Abstract
Vaccination of the global population against COVID-19 is a great scientific, logistical, and moral challenge. Despite the rapid development and authorization of several full-length Spike (S) protein vaccines, the global demand outweighs the current supply and there is a need for safe, potent, high-volume, affordable vaccines that can fill this gap, especially in low- and middle-income countries. Whether SARS-CoV-2 S-protein receptor-binding domain (RBD)-based vaccines could fill this gap has been debated, especially with regards to its suitability to protect against emerging viral variants of concern. Given a predominance for elicitation of neutralizing antibodies (nAbs) that target RBD following natural infection or vaccination, a key biomarker of protection, there is merit for selection of RBD as a sole vaccine immunogen. With its high-yielding production and manufacturing potential, RBD-based vaccines offer an abundance of temperature-stable doses at an affordable cost. In addition, as the RBD preferentially focuses the immune response to potent and recently recognized cross-protective determinants, this domain may be central to the development of future pan-sarbecovirus vaccines. In this study, we review the data supporting the non-inferiority of RBD as a vaccine immunogen compared to full-length S-protein vaccines with respect to humoral and cellular immune responses against both the prototype pandemic SARS-CoV-2 isolate and emerging variants of concern.
Subject terms: Biological sciences, Infectious diseases
Introduction
Unprecedented progress has been made in the design, development, production, and distribution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines to address the coronavirus disease 2019 (COVID-19) pandemic. COVAX, a global initiative aimed at equitable access to COVID-19 vaccines led by the World Health Organization (WHO), the Coalition for Epidemic Preparedness Innovations and Gavi, The Vaccine Alliance, has delivered over 370 million doses to 144 countries and territories (https://www.unicef.org/supply/covid-19-vaccine-market-dashboard). Despite these remarkable achievements, there are serious challenges threatening the initial target delivery of 2 billion doses in 2021, such as raw material shortages, manufacturing delays, safety signals, the emergence of multiple viral variants of concern and interest (VOC/VOI)1,2, and unequal procurement of vaccines by high-income countries. Ultimately, these challenges may result in a significant shortfall and delay in vaccine being made available to low- and middle-income countries (LMIC).
Most of the COVID-19 vaccines currently available or soon to be available to COVAX—including adenovirus-vectored, mRNA, and protein-subunit vaccines—use antigens based upon the SARS-CoV-2 full-length Spike (S) protein. Figure 1 illustrates the S-protein in its trimeric prefusion conformation showing the receptor-binding domain (RBD) on each protomer at the apex, the N-terminal domain (NTD) and the conserved S2 stalk region3,4. Vaccine approaches using just the RBD region as an antigen have the advantage of focusing immunity to key protective determinants. As an immunogen, the RBD shares multiple positive attributes with the parental full-length S-protein (Table 1), as well as potential production and manufacturing advantages such as delivery of abundant5, temperature-stable6,7 vaccine doses at an affordable cost, critical factors for distributing vaccine to LMIC. Following several successful preclinical proof-of-concept studies, RBD candidates have been advanced into clinical testing utilizing multiple vaccine platforms (Table 2). With the potential for making billions of doses of RBD vaccines available, its manufacture at single-site production facilities by experienced developing country vaccine manufacturers (DCVM) will obviate the need for numerous tech-transfers and minimize the risk of delays. This will be especially important in case there is a need to update the vaccine based on epidemiological trends, as is the case for seasonal influenza vaccines.
Fig. 1. Structure of the SARS-CoV-2 S-protein in the trimeric prefusion conformation.
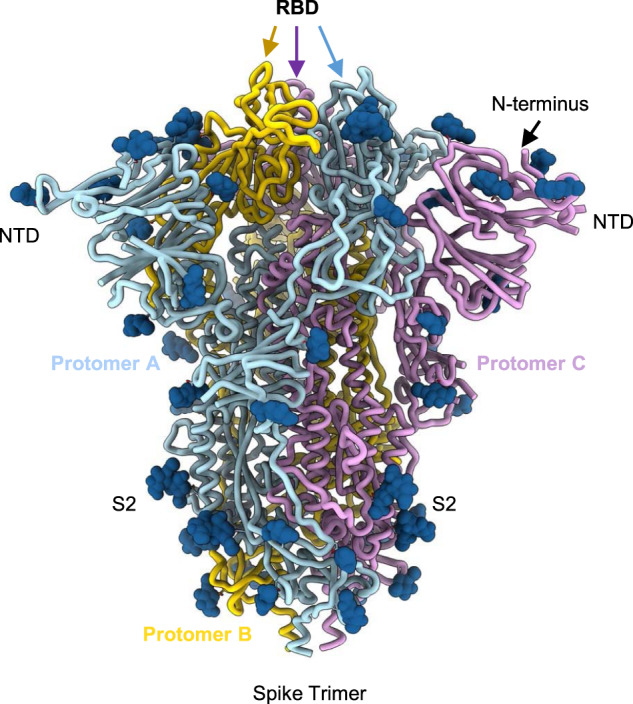
The structure of the S-protein trimer was modeled based on PDB 7LXY37. The three protomers (A, B, and C) are colored in cyan, yellow, and lilac, respectively. Structural components indicated include the NTD, the RBD, the N terminus, and the S2 domain.
Table 1.
Properties of RBD-based vaccine immunogens compared to full-length S-protein immunogens.
| Properties | RBD | S-protein | Comments |
|---|---|---|---|
| Structure | 3° | 4° | Tertiary structure recognized by conformational nAb |
| Neutralizing antibody titer | High | High | >90% Target RBD but Spike protein offers anti-NTD and anti-S2 nAb |
| RBD epitopes | Yes | Yes | RBD epitopes appear to undergo convergent evolution and cross-protect against CoV-2 variants and other sarbecoviruses. |
| NTD epitopes | No | Yes | NTD is showing deletions, insertions, and divergent substitutions |
| Neutralizing : binding antibody ratio | High | Medium | Induction of nAb that contribute to efficacy (CoP) favors an immune-focused strategy |
| CD4+ epitopes | Medium | High | RBD-specific immunity can be augmented by multimeric display (virus-like particles) and adjuvants |
| CD8+ epitopes | Low | Low | Subunit approaches (S-protein and RBD) appear to be devoid of CD8+ T-cell responses. S-protein offers better coverage for cell-mediated immunity (CMI). |
| B-cell memory | High | High | RBD-specific memory B cells show evidence of somatic hypermutation, which increases breadth of neutralization |
| Antibody persistence | Yes | Yes | Demonstrated >6 months (e.g., SK bioscience RBD-np and Pfizer-BioNTech S-protein vaccines) |
| NHP efficacy | High | High | Proof-of-concept demonstrated against upper and lower respiratory tract infection and disease |
| Clinical efficacy | Yes | Yes | 15 S-protein-based vaccines have reported ≥50% efficacy, with variations depending on the dominant viral variant. News reports of >90% for two Cuban RBD-based vaccines. |
| Supply volume | High | Medium | COVAX has negotiated favorable access terms for RBD vaccine candidates |
| Cost | Low | Medium | COGs low as produced by developing country vaccine manufacturers |
Table 2.
RBD-based vaccines in clinical development.
| Developer and vaccine | Manufacturing platform | Antigen and adjuvant | Dosing schedule (μg RBD) | Current clinical phase | Neutralizing antibody (vaccine/HCS) | References and Clinical Trial registration number |
|---|---|---|---|---|---|---|
| Adimmune AdCOVID | Baculovirus | RBDa + Alum | Phase 1 | NCT04522089 | ||
| Akston Biosciences AKS-452 | CHO | RBDa–Fc fusion protein | 1 Or 2 doses: 22.5, 45, or 90 μg | Phase 1/2 | NCT04681092 | |
| Biological E RBD219-N1C1 | Pichia | RBD331–549 + Alum/CpG | 2 Doses: 10 or 25 μg | Phase 2/3 | 23-Fold higher in mice | 5,88,89, CTRI/2021/06/034014 |
| Center for Genetic Engineering and Biotechnology Abdala CIGB 66 | Yeast | RBD331–529 + Alum | 3 Doses: 50 μg | Authorized in Cuba citing 92.28% clinical efficacy | ~8-Fold higher in NHP | 113,114, RPCEC00000359 |
| Center for Genetic Engineering and Biotechnology Mambisa CIGB 669 | Yeast | RBD331–529 + HBV nucleocapsid | 3 Doses: 50 μg (intranasal) or 2 doses in combination with CIGB 66 | Phase 2 | RPCEC00000345 | |
| Covaxx UB-612 | CHO | RBD340–359-Fc + 6 peptides + CpG/Alum | Phase 2 | ≥50-Fold higher in guinea pig (qNeu ELISA) | 86, NCT04773067 | |
| EuBiologics EuCorVac-19 | CHO | RBD319–541 + MPLA lipid nanospheres | 2 Doses 10 or 20 μg | Phase 1/2 | ~5-Fold higher in mice | 111, NCT04783311 |
| Finlay Vaccine Institute Soberana 02 | Baculovirus | Tetanus toxoid conjugated RBDa-Alum | 2 Doses 25 μg followed by 50 μg RBD-dimer with alum (FINLAY-FR-1A) | Authorized in Cuba citing 91.2% clinical efficacy | 115, RPCEC00000354 | |
| Hong Kong University LAIV | Hens’ eggs | RBDa (no adjuvant) | 2 Doses: 5 × 106– 5 × 107 | Phase 1 | NCT04809389 | |
| Pfizer/BioNTech BNT162b1 | mRNA | RBDa-trimer | 2 Doses: 30 μg | Phase 1 | 4.6-Fold higher in clinical study | 74,75,82,83, NCT04380701 |
| Serum Institute of India HBsAg-RBD-VLP | Pichia and Hansenula | RBD-VLP332–532 + Alum | 2 Doses: 1.8 or 2.1 μg | Phase 1/2 | >50-Fold higher for NHP, 4.6-fold higher for mice | 91,101,112, ACTRN12620000817943 |
| SK Bioscience GBP510 | CHO + E. coli | RBD328–531-nanoparticle + AS03 | 2 doses: 25 μg | Phase 3 | 5–8-Fold higher in clinical study | 26,78, NCT04750343 |
| Walvax/Abogen ARC0V-mRNA | mRNA | RBD319–541 | 15 μg | Phase 3 | 7, NCT04847102 | |
| West China Hospital SARS-Cov-2 RBD | Baculovirus | RBD319–545 + Alum | 2 Doses: 40 μg | Phase 3 | 87,117, NCT04904471 | |
| Anhui Zhifei Longcom Biopharmaceutical ZF2001 | CHO | RBD319–537 dimer + Alum | 3 Doses: 25 μg | Phase 3 | 2-Fold higher in clinical study | 76,77, NCT04646590 |
For current status of COVID-19 vaccine development, see https://covid19.trackvaccines.org/vaccines.
aRBD sequence data not available.
Available data are evaluated here from numerous preclinical and clinical studies, supporting the non-inferiority of RBD vaccine immunogens compared to full-length S-protein (Table 1), both with respect to eliciting homotypic and heterotypic cross-neutralizing antibody (nAb) responses and cross-protection against emerging variants.
Clinical data support nAb as a biomarker of protection
Vaccine-induced immunity and efficacy are being used to help establish a correlate of protection (CoP), a measurable immune response (often nAb titer) predictive of protection, as has been demonstrated for some licensed vaccines8,9. As of October 2021, both meta-analyses and prospective case–cohort sampling analyses have found a strong correlation between nAb titers and protection against COVID-1910–13. This is underpinned by passive immunization and preclinical vaccine efficacy studies in nonhuman primates (NHPs) that similarly support nAb as a CoP14–16.
Meta-analyses of numerous clinical candidates support the correlation between vaccine efficacy and nAb titer, regardless of vaccine platform. In seeking to establish a CoP for COVID-19 vaccines, both Earle et al.11 and Khoury et al.12 initially showed only a weak relationship between efficacy and reported nAb or binding antibody (bAb) titers11,12. This poor association could be explained by the high variation observed between the selected assays used by different studies17,18. This variability was partially mitigated by calibrating the assays against panels of human convalescent sera (HCS) run within the same study. When the titers were calibrated to HCS, a strong correlation between nAb and efficacy was observed, irrespective of the specific assay platform used (rank correlation ρ = 0.79). nAb calibrated against HCS accounted for up to 88.4% of the variation in efficacy observed across seven different vaccine studies and suggest that any vaccine, including RBD-based vaccines, which elicit significant amounts of nAb, is likely to afford protection. The correlation was further strengthened (rank correlation ρ = 0.86) when the analysis uncoupled the timing of the second dose and efficacy in the AstraZeneca ChAdOx trial13. A recent pre-publication investigating direct quantitative comparison of immune correlates results from the AZD12222 trial and the mRNA-1273 Coronavirus Efficacy (COVE) trial shows remarkably consistent results for nAb10. This direct comparison was enabled by calibrating assays and reporting results against the WHO International Standard17, steps that are essential for comparing clinical immunological data and by now should be the standard in the field19. The same pre-publication also noted that 68.5% of vaccine efficacy was mediated by the day 29 cID50 titer in the mRNA-1272 COVE trial10. The totality of the data support neutralization titer as a potential surrogate endpoint in future clinical trials of mRNA-1273. These statistical efforts to identify a CoP for COVID-19 vaccines are being increasingly recognized by vaccine manufacturers and regulatory agencies. The UK Medicines and Healthcare Products Regulatory Agency approved a pivotal Phase 3 study design to support vaccine authorization comparing the immunogenicity of a candidate vaccine to the immunogenicity of an approved vaccine with clinical evidence of efficacy, using nAb as a biomarker of protection20. Additional clinical trial-specific analyses expressed in International Units are awaited, as well as assessment of the durability of this correlation over time and impact of circulating VOC.
RBD is a key target of nAb
During the acute phase of natural infection, there is a rapid onset of protective immunity, typified by serum IgG nAb that targets RBD21. Functional RBD-specific responses at mucosal surfaces have also been noted, with secretory IgA shown to offer potent neutralization22, highlighting a preference for both systemic and mucosal responses. Depletion experiments demonstrate that 90% or more of the neutralizing activity present in the plasma of convalescent individuals is accounted for by RBD-specific nAb23–25. Although RBD-specific serum IgG titers were observed to wane following infection (with a half-life of 49 days), nAb titers and avidity increased over time for some individuals, consistent with affinity maturation23.
A second RBD depletion study of vaccinee sera from a Phase 1 clinical study, where participants were immunized with mRNA-1273, which includes a full prefusion S-protein trimer antigen, supported up to 99% of neutralizing activity to target RBD25. These results indicate that neutralizing activity in both natural and vaccine-induced immunity predominantly targets the RBD, even with a full spike trimer immunogen.
RBD is a key target for potent cross-neutralizing monoclonal antibodies
RBD exhibits tertiary structure and in its native conformation shows high affinity binding to the human ACE2 receptor (KD = 66 nM) and to conformational-dependent monoclonal antibodies (mAbs) such as CR3022 (KD = 56 nM) and S3094,26, a broadly cross-neutralizing sarbecoviruses mAb isolated from a convalescent patient27. Almost all antibodies with potent viral neutralizing activity (half maximal inhibitory concentration < 0.1 μg/ml) bind to RBD and many of them block interactions with the human ACE2 receptor23, which mediates viral entry into host cells (Fig. 2, ACE2-binding site of RBD outlined in black). In assessing convalescent patients, the most potent mAbs were found to bind to RBD and determined to block the RBD–ACE2 interaction at two specific sites, RBD-A (site Ia) and RBD-B (site Ib)28–31. Across several studies, the majority of the nAb polyclonal response (in some cases >90%) have been mapped to SARS-Cov-2 RBD, as has been the case with mAbs isolated from infected individuals23,28,32.
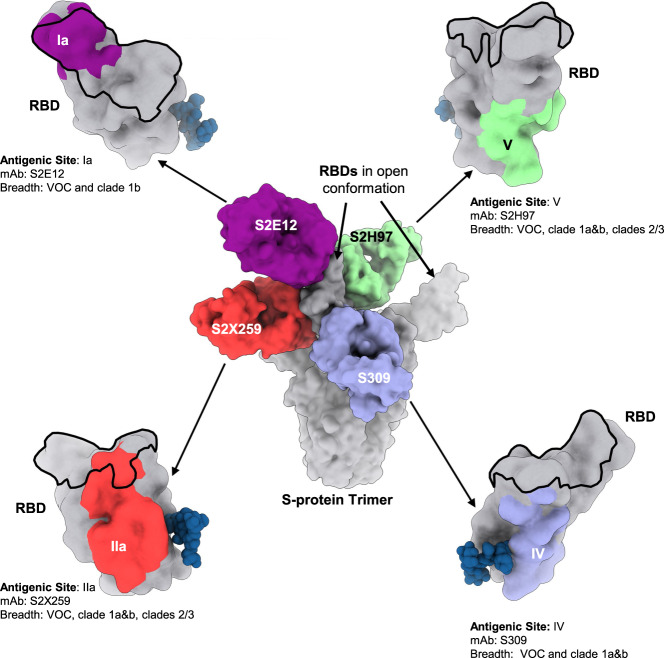
Fig. 2. Overview of SARS-CoV-2 RBD cross-neutralizing antigenic sites.
Center panel: composite model of the SARS-CoV-2 S-protein trimer (gray) with four distinct monoclonal antibodies (S2E12 in purple, S2X259 in red, S2H97 in green, and S309 in blue) bound to one RBD in the open conformation. Top and bottom, left and right panels: magnified model of RBD (gray) with the ACE2-binding site outlined in black and the respective mAb cognate epitopes indicated by color. In dark blue is a surface representation of the glycan at position N343, which is conserved across the sarbecovirus subgenus. Top left: antigenic site Ia (purple) is targeted by the S2E12 mAb, which neutralizes the VOC and clade 1b, SARS-CoV-2-related, sarbecoviruses29. Bottom left: antigenic site IIa (red) is targeted by the S2X259 mAb, which inhibits ACE2 binding, neutralizes the VOC and clades 1a&b SARS-CoV-2-related sarbecoviruses, and binds to clades 2/3 sarbecoviruses30. Bottom right: antigenic site IV (blue) is targeted by the S309 mAb, which neutralizes the VOC and clades 1a&b SARS-CoV-2-related sarbecoviruses27,35. Top right: antigenic site V (green) is targeted by S2H97, which neutralizes the VOC and clades 1a&b SARS-CoV-2-related sarbecoviruses, and binds to clades 2/3 sarbecoviruses34.
More recently, many protective epitopes have been footprinted to RBD, both within and outside of the ACE2 receptor-binding motif, with many recognized by mAbs able to cross-neutralize variants of the current pandemic, as well as other sarbecoviruses29,30,33,34 (Fig. 2). Of significance, is the identification of mAbs such as S309, newly identified S2H97 (determined to bind a novel RBD antigenic site designated site V), and S2E12 that bind RBD at discrete sites, and which are conserved across many clades of the sarbecoviruses27,34 (Fig. 2).
Of relevance to selection of the RBD as a target immunogen, the S309-neutralizing mAb recognizes an epitope containing a glycan at position N343, which is conserved across the sarbecovirus subgenus. It binds to multiple conformational states of the RBD presented from the S-protein (both open and closed) and mediates Fc-dependent effector functions such as antibody-dependent cellular cytotoxicity, supporting alternate mechanisms of protection. Its epitope resides on the opposite side of the ACE2 receptor-binding motif, which may explain its synergy with ACE2-inhibiting mAbs. S309 (evaluated in the clinic as VIR-7831) cross-neutralizes SARS-CoV-1 and SARS-CoV-2, and other sarbecoviruses27, and has been demonstrated to potently neutralize the Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1) and Epsilon (B.1.427/B.1.429) VOC/VOI, and protect Syrian hamsters against SARS-CoV-2 challenge in vivo35–37.
Based on results of a Phase 3 efficacy study, where the VIR-7831 mAb demonstrated an 85% reduction in hospitalization or death in at-risk individuals, the drug received Emergency Use Authorization from the US Food and Drug Admisnistration on 26 May 202138. Potent mAbs to other RBD-specific sites have also been in clinical development (REGN10889 and LY-COVV555), although at least one has been shown to significantly lose potency against the VOC following evolutionary changes in the RBD.
To date, several RBD bAbs with potent neutralizing activity have been demonstrated to be unaffected by the RBD mutations seen in the newly circulating Alpha (B.1.1.7) and Beta (B.1.351) viral variants29,39–41 (for a list of the mutations in RBD, see Table 3). These anti-RBD mAbs target conserved epitopes within the RBD and are potentially attractive therapeutics based on their resistance profile and will be important for novel immunogen design.
Table 3.
Amino acid mutations in the RBD region of SARS-CoV-2 variants of concern.
| WHO Label | Pango lineage | Mutations in RBD region |
|---|---|---|
| Alpha | B.1.1.7 | N501Y, (E484K*), (S494P*) |
| Beta | B.1.351, B.1.351.2, B.1.351.3 | K417N, E484K, N501Y |
| Delta | B.1.617.2, AY.1, AY.2, AY.3 | L452R, T478K, (K417N*) |
| Gamma | P.1, P.1.1, P.1.2 | K417T, E484K, N501Y |
Parentheses and * indicate the mutation is found in some sequences but not all. Information adapted from https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html#Concern, current as of 13 August 2021.
NTD: a functional neutralization target
Full S-protein vaccine antigens all contain the NTD, a second neutralization target. Several NTD-specific mAbs isolated from convalescent memory B cells, as well mAbs isolated from plasmablasts following mRNA vaccination, have demonstrated potent neutralizing capabilities against the original Wuhan-1 pandemic virus37,42. Many of these NTD-targeting mAbs are non-neutralizing and have been shown to inhibit cell-to-cell fusion, activate alternate effector functions, and have been demonstrated to be protective from SARS-CoV-2 challenge.
The NTD is a site of prevalent and recurrent deletion regions, which vary by length and location43. In addition, the currently designated VOCs harbor multiple mutations in the NTD, suggesting ongoing selective pressure, with NTD highly divergent across the different variants and other sarbecoviruses1. As such, an S-protein immunogen risks being mismatched at the NTD site with another variant. One example is the Beta variant (B.1.351) having a deletion in the NTD, compared to the Gamma variant (P.1), which contains point mutations.
RBD may be undergoing convergent evolution
Global sequencing efforts have documented virus evolution that includes mutations specifically in the S-protein1,2,44. Mutations and deletions in both the RBD (Table 3) and NTD of S-protein are of concern for ACE2 receptor-binding interactions and overall neutralizing activity1,2. As infectivity is reliant upon receptor binding, it has been suggested that mutation of the RBD sequence may be constrained to some degree at the ACE2-binding interface, limiting escape at this site39.
In support of this theory, co-evolution of separate virus lineages around the globe (Alpha: lineage B.1.1.7 or 501Y.V1 in the UK; Beta: lineage B.1.351 or 501Y.V2 in the Republic of South Africa; and Gamma: lineage P.1 or 501Y.V3 in Brazil) appear to carry one or more of the same mutations in RBD: K417N/T, E484K, and N501Y (Table 3). Similar mutations were observed to occur in forced viral evolution experiments using convalescent or vaccinee serum; passage of prototype virus with high-titer neutralizing antisera yielded a mutant virus carrying similar NTD deletions and the RBD-specific E484K mutation45. Mutation at the E484 site was shown to impact virus neutralization of the ancestral B.1 virus, which has been noted as a concern for emerging variants44,46–48 (Table 3), although recent epidemiological trends show prevalence for the Delta (B.1.617.2) variant that lacks this mutation (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/).
Many mutations in RBD appear to have enhanced receptor-binding affinity, resulting in increased infectivity and transmission. Viral variants that emerge harboring mutations in S-protein have the potential to evade nAb responses from previous infection, mAb therapies, or immunity from prior vaccination. The restricted antigenic changes observed in the SARS-COV-2 RBD support a convergent evolution theory, potentially a result of host adaptation leading to enhanced infectivity, and highlights the need for more broadly cross-neutralizing immunity49. As a result bivalent vaccine approaches targeting two or more variants are currently being explored for S-protein and RBD-based vaccines, in hopes of achieving broader coverage and protection against VOC48,50. although the utility of a bivalent vaccine formulation must be considered in the context of evolving epidemiology and additional booster shots with the prototype vaccine.
RBD-specific memory B cells
Investigation of convalescent donor samples have shown that memory B-cell responses against the SARS-CoV-2 S-protein increase between 1 and 8 months after infection51,52. Analysis of convalescent donors demonstrated that RBD-specific IgG memory B-cell responses were rich in recurrent and clonally expanded antibody sequences, with 10–90% of S-protein-specific memory B cells recognizing the RBD domain51,52. These clonal memory B cells were determined to express antibodies with evidence of somatic hypermutation, with some having increased neutralization potency and resistance to RBD mutations. Individuals with even modest plasma-neutralizing activity have also been shown to harbor rare IgG memory B cells that produce potent SARS-CoV-2 neutralization Ab53. For certain antibody lineages, maturation enabled neutralization of circulating SARS-CoV-2 VOC and heterologous sarbecoviruses54.
Recent work has shown that vaccination is similarly able to induce robust memory. The mRNA vaccines (Pfizer BNT162b2 or Moderna mRNA-1273) efficiently primed memory B cells specific for full-length S-protein and RBD, and these were detectable in all subjects after the second vaccine dose55.
CD4+ and CD8+ T-cell immune responses to RBD
Current clinical data suggest, but do not yet definitively prove, that T cells can contribute to vaccine-mediated protection against COVID-19. Providing CD4 T cell help to drive robust nAb responses is one possible mechanism of protection. It is also plausible that CD4 and CD8 T-cell responses play a larger role in modulating disease severity than in preventing asymptomatic or mild infections56. The role of T cells may become more pronounced in the absence of adequate nAb, such as when overall titers are low57, or when facing variants with mutations in nAb epitopes. For example, 50.8% vaccine efficacy was seen when nAbs were below the level of detection in the mRNA-1272 COVE trial10. Likewise, the Janssen Ad26.COV2.S vaccine was 52% protective in a South African clinical trial, with many cases of Beta variant infection, even though it did not elicit high antibodies against the Beta variant58,59. In contrast, CD8 T-cell depletion studies of convalescent primates prior to SARS-CoV-2 rechallenge only partly abrogated protection14 and in humans, Novavax has reported clinical efficacy of 86.3% against Alpha (B.1.1.7) and 50% against Beta (B.1.351) variants in the absence of any prominent CD8+ responses60–62. Sinovac63–65, Sinopharm66, and Bharat67 have also reported clinical efficacy without eliciting notable CD8+ responses. Thus, CD8 T-cell responses may be beneficial but are not absolutely required for protection.
Due to the anticipated benefit of vaccine-induced T-cell immunity to SARS-COV-2, CD4 and CD8 T-cell epitope mapping using overlapping peptides has been performed across the full length of the S-protein. An evaluation of CD4 T-cell responses in humans reported a frequency of at least 20% focused on discrete regions of the S-protein, including the NTD, the C-terminal domain, the neighboring fusion protein region, and the RBD68,69. It was also reported that the sequences of most T-cell epitopes are unaffected by mutations in the SARS-COV-2 VOC, including the Beta (B.1.351) variant70. Consistent with this finding, CD4 and CD8 T-cell reactivity to S-protein in vaccinees were robust against the SARS-COV-2 variant mutations.
The RBD contains fewer immunodominant CD4 T-cell epitopes than the full-length S-protein. Despite this fact, there is ample evidence that sufficient numbers of T-cell epitopes are presented by the RBD to generate good CD4 responses and elevated levels of functionally nAb16,71. The identification of multiple common CD4 T-cell epitopes within the RBD has been independently verified using different T-cell epitope mapping approaches69,72,73. Clinical studies with RBD antigens support the assertion that sufficient T-cell help is provided following immunization (Table 2). Immunization with an mRNA-encoded RBD vaccine candidate showed the induction of nAb titers ranging from 0.7-fold (1 μg dose) to 4.6-fold (30 μg dose) higher than HCS74,75. Notably, induction of robust nAb titers by RBD was not specific to the mRNA platform. A protein-subunit RBD-dimer vaccine co-administered with Alum, was similarly able to drive nAb responses in humans76,77, as was an RBD array displayed from a synthetic nanoparticle vaccine adjuvanted with AS0378. In summary, data on immune responses elicited by infection and vaccination with a S-protein or RBD antigen provide evidence that a potent RBD vaccine will recruit ample T-cell help.
CD8 T-cell epitopes may be less critical in subunit vaccines that protect primarily via humoral immunity. Epitope mapping of CD8 T-cell reactivity did not reveal an immunodominant region in S-protein, with epitopes being roughly equally distributed along the sequence68. Similar to CD4 epitopes, by definition, the RBD subdomain of S-protein contains fewer CD8 T-cell epitopes than the full-length S-protein. Therefore, it is not clear whether RBD would contain adequate CD8 T-cell epitopes for a vaccine whose protection was based on eliciting CD8 T cells. In this context, the broader number of epitopes in the full S-protein may be advantageous.
Due to large variations in T-cell assay methodology and resultant data, cross-study comparisons of T-cell responses are difficult, if not impossible, to interpret79,80. Full appreciation of the effect of vaccine platform and antigen on CD4 and CD8 T-cell responses will require standardized peripheral blood mononuclear cell collection and assay protocols, as was established within the COVID-19 Prevention Network and the COVE trial. Interpretable comparison of vaccine-induced cell-mediated immunity is expected within the year.
Preclinical studies support RBD as a potent vaccine antigen
Preclinical studies have evaluated the protective efficacy of several RBD-specific monoclonals, as well as RBD as a protective immunogen. Investigators evaluating RBD as a vaccine candidate have delivered the antigen either as DNA81, mRNA7,74,82–85, viral vector86, soluble monomer or dimer6,22,87–93, a fusion protein nanoparticle16,26,94–98, or as a virus-like particle (VLP)91,99–102. Additional RBD-based vaccine candidates in clinical trials (Table 2) have not yet published their preclinical work. The RBD sequences selected from the S-glycoprotein as candidate vaccines encode an ~25 kDa (>200 aa) glycosylated protein103, produced using either mammalian26, baculovirus-infected insect cells92, or yeast-based production platforms5,88,89,91,104. In general, vaccination strategies have focused on parenteral immunization to elicit high-titer serum nAb, although some preclinical studies support intranasal delivery of S-protein in NHPs and hamsters using a live vector approach105, and RBD on chitosan nanoparticles in mice106, with evidence for a role of mucosal immunity in reducing viral load in both the upper and lower respiratory tract.
Prototype RBD-np vaccine
The Institute for Protein Design (University of Washington) SARS-CoV-2 vaccine consists of a self-assembling, two-component nanoparticle that displays 60 copies of RBD per nanoparticle26. The RBD-np (nanoparticle) is produced in mammalian cells and maintains the two functional N-linked glycosylation sites (N331 and N343), important for proper folding of protein and for antibody recognition26. A comparison of the glycosylation patterns between the RBD-np and the S-2P trimer was very similar at the two sites, both exhibiting complex glycan that were heavily fucosylated. The RBD-np candidate administered intramuscularly proved safe, was as immunogenic as a recently described next-generation prefusion S-protein trimer107, and protected mice and rhesus macaques from live virus challenge. SK bioscience is developing the RBD-np vaccine technology from the University of Washington and is currently in Phase III clinical testing of the prototype variant B.1 vaccine (GPB510) adjuvanted with AS0378.
In initial mouse studies, the RBD-np vaccine was shown to induce nAb titers tenfold higher than the prefusion-stabilized S-protein trimer (S-2P) despite a fivefold lower dose26. The RBD-np vaccine induced antibodies targeting multiple distinct epitopes on the RBD, elicited a higher nAb : bAb ratio than HCS, and was fully protective against challenge with mouse-adapted SARS-CoV-2. Immunization of humanized mice transgenic for the non-rearranged human antibody variable and constant region germline repertoire confirmed the ability of this candidate to mount functional humanized nAbs, superior to that observed using S-2P prefusion trimer26.
NHP studies compared adjuvants (head-to-head) for advancement of the leading formulations for clinical development. Groups of rhesus macaques were immunized twice with RBD-np, plus one of five adjuvants including AS03, Alum-CpG, Alum, AS37, and another oil-in-water emulsion16. Both pseudovirus and live virus neutralization assays against the B.1 virus supported elevated titers with the AS03, Alum-CpG, and Alum adjuvants. Following intranasal/intratracheal challenge, significant protection was observed against viral burden in both the upper respiratory tract (URT) and in the lungs using an established sub-genomic mRNA assay with E-gene-specific primers, as well as protection against disease using positron emission tomography–computed tomography. Vaccination through intramuscular administration achieved a significant reduction in viral burden in the URT, potentially supporting the ability of RBD-based vaccines to also limit transmission, with evidence that greater serum nAb titers is inversely correlated with protection in the upper airways.
Animals similarly immunized with RBD-np adjuvanted with AS03 were followed for 5 months as part of a durability study and determined to maintain neutralizing titers (>1 : 1000) and ACE2-blocking activity16. The RBD-np-specific plasmablast response was also measured, 4 days after the second immunization, with the magnitude of antigen-specific IgG-secreting cells in blood correlating with the observed nAb responses. Immunized macaques exhibited an RBD-specific cell-mediated immune response, which was dominated by IL-2 and/or tumor necrosis factor-α (TNF-α)-secreting CD4+ T cells, an apparent Th1/Th2-balanced response, with little evidence of a CD8+ T-cell response.
A correlates analysis supported nAb, both using a wild-type and pseudovirus assay, as the top statistically significant CoP in both the nasal and pharyngeal compartments. Nanoparticle-specific IL2+ and TNF+ CD4 T-cell responses also emerged as a statistically significant CoP, suggesting the nanoparticle itself may contribute T-cell help16.
In a follow-on NHP study16, the RBD-np adjuvanted with AS03 was compared to the stable prefusion S-protein trimer HexaPro, where an additional four (4) Proline substitutions have been introduced to increase thermal stability and yield of production in mammalian cells107. Their evaluation, both as a soluble protein and displayed separately on a similar nanoparticle, supported near-equivalent neutralization titers after two doses.
Analysis of cross-neutralizing activity against VOC with sera from NHPs immunized with RBD-np adjuvanted with AS03 or Alum supported potent cross-neutralization against the Alpha (B.1.1.7) variant and favored AS03 as a preferred adjuvant for neutralization of the Beta (B.1.351) virus16. In pseudovirus assays, RBD cross-neutralization titers were equivalent to HexaPro immune sera for both VOC assessed, whereas RBD-np elicited higher titers of Abs against all RBD antigenic sites evaluated compared to HexaPro immune sera97. In addition, the data suggest that epitopes outside of the RBD do not significantly contribute to neutralization of VOC. Another NHP study evaluated the contribution of a third dose of RBD-np adjuvanted with Addavax. Primates boosted 6 months after primary immunization further elevated their cross-nAb titers against the Alpha (B.1.117) and Beta (B.1.351) variants, with GMT titers > 4e3 in a pseudovirus assay97.
Vaccine researchers around the world have demonstrated preclinical immunogenicity of multimeric RBD vaccine candidates utilizing a variety of presentation methods including the following: ferritin nanoparticles94,96,98,108,109, single-component protein nanoparticles95, two-component protein nanoparticles either self-assembling26 or assembled via SpyTag/Catcher technology91,99,110, and VLPs102,111. Together, these studies support the assertion that RBD displayed on a particle and co-administered with a suitable adjuvant represents a viable vaccine strategy for SARS-CoV-2, including against VOC.
RBD-based vaccines in the clinic
RBD as an immunogen has been advanced to the clinic independently by many developers (Table 2). Some large vaccine manufacturers, Serum Institute of India112, Biological E5, and SK Bioscience16,26, are developing RBD candidate vaccines scalable to hundreds of millions of doses at single-site production facilities, obviating the need for tech-transfer.
To date, two RBD vaccines developed in Cuba have reported favorable clinical efficacy data via news outlets113. The Center for Genetic Engineering and Biotechnology announced 92.28% efficacy following three doses of its Abdala vaccine114 and the Finlay Vaccine Institute reported 62% efficacy after two doses of its Soberana 02 vaccine and 91.2% efficacy following a third heterologous boost with the RBD-dimer vaccine115. Scientific reports of the efficacy studies are anticipated shortly. Three additional RBD vaccines have reported positive clinical immunogenicity and compared performance to a panel of HCS for benchmarking purposes: the SK Bioscience protein-subunit RBD-np78, the Zhifei Longcom protein-subunit RBD-dimer with Alum vaccine76, and the BioNTech mRNA RBD-trimer BNT162b1 vaccine74,75,82,83. In a press release, SK Bioscience reported interim results of a Phase 1/2 clinical trial, stating that the GBP510 vaccine, a two-component nanoparticle displaying 60 copies of RBD adjuvanted with AS03, was safe, well-tolerated, and demonstrated 100% nAb seroconversion with GMTs five to eight times higher than that in HCS after two doses78. Following three doses of the Zhifei RBD-dimer ZF2001 vaccine, 97% of recipients seroconverted with nAb GMTs twofold higher than the HCS panel tested76. In addition, Zhifei have reported a limited reduction in neutralization (1.6-fold) against the Beta (B.1.351) variant77.
BioNTech and Pfizer evaluated four mRNA candidate vaccines in Phase 1/2 clinical trials, including a lipid nanoparticle-formulated nucleoside-modified mRNA that encoded the RBD fused to a T4 fibritin-derived foldon, resulting in a trimeric RBD, BNT162b174,75,82,83. BNT162b1 elicited robust CD4+ and CD8+ T-cell responses, and strong humoral immunity after two doses (ranging from 1 to 50 μg per dose), with levels above that observed for HCS (1.9-fold higher at 10 μg dose; 4.6-fold higher at 30 μg dose) and equivalent to responses to the full S-protein candidate, BNT162b274,75,82,83. The BNT162b2 candidate vaccine, transcribing full-length S-protein mRNA, was associated with a lower incidence and severity of systemic reactions, particularly in older adults, and therefore the RBD vaccine (BNT162b1) did not move forward clinically. The authors speculated that the difference in observed reactogenicity might be explained by the number of RNA molecules in 30 μg of BNT162b1 being approximately five times as high as that in 30 μg of BNT162b275.
When we apply the ratio of the vaccine-induced vs. convalescent sera-neutralizing titer from these three RBD vaccines to the correlate analyses10–12, the predicted vaccine efficacy against the ancestral virus is well above 90% for both the SK Bioscience GPB510 and the BioNTech/Pfizer BNT162b1 RBD vaccines, and ~90% for Zhifei’s ZF2001.
The ability of RBD-based vaccines to impact viral burden in the upper airways and limit community transmission is still to be studied. As for S-protein-based vaccines already deployed116, it is likely that as immunity declines over time and variants arise, vaccinated individuals may be more likely to become transiently infected and transmit to others. This puts a spotlight on the required induction and duration of elevated nAb responses to maximize protection.
Conclusions
SARS-CoV-2-related data from multiple vaccine researchers support nAb as the key biomarker of protection irrespective of vaccine platform, as evaluated in mice, hamsters, NHPs, and humans. The RBD of the S-protein displays the major functionally neutralizing epitopes and, when used as an immunogen, may preferentially focus the immune response to highly potent and cross-protective determinants. Isolated mAbs to these protective determinants have been shown to cross-neutralize the VOC and support selection of RBD as a pan-sarbecovirus vaccine target49,110. Nonclinical and clinical vaccine studies demonstrate the exceptional properties and non-inferiority of RBD as an immunogen compared to full-length S-protein (Table 1). Focusing immunity to RBD through targeted vaccination strategies may have an important impact on eliciting elevated cross-nAb titers, with subsequent protection against viral variants and potentially limiting community transmission.
Given (1) the potential manufacturing and cost advantages of RBD as a vaccine immunogen across different platform technologies5,112, and (2) the engagement of experienced DCVM partners that can deliver billions of vaccine doses, RBD vaccine candidates with non-inferior safety and immunogenicity should continue to be evaluated, to address a critical medical need and ensure equitable access of vaccine.
With waning immunity and regulatory discussions on potential booster doses, we speculate that the availability of an RBD-based vaccine strategy that targets immunity to key cross-protective determinants may be a considerable advantage, a vaccination strategy potentially for both the previously immunized and those naturally infected.
Acknowledgements
We thank Drs. Lynda Stuart (Bill and Melinda Gates Foundation), David Veesler (University of Washington), Neil King (University of Washington), Bali Pulendran (Stanford University), Andrew Fiore-Gartland (Fred Hutchinson Cancer Research Center), and Galit Alter (Ragon Institute) for helpful discussions.
Author contributions
H.K., J.M.S., K.M., and D.W.V. contributed to the design and drafting of the manuscript. N.J. and I.K.Y. provided content and critical reviews.
Data availability
No data were generated for the review article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Harvey WT, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. Tracking SARS-CoV-2 Variants, www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (2021).
- 3.Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls AC, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J, et al. Process development and scale-up optimization of the SARS-CoV-2 receptor binding domain-based vaccine candidate, RBD219-N1C1. Appl. Microbiol. Biotechnol. 2021;105:4153–4165. doi: 10.1007/s00253-021-11281-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malladi SK, et al. Design of a highly thermotolerant, immunogenic SARS-CoV-2 spike fragment. J. Biol. Chem. 2021;296:100025. doi: 10.1074/jbc.RA120.016284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang NN, et al. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182:1271–1283 e1216. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plotkin SA. Updates on immunologic correlates of vaccine-induced protection. Vaccine. 2020;38:2250–2257. doi: 10.1016/j.vaccine.2019.10.046. [DOI] [PubMed] [Google Scholar]
- 9.Plotkin SA. Correlates of protection induced by vaccination. Clin. Vaccin. Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert, P. B. et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy trial. Preprint at medRxivhttps://www.medrxiv.org/content/10.1101/2021.08.09.21261290v4 (2021). [DOI] [PMC free article] [PubMed]
- 11.Earle KA, et al. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39:4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khoury DS, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 13.Voysey M, et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397:881–891. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMahan K, et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbett KS, et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arunachalam PS, et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021;594:253–258. doi: 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- 17.Mattiuzzo, G. Establishment of the WHO International Standard and Reference Panel for Anti-SARS-CoV-2 Antibody, https://cdn.who.int/media/docs/default-source/biologicals/ecbs/bs-2020-2403-sars-cov-2-ab-ik-17-nov-2020_4ef4fdae-e1ce-4ba7-b21a-d725c68b152b.pdf?sfvrsn=662b46ae_8&download=true (WHO, 2020).
- 18.Sholukh, A. M. et al. Evaluation of cell-based and surrogate SARS-CoV-2 neutralization assays. J. Clin. Microbiol. Jcm0052721, 10.1128/jcm.00527-21 (2021). [DOI] [PMC free article] [PubMed]
- 19.Kristiansen PA, et al. WHO International Standard for anti-SARS-CoV-2 immunoglobulin. Lancet. 2021;397:1347–1348. doi: 10.1016/S0140-6736(21)00527-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valneva. Valneva Initiates Phase 3 Clinical Trial for its Inactivated, Adjuvanted COVID-19 Vaccine Candidate, VLA2001, https://valneva.com/press-release/valneva-reports-positive-phase-1-2-data-for-its-inactivated-adjuvanted-covid-19-vaccine-candidate-vla2001/ (2021).
- 21.Suthar MS, et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep. Med. 2020;1:100040. doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021;13:eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piccoli L, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e1021. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greaney AJ, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e466. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greaney, A. J. et al. Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection. Sci. Transl. Med.13, 10.1126/scitranslmed.abi9915 (2021). [DOI] [PMC free article] [PubMed]
- 26.Walls AC, et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382 e1317. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto D, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 28.Rogers TF, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tortorici MA, et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tortorici, M. A. et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature, 10.1038/s41586-021-03817-4 (2021). [DOI] [PMC free article] [PubMed]
- 31.Barnes CO, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dejnirattisai W, et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200.e2122. doi: 10.1016/j.cell.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, et al. A combination of cross-neutralizing antibodies synergizes to prevent SARS-CoV-2 and SARS-CoV pseudovirus infection. Cell Host Microbe. 2021;29:806–818.e806. doi: 10.1016/j.chom.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starr, T. N. et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature, 10.1038/s41586-021-03807-6 (2021). [DOI] [PMC free article] [PubMed]
- 35.Cathcart, A. L. et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. Preprint at bioRxivhttps://www.biorxiv.org/content/10.1101/2021.03.09.434607v6.full (2021).
- 36.McCallum M, et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373:648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCallum M, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e2316. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vir&GSK. GSK and Vir Biotechnology announce sotrovimab (VIR-7831) receives Emergency Use Authorization from the US FDA for treatment of mild-to-moderate COVID-19 in high-risk adults and paediatric patients, https://www.gsk.com/en-gb/media/press-releases/gsk-and-vir-biotechnology-announce-sotrovimab-vir-7831-receives-emergency-use-authorization-from-the-us-fda/ (2021).
- 39.Cerutti G, et al. Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies. Structure. 2021;29:655–663.e654. doi: 10.1016/j.str.2021.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, L. et al. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science, 10.1126/science.abh1766 (2021). [DOI] [PMC free article] [PubMed]
- 41.Dejnirattisai W, et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954.e2939. doi: 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amanat F, et al. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell. 2021;184:3936–3948.e3910. doi: 10.1016/j.cell.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCarthy KR, et al. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Latif, A. A., et al. and the Center for Viral Systems Biology. SARS-CoV-2 (hCoV-19) Mutation Reports: Lineage Comparison, outbreak.info, https://outbreak.info/compare-lineages (2021).
- 45.Andreano, E. et al. SARS-CoV-2 escape from a highly neutralizing COVID-19 convalescent plasma. Proc. Natl Acad. Sci. USA118, 10.1073/pnas.2103154118 (2021). [DOI] [PMC free article] [PubMed]
- 46.Cele S, et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collier DA, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moyo-Gwete T, et al. Cross-reactive neutralizing antibody responses elicited by SARS-CoV-2 501Y.V2 (B.1.351) N. Engl. J. Med. 2021;384:2161–2163. doi: 10.1056/NEJMc2104192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams TC, Burgers WA. SARS-CoV-2 evolution and vaccines: cause for concern? Lancet Respir. Med. 2021;9:333–335. doi: 10.1016/S2213-2600(21)00075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu K, et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaebler C, et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dan, J. M. et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science371, 10.1126/science.abf4063 (2021). [DOI] [PMC free article] [PubMed]
- 53.Robbiani DF, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muecksch F, et al. Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity. 2021;54:1853–1868.e1857. doi: 10.1016/j.immuni.2021.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goel, R. R. et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naive and recovered individuals following mRNA vaccination. Sci. Immunol.6, 10.1126/sciimmunol.abi6950 (2021). [DOI] [PMC free article] [PubMed]
- 56.Peng Y, et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bange EM, et al. CD8(+) T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021;27:1280–1289. doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadoff J, et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alter G, et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021;596:268–272. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keech C, et al. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heath, P. T. et al. Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. N. Engl. J. Med. 10.1056/NEJMoa2107659 (2021). [DOI] [PMC free article] [PubMed]
- 62.Shinde V, et al. Efficacy of NVX-CoV2373 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021;384:1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palacios R, et al. Double-blind, randomized, placebo-controlled phase III clinical trial to evaluate the efficacy and safety of treating healthcare professionals with the adsorbed COVID-19 (inactivated) vaccine manufactured by Sinovac - PROFISCOV: a structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21:853. doi: 10.1186/s13063-020-04775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu Z, et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine (CoronaVac) in healthy adults aged 60 years and older: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021;21:803–812. doi: 10.1016/S1473-3099(20)30987-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18-59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021;21:181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jeewandara, C. et al. Antibody and T cell responses to Sinopharm/BBIBP-CorV in naive and previously infected individuals in Sri Lanka. Preprint at medRxivhttps://www.medrxiv.org/content/medrxiv/early/2021/07/19/2021.07.15.21260621.full.pdf (2021).
- 67.Ella R, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: interim results from a double-blind, randomised, multicentre, phase 2 trial, and 3-month follow-up of a double-blind, randomised phase 1 trial. Lancet Infect. Dis. 2021;21:950–961. doi: 10.1016/S1473-3099(21)00070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tarke A, et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Low JS, et al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science. 2021;372:1336–1341. doi: 10.1126/science.abg8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tarke A, et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poland GA, Ovsyannikova IG, Kennedy RB. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nolan, S. et al. A large-scale database of T-cell receptor beta (TCRbeta) sequences and binding associations from natural and synthetic exposure to SARS-CoV-2. Res. Sq. 10.21203/rs.3.rs-51964/v1 (2020).
- 73.Snyder, T. M. et al. Magnitude and dynamics of the T-cell response to SARS-CoV-2 infection at both individual and population levels. Preprint at medRxivhttps://www.ncbi.nlm.nih.gov/pubmed/32793919 (2020).
- 74.Sahin U, et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 75.Walsh EE, et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang S, et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021;21:1107–1119. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang B, et al. Serum sample neutralisation of BBIBP-CorV and ZF2001 vaccines to SARS-CoV-2 501Y.V2. Lancet Microbe. 2021;2:e285. doi: 10.1016/S2666-5247(21)00082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.SKbioscience. SK Bioscience’s COVID-19 Vaccine will enter Phase III clinical trial with Promising Interim Data, https://www.skbioscience.co.kr/en/news/news_01_01?mode=view&id=91& (2021).
- 79.Klasse, P. J., Nixon, D. F. & Moore, J. P. Immunogenicity of clinically relevant SARS-CoV-2 vaccines in nonhuman primates and humans. Sci. Adv.7, 10.1126/sciadv.abe8065 (2021). [DOI] [PMC free article] [PubMed]
- 80.Goletti, D. et al. The potential clinical utility of measuring severe acute respiratory syndrome coronavirus 2-specific T-cell responses. Clin. Microbiol. Infect. 10.1016/j.cmi.2021.07.005 (2021). [DOI] [PMC free article] [PubMed]
- 81.Jeong H, Choi YM, Seo H, Kim BJ. A novel DNA vaccine against SARS-CoV-2 encoding a chimeric protein of its receptor-binding domain (RBD) fused to the amino-terminal region of hepatitis B virus preS1 with a W4P mutation. Front. Immunol. 2021;12:637654. doi: 10.3389/fimmu.2021.637654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li J, et al. Safety and immunogenicity of the SARS-CoV-2 BNT162b1 mRNA vaccine in younger and older Chinese adults: a randomized, placebo-controlled, double-blind phase 1 study. Nat. Med. 2021;27:1062–1070. doi: 10.1038/s41591-021-01330-9. [DOI] [PubMed] [Google Scholar]
- 83.Mulligan MJ, et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 84.Elia U, et al. Design of SARS-CoV-2 hFc-conjugated receptor-binding domain mRNA vaccine delivered via lipid nanoparticles. ACS Nano. 2021;15:9627–9637. doi: 10.1021/acsnano.0c10180. [DOI] [PubMed] [Google Scholar]
- 85.Tai W, et al. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res. 2020;30:932–935. doi: 10.1038/s41422-020-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guirakhoo, F. et al. A novel SARS-CoV-2 multitope protein/peptide vaccine candidate is highly immunogenic and prevents lung infection in an adeno associated virus human angiotensin-converting enzyme 2 (AAV hACE2) mouse model. Preprint at bioRxivhttps://www.biorxiv.org/content/biorxiv/early/2020/11/30/2020.11.30.399154.full.pdf (2020).
- 87.Yang J, et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 88.Chen WH, et al. Genetic modification to design a stable yeast-expressed recombinant SARS-CoV-2 receptor binding domain as a COVID-19 vaccine candidate. Biochim. Biophys. Acta Gen. Subj. 2021;1865:129893. doi: 10.1016/j.bbagen.2021.129893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pollet, J. et al. SARSCoV-2 RBD219-N1C1: A yeast-expressed SARS-CoV-2 recombinant receptor-binding domain candidate vaccine stimulates virus neutralizing antibodies and T-cell immunity in mice. Hum. Vaccin. Immunother. 1–11, 10.1080/21645515.2021.1901545 (2021). [DOI] [PMC free article] [PubMed]
- 90.Malladi SK, et al. Immunogenicity and protective efficacy of a highly thermotolerant, trimeric SARS-CoV-2 receptor binding domain derivative. ACS Infect. Dis. 2021;7:2546–2564. doi: 10.1021/acsinfecdis.1c00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dalvie, N. C. et al. Engineered SARS-CoV-2 receptor binding domain improves manufacturability in yeast and immunogenicity in mice. Proc. Natl Acad. Sci. USA118, 10.1073/pnas.2106845118 (2021). [DOI] [PMC free article] [PubMed]
- 92.Dai L, et al. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020;182:722–733.e711. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun, S. et al. Interferon-armed RBD dimer enhances the immunogenicity of RBD for sterilizing immunity against SARS-CoV-2. Cell Res.10.1038/s41422-021-00531-8 (2021). [DOI] [PMC free article] [PubMed]
- 94.Kim, Y. I. et al. Development of spike receptor-binding domain nanoparticles as a vaccine candidate against SARS-CoV-2 infection in ferrets. mBio12, 10.1128/mBio.00230-21 (2021). [DOI] [PMC free article] [PubMed]
- 95.He, L. et al. Single-component, self-assembling, protein nanoparticles presenting the receptor binding domain and stabilized spike as SARS-CoV-2 vaccine candidates. Sci. Adv.7, 10.1126/sciadv.abf1591 (2021). [DOI] [PMC free article] [PubMed]
- 96.Ma X, et al. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity. 2020;53:1315–1330.0e1319. doi: 10.1016/j.immuni.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Walls AC, et al. Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell. 2021;184:5432–5447.e16. doi: 10.1016/j.cell.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.King, H. A. D. et al. Efficacy and breadth of adjuvanted SARS-CoV-2 receptor-binding domain nanoparticle vaccine in macaques. PNAS118, 10.1073/pnas.2106433118 (2021). [DOI] [PMC free article] [PubMed]
- 99.Kang YF, et al. Rapid development of SARS-CoV-2 spike protein receptor-binding domain self-assembled nanoparticle vaccine candidates. ACS Nano. 2021;15:2738–2752. doi: 10.1021/acsnano.0c08379. [DOI] [PubMed] [Google Scholar]
- 100.Tan TK, et al. A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 2021;12:542. doi: 10.1038/s41467-020-20654-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dalvie, N. C. et al. A modular protein subunit vaccine candidate produced in yeast confers protection against SARS-CoV-2 in non-human primates. Preprint at bioRxiv10.1101/2021.07.13.452251 (2021).
- 102.Zha, L. et al. Development of a vaccine against SARS-CoV-2 based on the receptor-binding domain displayed on virus-like particles. Vaccines9, 10.3390/vaccines9040395 (2021). [DOI] [PMC free article] [PubMed]
- 103.Lan J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 104.Zang J, et al. Yeast-produced RBD-based recombinant protein vaccines elicit broadly neutralizing antibodies and durable protective immunity against SARS-CoV-2 infection. Cell Discov. 2021;7:71. doi: 10.1038/s41421-021-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Doremalen, N. et al. Intranasal ChAdOx1 nCoV-19/AZD1222 vaccination reduces viral shedding after SARS-CoV-2 D614G challenge in preclinical models. Sci. Transl. Med. 10.1126/scitranslmed.abh0755 (2021). [DOI] [PMC free article] [PubMed]
- 106.Jearanaiwitayakul T, et al. Intranasal administration of RBD nanoparticles confers induction of mucosal and systemic immunity against SARS-CoV-2. Vaccines. 2021;9:768. doi: 10.3390/vaccines9070768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hsieh CL, et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Joyce, M. G. et al. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Preprint at bioRxivhttps://www.ncbi.nlm.nih.gov/pubmed/34013273 (2021). [DOI] [PMC free article] [PubMed]
- 109.Wang W, Huang B, Zhu Y, Tan W, Zhu M. Ferritin nanoparticle-based SARS-CoV-2 RBD vaccine induces a persistent antibody response and long-term memory in mice. Cell Mol. Immunol. 2021;18:749–751. doi: 10.1038/s41423-021-00643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cohen AA, et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang W-C, et al. SARS-CoV-2 RBD neutralizing antibody induction is enhanced by particulate vaccination. Adv. Mater. 2020;32:2005637. doi: 10.1002/adma.202005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dalvie, N. C. et al. Scalable, methanol-free manufacturing of the SARS-CoV-2 receptor binding domain in engineered Komagataella phaffii. Preprint at bioRxivhttps://www.ncbi.nlm.nih.gov/pubmed/33880471 (2021). [DOI] [PMC free article] [PubMed]
- 113.Frank, M. Cuba says second COVID-19 vaccine Soberana 2 boasts 91.2% efficacy, https://www.reuters.com/business/healthcare-pharmaceuticals/cuba-says-second-covid-vaccine-soberana-2-boasts-912-efficacy-2021-07-09/ (2021).
- 114.Limonta-Fernández, M. et al. The SARS-CoV-2 receptor-binding domain expressed in Pichia pastoris as a candidate vaccine antigen. Preprint at medRxivhttps://www.medrxiv.org/content/medrxiv/early/2021/07/03/2021.06.29.21259605.full.pdf (2021). [DOI] [PMC free article] [PubMed]
- 115.Chang-Monteagudo, A. et al. A single dose of SARS-CoV-2 FINLAY-FR-1A vaccine enhances neutralization response in COVID-19 convalescents, with a very good safety profile: An open-label phase 1 clinical trial. The Lancet Regional Health – Americas4, 10.1016/j.lana.2021.100079 (2021). [DOI] [PMC free article] [PubMed]
- 116.Brown CM, et al. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings - Barnstable County, Massachusetts, July 2021. MMWR Morb. Mortal. Wkly Rep. 2021;70:1059–1062. doi: 10.15585/mmwr.mm7031e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Meng F-Y, et al. Safety and immunogenicity of a recombinant COVID-19 vaccine (Sf9 cells) in healthy population aged 18 years or older: two single-center, randomised, double-blind, placebo-controlled, phase 1 and phase 2 trials. Signal Transduct. Target. Ther. 2021;6:271. doi: 10.1038/s41392-021-00692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were generated for the review article.