Abstract
Styrene-maleic acid (SMA) copolymers solubilize biological membranes to form lipid nanoparticles (SMALPs) that contain membrane proteins surrounded by native lipids, thus enabling the use of a variety of biophysical techniques for structural and functional studies. The question of whether SMALPs provide a truly natural environment or SMA solubilization affects the functional properties of membrane proteins, however, remains open. We address this question by comparing the photoactivation kinetics of rhodopsin, a G-protein-coupled receptor in the disk membranes of rod cells, in native membrane and SMALPs prepared at different molar ratios between SMA(3:1) and rhodopsin. Time-resolved absorption spectroscopy combined with complex kinetic analysis reveals kinetic and mechanistic differences between the native membrane and SMA-stabilized environment. The results suggest a range of molar ratios for nanoparticles suitable for kinetic studies.
Graphical abstract
Significance
Styrene-maleic acid (SMA) copolymers solubilize biological membranes to form nanoparticles with membrane proteins surrounded by native lipids. These have proven useful for structural studies of membrane proteins, but whether they provide an environment that does not affect their functional properties remains an open question. We show that solubilization of the membrane protein rhodopsin with relatively low SMA/rhodopsin molar ratios produces nanoparticles suitable for photokinetic studies. In such samples, we observe well-defined rhodopsin photoactivation intermediates. In contrast, using high SMA/protein molar ratios yields extremely slow photokinetics, the path becomes disrupted, and the active state is not reached. These results suggest that studies using proteins solubilized at high SMA concentrations should proceed with caution.
Introduction
Most membrane-bound proteins require a lipid membrane environment for function and lose their activity upon solubilization by detergents. Structural studies are difficult to conduct on membranes, especially in the presence of other proteins. Kinetic studies using optical techniques also face great difficulties because of enhanced light scattering in membrane suspensions. It is crucial to find solubilizing agents that do not alter the protein activation mechanism to circumvent these challenges.
Amphipathic copolymers such as styrene-maleic acid (SMA) have emerged as an important tool to solubilize membrane proteins for structural and functional studies. The addition of SMA to biological membranes results in the spontaneous formation of discoidal lipid nanoparticles (SMALPs) that contain active protein (1, 2, 3). Recently, we used SMA(3:1) to solubilize the G-protein-coupled receptor (GPCR) rhodopsin from its native membrane environment and compared the functionality of the solubilized protein to that observed in its native membrane (4).
In vertebrate rod cells, the photoreceptor rhodopsin initiates phototransduction. Rhodopsin consists of the apoprotein opsin and the chromophore ligand, 11-cis-retinal, covalently bound to lysine 296 through a protonated Schiff base linkage (5,6). Upon absorption of light, rhodopsin goes through a series of intermediates that first involve structural changes local to the chromophore but eventually result in global conformational changes that enable the binding of opsin to the G-protein transducin (7,8). The subsequent activation of transducin starts an enzyme cascade, which causes blockage of sodium channels in the rod cells, leading to a change in electrical current through the cells and thus detection of the light.
A great deal of information has been gathered over the years about the mechanism through which rhodopsin reaches its active state. Much of the understanding has been obtained on bovine rhodopsin with the use of time-resolved optical absorption (TROA), infrared and laser Raman spectroscopies (9, 10, 11, 12). Recent TROA studies have shown that the activation mechanism follows a square scheme (13) or a double-square scheme, depending on temperature and pH conditions (14).
In our earlier, complementary work (4), we determined the efficiency of rhodopsin membrane solubilization by SMA as a function of SMA/rhodopsin ratio and found that above the ratio of ∼20, practically complete solubilization is achieved, whereas below that only a fraction of the membranes is solubilized. A threshold SMA/lipid mass ratio of 1.2–1.3 has also been reported for complete solubilization of lipid vesicles by SMA(2:1) (15). Based on the size of dimyristoylphosphatidylcholine (DMPC) SMALPs (16), we estimated an SMA/rhodopsin ratio of around 10 in rhodopsin-containing SMALPs of the same size. This estimate gives an SMA/membrane mass ratio in the nanoparticles only slightly lower than the one mentioned above for lipid solubilization. Thus, SMA applied in solubilization above such minimal threshold value can be considered an excess and may or may not be desirable. We also monitored the formation of the photoinduced final state of rhodopsin in SMALPs. The results showed that the relative amount of SMA used in solubilization could dramatically impact rhodopsin’s photoresponse. During static photobleaching experiments at room temperature, particles formed at and below the SMA/rhodopsin ratio of 10 showed the known active-state intermediates, whereas rhodopsin solubilized in excess SMA, above the threshold ratio, did not. Thus, rhodopsin kinetics in SMA-stabilized nanoparticles may deviate from the kinetics observed in the native membrane. This idea prompted our study, in which we investigate the activation mechanism of rhodopsin in an SMA-stabilized environment using TROA spectroscopy.
The general aim of our work here is to test whether membrane-bound proteins solubilized into nanoparticles by SMA preserve the dynamic and kinetic properties they possess in their native membranes. Rhodopsin is uniquely suited for this test because its photokinetics has been studied extensively in native and artificial environments (17,18). In this comparative study, rhodopsin photokinetics is measured and evaluated in both membranes and SMALPs prepared at different SMA/rhodopsin ratios. The focus is on key reaction steps that follow the formation of the Lumi intermediate and lead to the active-state Meta II, which span the microsecond to second time interval, because these steps are known to be more sensitive to the protein environment than the steps leading to Lumi formation (19).
The results show that rhodopsin in SMALPs formed at relatively low ratios of SMA/rhodopsin follows a reaction mechanism that leads to the active state, whereas the mechanism in SMALPs made at high SMA/rhodopsin ratios is compromised, and thus, the active-state composition is not reached.
Materials and methods
Preparation of native membrane suspensions
Bovine rhodopsin in disk membrane was isolated from hypotonically washed rod outer segment (ROS) suspensions as reported previously (20) and resuspended in 10 mM Tris, 100 mM NaCl buffer (pH 8 at 30°C). The concentration of rhodopsin was determined spectrophotometrically (V750; JASCO, Easton, MD) by measuring the change in absorbance at 500 nm (ε = 40,600 M−1 cm−1) before and after irradiation in the presence of 3% lauramine oxide. The samples used to study rhodopsin in membrane were ∼25 μM and before use were mildly sonicated in an ice water bath under nitrogen using a Microson sonicator (Heat Systems Ultrasonics, Farmingdale, NY) at medium power output for three 30 s on-off cycles.
Preparation of SMA stock solution
SMA(3:1) copolymers, with three styrene units per one maleic acid unit, have aweight average molecular weight of 10 kg/mol (Polyscope Polymers B.V., Geleen, the Netherlands). The polymer was precipitated by the addition of 6 M HCl, rinsed with distilled water, and collected by filtration. The SMA stock solution (5% w/v) was made by dissolving SMA powder in Tris buffer, adjusted to pH 8, and stored at −20°C.
Solubilization of rhodopsin using SMA
Different volumes of the 5% (w/v) SMA stock solution were added directly to suspensions of native membrane that contained ∼50 μM rhodopsin to provide SMA/rhodopsin molar ratios of 5, 10, 20, and 100. Samples were centrifuged for 20 min at 10,000 × g, and the soluble fraction that contained rhodopsin-SMALPs was collected. Given the specific rhodopsin/lipid ratio in the ROS membrane, the SMA/rhodopsin molar ratio is approximately equal to 10 times the SMA/membrane mass ratios.
Time-resolved optical absorbance measurements
Activation kinetics of rhodopsin in native membrane and SMALPs (each ∼1 mg/mL rhodopsin) were investigated using TROA spectroscopy. The reaction was initiated by an ∼7 ns pulse of 477 nm light (80 μJ/mm2) generated by a dye laser pumped by the third harmonic of a neodymium-doped yttrium aluminum garnet (Nd:YAG) laser. After the laser excitation, post- minus prephotolysis difference spectra were recorded in the 330–630 nm wavelength range at selected delay times spaced on a logarithmic timescale using an apparatus described previously (21,22). Briefly, this system used an optical multichannel analyzer to obtain the absorption spectra of intermediates formed by laser photolysis. A pulse of white light from a flash lamp was used to probe the absorbance changes. The incident light was focused onto the sample, and the transmitted light was refocused onto the slit of a spectrograph that disperses the light across the channels of a gated charge-coupled device. A digital delay generator synchronized the gate pulses applied to the laser, flash lamp, and optical multichannel analyzer detector.
To enable minimal use of sample per photoflash, an insert was placed into a 4 mm × 4 mm cuvette (Starna Cells, Atascadero, CA), constructed to form a 2 μL sample chamber at the bottom, with a height of 1 mm and an actinic pathlength of 0.5 mm, contained between the cuvette walls (23). A l/2 cc glass syringe driven by a linear stepper motor was used to pump fresh sample after each measurement. 4 μL of fresh sample was delivered to the sample chamber after each photolysis pulse, and a channel on the insert allows for photolyzed material to leave and collect outside of the sample path in the cuvette. For efficient solubilization, the solution pH was set to 8, and because alkaline pH favors the forms with protonated Schiff base and higher temperature favors the forms with unprotonated Schiff base in rhodopsin kinetics (24), the experiments were carried out at 30°C to achieve a balance between the two forms.
Global exponential fitting
Assuming first-order kinetics, the time dependence of the post- minus prephotolysis absorption difference spectra enclosed in the data matrix, A(λ, t), was fitted to a sum of time-dependent exponential functions: A(λ, t) = Σ bi (λ) × exp(−ri × t) + D, where λ and t denote wavelength and time, respectively. The exponents ri are the apparent rates; bi (λ) are the amplitude spectra, called b-spectra; and D is the matrix of the residuals of the fit. Fitting the entire data matrix is technically challenging and can be avoided by separating the spectral and the temporal information in the data matrix using singular value decomposition: A = U × S × VT. Matrix U contains the orthonormal spectral vectors, V is the matrix of orthonormal temporal vectors, and matrix S is the measure of the U and V vector pair contributions to the data matrix. Only a limited number of time-dependent vectors, the significant V, were globally fitted to sums of exponentials yielding the apparent rates ri. The b-spectra were calculated by combining the results of the exponential fit with the significant U spectral vectors (25).
Spectral composition of sequential intermediates and time evolution of spectral forms
The experimental spectra in the data matrix are linear combinations of a limited number of spectral forms. The term “spectral form” refers to a model absorption spectrum characteristic of the state of the chromophore, such as unprotonated chromophore with λmax = 380 nm, called the unprotonated form, or protonated chromophore with λmax ≥ 420 nm, called the protonated form. These model spectra may represent a single intermediate or multiple, physically different isospectral intermediate states evolving in separate time intervals in the underlying reaction sequence. Because the spectral forms are not attached to intermediates in any specific kinetic mechanism, their time evolution can be assessed without knowing the reaction mechanism itself. The spectral forms that constitute the experimental spectra in the data matrix are more easily deduced from the spectra of the straight sequential intermediates than from the b-spectra. Thus, the b-spectra arranged in the B(λ, n) matrix were converted into the spectra of the sequential intermediates enclosed in the In(λ, n) matrix: In(λ, n) = B(λ, n) × W(n, n)−1. W(n, n) is the eigenvector matrix of the kinetic matrix constructed for the straight sequential scheme in which the apparent rates, ri, are assigned to the reaction rates between the intermediates. The intermediate spectra are interpreted as sums of the spectral forms: In(λ, n) = E(λ, m) × Q(m, n), where E(λ, m) is the matrix of the m spectral forms and Q(m,n) is the composition matrix of the n sequential intermediates. Note the number of sequential intermediates may be different from the number of spectral forms.
To obtain the time evolution of the spectral forms contained in the spectra of the straight sequential intermediates, first, the matrix of the time-dependent concentrations of the n sequential intermediates, Cin(n, t), was calculated using the kinetic matrix constructed for the straight sequential scheme. The product of Cin(n, t) and the composition matrix, Q(m, n), gives the time evolution of the m spectral forms: Cm(n, t) = Q(m,n) × Cin(n,t).
The Lumi, Meta480, Meta460, and Meta380 spectral forms used in the decomposition were obtained the following way. The Lumi spectral form is the experimental spectrum of the first sequential intermediate. The first sequential intermediate is not a single state, but an equilibrated mixture of the Lumi I and Lumi II states produced by an early microsecond transition not discussed in this work. The Meta480 and Meta380 and rhodopsin spectral forms were taken from static spectral records reported in our previous study (4), and the Meta460 spectral form is a blue-shifted variant of the Meta480 spectral form. The Meta480, Meta380, and Meta460 spectra were referenced against the rhodopsin spectrum and were scaled to yield the sum of all contributions, expressed as fraction numbers, equal to one for each intermediate of the straight sequential scheme.
Microscopic rates of kinetic models
For kinetic models more complex than the straight sequential scheme, the apparent rates from the global exponential fit cannot be assigned to microscopic rates in the model, and the spectra of the intermediates cannot be calculated from the experimental b-spectra directly. To validate a proposed complex kinetic model, we assume that the spectra of the spectral forms, E(λ, m), derived from the spectra of the sequential intermediates, represent the spectra of one or more intermediates of the complex model and thus can be used as intermediate model spectra for the complex scheme with l intermediate, E(λ, l). The microscopic rate constants of the reaction steps in the complex model are obtained by a fitting procedure designed to reproduce the experimental apparent rates by the eigenvalues and the experimental b-spectra by the product of the intermediate model spectra and the eigenvector matrix of the kinetic matrix for the proposed complex model: B(λ, n) = In(λ, n) × W(l, n). Models with l > n are considered degenerate and have multiple solutions (14).
Results
Rhodopsin in native membrane
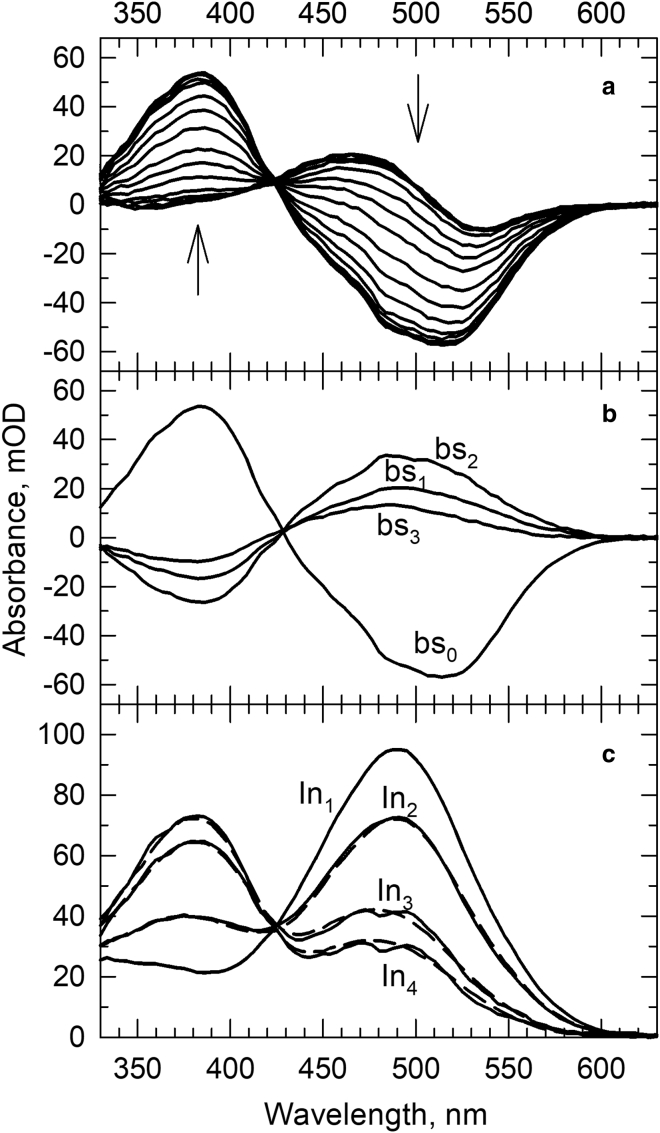
The time-resolved absorption difference spectra recorded for rhodopsin in its native membrane (rM) in the 330–630 nm wavelength range over a 1 μs–50 ms time window are shown in Fig. 1 a. Global exponential fit of the data matrix yielded 70 μs and 0.74 and 6.5 ms apparent lifetimes (τi = 1/ri), and the b-spectra corresponding to these lifetimes are shown in Fig. 1 b. The b-spectra in Fig. 1 b were converted into intermediate spectra of an unbranched, straight sequential scheme constructed using the apparent rates listed above. The intermediate spectra were decomposed into sums of spectral forms, as discussed above. For easier visualization of the experimental (Fig. 1 c, solid lines) and the reproduced sequential intermediate spectra (Fig. 1 c, dashed lines), the difference spectra were converted into absolute spectra by adding back the spectrum of the bleached rhodopsin, the scaled rhodopsin spectrum used as the reference spectrum for the spectral forms. The compositions of the sequential intermediate spectra expressed as fraction values of the different spectral forms present in the intermediates are listed in Table 1.
Figure 1.
(a) Light-induced absorption difference spectra of rhodopsin in membrane recorded at time delays from 1 μs to 50 ms. Arrows indicate the direction of change as time progresses. (b) The b-spectra corresponding to lifetimes 70 μs (bs1), 0.74 ms (bs2), and 6.5 ms (bs3) and the time-independent final spectrum (bs0) produced by global exponential fit to the time-dependent absorption spectra. (c) Solid lines: intermediate spectra of the sequential scheme calculated from the b-spectra. The scheme was constructed based on the lifetimes. Dashed lines: reproduction of the intermediate spectra by the model spectra using the spectral compositions obtained for the intermediates.
Table 1.
Fractional composition of sequential intermediates for membrane and nanoparticle samples
| Lumi | Meta480 | Meta380 | Meta460 | |
|---|---|---|---|---|
| rM | ||||
| In 1 | 1.00 | – | – | – |
| In 2 | 0.71 | 0.05 | 0.24 | – |
| In 3 | – | 0.45 | 0.55 | – |
| In 4 | – | 0.34 | 0.66 | – |
| rN10 | ||||
| In1 | 1.00 | – | – | – |
| In 2 | 0.92 | – | 0.08 | – |
| In 3 | 0.10 | 0.82 | 0.08 | – |
| In 4 | 0.10 | 0.65 | 0.25 | – |
| In 5 | – | 0.35 | 0.65 | – |
| rN20 | ||||
| In 1 | 1.00 | – | – | – |
| In 2 | 0.90 | 0.07 | 0.03 | – |
| In 3 | 0.33 | 0.65 | 0.02 | – |
| In 4 | 0.08 | 0.65 | 0.12 | 0.15 |
| In 5 | – | 0.10 | 0.30 | 0.60 |
| rN100 | ||||
| In 1 | 1.00 | – | – | – |
| In 2 | 0.45 | 0.55 | – | – |
| In 3 | 0.23 | 0.30 | 0.15 | 0.32 |
rM, membrane; rN, nanoparticle.
The classical sequential model of rhodopsin photoconversion, deduced from low-temperature experiments and used in many reports, considers only three sequential intermediates in the time interval covered in this study: Lumi, Meta I, and Meta II. We found four sequential intermediates, in agreement with an earlier study (13). Furthermore, the second and third sequential intermediate spectra display two well-separated absorption bands in Fig. 1 c, which can only be reconciled with the simultaneous presence of two molecular states in these intermediates. These clearly demonstrate that, despite its popularity, the classical sequential model is not a valid representation of kinetics at ambient temperatures.
Rhodopsin in SMALPs: SMA/rhodopsin molar ratio of 10
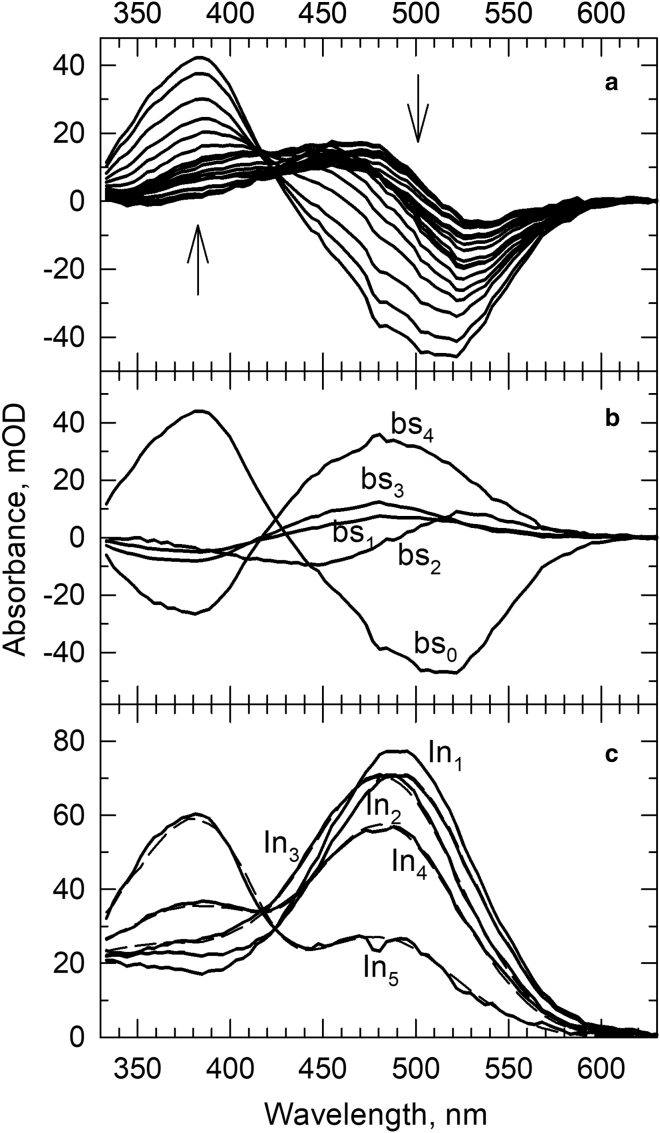
The time-resolved absorption difference spectra were recorded at delay times from 1 μs to 2 s for the nanoparticle samples made with an SMA/rhodopsin molar ratio of 10 (rN10). These are shown in Fig. 2 a. Global exponential fitting resolved 85 μs, 2.5 ms, 69 ms, and 0.73 s apparent lifetimes and the b-spectra are displayed in Fig. 2 b. Fitting only three apparent lifetimes, which was sufficient for the membrane sample, produced unacceptable residuals with spectral structures above the noise level. The long lifetimes suggest that rhodopsin kinetics in the rN10 nanoparticle is much slower than in the membrane. The sequential intermediate spectra calculated from the b-spectra and the spectra reproduced by the spectral forms according to the composition matrix in Table 1 are displayed in Fig. 2 c (solid lines, calculated spectra, and dashed lines, composition matrix spectra).
Figure 2.
(a) Light-induced absorption difference spectra of rhodopsin in the rN10 sample recorded at time delays from 1 μs to 2 s. Arrows indicate the direction of change as time progresses. (b) The b-spectra corresponding to lifetimes 85 μs (bs1), 2.5 ms (bs2), 69 ms (bs3), and 0.73 s (bs4) and the time-independent final spectrum (bs0) produced by global exponential fit to the time-dependent absorption spectra. (c) Solid lines: intermediate spectra of the sequential scheme calculated from the b-spectra. The scheme was constructed based on the lifetimes defined above. Dashed lines: reproduction of the intermediate spectra by the model spectra using the spectral compositions obtained for the intermediates.
Although the membrane and the rN10 nanoparticle sample display very different apparent rates, the compositions of their last intermediate spectra are surprisingly similar, as both final states contain the Meta480 and Meta380 forms in a roughly 1:2 ratio. This final composition, characteristic of the equilibrium between Meta I480 and the active-state Meta II at 30°C (13), however, must have been reached in the rN10 nanoparticle differently from the membrane because the number of apparent rates in the rN10 nanoparticle is one more than in the membrane.
Rhodopsin in SMALPs: SMA/rhodopsin molar ratios of 20 and 100
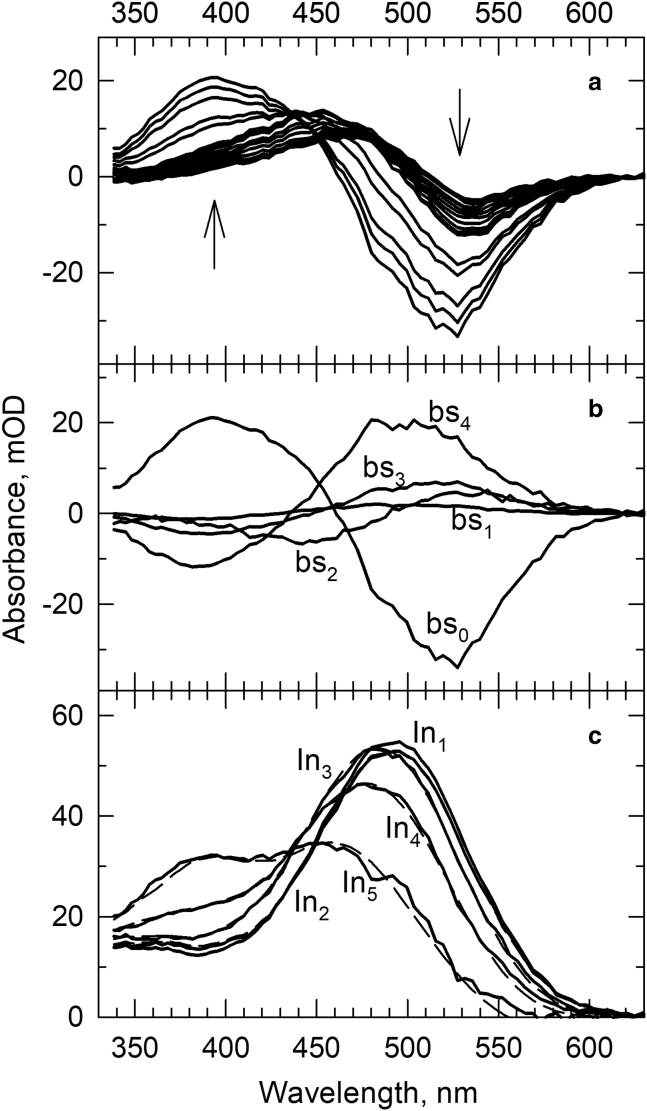
The time-resolved absorption difference spectra for the nanoparticle samples prepared at SMA/rhodopsin molar ratio of 20 (rN20) were recorded at delay times from 10 μs to 20 s and are shown in Fig. 3 a. Global exponential fitting revealed four lifetimes: 0.26 ms, 12 ms, 0.78 s, and 6.4 s. The corresponding b-spectra are shown in Fig. 3 b, and the sequential intermediate spectra calculated from them are in Fig. 3 c, solid lines, together with the spectra reproduced by the spectral forms according to the composition matrix, dashed lines. The b-spectra and the absolute spectra of the sequential intermediates, and particularly the composition matrix in Table 1, indicate that only very limited amounts of the unprotonated intermediates, absorbing at 380 nm, are produced during Lumi decay.
Figure 3.
(a) Light-induced absorption difference spectra of rhodopsin in the rN20 sample recorded at time delays from 10 μs to 20 s. Arrows indicate the direction of change as time progresses. (b) The b-spectra corresponding to lifetimes 0.26 ms (bs1), 12 ms (bs2), 0.78 s (bs3), and 6.4 s (bs4) and the time-independent final spectrum (bs0) produced by global exponential fit to the time-dependent absorption spectra. (c) Solid lines: intermediate spectra of the sequential scheme calculated from the b-spectra. The scheme was constructed based on the lifetimes. Dashed lines: reproduction of the intermediate spectra by the model spectra using the spectral compositions obtained for the intermediates.
The final stage detected is also different from that of the membrane and the rN10 samples because no combination of Meta480 and Meta380 spectral forms could reproduce the last intermediate spectrum; it required an additional form having λmax at 460 nm, Meta460. This blue-shifted protonated form is a characteristic component of the spectral forms detected on the seconds-to-minutes timescale during the deactivation of rhodopsin (26). The tendency to deviate from the kinetic path followed by rhodopsin in the native membrane, as observed in the rN20 sample, is even stronger in the rN100 nanoparticle. The time-resolved absorption difference spectra recorded in the 10 μs–10 s time interval are shown in Fig. 4 a. Regardless of the lower accuracy of the data, as compared to the other samples presented, it is quite obvious that significant spectral changes occur only at very late times. Two exponentials with 56 ms and 3.2 s lifetimes were fitted to the data. The faster (56 ms) b-spectrum in Fig. 4 b looks like a Lumi minus Meta480 spectral change, and only the slower b-spectrum (3.2 s) shows the conversion of protonated forms into unprotonated ones. The absolute spectra in Fig. 4 c and the composition matrix in Table 1 show that the 380 nm absorbing form appears together with the 460 nm absorbing form. The latter is known to be part of the deactivation process, as already mentioned for the rN20 sample.
Figure 4.
(a) Light-induced absorption difference spectra of rhodopsin in the rN100 sample recorded at time delays from 10 μs to 10 s. Arrows indicate the direction of change as time progresses. (b) The b-spectra corresponding to lifetimes 56 ms (bs1) and 3.2 s (bs2) and the time-independent final spectrum (bs0) produced by global exponential fit to the time-dependent absorption spectra. (c) Solid lines: intermediate spectra of the sequential scheme calculated from the b-spectra. The scheme was constructed based on the lifetimes. Dashed lines: reproduction of the intermediate spectra by the model spectra using the spectral compositions obtained for the intermediates.
Because 380 nm absorbing forms are present past the active state (26), it is difficult to state what portion of the Meta380 spectral component in the final sequential intermediate belongs to the true Meta II intermediate of the active state. This reasoning also applies to the final state observed in the rN20 sample. The Meta380 spectral form has a bigger share in the final spectrum of the rN20 sample, making it more probable that the true Meta II intermediate of the active state is present. Fluorescence measurements reported in our earlier work (4) showed that the retinylidene Schiff base linkage is intact in the late forms.
The data for rN20 and rN100 nanoparticles presented above show very distorted rhodopsin photokinetics that do not lead to the active state. We tested whether removal of the excess SMA from the SMALPs formed could restore the rhodopsin photokinetics and whether the active-state composition seen in SMALPs prepared at low SMA/rhodopsin ratios could be reached. We carried out buffer exchange on the rN100 sample and replaced the SMA solution with an SMA-free buffer. A comparison of the photolysis products before and after excess SMA removal showed no spectral difference, the active state was not restored. This indicates that when SMA is used in excess of the minimal amount required for solubilization, it binds to the membrane in a way that causes further structural changes in the nanoparticles formed, thus changing the rhodopsin environment, and these changes are not reversible.
Time evolution of spectral forms in native membrane and nanoparticles
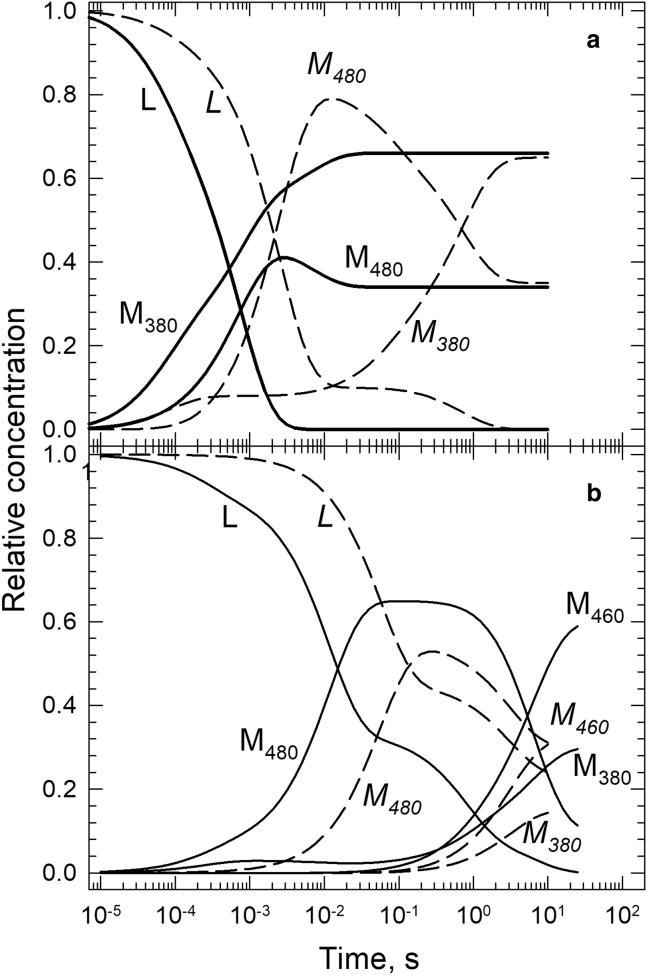
The time evolution of spectral forms gives a better picture of the kinetics than the composition of the sequential intermediates can provide. It is calculated as the product of the composition matrix and the matrix of the time-dependent concentrations of the sequential intermediates, as discussed above. Fig. 5 shows the time courses of the Lumi, Meta480, Meta380, and Meta460 spectral forms for the membrane (Fig. 5 a, solid lines), rN10 (Fig. 5 a, dashed lines and italicized labels), rN20 (Fig. 5 b, solid lines), and rN100 (Fig. 5 b, dashed lines and italicized labels) samples.
Figure 5.
(a) Time evolution of Lumi, Meta480, and Meta380 spectral forms during rhodopsin photoreaction in membrane (L, M480, and M380, solid lines) and in rN10 nanoparticles (L, M480, and M380, dashed lines). (b) Time evolution of Lumi, Meta480, Meta460, and Meta380 spectral forms during rhodopsin photoreaction in rN20 (L, M480, M460, and M380, solid lines) and in rN100 nanoparticles (L, M480, M460, and M380, dashed lines).
The time courses of the spectral forms for the membrane (Fig. 5 a, solid lines) and for the rN10 sample (Fig. 5 a, dashed lines) differ significantly. The membrane sample shows a branched decay of the Lumi form into the Meta480 and Meta380 forms at early times, followed by equilibration of the latter two forms on the millisecond timescale. These are in agreement with an earlier report (13). Rhodopsin in the rN10 sample completes a similar but slower kinetic path and reaches the final active state observed in the native membrane. Whereas it takes rhodopsin less than 10 ms to reach the active state in the membrane, it takes one more step and almost a second to do so in the rN10 nanoparticle. Furthermore, in the rN10 nanoparticle the protonated Meta480 is the dominant spectral form and not the unprotonated Meta380 form. The latter builds up gradually in the slow transitions. Interestingly, the Lumi spectral form is present for a long time; a fraction of it follows the time dependence of the Meta480 form. The simultaneous presence of two spectral forms indicates a reversible step in the kinetics; thus, the kinetic scheme for rN10 may contain a Lumi-to-Meta480 equilibrium step as part of the Lumi decay. The time courses of the spectral forms also suggest that two isospectral intermediates may be responsible for the long-lasting decay of the Meta480 spectral form.
The time courses for the rN20 and the rN100 nanoparticles in Fig. 5 b show a drastically different picture from the one just discussed. The photoactivation pathway for rhodopsin in these nanoparticles exhibits very slow kinetics, and the compositions of their final intermediates are very different from the ones seen in the membrane and the rN10 samples. In rN20 (Fig. 5 b, solid lines), around 65% of the Lumi spectral form decays into Meta480 reversibly and only around 2–3% of it into Meta380. Only at late times, around 1 s, do we see more Meta380 formation from both Lumi and Meta480. However, the buildup of Meta380 is accompanied by the formation of twice as much Meta460. The Meta460 spectral form, and probably a big part of the Meta380, is not part of the kinetic chain leading to the active state. The rN100 sample (Fig. 5 b, dashed lines) displays even slower kinetics and practically a single, and even more reversible, transition between Lumi and Meta480 before the onset of spectral changes characteristic of deactivation after the active state in rhodopsin kinetics.
Discussion
The description of kinetics on the molecular level involves identifying the intermediate states, presenting a kinetic model that contains the interconnections between states, and defining the microscopic reaction rate constants of the connecting steps. The kinetic picture presented above deals with apparent rates and b-spectra, spectral forms, and their time evolution. None of these actually needed the introduction of kinetic schemes; we used the intermediates of the sequential scheme instead of the b-spectra only for convenience and better visualization of the spectral forms present at the different stages of the reaction. The aim of the analysis above was to get hints on the structure of the possible kinetic schemes. Based on these hints, we now connect the experimental observations to feasible kinetic models that contain intermediates with spectral characteristics of molecular states.
We showed that the kinetic path of rhodopsin solubilized at SMA/rhodopsin molar ratios of 20 and above is compromised by the onset of processes that do not belong to the path leading to the active state. Analyzing distorted or fragmented kinetic models is outside the scope of this work. Therefore, kinetic analysis of the rN20 and the rN100 samples will not be pursued further. Instead, we focus on rhodopsin kinetics in the rN10 sample, in which the influence of SMA can be fully explored.
The kinetic picture we deduce for the rN10 sample also applies to nanoparticles made at lower SMA/rhodopsin molar ratios. Our previous work showed that they all have the same spectral composition seconds after photobleaching (4). To get more insight into the kinetics at lower molar ratios, we tested the kinetics in the nanoparticle made at SMA/rhodopsin ratio of five (rN5), the results of which are not presented here because we found only slight deviations, bordering the limits of reproducibility, from the kinetics observed and reported here for the rN10 sample. It is very likely that nanoparticles made at SMA/rhodopsin molar ratios that are not exceeding 10–15 have similar structures, resembling, perhaps, the reported disk-shaped SMALP structure (16), whereas nanoparticles made at higher molar ratios are significantly different from these. The threshold molar ratio of 10–15 corresponds roughly to the minimal SMA/lipid mass ratio of 1.2:1 needed for complete solubilization of lipid membranes (15).
Rhodopsin in membrane follows the square kinetic model
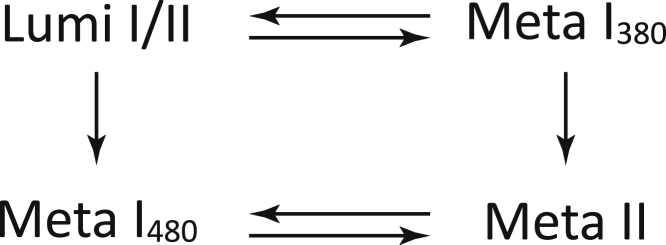
Rhodopsin in native membranes showed three apparent lifetimes and four b-spectra, which is typically seen at ambient temperatures and neutral pH and adequately described by the square model (Fig. 6; (13)).
Figure 6.
Transitions between the late intermediates in the square model of the rhodopsin activation mechanism.
The shapes of the b-spectra are intimately connected to the underlying kinetic scheme, the elements of which can be deduced following a few simple rules (27). The b-spectra that correspond to well-separated lifetimes are viewed as difference spectra produced by intermediates participating in a transition. The positive lobe represents the spectra of the decaying and the negative lobe the spectra of the emerging intermediates in the transition. Reversible transitions display b-spectra with reduced amplitudes, and branched decays of intermediates produce more than one spectral component in the negative lobe. Each transition is considered a fragment of the kinetic scheme.
The b-spectrum with 70 μs lifetime in Fig. 7 a, bs1, has the shape of a Lumi minus Meta380 difference spectrum with reduced amplitude. According to the rules of b-spectra interpretation, it represents the partial decay of the Lumi intermediate into the Meta I380 intermediate via a reversible step in the scheme. The b-spectrum also has a small Lumi-to-Meta I480 contribution, suggesting a branched Lumi decay mechanism. These are in agreement with the square model. The b-spectrum with 0.74 ms lifetime, bs2, shows the decay of the equilibrated Lumi/Meta I380 mixture into both Meta I480 and Meta II via irreversible steps, as the square model predicts. The 6.5 ms b-spectrum, bs3, is a Meta480 minus Meta380 difference spectrum and represents the establishment of the final equilibrium between the Meta I480 and Meta II intermediates. The spectrum of the equilibrated mixture of the Meta I480 and the Meta II active state is the last b-spectrum in Fig. 7 a, bs0.
Figure 7.
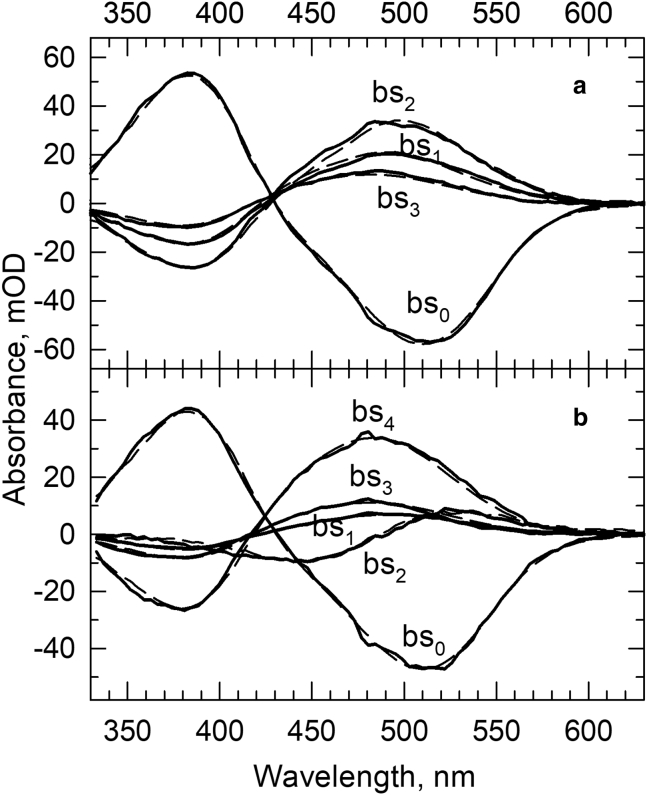
(a) Fitting the square model to the experimental b-spectra (solid lines) and lifetimes obtained for rhodopsin in membrane. The b-spectra corresponding to 73 μs (bs1), 0.77 ms (bs2), and 6.6 ms (bs3) lifetimes and the time-independent final spectrum (bs0) reproduced by the kinetic matrix are shown by dashed lines. (b) Fitting the double-square model to the experimental b-spectra (solid lines) and lifetimes obtained for rhodopsin in rN10 nanoparticles. The b-spectra corresponding to 93 μs (bs1), 2.7 ms (bs2), 68 ms (bs3), and 0.74 and 0.75 s (degenerate, bs4) lifetimes and the time-independent final spectrum (bs0) reproduced by the kinetic matrix are shown by dashed lines.
The validity of the square model was tested and the microscopic rate constants of the transitions between the intermediate states in the model were obtained by a fitting procedure designed to reproduce the experimental b-spectra and the lifetimes. The reproduced b-spectra corresponding to the reproduced lifetimes of 73 μs and 0.77 and 6.6 ms are shown in Fig. 7 a, dashed lines. The microscopic rate constants are presented in Table 2. The rate constants are similar to those published for rhodopsin at 30°C and pH 7 (13). We can conclude that the reaction kinetics of rhodopsin in the native membrane at 30°C and pH 8 is adequately described by the square model (Fig. 6), the intermediates of which represent molecular states of rhodopsin.
Table 2.
Microscopic reaction rate constants, s−1, for membrane in the square model and for rN10 nanoparticles in the double-square model
| rM (k/−k) | rN10 (k/−k) | |
|---|---|---|
| Lumi II ↔ Meta I380 | 3.3 × 103/8.0 × 103 | 8.9 × 102/9.6 × 103 |
| Lumi II ↔ Meta Ia480 | 7.4 × 102/0 | 3.4 × 102/4.1 × 101 |
| Meta I380 ↔ Meta IIa | 3.0 × 103/0 | 2.9 × 102/1.0 × 101 |
| Meta Ia480 ↔ Meta IIa | 1.0 × 102/5.1 × 101 | 3.2 × 10−1/1.0 |
| Meta Ia480 → Meta Ib480 | N/A | 1.6 |
| Meta Ib480 → Meta IIb | N/A | 1.2 |
| Meta IIa ↔ Meta IIb | N/A | 8.6 × 10−1/4.6 × 10−1 |
N/A, not applicable; rM, membrane.
Rhodopsin in rN10 nanoparticle follows the double-square kinetic model
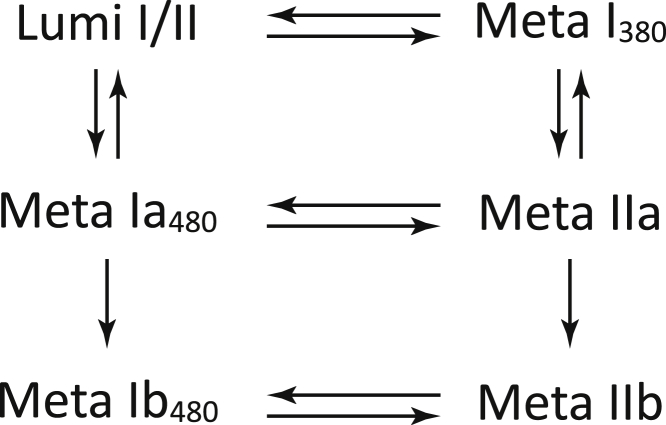
Rhodopsin in both the native membrane and the rN10 nanoparticle completes the kinetic path and displays the spectrum characteristic for the active state at the end of the reaction chain. However, in the nanoparticle, it takes rhodopsin one more step to cover the path from Lumi to the active state, and there are likely two Meta I480 states in the reaction sequence. To accommodate the extra step and intermediate, the single-square model was extended by adding additional intermediates. The extended kinetic model is the double-square model (Fig. 8) introduced by us recently to describe the kinetics at moderately low temperatures and elevated pH values of bovine rhodopsin in membrane (14) and lipid nanodisks (18), as well as human rhodopsin kinetics in membrane (17). The model contains a pair of isospectral Meta Ia480/Meta Ib480 protonated intermediates and their unprotonated Meta IIa/Meta IIb counterparts.
Figure 8.
Transitions between the late intermediates in the double-square model of the rhodopsin activation mechanism.
The 85 μs first b-spectrum, Fig. 7 b, bs1, is similar in shape but somewhat smaller in amplitude than the 70 μs first b-spectrum observed in the membrane. It shows the partial decay of the Lumi intermediate into the Meta I380 intermediate in the scheme via an equilibrium step, which is more back shifted than the analogous step in the membrane. The 2.5 ms second lifetime is 3.4 times longer than the 0.74 ms second lifetime detected in the membrane, and the corresponding b-spectrum, Fig. 7 b, bs2, has the shape of a nearly perfect Lumi minus Meta480 difference spectrum. The long-lasting and simultaneous presence of Lumi and Meta480 spectral forms in Fig. 5 a is consistent with the Lumi-to-Meta Ia480 step in the double-square model being reversible, which makes the Meta I380-to-Meta IIa step reversible as well. The lack of a Meta380 spectral component in the negative lobe of the 2.5 ms b-spectrum indicates that the equilibrated Lumi/Meta I380 mixture decays preferentially through the protonated Meta Ia480 intermediate and not through the unprotonated Meta IIa. The assignment of the first and second b-spectra to the steps in the upper part of the double-square model (Fig. 8) thus seems quite straightforward and similar to the analogous assignment of the corresponding membrane b-spectra to the square model above (Fig. 6).
The third and fourth b-spectra are more problematic to assign to specific steps in the scheme. The 69 ms third and the 0.73 s fourth b-spectra, Fig. 7 b, bs3 and bs4, respectively, both have the shape of a Meta480 minus Meta380 difference spectrum. The higher amplitude of the 0.73 s b-spectrum shows that most of the Schiff base deprotonation needed to reach the active state occurs in the slowest step, which is more than a hundred times slower than the last step in the membrane. The information provided by the two b-spectra, however, is not sufficient for the assignment of the last two b-spectra to specific steps in the lower half of the model for the very simple reason that the double-square model requires five lifetimes and six b-spectra for unambiguous assignment. We have one less, and thus, our model is degenerate and there are multiple ways to fit it to the b-spectra and the lifetimes (14). The ambiguous microscopic rate constants belong to the steps in the lower half, the slow part of the model, and thus have small and restricted values in any of the possible solutions. The actual values have no relevance regarding the conclusions reached in this study. The fit we present assumes temporal degeneracy (14). The longest, 0.73 s experimental lifetime represents two degenerate eigenvalues, and the corresponding experimental b-spectrum is the result of two degenerate eigenvectors of the kinetic matrix.
The reproduction of the b-spectra by the model is shown in Fig. 7 b, dashed lines; the microscopic rate constants that produced the 93 μs, the 2.7 ms, the 68 ms, and the degenerate 0.74 and 0.75 s lifetimes are listed in Table 2. The double-square model provides an adequate description of the kinetics in the rN10 nanoparticle, and the intermediates in the model can be regarded as molecular states of rhodopsin.
Time evolution of intermediates present in the membrane and the rN10 nanoparticle
The table of microscopic rate constants contains all the essential information regarding the kinetic mechanism. For a complex mechanism, however, inspection of the rate constants is not the most convenient way to visualize how the reaction progresses in time. The time dependence of the intermediate concentrations gives a better picture. The latter can be calculated from the kinetic matrices obtained in the fit to the square and double-square models.
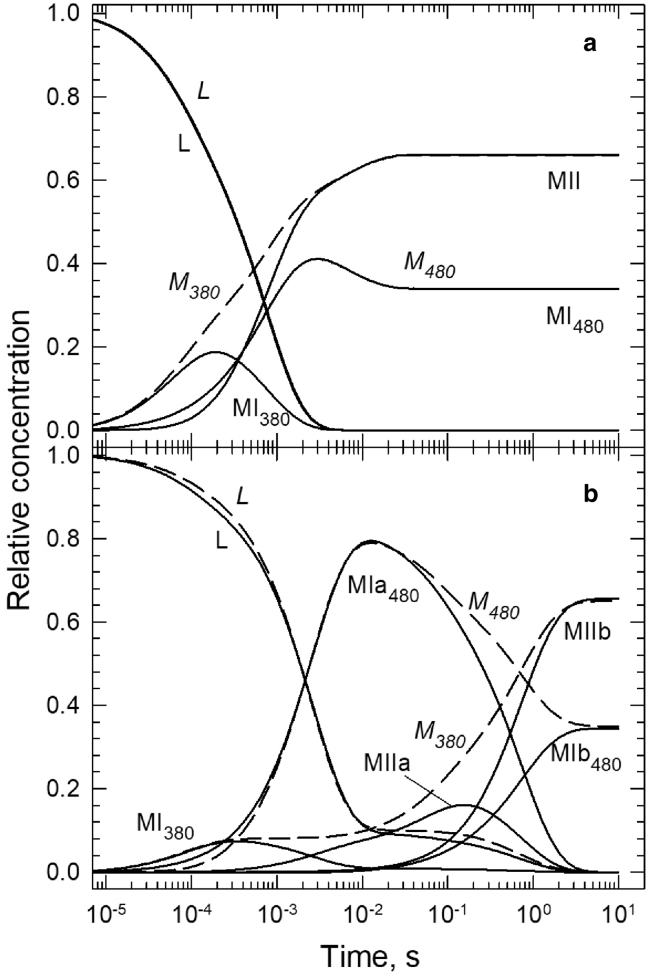
In Fig. 9 a, the solid lines show the time courses of the Lumi, Meta I480, Meta I380, and Meta II intermediate states calculated from the microscopic rate constants in the square model deduced for rhodopsin in the membrane. For comparison, the time courses of the spectral forms calculated based on the spectral compositions of the sequential intermediates, as discussed above in detail and depicted in Fig. 5 a, are also shown by dashed lines and italicized labels. Fig. 9 a illustrates the splitting of the time course of the Meta380 spectral form into two, the time courses of the isospectral Meta I380 (early times) and Meta II (late times) intermediates occurring in consecutive time intervals.
Figure 9.
(a) Time evolution of rhodopsin photoreaction in membrane (solid lines), showing the Lumi (L), Meta I380 (MI380), Meta I480 (MI480), and Meta II (MII) intermediate states derived from the square model. The time courses of the corresponding spectral forms (L, M380, and M480) from Fig. 5a are shown for comparison (dashed line). (b) Time evolution of rhodopsin photoreaction in rN10 nanoparticles (solid lines), showing the Lumi (L), Meta I380 (MI380), Meta Ia480 (MIa480), Meta IIa (MIIa), Meta Ib480 (MIb480), and Meta IIb (MIIb) intermediate states derived from the double-square model. The dashed lines show the time courses of the corresponding spectral forms (L, M380, and M480) from Fig. 5a.
The double-square model for the rN10 sample has six intermediates, the time courses of which were calculated based on the microscopic rate constants and are plotted in Fig. 9 b, solid lines. Dashed lines and italicized labels show the time courses of the spectral forms taken from Fig. 5 a. Here, the time course of the Meta480 spectral form is split into those of the isospectral Meta Ia480 (early times) and Meta Ib480 (late times) intermediates. In a similar manner, the time course of the Meta380 spectral form is split into three, the time courses of the isospectral Meta I380 (early times), Meta IIa (middle times), and Meta IIb (late times) intermediate states present in separate consecutive time intervals. Fig. 9, a and b demonstrate in a simple way the difference between the spectral forms detected in a reaction and the intermediate states responsible for the spectral forms. It is interesting to note that absorption transients recorded at single wavelengths always represent the time evolution of spectral forms, which may or may not belong to single intermediate states.
The effect of SMA on rhodopsin kinetics
A comparison of the intermediate states and their time evolutions between the membrane (Fig. 9 a) and the rN10 nanoparticle (Fig. 9 b) gives a clear picture of the SMA influence on rhodopsin kinetics. The early part of the reaction sequence, the branched decay of Lumi into Meta I380 and Meta I480, is not very different in the membrane and in the rN10 nanoparticle. The first lifetimes are similar, 70 μs in membrane and 85 μs in rN10. However, the equilibrium between Lumi and Meta I380 is somewhat more back shifted in rN10 than in membrane. The backshift slows down the Meta I380-to-Meta IIa conversion and reduces its contribution to the second b-spectrum. The 2.5 ms second lifetime in the rN10 sample is longer by a factor of 3.4 than the analogous 0.74 ms lifetime in the membrane, which is a significant but not a dramatic change. Because the second b-spectrum in rN10 shows only Lumi-to-Meta Ia480 conversion, the fraction of the unprotonated intermediates present in the reaction mixture stays constant during the second transition, around 0.1, the same low value as after the first transition. In the membrane, the equilibrium between Lumi and Meta I380 after the first transition is less back shifted than in the nanoparticle, and the concentration ratio between unprotonated and protonated states is above 1 after the second transition.
This is a very significant difference in rhodopsin kinetics between the rN10 and the membrane samples and shows that photoactivated rhodopsin in SMA more strongly follows the protonated pathway, giving preference to the path through Meta Ia480 over the path through Meta IIa. Another significant difference between the two samples is the drastic slowdown of the reaction as it progresses in the rN10 sample. In membranes, lowering the temperature is known to affect both the kinetic rates and the preference between pathways connecting Lumi to the active state (28). However, the effect of temperature is consistent with hindering the deprotonation of the Schiff base all along the reaction chain, starting with the equilibrium between Lumi and Meta I380, which becomes very back shifted and thus undetectable below 20°C (13), and ending with the active state, which is dominated by the Meta I480 intermediate. Lowering the temperature slows down the kinetics uniformly along the reaction chain, and it is far less drastic than observed in the SMALP samples at the late stages.
Potential implications for kinetic effects of SMA on other GPCRs
The types of general kinetic pathways and conclusions reached here for rhodopsin can be extended to other proteins of the GPCR family, as the proteins all have inactive and active conformations to control their interaction with the G-protein. The conversion from the inactive state to the active state is triggered by light in rhodopsin and by ligand binding in other receptor proteins. To reach the active state, some helices in rhodopsin change their physical positions to accommodate the G-protein (29). This physical rearrangement causes a relatively large volume change, especially in the cytoplasmic face, which requires free space provided by the polyunsaturated alkyl chains of the membrane lipids and mobility of the surrounding lipid molecules. The details of the reaction chain connecting the triggered initial state to the active state are best known for rhodopsin. Structural, infrared, and electron paramagnetic resonance spectroscopy studies have shown that the protein backbone of rhodopsin maintains the inactive form in all states with a protonated chromophore, including Meta I480. Only upon the Meta I480-to-Meta II transition does the protein undergo the big conformational change necessary to accommodate the G-protein (12,30,31).
In the early steps of the activation mechanism, the structural changes are restricted predominantly to the retinal binding pocket, which is equivalent to the ligand binding site in other receptor proteins. This part of the kinetics is not very sensitive to the fluidity of the lipid phase of the surrounding membrane.
The effect of SMA on the early part of the kinetics is thus not significant. It becomes somewhat more pronounced in the next steps, which occur on the early millisecond timescale in rhodopsin kinetics and are accompanied by bigger changes in or near the binding pocket and, perhaps, by rearrangements of bound lipids surrounding the helices. These structural changes allow partial deprotonation of the retinal Schiff base, producing Meta II in the square model and Meta IIa in the double-square model through Meta I380. These steps likely communicate the trigger pulse to the helical backbone, and steps analogous to those detected in rhodopsin kinetics are expected for other receptor proteins.
The dramatic changes in kinetics occur in the late steps, indicating that SMA mainly affects the steps associated with movements of helices in the protein backbone. Forcing rhodopsin to follow the protonated path through Meta I480 means that SMA locks and holds the protein in the inactive conformational state strongly enough to cause a dramatic slowdown of the late kinetic steps.
The slow rates can be explained in a number of ways. They could be caused by increased rigidity of the lipid environment because of the insertion of the hydrophobic styrene blocks of the copolymer between the unsaturated alkyl chains of lipids or displacing the entire lipid molecule, thus slowing down or even preventing the protein conformation changes necessary for the reaction to progress. The aromatic ring of styrene may also bind to the proposed cholesterol binding site of the inactive conformation (32) and slow down or freeze the movements of the helices needed to reach the active state. At very high SMA/protein ratios, the receptor may stay in the inactive state permanently. The results and the reasoning presented here may apply to many membrane-bound GPCR proteins. Thus, the effects of SMA on the kinetics reported here may be quite ubiquitous.
Lipid nanodisks enclosing rhodopsin can also be made the traditional way using detergent-solubilized protein, lipids, and appropriate edge proteins. The kinetics in these particles deviates from membrane kinetics only slightly (18), to a much smaller extent than presented here for the lipid nanoparticles made with SMA. The use of SMALPs has the big and sometimes crucial advantage of avoiding the application of detergents harmful to many membrane proteins. However, our work suggests that until optimal SMA conditions are first established, lipid nanodisks may be more appropriate for kinetic and dynamic membrane protein studies.
Conclusions
Solubilization of the membrane protein rhodopsin with SMA at SMA/protein molar ratios not exceeding 10–15, at or below the threshold SMA/membrane mass ratio needed for complete solubilization, produces nanoparticles that are suitable for kinetic studies. The effects of SMA at low SMA/protein molar ratios do not prevent intermediates involved in rhodopsin activation from forming, and the mechanism follows the general path leading to the active state in membrane. The details of the kinetics, however, do differ significantly from those seen in the native membrane. The main differences include revealing additional kinetic steps not resolved in the membrane, giving preference to the pathway through the protonated inactive forms over the unprotonated ones, and increasingly slowing down the reaction steps at the late stages at which big conformational and volume changes occur. In contrast, for SMALPs made at high SMA/protein molar ratios, the kinetics of rhodopsin is extremely slow, its path becomes disrupted, and the active state is not reached, thus rendering the solubilized protein unsuitable for kinetic studies. These results suggest that the interpretation of membrane protein dynamics studies carried out at high SMA concentrations should proceed only with caution.
Acknowledgments
This work was supported by National Institutes of Health Grant R01EY029343 (D.S.K. and D.L.F.).
Editor: John Conboy.
References
- 1.Hall S.C.L., Tognoloni C., Arnold T. Influence of poly(styrene- co-maleic acid) copolymer structure on the properties and self-assembly of SMALP nanodiscs. Biomacromolecules. 2018;19:761–772. doi: 10.1021/acs.biomac.7b01539. [DOI] [PubMed] [Google Scholar]
- 2.Kopf A.H., Dörr J.M., Killian J.A. Factors influencing the solubilization of membrane proteins from Escherichia coli membranes by styrene-maleic acid copolymers. Biochim. Biophys. Acta Biomembr. 2020;1862:183125. doi: 10.1016/j.bbamem.2019.183125. [DOI] [PubMed] [Google Scholar]
- 3.Xue M., Cheng L., Marrink S.J. Molecular mechanism of lipid nanodisk formation by styrene-maleic acid copolymers. Biophys. J. 2018;115:494–502. doi: 10.1016/j.bpj.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitch S.G., Yao W., Farrens D.L. Functional integrity of membrane protein rhodopsin solubilized by styrene-maleic acid copolymer. Biophys. J. 2021;120:3508–3515. doi: 10.1016/j.bpj.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis J.W., Kliger D.S. Photointermediates of visual pigments. J. Bioenerg. Biomembr. 1992;24:201–210. doi: 10.1007/BF00762678. [DOI] [PubMed] [Google Scholar]
- 6.Ostroy S.E. Rhodopsin and the visual process. Biochim. Biophys. Acta. 1977;463:91–125. doi: 10.1016/0304-4173(77)90004-0. [DOI] [PubMed] [Google Scholar]
- 7.Molday R.S., Moritz O.L. Photoreceptors at a glance. J. Cell Sci. 2015;128:4039–4045. doi: 10.1242/jcs.175687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janz J.M., Farrens D.L. Rhodopsin activation exposes a key hydrophobic binding site for the transducin α-subunit C terminus. J. Biol. Chem. 2004;279:29767–29773. doi: 10.1074/jbc.M402567200. [DOI] [PubMed] [Google Scholar]
- 9.Lewis A., Fager R.S., Abrahamson E.W. Tunable laser resonance Raman spectroscopy of the visual process. I: The spectrum of rhodopsin. J. Raman Spectrosc. 1973;1:465–470. [Google Scholar]
- 10.Lehmann N., Alexiev U., Fahmy K. Linkage between the intramembrane H-bond network around aspartic acid 83 and the cytosolic environment of helix 8 in photoactivated rhodopsin. J. Mol. Biol. 2007;366:1129–1141. doi: 10.1016/j.jmb.2006.11.098. [DOI] [PubMed] [Google Scholar]
- 11.Tsukamoto H., Szundi I., Kliger D.S. Rhodopsin in nanodiscs has native membrane-like photointermediates. Biochemistry. 2011;50:5086–5091. doi: 10.1021/bi200391a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogel R., Siebert F. Fourier transform IR spectroscopy study for new insights into molecular properties and activation mechanisms of visual pigment rhodopsin. Biopolymers. 2003;72:133–148. doi: 10.1002/bip.10407. [DOI] [PubMed] [Google Scholar]
- 13.Thorgeirsson T.E., Lewis J.W., Kliger D.S. Effects of temperature on rhodopsin photointermediates from lumirhodopsin to metarhodopsin II. Biochemistry. 1993;32:13861–13872. doi: 10.1021/bi00213a015. [DOI] [PubMed] [Google Scholar]
- 14.Szundi I., Funatogawa C., Kliger D.S. Complexity of bovine rhodopsin activation revealed at low temperature and alkaline pH. Biochemistry. 2016;55:5095–5105. doi: 10.1021/acs.biochem.6b00687. [DOI] [PubMed] [Google Scholar]
- 15.Zhang R., Sahu I.D., Lorigan G.A. Characterizing the structure of lipodisq nanoparticles for membrane protein spectroscopic studies. Biochim. Biophys. Acta. 2015;1848:329–333. doi: 10.1016/j.bbamem.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamshad M., Grimard V., Dafforn T.R. Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res. 2015;8:774–789. doi: 10.1007/s12274-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funatogawa C., Szundi I., Kliger D.S. A comparison between the photoactivation kinetics of human and bovine rhodopsins. Biochemistry. 2016;55:7005–7013. doi: 10.1021/acs.biochem.6b00953. [DOI] [PubMed] [Google Scholar]
- 18.Szundi I., Funatogawa C., Kliger D.S. Protein sequence and membrane lipid roles in the activation kinetics of bovine and human rhodopsins. Biophys. J. 2017;113:1934–1944. doi: 10.1016/j.bpj.2017.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epps J., Lewis J.W., Kliger D.S. Lumi I--> Lumi II: the last detergent independent process in rhodopsin photoexcitationt. Photochem. Photobiol. 2006;82:1436–1441. doi: 10.1562/2006-02-01-RA-792. [DOI] [PubMed] [Google Scholar]
- 20.Thorgeirsson T.E., Lewis J.W., Kliger D.S. Photolysis of rhodopsin results in deprotonation of its retinal Schiff’s base prior to formation of metarhodopsin II. Photochem. Photobiol. 1992;56:1135–1144. doi: 10.1111/j.1751-1097.1992.tb09738.x. [DOI] [PubMed] [Google Scholar]
- 21.Lewis J.W., Warner J., Kliger D.S. Noise reduction in laser photolysis studies of photolabile samples using an optical multichannel analyzer. Rev. Sci. Instrum. 1987;58:945–949. [Google Scholar]
- 22.Lewis J.W., Kliger D.S. Absorption spectroscopy in studies of visual pigments: spectral and kinetic characterization of intermediates. Methods Enzymol. 2000;315:164–178. doi: 10.1016/s0076-6879(00)15842-2. [DOI] [PubMed] [Google Scholar]
- 23.Lewis J.W., Kliger D.S. Microliter flow cell for measurement of irreversible optical absorbance transients. Rev. Sci. Instrum. 1993;64:2828–2833. [Google Scholar]
- 24.Williams T.P. An isochromic change in the bleaching of rhodopsin. Vision Res. 1970;10:525–533. doi: 10.1016/0042-6989(70)90046-5. [DOI] [PubMed] [Google Scholar]
- 25.Henry E.R., Hofrichter J. Singular value decomposition: application to analysis of experimental data. Methods Enzymol. 1992;210:129–192. [Google Scholar]
- 26.Szundi I., Lewis J.W., Kliger D.S. Effect of NADPH on formation and decay of human metarhodopsin III at physiological temperatures. Vision Res. 2000;40:3039–3048. doi: 10.1016/s0042-6989(00)00148-6. [DOI] [PubMed] [Google Scholar]
- 27.Szundi I., Lewis J.W., Kliger D.S. Deriving reaction mechanisms from kinetic spectroscopy. Application to late rhodopsin intermediates. Biophys. J. 1997;73:688–702. doi: 10.1016/S0006-3495(97)78103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schleicher A., Hofmann K.P. Kinetic study on the equilibrium between membrane-bound and free photoreceptor G-protein. J. Membr. Biol. 1987;95:271–281. doi: 10.1007/BF01869489. [DOI] [PubMed] [Google Scholar]
- 29.Marr K., Peters K.S. Photoacoustic calorimetric study of the conversion of rhodopsin and isorhodopsin to lumirhodopsin. Biochemistry. 1991;30:1254–1258. doi: 10.1021/bi00219a013. [DOI] [PubMed] [Google Scholar]
- 30.Farrens D.L., Altenbach C., Khorana H.G. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 31.Ruprecht J.J., Mielke T., Schertler G.F. Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 2004;23:3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sejdiu B.I., Tieleman D.P. Lipid-protein interactions are a unique property and defining feature of G protein-coupled receptors. Biophys. J. 2020;118:1887–1900. doi: 10.1016/j.bpj.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]