Main text
Our understanding of the molecular basis of G protein-coupled receptors (GPCRs) activation and signaling has exploded over the last few years. X-ray crystallography and single-particle cryo-electron microscopy have provided static structural snapshots for a range of receptor conformational states and receptor-signaling partner complexes (1). In parallel, a combination of biophysical techniques and computational studies have revealed that GPCRs are highly dynamic allosteric proteins that sample complex conformational landscapes (2). Altogether, these approaches have described how receptor activation and signaling pathways relate to such landscapes in a ligand-dependent manner and offered penetrating glimpses into the inner workings of GPCRs.
Whereas most of the early studies made use of detergent-solubilized receptors, there has been an increasing appreciation for the roles played by the lipid bilayer and specific lipids in modulating the thermodynamic and functional equilibrium between GPCR conformational states. Many studies have demonstrated that mechanical properties of the bilayer (e.g., bilayer thickness, bending stiffness, and monolayer spontaneous curvature) can affect the function of GPCRs (3,4). However, biophysical studies are difficult to conduct on vesicle membranes. In response, lipid nanoparticle systems, such as nanodisks (5), styrene maleic acid (SMA), SMA lipid particles (SMALPs) (6), and others (7) have been developed to study GPCRs in lipidic environments in a form that is soluble and amenable to a wide range of structural and biophysical experiments. Nanodisk and SMALPs consist of a lipid disk that is surrounded by amphipathic scaffolds interacting with the hydrophobic portion of lipid bilayers and with the aqueous solution. Many biophysicists are familiar with the two helical protein belts termed membrane scaffold proteins (MSPs) derived from the ApoA-1 apolipoprotein of high-density lipoproteins that form the scaffold for the nanodisk system. In SMALPs, this is an SMA copolymer that plays a role analogous to the MSPs. A GPCR reconstituted in a nanoscale lipid particle, therefore, consists of the receptor surrounded by a few layers of lipids, the number of which is determined by the type of ApoA-1 variant in nanodisks (8) or by the SMA/protein ratio in SMALPs. Typically, nanodisks are formed by mixing a detergent-solubilized membrane protein with lipids before the addition of MSP and removal of detergent by adsorbent polystyrene beads. On the other hand, SMALPs are formed directly from biomembranes (think of a molecular cookie cutter) in a process that does not require detergent. This may be advantageous for receptors with low stability in detergents, but whether SMALPs provide an environment that does not affect their functional properties remains a fundamental yet open question.
In this issue of Biophysical Journal (9), Szundi and colleagues use time-resolved optical absorption spectroscopy to rigorously compare the photoactivation kinetics of the prototypical GPCR rhodopsin measured in SMALPs with kinetics obtained in native membranes. Rhodopsin is uniquely suited for this test because its photokinetics has been studied extensively in native and model membranes and in lipid nanodisks (10). Here, the authors demonstrate that the active state of rhodopsin is formed in SMALPs when the native membranes are solubilized with relatively low ratios of SMA to rhodopsin (≤10–15). However, rhodopsin reaches its active state much slower in SMALPs (1 s) than in native membranes (10 ms) (Fig. 1). The strength of the study relies on the ability of time-resolved optical absorption spectroscopy, cleverly exploited by the authors, to follow the time dependence of rhodopsin photointermediates concentration (conformational states in other GPCRs) after light activation (ligand binding in other GPCRs) over a wide range of timescales, from microseconds to seconds. A key finding is that the dramatic changes in kinetics occur in the late steps of the kinetic pathway, indicating that SMA mainly affects the steps associated with the movements of helices in the protein backbone (i.e., shape change of the protein necessary to accommodate the G protein) and not the early steps of rhodopsin activation (i.e., when conformational changes are restricted to the retinal pocket). With higher SMA/rhodopsin ratios, the kinetic pathway is disrupted so that the active photoproduct is not formed (see also (11)), most likely because most lipids have been lost.
Figure 1.
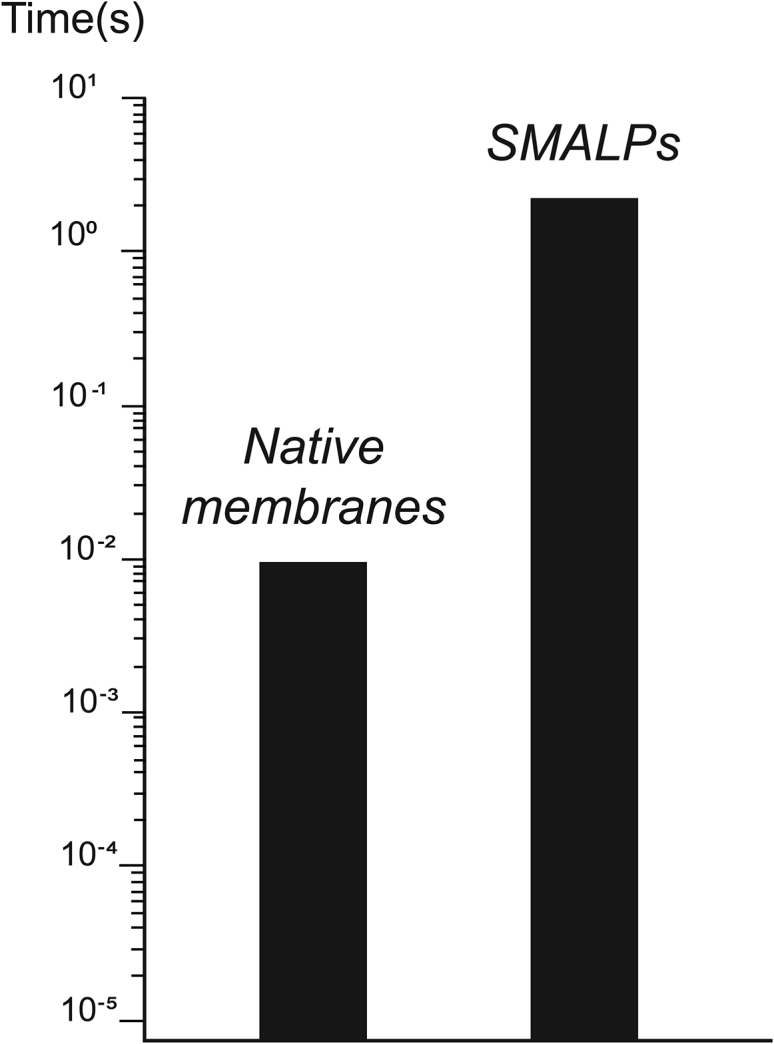
Time it takes rhodopsin to reach the active MII state in native membranes (left) and SMALPs (right) as measured by time-resolved optical absorption spectroscopy.
Recent developments in structural biology are allowing researchers to quickly determine structures of GPCRs (1,12), and lipid nanoscale bilayers are playing a supporting role (7). The study by Szundi et al. (9) indicates that optimal SMA conditions must be established first if subsequent structural studies of GPCRs in SMALPs are meant to be functionally relevant. It is also a reminder that the thermodynamic and kinetic equilibrium between functional states of GPCRs is linked to properties of the lipid bilayer via lipid-protein interactions. Key receptor conformational changes are coupled energetically to membrane deformation that extends further than the first shell of lipids surrounding the protein. The key for optimal GPCR functioning in nanoscale lipid bilayers may then be to find the right compromise between the size of the nanoparticle amenable to experiments and the number of layers of lipids surrounding the receptor.
There is no doubt that polymer nanoparticles will play an important role in our understanding of the molecular basis of membrane protein function. The limitations of the SMALPs technology highlighted in Szundi et al. (9) for GPCRs may not apply to other membrane proteins that offer a nondeformable interface to the lipid membrane or to proteins that show high affinity for a particular lipid. It is anticipated that the development of newer polymers and a deeper understanding of lipid behavior in nanoscale lipid bilayer, together with detailed functional studies such as the one by Szundi et al. (9), will allow SMALPs to be applied to a wide range of membrane proteins and optimized for GPCRs.
Acknowledgments
The author is supported by the Intramural Research Program of the National Cancer Institute.
Editor: John Conboy.
References
- 1.Weis W.I., Kobilka B.K. The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 2018;87:897–919. doi: 10.1146/annurev-biochem-060614-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Latorraca N.R., Venkatakrishnan A.J., Dror R.O. GPCR dynamics: structures in motion. Chem. Rev. 2017;117:139–155. doi: 10.1021/acs.chemrev.6b00177. [DOI] [PubMed] [Google Scholar]
- 3.Brown M.F. Soft matter in lipid-protein interactions. Annu. Rev. Biophys. 2017;46:379–410. doi: 10.1146/annurev-biophys-070816-033843. [DOI] [PubMed] [Google Scholar]
- 4.Soubias O., Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim. Biophys. Acta. 2012;1818:234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLean M.A., Gregory M.C., Sligar S.G. Nanodiscs: a controlled bilayer surface for the study of membrane proteins. Annu. Rev. Biophys. 2018;47:107–124. doi: 10.1146/annurev-biophys-070816-033620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knowles T.J., Finka R., Overduin M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc. 2009;131:7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 7.Lavington S., Watts A. Lipid nanoparticle technologies for the study of G protein-coupled receptors in lipid environments. Biophys. Rev. 2020;12:1287–1302. doi: 10.1007/s12551-020-00775-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagn F., Etzkorn M., Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szundi I., Pitch S.G., Kliger D.S. Styrene-maleic acid copolymer effects on the function of the GPCR rhodopsin in lipid nanoparticles. Biophys. J. 2021;120:4337–4348. doi: 10.1016/j.bpj.2021.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsukamoto H., Szundi I., Kliger D.S. Rhodopsin in nanodiscs has native membrane-like photointermediates. Biochemistry. 2011;50:5086–5091. doi: 10.1021/bi200391a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitch S.G., Yao W., Farrens D.L. Functional integrity of membrane protein rhodopsin solubilized by styrene-maleic acid copolymer. Biophys J. 2021;120:3508–3515. doi: 10.1016/j.bpj.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin J., Chen K.M., Rosenbaum D.M. Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane. Nature. 2020;584:125–129. doi: 10.1038/s41586-020-2379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


