Abstract
The transcription factor NFATC2 induces β cell proliferation in mouse and human islets. However, the genomic targets that mediate these effects have not been identified. We expressed active forms of Nfatc2 and Nfatc1 in human islets. By integrating changes in gene expression with genomic binding sites for NFATC2, we identified approximately 2200 transcriptional targets of NFATC2. Genes induced by NFATC2 were enriched for transcripts that regulate the cell cycle and for DNA motifs associated with the transcription factor FOXP. Islets from an endocrine-specific Foxp1, Foxp2, and Foxp4 triple-knockout mouse were less responsive to NFATC2-induced β cell proliferation, suggesting the FOXP family works to regulate β cell proliferation in concert with NFATC2. NFATC2 induced β cell proliferation in both mouse and human islets, whereas NFATC1 did so only in human islets. Exploiting this species difference, we identified approximately 250 direct transcriptional targets of NFAT in human islets. This gene set enriches for cell cycle–associated transcripts and includes Nr4a1. Deletion of Nr4a1 reduced the capacity of NFATC2 to induce β cell proliferation, suggesting that much of the effect of NFATC2 occurs through its induction of Nr4a1. Integration of noncoding RNA expression, chromatin accessibility, and NFATC2 binding sites enabled us to identify NFATC2-dependent enhancer loci that mediate β cell proliferation.
Keywords: Cell Biology, Endocrinology
Keywords: Beta cells, Cell cycle, Diabetes
Introduction
Genetic association and linkage studies are designed to investigate the relationship between genetic variation at a gene locus and phenotypes, including those with disease relevance. GWAS in humans have identified more than 200 loci associated with type 2 diabetes (T2D) (1). We previously asked whether variations in the expression of candidate genes located at GWAS loci are regulated by one or more transcription factors (TFs). We analyzed a rich data set consisting of islet mRNA abundance measurements in 500 mice derived from an F2 intercross between a diabetes-resistant and a diabetes-susceptible obese mouse strain (2).
The data set provided numerous instances in which the abundance of mRNA transcripts mapped to locations coinciding with the physical location of their corresponding genes (cis-eQTL). However, even more numerous were instances in which the abundance of multiple mRNAs comapped to a common locus (trans-eQTL), suggesting that the locus contains a master regulator of the comapping genes. We asked whether the abundance of mRNA transcripts encoding genes for T2D identified in human GWAS map to a similar “hot spot” in the mouse genome. We discovered that approximately 40% of these mRNA abundance phenotypes mapped to a broad region on mouse chromosome 2. Using mediation analysis, we identified Nfatc2 as a regulator of expression of these genes. Further, we demonstrated that NFATC2 potently stimulates β cell proliferation in mouse and human islets (2).
We sought to better understand how such a large proportion of GWAS genes are under the regulation of NFATC2 and to identify its direct transcriptional targets that mediate enhanced β cell proliferation. One possibility is that the binding sites for NFAT proteins are proximal to binding sites for other TFs that affect β cell gene expression and proliferation. This would also predict that some of these other TFs interact and act in concert with NFATC2. We performed ChIP studies to identify genomic binding sites of NFATC2 in human islets. We found that NFATC2 binding sites are proximal to binding sites for several key islet TFs (e.g., NKX6-1, FOXA2, and PDX1). By combining our ChIP data with RNA-Seq, we were able to infer which regulatory sites are stimulatory and which are inhibitory. Our studies establish FOXP proteins as essential binding partners for NFATC2 in the context of β cell proliferation. We identify Nr4a1 as a target of NFATC2 that mediates a major part of the ability of NFATC2 to stimulate β cell proliferation. Finally, we integrated noncoding RNA sequencing (ncRNA-Seq), chromatin accessibility, and NFATC2 genomic binding to identify NFATC2-dependent enhancer loci in human islets. Our results provide a catalog of direct and indirect transcriptional targets of NFATC2 that mediate enhanced β cell proliferation in human islets.
Results
NFAT regulates gene expression in human islets.
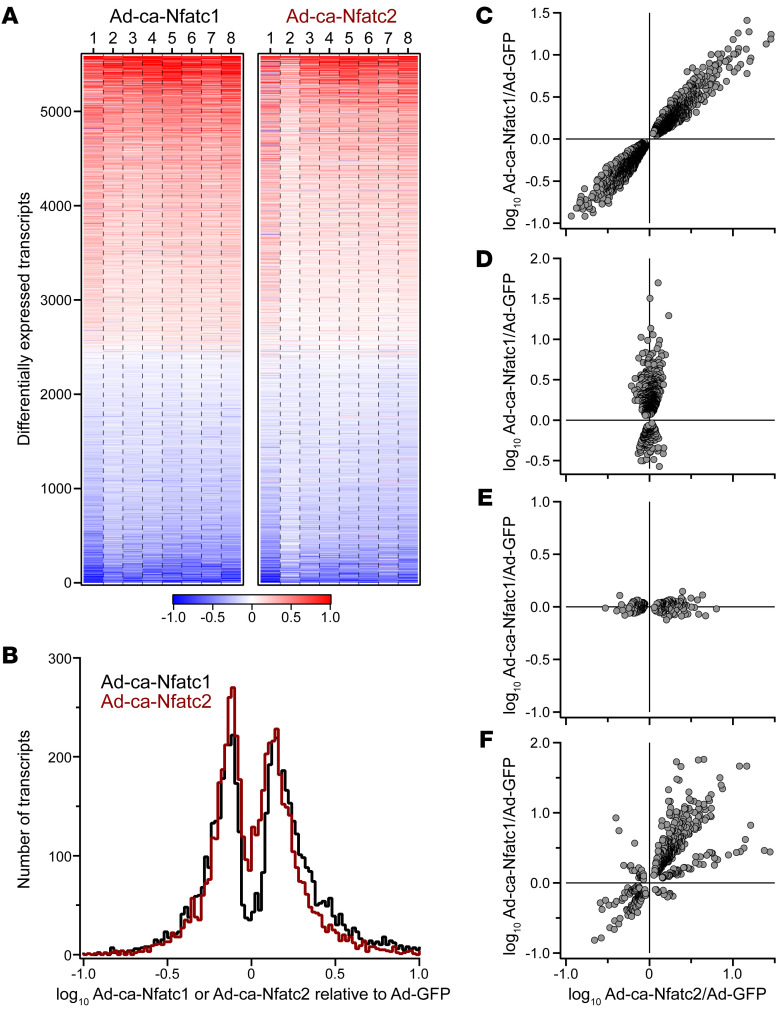
To explore the mechanism by which NFAT stimulates β cell proliferation in human islets, we profiled the human islet transcriptome following overexpression of constitutively active (CA) forms of Nfatc1 and Nfatc2 (3). From 8 distinct human islet donors, a total of approximately 19,000 transcripts were identified, of which 5589 were differentially expressed (DE) in response to NFATC1 and/or NFATC2 (Figure 1A and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI144833DS1). Among the DE genes, approximately 20% and approximately 12% were induced more than 2-fold by NFATC1 and NFATC2, respectively (Figure 1B). Similarly, approximately 9% and approximately 7% of the DE genes were suppressed more than 50% by NFATC1 and NFATC2, respectively.
Figure 1. NFATC1 and NFATC2 regulate gene expression in human islets.
Whole-islet RNA was sequenced from 8 separate human donors (donors 1–8) 48 hours after Ad-mediated expression of CA Nfatc1 or Nfatc2; Ad-GFP was used as the negative control. (A) Heatmap shows the expression of transcripts among the individual samples that were differentially regulated in response to NFATC1 and/or NFATC2 relative to GFP (5589 transcripts). Transcripts are ordered by the average increase in expression in response to NFATC2. Red, increased expression; blue, decreased. A small number of transcripts showed a greater than 10-fold change in expression. For illustration purposes, z scale for heatmap is truncated at a 10-fold change. A complete list of all transcripts and their regulation by NFATC1 and NFATC2 is provided in Supplemental Table 1. (B) Distribution of log10 fold-change in expression for NFATC1- and NFATC2-regulated transcripts. Transcripts were further classified as those that were regulated equally in response to NFATC1 and NFATC2 (C, 3141 transcripts), regulated by NFATC1 only (D, 794 transcripts), regulated by NFATC2 only (E, 226 transcripts), or differentially regulated by NFATC1 and NFATC2 in magnitude or direction (F, 631 transcripts).
As we reported in mouse islets, identifying transcripts that demonstrate differential versus common regulation by NFATC1 and NFATC2 is one approach to linking their effects on β cell proliferation and insulin secretion (2). More than 50% of the DE genes (3141 genes) tend to be equally modulated by NFATC1 and NFATC2 (Figure 1C). This may reflect a homologous protein structure between NFAT2 and NFAT1, the protein products of Nfatc1 and Nfatc2, respectively, including the highly conserved REL homology domain, which confers a nearly identical consensus binding motif (4, 5). Overall, these results demonstrate that NFATC1 and NFATC2 mediate similar changes in gene expression in human islets.
Among the approximately 1700 human genes that are annotated as being associated with regulation of the cell cycle (Gene Ontology GO:0007049), 1642 are expressed in human islets, of which 723 (~44%) are DE in response to NFATC1 or NFATC2. Interestingly, more than half of the DE cell cycle genes are regulated similarly by NFATC1 and NFATC2, including ASF1B and CDKN1A (Supplemental Table 2). Previously, we showed that overexpression of ASF1B was sufficient to induce human β cell proliferation (6). Others have shown that CDKN1A decreases β cell proliferation (7, 8). NFATC1 and NFATC2 induce the expression of ASF1B while suppressing CDKN1A expression, resulting in increased β cell proliferation. These results demonstrate that NFATC1 and NFATC2 directly or indirectly regulate the expression of genes that themselves are sufficient to induce human β cell proliferation, suggesting that NFAT may be a master regulator of β cell proliferation. In contrast to the number of genes that were equally regulated by NFATC1 and NFATC2, many fewer were solely regulated by one of the NFAT isoforms. For example, 889 and 315 genes were regulated exclusively by NFATC1 (Figure 1D) or NFATC2 (Figure 1E), respectively or differentially regulated by NFATC1 and NFATC2 in magnitude or direction (Figure 1F).
Several physiological pathways were significantly enriched (z score > 3) within the gene sets that were transcriptionally regulated by NFATC1 and/or NFATC2 in human islets (Supplemental Figure 1 and Supplemental Table 3). For example, mitochondria-associated genes were enriched (z > 9) in the transcripts that NFATC1 and NFATC2 induced, whereas a number of pathways associated with the nucleus were enriched (z > 12) in transcripts that were suppressed by both TFs. Several pathways were also enriched in gene sets that showed selective regulation by NFATC1 or NFATC2. For example, regulation of insulin secretion was enriched (z ≈ 4.5) in transcripts that were uniquely induced by NFATC2, including IRS1, CDK16, SFRP1, NPFF, and NR1D1. Genes induced exclusively by NFATC1 enriched for histone ubiquitination (z ≈ 4.8), whereas terms associated with mRNA processing were suppressed by NFATC1. Taken together, these results suggest that NFATC1 and NFATC2 positively regulate expression of genes encoding mitochondrial proteins and negatively regulate genes encoding nuclear proteins. Less robust enrichment was observed for smaller gene sets that showed selective regulation by NFATC1 versus NFATC2.
We showed that, in addition to inducing β cell proliferation, NFATC2 is a key driver of genes in mouse islets whose orthologs have been associated with diabetes in human GWAS (2). Among approximately 242 genes that have been associated with diabetes in humans, the expression of 82 was regulated by NFATC1 and/or NFATC2 in human islets (Supplemental Figure 2). The majority of these were equally regulated by NFATC1 and NFATC2, including HNF4A, CDC123, HHEX, GLIS3, and SLC30A8. GIPR and MACF1 were exclusively regulated by NFATC1, whereas the expression of IRS1 was regulated by NFATC2. These results extend our earlier observations in the mouse by showing that in human islets, NFATC2 is a key regulator of genes associated with diabetes. Further, the association of T2D susceptibility to a locus containing NFATC2 in human GWAS (Supplemental Figure 3) may in part reflect NFATC2-dependent regulation of these diabetes-associated effector genes.
NFATC2 preferentially binds sites proximal to genes in human islets.
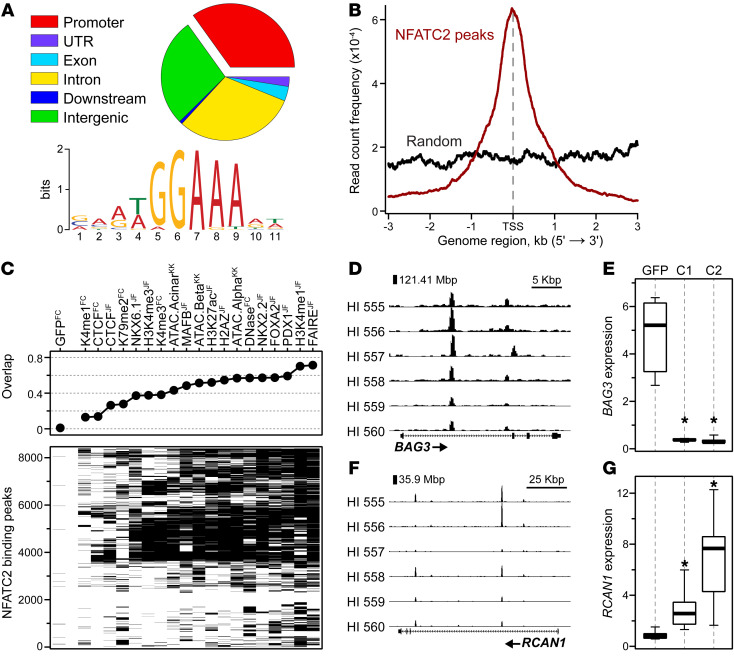
To identify direct transcriptional targets of NFATC2, we performed genome-wide ChIP-Seq for NFATC2 in 6 distinct human islet donors. We identified a total of approximately 8600 binding sites, approximately 35% of which were within the promoter region (±3 Kbp from the transcription start site [TSS]) of known genes (Figure 2A). The NFAT consensus motif (TGGAAA) was the most common DNA sequence present at the NFATC2 binding sites (Fisher’s exact test, P = 2 × 10–668). Most of these binding sites were very close to the TSS of the nearest gene (Figure 2B), suggesting these genes may be direct transcriptional targets of NFATC2.
Figure 2. NFATC2 preferentially binds DNA proximal to gene loci in human islets.
Islets from 6 separate human donors were transduced with Ad-ca-Nfatc2 or Ad-GFP as the negative control. Forty-eight hours later, ca-Nfatc2–bound chromatin was isolated and sequenced, resulting in the identification of 8635 genome-wide NFATC2 binding sites. (A) Genomic distribution is illustrated as a percentage of all sites, as well as the position weight matrix for the most common DNA sequence present at NFATC2 binding sites. (B) Average read density for all NFATC2 binding sites or an equal number of randomly selected sites ±3 kb from all known TSSs. (C) Enrichment scores for alignment of NFATC2 sites with previously published ChIP-Seq for CTCF, several TFs (MAFB, NKX6.1, FOXA2, PDX1), histone modifications, and measures of chromatin accessibility (ATAC-Seq, DNase, FAIRE-Seq) in human islets. Published data from the labs of Francis Collins (10), Klaus Kaestner (11), and Jorge Ferrer (12, 13). Genome browser views for NFATC2 binding at the BAG3 (D) and RCAN1 (F) gene loci. Expression of BAG3 (E) and RCAN1 (G) in human islets 48 hours after transduction with Ad-GFP (GFP), Ad-ca-Nfatc1 (C1), or Ad-ca-Nfatc2 (C2). Data are represented as mean ± SEM. *P < 0.05, Student’s 2-tailed t test for C1 or C2 versus GFP.
Previous studies have identified binding sites for key TFs in human islets, including NKX6-1, NKX2-2, FOXA2, and PDX1 (9). In addition, various histone modifications (e.g., H3K27ac, H3K4me1) and chromatin accessibility (e.g., ATAC, FAIRE, DNase) measurements have been published for human islets (9–13). We investigated the extent to which NFATC2 binding sites overlap with these other TFs and epigenome modifications. All ChIP sets overlapped significantly with NFATC2 binding, except for H3K79me2 (Supplemental Figure 4). Interestingly, more than 60% of the NFATC2 binding sites also bound NKX2-2, FOXA2, and PDX1 (Figure 2C). Furthermore, more than 70% of the NFATC2 sites also contained strong signals for H3K4me1, a feature of active and primed enhancer loci (14). These results suggest that NFATC2 may work in concert or compete with other key TFs at critical regulatory loci in human islets. Finally, approximately 45% of the NFATC2 binding sites cooccurred with previously identified β cell assay for transposase-accessible chromatin–sequencing (ATAC-Seq) peaks, indicating that NFATC2 commonly binds to open chromatin sites, but may also pioneer less accessible chromatin regions.
NFAT can directly activate or suppress the expression of target genes (5). We integrated our NFATC2 binding data with the RNA-Seq data to identify examples of both induction and repression in human islets. For example, NFATC2 binds at 2 sites within the first intron of BCL2-associated athanogene 3 (BAG3) at approximately 121.42 and approximately 121.43 Mbp (Figure 2D). These 2 NFATC2 binding sites likely function as negative regulators, as both NFATC1 and NFATC2 strongly suppress the expression of BAG3 in human islets (Figure 2E). Previous work has shown that BAG3 protein is abundant in human β cells (15), consistent with our measure of BAG3 expression in human islets treated with the control adenovirus Ad-GFP (Figure 2E). Silencing Bag3 expression in a mouse β cell line leads to enhanced glucose-stimulated insulin secretion (15). In contrast to the negative regulation of BAG3, NFATC2 positively regulates the expression of RCAN1 (regulator of calcineurin 1). RCAN1 binds to the catalytic domain of calcineurin A and decreases its phosphatase activity (16), thereby negatively regulating NFAT transcriptional activity. NFATC2 binds to the promoter of a short isoform of RCAN1 at approximately 35.9 Mbp and the first intron of a long isoform at approximately 35.95 Mbp on Chr 21 (Figure 2F), leading to an approximately 3-fold increase in RCAN1 expression in human islets (Figure 2G). Previous studies have identified NFATC1 binding sites in the promoter of RCAN1 (17). Our ChIP data suggest that NFATC2 binds to cis regulatory elements, resulting in increased RCAN1 expression, forming a negative feedback loop on endogenous NFATC2 activity. The significant induction of RCAN1 expression likely reflects our use of the CA form of NFATC2, which is not subject to calcineurin-dependent regulation (3). A further direct target of NFATC2 appears to be VEGFA. VEGFA is a growth factor that is important for islet vascularization. NFATC2 binds within the first 2 introns of VEGFA on Chr 6 at approximately 43.74 Mbp, leading to a 40% decrease in expression. These results demonstrate that by linking our NFATC2-binding data with NFATC2-dependent gene regulation in human islets, we can identify genes directly targeted by NFAT that play key roles in islet function.
Identification of direct transcriptional targets of NFATC2 in human islets.
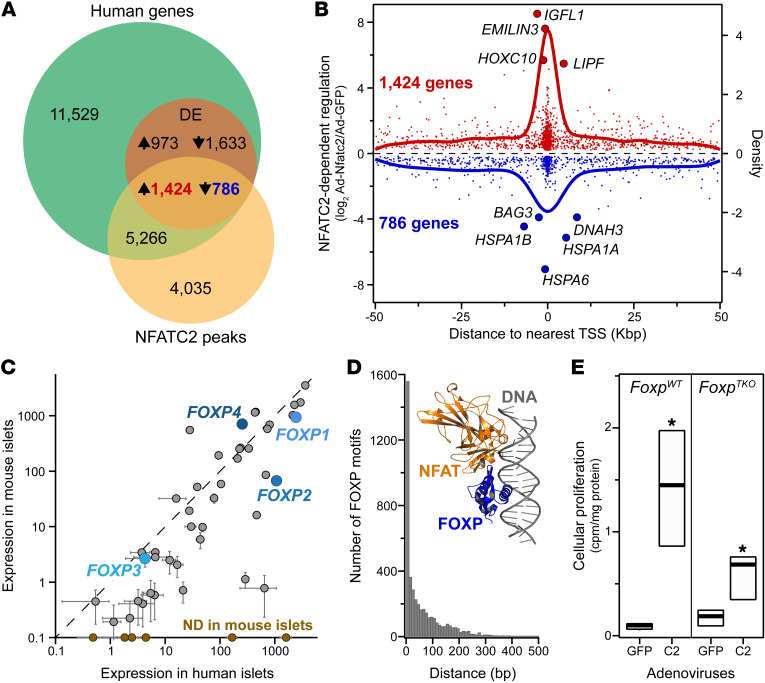
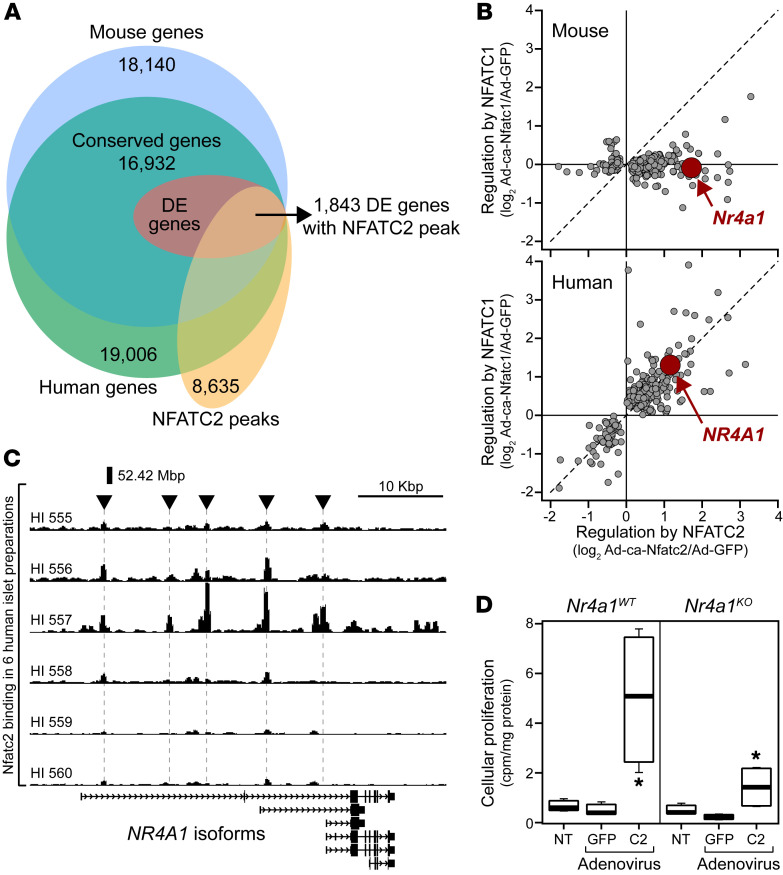
To elucidate the direct transcriptional networks initiated by NFAT that lead to enhanced β cell proliferation, we integrated our NFATC2 human RNA-Seq and ChIP-Seq data sets. Among the approximately 19,000 transcripts and approximately 8600 NFATC2 binding sites, we identified approximately 2200 DE genes that had one or more NFATC2 binding peaks within 50 Kbp of the TSS (Figure 3A). A 50 Kbp window was chosen, based on the performance of different window sizes in determining target genes by ChIP-Seq and RNA-Seq (18). Previous studies have used this approach to define possible direct transcriptional targets of TFs (19). Of these DE genes, the expression of 1425 was induced, while 786 were suppressed by NFATC2 (Supplemental Table 4). There is a strong relationship between proximity of NFATC2 binding to the TSS and the magnitude of gene regulation; binding sites closer to the TSS tended to result in larger fold changes in expression (Figure 3B).
Figure 3. NFATC2 complexes with FOXP to regulate cellular proliferation in mouse islets.
(A) Whole islet RNA-Seq (19,006 transcripts) and NFATC2 genomic binding (8635 sites) were integrated to identify direct NFATC2 transcriptional targets in human islets. 4817 Genes were DE in response to NFATC2 overexpression; 2211 of these DE transcripts contained an NFATC2 binding site within 50 Kbp of the TSS, suggesting they are directly regulated by NFATC2. (B) Relationship between the magnitude of NFATC2-dependent gene regulation and distance from the NFATC2 peak to the TSS is illustrated for the direct target genes. (C) Expression levels for approximately 50 known NFAT binding partners in mouse versus human islets; FOXP family is highlighted. Six genes were expressed in human, but not mouse islets (ND in mouse islets): 3 members of the POU family (POU4F3, POU4F2, and POU5F1), GATA1, EGR4, and MMP1, in increasing expression in human islets. (D) Frequency distribution between NFATC2 binding sites and known FOXP motif (MA0481.2, GTAAACA). Inset: crystal structure (PDB: 2AS5) of DNA binding domains for FOXP and NFAT in complex with DNA segment for the murine Il2 promoter ARRE2 site gene. (E) NFATC2-induced cellular proliferation in islets from WT(FoxpWT) and Foxp1, Foxp2, and Foxp4 triple-knockout mice (FoxpTKO). Data are represented as mean ± SEM. *P < 0.05 for NFATC2 versus GFP, Student’s 2-tailed t test. FoxpWT, n = 3; FoxpTKO, n = 4.
To gain insight into the physiological pathways that may be regulated by NFAT, we asked whether the DE genes directly targeted by NFATC2 were enriched for GO or KEGG annotations (Supplemental Figure 5 and Supplemental Table 5 and refs. 20, 21). Genes that play a key role in the unfolded protein response (UPR) were significantly enriched (z > 8) among the 786 genes that had an NFATC2 peak proximal to their promoter and were suppressed upon NFATC2 overexpression, including DDIT3 (CHOP) and EIF2AK3 (PERK) as well as several heat shock protein chaperones (e.g., HSPA1A, HSPA6, HSPA1B; see also Figure 3B). In contrast, direct transcriptional targets that were induced by NFATC2 were enriched for regulation of cellular communication by electrical coupling (z ≈ 6), histone methyl-transferase complex (z ≈ 5), and pathways associated with ciliary transport (z ≈ 5). Genes that did not have an NFATC2 peak, but were DE in response to NFATC1 and NFATC2 in mouse and human islets (i.e., were indirect targets), were enriched for pathways distinct from those with NFATC2 peaks. For example, indirect targets that were suppressed were enriched for ubiquitin and proteosomal degradation processes, whereas genes that were induced were enriched for chemokine-associated pathways.
NFAT has several known binding partners, including FOS, JUN, and FOXP3 (22, 23). In rat islets, overexpression of Fos leads to increased Nr4a1 expression in parallel with enhanced β cell proliferation (24). Several of the NFAT binding partners are abundantly expressed in human and mouse islets (e.g., 3 members of the FOXP family) and thus could modulate the effect of NFAT on gene expression and β cell proliferation in human islets (Figure 3C).
We performed 2 separate analyses to determine whether one or more of these binding partners are involved in NFAT-dependent gene regulation in human islets. First we performed a ChIP enrichment analysis (ChEA) to determine whether other transcription factors bound near, and co-regulated the same DE genes as NFATC2 (25). Among the 1425 genes whose expression increases in response to NFATC2 binding, FOXP1 binding was found to be significantly enriched (P < 10–11) in human embryonic stem cells (Supplemental Figure 6). Second, we asked, among all NFATC2 binding sites, whether there are motifs for other known TFs that essentially cooccur, i.e., are within 500 bp of the NFATC2 binding locus. We found highly significant cooccurrence of FOXP1 motifs (P < 10–15), where approximately 50% of all NFATC2 sites associated with a FOXP1 motif (MA0481.2 or GTAAACA). Further, the distance between NFATC2 binding and the FOXP1 motif was strongly skewed to less than 50 base pairs (Figure 3D), suggesting that NFAT and FOXP proteins form a heterodimer, consistent with a crystal structure of the DNA-binding domains of NFAT and FOXP bound to DNA (Figure 3D; ref. 23). To evaluate a direct interaction between NFATC2 and FOXP proteins in human β cells, we used the HA-tag on ca-NFATC2 to perform immunoprecipitation (IP) in EndoC-βH2 β cells. Cells were transduced with either Ad-GFP or Ad-ca-Nfatc2, followed by HA-IP and Western blot for FOXP4. To minimize coimmunoprecipitation of FOXP4 and ca-Nfatc2 proteins on large DNA fragments, cells were treated with DNase1 prior to ca-Nfatc2 IP. FOXP4 was enriched (~3.4-fold) in β cells expressing ca-Nfatc2 versus GFP (Supplemental Figure 7; see complete unedited blots in the supplemental material). These results suggest that NFAT and FOXP4 form a physical complex in human β cells.
We sought to experimentally validate the requirement of FOXP for NFAT to induce β cell proliferation. Three of the four FOXP isoforms (FOXP1, FOXP2, and FOXP4) are abundantly expressed in mouse and human islets, whereas FOXP3 is normally expressed in T cells (Figure 3C and ref. 26). We asked whether our CA form of NFATC2 retained the ability to drive β cell proliferation in islets from WT or a mutant mouse lacking Foxp1, Foxp2, and Foxp4; FoxpTKO (27). Whereas NFATC2 yielded an approximately 10-fold increase in β cell proliferation in WT islets, an approximately 6-fold increase was observed in islets from FoxpTKO mice (Figure 3E). These results suggest that one or more of the FOXP proteins are required for NFATC2 to fully stimulate β cell proliferation in mouse islets.
Identification of NR4A1 as a key NFATC2 target gene.
Since NFATC2 stimulates β cell proliferation in both human and mouse islets, we hypothesized that a common set of target genes in both mouse and human is targeted by NFATC2. In striking contrast to NFATC2, NFATC1 is capable of inducing β cell proliferation only in human islets (2). This species difference enabled us to identify a small set of genes that are direct targets of NFATC2 and follow a β cell proliferation signature.
First, we identified approximately 17,000 islet transcripts that are conserved between mouse and human (Figure 4A). Among these transcripts, approximately 3800 were DE (FDR < 0.05) in response to NFATC1 and/or NFATC2 in mouse and human islets. To focus on genes directly regulated by NFATC2, we further narrowed this to 1844 by requiring that an NFATC2 binding site be within 50 Kbp of the TSS. We then exploited our earlier observation that NFATC2 stimulates β cell proliferation in both mouse and human, whereas NFATC1 does so only in human (2). Among the 1844 DE transcripts directly targeted by NFATC2, only 254 followed the same pattern as that seen in the effect of NFATC1 versus NFATC2 on β cell proliferation in mouse and human islets (Figure 4B and Supplemental Table 6). Pathways significantly enriched (z > 5) in this relatively small set of genes included DNA replication initiation (z > 10), DNA replication (z > 8), nuclear replication fork, and translation factor activity (z > 7) and included NR4A1, CDK2, CCNE2, CCNA2, POLA2, PCNA, and several of the mini-chromosome maintenance genes (Supplemental Figure 8). Further, the promoters of these β cell proliferation signature genes were enriched for binding motifs associated with the cell cycle–related TF E2F1 (Supplemental Table 7).
Figure 4. Identification of NR4A1 as a key target gene for NFATC2 to induce cellular proliferation in human islets.
(A) Integration of mouse and human islet transcriptomic data with NFATC2 binding data identifies NFAT direct transcriptional targets common to mouse and human. 1844 Genes were regulated by NFATC1 and/or NFATC2 and have an NFATC2 binding site within 50 Kbp of the gene’s TSS. (B) Identification of 254 direct transcriptional targets that followed a β cell proliferation signature regulated only by NFATC2 in mouse (top panel) and by NFATC1 and NFATC2 in human (bottom panel). The gene NR4A1 is indicated. (C) Genome browser view of the NFATC2 binding sites at the NR4A1 locus in 6 separate human islet donors. (D) Cellular proliferation in islets from WT (Nr4a1WT) and Nr4a1 knockout (Nr4a1KO) mice 48 hours following overexpression of GFP, ca-Nfatc2, or with no treatment (NT). Data are represented as mean ± SEM. *P < 0.05 for NFATC2 versus GFP, Student’s 2-tailed t test. n = 4 for each genotype.
NR4A1 (Nur77) has previously been shown to regulate β cell proliferation in mouse and human islets (28). The islet expression of NR4A1 follows the β cell proliferation signature. It is induced by NFATC2 in both mouse and human islets, but only by NFATC1 in human islets (Figure 4B). Interestingly, among the NR4A family of TFs, only NR4A1 showed this pattern of NFAT-dependent regulation (Supplemental Figure 9). The expression of Nr4a2 was exclusively induced by Nfatc2 in mouse islets and not at all in human islets (NR4A2), whereas Nr4a3 was induced by both NFATC1 and NFATC2 in mouse and human (NR4A3) islets. These results suggest that NR4A1 is a key transcriptional target of NFAT that underlies the ability of NFATC1 and NFATC2 to induce β cell proliferation in mouse and human islets. We identified several NFATC2 binding sites upstream of the NR4A1 gene locus (Figure 4C). This locus has been previously demonstrated to operate as a super enhancer for NR4A1 expression (29). One or more of these binding sites may function as key regulatory sites that mediate NFAT-induced induction of NR4A1 in human islets.
To experimentally validate a requirement of NR4A1 for NFATC2-induced β cell proliferation, we asked whether NFATC2 can induce β cell proliferation in islets from Nr4a1-knockout (Nr4a1KO) mice. As with our observations of the FoxpTKO mice (Figure 3E), islets from Nr4a1KO mice yielded significantly reduced β cell proliferation in response to NFATC2 compared with islets from WT mice, as judged by the incorporation of [3H]-thymidine into newly synthesized DNA (Figure 4D). To extend these observations, we used the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) to measure cellular proliferation, coupled with immunostains for insulin, glucagon, and somatostatin to discriminate the major islet cell types stimulated to proliferate in WT and Nr4a1KO mice. Islets from both mice were untreated or transduced with Ad-LacZ or Ad-ca-Nfatc2 and labeled with EdU for 48 hours in culture. Islets were then dispersed and immunostained for EdU and either insulin or glucagon and somatostatin to identify β cells or α cells and δ cells, respectively; DAPI was used to mark nuclei. In WT islets, 79% of cells that were induced to proliferate in response to ca-Nfatc2 were β cells, while 15% were α cells or δ cells (Supplemental Figure 10A). In Nr4a1KO islets, 77% of replicating cells were β cells, while only 3% were α cells or δ cells. These results suggest regardless of the presence of NR4A1, β cells are the primary cell type induced to proliferate in response to ca-Nfatc2. However, this does not provide information about the relative requirement of NR4A1 for NFATC2-dependent β cell proliferation.
To determine the effect of Nr4a1 loss on the ability of ca-Nfatc2 to stimulate β cell proliferation, we computed the percentages of β cells and non-β cells induced to proliferate in response to ca-Nfatc2 in WT and Nr4a1KO islets. We found that 10.9% percent of β cells were positive for EdU in WT islets, whereas in Nr4a1KO islets, this was reduced to 6.6% (Supplemental Figure 10B). While the loss of Nr4a1 blunted NFATC2-induced β cell proliferation, it did not affect the small amount of α cell and δ cell proliferation (Supplemental Figure 10B). Representative images of proliferating β cells are shown in Supplemental Figure 11 (WT mice) and Supplemental Figure 12 (Nr4a1KO mice) and of proliferating α cells and δ cells in Supplemental Figure 13 (WT mice) and Supplemental Figure 14 (Nr4a1KO mice). High-resolution images of proliferating cells from WT and Nr4a1KO islets are provided in Supplemental Figure 15.
Identification of NFATC2-dependent enhancer loci in human islets.
Our results strongly suggest that NFATC2 directly targets a small set of genes that includes NR4A1, leading to enhanced β cell proliferation. In addition, NFAT regulates the islet expression of approximately 34% of genes that have been associated with diabetes in human GWAS. NFATC2 has been shown to regulate transcription by binding to the promoter as well as enhancer loci of target genes (30). To identify the NFATC2-dependent enhancers, we characterized ncRNA originating from open chromatin bound with NFATC2. Also characterized as enhancer RNA (eRNA), ncRNAs emerging from distal regulatory regions with open chromatin are hallmark features of long-distance enhancer regulation of target gene expression (31, 32). The abundance of eRNA is proportional to enhancer activity (33–35). In some cases, eRNAs are better indicators of an active enhancer activity than H3K27ac (36). To characterize which NFATC2-dependent enhancers are critical for regulation of GWAS genes and β cell proliferation, we performed ATAC-Seq and ncRNA-Seq on human islets treated with Ad-GFP or Ad-ca-Nfatc2. Thus, by intersecting NFATC2 binding sites with NFATC2-dependent regulation of ncRNA, chromatin accessibility, and protein-coding transcripts (mRNA), we could nominate key regulatory loci for GWAS genes and β cell proliferation.
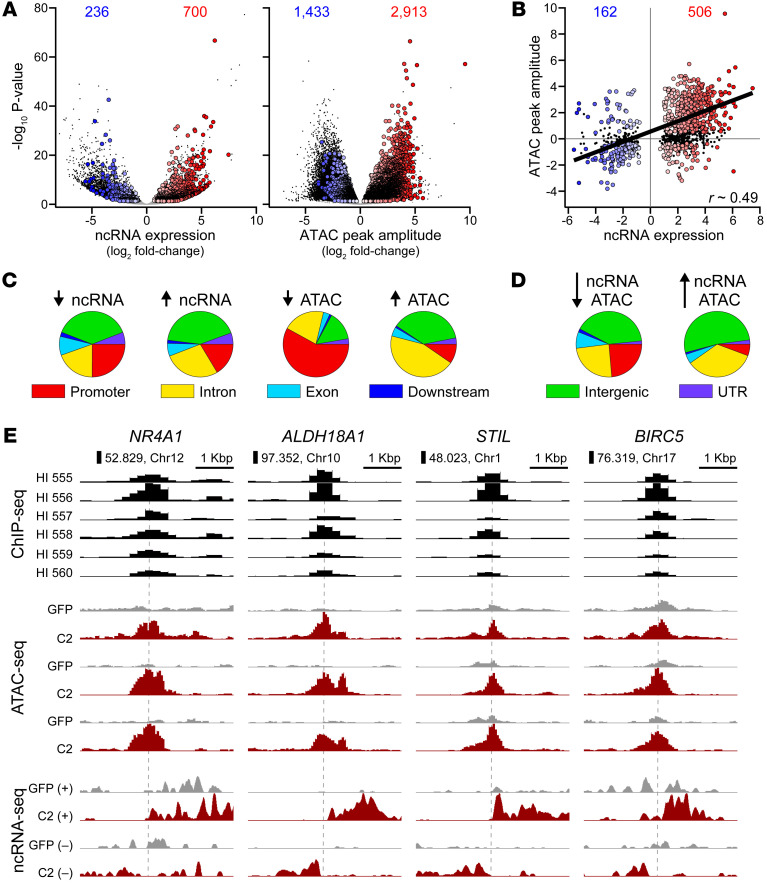
In parallel with mRNA profiling (RNA-Seq), we performed ncRNA-Seq on islets from 5 separate human donors, treated with Ad-GFP or Ad-ca-Nfatc2. Total RNA was depleted of polyadenylated transcripts and sequenced. ncRNAs were defined by excluding coding-gene transcripts, ncRNA that overlapped coding genes in the same strand, and RNA shorter than 100 base pairs. Among the approximately 92,000 ncRNAs identified, 12,877 were DE (adjusted P < 0.05 and log2 fold change > |0.5|) in response to ca-Nfatc2 (Supplemental Figure 16A). Of these, 936 were located within 1 Kbp of an NFATC2 binding site, 700 were induced, and 236 were suppressed (Figure 5A). Thus, approximately 7% of the DE ncRNAs were directly regulated by NFATC2. The distribution of NFATC2-dependent ncRNAs across genomic compartments (i.e., promoter, intron, exon, downstream, distal intergenic, or UTR) was similar whether their expression increased or decreased in response to NFATC2 overexpression (Figure 5C). A greater proportion of all DE ncRNAs occurred in intergenic regions (Supplemental Figure 16C) rather than those with a proximal NFATC2 binding site, which were biased for promoters (Figure 5C).
Figure 5. Identification of NFATC2-dependent enhancer loci in human islets.
ncRNA expression was measured in 5 separate human islet samples. (A) Volcano plots (–log10 P value versus log2 fold-change) for expression of 91,712 ncRNAs (left panel) and 231,232 ATAC peaks (right panel) in human islets in response to Ad-ca-Nfatc2 versus Ad-GFP. DE ncRNAs or DA ATAC peaks (P < 0.05, DESeq2) that contain an NFATC2 binding peak within 1 Kbp are shown as colored circles. Blue, suppressed; red, increased. Numbers are shown for each. Chromatin accessibility was measured in 3 separate human islet samples. (B) The log2 fold change in the ATAC peak amplitude versus ncRNA expression for 1105 combined loci containing an NFATC2 binding peak within 1 Kbp. Colored circles designate loci where both ATAC peak and ncRNA expression were significantly different in response to ca-Nfatc2 (P < 0.05, DESeq2). Black dots indicate loci where ATAC peak did not change but yielded a DE ncRNA. Numbers are shown for each. Distribution of NFATC2-dependent ncRNAs and ATAC peaks (C) and colocalized ncRNA-ATAC peaks (D) among annotated genomic compartments. (E) Browser views of NFATC2-dependent enhancer loci proximal to NR4A1, ALDH18A1, STIL, and BIRC5 genes. NFATC2 ChIP-Seq in 8 separate human islet donors. ATAC-Seq in 3 separate human islet donors in response to Ad-GFP or Ad-ca-Nfatc2. Aggregate ncRNA-Seq in 5 separate human islet donors for transcripts identified on forward (+) or reverse (–) strands (see Methods) in response to Ad-GFP or Ad-ca-Nfatc2 treatments.
A typical enhancer is associated with open or accessible chromatin (37). To identify NFATC2-dependent changes in chromatin accessibility, we performed ATAC-Seq on islets from 3 separate human donors, each treated with either Ad-GFP or Ad-ca-Nfatc2. Of the more than 230,000 ATAC peaks we identified, approximately 26% (59,597 peaks) were differentially accessible (DA) (P < 0.05) in response to NFATC2, with approximately twice the number becoming less accessible, i.e., closing, than more accessible, i.e., opening (Supplemental Figure 16A). Among these differential ATAC peaks, 4346 had an NFATC2 peak within 1 Kbp. An additional approximately 3678 peaks did not change yet had an NFATC2 binding peak within 1 Kbp (Figure 5A). Thus, as with our observations for NFATC2-dependent regulation of ncRNAs, NFATC2 directly regulates approximately 7% of ATAC peaks that show differential accessibility. In contrast to the ncRNAs, the genomic compartments for NFATC2-dependent ATAC peaks were different depending on whether they closed or opened in response to NFATC2. Peaks that closed tended to reside in gene promoters, whereas peaks that opened were present within intergenic or intronic regions, indicating a possible shift between proximal and distal regulation (Figure 5C). A similar trend was observed for all DA ATAC peaks (Supplemental Figure 16C). Interestingly, opening peaks strongly enriched for NFAT motifs, while closing peaks enriched for motifs associated with FOS and JUN (Supplemental Table 8).
Integration of the ATAC, ncRNA, and ChIP data sets yielded approximately 977 NFATC2-dependent enhancer loci in human islets (Figure 5B). There was a positive correlation (r ≈ 0.49, P < 10–4) between changes in ncRNA expression and chromatin accessibility evoked by NFATC2. This relationship was even stronger for all ncRNA/ATAC pairs regardless of whether they were proximal to NFATC2 binding sites (Supplemental Figure 16B). The NFATC2-dependent enhancers tended to occur in genomic compartments that reflected the combination of NFATC2-dependent ncRNAs and ATAC peaks (Figure 5D). The distribution of the genomic compartment for all ncRNA/ATAC pairs was similar to what we observed for DE ncRNAs (>50% in intergenic regions) and was not different depending on whether there was increased or decreased ncRNA expression in response to NFATC2 (Supplemental Figure 16D).
To identify candidate effector genes for the NFATC2-dependent enhancer loci, we relied on gene membership within topologically associating domains (TADs). A regulatory locus within a TAD is more likely to target genes within that TAD than genes outside the TAD (38, 39). Among the approximately 2600 TADs identified in human islets (40), 587 contained one or more of the NFATC2-dependent enhancers (Supplemental Table 9). To associate NFATC2 enhancer loci to putative effector genes, we focused on (a) our β cell proliferation signature gene set and (b) genes associated with T2D in human GWAS.
Among the 254 direct target genes that manifested our β cell proliferation signature (Figure 4B), 84 were within a TAD that had an NFATC2 enhancer locus (Supplemental Table 10). For example, an NFATC2-dependent enhancer is located on Chr 12 at approximately 52.8 Mbp, which is in the same TAD and approximately 383 Kbp from the NR4A1 gene locus (Figure 5C). In response to NFATC2, the expression of an ncRNA, MSTRG.93445, increased approximately 3-fold in parallel with an approximately 2-fold increase in NR4A1 expression. Proximal to ALDH18A1, another β cell proliferation signature gene, was an NFATC2 enhancer on Chr 10 at approximately 97 Mbp that resulted in a greater than 40-fold increase in the expression of two flanking ncRNAs, MSTRG.54591 and MSTRG.54592. ALDH18A1 has been associated with β cell survival in diabetes-resistant versus diabetes-sensitive mouse strains (41).
A third example is an NFATC2 enhancer locus located within an islet stretch enhancer on Chr 1 at approximately 48 Mbp, which is approximately 245 Kbp from Scl/Tal1 interrupting locus (STIL). At this locus, NFATC2 induced the expression of 2 ncRNAs flanking the enhancer, MSTRG.9208 and MSTRG.9209, approximately 7-fold and approximately 6-fold, respectively. This eRNA regulation was in parallel with an approximately 2-fold induction of STIL. STIL is a key regulator of the mitotic spindle checkpoint and is essential for centriole replication (42). Overexpression of STIL promotes proliferation of dopaminergic neuronal cells (43). Taken together, our data suggest that NFATC2 binds to long-distance enhancers to regulate the expression of genes that promote β cell proliferation.
Earlier, we defined direct NFATC2 target genes as those that change expression in response to NFATC2 overexpression and have an NFATC2 binding site within 50 Kbp. However, as illustrated in the above examples, enhancers can operate from distances much farther than 50 Kbp, yet function as transcriptional regulators. An example of a gene that does not have a proximal NFATC2 binding site but is within a TAD with an NFATC2-dependent enhancer locus is BIRC5. An NFATC2-dependent enhancer is located within an islet-specific stretch enhancer (44) that is approximately 110 Kb from BIRC5 (Figure 5E). NFATC2 induces the expression of an ncRNA, MSTRG.178739 (~4-fold), and BIRC5 (~3-fold) in human islets. Previous work has shown that BIRC5 is sufficient to regulate β cell proliferation (45–47).
Given our observations that FOXP is a key binding partner for NFATC2-induced β cell proliferation, we asked whether NFATC2-dependent enhancers show evidence of FOXP binding. Among 176 NFATC2-dependent enhancer loci that occurred within a TAD that contained one or more of the β cell proliferation signature genes, approximately 40% had a proximal FOXP motif (Supplemental Table 11). These results suggest that FOXP may be a binding partner for a large portion of the NFATC2-dependent enhancer loci for effector genes that regulate β cell proliferation.
Previously, we showed that NFATC2 regulates the expression of many putative effector genes of regulatory loci associated with T2D in human GWAS (2). However, these studies did not discriminate between direct and indirect regulation or promoter and enhancer-mediated regulation. Here, we identified 156 SNPs that are strongly associated with T2D in human GWAS (48) and are present within 1 Kbp of an NFATC2-dependent enhancer locus (Supplemental Table 12). These GWAS-associated loci are located within 13 TADs that together contain 22 genes previously linked to T2D. Of these nominated T2D effector genes, 7 are regulated by NFATC2.
An example of a GWAS locus where NFATC2 regulates the reported T2D GWAS effector gene occurs at the ARAP1/STARD10 gene locus. NFATC2 binds to and induces the expression of an ncRNA, MSTRG.73719, approximately 9.5-fold that is located within an intron of ARAP1 at approximately 72.43 Mbp. Both genes have been predicted as the targets of the diabetes risk variants (49, 50). Our results support both genes as targets of the regulatory locus, as ca-Nfatc2 induces the expression of ARAP1 and STARD10 approximately 1.5-fold and approximately 1.4-fold, respectively.
Discussion
The laboratories of Crabtree and Kim first established that the Calcineurin/NFAT pathway regulates β cell mass (51, 52). This finding prompted several laboratories to identify small molecule inhibitors of NFAT-regulating kinases as potential diabetes therapeutics (53–57). There are 4 canonical NFAT proteins, all of which are expressed in β cells (51). Our study focused on NFATC1 and NFATC2. We previously identified NFATC2 as an islet regulator of genes associated with diabetes risk in human GWAS. We included NFATC1 in studies reported here because of extensive literature describing its function in human β cells (51, 52, 58). Future studies are required to explore possible links between NFATC3 and NFATC4 in effects on β cell proliferation in human islets. Previously, we showed that overexpression of CA forms of NFATC1 and NFATC2 drives human β cells to proliferate (2). Surprisingly, NFATC1 was only effective in humans, whereas NFATC2 was effective in both mouse and human islets. While substantial differences exist in the pathways that govern β cell proliferation between mouse and human, we sought to identify the conserved elements of NFAT-dependent regulation of β cell proliferation.
We identified a small set of genes, the β cell proliferation signature gene set, based on 2 properties: (a) an expression pattern that matched the ability of NFATC1 and NFATC2 to induce β cell proliferation in mouse and human islets and (b) the presence of an NFATC2 peak within 50 Kbp of the TSS. The species selectivity of NFATC1 in driving β cell proliferation coupled with the ability of NFATC2 to do so in both mouse and human islets proved to be a strong filter, as only approximately 500 genes followed this pattern. Of these, 254 had an NFATC2 peak proximal to the TSS, suggesting they are direct transcriptional targets that mediate the effect of NFAT on β cell proliferation. This set of 254 genes was enriched for cell cycle, DNA replication, and related pathways and included CCNA2, CCNE2, TFDP1, PCNA, GADD45B, ESPL1, CDK2, MCM2, MCM3, MCM6, E2F1, and CDC7, all well-known players in cell cycle control. However, a few genes in this β cell proliferation signature gene set are virtually uncharacterized, including C10orf88 (2310057M21Rik), C5orf52 (4921536K21Rik), ZNF438 (Zfp438), and ZNF556. That these uncharacterized genes segregate with the β cell proliferation signature gene set suggests that they too play a critical role in β cell proliferation. The remaining 246 genes that did not have a proximal NFATC2 peak were enriched for cell cycle processes (e.g., DNA replication, nuclear replication fork), although less strongly than the direct transcriptional target gene list. These results suggest that NFAT directly initiates cellular proliferation by targeting genes in the β cell proliferation signature gene set (e.g., ASF1B, NR4A1), which results in downstream transcriptional changes for other genes associated with cellular replication (e.g., CDK1, MCM10, POLB).
Our approach led us to NR4A1 as a direct transcriptional target of NFATC2 that mediates the effect of NFAT on β cell proliferation. NR4A1 is a nuclear receptor with no known endogenous ligands (59). We demonstrated that NFATC2-induced β cell proliferation was blunted in Nr4a1KO mice by measuring incorporation of tritiated thymidine in whole islets and incorporation of EdU in dispersed islet cells costained with insulin, or glucagon and somatostatin, to identify β cells or α cells and δ cells, respectively. The EdU studies suggest that NFATC2 primarily induces β cell proliferation in mouse islets. Interestingly, Nr4a1KO islets showed reduced β cell proliferation under basal conditions (Ad-LacZ), consistent with reduced β cell mass that has been reported for Nr4a1KO mice (28). Newgard and colleagues previously showed that NR4A1 mediates the stimulatory effect of NKX6.1 on β cell proliferation in rat and human islets (28). In addition to NR4A1, NKX6.1 induced the expression of E2f1, as well as Ube2c and Ube2s, which are E3 ubiquitin conjugating enzymes that target the cell cycle inhibitor p21 for proteosomal degradation. In human islets, NFAT induces the expression of E2F1 without changes in NKX6.1, UBE2C, or UBE2S expression, suggesting the 2 TFs mediate parallel pathways that regulate β cell proliferation. Interestingly, approximately 40% of NFATC2 binding sites overlap with NKX6.1 binding sites, suggesting that some genes may be coregulated by NFATC2 and NKX6.1. Finally, we identified 2 E3 ligases in our β cell proliferation signature gene set: UBE2T and UBE2M. Further work is required to determine whether p21 or other key negative regulators of the cell cycle are substrates of the E3 ligases.
We asked whether other TFs regulated a set of genes similar to those in our β cell proliferation gene set. Not only do E2F1, FOXM1, and TFDP1 target a similar gene set, they are included in the β cell proliferation gene set, suggesting they may work in concert with NFATC2 to stimulate β cell proliferation. We previously reported that overexpression of FOXM1 is sufficient to drive human β cell proliferation (60). Shirakawa et al. showed that FOXM1, PLK1, and CENPA function in a pathway of adaptive β cell proliferation (61). Further, our analysis shows that the β cell proliferation signature gene set is enriched for CENPA transcriptional targets. These results suggest that NFAT, in tandem or through FOXM1 and CENPA, is utilizing a similar pathway to induce β cell proliferation. Finally, the DNA binding motif associated with E2F1 is enriched in the promoters of the β cell proliferation signature genes. E2F1 has previously been implicated in β cell proliferation (62, 63).
We identified a significant enrichment for mitochondria-related genes among those induced by NFATC1 and NFATC2 (Supplemental Figure 1). However, when restricted to only those genes directly targeted by NFATC2 (i.e., having an NFATC2 peak proximal to the TSS), this mitochondrial enrichment was lost, suggesting that bulk mitochondrial gene changes are likely not direct transcriptional targets of NFATC2. In contrast, direct transcriptional targets that are induced by NFATC2 enrich for cilia-related genes. Cilia are found on every β cell and are required for proper β cell function (64). Furthermore, cilia-related genes are associated with T2D (65, 66).
Direct transcriptional targets that were suppressed by NFATC2 were enriched for GO terms involving the UPR and regulation of ER stress. Genes found in these sets include the protein chaperones HSPA1A, HSPA1B, HSPA5, and HSPA6 and the UPR genes CHOP and PERK (Supplemental Figure 5). NFAT has previously been associated with ER stress. The ER stress-inducing agents thapsigargin (TG) and tunicamycin (TM) induced the expression of NFAT in MIA PaCa-2 cells, a human pancreas epithelial cell line (67). Our results suggest that NFAT may function as a negative regulator of ER stress and the UPR and that the induction observed in MIA PaCa-2 cells in response to TG and TM may reflect a compensatory upregulation of NFAT.
Several direct target genes of NFATC2 are negatively regulated and play a role in the ER stress response and protein folding, including DDIT3 (CHOP), EIF2AK3 (PERK), HSPA1A, HSPA6, and HSPA1B. UPR and ER stress have been linked to β cell proliferation and as a mechanism with which to gauge insulin demand (68). It is possible that NFATC2 may decrease the expression of these ER stress-regulating genes to induce a state conducive for β cell proliferation.
We have also identified direct transcriptional targets of NFAT that are known to play key roles in islet function. For example, NFATC1 and NFATC2 suppress BAG3 expression approximately 14-fold in human islets, which may be mediated by a strong NFATC2 binding site within the first intron of BAG3 (Figure 2, D and E). BAG3 has been shown to negatively regulate insulin secretion (15).
Powers and colleagues have evaluated the effects of β cell–derived VEGFA on islet function in mice (69). VEGFA plays a critical role during pancreas development by recruiting endothelial cells necessary for islet vasculature (70). However, excessive VEGFA signaling results in diminished β cell proliferation (71). Our data show that NFATC2 suppresses VEGFA expression in human islets. It is possible that NFATC2-dependent suppression of VEGFA in part underlies enhanced β cell proliferation.
The revascularization of transplanted islets is a crucial step for islet graft survival and function (72). Elevated islet expression of VEGFA has been shown to improve the revascularization process (73). Tacrolimus (or FK506), an immunosuppressant that is commonly used to prevent organ rejection, elevates VEGFA expression in ingrafted islets (74). Tacrolimus is also a potent inhibitor of the calcineurin-dependent NFAT pathway (75). Paradoxically, however, in parallel with increased islet VEGFA expression, tacrolimus blunted islet revascularization, suggesting it may have pleiotropic effects in vivo. It is possible that tacrolimus-induced VEGFA expression may lead to enhanced islet revascularization if the islets are treated prior to transplantation. Indeed, pancreatic islets codelivered with FK506 loaded beads into the transplant site of diabetic mice result in better glycemic outcomes than those in mice treated with FK506 systemically (76).
Most T2D GWAS variants do not align with protein-coding regions of the genome, but rather reside in noncoding, regulatory regions: intronic and intergenic (77). Thus, diabetes risk is strongly linked to altered gene regulation rather than alterations in gene products. Establishing which genes are regulated by these loci is critical to understand their disease risk. We have previously shown that NFATC2 regulates the expression of several genes associated with diabetes in human GWAS (2). Here, we extend these observations by showing that 82 of 242 genes associated with diabetes are DE with respect to NFATC1 and/or NFATC2 in human islets (Supplemental Figure 2). Of these, 47 genes have an NFATC2 peak within 50 Kbp of the TSS, suggesting they may be direct transcriptional targets of NFATC2. Interestingly, the NFATC2 locus is strongly associated with T2D diabetes (Supplemental Figure 3), which may in part reflect NFATC2-dependent regulation of the GWAS genes.
Our goal was to integrate chromatin binding and gene regulation to identify direct transcriptional targets of NFATC2 that regulate β cell proliferation in human islets. Several methods have been used to define direct targets of TFs, including ChIP-Seq, RNA-Seq, 3D chromatin architecture, and positional data (18, 78–80). A relatively novel approach utilized ncRNAs originating from enhancers, eRNA, to nominate direct targets of enhancers in a TF perturbed system (32, 81). We have endeavored to identify direct targets of NFATC2 in human islets using both the traditional ChIP-based approach and an eRNA-based method. Integration of ncRNA expression, chromatin accessibility, and NFATC2 binding allowed us to identify 977 NFATC2-dependent enhancer loci. Among these enhancers, chromatin accessibility increased in approximately 42%, decreased in approximately 19%, and remained unchanged in approximately 39%. Thus, NFATC2 functions as a pioneering factor or utilizes existing open chromatin sites at more than 80% of the NFATC2-dependent enhancer loci.
The NFATC2 gene locus contains SNPs that are associated with T2D in human GWAS (Supplemental Figure 3). These local SNPs may influence NFATC2 expression. In addition, T2D-associated SNPs may interfere with NFATC2 binding at distal enhancer loci. We identified a small set of the NFATC2-dependent enhancers (~2%) that harbor 156 SNPs associated with T2D in human GWAS. To nominate possible effector genes at these loci, we relied on gene membership within TADs (40) and NFATC2-dependent regulation in mRNA expression in human islets. We identified 13 TADs that contain both a T2D-associated NFATC2 enhancer locus and one or more of 22 nominated GWAS effector genes. Among these GWAS genes, 7 were regulated by NFATC2, suggesting they may be direct transcriptional targets of the enhancer locus. It is possible that their association with T2D may reflect disruption of this NFATC2-dependent regulation. Interestingly, 15 putative GWAS effector genes at the other enhancer loci were not regulated by NFATC2, suggesting that other genes may be the true target for these loci. As only 7 of 242 T2D GWAS effector genes with an NFATC2-dependent enhancer in the same TAD were DE, it appears that NFATC2-dependent enhancers regulate a very small proportion of nominated T2D effectors. In contrast, approximately 20% of the proposed GWAS genes that are DE in response to NFATC2, have an NFATC2 binding site within 50 Kbp of the gene’s TSS, which may reflect direct transcriptional regulation.
We identified NFATC2-dependent enhancers located within TADs that contain genes following our β cell proliferation signature, enhanced β cell proliferation in mouse and human islets in response to NFATC2, but for NFATC1 only in human islets. These included NR4A1, STIL, ALDH18A1, and BIRC5 (Figure 5E). NR4A1, STIL, and ALDH18A1 all had proximal NFATC2 binding sites in addition to an NFATC2-dependent enhancer, suggesting they may be subject to both proximal and distal regulation by NFATC2. In contrast, BIRC5 may be exclusively regulated by a distal enhancer locus, highlighting the importance of identifying the enhancer loci via the ncRNA, ATAC, and NFATC2 binding profiles for this class of effector genes. Our results suggest that as much as 25% of the β cell proliferation signature gene set may be regulated exclusively by enhancer loci and 33% by a combination of distal enhancer and proximal regulatory sites. Thus, identifying TF-dependent enhancers through eRNA profiling can establish direct regulatory relationships that arbitrary distances to TSSs may not capture.
In summary, we have demonstrated that NFATC2 is a key TF for genes that regulate β cell proliferation and function. We show that FOXP TFs are key binding partners of NFATC2 and mediate its effect on proliferation. Downstream of NFATC2, we determined that Nr4a1 is an NFATC2 target gene that mediates much of its ability to stimulate β cell proliferation. In addition, NFATC2 can act as an enhancer to direct gene expression and affect target genes that mediate β cell proliferation and T2D susceptibility.
Limitations.
We acknowledge certain limitations in our study of NFAT-mediated β cell proliferation in human islets. The NFAT proteins used in this study were CA, due to several regulatory serine residues mutated to alanine residues. These mutants do not require activation by the phosphatase calcineurin and are not subject to inactivation by regulatory kinases. These regulatory steps likely play important roles in β cell proliferation mediated by the NFAT signaling pathway. In addition to the Ser-to-Ala mutations, our Nfatc2 construct has 3 tandem HA tags at the amino-terminus, which we used for the ChIP-Seq and IP studies. It is possible these HA tags interfere with endogenous protein interactions. The adenoviruses employed to overexpress ca-Nfatc1 and ca-Nfatc2 used the CMV promoter, yielding approximately 900- and approximately 200-fold greater expression than endogenous NFATC1 and NFATC2 expression, respectively. Our previous results strongly suggest that ca-Nfatc1 and ca-Nfatc2 stimulate proliferation of human β cells (2). However, rigorous quantification of islets from multiple human donors for insulin-positive cells that costain for Ki67, BrdU/EdU, or PHH3 under control versus NFAT overexpression conditions is required to firmly establish NFAT as a potent mitogen of human β cells.
Methods
A detailed description is found in the Supplemental Methods.
Human islet procurement.
All human islets were obtained through the Integrated Islet Distribution Program (IIDP). Upon receipt, all human islets were incubated overnight in RPMI 1640 supplemented with 8.0 mM glucose, 1% antibiotic/antimycotic (Gibco, Thermo Fisher Scientific), 5 mM HEPES, and 10% FBS before experiments were performed. All human islet preparations were greater than 80% islet purity and 90% viability. Supplemental Table 13 lists the human islet preparations used for this study, along with donor demographics.
Mouse studies.
Mice were housed in groups of 2 to 5 mice per cage in temperature- and humidity-controlled rooms with a 12-hour light/12-hour dark cycle (6:00 am–6:00 pm). After weaning, mice were maintained on chow diet (Purina 5008).
Statistics.
For several experiments that compared data between 2 groups, Student’s 2-tailed t test was used. When more than 2 groups were compared, 2-way ANOVA was used. In general, P <0.05 was considered statistically significant. When defining enrichments using ChEA, Fisher’s exact test was used, and an adjusted P value of less than 0.05 was defined as statistically significant. Posterior probabilities (PP) were used to define statistical significance for the RNA-Seq data. In this manner, PP1 < 0.01 indicates differential expression relative to Ad-GFP treatment.
Study approval.
All studies performed on mice were approved by the Animal Care and Use Committees at University of Wisconsin–Madison (protocol A005821) and Brigham Young University (protocol 17-1202). Human donors were deidentified in accordance with HIPAA regulations. The human islets that are distributed by the IIDP are from approved cadaveric organ donors from which at least one other organ has been approved for transplantation. Because the donors are brain dead, the IRB’s from the institutions that isolate the islets consider the tissue as “Exempt” from Human Studies Approval.
Data availability.
All sequencing data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE158499). A browser session that contains the Nfatc2-dependent ChIP-Seq, ATAC-Seq, and ncRNA-Seq data is available here: http://genome.ucsc.edu/s/ssimonett/hg19_Nfatc2_human_islet_genome_browser
Author contributions
SPS, LAS, MER, DSS, KLS, JAH, and DRT conducted the experiments. SPS, SS, RB, CD, LAS, JC, MNB, CPC, JAH, CK, SK, and MPK performed analyses. SPS, JS, RS, JST, IPM, ADA, and MPK conceived of the study. SPS, ADA, and MPK wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the JDRF (17-2011-261, 2-SRA-2015-57-Q-R to ADA), the NIH (DK058037, DK101573, DK102948, DK066369 to ADA; GM102756 to CK; DK090570, DK050203 to RS; HL147571, HL148719, HL126509 to IPM; HG011371, HG003747 to SK; 5T32GM007197-43, T32HL007381 to LAS), the USDA (2020-67017-30846), and the Beatson Foundation (2019-003 to JST), and from the University of Wisconsin–Madison, Department of Biochemistry and Office of the Vice Chancellor for Research and Graduate Education, with funding from the Wisconsin Alumni Research Foundation (to MPK). Human pancreatic islets were provided by the National Institute of Diabetes and Digestive and Kidney Diseases–funded IIDP at City of Hope (NIH grant 2UC4DK098085). We are grateful for graphical expertise provided by Laura Vanderploeg within the Media Center of the Department of Biochemistry, University of Wisconsin–Madison, and to Nikolina Nakic, Imperial College London, for guidance on our pancreatic islet ATAC-Seq studies.
Version 1. 09/07/2021
In-Press Preview
Version 2. 11/01/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(21):e144833.https://doi.org/10.1172/JCI144833.
Contributor Information
Shane P. Simonett, Email: ssimonett@wisc.edu.
Sunyoung Shin, Email: Sunyoung.Shin@utdallas.edu.
Jacob A. Herring, Email: herrin06@gmail.com.
Rhonda Bacher, Email: rbacher@ufl.edu.
Linsin A. Smith, Email: linsin@uchicago.edu.
Chenyang Dong, Email: cdong39@wisc.edu.
Mary E. Rabaglia, Email: buffalum77@gmail.com.
Donnie S. Stapleton, Email: dsstaple@biochem.wisc.edu.
Kathryn L. Schueler, Email: schueler@biochem.wisc.edu.
Jeea Choi, Email: angeljjjj@gmail.com.
Matthew N. Bernstein, Email: mnbernstein@wisc.edu.
Daniel R. Turkewitz, Email: danielturkewitz@gmail.com.
Carlos Perez-Cervantes, Email: cperez5@uchicago.edu.
Jason Spaeth, Email: jspaeth@iu.edu.
Roland Stein, Email: roland.stein@vanderbilt.edu.
Jeffery S. Tessem, Email: jeffery_tessem@byu.edu.
Christina Kendziorski, Email: kendzior@biostat.wisc.edu.
Ivan P. Moskowitz, Email: imoskowitz@peds.bsd.uchicago.edu.
Mark P. Keller, Email: mark.keller@wisc.edu.
Alan D. Attie, Email: adattie@wisc.edu.
References
- 1.Mahajan A, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keller MP, et al. The transcription factor Nfatc2 regulates β-cell proliferation and genes associated with type 2 diabetes in mouse and human islets. PLoS Genet. 2016;12(12):e1006466. doi: 10.1371/journal.pgen.1006466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monticelli S, Rao A. NFAT1 and NFAT2 are positive regulators of IL-4 gene transcription. Eur J Immunol. 2002;32(10):2971–2978. doi: 10.1002/1521-4141(2002010)32:10<2971::AID-IMMU2971>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 4.Rao A, et al. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 5.Mognol GP, et al. Cell cycle and apoptosis regulation by NFAT transcription factors: new roles for an old player. Cell Death Dis. 2016;7:e2199. doi: 10.1038/cddis.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul PK, et al. Histone chaperone ASF1B promotes human β-cell proliferation via recruitment of histone H3.3. Cell Cycle. 2016;15(23):3191–3202. doi: 10.1080/15384101.2016.1241914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu EE, et al. SOX4 allows facultative β-cell proliferation through repression of Cdkn1a. Diabetes. 2017;66(8):2213–2219. doi: 10.2337/db16-1074. [DOI] [PubMed] [Google Scholar]
- 8.Robitaille K, et al. High-throughput functional genomics identifies regulators of primary human beta cell proliferation. J Biol Chem. 2016;291(9):4614–4625. doi: 10.1074/jbc.M115.683912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46(2):136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stitzel ML, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12(5):443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ackermann AM, et al. Integration of ATAC-Seq and RNA-Seq identifies human alpha cell and beta cell signature genes. Mol Metab. 2016;5(3):233–244. doi: 10.1016/j.molmet.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42(3):255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran I, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16(4):435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iorio V, et al. BAG3 regulates formation of the SNARE complex and insulin secretion. Cell Death Dis. 2015;6:e1684. doi: 10.1038/cddis.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genesca L, et al. Phosphorylation of calcipressin 1 increases its ability to inhibit calcineurin and decreases calcipressin half-life. Biochem J. 2003;374(pt 2):567–575. doi: 10.1042/BJ20030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh M, et al. The CCAAT/enhancer binding protein beta (C/EBPbeta) cooperates with NFAT to control expression of the calcineurin regulatory protein RCAN1-4. J Biol Chem. 2010;285(22):16623–16631. doi: 10.1074/jbc.M109.098236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sikora-Wohlfeld W, et al. Assessing computational methods for transcription factor target gene identification based on ChIP-Seq data. PLoS Comput Biol. 2013;9(11):e1003342. doi: 10.1371/journal.pcbi.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gertz J, et al. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013;52(1):25–36. doi: 10.1016/j.molcel.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(d1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, et al. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392(6671):42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 23.Wu Y, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126(2):375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 24.Ray JD, et al. Nkx6.1-mediated insulin secretion and β-cell proliferation is dependent on upregulation of c-Fos. FEBS Lett. 2016;590(12):1791–1803. doi: 10.1002/1873-3468.12208. [DOI] [PubMed] [Google Scholar]
- 25.Lachmann A, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devaud C, et al. Foxp3 expression in T regulatory cells and other cell lineages. Cancer Immunol Immunother. 2014;63(9):869–876. doi: 10.1007/s00262-014-1581-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spaeth JM, et al. The FOXP1, FOXP2 and FOXP4 transcription factors are required for islet alpha cell proliferation and function in mice. Diabetologia. 2015;58(8):1836–1844. doi: 10.1007/s00125-015-3635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tessem JS, et al. Nkx6.1 regulates islet β-cell proliferation via Nr4a1 and Nr4a3 nuclear receptors. Proc Natl Acad Sci U S A. 2014;111(14):5242–5247. doi: 10.1073/pnas.1320953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas GD, et al. Deleting an Nr4a1 super-enhancer subdomain ablates Ly6C(low) monocytes while preserving macrophage gene function. Immunity. 2016;45(5):975–987. doi: 10.1016/j.immuni.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao ZJ, et al. NFATc2 enhances tumor-initiating phenotypes through the NFATc2/SOX2/ALDH axis in lung adenocarcinoma. Elife. 2017;6:e26733. doi: 10.7554/eLife.26733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold PR, et al. Diversity and emerging roles of enhancer RNA in regulation of gene expression and cell fate. Front Cell Dev Biol. 2019;7:377. doi: 10.3389/fcell.2019.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XH, et al. Transcription-factor-dependent enhancer transcription defines a gene regulatory network for cardiac rhythm. Elife. 2017;6:e31683. doi: 10.7554/eLife.31683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azofeifa JG, et al. Enhancer RNA profiling predicts transcription factor activity. Genome Res. doi: 10.1101/gr.225755.117. [published online February 15, 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danko CG, et al. Identification of active transcriptional regulatory elements from GRO-Seq data. Nat Methods. 2015;12(5):433–438. doi: 10.1038/nmeth.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507(7493):455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tyssowski KM, et al. Different neuronal activity patterns induce different gene expression programs. Neuron. 2018;98(3):530–546. doi: 10.1016/j.neuron.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49(5):825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dowen JM, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159(2):374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji X, et al. 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell. 2016;18(2):262–275. doi: 10.1016/j.stem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenwald WW, et al. Pancreatic islet chromatin accessibility and conformation reveals distal enhancer networks of type 2 diabetes risk. Nat Commun. 2019;10(1):2078. doi: 10.1038/s41467-019-09975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh H, et al. Novel genes underlying beta cell survival in metabolic stress. Bioinformation. 2013;9(1):37–41. doi: 10.6026/97320630009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.David A, et al. Lack of centrioles and primary cilia in STIL(-/-) mouse embryos. Cell Cycle. 2014;13(18):2859–2868. doi: 10.4161/15384101.2014.946830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, et al. STIL: a multi-function protein required for dopaminergic neural proliferation, protection, and regeneration. Cell Death Discov. 2019;5:90. doi: 10.1038/s41420-019-0172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker SC, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110(44):17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, et al. Prolactin-stimulated survivin induction is required for beta cell mass expansion during pregnancy in mice. Diabetologia. 2015;58(9):2064–2073. doi: 10.1007/s00125-015-3670-0. [DOI] [PubMed] [Google Scholar]
- 46.Wu X, et al. Survivin is required for beta-cell mass expansion in the pancreatic duct-ligated mouse model. PLoS One. 2012;7(8):e41976. doi: 10.1371/journal.pone.0041976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, et al. Perinatal survivin is essential for the establishment of pancreatic beta cell mass in mice. Diabetologia. 2009;52(10):2130–2141. doi: 10.1007/s00125-009-1469-6. [DOI] [PubMed] [Google Scholar]
- 48.Gaulton KJ, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47(12):1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kulzer JR, et al. A common functional regulatory variant at a type 2 diabetes locus upregulates ARAP1 expression in the pancreatic beta cell. Am J Hum Genet. 2014;94(2):186–197. doi: 10.1016/j.ajhg.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carrat GR, et al. Decreased STARD10 expression is associated with defective insulin secretion in humans and mice. Am J Hum Genet. 2017;100(2):238–256. doi: 10.1016/j.ajhg.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heit JJ, et al. Calcineurin/NFAT signalling regulates pancreatic beta-cell growth and function. Nature. 2006;443(7109):345–349. doi: 10.1038/nature05097. [DOI] [PubMed] [Google Scholar]
- 52.Goodyer WR, et al. Neonatal β cell development in mice and humans is regulated by calcineurin/NFAT. Dev Cell. 2012;23(1):21–34. doi: 10.1016/j.devcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aamodt KI, et al. Development of a reliable automated screening system to identify small molecules and biologics that promote human β-cell regeneration. Am J Physiol Endocrinol Metab. 2016;311(5):E859–E868. doi: 10.1152/ajpendo.00515.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdolazimi Y, et al. CC-401 promotes β-cell replication via pleiotropic consequences of DYRK1A/B inhibition. Endocrinology. 2018;159(9):3143–3157. doi: 10.1210/en.2018-00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dirice E, et al. Inhibition of DYRK1A stimulates human β-cell proliferation. Diabetes. 2016;65(6):1660–1671. doi: 10.2337/db15-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen W, et al. Inhibition of DYRK1A and GSK3B induces human β-cell proliferation. Nat Commun. 2015;6:8372. doi: 10.1038/ncomms9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang P, et al. A high-throughput chemical screen reveals that harmine-mediated inhibition of DYRK1A increases human pancreatic beta cell replication. Nat Med. 2015;21(4):383–388. doi: 10.1038/nm.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai C, et al. Age-dependent human β cell proliferation induced by glucagon-like peptide 1 and calcineurin signaling. J Clin Invest. 2017;127(10):3835–3844. doi: 10.1172/JCI91761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu L, Chen L. Characteristics of Nur77 and its ligands as potential anticancer compounds (Review) Mol Med Rep. 2018;18(6):4793–4801. doi: 10.3892/mmr.2018.9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davis DB, et al. FoxM1 is up-regulated by obesity and stimulates beta-cell proliferation. Mol Endocrinol. 2010;24(9):1822–1834. doi: 10.1210/me.2010-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shirakawa J, et al. Insulin signaling regulates the FoxM1/PLK1/CENP-A pathway to promote adaptive pancreatic β cell proliferation. Cell Metab. 2017;25(4):868–882. doi: 10.1016/j.cmet.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fajas L, et al. Impaired pancreatic growth, beta cell mass, and beta cell function in E2F1 (-/- )mice. J Clin Invest. 2004;113(9):1288–1295. doi: 10.1172/JCI18555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grouwels G, et al. Ectopic expression of E2F1 stimulates beta-cell proliferation and function. Diabetes. 2010;59(6):1435–1444. doi: 10.2337/db09-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hughes JW, et al. Primary cilia control glucose homeostasis via islet paracrine interactions. Proc Natl Acad Sci U S A. 2020;117(16):8912–8923. doi: 10.1073/pnas.2001936117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerdes JM, et al. Ciliary dysfunction impairs beta-cell insulin secretion and promotes development of type 2 diabetes in rodents. Nat Commun. 2014;5:5308. doi: 10.1038/ncomms6308. [DOI] [PubMed] [Google Scholar]
- 66.Kluth O, et al. Decreased expression of cilia genes in pancreatic islets as a risk factor for type 2 diabetes in mice and humans. Cell Rep. 2019;26(11):3027–3036. doi: 10.1016/j.celrep.2019.02.056. [DOI] [PubMed] [Google Scholar]
- 67.Jiang S, et al. Altered activity patterns of transcription factors induced by endoplasmic reticulum stress. BMC Biochem. 2016;17:8. doi: 10.1186/s12858-016-0060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma RB, et al. Insulin demand regulates β cell number via the unfolded protein response. J Clin Invest. 2015;125(10):3831–3846. doi: 10.1172/JCI79264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brissova M, et al. Pancreatic islet production of vascular endothelial growth factor--a is essential for islet vascularization, revascularization, and function. Diabetes. 2006;55(11):2974–2985. doi: 10.2337/db06-0690. [DOI] [PubMed] [Google Scholar]
- 70.Cai Q, et al. Enhanced expression of VEGF-A in β cells increases endothelial cell number but impairs islet morphogenesis and β cell proliferation. Dev Biol. 2012;367(1):40–54. doi: 10.1016/j.ydbio.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brissova M, et al. Islet microenvironment, modulated by vascular endothelial growth factor-A signaling, promotes β cell regeneration. Cell Metab. 2014;19(3):498–511. doi: 10.1016/j.cmet.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brissova M, Powers AC. Revascularization of transplanted islets: can it be improved? Diabetes. 2008;57(9):2269–2271. doi: 10.2337/db08-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang N, et al. Elevated vascular endothelial growth factor production in islets improves islet graft vascularization. Diabetes. 2004;53(4):963–970. doi: 10.2337/diabetes.53.4.963. [DOI] [PubMed] [Google Scholar]
- 74.Nishimura R, et al. Tacrolimus inhibits the revascularization of isolated pancreatic islets. PLoS One. 2013;8(4):e56799. doi: 10.1371/journal.pone.0056799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hogan PG, et al. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17(18):2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 76.Pathak S, et al. Single synchronous delivery of FK506-loaded polymeric microspheres with pancreatic islets for the successful treatment of streptozocin-induced diabetes in mice. Drug Deliv. 2017;24(1):1350–1359. doi: 10.1080/10717544.2017.1377317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536(7614):41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang S, et al. Target analysis by integration of transcriptome and ChIP-Seq data with BETA. Nat Protoc. 2013;8(12):2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ron G, et al. Promoter-enhancer interactions identified from Hi-C data using probabilistic models and hierarchical topological domains. Nat Commun. 2017;8(1):2237. doi: 10.1038/s41467-017-02386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baxter JS, et al. Capture Hi-C identifies putative target genes at 33 breast cancer risk loci. Nat Commun. 2018;9(1):1028. doi: 10.1038/s41467-018-03411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Perez-Cervantes C, et al. Enhancer transcription identifies cis-regulatory elements for photoreceptor cell types. Development. 2020;147(3):dev184432. doi: 10.1242/dev.184432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE158499). A browser session that contains the Nfatc2-dependent ChIP-Seq, ATAC-Seq, and ncRNA-Seq data is available here: http://genome.ucsc.edu/s/ssimonett/hg19_Nfatc2_human_islet_genome_browser