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levental I, et al. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry. 2010;49(30):6305–6316. doi: 10.1021/bi100882y. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, et al. Protein depalmitoylation is induced by Wnt5a and promotes polarized cell behavior. J Biol Chem. 2015;290(25):15707–15716. doi: 10.1074/jbc.M115.639609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tani-ichi S, et al. Structure and function of lipid rafts in human activated T cells. Int Immunol. 2005;17(6):749–758. doi: 10.1093/intimm/dxh257. [DOI] [PubMed] [Google Scholar]
- 44.Anfosso F, et al. Outside-in signaling pathway linked to CD146 engagement in human endothelial cells. J Biol Chem. 2001;276(2):1564–1569. doi: 10.1074/jbc.M007065200. [DOI] [PubMed] [Google Scholar]
- 45.Dagur PK, et al. Endothelial-binding, proinflammatory T cells identified by MCAM (CD146) expression: characterization and role in human autoimmune diseases. Autoimmun Rev. 2015;14(5):415–422. doi: 10.1016/j.autrev.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larochelle C, et al. Melanoma cell adhesion molecule identifies encephalitogenic T lymphocytes and promotes their recruitment to the central nervous system. Brain. 2012;135(pt 10):2906–2924. doi: 10.1093/brain/aws212. [DOI] [PubMed] [Google Scholar]
- 47.Flanagan K, et al. Laminin-411 is a vascular ligand for MCAM and facilitates TH17 cell entry into the CNS. PLoS One. 2012;7(7):e40443. doi: 10.1371/journal.pone.0040443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider-Hohendorf T, et al. VLA-4 blockade promotes differential routes into human CNS involving PSGL-1 rolling of T cells and MCAM-adhesion of TH17 cells. J Exp Med. 2014;211(9):1833–1846. doi: 10.1084/jem.20140540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larochelle C, et al. Melanoma cell adhesion molecule-positive CD8 T lymphocytes mediate central nervous system inflammation. Ann Neurol. 2015;78(1):39–53. doi: 10.1002/ana.24415. [DOI] [PubMed] [Google Scholar]
- 50.Hartl FA, et al. Noncanonical binding of Lck to CD3epsilon promotes TCR signaling and CAR function. Nat Immunol. 2020;21(8):902–913. doi: 10.1038/s41590-020-0732-3. [DOI] [PubMed] [Google Scholar]
- 51.Yan X, et al. A novel anti-CD146 monoclonal antibody, AA98, inhibits angiogenesis and tumor growth. Blood. 2003;102(1):184–191. doi: 10.1182/blood-2002-04-1004. [DOI] [PubMed] [Google Scholar]
- 52.Labib M, et al. Electrochemical methods for the analysis of clinically relevant biomolecules. Chem Rev. 2016;116(16):9001–9090. doi: 10.1021/acs.chemrev.6b00220. [DOI] [PubMed] [Google Scholar]
- 53.Egawa T, et al. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204(8):1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.