Abstract
Amyloids are proteinaceous deposits considered an underlying pathological hallmark of several degenerative diseases. The mechanism of amyloid formation and its inhibition still represent challenging issues, especially when protein structure cannot be investigated by classical biophysical techniques as for the intrinsically disordered proteins (IDPs). In this view, the need to find an alternative way for providing molecular and structural information regarding IDPs prompted us to set a novel, to our knowledge, approach focused on UV Resonance Raman (UVRR) spectroscopy. To test its applicability, we study the fibrillation of hen-egg white lysozyme (HEWL) and insulin as well as their interaction with resveratrol, employing also intrinsic fluorescence spectroscopy, Fourier transform infrared (FTIR) spectroscopy, and atomic force microscopy (AFM). The increasing of the β-sheet structure content at the end of protein fibrillation probed by FTIR occurs simultaneously with a major solvent exposure of tryptophan (Trp) and tyrosine (Tyr) residues of HEWL and insulin, respectively, as revealed by UVRR and intrinsic fluorescence spectroscopy. However, because the latter technique is successfully used when proteins naturally contain Trp residues, it shows poor performances in the case of insulin, and the information regarding its tertiary structure is exclusively provided by UVRR spectroscopy. The presence of an increased concentration of resveratrol induces mild changes in the secondary structure of both protein fibrils while remodeling HEWL fibril length and promoting the formation of amorphous aggregates in the case of insulin. Although the intrinsic fluorescence spectra of proteins are hidden by resveratrol signal, UVRR Trp and Tyr bands are resonantly enhanced, showing a good sensitivity to the presence of resveratrol and marking a modification in the noncovalent interactions in which they are involved. Our findings demonstrate that UVRR is successfully employed in the study of aggregation-prone proteins and of their interaction with ligands, especially in the case of Trp-lacking proteins.
Significance
The mechanisms underlying protein aggregation and protein-drug interaction are still elusive. Ultraviolet resonance Raman spectroscopy (UVRR) is employed to investigate both phenomena by enhancing specific aromatic amino acid vibrations generally involved in protein self-assembly and in ligand interaction. UVRR has been exploited together with FTIR, fluorescence spectroscopy, and AFM to obtain a robust interpretation of the data, showing that the ligand remodels the morphology of protein fibrils and that aromatic residues are sensitive markers of the presence of the ligand, allowing us to speculate about their involvement as ligand binding sites. UVRR demonstrates itself to be more informative on Trp-lacking proteins compared with fluorescence spectroscopy and in general when the interaction between proteins and fluorescent ligand is studied.
Introduction
Amyloids are extended aggregates formed by misfolded proteins, whose secondary structure is rich in β-sheets. Despite their presence in tissues and organs being considered a pathological hallmark of several human disorders such as Parkinson’s disease, Alzheimer’s disease, type II diabetes, and gestational diabetes (1, 2, 3), protein aggregates can also be physiological for cells (4).
It is worthwhile to mention that several proteins undergoing toxic fibrillation in living organisms belong to the class of intrinsically disordered proteins (IDPs). The aggregation of IDPs, as well as of other globular proteins, may be slowed down or inhibited by the interaction with small ligands such as polyphenols, the presence of which sometimes turned out into a reduced cytotoxicity of the aggregates (5, 6, 7, 8). Although the mechanism of inhibition is still not fully understood, polyphenols’ aromaticity, hydrophobicity, and low water solubility promote the binding with biomolecules, generally involving protein hydrophobic and aromatic portions (9, 10, 11, 12, 13, 14, 15, 16). However, differently from globular proteins, IDPs present an outstanding structural flexibility, which enables them to populate an ensemble of different, reversible, and thermodynamically stable conformations, making them suitable for performing different biological functions (17,18). This aspect not only hampers the study of their structure but also complicates the possibility of making molecular predictions regarding protein-ligand binding.
Although the investigation of IDPs’ native and fibrillar structures cannot be done by x-ray diffraction because of the lack of a well-defined electron density (19,20), cryogenic electron microscopy (cryo-EM) and solid-state NMR could provide structural insights on IDPs. However, these techniques require the use of cryogenic temperature, isotope labeling, large amounts of sample, and so on (2,21, 22, 23). In addition, resolving by cryo-EM the structure of proteins smaller than 50 kDa is still challenging (24).
In this respect, Ultraviolet Resonance Raman (UVRR) spectroscopy can be considered as a valuable alternative to shed new light on protein structure. Differently from cryo-EM and solid-state NMR, proteins can be studied in their aqueous-like environment at a physiological temperature, without chemical manipulation and protein size limitation (21,24,25). Thanks to the resonance condition, UVRR opens up the possibility of studying complex biological systems, as well as their interaction with ligands, by selectively enhancing only the vibrational signals arising from specific biomolecular chromophores (26,27). In the case of protein-based systems, the selective enhancement provided by UVRR allows the direct investigation of aromatic amino acids (14), especially tyrosine (Tyr) and tryptophan (Trp) residues, which play an active role in the self-assembly process involved in fibrils’ formation (15, 16, 17, 18). As a matter of fact, another classical biophysical technique has been widely used for the purpose of fibrillation studies as well as for the investigation of Tyr and Trp residues microenvironments and solvation, namely intrinsic fluorescence spectroscopy. However, the actual efficacy of this technique is strictly dependent on the presence of Trp residues in proteins, as Tyr residues are less sensitive to solvent exposure and binding with ligands, possibly leading to misleading considerations (28,29).
In this context, UVRR demonstrates that it can overcome the limitations of intrinsic fluorescence spectroscopy, especially dealing with Trp-lacking proteins and when the interaction with a fluorescent ligand is probed, because only the protein contributions are enhanced. In fact, although UVRR Tyr markers are sensitive to the modification of noncovalent interactions and to the change in microenvironments’ hydrophobicity, intrinsic Tyr fluorescence manifests its insensitiveness to both phenomena, hence resulting in being less applicable in these cases (30,31).
The aim of this work is to verify the potential of UVRR combined with complementary techniques, such as Fourier transform infrared (FTIR) spectroscopy, intrinsic fluorescence spectroscopy, and atomic force microscopy (AFM), for exploring the secondary and tertiary structures of aggregation-prone proteins during fibrillation and upon their interaction with an antioxidant.
To this purpose, we explored the fibrillation of two model proteins, namely hen-egg white lysozyme (HEWL) and human insulin, and their interaction with a simple hydrophobic ligand, belonging to polyphenolic compounds with antioxidant properties: resveratrol. Indeed, because HEWL is characterized by the presence of both Tyr and Trp residues, whereas insulin has no Trp residues, they are ideal candidates for our study. Thanks also to the support of FTIR spectroscopy, the complexity of the UVRR spectra of HEWL and insulin has been disentangled, and, by observing the modifications of the Trp and Tyr vibrational bands in UVRR spectra, we could target the bands mainly affected by fibrillation as well as those affected by the interaction with resveratrol. In particular, we demonstrated that resveratrol mildly modifies protein fibril secondary structure while relevantly remodeling their morphology. In particular, the presence of resveratrol during HEWL and insulin fibrillation drives the formation of different fibrillar polymorphs, with a modified solvent exposure of aromatic amino acids. In this respect, the use of UVRR spectroscopy and the possibility of enhancing only peculiar chromophores’ vibrational modes are essential to emphasize the modification occurring exclusively in protein tertiary structure, avoiding the interference of resveratrol signals. Differently, FTIR is scarcely sensitive to protein tertiary structure modification, and successful fluorescence spectroscopy data interpretation depends on the presence of Trp residues in the amino acid sequence and on the interaction between a protein and a nonfluorescent ligand. In our case, the fluorescent contribution of resveratrol overlaps with both protein spectra, and no insights on protein tertiary structure could be deduced by fluorescence spectroscopy.
On the contrary, the outstanding ability to enhance only peculiar protein vibrational modes and also the possibility of distinguishing the effect of water exposure and H-bonding make UVRR spectroscopy a valuable experimental technique to discern the nature of intermolecular interactions established between proteins and ligands (32,33). Finally, our UVRR results, complemented with computational evidence reported in literature analyzing the interaction between HEWL, insulin and other polyphenolic ligands (10,12,34) pointed out the involvement of Trp (in the case of HEWL) and Tyr (in the case of insulin) residues as resveratrol binding sites.
Overall, our results show that a multitechnique approach is mandatory to infer about protein fibrillation and their interaction with ligands and that UVRR may play a pivotal role in this type of study.
Materials and methods
Preparation of protein samples
HEWL (L6876), human recombinant insulin (91077C), and resveratrol (R5010) were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification.
Resveratrol was dissolved in 100% ethanol and further diluted in phosphate-buffered saline (PBS (pH 1.8)). HEWL and insulin solutions were prepared in acidic conditions (PBS at pH 1.8, 1.02 mM adjusted by a solution of HCl 37%). The samples were then diluted to the final concentration of 1 mM using the solution of resveratrol, reaching (protein/resveratrol) molar ratios of (1:0), (1:1), and (1:2). HEWL/insulin (1% EtOH) were defined as “HEWL” and “insulin,” HEWL/insulin + resveratrol (1:1) as “HR1”and “IR1,” and HEWL/insulin + resveratrol (1:2) as “HR2” and “IR2,” respectively. Part of these solutions were measured as freshly prepared solutions, and the remaining parts were kept at 65°C and shacked at 750 rpm in a dark place to induce fibrillation. The fibrillation was stopped when the gelatinous phase was achieved, i.e., after 7 days for HEWL and after 1 day for insulin. Then, the solutions were centrifuged at 16,000 × g for 40 min at room temperature. The supernatant solutions were used to estimate the concentration of the gelatinous fraction. Finally, the pellet was resuspended in PBS before measurement, achieving a final ethanol concentration of 1% EtOH.
UVRR spectroscopy
UVRR measurements were carried out at the BL10.2-IUVS (Inelastic UltraViolet Scattering) beamline at Elettra Sincrotrone Trieste using the experimental setup reported elsewhere (35). Samples spectra were measured using two different wavelengths, namely 228 and 244 nm. Synchrotron radiation was used to get the 228 nm source, and the measurements taken at 244 nm were carried out using an Ar + laser (Lexel Laser, Fremont, CA). Protein spectra were acquired at room temperature. The diffused radiation was collected in backscattering geometry and energetically dispersed using a spectrometer with 1800 g/mm (Trivista; Princeton Instruments, Trenton, NJ). The final spectral bandwidth is 20 cm−1 at 228 nm and is 8 cm−1 at 244 nm. Samples excited at 228 nm were irradiated for 3 h, accumulating one spectrum every 15 min with an incoming radiation of 16 μW. Differently, with 244 nm of excitation wavelength, samples were irradiated for 10 s at 140 μW. The final spectrum was the average of 40 spectra. Spectra of buffer and of resveratrol solutions were taken as well, and their contributions were carefully removed from the measured spectra of proteins. Samples were horizontally oscillated during the measurement to avoid photodamage.
Infrared spectroscopy
Infrared spectra of the samples were collected at the infrared beamline SISSI (Synchrotron Infrared Source for Spectroscopic and Imaging), Elettra Sincrotrone Trieste (Trieste, Italy) (36).
Attenuated total reflectance (ATR) spectra of the samples were collected using a MIRacle Single Reflection ATR box (PIKE Technologies, Fitchburg, WI) with a germanium (Ge) internal reflective element. Spectra were acquired using the Vertex 70 interferometer (Bruker Optik, Ettlingen, Germany) equipped with an MIR (Mid-InfraRed) DLaTGS detector and a KBr beam splitter. A 2 μL drop of each sample was deposited onto the internal reflective element, and the measurements were performed during drop dehydration at ambient temperature under a gentle nitrogen flux to prevent water vapor spectral interference. For the same purpose, the interferometric compartment and ATR accessory were also purged with nitrogen. For each spectrum, 15 × 128-scan interferograms were collected with a scanner velocity of 7 kHz and 4 cm−1 of resolution. The spectral region explored spanned a range within 5000–500 cm−1. A 256-scan interferogram of the background was collected under identical external conditions.
AFM
A 15 μL drop of the diluted solution containing fibrils was spotted on freshly cleaved mica and let absorb for 15 min (dilutions were optimized to obtain a sufficient number of fibrils on the surface). Then, the deposited samples were washed with 1 mL of deionized water and we dried with a gentle stream of nitrogen. To avoid sticking effects due to the residual water on the surface, we kept the samples overnight in a N2 chamber. We performed topographic measurements in dynamic mode in air with commercial cantilevers (details in Supporting materials and methods) on MFP-3D (Oxford Instruments/Asylum Research, Goleta, CA) and Solver Pro (NT-MDT, Moscow, Russia) instruments at the Nano Innovation Laboratory at Elettra. Images were acquired with a pixel resolution of roughly 5 nm, acquiring 5 × 5 μm2 images at 1024 × 1024 square pixels, and then analyzed with Gwyddion software to remove the background with standard flattening processes (37). To obtain the length distribution of fibrils, we performed a statistical analysis using the Persistence software, developed by De La Cruz lab (38).
Intrinsic fluorescence spectroscopy
Fluorescence spectra were collected at 24°C using a Perkin Elmer LS-55 spectrofluorometer (Perkin Elmer, Waltham, MA). Solutions of HEWL (5 μM) were excited at 280 nm and insulin ones (20 μM) at 276 nm, recording the emission in the range of 200–500 nm. 500 μL of each solution was loaded inside a quartz cell and measured at different times to follow the aggregation kinetics.
Results and discussion
Modifications of HEWL and insulin secondary structure change upon fibrillation
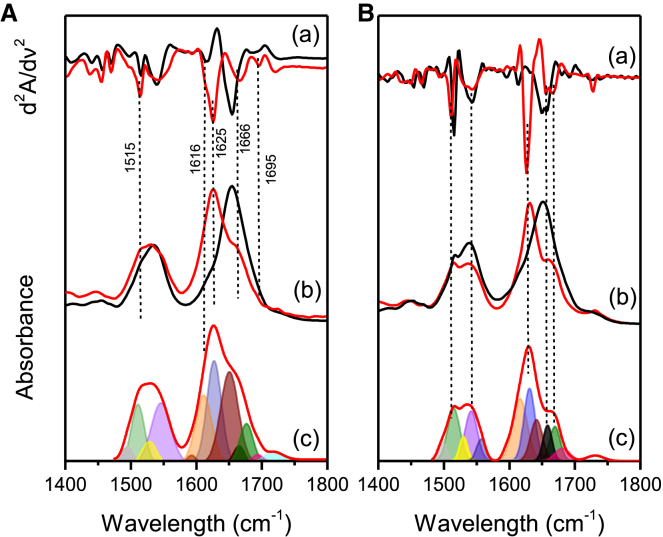
Protein secondary structure is conventionally investigated by recording protein FTIR spectra, focusing on amide I (1600–1700 cm−1) and amide II (1490–1580 cm−1) bands (see Fig. 1). This spectral range is dominated by specific normal modes of vibrations related to peptide linkage (39, 40, 41). In particular, because of the high sensitivity of C=O stretching to its own H-bond state, amide I is commonly used to discern protein secondary structure composition (39,42). The amide bands of freshly prepared and fibril solution of HEWL and insulin are shown in Fig. 1, A (b) and B (b), respectively. To resolve the overlapping amide I band components arising from different secondary structural elements, the second-derivative analysis of the FTIR absorbance spectra was performed (40,41). The second-derivative spectra of HEWL and insulin as freshly prepared and fibril solutions are shown and in Fig. 1, A (a) and B (a), respectively, and the positions of the minima are reported in Table 1, accompanied by a complete assignment. Additionally, to follow the spectral modification of protein secondary structure content, FTIR spectra were fitted employing Gaussian curves, the centers of which were initialized using the minima found in the second-derivative spectra (indicated by the dashed lines in Fig. 1, A (c) and B (c). The fitting results, also reported in Table 1, highlight that the secondary structure of both freshly prepared HEWL and insulin is mostly characterized by a predominant content of α-helix (58 and 40% of the amide I band area, respectively), in agreement with the literature (43,44). The remaining band areas (42% for HEWL and 60% for insulin) account for other vibrational contributions arising from side chains, unordered structures, and β-sheets. As reported in Table 1, the 1616 cm−1 band has been assigned to a mixed contribution arising from side chains, in particular to Tyr-OH vibrational modes, and intermolecular β-sheets. In particular, depending on the protein investigated, a plethora of studies assigned the sub-band below 1620 cm−1 to intermolecular β-sheets, especially when amyloid fibrils are detected (43,45,46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56), whereas many others attributed this band to side chain vibrations (39,42,43). Additionally, the presence of β-sheet signals below 1620 cm−1 is not restricted to the formation of amyloid fibrils, but it has been observed during the formation of small oligomeric species or the presence of small aggregates, especially when proteins are kept under harsh conditions (45,47,57).
Figure 1.
(A) (a) Second derivative spectra of HEWL as freshly prepared and fibril solutions (depicted in black and in red, respectively), (b) FTIR spectra of the HEWL freshly prepared and fibril solutions (depicted in black and in red, respectively), and (c) the fit of the HEWL fibrils spectrum with amide I and II sub-bands highlighted with colors. Dashed lines are used to distinguish different secondary structure components. (B) (a) Second derivative of insulin as freshly prepared and fibril solutions (depicted in black and in red, respectively), (b) FTIR spectra of the second-derivative spectra of insulin freshly prepared and fibril solutions (depicted in black and in red, respectively), and (c) the fit of the insulin fibrils spectrum with amide I and II sub-bands highlighted with colors. Dashed lines are used to discern the secondary structure components.
Table 1.
Assignment, spectral positions, and content of amide I secondary structures probed by FTIR spectroscopy
| HEWL (1% EtOH) |
Insulin (1% EtOH) |
Assignment (39, 40, 41, 42, 43,45,58) | ||||||
|---|---|---|---|---|---|---|---|---|
| Freshly prepared |
Fibril |
Freshly prepared |
Fibril |
|||||
| (±2 cm−1) | (%) | (±2 cm−1) | (%) | (±2 cm−1) | (%) | (±2 cm−1) | (%) | |
| 1616 | 10.5 | 1616 | 20 | 1615 | 10 | 1616 | 31 | side chains/β-sheets |
| – | – | 1626 | 31 | 1631 | 8 | 1630 | 28 | intermolecular β-sheets |
| 1641 | 12.5 | – | – | 1648 | 32 | 1645 | 16 | unordered |
| 1654 | 58 | 1651 | 3 | 1656 | 40 | 1654 | 8 | α-helix |
| – | – | 1666 | 30 | – | – | 1668 | 12 | irregular helix/loops |
| – | – | 1677 | 10 | 1678 | 3 | – | – | turns |
| 1683 | 19 | 1695 | 2 | 1691 | 6 | 1695 | 5 | β-sheets |
The assignment of amide I secondary structures is made on the basis of the minima found in the second-derivative spectra of the freshly prepared and of the fibril solutions of HEWL and insulin (1% EtOH). The spectral position and the content of each protein secondary structure is reported as well. The error estimated for each area is approximately of 2%.
In our case, multiple factors such as high protein concentration and acidic pH may induce fast modification on protein native structure (45,57). Consequently, we could not exclude that our experimental conditions may have induced the formation of a small percentage of additional intermolecular β-sheets, which may contribute to the 1616 cm−1 band intensity, conventionally solely attributed to Tyr-OH vibrations (i.e., a mixture of the in-plane C-C stretching νCC and the deformation of the ring (δHCC)).
As shown in Fig. 1 and Table 1, the fibrillation of HEWL and insulin is accompanied by a significant change in the FTIR spectra line shapes, compared to the freshly prepared protein solutions. By following the modification of the amide I shape, we observed that both fibrils present two intense β-sheet bands located approximately in the regions (1626–1631) cm−1 and a little one at 1695 cm−1, which are typical of intermolecular β-sheets. The position of the Am-I maximum when mature fibrils are formed is informative on the β-sheet organization, namely whether they have a parallel or an antiparallel orientation. Generally, when an Am-I maximal peak is located below 1630 cm−1 and a minor contribution is detected in the region 1686–1695 cm−1, the intermolecular β-sheets are antiparallel, whereas when the Am-I maximum is located at 1630 cm−1 or higher wavelengths and the high-energy band is no longer detected, β-sheets have a parallel orientation (45,59,60). Anyway, small deviations to this rule are commonly observed; in fact, as reported for transthyretin and other systems, the lower energy position of the β-sheet band can shift depending on their length, namely on the number of strands composing the sheets (56). Because of this evidence and the fact that proteins could also be composed by a mixed composition of parallel and antiparallel β-sheets (61), it is preferable to refer to those bands as generic intermolecular β-sheets bands. Moreover, in both cases, fibrillation also induces the rise of loops and irregular helical structures located at 1666–1668 cm−1 and the increasing of the 1616 cm−1 (see Table 1) and 1515 cm−1 band intensities, together with a frequency red shift of the latter band. Both the 1616 cm−1 and the 1515 cm−1 bands arise from a mixed contribution of the in-plane C-C stretching and from the deformation of the Tyr ring (δHCC). Although the 1616 cm−1 band has not been extensively studied because its contribution rarely appears as a well-resolved peak, the intensity and the band frequency modifications of the 1515 cm−1 are easier to monitor because they are generally more distinguishable from the contribution of amide II (62). This band is considered as a local reporter of the tertiary structure modification and on the H-bond network where Tyr residues are on average involved (63,64). In particular, as reported also for other systems (62,65,66), the 1515 cm−1 band increases its intensity (see Fig. 1, A (b) and B (b)) and red shifts of ∼2 cm−1 upon fibrillation. A similar red shift could be thought of as meaningless, but it is actually larger than others already reported in literature (66,67). It accounts for a change in the hydrogen bond network in which Tyr residues are involved, and it has been reported many times working with aggregated proteins (62,68) indicating the formation of a stronger H-bond network in which tyrosine OH groups are involved as H-donors (62,69).
The secondary structure arrangement predicted by FTIR spectroscopy is confirmed by UVRR spectra obtained at 244 nm. Generally, proteins’ structure modifications are highlighted by spectral modifications normalized to the peak intensity of an internal standard. In our work, we avoid the use of an internal standard for spectra normalization because the concentration of an internal standard could also reach 1700 times that of the protein studied. Depending on the protein investigated, its structure could change significantly for the presence of these substances. To find a protocol that does not depend on the presence of an internal standard, we used the FTIR technique as a gold standard for the study of protein structure, and we compared the results obtained with those of UVRR spectroscopy. Peptide bond vibrational modes are resonantly enhanced by a 190–200 nm excitation wavelength, inducing a strong expansion of amide I, II, and III signals. Similarly to FTIR vibrational amide bands, amide I (1640–1680 cm−1) consists mainly of the C=O stretching, whereas amide II (∼1550 cm−1) and III (1200–1340 cm−1) consist of a combination between CN stretching and NH bending (39). Protein secondary structure is usually derived from the spectral position of Am-I and Am-III (70). The correlation between the position of amide bands and the composition of the protein secondary structure was made using chemometrics. Hence, UVRR spectra of purely α-helix, β-sheet, and unordered secondary structure-like proteins with a well-known x-ray structure were collected using an incoming radiation energy of 193–206 nm, and their amide I, II, III, and S positions were used to directly unravel the unknown secondary structure of other proteins (71). A complete overview of protein secondary structure components based on the analysis of the UVRR protein spectra can be found in (26,72, 73, 74). Because the radiation energies used in this work do not satisfy amide band resonance conditions, those bands were decomposed by a linear fitting using a routine based on the Levenberg-Marquardt nonlinear least-squares method as proposed in (73).
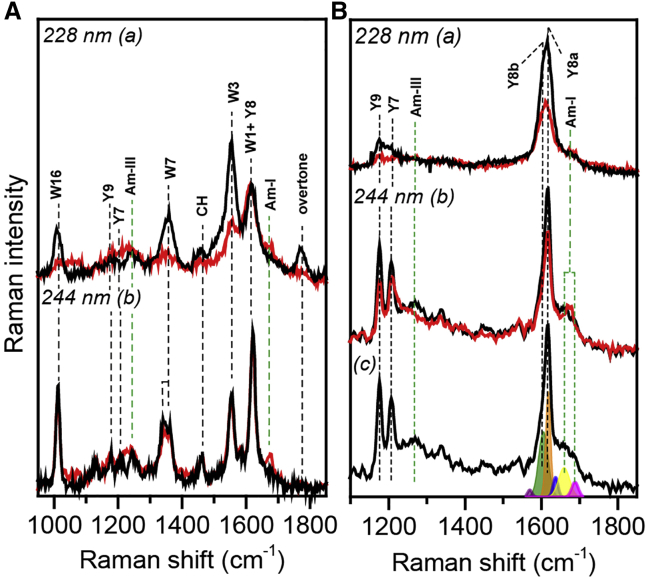
Because of the poor signal/noise ratio of the UVRR spectra obtained using synchrotron radiation at excitation wavelength lower than 220 nm, we monitored the modification of protein secondary structure using a 244 nm excitation wavelength, at which aromatic amino acid vibrational modes are less enhanced. In particular, in Fig. 2, A (b) and B (b), the 244 nm UVRR spectra of freshly prepared and fibrils solutions of HEWL and insulin are presented, and the vibrational peaks are assigned using the same nomenclature reported in (32). In both cases, the modification of the secondary structure could be easily observed by the shape variation and by the energy shift of amide I and III bands (Am-I and Am-III in Fig. 2, A (b) and B (b), indicated by green dashed lines). Similar to FTIR data analysis, a fitting procedure has been applied to the 244 nm HEWL and insulin freshly prepared and fibril spectra (see Fig. 2 B (c)) to follow the modification of those bands upon fibrillation (see results in Table 2). Further details on the UVRR spectra fitting procedure are reported in the Supporting materials and methods. From Table 2, we can deduce that both freshly prepared HEWL and insulin solutions have Am-I and Am-III typical of a mixed α-helix/unordered populated protein (73, 74, 75), whereas fibril solutions are characterized by a red shifting of Am-III and an Am-I located approximately at ∼1670 cm−1, which is typically found in a β-sheet-rich protein and in amyloid fibrils (19,32,74,76,77). In particular, the Am-III band position located at 1242 cm−1 corresponds to a Ramachandran angle of 150°, associated with an antiparallel β-sheet structure. This observation further justifies our choice to not define the orientation of β-sheets, as this evidence can be interpreted in contrast with our FTIR results (78).
Figure 2.
UVRR spectra of freshly prepared (depicted in black) and fibrils (depicted in red) solutions of (A) HEWL and (B) insulin taken at (a) 228 nm and (b) 244 nm excitation wavelength. Peak assignment has been made according to the nomenclature reported in (32); Trp, Tyr, and amide bands are indicated with the symbols W∗, Y∗, and Am-∗, respectively. (c) The fitting of freshly prepared insulin spectrum taken at 244 nm.
Table 2.
Assignment of protein secondary structure made by UVRR spectra analysis through amide I and III spectral positions
| HEWL |
Insulin |
|||
|---|---|---|---|---|
| Am-I (cm−1) | Am-III (cm−1) | Am-I (cm−1) | Am-III (cm−1) | |
| Freshly prepared | (1656, 1674) ± 2 (α-helix, β-sheets) | 1246 ± 1 | (1656, 1683) ± 2 (α-helix, unordered) | 1257 ± 2 |
| Fibrils | 1672 ± 3 (β-sheets) | 1229 ± 3 | 1671 ± 1 (β-sheets) | 1242 ± 2 |
Am-I and Am-III positions evaluated by the fit of the 244 nm UVRR spectra of freshly prepared and fibril solutions of HEWL and insulin. A complete attribution of the secondary structure has been made according to (74).
Tertiary structure changes upon HEWL and insulin fibrillation
UVRR spectra of freshly prepared and fibril solutions of HEWL and insulin could be used to evaluate proteins’ tertiary structure modification upon fibrillation (see Fig. 2, A and B). Differently from the 244 nm spectra, the 228 nm ones present a furtherly enhanced signal of Trp (indicated with W∗ in Fig. 2 A) and Tyr bands (i.e., Y∗ in Fig. 2, A and B), letting this wavelength be particularly suitable for the study of protein tertiary structure. In fact, the spectrum in Fig. 2 A (a) is characterized by an overtone band at ∼1760 cm−1 arising from the sum of two Trp bands located at 1009 and 757 cm−1. The presence of the overtone is rarely observed working in off-resonance conditions, explaining why it is absent in the 244 nm spectra (Fig. 2 A b) (27). Thus, the 228 nm spectra of HEWL are dominated by the vibrational features assigned to Trp bands (32,76,79,80), whereas the insulin ones by those attributed to Tyr bands because no tryptophan residues are present in the amino acid sequence. To highlight the structural modifications, we normalized the spectra to the CH band at 1450 cm−1. In fact, such a band is expected not to be resonantly enhanced like the Trp and Tyr bands and therefore is expected to not change in intensity during the fibrilization process. Both the 228 nm UVRR spectra of insulin and HEWL solutions are characterized by the damping of the Trp and Tyr residues peak intensities when mature fibrils are formed (see Fig. 2, A (a) and B (a), respectively). This evidence confirms previously obtained UVRR results concerning HEWL and insulin fibrillation (81, 82, 83, 84), in which the Phe UVRR band cross section (at 1003 cm−1) decreases its intensity upon HEWL fibrillation (76,85), and similarly, both Phe and Tyr bands decrease their Raman cross sections during insulin aggregation (81, 82, 83). Despite the intensity damping of aromatic residues bands being correlated to the establishment of a strong H-bonding network in which aromatic residues are involved (86, 87, 88, 89, 90), a quantitative evaluation of the H-bond sensitivity of these bands is complicated by environmental influences and only the energy shift of specific UVRR bands has been straightforwardly associated to the formation of H-bond (84). Indeed, modified solvent exposure, hydrophobicity, and solvent polarity are considered the main causes of aromatic amino acid UVRR band cross section modification (76,79,84,85,91,92). In fact, Xu and colleagues demonstrated that the Phe peak at 1003 cm−1 is particularly sensitive to water exposure and solvent polarity (76), and Asher et al. explained the correlation between the intensity increasing of both Trp and Tyr UVRR bands and the increased solvation of their microenvironments as the result of a blue shift of their absorbance spectra upon hydration (91). In our case, the UV-Vis spectra of both HEWL and insulin fibrils are characterized by a blue shift of the main absorption peak while maintaining a comparable or even a reduced absorption degree (see Fig. S1). This implies that self-absorption plays a minor role in the modification of the aromatic amino acid band, which is actually due to water exposure and hydrophobicity variation of Trp and Tyr residues.
Thus, the intensity damping of these UVRR bands upon fibrillation suggests that part of the Trp and Tyr residues in HEWL and insulin surely reside in a less hydrophobic and water-exposed environment when mature fibrils are formed and, eventually, could form hydrogen bonds with solvent molecules.
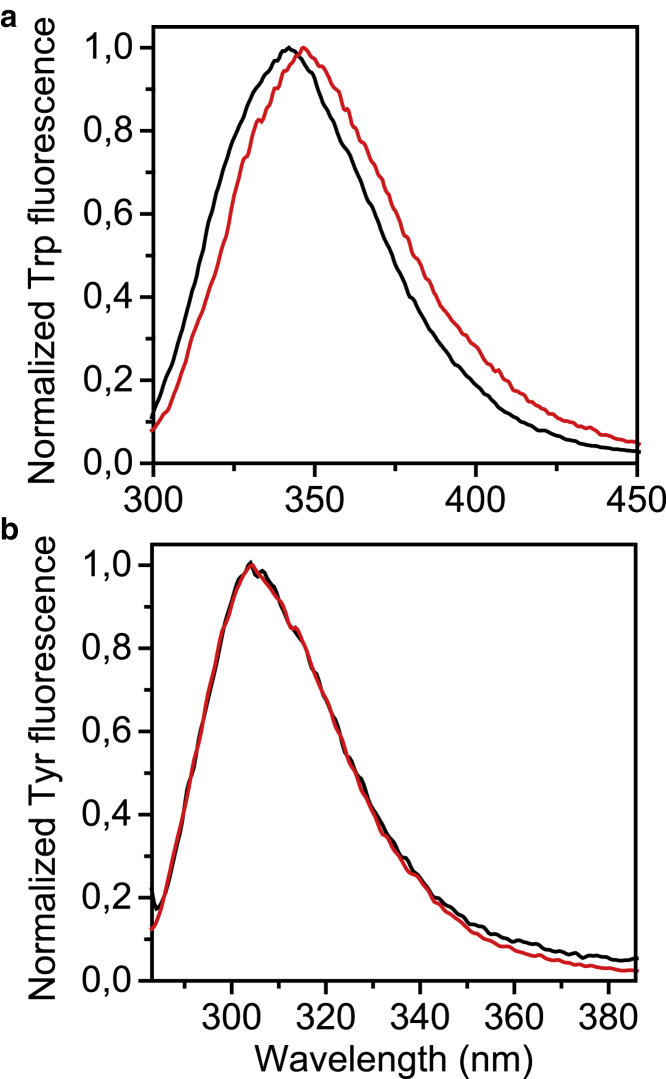
In the case of HEWL fibrils, we can deduce similar conclusions by using intrinsic fluorescence spectroscopy. Among the six Trp residues of HEWL, 80% of its fluorescence spectrum arises from Trp62, situated in the enzymatic site, and Trp108 (93). During fibrillation, we observed a quenching of the Trp maximal peak (data not shown) and a red shift of the Trp peak from 342 to 345 nm when mature fibrils are formed (see Fig. 3 (a)). Both the Trp fluorescence quenching and the red shift of the peak suggest an increased solvent exposure of part of the Trp residues and/or the establishment of a H-bond network in which some of them are involved (29,94). Nevertheless, when mature fibrils are formed in similar fibrillation conditions, the sequence 57–107 was found to be inaccessible to pepsin digestion (95). Because Trp62 is buried in the hydrophobic core of the fibrils, we can deduce that in respect to the freshly prepared solution, more Trp residues are solvent exposed when fibrillation ends with the exception of Trp62 and Trp63, thus confirming our UVRR findings.
Figure 3.
(a) Normalized Trp fluorescence intensity of HEWL freshly prepared and fibril solutions (depicted in black and in red, respectively). Trp peak maximum red shifts from 342 nm toward 345 nm during fibrillation. (b) Normalized Tyr fluorescence intensity of insulin freshly prepared and fibril solutions (depicted in black and in red, respectively). Tyr maximal peak does not shift in energy upon fibrillation.
Unfortunately, in the case of Trp-lacking proteins, intrinsic fluorescence spectroscopy cannot infer protein residue solvent exposure (30,31). In fact, in Fig. 3 (b) we do not observe any red shift of insulin Tyr fluorescence peak upon fibrillation and/or upon their solvent exposure. During insulin fibrillation, we observed only an intensity quenching of the Tyr peak located at 305 nm (see Fig. S2). Nevertheless, this quenching phenomenon could not be traced back straightforwardly only to a possible Tyr solvent exposure, in accordance with the findings reported by Bekard (30) on bovine insulin and by Dusa (31) on α-synuclein.
In light of this limitation, UVRR is an alternative and valuable approach to get important insights regarding protein tertiary structure and aromatic residues microenvironments’ hydrophobicity, even in the case of Trp-lacking proteins. Noteworthy, in Fig. 2 B (a) (freshly prepared and fibril solutions), we could easily observe that not only do Tyr peak intensities undergo quenching (especially the Y8 band, which is actually a doublet, Y8a and Y8b), but we could also consider other markers to gain additional information on the effect of fibrillation (96). As already reported in literature, the Y8b band position has been used as an indicator of the strength of an H-bond in which Tyr is involved as the H-donor, and the modification in the relative intensity of Tyr Fermi doublet R = I853/I828 has been used as a marker of Tyr average microenvironment hydrophobicity variation (84,96, 97, 98). We verified the presence of these markers analyzing the 244 nm spectra because of its higher spectral resolution (see Fig. 2 B (b)): the position of the Y8b band in freshly prepared and fibril solutions is located at (1606 ± 3) cm−1 and (1597 ± 2) cm−1, respectively. The red-shifted position of the Y8b band suggests that some Tyr residues act as H-donors when mature fibrils are formed (84). Additionally, from the fitting of the Tyr Fermi doublet 828–853 cm−1 (see Fig. S3), we estimated how the area modification of these bands influenced the R ratio. In particular, R is equal to 1.6 in the case of freshly prepared insulin solution, whereas in the case of fibril solution, it is equal to 2.2. Phenomenologically, the increase of the 853 cm−1 component is associated with a higher hydrophobicity of Tyr environment, or it is observed when the hydroxyl group acts as a strong H-bond acceptor (96). In light of the previous considerations, we deduce that the overall intensity decreasing of Tyr bands, the red-shifted position of the Y8b band, and the value of the R ratio are all clear hints of Tyr-increased solvent exposure and of the phenolic ring’s involvement in an H-bond network when mature fibrils are formed (88,89). These assumptions are in agreement with previously reported ANS (8-Anilinonaphthalene-1-sulfonic acid) fluorescence, crystallographic, and proteolysis results as well as with molecular dynamics (MD) simulations carried out on insulin (11,84,96,98, 99, 100, 101), confirming the ability of UVRR to infer solvent exposure and change in the hydrophilicity of the aromatic residue microenvironment.
Interaction with resveratrol
The multitechnique approach presented for the study of protein fibrillation has also been employed for the study of protein-ligand interaction, which is generally mediated by intermolecular forces such as H-bonding, cation-π interaction, van der Waals forces, and so on. In the next sections, we further elucidate the advantages of the use of UVRR spectroscopy compared to fluorescence spectroscopy for the investigation of protein tertiary structure modification, especially when the ligand studied exhibits a large fluorescent signal and the protein lacks Trp residues.
Fibril morphology modification induced by protein-resveratrol interaction
Polyphenols and natural compounds, in general, have been demonstrated to act as antiamyloidogenic compounds (102). In light of these results, the effect induced by resveratrol during HEWL and insulin fibrillation has been evaluated. Because the interaction between proteins and ligands occurs via noncovalent interaction (H-bonding, van der Waals, aromatic, and hydrophobic interactions), resveratrol has been used as one of the smallest aromatic model drugs to infer the molecular mechanism of interaction.
Resveratrol has been incubated with HEWL and insulin in a concentration-dependent manner and in environmental conditions prone to induce aggregation. As anticipated in the Materials and methods, we named (H/I)R1 and (H/I)R2 the mixture (protein/resveratrol) (1:1) and (1:2), respectively. HEWL and insulin were replaced with “HE” and “IE”, respectively, only in Fig. 4 to ensure a good visibility of the image.
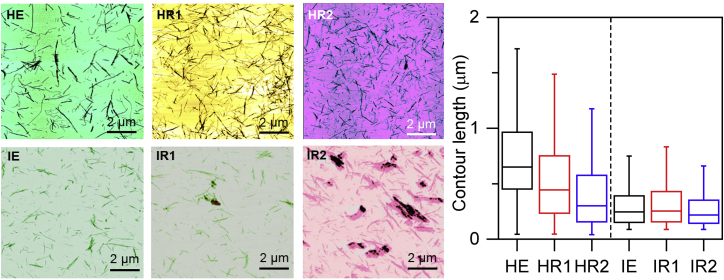
Figure 4.
(left) 10 × 10 μm2 AFM topographic images of HEWL (HE in black, HR1 in red, and HR2 in blue) and insulin (IE in black, IR1 in red, IR2 in blue) fibrils. False color scales represent the three-dimensional topographic height. (Right) The distributions of fibrils contour length are presented. The error bands represent the 25th and the 75th percentiles of the contour length distribution.
AFM images reported in Fig. 4 show that resveratrol actually remodels the structure of both HEWL and insulin fibrils in a dose-dependent manner. To quantify this qualitative behavior, AFM images were statistically analyzed to estimate the distribution of fibril contour lengths with and without resveratrol. The results of AFM analysis are summarized in Fig. 4, in which the most probable value for fibril contour length corresponds to the 50th percentile of the distribution. In particular, the long and well-separated HEWL fibrils obtained in the absence of resveratrol appear gradually shorter and straighter by increasing the antioxidant concentration, whereas the contour length of the insulin fibrils is mildly modified by the presence of resveratrol, which actually induces the formation of amorphous aggregates by increasing resveratrol concentration (see Fig. 4).
Interestingly, Frare and colleagues indicated that HEWL could undergo hydrolysis when incubated at pH < 3, producing fragments with a high tendency to self-aggregate (95). However, this evidence has been widely discussed in other works, suggesting that hydrolysis does not occur in all the cases, but it strongly depends on the preparation methods (93,103). In fact, Arnaudov and colleagues reported that the incubation of HEWL at pH 2 and high temperature mostly consists in the production of full-length fibrils (103), and similar results were obtained also by Chaari et al., which indicates that agitation at 750 rpm could counteract hydrolysis, giving rise to full-length proteins (93). Our sample preparation followed a protocol similar to that of Chaari and colleagues, willing to reduce the possibility of chemical degradation. Noteworthy, the reduction of HEWL fibrils length in presence of resveratrol does not depend on hydrolysis, as HR1 and HR2 were obtained in the same acidic conditions. Therefore, the reduction observed appears to be the result of a concentration-dependent interaction with the antioxidant.
Similarly, in the case of insulin, IR1, and IR2, we observed the formation of nonfibrillar spherical aggregates coexisting with mature fibrils of comparable length with increasing the concentration of resveratrol, as observed also elsewhere (104,105). These findings are in agreement with scientific literature, which reports how the effects of antioxidants on protein fibrils are their morphological remodeling and how the modification of the fibrillation pathway lead to the production of amorphous aggregates (5,6).
FTIR spectroscopy probes secondary and tertiary structure modification upon the interaction between HEWL, insulin, and resveratrol
The relevant shrinking of HEWL fibrils induced by an increased concentration of resveratrol is not accompanied by significant secondary structure modifications, as evidenced by FTIR analysis performed on HEWL, HR1, and HR2 samples reported in Fig. 5 (a). The results of FTIR band fitting of HEWL, HR1, and HR2 spectra reported in Table 3 indicate that resveratrol induces an increased concentration of β-sheet structure (from 29 to 40%) and a reduced contribution of α-helix (from 3 to 10%) upon increasing the content of resveratrol. Differently, resveratrol induces a modification in insulin secondary structure. In fact, as clearly visible from the second-derivative spectra reported in Fig. 5 (b), the presence of resveratrol induces the increasing of the α-helix contribution (1658 cm−1) instead of the unordered and turn populations (1666 and 1670 cm−1, respectively). Additionally, band fitting of insulin spectra highlights that the low-energy β-sheet band (at ∼1630 cm−1 in the insulin case), blue shifts to 1627 cm−1 upon increasing the concentration of resveratrol as reported in Table 3. This evidence indicates a reorganization of the β-sheet disposition, passing from shorter to longer ones when resveratrol concentration is double that of insulin.
Figure 5.
FTIR spectra of amide I and II bands (1450–1750 cm−1) of (a, top) HEWL fibrils in absence (in black) and presence of resveratrol (HR1 in red and HR2 in blue) and (b) insulin fibrils in absence (in black) and presence of resveratrol (IR1 in red and IR2 in blue) with their second-derivative spectra in the corresponding panel (on the bottom).
Table 3.
The impact of resveratrol on fibril contour lengths and secondary structure probed by FTIR
| Contour length (nm) | FTIR β-sheets position (±2 cm−1) and content (%) | FTIR α-helix position (±2 cm−1) and content (%) | |
|---|---|---|---|
| HEWL | 640 [421:964] | 1626 (29%) | 1656 (3%) |
| HR1 | 445 [234:755] | 1626 (39%) | 1656 (9%) |
| HR2 | 300 [155:577] | 1624 (40%) | 1656 (10%) |
| Insulin | 247 [151:391] | 1630 (32%) | 1658 (7.5%) |
| IR1 | 242 [153:412] | 1627, 1631 (28%) | 1658 (18%) |
| IR2 | 218 [143:352] | 1627 (34%) | 1658 (18%) |
The positions of the β-sheets bands extracted from FTIR spectra of HEWL, HR1, HR2, insulin, IR1, IR2 fibrils solutions together with the associated contour length distribution. The 25th and the 75th percentiles of the distribution are indicated in squared brackets.
More interestingly, from the FTIR spectra of insulin in Fig. 5 (b), we observe a further 2 cm−1 of the Tyr-OH band located in the region 1510–1520 cm−1 upon interaction with resveratrol, indicating a further strengthening of the H-bond network in which the OH group of tyrosine residues are involved when the antioxidant is present (62,63,66, 67, 68). Additionally, this band increases its intensity with increasing the concentration of resveratrol. In the literature, there is no clear evidence of the correlation between intensity modification of the ∼1515 cm−1 band and a possible mechanism of interaction; anyway, we observed that an intensity increasing of this band is always accompanied by its energy red shifting, consequent to a modification of the H-bond network around tyrosine residues. Interestingly, the low-energy flank of amide I present a resveratrol-concentration-dependent variation; the tyrosine doublet band at 1594–1606 cm−1 (ν8b, ν8a) modifies its relative intensity by increasing the 1606 cm−1 band and by decreasing the 1594 cm−1 one (see Figure 5 (b)) (67). Noteworthily, those bands are sensitive to tyrosine ring deformations and symmetry losses, mainly because of intermolecular interactions, confirming that tyrosine residues could be involved in a direct interaction with the antioxidant or could reflect structural modifications resulting from the interaction between insulin and resveratrol.
Tertiary structure of HEWL and insulin fibrils in presence of resveratrol
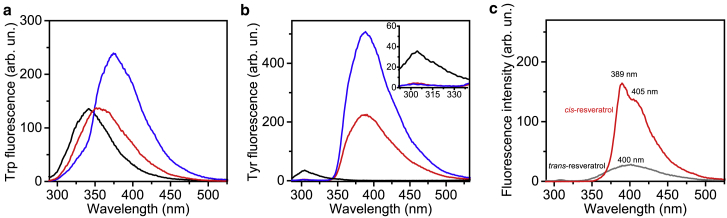
The modification of the protein tertiary structure upon interaction with a ligand is usually exploited in literature by fluorescence spectroscopy. In this case, intrinsic fluorescence spectroscopy has been used to indirectly investigate the effect of resveratrol during the fibrillation of both HEWL and insulin. Fig. 6 shows the fluorescence data obtained for HEWL (Fig. 6 (a)) and insulin (Fig. 6 (b)) at different concentrations of resveratrol, namely when the protein/antioxidant ratio is (1:0) (in black), (1:1) (in red), and (1:2) (in blue). In Fig. 6, (a) and (b), the Trp and Tyr intrinsic fluorescence peaks of HEWL and insulin fibrils obtained in the absence and in the presence of resveratrol have been presented, respectively.
Figure 6.
(a) Trp fluorescence of HEWL fibrils solution in absence (in black) and in presence of resveratrol, i.e., HR1 (in red) and HR2 (in blue). (b) Tyr fluorescence of freshly prepared solution of insulin in absence (in black) and in presence of resveratrol, i.e., IR1 (in red) and IR2 (in blue). (c) Trans- (in gray) and cis- (in red) resveratrol spectra. Trans isoform is characterized by a low quantum yield symmetric peak with a maximum at 400 nm, whereas cis isoform by a high quantum yield contribution characterized by two peaks located at 389 and 405 nm.
As clearly shown in Fig. 6 (a), once HEWL fibrils are formed in absence of resveratrol, the Trp maximal peak is located at 345 ± 0.5 nm, indicating that part of the Trp residues are solvent exposed (29). Because Trp62 and Trp108 are the dominant fluorescent emitters of HEWL and Trp62 has been found buried in the hydrophobic core of the HEWL fibrils (95,106), it is likely that Trp108 results in greater solvent exposure when HEWL fibrils are formed. Thus, the fluorescent signal we detected measuring HEWL fibrils arises mainly from Trp62. The presence of resveratrol in the case of HR1 promotes the red shifting of Trp emission peak to 352 ± 0.5 nm, indicating that the interaction with resveratrol during fibrillation actually stabilizes HEWL fibrils in a different structural conformational, inducing a further solvent exposure of Trp residues. Docking studies reported that resveratrol could bind to specific portions involving Trp62 (10). In light of this consideration, we could speculate on two possible effects induced by the presence of resveratrol at this specific concentration; one possible scenario could suggest that resveratrol provokes the complete solvent exposure of Trp108, and the other possibility is that resveratrol binds to Trp62 during fibrillation and stabilizes protein in a different fibrillar conformation, with a more solvent-exposed hydrophobic core. In the case of HR2, the emission peak results in a further red shift to 376 ± 0.5 nm and increases its quantum yield, which is actually not compatible with a typical Trp signal. In fact, in this case, the protein contribution is completely hidden by that of resveratrol, which is no more in a totally trans conformation as reported in Fig. 6 (c). In fact, the trans to cis isomerization of resveratrol provokes the observed hyperchromic effect with a modification of the emission peak shape as reported in Fig. 6 (c). However, in the case of HR2, the emission peak does not resemble that of a trans- nor that of a cis-resveratrol one, and it is also different from the superposition of the HEWL and resveratrol fluorescence signals (data not shown). Hence, we can conclude that the peak detected in the HR2 spectrum may arise from a mixed population of free and bound resveratrol in both the conformations or may arise from resveratrol blocked in an alternative conformation by the binding with HEWL. The interaction at this concentration of resveratrol actually hides the fluorescence signal arising from HEWL, indicating the need to find an alternative technique for studying the interaction. It is also important to mention that the trans to cis isomerization is not induced by UV light in this case, as proteins underwent fibrillation stored in a dark place. The effect of resveratrol concentration, incubation temperature, and low pH could surely affect the stabilization of the trans population (107). However, without UV exposure for days at the resveratrol concentration explored, the trans conformation remains stable (107). Additionally, when the protein/resveratrol concentration ratio is (1:1), we observed two different fluorescence spectra, indicating that the interaction with HEWL and insulin could differently affect the stabilization of the trans conformation (108).
At the same time, Fig. 6 (b) shows that insulin fibrils obtained in presence of resveratrol (IR1 and IR2) present a strong intensity damping of the Tyr emission peak compared to insulin fibrils. More importantly, the fluorescence spectra of IR1 and IR2 are completely dominated by the signal arising from resveratrol, showing a single broad peak at 388 nm. The hyperchromic effect observed even in these cases suggests that the interaction between insulin and the antioxidant promotes the emergence of a resveratrol emission peak, whose shape does not resemble that of its trans nor that of its cis conformations. Differently from HR2, the maximal peak of the resveratrol contribution when interacting with insulin appears furtherly red shifted (109). Thus, because of the hyperchromic effect of resveratrol and to the insensitiveness of Tyr bands to insulin fibrillation and interaction with ligands, an alternative tool has to be used to shed light into protein tertiary structure when fibrils are formed, especially in presence of a fluorescent ligand.
Because of its exclusive sensitiveness to protein chromophores, UVRR spectroscopy has been used to discern about the structural modifications provoked by resveratrol on both proteins without observing interference from the antioxidant signal and being able to register Trp and Tyr bands modification upon interaction with the ligand.
In particular, Fig. 7 (a) and (b) show the 228 nm UVRR spectra HEWL, HR1, and HR2 and insulin, IR1, and IR2, respectively. In the case of HEWL (Fig. 7 (a)), the 228 nm UVRR spectra show a strong damping of the symmetric phenyl ring mode of Trp (W3 band) in a resveratrol-concentration-dependent manner. To better investigate this peculiar behavior, the spectra were fitted to quantify the relative area variation of W3 band with respect to the W1 one and to evaluate a possible W3 band shift. The results reported in Table 4 indicate that in the HR2 case, the W3 band of Trp residues is quenched and red shifted. As reported elsewhere (88), W3 band position is mostly sensitive to the variations occurring in the torsional angle χ2,1 of proteins. The observed modification of W3 peak intensity and position indicates the interaction with resveratrol affects and modifies HEWL torsional angles (86,88).
Figure 7.

UVRR spectra of (a) HEWL (in black), HR1 (in red), and HR2 (in blue) and (b) of insulin (black), IR1 (red), and IR2 (blue) obtained using 228 nm of excitation wavelength and presented in the region 1400–1800 cm−1.
Table 4.
Resveratrol changes the relative intensity of Trp peaks in a concentration-dependent manner
| W3 area/W1 area | W3 position (cm−1) | |
|---|---|---|
| HEWL | 0.63 ± 0.04 | 1557 ± 2 |
| HR1 | 0.6 ± 0.03 | 1557 ± 1 |
| HR2 | 0.33 ± 0.03 | 1554 ± 1 |
W3/W1 area ratio and W3 position of 228 nm UVRR spectra of HEWL, HR1, and HR2.
Interestingly, the modification of peptide torsion angle occurs only in the HR2 case, when possibly a mixed population of trans- and cis-resveratrol occurs, as evidenced by our intrinsic fluorescence results. Because the UVRR spectra of HR1 and HR2 fibrils are mainly dominated by proteins’ signal, we could deduce that the effects observed on the W3 band are direct consequences of the Trp-resveratrol interaction in a concentration-dependent manner (86,88). As observed by molecular dynamics simulation, the modification of dihedral angle χ2 of protein residues actually is crucial for binding affinity and can be strongly modified by the interaction with small aromatic compounds (110).
Because the docking study suggests that Trp62 is involved in the interaction with resveratrol and the number of estimated binding sites is ∼1, we speculate that the UVRR and fluorescence Trp modifications are induced by the involvement of Trp62 in the interaction with resveratrol.
Conversely, the 228 nm UVRR spectra of insulin show that the Y8 band (in-plane stretching of the ring) of the fibrils increases with increasing the concentration of resveratrol. Similar results have been obtained by Wang and colleagues studying the interaction between Aβ proteins and myricetin (111). In fact, they reported that the intensity of both Phe and Tyr bands increased upon addition of myricetin, which actually inhibits the formation of Aβ fibrils, concluding that the inhibition occurs consequently to the ligand-aromatic amino acid interaction (111). Therefore, the formation of insulin amorphous aggregates and the increasing of the UVRR Tyr band intensity in a resveratrol-concentration-dependent manner could be the results of the direct interaction between tyrosine residues and resveratrol. The interaction with resveratrol could increase the hydrophobicity of Tyr microenvironments or could underlie the formation of a strong H-bond in which Tyr is involved (79). Hence, the formation of nonfibrillar aggregates in a resveratrol-concentration-dependent manner, together with the modification of the Tyr-OH band at 1515 and 1594–1606 cm−1 detected by FTIR spectroscopy, which is sensitive to ring deformation induced by the establishment of intermolecular interactions in which Tyr residues are involved (such as H-bonding), leads us to infer the involvement of tyrosine residues as direct resveratrol binding sites via intermolecular forces. Docking studies exploiting the interaction between insulin and other simple aromatic compounds confirm our interpretation, highlighting that they actually bind to a specific portion of the insulin B domain, composed of (Tyr26, Phe24, Phe25, Tyr16) (11,112, 113, 114), which is the same portion able to self-assemble independently (99).
Conclusions
The potential of UVRR spectroscopy in monitoring modifications occurring in protein secondary and tertiary structures upon fibrillation and interaction with a hydrophobic ligand has been exploited in combination with FTIR spectroscopy, fluorescence spectroscopy, and AFM. Specifically, because of its high sensitivity to hydrophobic modifications of aromatic residues microenvironment, UVRR is more informative than intrinsic fluorescence spectroscopy when proteins do not contain Trp residues. Additionally, UVRR exploiting electronic resonances allows enhancing only selected proteins’ vibrational modes peculiar to the protein-ligand interaction, thus decreasing the background caused by unwanted spectral contributions. FTIR spectroscopy, effective at probing protein secondary structures and complemented with morphological information that can be extracted from AFM measurements, allows for a more comprehensive interpretation of the UVRR data.
To benchmark the potential of UVRR, this multitechnique approach has been employed on two model proteins prone to fibrillation (HEWL and insulin), also studying their interaction with a model ligand: resveratrol. We observed that HEWL and insulin fibril morphology is completely remodeled by the incubation with resveratrol in a concentration-dependent manner; in the case of HEWL, fibrils become shorter and straighter, whereas in the case of insulin, resveratrol induces the formation of nonfibrillar aggregates. Differently from fluorescence spectra in which the resveratrol completely hides the proteins’ contribution, UVRR offers the possibility of following the proteins’ tertiary structure modification due to a different concentration of resveratrol by completely removing its contribution by selectively probe-peculiar protein vibrations. However, UVRR manifests its powerfulness in the case of Trp-lacking proteins, as the Tyr fluorescence peak is completely insensitive to the solvent effect and to noncovalent interactions. Together with FTIR results, UVRR demonstrates that part of the Trp and Tyr residues of HEWL and insulin could be considered close to the resveratrol binding sites.
Indeed, this work proposes a reliable interpretation of UVRR spectra of proteins undergoing fibrillation and provides a firm multitechnique experimental approach that can be followed for the investigation of biologically more relevant systems such as IDPs and of their interaction with drugs.
From a general point of view, the characterization of fibrillar molecular architecture of model proteins by UVRR spectroscopy is important not only to answer fundamental questions related to protein folding, but it is also crucial for the development of therapeutics interventions and for diagnostics goals.
Author contributions
M.P. conceived the experiments. M.P., F.D., L.V., and C.M. designed the experimental plan. M.P. performed UVRR and FTIR measurements. M.P. and S.P. performed the fluorescence spectroscopy measurements, and P.P. performed the AFM measurements and their analysis. A.G. contributes to the Raman setup optimization. M.P. analyzed all the experimental results and wrote the manuscript. F.D., L.P., P.P., S.P., L.V., and C.M. revised the manuscript. L.V. and C.M. supervised the entire work.
Editor: Wendy Shaw.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.08.032.
Contributor Information
Maria Pachetti, Email: maria.pachetti@burlo.trieste.it.
Lisa Vaccari, Email: lisa.vaccari@elettra.eu.
Claudio Masciovecchio, Email: claudio.masciovecchio@elettra.eu.
Supporting material
References
- 1.Moran S.D., Zanni M.T. How to get insight into amyloid structure and formation from infrared spectroscopy. J. Phys. Chem. Lett. 2014;5:1984–1993. doi: 10.1021/jz500794d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iadanza M.G., Jackson M.P., Radford S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018;19:755–773. doi: 10.1038/s41580-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fusco G., Chen S.W., De Simone A. Structural basis of membrane disruption and cellular toxicity by α-synuclein oligomers. Science. 2017;358:1440–1443. doi: 10.1126/science.aan6160. [DOI] [PubMed] [Google Scholar]
- 4.Watt B., van Niel G., Marks M.S. PMEL: a pigment cell-specific model for functional amyloid formation. Pigment Cell Melanoma Res. 2013;26:300–315. doi: 10.1111/pcmr.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ono K., Yamada M. Antioxidant compounds have potent anti-fibrillogenic and fibril-destabilizing effects for alpha-synuclein fibrils in vitro. J. Neurochem. 2006;97:105–115. doi: 10.1111/j.1471-4159.2006.03707.x. [DOI] [PubMed] [Google Scholar]
- 6.He J., Xing Y.-F., Zeng C.-M. Tea catechins induce the conversion of preformed lysozyme amyloid fibrils to amorphous aggregates. J. Agric. Food Chem. 2009;57:11391–11396. doi: 10.1021/jf902664f. [DOI] [PubMed] [Google Scholar]
- 7.Kim C.-Y., Lee C., Jang J.-H. Neuroprotective effect of epigallocatechin-3-gallate against β-amyloid-induced oxidative and nitrosative cell death via augmentation of antioxidant defense capacity. Arch. Pharm. Res. 2009;32:869–881. doi: 10.1007/s12272-009-1609-z. [DOI] [PubMed] [Google Scholar]
- 8.Feng Y., Wang X.P., Liu R.T. Resveratrol inhibits beta-amyloid oligomeric cytotoxicity but does not prevent oligomer formation. Neurotoxicology. 2009;30:986–995. doi: 10.1016/j.neuro.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Stirpe A., Pantusa M., Guzzi R. Resveratrol induces thermal stabilization of human serum albumin and modulates the early aggregation stage. Int. J. Biol. Macromol. 2016;92:1049–1056. doi: 10.1016/j.ijbiomac.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Gazova Z., Siposova K., Nagy M. Amyloid aggregation of lysozyme: the synergy study of red wine polyphenols. Proteins. 2013;81:994–1004. doi: 10.1002/prot.24250. [DOI] [PubMed] [Google Scholar]
- 11.Alijanvand S.H., Christensen M.H., Otzen D.E. Novel noscapine derivatives stabilize the native state of insulin against fibrillation. Int. J. Biol. Macromol. 2020;147:98–108. doi: 10.1016/j.ijbiomac.2020.01.061. [DOI] [PubMed] [Google Scholar]
- 12.Wang J.-B., Wang Y.-M., Zeng C.-M. Quercetin inhibits amyloid fibrillation of bovine insulin and destabilizes preformed fibrils. Biochem. Biophys. Res. Commun. 2011;415:675–679. doi: 10.1016/j.bbrc.2011.10.135. [DOI] [PubMed] [Google Scholar]
- 13.Lemkul J.A., Bevan D.R. Destabilizing Alzheimer’s Abeta(42) protofibrils with morin: mechanistic insights from molecular dynamics simulations. Biochemistry. 2010;49:3935–3946. doi: 10.1021/bi1000855. [DOI] [PubMed] [Google Scholar]
- 14.Porat Y., Abramowitz A., Gazit E. Inhibition of amyloid fibril formation by polyphenols: structural similarity and aromatic interactions as a common inhibition mechanism. Chem. Biol. Drug Des. 2006;67:27–37. doi: 10.1111/j.1747-0285.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 15.Levy-Sakin M., Shreberk M., Gazit E. Targeting insulin amyloid assembly by small aromatic molecules: toward rational design of aggregation inhibitors. Islets. 2009;1:210–215. doi: 10.4161/isl.1.3.9609. [DOI] [PubMed] [Google Scholar]
- 16.Hirohata M., Hasegawa K., Naiki H. The anti-amyloidogenic effect is exerted against Alzheimer’s beta-amyloid fibrils in vitro by preferential and reversible binding of flavonoids to the amyloid fibril structure. Biochemistry. 2007;46:1888–1899. doi: 10.1021/bi061540x. [DOI] [PubMed] [Google Scholar]
- 17.Salvi N., Abyzov A., Blackledge M. Solvent-dependent segmental dynamics in intrinsically disordered proteins. Sci. Adv. 2019;5:eaax2348. doi: 10.1126/sciadv.aax2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright P.E., Dyson H.J. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 19.Shashilov V.A., Sikirzhytski V., Lednev I.K. Quantitative methods for structural characterization of proteins based on deep UV resonance Raman spectroscopy. Methods. 2010;52:23–37. doi: 10.1016/j.ymeth.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 21.Tuttle M.D., Comellas G., Rienstra C.M. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B., Ge P., Jiang L. Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 2018;9:3609. doi: 10.1038/s41467-018-05971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerrero-Ferreira R., Taylor N.M., Stahlberg H. Cryo-EM structure of alpha-synuclein fibrils. eLife. 2018;7:e36402. doi: 10.7554/eLife.36402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Huynh D.T., Yeates T.O. A 3.8 Å resolution cryo-EM structure of a small protein bound to an imaging scaffold. Nat. Commun. 2019;10:1864. doi: 10.1038/s41467-019-09836-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heise H., Hoyer W., Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci. USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oladepo S.A., Xiong K., Lednev I.K. UV resonance Raman investigations of peptide and protein structure and dynamics. Chem. Rev. 2012;112:2604–2628. doi: 10.1021/cr200198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asher S.A., Ludwig M., Johnson C.R. UV resonance Raman excitation profiles of the aromatic amino acids. J. Am. Chem. Soc. 1986;108:3186–3197. [Google Scholar]
- 28.Vivian J.T., Callis P.R. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakowicz J.R., Masters B.R. Principles of fluorescence spectroscopy, third edition. J. Biomed. Op. 2008;13:029901. [Google Scholar]
- 30.Bekard I.B., Dunstan D.E. Tyrosine autofluorescence as a measure of bovine insulin fibrillation. Biophys. J. 2009;97:2521–2531. doi: 10.1016/j.bpj.2009.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dusa A., Kaylor J., Fink A.L. Characterization of oligomers during α-synuclein aggregation using intrinsic tryptophan fluorescence. Biochemistry. 2006;45:2752–2760. doi: 10.1021/bi051426z. [DOI] [PubMed] [Google Scholar]
- 32.Schlamadinger D.E., Gable J.E., Kim J.E. Hydrogen bonding and solvent polarity markers in the UV resonance Raman spectrum of tryptophan: application to membrane proteins. J. Phys. Chem. B. 2009;113:14769–14778. doi: 10.1021/jp905473y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López-Peña I., Leigh B.S., Kim J.E. Insights into protein structure and dynamics by ultraviolet and visible resonance Raman spectroscopy. Biochemistry. 2015;54:4770–4783. doi: 10.1021/acs.biochem.5b00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabiee A., Ebrahim-Habibi A., Nemat-Gorgani M. How curcumin affords effective protection against amyloid fibrillation in insulin. Food Funct. 2013;4:1474–1480. doi: 10.1039/c3fo00019b. [DOI] [PubMed] [Google Scholar]
- 35.D’Amico F., Saito M., Masciovecchio C. UV resonant Raman scattering facility at Elettra. Nucl. Instrum. Methods Phys. Res. A. 2013;703:33–37. [Google Scholar]
- 36.Lupi S., Nucara A., Kiskinova M. Performance of SISSI, the infrared beamline of the ELETTRA storage ring. J. Opt. Soc. Am. B. 2007;24:959–964. [Google Scholar]
- 37.Nečas D., Klapetek P. Gwyddion: an open-source software for SPM data analysis. Cent. Eur. J. Phys. 2012;10:181–188. [Google Scholar]
- 38.Graham J.S., McCullough B.R., De La Cruz E.M. Multi-platform compatible software for analysis of polymer bending mechanics. PLoS One. 2014;9:e94766. doi: 10.1371/journal.pone.0094766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barth A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta. 2007;1767:1073–1101. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Byler D.M., Susi H. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers. 1986;25:469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- 41.Yang H., Yang S., Yu S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015;10:382–396. doi: 10.1038/nprot.2015.024. [DOI] [PubMed] [Google Scholar]
- 42.Barth A., Zscherp C. What vibrations tell us about proteins. Q. Rev. Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 43.Zako T., Sakono M., Maeda M. Bovine insulin filaments induced by reducing disulfide bonds show a different morphology, secondary structure, and cell toxicity from intact insulin amyloid fibrils. Biophys. J. 2009;96:3331–3340. doi: 10.1016/j.bpj.2008.12.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mossuto M.F., Dhulesia A., Salvatella X. The non-core regions of human lysozyme amyloid fibrils influence cytotoxicity. J. Mol. Biol. 2010;402:783–796. doi: 10.1016/j.jmb.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou Y., Li Y., Ma G. Parallel β-sheet fibril and antiparallel β-sheet oligomer: new insights into amyloid formation of hen egg white lysozyme under heat and acidic condition from FTIR spectroscopy. J. Phys. Chem. B. 2013;117:4003–4013. doi: 10.1021/jp4003559. [DOI] [PubMed] [Google Scholar]
- 46.Konar M., Mathew A., Dasgupta S. Effect of silica nanoparticles on the amyloid fibrillation of lysozyme. ACS Omega. 2019;4:1015–1026. doi: 10.1021/acsomega.8b03169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulaj M., Foley J., Muschol M. Amyloid oligomers and protofibrils, but not filaments, self-replicate from native lysozyme. J. Am. Chem. Soc. 2014;136:8947–8956. doi: 10.1021/ja502529m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkataramani S., Truntzer J., Coleman D.R. Thermal stability of high concentration lysozyme across varying pH: a Fourier Transform Infrared study. J. Pharm. Bioallied Sci. 2013;5:148–153. doi: 10.4103/0975-7406.111821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matheus S., Friess W., Mahler H.-C. FTIR and nDSC as analytical tools for high-concentration protein formulations. Pharm. Res. 2006;23:1350–1363. doi: 10.1007/s11095-006-0142-8. [DOI] [PubMed] [Google Scholar]
- 50.Meersman F., Heremans K. Temperature-induced dissociation of protein aggregates: accessing the denatured state. Biochemistry. 2003;42:14234–14241. doi: 10.1021/bi035623e. [DOI] [PubMed] [Google Scholar]
- 51.Piccirilli F., Plotegher N., Bubacco L. High-pressure-driven reversible dissociation of α-synuclein fibrils reveals structural hierarchy. Biophys. J. 2017;113:1685–1696. doi: 10.1016/j.bpj.2017.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiti F., Webster P., Dobson C.M. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl. Acad. Sci. USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villegas V., Zurdo J., Serrano L. Protein engineering as a strategy to avoid formation of amyloid fibrils. Protein Sci. 2000;9:1700–1708. doi: 10.1110/ps.9.9.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fändrich M., Forge V., Diekmann S. Myoglobin forms amyloid fibrils by association of unfolded polypeptide segments. Proc. Natl. Acad. Sci. USA. 2003;100:15463–15468. doi: 10.1073/pnas.0303758100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fändrich M., Dobson C.M. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 2002;21:5682–5690. doi: 10.1093/emboj/cdf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zandomeneghi G., Krebs M.R.H., Fändrich M. FTIR reveals structural differences between native beta-sheet proteins and amyloid fibrils. Protein Sci. 2004;13:3314–3321. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez C., Miti T., Ullah G. Mechanism of fibril and soluble oligomer formation in amyloid beta and hen egg white lysozyme proteins. J. Phys. Chem. B. 2019;123:5678–5689. doi: 10.1021/acs.jpcb.9b02338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nielsen L., Frokjaer S., Brange J. Studies of the structure of insulin fibrils by Fourier transform infrared (FTIR) spectroscopy and electron microscopy. J. Pharm. Sci. 2001;90:29–37. doi: 10.1002/1520-6017(200101)90:1<29::aid-jps4>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 59.Bouchard M., Zurdo J., Robinson C.V. Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 2000;9:1960–1967. doi: 10.1110/ps.9.10.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lomont J.P., Ostrander J.S., Zanni M.T. Not all β-sheets are the same: amyloid infrared spectra, transition dipole strengths, and couplings investigated by 2D IR spectroscopy. J. Phys. Chem. B. 2017;121:8935–8945. doi: 10.1021/acs.jpcb.7b06826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmick S.D., Weliky D.P. Major antiparallel and minor parallel β sheet populations detected in the membrane-associated human immunodeficiency virus fusion peptide. Biochemistry. 2010;49:10623–10635. doi: 10.1021/bi101389r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tremmel S., Beyermann M., Fabian H. 13C-labeled tyrosine residues as local IR probes for monitoring conformational changes in peptides and proteins. Angew. Chem. Int.Engl. 2005;44:4631–4635. doi: 10.1002/anie.200500547. [DOI] [PubMed] [Google Scholar]
- 63.Reinstädler D., Fabian H., Naumann D. Refolding of thermally and urea-denatured ribonuclease A monitored by time-resolved FTIR spectroscopy. Biochemistry. 1996;35:15822–15830. doi: 10.1021/bi961810j. [DOI] [PubMed] [Google Scholar]
- 64.Berthomieu C., Hienerwadel R. Vibrational spectroscopy to study the properties of redox-active tyrosines in photosystem II and other proteins. Biochim. Biophys. Acta. 2005;1707:51–66. doi: 10.1016/j.bbabio.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 65.Chirgadze Y.N., Fedorov O.V., Trushina N.P. Estimation of amino acid residue side-chain absorption in the infrared spectra of protein solutions in heavy water. Biopolymers. 1975;14:679–694. doi: 10.1002/bip.1975.360140402. [DOI] [PubMed] [Google Scholar]
- 66.Zscherp C., Aygün H., Mäntele W. Effect of proline to alanine mutation on the thermal stability of the all-β-sheet protein tendamistat. Biochim. Biophys. Acta. 2003;1651:139–145. doi: 10.1016/s1570-9639(03)00263-2. [DOI] [PubMed] [Google Scholar]
- 67.Gabashvili I.S., Menikh A., Fragata M. Protein structure of photosystem II studied by FT-IR spectroscopy. Effect of digalactosyldiacylglycerol on the tyrosine side chain residues. J. Mol. Struct. 1998;444:123–133. [Google Scholar]
- 68.Fabian H., Schultz C., Naumann D. Impact of point mutations on the structure and thermal stability of ribonuclease T1 in aqueous solution probed by Fourier transform infrared spectroscopy. Biochemistry. 1994;33:10725–10730. doi: 10.1021/bi00201a021. [DOI] [PubMed] [Google Scholar]
- 69.Arrondo J.L.R., Muga A., Goñi F.M. Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog. Biophys. Mol. Biol. 1993;59:23–56. doi: 10.1016/0079-6107(93)90006-6. [DOI] [PubMed] [Google Scholar]
- 70.Shashilov V., Xu M., Lednev I.K. Probing a fibrillation nucleus directly by deep ultraviolet Raman spectroscopy. J. Am. Chem. Soc. 2007;129:6972–6973. doi: 10.1021/ja070038c. [DOI] [PubMed] [Google Scholar]
- 71.Chi Z., Asher S.A. UV resonance Raman determination of protein acid denaturation: selective unfolding of helical segments of horse myoglobin. Biochemistry. 1998;37:2865–2872. doi: 10.1021/bi971161r. [DOI] [PubMed] [Google Scholar]
- 72.Kurouski D., Van Duyne R.P., Lednev I.K. Exploring the structure and formation mechanism of amyloid fibrils by Raman spectroscopy: a review. Analyst (Lond.) 2015;140:4967–4980. doi: 10.1039/c5an00342c. [DOI] [PubMed] [Google Scholar]
- 73.Maiti N.C., Apetri M.M., Anderson V.E. Raman spectroscopic characterization of secondary structure in natively unfolded proteins: α-synuclein. J. Am. Chem. Soc. 2004;126:2399–2408. doi: 10.1021/ja0356176. [DOI] [PubMed] [Google Scholar]
- 74.JiJi R.D., Balakrishnan G., Spiro T.G. Intermediacy of poly(L-proline) II and beta-strand conformations in poly(L-lysine) beta-sheet formation probed by temperature-jump/UV resonance Raman spectroscopy. Biochemistry. 2006;45:34–41. doi: 10.1021/bi051507v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simpson J.V., Balakrishnan G., Jiji R.D. MCR-ALS analysis of two-way UV resonance Raman spectra to resolve discrete protein secondary structural motifs. Analyst (Lond.) 2009;134:138–147. doi: 10.1039/b814392g. [DOI] [PubMed] [Google Scholar]
- 76.Xu M., Ermolenkov V.V., Lednev I.K. Hen egg white lysozyme fibrillation: a deep-UV resonance Raman spectroscopic study. J. Biophotonics. 2008;1:215–229. doi: 10.1002/jbio.200710013. [DOI] [PubMed] [Google Scholar]
- 77.Xu M., Shashilov V., Lednev I.K. Probing the cross-β core structure of amyloid fibrils by hydrogen-deuterium exchange deep ultraviolet resonance Raman spectroscopy. J. Am. Chem. Soc. 2007;129:11002–11003. doi: 10.1021/ja073798w. [DOI] [PubMed] [Google Scholar]
- 78.Sikirzhytski V., Topilina N.I., Lednev I.K. Fibrillation mechanism of a model intrinsically disordered protein revealed by 2D correlation deep UV resonance Raman spectroscopy. Biomacromolecules. 2012;13:1503–1509. doi: 10.1021/bm300193f. [DOI] [PubMed] [Google Scholar]
- 79.Chi Z., Chen X.G., Asher S.A. UV resonance Raman-selective amide vibrational enhancement: quantitative methodology for determining protein secondary structure. Biochemistry. 1998;37:2854–2864. doi: 10.1021/bi971160z. [DOI] [PubMed] [Google Scholar]
- 80.Chi Z., Asher S.A. Ultraviolet resonance Raman examination of horse apomyoglobin acid unfolding intermediates. Biochemistry. 1999;38:8196–8203. doi: 10.1021/bi982654e. [DOI] [PubMed] [Google Scholar]
- 81.Kurouski D., Washington J., Lednev I.K. Disulfide bridges remain intact while native insulin converts into amyloid fibrils. PLoS One. 2012;7:e36989. doi: 10.1371/journal.pone.0036989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kurouski D., Deckert-Gaudig T., Lednev I.K. Structure and composition of insulin fibril surfaces probed by TERS. J. Am. Chem. Soc. 2012;134:13323–13329. doi: 10.1021/ja303263y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kurouski D., Lauro W., Lednev I.K. Amyloid fibrils are “alive”: spontaneous refolding from one polymorph to another. Chem. Commun. (Camb.) 2010;46:4249–4251. doi: 10.1039/b926758a. [DOI] [PubMed] [Google Scholar]
- 84.Hildebrandt P.G., Copeland R.A., Prendergast F.G. Tyrosine hydrogen-bonding and environmental effects in proteins probed by ultraviolet resonance Raman spectroscopy. Biochemistry. 1988;27:5426–5433. doi: 10.1021/bi00415a007. [DOI] [PubMed] [Google Scholar]
- 85.Xu M., Ermolenkov V.V., Lednev I.K. Lysozyme fibrillation: deep UV Raman spectroscopic characterization of protein structural transformation. Biopolymers. 2005;79:58–61. doi: 10.1002/bip.20330. [DOI] [PubMed] [Google Scholar]
- 86.Miura T., Takeuchi H., Harada I. Characterization of individual tryptophan side chains in proteins using Raman spectroscopy and hydrogen-deuterium exchange kinetics. Biochemistry. 1988;27:88–94. doi: 10.1021/bi00401a015. [DOI] [PubMed] [Google Scholar]
- 87.Walsh M.A., Schneider T.R., Wilson K.S. Refinement of triclinic hen egg-white lysozyme at atomic resolution. Acta Crystallogr. D Biol. Crystallogr. 1998;54:522–546. doi: 10.1107/s0907444997013656. [DOI] [PubMed] [Google Scholar]
- 88.Takeuchi H. Raman structural markers of tryptophan and histidine side chains in proteins. Biopolymers. 2003;72:305–317. doi: 10.1002/bip.10440. [DOI] [PubMed] [Google Scholar]
- 89.Takeuchi H., Ohtsuka Y., Harada I. Ultraviolet resonance Raman study on the binding mode of enkephalin to phospholipid membranes. J. Am. Chem. Soc. 1992;114:5321–5328. [Google Scholar]
- 90.Jakubek R.S., Handen J., Lednev I.K. Ultraviolet resonance Raman spectroscopic markers for protein structure and dynamics. Trends Analyt. Chem. 2018;103:223–229. doi: 10.1016/j.trac.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Asher S.A., Larkin P.J., Teraoka J. Ultraviolet resonance Raman and absorption difference spectroscopy of myoglobins: titration behavior of individual tyrosine residues. Biochemistry. 1991;30:5944–5954. doi: 10.1021/bi00238a019. [DOI] [PubMed] [Google Scholar]
- 92.Chi Z., Asher S.A. UV Raman determination of the environment and solvent exposure of Tyr and Trp residues. J. Phys. Chem. B. 1998;102:9595–9602. [Google Scholar]
- 93.Chaari A., Fahy C., Rholam M. Insights into kinetics of agitation-induced aggregation of hen lysozyme under heat and acidic conditions from various spectroscopic methods. PLoS One. 2015;10:e0142095. doi: 10.1371/journal.pone.0142095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rosario-Alomar M.F., Quiñones-Ruiz T., Lednev I.K. Hydrogen sulfide inhibits amyloid formation. J. Phys. Chem. B. 2015;119:1265–1274. doi: 10.1021/jp508471v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Frare E., Polverino De Laureto P., Fontana A. A highly amyloidogenic region of hen lysozyme. J. Mol. Biol. 2004;340:1153–1165. doi: 10.1016/j.jmb.2004.05.056. [DOI] [PubMed] [Google Scholar]
- 96.Hernández B., Coïc Y.M., Ghomi M. All characteristic Raman markers of tyrosine and tyrosinate originate from phenol ring fundamental vibrations. J. Raman Spectrosc. 2016;47:210–220. [Google Scholar]
- 97.Hernández B., Pflüger F., Ghomi M. Characteristic Raman lines of phenylalanine analyzed by a multiconformational approach. J. Raman Spectrosc. 2013;44:827–833. [Google Scholar]
- 98.Rava R.P., Spiro T.G. Resonance enhancement in the ultraviolet Raman spectra of aromatic amino acids. J. Phys. Chem. 1985;89:1856–1861. [Google Scholar]
- 99.Ivanova M.I., Sievers S.A., Eisenberg D. Molecular basis for insulin fibril assembly. Proc. Natl. Acad. Sci. USA. 2009;106:18990–18995. doi: 10.1073/pnas.0910080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Whittingham J.L., Scott D.J., Guy Dodson G. Insulin at pH 2: structural analysis of the conditions promoting insulin fibre formation. J. Mol. Biol. 2002;318:479–490. doi: 10.1016/S0022-2836(02)00021-9. [DOI] [PubMed] [Google Scholar]
- 101.Tito P., Nettleton E.J., Robinson C.V. Dissecting the hydrogen exchange properties of insulin under amyloid fibril forming conditions: a site-specific investigation by mass spectrometry. J. Mol. Biol. 2000;303:267–278. doi: 10.1006/jmbi.2000.4142. [DOI] [PubMed] [Google Scholar]
- 102.Porzoor A., Alford B., Macreadie I. Anti-amyloidogenic properties of some phenolic compounds. Biomolecules. 2015;5:505–527. doi: 10.3390/biom5020505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arnaudov L.N., de Vries R. Thermally induced fibrillar aggregation of hen egg white lysozyme. Biophys. J. 2005;88:515–526. doi: 10.1529/biophysj.104.048819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ehrnhoefer D.E., Bieschke J., Wanker E.E. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 105.Woods L.A., Platt G.W., Radford S.E. Ligand binding to distinct states diverts aggregation of an amyloid-forming protein. Nat. Chem. Biol. 2011;7:730–739. doi: 10.1038/nchembio.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tokunaga Y., Sakakibara Y., Sugimoto Y. Analysis of core region from egg white lysozyme forming amyloid fibrils. Int. J. Biol. Sci. 2013;9:219–227. doi: 10.7150/ijbs.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Flieger J., Tatarczak-Michalewska M., Blicharska E. Characterization of the cis/trans isomerization of resveratrol by high-performance liquid chromatography. Anal. Lett. 2017;50:294–303. [Google Scholar]
- 108.Hong Z., Asher S.A. Dependence of Raman and resonance Raman intensities on sample self-absorption. Appl. Spectrosc. 2015;69:75–83. doi: 10.1366/14-07531. [DOI] [PubMed] [Google Scholar]
- 109.Chen Y.-Y., Xiao L., Wang P. Biological relevance of the interaction between resveratrol and insulin. Food Biophys. 2013;8:282–289. [Google Scholar]
- 110.Wang L., Berne B.J., Friesner R.A. On achieving high accuracy and reliability in the calculation of relative protein-ligand binding affinities. Proc. Natl. Acad. Sci. USA. 2012;109:1937–1942. doi: 10.1073/pnas.1114017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang M., Jiji R.D. Resolution of localized small molecule-Aβ interactions by deep-ultraviolet resonance Raman spectroscopy. Biophys. Chem. 2011;158:96–103. doi: 10.1016/j.bpc.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 112.Mishra N.K., Krishna Deepak R.N.V., Verma S. Controlling in vitro insulin amyloidosis with stable peptide conjugates: a combined experimental and computational study. J. Phys. Chem. B. 2015;119:15395–15406. doi: 10.1021/acs.jpcb.5b08215. [DOI] [PubMed] [Google Scholar]
- 113.Gong H., He Z., Huang K. Effects of several quinones on insulin aggregation. Sci. Rep. 2014;4:5648. doi: 10.1038/srep05648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carbonaro M., Ripanti F., Nucara A. Human insulin fibrillogenesis in the presence of epigallocatechin gallate and melatonin: structural insights from a biophysical approach. Int. J. Biol. Macromol. 2018;115:1157–1164. doi: 10.1016/j.ijbiomac.2018.04.134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.