Abstract
Repeat-induced point mutation is a genetic process that creates cytosine-to-thymine (C-to-T) transitions in duplicated genomic sequences in fungi. Repeat-induced point mutation detects duplications (irrespective of their origin, specific sequence, coding capacity, and genomic positions) by a recombination-independent mechanism that likely matches intact DNA double helices directly, without relying on the annealing of complementary single strands. In the fungus Neurospora crassa, closely positioned repeats can induce mutation of the adjoining nonrepetitive regions. This process is related to heterochromatin assembly and requires the cytosine methyltransferase DIM-2. Using DIM-2-dependent mutation as a readout of homologous pairing, we find that GC-rich repeats produce a much stronger response than AT-rich repeats, independently of their intrinsic propensity to become mutated. We also report that direct repeats trigger much stronger DIM-2-dependent mutation than inverted repeats. These results can be rationalized in the light of a recently proposed model of homologous DNA pairing, in which DNA double helices associate by forming sequence-specific quadruplex-based contacts with a concomitant release of supercoiling. A similar process featuring pairing-induced supercoiling may initiate epigenetic silencing of repetitive DNA in other organisms, including humans.
Significance
There exists a large repertoire of homology-directed processes that apparently involve interactions between intact chromosomal regions. Neurospora crassa possesses one such process, known as repeat-induced point mutation (RIP). RIP involves a number of conserved epigenetic factors and is also being extremely sensitive to DNA homology. By taking advantage of this unique system, we show that recognition of repetitive DNA is driven mainly by GC basepairs. We also show that the relative orientation of closely positioned repeats plays an important role in modulating the activity of the heterochromatin-related pathway of RIP. Our results support a model in which homologous intact double-stranded DNAs (dsDNAs) can associate by forming short interspersed quadruplexes and, furthermore, suggest a role for this process in initiating heterochromatin formation on repetitive DNA.
Introduction
The existence of recombination-independent pairing is well documented (1). Mammals feature a particularly rich repertoire of such phenomena (2,3). For example, during early meiosis in mice, homologous chromosomes can pair transiently in the absence of programmed DNA breaks (4). During early mammalian development, a number of homologous loci engage in extensive pairing, presumably to establish appropriate patterns of gene expression (5). The transient association of two X chromosomes before the onset of random X-chromosome inactivation provides arguably the most well-known instance of such a process (6). Recombination-independent pairing has also been described in flies (7,8), worms (9), and yeast (10, 11, 12).
Mammalian genomes also contain large amounts of highly repetitive (“self-homologous”) DNA normally silenced in the form of constitutive heterochromatin (13). Importantly, in mammals, the formation of heterochromatin on tandemly repeated DNA does not require RNA interference (14,15). Pathological misregulation of this process may be associated with several types of cancer (16,17) and other disease, such as type I facioscapulohumeral muscular dystrophy (18). The nature of the mechanism(s) responsible for the identification of repetitive DNA sequences remains largely unknown, but it has been proposed to involve pairwise interactions between repeat units (19).
The strongest evidence for the existence of homologous dsDNA-dsDNA pairing in vivo is obtained by studying two gene silencing processes in the fungus Neurospora crassa. Both processes detect DNA sequence homology by a yet unidentified mechanism that does not require the RecA-like recombinases and, instead, matches intact DNA double helices directly. These processes are known as “repeat-induced point mutation” (RIP, (20)) and “meiotic silencing by unpaired DNA” (MSUD, (21)). RIP introduces cytosine-to-thymine (C-to-T) transitions in duplicated genomic sequences, whereas MSUD induces transient RNA interference against dissimilar DNA sequences present at the allelic positions on a pair of homologous chromosomes (21). Since its discovery in N. crassa (22), RIP was demonstrated experimentally in several filamentous fungi, and signatures of RIP were detected computationally in the genomes of most Pezizomycotina (23) and some Basidiomycota species (24). A process called "methylation induced premeiotically" was also described in the fungus Ascobolus immersus, in which duplications undergo cytosine methylation instead of mutation (25).
RIP takes place after fertilization but before karyogamy in cells that harbor haploid nuclei of both parental types. This period is known as the premeiotic stage. In Neurospora, RIP can accurately identify segments of chromosomal DNA that share only several hundred basepairs (bp) of homology (22,26). Duplications are recognized irrespective of their origin, particular sequence, coding capacity, or genomic positions. The ability of RIP to detect two identical gene-sized DNA sequences, even when present on different chromosomes, suggests that an efficient and global homology search is involved. The corresponding mechanism should solve two problems inherent in eukaryotic genomes, namely the relatively slow diffusion of chromatin and the occlusion of DNA sequences by tightly bound proteins (e.g., histones). The same problems are also faced by the homology search during break-induced interchromosomal recombination (27). In yeast, the latter takes a few hours and involves global ATP-dependent degradation of histones, which increases the overall chromosome mobility and recombination rates (28). In its turn, RIP, which progresses over the course of several days, may also require global chromatin remodeling to promote mobility and accessibility of genomic DNA.
By analyzing the occurrence of mutations in strategically designed synthetic repeats in N. crassa, it was discovered that RIP could still detect the presence of homologous trinucleotides (triplets) interspersed with a periodicity of 11 bp along the participating DNA segments, which corresponds to the overall sequence identity of only 27% (20). Further studies revealed that some specific triplets (such as GAC) were particularly effective at promoting RIP (29). Taken together, these results suggested a possibility that RIP involved direct dsDNA-dsDNA pairing in which sequence-specific contacts between homologous DNA segments could only be established in register with their double-helical structure (20,26).
A molecular mechanism of the direct dsDNA-dsDNA pairing that consistently explained the above results was subsequently proposed (30). This model is based on the fact that canonical Watson-Crick (WC) basepairs have unique, yet self-complementary, electrostatic patterns along major groove edges, thus permitting, in principle, binding of two complementary double-stranded stacks without disturbing the WC pairing. This molecular property was already implicated in the early theories of DNA replication (31) and homologous recombination (32). According to the dsDNA-dsDNA pairing model, a sequence-specific contact between two dsDNAs corresponds to a short quadruplex stack of three to four planar quartets formed by identical WC basepairs (30). The energy of quartet formation includes a large nonspecific contribution of ionic interactions and a hydrogen bonding term. As predicted, strong polarization of hydrogen bonds in GC quartets may provide additional stabilization energy compared with AT quartets (30). Because quadruplexes can only be formed at intervals corresponding to half-integral numbers of helical turns, the observed periodicity of 11 bp (or 22 bp) suggests that DNA pairing is accompanied by a concomitant change in the linking number of the participating DNAs and results in the accumulation of supercoiling in the adjacent regions. Although this mechanism remains hypothetical, it currently represents the only detailed model that can consistently explain the unusual homology recognition patterns of RIP (33,34) and MSUD (21).
In N. crassa, RIP can be executed by two largely independent pathways. The first pathway relies on the putative C5-cytosine methyltransferase (CMT) RID (35). The second pathway requires DIM-5 (a histone H3 lysine-9 (H3K9) methyltransferase), DIM-2 (a canonical CMT) and heterochromatin protein 1 (HP1) (36,37). The two pathways feature opposite substrate preferences. Whereas RID-dependent RIP is largely limited to the repeats, DIM-2-dependent RIP tends to mutate the adjoining nonrepetitive regions. Whereas RID-like proteins form a group that is likely endemic to filamentous fungi (38), DIM-5 belongs to a conserved SUV39 family of lysine methyltransferases that participate in silencing of repetitive DNA in the context of constitutive heterochromatin (13). The uncovered role of DIM-5 in RIP suggested a possibility that SUV39 proteins can be recruited and/or activated by homologous dsDNA-dsDNA interactions (39).
One aspect of Neurospora RIP that makes it particularly useful for understanding other putative recombination-independent homology-directed phenomena is its ability to provide an accurate readout of DNA homology (26). More specifically, in N. crassa, the expected number of RIP appears to be accurately related to the amount of inducing homology, provided that the levels of RIP are not saturated and a sufficiently large number of RIP products are sampled. This property holds true for DNA sequences that are short enough to be manipulated with single-basepair precision, which allows addressing a number of biophysical questions concerning the mechanism of homologous dsDNA-dsDNA pairing. Furthermore, the presence of two distinct RIP pathways in N. crassa brings an important advantage to studying molecular events involved in recombination-independent DNA homology recognition because these pathways can be switched on and off independently, allowing a wider spectrum of questions to be pursued.
To quantify the magnitude of RIP along a given DNA region, a new computational approach named the partitioned RIP propensity (PRP) was developed (33). PRP takes as an input the occurrence of individual mutations and estimates the probability of mutation for a short DNA segment rather than for a particular site or a sequence motif (33). In doing so, PRP is designed to avoid complications associated with nonuniform distributions of RIP substrates in natural sequences, thus permitting one to distinguish regions that may be intrinsically different with respect to being affected by RIP. Initial application of the PRP approach to reanalyze the earlier data (20) led to the idea of mechanical coupling between DNA paring and DNA supercoiling, with a number of implications for the function of repetitive DNA (33).
Here, we use the DIM-2-dependent RIP as a readout of repeat recognition to test two predictions made by the quadruplex-based pairing model (30). We have found that GC-rich repeats indeed promote much stronger RIP compared with AT-rich repeats, with the relative contribution of AT basepairs being close to zero. We have also found that direct repeats trigger stronger RIP in the adjacent nonrepetitive regions compared with inverted repeats; both the spacer between the duplicated sequences (the linker) and the flanks are similarly affected by this process. These and other results further corroborate the idea that the homologous pairing for RIP involves formation of interspersed quadruplexes and produces local DNA supercoiling stress. In the case of inverted repeats, this stress would favor the formation of plectonemic structures on the linker and the flanks, antagonizing nucleosome assembly and therefore reducing the amount of substrate for DIM-5 to produce H3K9me3 and initiate DIM-2-dependent RIP.
Materials and methods
Plasmids
Plasmids were constructed using standard molecular cloning techniques as previously described (20,36). All inserts were verified by sequencing. All plasmids used in this study are listed in Table 1. Plasmid maps are provided in EMBL format in the Supporting material, Data S1.
Table 1.
Repeat constructs analyzed in this study
| Sequence identifier | Repeat identifier | Repeat type | Homology length | Homology GC% | Linker length | Repeat source (GenBank accession number) | Plasmid |
|---|---|---|---|---|---|---|---|
| S1 | R1 | direct | 802 | 28.1 | 729 | E. coli (CP000948.1: 3900805-3900009) | pFOC100J |
| S2 | R2 | direct | 802 | 32.2 | 729 | A. vaga (EU637017.1: 33915-34711) | pEAG238B |
| S3 | R3 | direct | 802 | 53.5 | 729 | E. coli (CP000948.1: 1637844-1637049) | pFOC100B |
| S3 | R4 | inverted | 802 | 53.5 | 729 | E. coli (CP000948.1: 1637844-1637049) | pFOC100E |
| S4 | R5 | direct | 802 | 66.2 | 729 | E. coli (CP000948.1: 257444-258240) | pFOC100G |
| S4 | R6 | inverted | 802 | 66.2 | 729 | E. coli (CP000948.1: 257444-258240) | pFOC100H |
| S4 | R7 | direct | 802 | 66.2 | 2187 | E. coli (CP000948.1: 257444-258240) | pFOC100R |
| S4 | R8 | direct | 401 | 64.6 | 365 | E. coli (CP000948.1: 257845-258240) | pFOC100N |
| S4 | R9 | direct | 401 | 64.6 | 729 | E. coli (CP000948.1: 257845-258240) | pFOC100M |
Manipulation of Neurospora strains
Linearized plasmids were transformed into his-3 strains as previously described (20,36). Homokaryotic repeat-carrying strains were obtained by macroconidiation of the primary his-3+ transformants. The integrity of transformed DNA was verified by sequencing. All strains created in this study are listed in Table 2. Crosses were setup as previously described (20,36). All crosses analyzed in this study are listed in Table 3.
Table 2.
Strains used in this study
| Strain identifier | Mating type | Genotype | Source | Reference |
|---|---|---|---|---|
| FGSC#8594 | a | dim-2Δ, his-3 | FGSC1 | (40) |
| FGSC#12354 | A | ridΔ | FGSC | (40) |
| C02.1 | A | ridΔ, mus-52Δ, his-3 | cross progeny of FGSC#9539 and FGSC#12354 | (36) |
| C03.1 | a | ridΔ, mus-52Δ, his-3 | cross progeny of FGSC#9720 and FGSC#12353 | (36) |
| C96.1 | A | dim-2Δ | cross progeny of FGSC#8594 and FGSC#2489 | (36) |
| T465.4h | A | ridΔ, mus-52Δ | C02.1 transformed with pEAG238B | (33) |
| T618.1h | a | ridΔ, mus-52Δ | C03.1 transformed with pFOC100B | this study |
| T645.2h | a | ridΔ, mus-52Δ | C03.1 transformed with pFOC100E | this study |
| T646.9h | a | ridΔ, mus-52Δ | C03.1 transformed with pFOC100J | this study |
| T648.4h | A | ridΔ, mus-52Δ | C02.1 transformed with pFOC100J | this study |
| T649.4h | a | ridΔ, mus-52Δ | C03.1 transformed with pFOC100G | this study |
| T650.4h | a | ridΔ, mus-52Δ | C03.1 transformed with pFOC100H | this study |
| T660.5h | a | dim-2Δ | FGSC#8594 transformed with pFOC100H | this study |
| T672.4h | a | ridΔ, mus-52Δ | C03.1 transformed with pFOC100N | this study |
| T674.1h | a | ridΔ, mus-52Δ | C03.1 transformed with pFOC100M | this study |
| T676.1h | a | ridΔ, mus-52Δ | C03.1 transformed with pFOC100R | this study |
The Fungal Genetics Stock Center
Table 3.
Crosses analyzed in this study
| Cross identifier | Female parent | Tested repeat | Male parent | Tested repeat |
|---|---|---|---|---|
| X1 | T618.1h | R3 | T465.4h | R2 |
| X2 | T645.2h | R4 | T465.4h | R2 |
| X3 | T646.9h | R1 | T465.4h | R2 |
| X4 | T649.4h | R5 | T648.4h | R1 |
| X5 | T650.4h | R6 | T648.4h | R1 |
| X6 | C96.1 | NA | T660.5h | R5 |
| X7 | FGSC#12354 | NA | T649.4h | R5 |
| X8 | FGSC#12354 | NA | T676.1h | R7 |
| X9 | FGSC#12354 | NA | T672.4h | R8 |
| X10 | FGSC#12354 | NA | T674.1h | R9 |
Genomic DNA extraction, PCR amplification, and sequencing
Ascospores were sampled as previously described (20,36). For each cross, up to 150 ascospore clones were first genotyped and sorted by the corresponding repeat allele. 50 clones (per repeat construct per each dim-2+/+, ridΔ/Δ cross) and 25 clones (per repeat construct per each dim-2Δ/Δ, rid+/+ cross) were chosen for analysis. PCR products were sequenced at Eurofins Genomics (Cologne, Germany). Chromatograms were assembled into contigs with Phred-Phrap (41). Contigs were validated manually using Consed (42).
Sequence and statistical analysis
For each repeat construct for each cross, assembled contigs were aligned with the reference using ClustalW (43). Mutations were analyzed as previously described (20,33,36). Graphs were plotted using the ggplot2 package in R (44). All sequence alignments generated in this study are provided in CLUSTAL format in the Supporting material, Data S2.
Results
The quadruplex-based pairing model (30) predicts that homologous GC-rich sequences should engage in stronger pairing for RIP than AT-rich sequences. This prediction is based on the intrinsic property of GC basepairs to form more stable quartets than AT basepairs (30). However, testing this prediction with RIP runs into an obvious problem; GC-rich repeats have more cytosines, thus they are a priori expected to mutate more strongly. In addition, DNA homology becomes reduced in successive cycles of RIP in the same cross, and the rate of reduction is also coupled to GC content. In Neurospora, these problems can be avoided by using DIM-2-dependent RIP as the only pathway because it is induced by closely positioned repeats but produces mutations on adjacent nonrepetitive regions, which separates the inducer and the substrate of mutation (26,33). Furthermore, because DIM-2-dependent RIP is relatively weak, it also fulfills the requirement for not being saturated, thus improving the signal range.
DIM-2-dependent RIP is modulated by GC content and orientation of closely positioned perfect repeat units
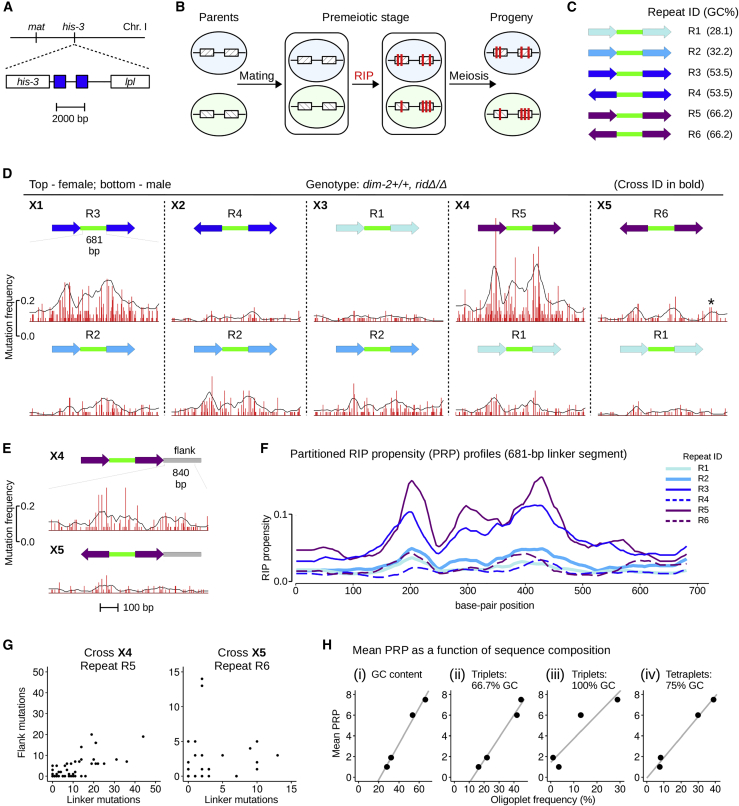
Four natural DNA sequences were used to create all repeat constructs in this study (Fig. 1 C; Table 1). Sequences S1, S3, and S4 originated in the bacterium Escherichia coli, whereas sequence S2 was obtained from the bdelloid rotifer Adineta vaga (Table 1). The sequences were chosen to represent different levels of GC content, from 28.1 to 66.2% (Fig. 1 C). Repeat constructs R1–R6 have exactly the same linker and the flanks. These constructs also have the same repeat unit length (802 bp). The constructs differ with respect to the sequence and/or orientations of the repeat units (Fig. 1 C; Table 1). All constructs are integrated into the same locus, between his-3 and lpl on chromosome 1 (Fig. 1 A). Two different constructs, each carried by one parental strain, were tested simultaneously in each cross (Fig. 1 B). Five isogenic dim-2+/+, ridΔ/Δ crosses with different combinations of repeat constructs were analyzed (Fig. 1 D; Table 3). 50 randomly sampled spore clones (per construct per cross) were assayed for RIP. Repeats R1 and R2 were each tested in three different crosses (Fig. 1 D; Table 3). The occurrence of mutations was analyzed as mutation frequency and as the associated PRP profile (Fig. 1 D).
Figure 1.
DIM-2-dependent RIP is affected by the GC content and the relative orientation of closely positioned perfect repeat units. (A) A pair of closely positioned perfect repeat units is integrated between his-3 and lpl genes on chromosome 1. The same integration site is used to create all repeat-carrying strains in this study (Table 2). (B) Mutation of two constructs can be assayed simultaneously in one cross. (C) Analyzed perfect repeats R1–R6 have the same overall length and the same linker sequence (green). Repeats differ with respect to GC content and relative orientation (Table 1). (D) Mutation of R1–R6 by DIM-2-dependent RIP was analyzed in five crosses (Table 3). Cross and repeat identifiers are indicated. 50 ascospore clones (per repeat construct per cross) were analyzed for RIP in the linker region. A 681-bp segment of the 729-bp linker was sequenced. The level of RIP is expressed as the number of mutations per site (“mutation frequency,” red) and as “partitioned RIP propensity” (PRP) profile (black). Data in (D and E) are plotted at the same scale (both x axis and y axis). (E) DIM-2-dependent RIP in the lpl-proximal flank was analyzed for the two GC-rich repeats R5 and R6 (crosses X4 and X5, respectively). (F) DIM-2-dependent PRP profiles for the linker region. Repeat identifiers are indicated. PRP profiles for repeats R1 and R2 are based on the combined data (crosses X3–X5 and X1–X3, respectively). (G) The relationship between the number of mutations in the linker versus the flank in individual spore clones for the GC-rich repeats R4 and R5. (H) Mean PRP as a function of sequence composition expressed as the overall GC content (i), the frequency of two triplet types (ii and iii) and the frequency of one tetraplet type (iv). To see this figure in color, go online.
Overall, our results show that perfect direct repeats with high GC contents promote the strongest DIM-2-dependent RIP in the linker region (Fig. 1, D and F; repeats R3 and R5). Strikingly, simply flipping one repeat unit in R3 and R5 (to produce inverted repeats R4 and R6, respectively) reduced DIM-2-dependent RIP to the levels observed for AT-rich direct repeats R1 and R2 (Fig. 1, D and F). Despite these dramatic differences in RIP levels, however, the positions of local minima and maxima in the linker PRP profiles remained similar (Fig. 1 F).
Inverted repeats are readily mutated by RID-dependent RIP
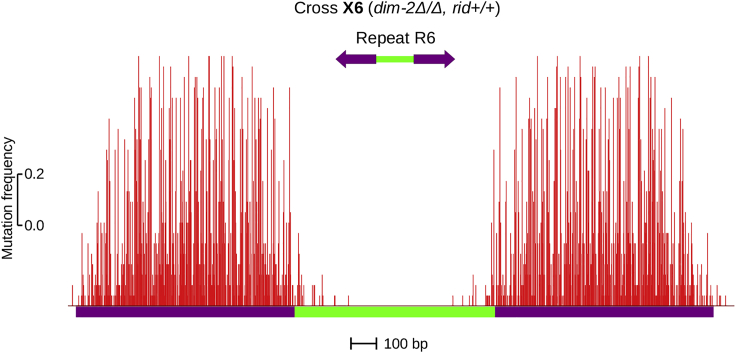
Because inverted repeats appeared to trigger fewer DIM-2-dependent RIP than direct repeats (Fig. 1 D), it was important to exclude the possibility that the inversion of a repeat unit in this particular construct somehow disturbed the homologous pairing. In this case, the inversion should similarly hinder both RIP pathways, which, in principle, should be noticeable in the wild-type background. Earlier studies argued against this interpretation (20,29,45). Nevertheless, to address this issue formally, we have now assayed mutation of repeat R6 by RID-dependent RIP (Fig. 2; Table 3). Very strong RIP was observed, with mutation frequency approaching saturation at many sites (Fig. 2). Also notable is the very low level of RID-dependent mutation in the linker region (Fig. 2). These results show that 1) the inversion of a repeat unit in repeats R5 and R6 has no apparent effect on the efficiency of homologous pairing and 2) RID- and DIM-2-dependent RIP pathways are affected differently by the relative orientation of the repeat units.
Figure 2.
Perfect GC-rich inverted repeats trigger strong RID-dependent RIP. 25 random progeny clones carrying repeat R6 were analyzed. The magnitude of RIP is expressed as the number of mutations per site (“mutation frequency,” red). Data are plotted at the same scale as in Fig. 1D (both x axis and y axis). To see this figure in color, go online.
DIM-2-dependent RIP is regulated similarly in the linker and in the flanks
It was important to determine whether DIM-2-dependent RIP could only mutate the linker region of these particular repeat constructs or whether it could also mutate the flanks, as suggested by the previous study (36). Focusing on the two GC-rich repeats R5 and R6, the “right” lpl-proximal flank was sequenced in the same 50 spore clones that have been assayed for RIP in the linker. A moderate level of RIP was found in the flank region adjacent to the direct repeat, and a substantially lower level of mutation was found in the same region adjacent to the inverted repeat (Fig. 1 E). The relative difference in “flank” RIP between repeats R5 vs. R6 is similar to the relative difference in “linker” RIP for the same repeats (Fig. 1 D). Taken together, these results suggest that DIM-2-dependent RIP is controlled in the linker and the flanks by the same or tightly related processes. Note, however, that in the case of direct repeat R5, there is a positive correlation between the “linker” and “flank” RIP on the per spore basis (Fig. 1 G). No such correlation is noticeable for the inverted repeat R6, for which the two spore clones with the strongest “flank” RIP (corresponding to 13 and 14 mutations) had only two mutations in the linker (Fig. 1 G).
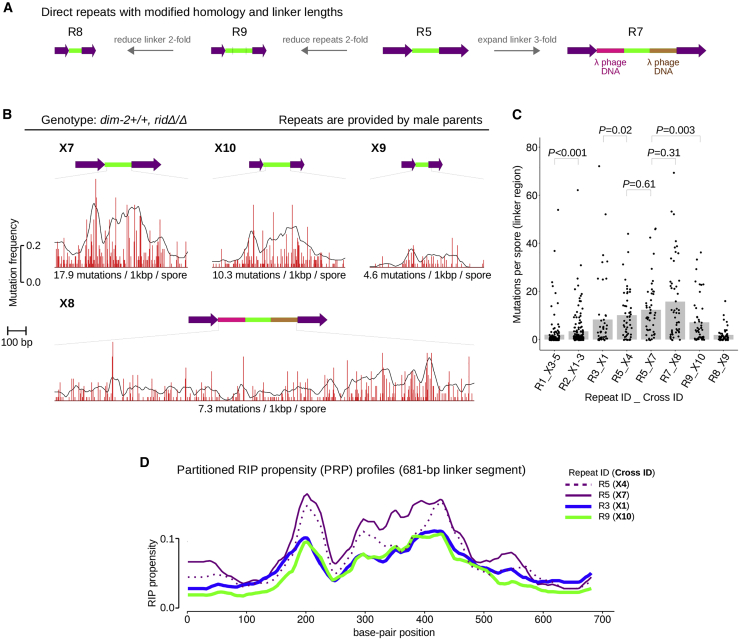
DIM-2-dependent RIP of closely positioned repeats is modulated by the relative lengths of constituent segments
The above results suggest that direct repeats promote stronger RIP by the DIM-2 pathway. To learn more about the relationship between the length parameters of direct repeats and ensuing DIM-2-dependent RIP, we have altered the linker length or the repeat length or both (Fig. 3 A). All crosses had the same female parent, whereas repeats were provided by the otherwise isogenic male parents (Tables 2 and 3). We first confirmed that our “standard” GC-rich direct repeat R5 was mutated similarly in this modified situation (Fig. 3, B–D: compare repeat R5 in crosses X4 and X7). We then proceeded to test three additional repeats derived from R5. Repeat R7 has exactly the same homology units as R5, whereas its linker has been expanded threefold, from 729 to 2187 bp (Fig. 3 A; Table 1). Repeat R9 has exactly the same linker as R5, but its homology units have been reduced twofold, from 802 to 401 bp (Fig. 3 A; Table 1). And, last, repeat R8 has the same homology units as R9, whereas its linker has been reduced twofold (Fig. 3 A; Table 1).
Figure 3.
DIM-2-dependent RIP of closely positioned repeats is modulated by the relative lengths of constituent segments. (A) Three additional repeats R7, R8, and R9 were constructed based on the GC-rich direct repeat R5. (B) Repeats were always provided by male parents (Table 3). 50 random ascospore clones were analyzed (per repeat per cross). The magnitude of RIP is expressed as in Fig. 1D. Data are plotted at the same scale as in Fig. 1D (both x axis and y axis). (C) Per spore numbers of DIM-2-dependent mutations in the sequenced linker segment. For repeats R1 and R2, the combined data sets are used. The mean number of RIP is indicated by a gray bar. The difference between empirical distributions of mutation counts is evaluated for significance by the Kolmogorov-Smirnov test. (D) DIM-2-dependent PRP profiles for the linker region. Repeat identifiers are indicated. To see this figure in color, go online.
Remarkably, the threefold expansion of the linker had no significant effect on the total number of mutations observed in this region; essentially the same (or apparently somewhat larger) number of mutations became distributed more or less evenly across the longer region (Fig. 3, B and C: compare R5 and R7). Mutation of the linker was significantly decreased upon halving the repeat length (Fig. 3, B and C: compare R5 and R9). Finally, the twofold reduction of the linker decreased its mutation even further (Fig. 3, B and C).
Discussion
Homologous pairing for RIP is likely driven by GC-rich but not GC-pure oligoplets
Our results show that perfect repeats with higher GC content promote stronger DIM-2-dependent RIP (Fig. 1 D). Assuming that the latter reflects the ability of repeats to engage in homologous pairing, these results suggest that perfect GC-rich repeats can pair more efficiently than perfect AT-rich repeats. For the assayed direct repeats R1, R2, R3, and R5, the dependence of the mean PRP on the overall GC content is approximated by a linear regression (Fig. 1 Hi). Interestingly, extrapolation to zero-RIP indicates that the process should stop when the overall GC content falls below ∼20%, which might suggest that only GC basepairs increase the energy of pairing, whereas AT pairs actually reduce it. In reality, this simplistic analysis is not entirely correct because, according to the earlier data, pairing for RIP should involve a minimum of three and, perhaps, four consecutive basepairs (20,29,30). Fig. 1 Hii–iv displays the results of a similar analysis for representative oligoplet patterns with GC contents above 50%. The pairing evidently is not driven by GC-pure oligoplets, because it occurs without such GC-pure triplets (Fig. 1 Hii). In contrast, the content of tetraplets with a single AT-pair yields a good linear fit convergent at zero (Fig. 1 Hiv). These results agree with the earlier predictions concerning the mechanism of pairing via short oligoplets and the associated contrasting roles of AT and GC basepairs. In addition, these results suggest that efficient homologous pairing for RIP likely requires mixed GC-rich sequences. One such motif, 5′-GAC-3′, was already implicated in stimulating RIP of repeats with interspersed rather than perfect homology (29).
Per spore correlations between RIP in flank versus linker segments
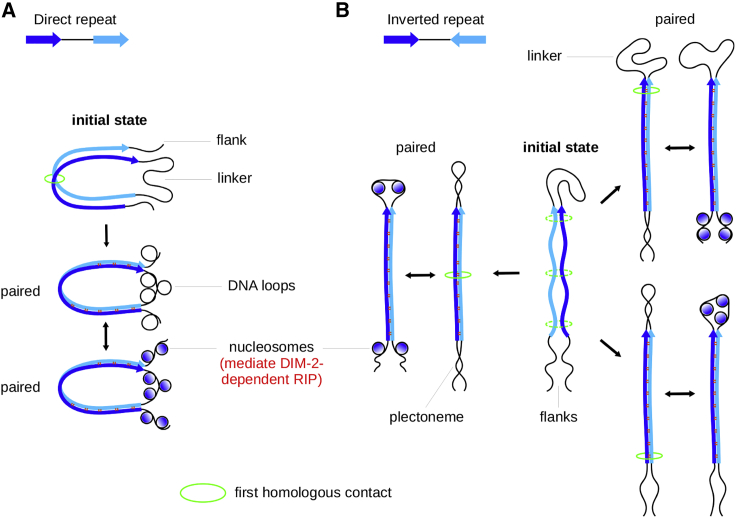
To our knowledge, the statistics of per spore correlations of RIP were never studied in the earlier literature. Our results suggest that mutations of regions adjacent to the repeat units are not statistically independent (Fig. 1 G) and that their correlations are qualitatively different for direct versus inverted orientations of the same homologous sequences. These differences support the supercoiling-driven model of DIM-2-dependent RIP (Fig. 4) explained below.
Figure 4.
Proposed role of pairing-induced DNA supercoiling in regulating DIM-2-dependent RIP. Recombination-independent pairing of direct (A) versus inverted (B) repeats is expected to create the same amount of DNA supercoiling. The way by which this supercoiling is utilized can be strongly influenced by the relative orientation of the closely positioned repeat units. To see this figure in color, go online.
Both direct and inverted repeats can start pairing, with some probability, at any position along the repeat units (Fig. 4). The first homologous contact creates a double-stranded contour that contains the linker and two adjacent segments of the repeat units. Any closed DNA contour is characterized by a certain linking number that cannot be changed without breaking at least one of the strands (46). This number is set by the first contact (Fig. 4). The linking number can be qualitatively interpreted as an algebraic sum of DNA twisting and supercoiling (46). Whereas the contour is closed, these two components are coupled. As the pairing proceeds further along the homology length, modulations of the average twisting and supercoiling should be mutually compensating. The pairing is expected to significantly affect the conformations of the participating dsDNAs (30). The ability of only certain interspersed homologies with 11- to 12-bp periodicities to promote RIP (20) suggests that the helical twist in the paired DNAs differs from that of free DNA. A compensatory change in supercoiling should be induced mainly on the linker because it remains flexible. Similar considerations apply to pairing of the repeat segments adjacent to flanking DNA (Fig. 4). Because they are outside the contour, their twisting should be compensated by supercoiling in the flanks (instead of the linker). Eventually, this supercoiling should be absorbed by surrounding bulk DNA. Taken together, the model suggests that as the pairing progresses both the linker and the flanks become transiently supercoiled. The presence of this supercoiling can provide a physical mark that targets them for DIM-2-dependent RIP, in part by facilitating the association of histone cores (33).
The closed contours in Fig. 4, A and B are also responsible for the partitioning of the induced DNA supercoiling. The length and topology of these contours are inherently different for direct and inverted repeats, which is crucial for all of the subsequent events and the outcome of the DIM-2-dependent RIP. In the case of direct repeats, the overall contour length does not depend on the position of the first contact; the total length of the included paired DNA will be always equal to that of one repeat unit (Fig. 4 A). The corresponding amount of supercoiling will be transferred to the linker, and the same amount will also be transferred to the flanks. The rotational orientation of the paired repeat units may affect the quality of pairing and the amount of supercoiling, but this value is always the same for the flanks and the linker. As a result, the amplitudes of DIM-2-dependent RIP on the linker and the flanks should be positively correlated, and this is indeed seen in the left panel of Fig. 1 G. Only one of the two flanks was assayed in our experiment; therefore, the average numbers of mutations on the x and y axes are different. Nevertheless, the positive correlation is evident.
Inverting one of the two closely positioned repeat units qualitatively changes the partitioning of pairing-induced supercoiling (Fig. 4 B). For the inverted repeats, the overall length of the double-stranded contour will depend on the position of the first homologous contact (Fig. 4 B). If the first such contact occurs near the linker, most homologous DNA will be excluded from the contour, and all supercoiling will be transferred to the flanks. In the opposite situation, when the initial contact occurs near the flanks, the contour will include both repeat units, and therefore, all supercoiling will be transferred to the linker. As a result, the strongest amplitudes of DIM-2-dependent RIP for the linker and the flanks should be found in different spore clones; in other words, they should be anticorrelated. However, this idea concerns only the two extreme cases. Intermediate situations will produce supercoiling on both the linker and the flanks. In such situations, hardly any linker-flank correlations of RIP signals might be detectable because the partitioning of supercoiling and the corresponding amplitudes of RIP are coupled. The experimental pattern revealed in the right panel of Fig. 1 G agrees with the above expectations. Indeed, overall, the plotted points look randomly scattered, but there are a few spores in which the maximal amplitudes of DIM-2-dependent RIP were observed for either the flank or the linker but not both of them.
DIM-2-dependent RIP of direct and inverted repeats is regulated by the same mechanism
As shown previously (20,33), the wild-type RIP of closely positioned repeats is altered dramatically by flipping one of the two repeat units. For inverted orientations, linker PRP profiles typically look dominated by RIP “leaking” from the repeats. However, observations made in the wild-type genetic backgrounds could not disentangle individual contributions of the two RIP pathways. Our current results show that, in the case of inverted repeats, RID-dependent RIP spreads into the linker for only ∼150 bp from each repeat border (Fig. 2). In contrast, DIM-2-dependent RIP occurs throughout the entire linker. The absolute level of mutation is much higher for direct repeats (Fig. 1 D), but despite this difference, the linker PRP profiles for direct and inverted repeats tend to have major peaks at the same positions (Fig. 1, D and F), suggesting that the mechanism of DIM-2-dependent RIP is in both cases similar and potentially related with the pairing-induced DNA supercoiling.
According to the proposed model (33), the characteristic shapes of PRP profiles for spacers between closely positioned repeats likely reflect the nucleosome-dependent accessibility of this DNA to DIM-2. The evident similarity of such profiles for the same repeats with direct and inverted orientations of (profiles R3 vs. R4, and R5 vs. R6 in Fig. 1, D and F) suggests that the principal peaks have similar origin and that inversion of orientations has little effect on the positioning of the nucleosomes. A closer inspection reveals an interesting effect that may be relevant to the mechanism of DIM-2-dependent RIP. Specifically, the linker profiles of repeats R3 and R4 have prominent peaks around 200, 300, and 430 bp, with no significant peaks near the edges (Fig. 1 F; individual profiles are provided in Fig. 1 D). These three peaks can also be found at the same positions in the linker profile of repeat R5 (Fig. 1, D and F). However, in the linker profile of repeat R6, the third peak is shifted to 400 bp, whereas a new peak can be seen at 600 bp, making this profile overall less symmetrical (Fig. 1 F; the new peak is marked with an asterisk in Fig. 1 D).
This effect can be explained by the proposed model (Fig. 4). Repeats with higher GC content will engage in more efficient homologous pairing, thus producing stronger supercoiling and allowing the maximal number of nucleosomes on the linker. Such dense packing of nucleosomes between inverted repeat units will be asymmetrical because only one nucleosome may likely be placed near the linker-repeat junction at any given time (Fig. 4 B; also discussed below).
Overall, the model (Fig. 4) suggests that DIM-2-dependent RIP of direct versus inverted repeats is triggered by the supercoiling stress of the same sign that is likely produced by the same molecular mechanism.
Contrasting amplitudes of DIM-2-dependent RIP on direct and inverted repeats
In the wild-type genetic backgrounds, PRP profiles of closely positioned repeats are characterized by contrasting RIP propensities inside the linkers for direct and inverted orientations, respectively (20). As suggested earlier, inverted repeats could feature drastically weaker DIM-2-dependent RIP because establishing the first homologous contact near the linker (as opposed to near the flanks, Fig. 4 B) is preferred energetically (33). The results in this study, notably, the PRP profiles in Fig. 1 F and the patterns of per spore correlations in Fig. 1 G, indicate the following: 1) DIM-2-dependent RIP works qualitatively similarly on both types of repeats, and 2) all types of pairing scenarios (Fig. 4 B) are admissible. On the other hand, the above mentioned difference in the amplitudes of RIP can be due to specific conditions found on DNA segments adjacent to repeat units just after the pairing, as discussed below.
During homologous pairing of direct repeats, the supercoiling stress that accumulates on the linker is first expected to produce separate loops because the linker itself needs to remain extended to bridge the opposite ends of the paired direct repeat units (Fig. 4 A). The high bending rigidity of DNA will tend to increase the diameters of these loops, thus reducing the end-to-end distance of the linker, which will be opposed by yet higher bending rigidity of the paired repeat units. In this situation, the assembly of nucleosomes is expected to relieve the strain by stabilizing smaller DNA loops and thus allowing the linker to accommodate a larger number of supercoiling turns favoring the complete pairing of long repeats. The supercoiling stress on the flanks should initially induce DNA loops similar to those in the linker (Fig. 4 A). These loops can be absorbed by surrounding bulk DNA or they can be stabilized by the formation of nucleosomes that will mediate DIM-2-dependent RIP by the same mechanism that operates on the linker (Fig. 4 A). Some additional steps will then be required to produce H3K9me3 on these newly formed nucleosomes.
In contrast, in the case of inverted repeats, the two ends of the linker are parallel and thus remain in contact when the homologous units are paired (Fig. 4 B). In this situation, the linking number of the contour can be maintained by forming a plectoneme rather than separate loops. As a result, the formation of nucleosomes should be less favored because the plectonemes are not strained and can have sufficiently low energy, even if remaining nucleosome-free. The same process may also work in the flanks, where the plectoneme can be formed by rotating the paired repeat units. Thus, there could be a competition (thermodynamic or/and kinetic) between these mutually exclusive processes, namely, the folding of plectonemes and the formation nucleosomes. Overall, compared with the direct repeats, the relatively high stability of plectonemes to the either side of the paired inverted repeats should make nucleosome assembly less favorable, thus decreasing RIP on both the linker and the flanks.
DIM-2-dependent RIP can be influenced by the length parameters
According to our model, the double-helical twisting of the paired homologous segments produces DNA supercoiling that controls DIM-2-dependent RIP (Fig. 4). Only DNA supercoiling that leads to the formation of H3K9me3-containing repeat-proximal nucleosomes will result in RIP. In general, whereas the overall change in the linking number should be the same regardless of repeat orientation, a larger fraction of DNA supercoiling can be used for nucleosome assembly for direct repeats. This reasoning is consistent with DIM-2-dependent mutation of repeats R3–R6 (Fig. 1). However, there is at least one additional layer of complexity that can affect DIM-2-dependent RIP on the linker, specifically, the need to accommodate a discrete number of nucleosomes. To better understand the relationship between the linker size and DIM-2-dependent RIP, we designed several additional constructs (Fig. 3 A; Table 1). Repeats R7 and R9 were derived from R5 by either tripling the linker length (R7) or halving the repeat unit length (R9). Repeat R8 was derived from R9 by halving the linker length (Fig. 3 A).
To assay mutation of these constructs in an isogenic experimental system, we used a standard ridΔ strain (FGSC#12354) as a female parent. Another ridΔ strain of an opposite mating type was used as a recipient for all the constructs. RIP of only one repeat construct per cross could be assayed in this situation. We also tested repeat R5 to ensure that RIP outcomes are compatible between the two crossing strategies. Indeed, the levels of mutation of R5 in crosses X4 vs. X7 were very similar (Fig. 3, B and C). Interestingly, the threefold expansion of the linker had no significant effect on its mutation by DIM-2-mediated RIP (Fig. 3, B and C: R5 vs. R7). Because the total number of mutations remained nearly the same, the per site frequency of mutation decreased threefold (Fig. 3 B). In contrast, focusing on repeats R8 and R9, doubling the linker length strongly increased both the total number of mutations and mutation frequency (by 4.5-fold and 2.2-fold, respectively). Finally, focusing on repeats R5 and R9, halving the length of homology reduced the frequency of mutation almost proportionally, by 42.5% (Fig. 3, B and C). These nontrivial results are explained below.
First, the effect of shortening only the repeat unit length can be understood by comparing the PRP profiles of repeats R5, R9, and R3 (which differ from R5 by GC content rather than the homology length). The PRP profiles of repeats R3 and R9 are very similar (Fig. 4 D). This result suggests that decreasing (or increasing) the GC content produces the same effect as decreasing (or increasing) the homology length. This result supports the idea that repeats with lower GC content induce less DNA supercoiling, potentially because they fail to pair along their entire length. On the other hand, R9 pairing appears to be robust (for its size) and unconstrained by the relatively long linker.
Second, our results suggest that repeat R5 can engage in very strong pairing, possibly inducing excessive supercoiling stress on the linker. In this situation, the threefold expansion of the linker can only be beneficial for DIM-2-dependent RIP because it permits the same amount of supercoiling to be distributed over a larger region.
Finally, repeat R8 represents a scaled version of repeat R5, for which both the linker and the repeat units were reduced twofold. However, this scaling operation produces a linker that can carry no more than two canonical nucleosomes, which must be packed very tightly. In reality, perhaps only one nucleosome can be readily assembled on this linker, whereas the assembly of the second one is far less likely. It is possible that two or more nucleosomes need to be assembled together to be cross-linked by HP1 to promote robust DIM-2-dependent RIP. In this situation, doubling the linker length (Fig. 3: compare repeats R8 and R9) relieves the restriction on nucleosome assembly and allows the homologous segments to pair completely.
Conclusion
In summary, our current results support and advance the idea that homologous pairing for RIP involves the formation of short interspersed quadruplexes between double-stranded DNA molecules. This process is likely accompanied by a change in the twist of the participating dsDNAs. The resultant accumulation of the local supercoiling stress may serve as a signal for activating DIM-2-dependent RIP. Because the latter requires the conserved SUV39 methyltransferase DIM-5, our results further suggest that SUV39 proteins can be recruited (or activated) by the supercoiling stress in general. If so, this process may be responsible for the initiation of heterochromatin assembly on repetitive DNA in other organisms, including humans.
Author contributions
E.G. and A.K.M. designed the study. F.C. and T.-S.N. performed the experiments. E.G. and A.K.M. wrote the manuscript.
Acknowledgments
This work was supported by Agence Nationale de la Recherche (grants 11-LABX-0011, 10-LABX-0062 and ANR-19-CE12-0002), Fondation pour la Recherche Médicale (grant AJE20180539525), Centre National de la Recherche Scientifique (CNRS), and Institut Pasteur.
Editor: Helmut Schiessel.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.09.014.
Contributor Information
Alexey K. Mazur, Email: alexey@ibpc.fr.
Eugene Gladyshev, Email: eugene.gladyshev@gmail.com.
Supporting material
References
- 1.Joyce E.F., Erceg J., Wu C.-T. Pairing and anti-pairing: a balancing act in the diploid genome. Curr. Opin. Genet. Dev. 2016;37:119–128. doi: 10.1016/j.gde.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joyce E.F., Williams B.R., Wu C.-T. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 2012;8:e1002667. doi: 10.1371/journal.pgen.1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apte M.S., Meller V.H. Homologue pairing in flies and mammals: gene regulation when two are involved. Genet. Res. Int. 2012;2012:430587. doi: 10.1155/2012/430587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boateng K.A., Bellani M.A., Camerini-Otero R.D. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev. Cell. 2013;24:196–205. doi: 10.1016/j.devcel.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogan M.S., Parfitt D.-E., Spector D.L. Transient pairing of homologous Oct4 alleles accompanies the onset of embryonic stem cell differentiation. Cell Stem Cell. 2015;16:275–288. doi: 10.1016/j.stem.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu N., Tsai C.-L., Lee J.T. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 7.McKim K.S., Green-Marroquin B.L., Hawley R.S. Meiotic synapsis in the absence of recombination. Science. 1998;279:876–878. doi: 10.1126/science.279.5352.876. [DOI] [PubMed] [Google Scholar]
- 8.Duncan I.W. Transvection effects in Drosophila. Annu. Rev. Genet. 2002;36:521–556. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- 9.Dernburg A.F., McDonald K., Villeneuve A.M. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 10.Weiner B.M., Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 11.Ding D.-Q., Yamamoto A., Hiraoka Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 2004;6:329–341. doi: 10.1016/s1534-5807(04)00059-0. [DOI] [PubMed] [Google Scholar]
- 12.Kim S., Liachko I., Dunham M.J. The dynamic three-dimensional organization of the diploid yeast genome. eLife. 2017;6:e23623. doi: 10.7554/eLife.23623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saksouk N., Simboeck E., Déjardin J. Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin. 2015;8:3. doi: 10.1186/1756-8935-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrick D., Fiering S., Whitelaw E. Repeat-induced gene silencing in mammals. Nat. Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 15.Wang F., Koyama N., Tsukamoto T. The assembly and maintenance of heterochromatin initiated by transgene repeats are independent of the RNA interference pathway in mammalian cells. Mol. Cell. Biol. 2006;26:4028–4040. doi: 10.1128/MCB.02189-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Déjardin J. Switching between epigenetic states at pericentromeric heterochromatin. Trends Genet. 2015;31:661–672. doi: 10.1016/j.tig.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Kadoch C., Copeland R.A., Keilhack H. PRC2 and SWI/SNF chromatin remodeling complexes in health and disease. Biochemistry. 2016;55:1600–1614. doi: 10.1021/acs.biochem.5b01191. [DOI] [PubMed] [Google Scholar]
- 18.Hamel J., Tawil R. Facioscapulohumeral muscular dystrophy: update on pathogenesis and future treatments. Neurotherapeutics. 2018;15:863–871. doi: 10.1007/s13311-018-00675-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorer D.R., Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 20.Gladyshev E., Kleckner N. Direct recognition of homology between double helices of DNA in Neurospora crassa. Nat. Commun. 2014;5:3509. doi: 10.1038/ncomms4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhoades N., Nguyen T.-S., Gladyshev E. Recombination-independent recognition of DNA homology for meiotic silencing in Neurospora crassa. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2108664118. e2108664118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selker E.U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- 23.Hane J.K., Williams A.H., Oliver R.P. Repeat-induced point mutation: a fungal-specific, endogenous mutagenesis process. Genet. Transform. Syst. Fungi. 2015;2:55–68. [Google Scholar]
- 24.Horns F., Petit E., Hood M.E. Patterns of repeat-induced point mutation in transposable elements of basidiomycete fungi. Genome Biol. Evol. 2012;4:240–247. doi: 10.1093/gbe/evs005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossignol J.L., Faugeron G. Gene inactivation triggered by recognition between DNA repeats. Experientia. 1994;50:307–317. doi: 10.1007/BF01924014. [DOI] [PubMed] [Google Scholar]
- 26.Gladyshev E., Kleckner N. Recombination-independent recognition of DNA homology for repeat-induced point mutation. Curr. Genet. 2017;63:389–400. doi: 10.1007/s00294-016-0649-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barzel A., Kupiec M. Finding a match: how do homologous sequences get together for recombination? Nat. Rev. Genet. 2008;9:27–37. doi: 10.1038/nrg2224. [DOI] [PubMed] [Google Scholar]
- 28.Hauer M.H., Gasser S.M. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 2017;31:2204–2221. doi: 10.1101/gad.307702.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gladyshev E., Kleckner N. Recombination-independent recognition of DNA homology for repeat-induced point mutation (RIP) is modulated by the underlying nucleotide sequence. PLoS Genet. 2016;12:e1006015. doi: 10.1371/journal.pgen.1006015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazur A.K. Homologous pairing between long DNA double helices. Phys. Rev. Lett. 2016;116:158101. doi: 10.1103/PhysRevLett.116.158101. [DOI] [PubMed] [Google Scholar]
- 31.Löwdin P.O. In: Electronic Aspects of Biochemistry. Pullman B., editor. Academic Press; 1964. Some aspects of DNA replication: Incorporation errors and proton transfer; pp. 167–201. [Google Scholar]
- 32.McGavin S. A model for the specific pairing of homologous double-stranded nucleic acid molecules during genetic recombination. Heredity. 1977;39:15–25. doi: 10.1038/hdy.1977.39. [DOI] [PubMed] [Google Scholar]
- 33.Mazur A.K., Gladyshev E. Partition of repeat-induced point mutations reveals structural aspects of homologous DNA-DNA pairing. Biophys. J. 2018;115:605–615. doi: 10.1016/j.bpj.2018.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazur A.K., Nguyen T.-S., Gladyshev E. Direct homologous dsDNA-dsDNA pairing: how, where, and why? J. Mol. Biol. 2020;432:737–744. doi: 10.1016/j.jmb.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Freitag M., Williams R.L., Selker E.U. A cytosine methyltransferase homologue is essential for repeat-induced point mutation in Neurospora crassa. Proc. Natl. Acad. Sci. USA. 2002;99:8802–8807. doi: 10.1073/pnas.132212899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gladyshev E., Kleckner N. DNA sequence homology induces cytosine-to-thymine mutation by a heterochromatin-related pathway in Neurospora. Nat. Genet. 2017;49:887–894. doi: 10.1038/ng.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aramayo R., Selker E.U. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb. Perspect. Biol. 2013;5:a017921. doi: 10.1101/cshperspect.a017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 39.Gladyshev E. Repeat-induced point mutation and other genome defense mechanisms in fungi. Microbiol. Spectr. 2017;5 doi: 10.1128/microbiolspec.FUNK-0042-2017. FUNK 0042-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCluskey K., Wiest A., Plamann M. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J. Biosci. 2010;35:119–126. doi: 10.1007/s12038-010-0014-6. [DOI] [PubMed] [Google Scholar]
- 41.Ewing B., Hillier L., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 42.Gordon D., Abajian C., Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 43.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 45.Irelan J.T., Hagemann A.T., Selker E.U. High frequency repeat-induced point mutation (RIP) is not associated with efficient recombination in Neurospora. Genetics. 1994;138:1093–1103. doi: 10.1093/genetics/138.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cantor C.R., Schimmel P.R. W. H. Freeman; San Francisco, CA: 1980. Biophysical Chemistry. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.