Abstract
Background:
Inflammatory cells in previous human skin single-cell data constituted only a small fraction of the overall cell population, such that functional subsets were difficult to ascertain.
Objective:
Our aims were to overcome the limitation by applying single-cell transcriptomics to emigrating cells from skin and elucidate ex vivo gene expression profiles of pathogenic vs. regulatory immune cell subsets in psoriasis skin.
Methods:
We harvested emigrating cells from human psoriasis skin after incubation in culture medium without enzyme digestion or cell sorting and analyzed cells with single-cell RNA sequencing and flow cytometry simultaneously.
Results:
Unsupervised clustering of harvested cells from psoriasis and control skin identified NK cells, T-cell subsets, dendritic cell subsets, melanocytes, and keratinocytes in different layers. Comparison between psoriasis vs. control cells within each cluster identified: 1) Cutaneous Type 17 T-cells display highly differing transcriptome profiles depending on IL-17A vs. IL-17F expression and IFNγ vs. IL-10 expression, 2) Semimature dendritic cells are regulatory dendritic cells with high IL-10 expression but a subset of semimature dendritic cells expresses IL-23A and IL-36G in psoriasis, and 3) CCL27-CCR10 interaction is potentially impaired in psoriasis due to decreased CCL27 expression in basal keratinocytes.
Conclusion:
We propose that single-cell transcriptomics applied to emigrating cells from human skin provide an innovative study platform to compare gene expression profiles of heterogenous immune cells in various inflammatory skin diseases.
Clinical Implication:
Our method can readily be extended to study modulation of leukocyte transcripts by immune-targeted treatments.
Keywords: Psoriasis, single-cell RNA sequencing, T-cells, dendritic cells, keratinocytes, emigrating cells
Capsule Summary:
Single-cell transcriptomics applied to emigrating cells from psoriasis skin elucidate ex vivo gene expression profiles of cutaneous type 17 T-cell and regulatory T-cell subsets and mature vs. semimature dendritic cells.
Introduction
Psoriasis vulgaris is the most common inflammatory disease in humans that is mediated by IL-17 producing Type 17 T-cells (T17 cells), which includes CD4+ (Th17) and CD8+ (Tc17) subsets1. The heterogeneity in T17 cells is suggested by the finding that individual T-cells can synthesize only IL-17A or IL-17F (homodimers) or they can synthesize IL-17A/F (heterodimer), while each of these IL-17 isoforms has differing ability to activate IL-17 receptors on target cells2. The expression of pathogenic vs. regulatory markers in IL-17A vs. IL-17F-producing T-cells is presently unknown.
The chronicity of psoriatic lesions may be caused by defective negative regulatory pathways, such as decreased function of regulatory T-cells (Tregs)3, reduced Type 1 regulatory T-cells (Tr1)4 and low expression of negative regulatory factors that are highly expressed in resolving delayed-hypersensitivity skin reaction lesions5. However, the function of Tregs has been assessed only in peripheral blood3, and the properties of Tregs and other regulatory immune cells in human skin is largely unexplored6.
Normal skin contains a subset of myeloid dendritic cells (DCs) that are characterized by BDCA-3 expression, IL-10 production and expression of negative immune regulators, so this population is considered to be regulatory contributing to immune homeostasis/tolerance under non-inflammatory conditions7. In contrast, psoriasis lesions contain an increased number of “inflammatory” CD11c+ DCs and mature populations marked by DC-LAMP expression8. The relative expression of inflammatory vs. regulatory molecules in DC populations present in psoriasis lesions has not been widely explored.
Single-cell RNA sequencing (scRNA-seq) is a powerful technique to characterize gene expression in individual leukocytes or other tissue-resident cells in human skin9–19. However, leukocyte subsets were difficult to study in depth in human skin, as other skin cells such as fibroblasts and keratinocytes dominated the overall cell mixture. In this study, we have performed scRNA-seq in psoriasis and control skin using emigrating cells from skin biopsies to enrich leukocyte subsets without cell sorting or activation. We find novel and distinct patterns of gene expression within different T17 cell subsets, Tregs, mature and semimature DC subsets and keratinocytes in different layers that are disease-related and may drive key inflammatory pathways in psoriasis.
Methods
Please see the supplementary experiment section and Fig E1, E2 and E3 in the article’s Online Repository for validation of our new method - scRNA-seq of emigrating cells from skin. With skin biopsies from the same sample, we compared our new method with our group’s previous methods – 1) scRNA-seq of isolated cells from cryopreserved tissues by conventional enzyme digestion9–11, and 2) flow cytometry analysis of emigrating cells20 (Fig E1 and Fig E2). In addition, we compared our new method with existing scRNA-seq data of isolated cells from fresh skin tissues by conventional enzyme digestion (Fig E3). For extended methods, please see the supplemental materials section in the article’s Online Repository. The scRNA-seq data have been deposited in NCBI′s Gene Expression Omnibus and are publicly accessible through GEO Series accession number GSE151177. All the scripts used to present and analyze the data are publicly available from https://github.com/jaehwan79/KruegerLab_scRNAseq_emigrating_cells_human_skin. There is no restriction on the use of the data or code.
Patients
Human subject research was performed in accordance with the Helsinki Declaration and approved by the Institutional Review Board of the Rockefeller University, New York, NY, USA. Demographics and disease severity of the study cohort is presented in Table E1 in this article’s Online Repository.
Harvesting emigrating cells from skin biopsy tissues
6 mm punch skin biopsy tissues from 13 psoriasis patients and 5 healthy volunteers were bisected, and one of the two bisected skin tissues was incubated in culture medium for harvesting emigrating cells. To split epidermis and dermis (Video E1), harvested skin tissue was immediately placed in 0.2% Dispase II (Sigma-Aldrich) and incubated in a humidified incubator at 37°C and 5% CO2 for 3 hours. Then, epidermis and dermis were separated with forceps and sliced into small pieces with #10 blade scalpels (Video E2). Epidermis and dermis were separately incubated in RPMI-1640 medium with L-glutamine (Cytiva) supplemented with 10% human albumin serum (Sigma-Aldrich) without any enzyme in a humidified incubator at 37°C and 5% CO2. Nonplastic adherent cells that had emigrated out of epidermis and dermis were harvested after 48 hours (Video E3). The harvested cells from epidermis and dermis were combined and filtered through a 40-μm cell strainer (Corning) (Video E4) and stored on ice. The cell numbers and viability were determined using a Countess automated cell counter (Invitrogen) and trypan blue staining (BioRad). The cell viability of individual samples is presented in Table E1.
Single-cell RNA sequencing experiments
The 10x Genomics Chromium Single Cell 3’ Reagents Kit user guide (https://support.10xgenomics.com) was used to prepare the single cell suspension. All the samples were sequenced by a single NextSeq 500 sequencer (Illumina) with the identical run parameters: read 1 for 28 cycles, read 2 for 55 cycles, and index for 8 cycles. Sequencing information including the sequencing saturation and depth is presented in Table E1.
Analysis of single-cell RNA sequencing data
We used Seurat R package (version 3.0) installed in R (version 3.6.2) for the downstream single-cell RNA sequencing data analysis21. Before data integration, single-cell data quality control was performed separately for each individual sample as previously described for human skin single-cell RNA sequencing analyses9–11.
Principal component analysis and graph-based clustering analysis were performed, and differentially expressed genes of each cluster compared to all other cells were found. The average gene expression of psoriasis vs. control cells within a cluster was calculated. To compare psoriasis vs. control cells within clusters representing each type of skin immune cells, we merged adjacent clusters of common immune cell subsets (Fig E4). For immune cell cluster comparison, cells expressing target genes within each cluster were quantified by (1) number of cells expressing the target gene and (2) proportion of cells expressing the target gene.
We used iTALK R package (version 0.1.0) for the receptor-ligand interaction analysis. Differentially expressed genes between psoriasis vs. control in each cluster were found, and then the fold changes of differentially expressed genes were used to calculate receptor-ligand interaction between different cell clusters.
Multiparameter flow cytometry experiments
When emigrating cells from skin biopsy tissues were harvested and 5,000-10,000 harvested skin cells were entered into the 10X Genomics single-cell chip, the rest of harvested skin cells were simultaneously analyzed by multiparameter flow cytometry with dendritic cell panels. The harvested skin cells were washed and incubated in ice for 30 minutes with fluorochrome-conjugated monoclonal antibodies to cell-surface markers, and the cells were acquired with the BD LSR II flow cytometer (BD Biosciences) and analyzed with FlowJo software (TreeStar).
Results
Microfluidic partitioning of emigrating cells from human skin empowers single-cell transcriptomic profiling of heterogeneous immune cells under ex vivo conditions
Dimensionality reduction analysis of 23,220 single-cell data identified clusters of NK cells, CD161+ T-cells, CD8+ T-cells, CD4+ T-cells, Tregs, mature DCs, semimature DCs, macrophages, melanocytes, and KCs in different layers of Stratum (S.) corneum, S. granulosum, S. spinosum, and S. basale without subclustering (Fig 1A–E). Leukocytes (NK cells, T-cells, dendritic cells and macrophages) constituted 53.0% of scRNA-seq data of psoriasis skin and 15.5% of control skin (Fig 1F and Fig E5B). The average number of T-cells per skin biopsy sample increased 7.0 times from control (67.4 ± 19.6) to psoriasis (472.9 ± 108.5), and the average number of DCs per skin biopsy sample increased 3.4 times from control (70.8 ± 33.4) to psoriasis (241.7 ± 63.7) (p < 0.05, Fig E5C). The top 10 most differentially expressed genes in each cluster are presented in Fig E6.
Figure 1. Single-cell transcriptomic profiling of leukocytes and keratinocytes in human psoriasis and control skin.
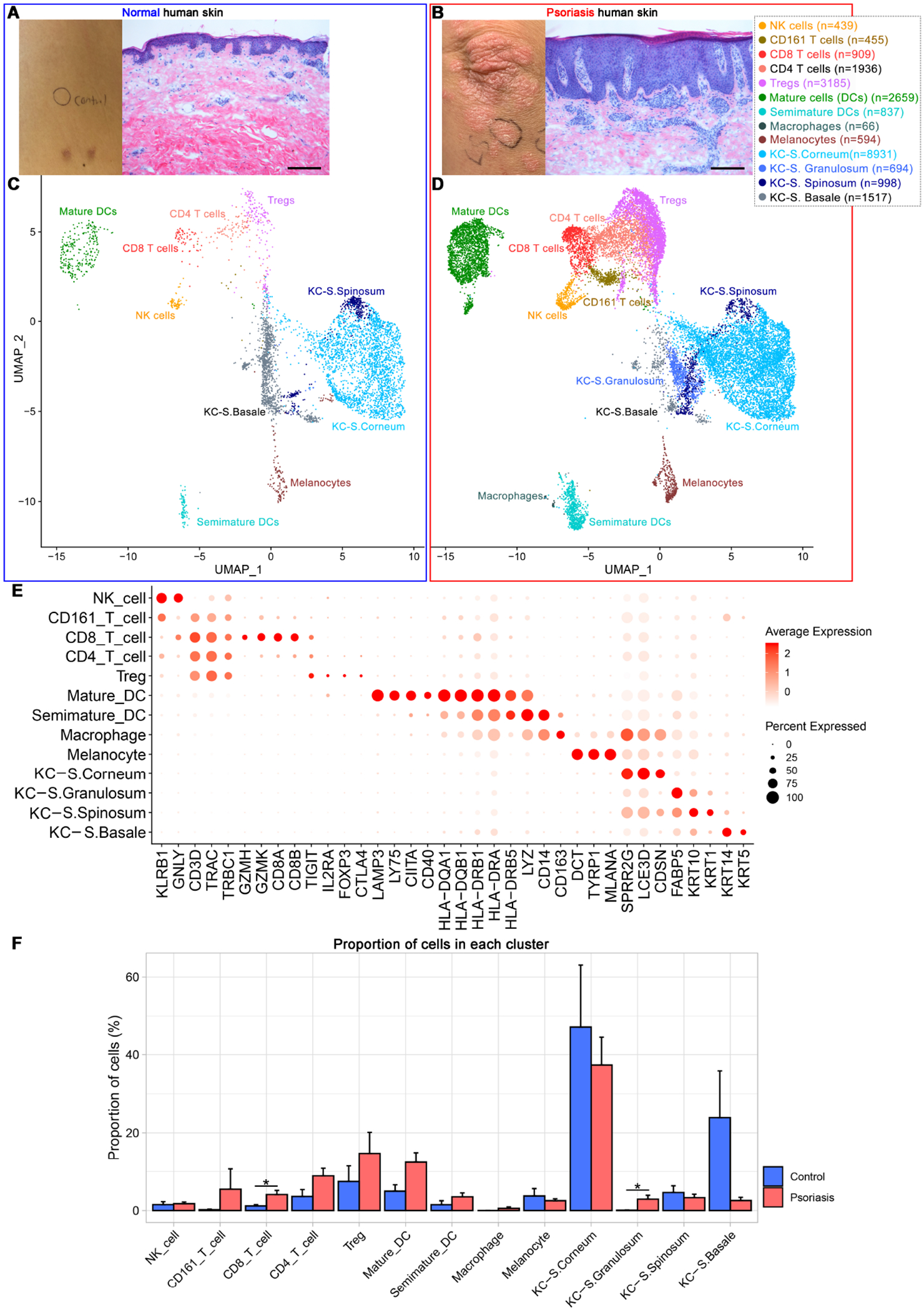
Clinical & microscopic figures of control (A) and psoriasis skin (B) and their leukocyte & keratinocyte scRNA-seq data presented in the Uniform Manifold Approximation and Projection plot (C and D). (E) Dot plot displaying expression levels of cluster-defining genes. (F) Cell composition of individual samples. Treg, regulatory T-cell; DCs, dendritic cells; KC, keratinocytes, S, Stratum. Scale bar in (A) and (B) = 200 μm, Proportion of cells in (F) = average number of cells in cluster within individual sample / total number of cells within individual sample x 100 (%), Error bar in (F) = Standard Error of Mean.
CD161+ T-cell and CD8+ T-cell clusters were segregated from other T-cell clusters (Figure 1C and 1D), but CD4+ T-cell and Treg clusters were not segregated and the designations were nominal (Fig 1E). Compared to CD4+ T-cell cluster, Treg cluster was characterized by a higher proportion of cells expressing CD25 (IL2RA) (16.1% in Treg vs. 1.7% in CD4+ T-cell clusters), FoxP3 (16.2% in Treg vs. 0.4% in CD4+ T-cell clusters) and CTLA4 (14.6% in Treg vs 1.7% in CD4+ T-cell clusters) (Fig E7). When we applied heuristic cut-off value of 1 to FoxP3 expression, FoxP3High subsets in the Treg cluster expressed high levels of CD25 (IL2RA) with low levels of CD127 (IL7R) together with high levels of OX40 (TNFRSF4), TIGIT, CTLA4, and CCR10 (Fig 2 and Fig E6). When T-cell clusters were subset by cytokine mRNA expression, numerous cytokines were expressed by T-cells in psoriasis skin (Fig 2 and Fig E7) - TNFSF10 (23.9%), TNFSF12 (15.6%), CCL20 (10.1%), IL-36G (5.9%), TNF (4.2%), IL-26 (3.0%), IFNγ (2.8%), IL-17F (0.9%), IL-17A (0.6%), IL-22 (0.3%), and IL-13 (0.1%).
Figure 2. The average gene expression within clusters of NK cells and T-cell subsets.
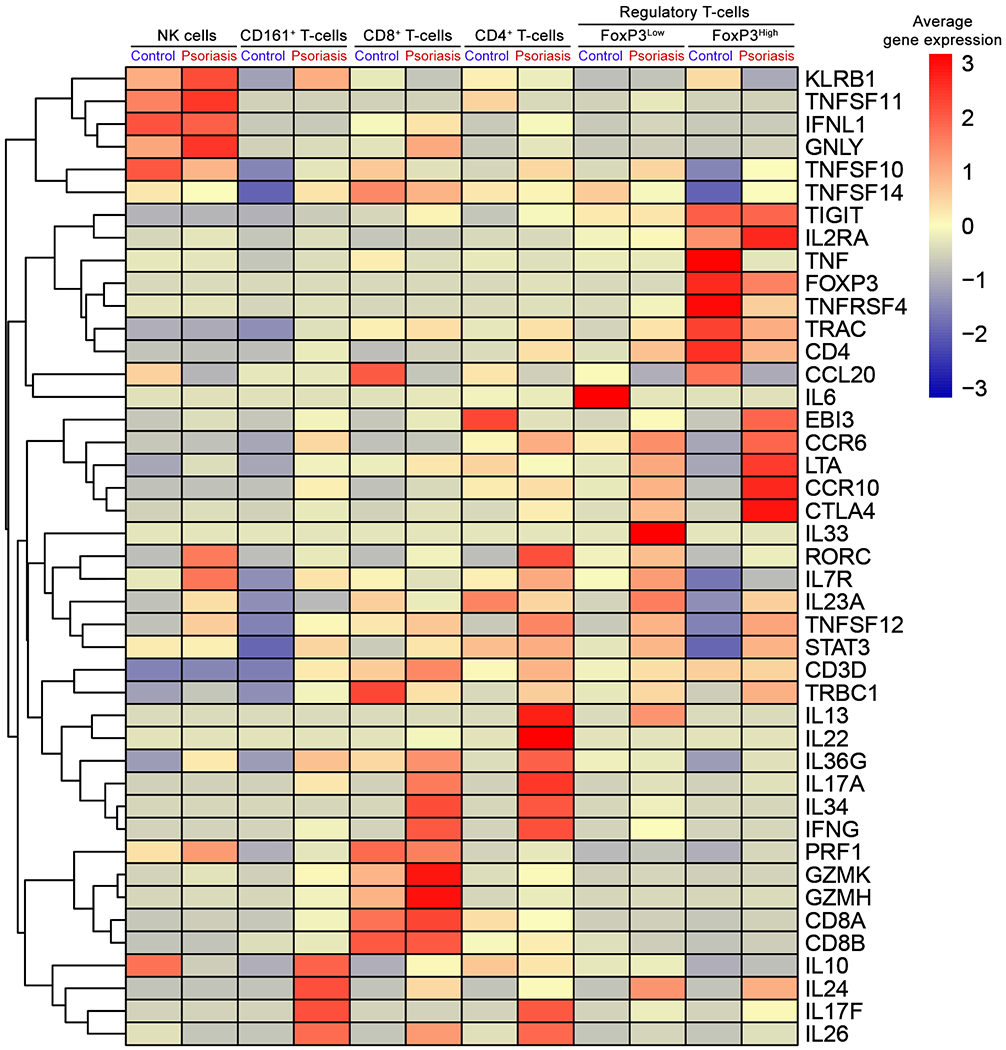
Heatmap of scRNA-seq analysis illustrates the average gene expression within clusters of NK cell and T-cell subsets, split by psoriasis and control. Regulatory T-cell cluster is divided into cells with FoxP3 expression high (FoxP3High) and cells with FoxP3 expression low (FoxP3Low) using a cut-off value of 1. Genes with similar expression patterns are linked by a complete linkage method.
Cutaneous T17 cells display highly differing transcriptomes depending on IL-17A vs. IL-17F expression and IFNγ vs. IL-10 expression
T17 cells constituted 2.7% of CD4+ T-cell cluster, 1.3% of CD8+ T-cell cluster, 2.4% of CD161+ T-cell cluster and 0.5% of Treg cluster (Fig E7). When T17 cells were subdivided into Th17 (CD4+ T17) cells, Tc17 (CD8+ T17) cells, CD161+ T17 cells, and T17 cells in a Treg cluster (Fig E8), Th17 (CD4+ T17) cells expressed IL-17A (32.7%) or IL-17F (76.9%) (Fig E9). In contrast, Tc17 (CD8+ T17) cells expressed only IL-17A, and IL-17F was undetected in Tc17 cells with a cut-off value of 1. 50% of Tc17 cells expressed IFNγ, while only 13.5% of Th17 cells expressed IFNγ. 5.8% of Th17 cells expressed IL-10, while IL-10 was undetected in Tc17 cells with a cut-off value of 1. CD161+ T17 cells co-expressed CD8B (36.4%) or CD4 (9.1%) and expressed IL-17A (36.4%), IL-17F (72.7%) and IL-10 (9.1%).
Majority of T17 cells expressed either IL-17A or IL-17F and only 7.8% of T17 cells co-expressed IL-17A/IL-17F (Fig 3A). Excluding IL-17A/IL-17F co-producing T17 cells, IFNγ was expressed by 25.6% of IL-17A producing T17 cells, while only 3.9% of IL-17F producing T17 cells expressed IFNγ. In contrast, IL-10 was undetected by IL-17A producing T17 cells with a cut-off value of 1, but 5.9% of IL-17F producing T17 cells expressed IL-10.
Figure 3. The average gene expression of cutaneous Type 17 T-cell (T17 cell) subsets.
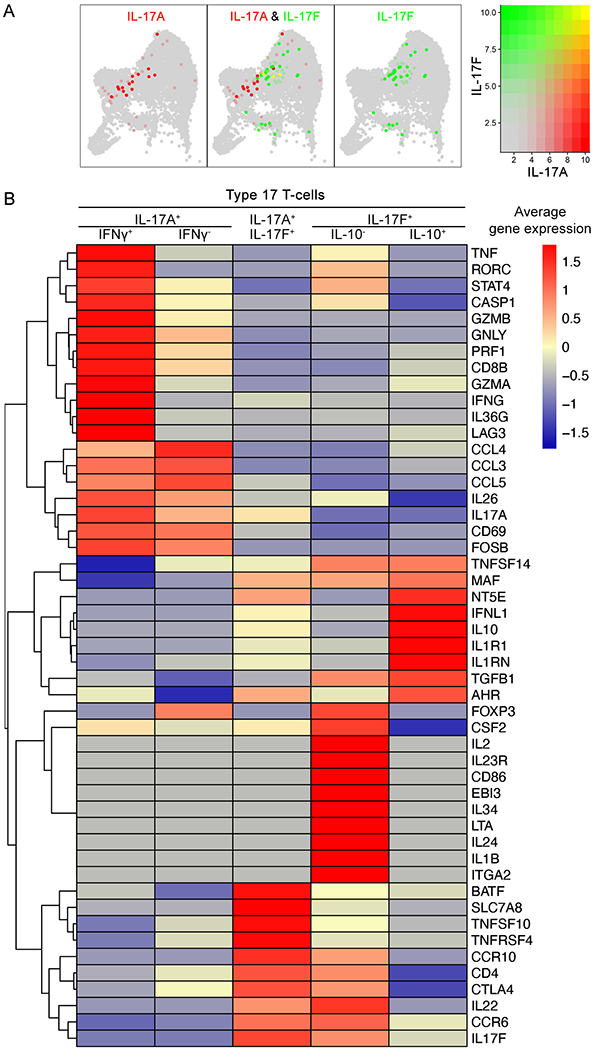
(A) IL-17A (red), IL-17F (green) and IL-17A/IL17F co-expression (yellow) within T-cell subset clusters visualized in low-dimensional space. (B) Heatmap of scRNA-seq analysis illustrates the average gene expression of T17 cell subsets. Genes with similar expression patterns are linked by a complete linkage method.
IL−17A+ T17 cells in psoriasis skin expressed high levels of inflammatory cytokines, such as IL-26, CCL3, CCL4 and CCL5, which have been reported in pathogenic T17 cell transcripts22, 23 (Fig 3B). In addition, IL-17A+ IFNγ+ T17 cells expressed high levels of transcription factors, such as RORC and STAT46, 23, inflammatory cytokines including IL-36G and TNF1, cytotoxic transcripts such as GZMA, GZMB, GNLY, CD8B and PRF1, and other pathogenic T17 cell transcripts23, 24, such as CASP1 and LAG3.
IL-17F+ IL-10+ T17 cells expressed high levels of MAF, AHR, CD73 (NT5E) and IL1RN, which have been reported in non-pathogenic IL-10-producing T17 cell transcripts23, 25–30 (Fig 3B). In contrast, IL-17F+ IL-10− T17 cells expressed high levels of inflammatory cytokines, such as IL-1β, IL-2, IL-24, IL-34, EBI3 and LTA, and IL23R. Both IL-17F+ IL-10− T17 cells and IL-17A+ IL-17F+ T17 cells expressed high levels of IL-22 and CCR6. In addition, IL-17A+ IL-17F+ T17 cells expressed high levels of BATF, SLC7A8 and TNFSF10.
Mature DCs in psoriasis skin express high levels of IL-23A & IL-36G and low levels of KYNU compared to mature DCs in control skin
When the average gene expression was compared between mature DC and semimature DC clusters split by psoriasis vs. control in the scRNA-seq data, mature DCs were characterized by 1) high expression of MHC class II molecules, 2) skin DC marker8 expression of CD86, DC-LAMP (LAMP3), CD205 (LY75), CD40, CIITA, CD80, PD1-L1 (CD274) and PD1-L2 (PDCD1LG2), and 3) expression of DC regulatory tryptophan metabolism enzyme31 KYNU (Fig 4A and Fig E10C).
Figure 4. Simultaneous scRNA-seq and flow cytometry analyses define transcriptomic profiles of mature vs. semimature dendritic cells.
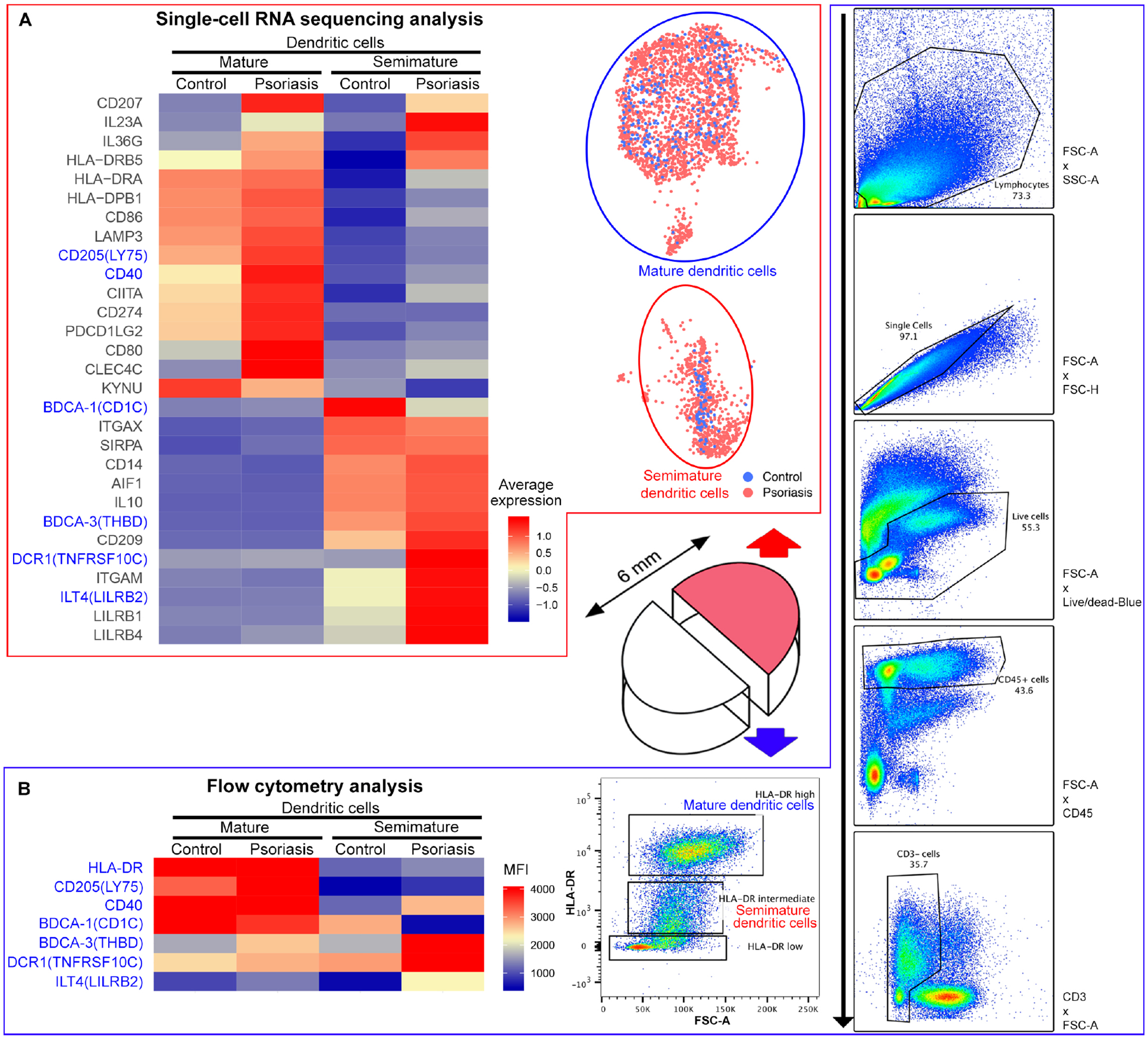
(A) Heatmap of scRNA-seq analysis illustrates the average gene expression within mature vs. semimature DC clusters, split by psoriasis and control. (B) Heatmap of flow cytometry analysis illustrates Median Fluorescence Intensity (MFI) of HLA-DRHigh (mature) or HLA-DRLow (semimature) CD45+ CD3− HLA-DR+ dendritic cells in control and psoriasis skin. Consistent findings between scRNA-seq and flow cytometry analyses are marked in blue.
Simultaneous flow cytometry analysis confirmed scRNA-seq findings of mature DCs (Fig 4B and Figure E10A). The Median fluorescence intensity (MFI) of HLA-DR, CD205 (LY75) and CD40 in mature DCs was higher than the MFI of HLA-DR, CD205 and CD40 in semimature DCs (Fig 4B). In addition, the proportions of CD205+ and CD40+ cells in mature DCs were higher than the proportion of CD205+ and CD40+ cells in semimature DCs (Fig E10A).
Mature DCs in psoriasis skin expressed more IL-23A and IL-36G compared to mature DCs in control skin. IL-23A or IL-36G expression in mature DCs increased 4.2 times from control skin (1.5%, 4/269) to psoriasis skin (6.2%, 148/2390) (Fig E11A and E11C). In contrast, mature DCs in psoriasis skin expressed less KYNU than mature DCs in control skin. KYNU expression in mature DCs decreased 2.1 times from control skin (21.6%, 58/269) to psoriasis skin (10.4%, 249/2390) (Fig E10C and Fig E11D).
Semimature DCs in psoriasis skin express high levels of IL-10, BDCA-3 and LILRB2, but a subset of semimature DCs also expresses IL-23A and IL-36G
Semimature DCs were characterized by 1) intermediate expression of MHC class II molecules (HLA-DRB5), 2) skin DC marker8 expression of CD11c (ITGAX), SIRPA, CD14, AIF1 and CD209 (DC-SIGN) and 3) High IL-10 expression (Fig 4A and Fig E10B). Among all IL-10 expressing cells in total scRNA-seq data, 53.5% (145/271) of IL-10 expressing cells were semimature DCs (Fig 5A and 5B). IL-10 expressing semimature DCs in psoriasis skin highly co-expressed BDCA-3 (THBD) and LILRB2 (ILT4). 46.5% (60/129) of IL-10 expressing semimature DCs in psoriasis skin co-expressed BDCA-3 (Fig 5C, 7D and 7F). 41.9% (54/129) of IL-10 expressing semimature DCs in psoriasis skin co-expressed LILRB2.
Figure 5. IL-10, BDCA-1 (CD1C), BDCA-3 (THBD), LILRB2 (ILT4) and IL-23 expression in mature dendritic cells (DCs) and semimature DCs.
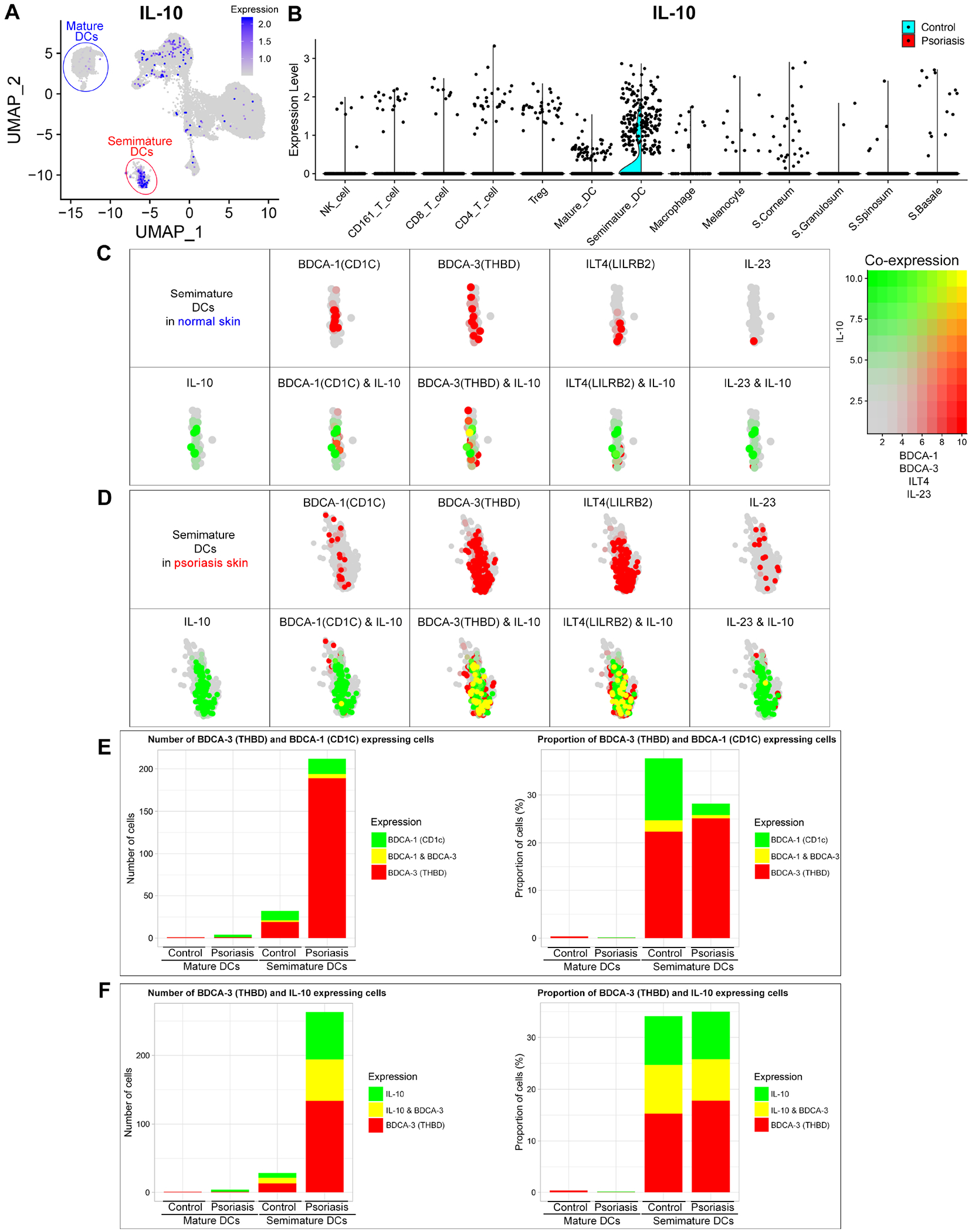
IL-10 expression in total scRNA-seq data is visualized in low-dimensional space (A) and violin plot of each immune cell cluster (B). Co-expression of IL-10 & BDCA-1, BDCA-3, LILRB2 or IL-23 in normal skin semimature DCs (C) and psoriasis skin semimature DCs (D) is visualized in low-dimensional space. Cells with expression of BDCA-3 & BDCA-1 (E) and BDCA-3 & IL-10 (F) within each cluster of mature and semimature DCs in normal and psoriasis skin are quantified by number of cells and proportion of cells. Proportion of cells = number of target gene expressing cells within cluster / total number of cells within cluster x 100 (%).
Psoriasis semimature DCs expressed more BDCA-3, DCR1 (TNFRSF10C), ITGAM, LILRB2, LILRB1 and LILRB4, and less BDCA-1 (CD1c)8 than semimature DCs in control skin (Fig 4A, 5C, 5D and Fig E10D). The BDCA-3 to BDCA-1 ratio increased 5.2 times from control (1.6) to psoriasis skin (8.4) (Fig 5E).
Simultaneous flow cytometry analysis confirmed scRNA-seq findings of semimature DCs (Fig 4B and Fig E10A). The MFI of BDCA-3, DCR1 and LILRB2 in psoriasis semimature DCs was higher than the MFI of BDCA-3, DCR1 and LILRB2 in control semimature DCs (Fig 4B). The MFI of BDCA-1 in psoriasis semimature DCs was lower than the MFI of BDCA-1 in control semimature DCs. In addition, the proportion of BDCA-3+ and LILRB2+ cells in psoriasis semimature DCs was higher than the proportion of BDCA-3+ and LILRB2+ cells in control semimature DCs (Fig E10A). The proportion of BDCA-1+ cells in psoriasis semimature DCs was lower than the proportion of BDCA-1+ cells in control semimature DCs.
When we analyzed the receptor-ligand interaction with the scRNA-seq data, inhibitory receptors LILRB2 and LILRB1 in psoriasis semimature DCs were potentially stimulated by a key component of MHC class I molecules (B2M) in CD161+ T-cells, CD4+ T-cells, melanocytes and KCs in stratum corneum32 (Fig E12A).
Like mature DCs, semimature DCs in psoriasis skin expressed more IL-23A and IL-36G than semimature DCs in control skin (Fig 4A and Fig E11). The number of cells expressing IL-23A or IL-36G in semimature DCs increased 9.8 times from control (1.2%, 1/85) to psoriasis skin (11.6%, 87/752). Semimature DCs expressing IL-23A or IL-36G and semimature DCs expressing IL-10 were independent – only 0.6% (1/157) of semimature DCs expressing IL-23A or IL-10 co-expressed IL-23A and IL-10.
scRNA-seq of emigrating cells from human skin identifies locations of keratinocyte transcriptome changes implicated in psoriasis pathogenesis
Our scRNA-seq data showed that the decrease of FLG expression in psoriasis epidermis occurs in stratum corneum33 (Fig 6A). 40.7% (700/1720) of FLG expressing KCs were located in S. corneum (Fig 6B), and the number of KCs expressing FLG in S. corneum decreased 2.1 times from control (18.8%, 473/2513) to psoriasis (3.5%, 227/6418).
Figure 6. Gene expression within keratinocyte clusters and CCL27 (keratinocyte)–CCR10 (Treg) interactions.
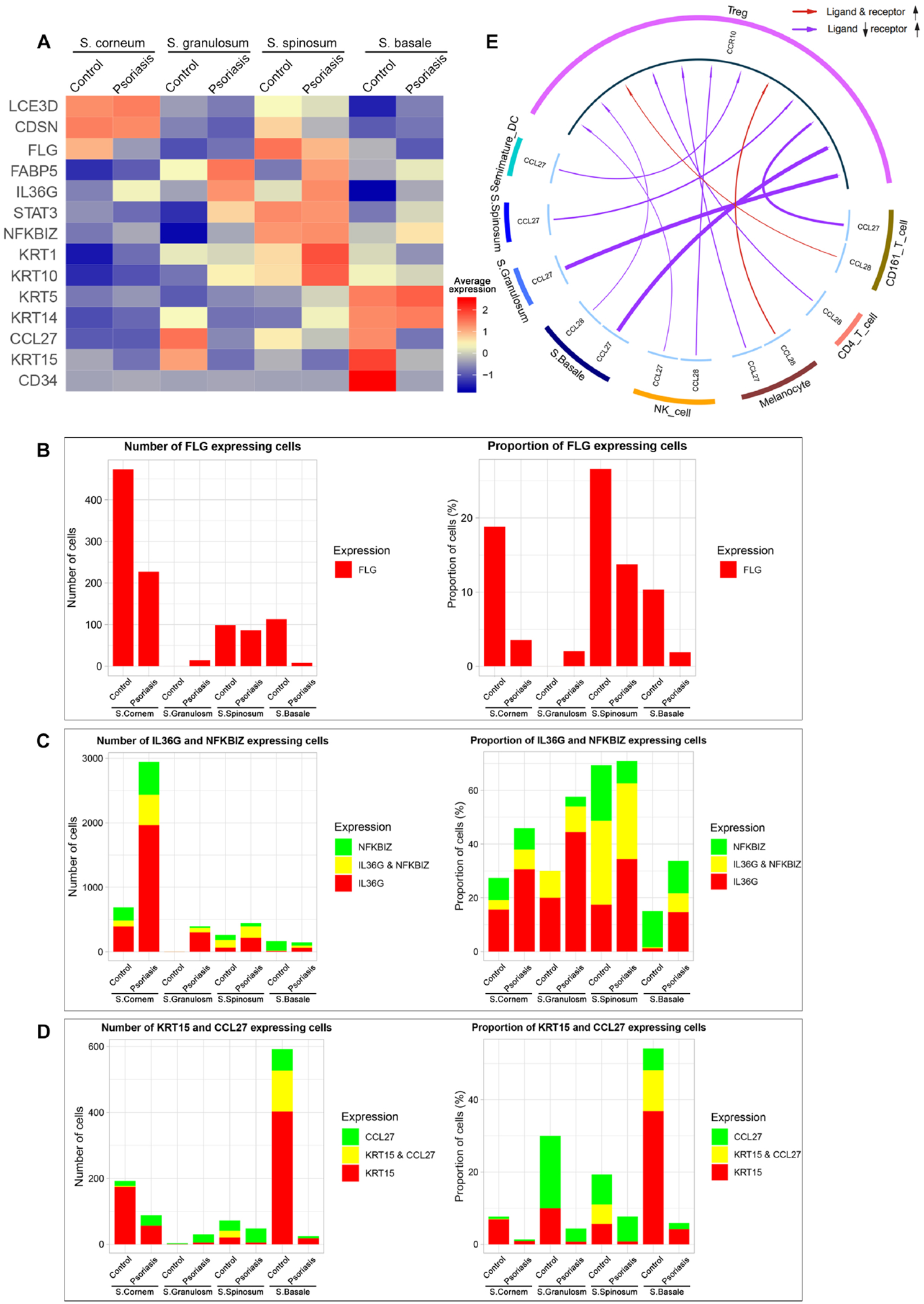
(A) Heatmap of scRNA-seq analysis illustrates the average gene expression within clusters of keratinocytes (KCs) in Stratum (S.) corneum, S. granulosum, S. spinosum and S. basale, split by psoriasis and control. Cells with expression of FLG (B), IL-36G & NFKBIZ (C) and KRT15 & CCL27 (D) within each layer of KCs in control and psoriasis epidermis are quantified by number of cells and proportion of cells. Proportion of cells = number of target gene expressing cells within cluster / total number of cells within cluster x 100 (%). (E) CCL27/CCL28-CCR10 interaction in psoriasis compared to control highlights decreased CCL27 in basal keratinocytes interacting with CCR10 in Tregs.
Our data showed that the increase of IL-36G expression in psoriasis epidermis occurs in suprabasal layers (S. corneum, S. granulosum and S. spinosum) and NFKBIZ, a transcriptional regulator of IL-36–driven gene expression34 is co-expressed with IL-36G in stratum spinosum. 97.2% (3196/3288) of KCs expressing IL-36G in psoriasis skin was located in suprabasal layers, and 51.0% (292/573) of KCs expressing IL-36G in S. spinosum of psoriasis and control skin co-expressed NFKBIZ (Fig 6C).
Our data showed that CCL27 (chemotactic ligand for skin-associated memory T lymphocytes binding to CCR10)35 and KRT15 (keratinocyte stem marker of quiescence)36 were decreased in stratum basale (Fig 6D). CCL27 expression in KCs in S. basale decreased 10.5 times from control (17.3%, 189/1093) to psoriasis (1.7%, 7/424). KRT15 expression in KCs in S. basale decreased 11.3 times from control (48.1%, 526/1093) to psoriasis (4.2%, 18/424). When we analyzed the CCL27/CCL28-CCR10 interaction with the scRNA-seq data, CCL27-CCR10 interaction was potentially impaired in psoriasis due to decreased CCL27 expression in basal KCs37, 38 (Fig 6E).
Discussion
Recent scRNA-seq technology provided the opportunity to compare gene expression profiles of heterogenous immune cells in the skin without the need to predetermine markers for various cell subsets. Co-authors of this paper pioneered single-cell transcriptome profiling of keratinocytes in lupus nephritis patients’ skin10, 11 and single-cell transcriptome profiling of fibroblasts in atopic dermatitis patients’ skin9, and many researches are now utilizing scRNA-seq for studying human skin to further understand inflammatory skin diseases12–17. However, scRNA-seq assessment of immune cells in human skin has been challenging with whole tissue enzyme dissociation where most cells are keratinocytes and fibroblasts, and immune cells constitute less than 5% of the isolated cells. To study immune cell subsets in the skin with scRNA-seq, skin immune cells isolated by enzyme digestion are often required to be sorted, activated and enriched before single-cell library construction, which may lead to changes in their gene expression or even function39, 40.
In an attempt to overcome the limitation of enzyme dissociation for isolation of skin immune cells and to obtain single-cell transcriptome of human immune cells with minimal manipulation, we harvested emigrating cells from skin biopsy tissues after 48-hour incubation in culture medium without enzyme digestion. Our single-cell approach was established for flow cytometry analysis of cutaneous DCs7, 20, 41, 42 and T-cells43, 44, but it has not been linked to single-cell transcriptomics to study heterogeneous immune cells in human skin.
With our new single-cell approach, we found cutaneous T17 cell subsets with highly differing transcriptomes depending on IL-17A vs. IL-17F expression and IFNγ vs. IL-10 expression: 1) IL-17A+ IFNγ+ T17 cells, 2) IL-17A+ IFNγ− T17 cells, 3) IL-17A+ IL-17F+ T17 cells, 4) IL-17F+ IL-10− T17 cells and 5) IL-17F+ IL-10+ T17 cells (Fig 3B). The T17 cell subset that most conforms with current pathogenic subsets6, 22–24 is the IL-17A+ IFNγ+ population, synthesizing high levels of TNF, IL-26 and IL-36G and mostly within CD8+ T-cells that co-express cytotoxic markers (Tc17 T-cells).
In contrast, the largest subset of IL-17-producing T-cells isolated from psoriasis lesions are those that are IL-17F+ IL-10−. This population (presumptive IL-17F/F producers) constitutes 53% of T17 T-cells and is about 5-fold more frequent than cells co-producing IL-17A and IFNγ. High expression of cytokines such as IL-1B, CSF-2, LTA, IL-24 and IL-34 likely identifies a different inflammatory potential from cells exposing IL-17A and IFNγ. Interestingly, this subset has the highest expression of the IL-23 receptor, so it might be the most strongly affected by therapeutic IL-23 antagonists. Perhaps this subset also stems from inflammatory conversion of Tregs, as FoxP3 has the highest expression among all T17 cells. While a recent report found some human blood T-cells synthesized only IL-17F after culture in polarizing conditions45, we believe this is the first report of a unique and sizable IL-17F+ T-cell population in the context of an organ affected by an IL-17-driven autoimmune condition.
The population that best fits the description of non-pathogenic T17 cells6, 23–26, 30, 46 is the IL-17F+ IL-10+ population, but this population constitutes only 4% of overall T17 cells. IL-17F+ IL10+ T17 cells expressed low levels of FoxP3 and high levels of AHR and MAF. Since Aryl hydrocarbon Receptor (AHR) interacts with c-Maf (MAF) to promote the differentiation of Tr1 cells47, cutaneous IL-17F+ T17 cells may have plasticity to differentiate into Tr1 cells rather than Tregs.
We found distinct DC subsets in human skin, including mature DCs and semimature DCs (Fig 4). High expression of MHC and co-stimulatory molecules by mature DCs in psoriasis may be the reason for the highly T-cell stimulatory nature of these cells8. Semimature DCs are regulatory DCs expressing high levels of IL-10, BDCA-3 and LILRB2. Single-cell transcriptome of semimature DCs is consistent with gene expression profiles of human skin resident BDCA3+ regulatory DCs described by Chu, C.C. et al.7.
Since mature DCs in psoriasis skin express high levels of IL-23A & IL-36G and low levels of KYNU compared to mature DCs in control skin, mature DCs in psoriasis skin may lose tolerogenicity that is maintained by IDO1-KYNU-AHR loop31, 48, 49 in normal condition and express IL-23A and IL-36G (Fig E11). A subset of regulatory semimature DCs in psoriasis skin expresses IL-23A and IL-36G; dysfunctional regulatory DCs in psoriasis skin may contribute to the immune tolerance impairment implicated in psoriasis.
Our approach has some limitations50: 1) 48 hour in vitro culture and cell emigration may change the status of cells and cause the change of gene expression, 2) we observed that the number of isolated skin cells and the proportion of different cell types were not consistent between samples, 3) heterogeneous skin cells with variable sizes were not reliably counted by automated cell counters. As a result, the number of cells submitted to a microfluidic platform was not consistent between samples and sequencing depth per cells became variable (Table E1), 4) some skin resident immune cells may not be migratory and might require other isolation techniques.
With novel findings of distinct immune cell transcriptome changes in psoriasis, we propose that single-cell transcriptomics applied to emigrating cells from skin provide an innovative study platform to compare gene expression profiles of heterogenous immune cells in human skin. This method can also readily be extended to study other inflammatory skin diseases or modulation of leukocyte transcripts by immune-targeted treatments.
Supplementary Material
Acknowledgments:
We thank Rockefeller University Research Facilitation Office for regulatory and administrative assistance. We thank Rockefeller University Genomics Resource Center for providing access to Illumina sequencing.
Funding:
This work was supported by National Psoriasis Foundation Discovery Grant 2018. This work was supported by in part grant no. UL1TR001866 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program.
Disclosure of potential conflict of interest:
J.K. has received research funds from AbbVie. E.G. has received research funds from AbbVie, Celgene, Eli Lilly, Janssen, Medimmune/Astra Zeneca, Novartis, Pfizer, Regeneron, Vitae, Glenmark, Galderma, Asana, Innovaderm, Dermira, and UCB and is also a consultant for Sanofi Aventis, Regeneron, Stiefel/GlaxoSmithKline, MedImmune, Celgene, Anacor, AnaptysBio, Dermira, Galderma, Glenmark, Novartis, Pfizer, Vitae, Leo Pharma, AbbVie, Eli Lilly, Kyowa, Mitsubishi Tanabe, Asana Biosciences, and Promius. J.G.K. has received research support from Pfizer, Amgen, Janssen, Lilly, Merck, Novartis, Kadmon, Dermira, Boehringer, Innovaderm, Kyowa, BMS, Serono, BiogenIdec, Delenex, AbbVie, Sanofi, Baxter, Paraxel, Xenoport, and Kineta. The rest of the authors declare that they have no relevant conflict of interest.
Abbreviations:
- AHR
Aryl hydrocarbon Receptor
- CCL
CC chemokine ligand
- DC
Dendritic cell
- FLG
Filaggrin
- KC
Keratinocyte
- MFI
Median fluorescence intensity
- NK
Natural Killer
- S.
Stratum
- scRNA-seq
Single-cell RNA sequencing
- T17
Type 17 T-cell
- Tc17
Cytotoxic type 17 T-cell
- Th17
T helper 17 T-cell
- Treg
Regulatory T-cell
- Tr1
Type 1 regulatory T-cell
- UMAP
Uniform Manifold Approximation and Projection
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kim J, Krueger JG. Highly Effective New Treatments for Psoriasis Target the IL-23/Type 17 T Cell Autoimmune Axis. Annu Rev Med 2017; 68:255–69. [DOI] [PubMed] [Google Scholar]
- 2.Wright JF, Guo Y, Quazi A, Luxenberg DP, Bennett F, Ross JF, et al. Identification of an interleukin 17F/17A heterodimer in activated human CD4+ T cells. Journal of Biological Chemistry 2007; 282:13447–55. [DOI] [PubMed] [Google Scholar]
- 3.Sugiyama H, Gyulai R, Toichi E, Garaczi E, Shimada S, Stevens SR, et al. Dysfunctional Blood and Target Tissue CD4+CD25high Regulatory T Cells in Psoriasis: Mechanism Underlying Unrestrained Pathogenic Effector T Cell Proliferation. The Journal of Immunology 2005; 174:164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J, Lee J, Gonzalez J, Fuentes-Duculan J, Garcet S, Krueger JG. Proportion of CD4(+)CD49b(+)LAG-3(+) Type 1 Regulatory T Cells in the Blood of Psoriasis Patients Inversely Correlates with Psoriasis Area and Severity Index. J Invest Dermatol 2018. [DOI] [PubMed] [Google Scholar]
- 5.Gulati N, Suárez-Fariñas M, da Rosa JC, Krueger JG. Psoriasis is characterized by deficient negative immune regulation compared to transient delayed-type hypersensitivity reactions. F1000Research 2015; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu X, Tian J, Wang S. Insight Into Non-Pathogenic Th17 Cells in Autoimmune Diseases. Front Immunol 2018; 9:1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu CC, Ali N, Karagiannis P, Di Meglio P, Skowera A, Napolitano L, et al. Resident CD141 (BDCA3)+ dendritic cells in human skin produce IL-10 and induce regulatory T cells that suppress skin inflammation. J Exp Med 2012; 209:935–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaba LC, Fuentes-Duculan J, Eungdamrong NJ, Abello MV, Novitskaya I, Pierson KC, et al. Psoriasis is characterized by accumulation of immunostimulatory and Th1/Th17 cell-polarizing myeloid dendritic cells. J Invest Dermatol 2009; 129:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He H, Suryawanshi H, Morozov P, Gay-Mimbrera J, Del Duca E, Kim HJ, et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J Allergy Clin Immunol 2020. [DOI] [PubMed] [Google Scholar]
- 10.Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, et al. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight 2017; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Der E, Suryawanshi H, Morozov P, Kustagi M, Goilav B, Ranabothu S, et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol 2019; 20:915–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D, Kobayashi T, Voisin B, Jo JH, Sakamoto K, Jin SP, et al. Targeted therapy guided by single-cell transcriptomic analysis in drug-induced hypersensitivity syndrome: a case report. Nat Med 2020; 26:236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devitt K, Hanson SJ, Tuong ZK, McMeniman E, Soyer HP, Frazer IH, et al. Single-cell RNA sequencing reveals cell type-specific HPV expression in hyperplastic skin lesions. Virology 2019; 537:14–9. [DOI] [PubMed] [Google Scholar]
- 14.Sole-Boldo L, Raddatz G, Schutz S, Mallm JP, Rippe K, Lonsdorf AS, et al. Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun Biol 2020; 3:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi R, Grzenda A, Allison TF, Rawnsley J, Balin SJ, Sabri S, et al. Defining Transcriptional Signatures of Human Hair Follicle Cell States. J Invest Dermatol 2020; 140:764–73 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue D, Tabib T, Morse C, Lafyatis R. Transcriptome landscape of myeloid cells in human skin reveals diversity, rare populations and putative DC progenitors. J Dermatol Sci 2020; 97:41–9. [DOI] [PubMed] [Google Scholar]
- 17.Rojahn TB, Vorstandlechner V, Krausgruber T, Bauer WM, Alkon N, Bangert C, et al. Single-cell transcriptomics combined with interstitial fluid proteomics defines cell type-specific immune regulation in atopic dermatitis. J Allergy Clin Immunol 2020. [DOI] [PubMed] [Google Scholar]
- 18.Lowe MM, Naik HB, Clancy S, Pauli M, Smith KM, Bi Y, et al. Immunopathogenesis of hidradenitis suppurativa and response to anti-TNFα therapy. JCI insight 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudjonsson JE, Tsoi LC, Ma F, Billi AC, van Straalen KR, Vossen AR, et al. Contribution of plasma cells and B-cells to hidradenitis suppurativa pathogenesis. JCI insight 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita H, Nograles KE, Kikuchi T, Gonzalez J, Carucci JA, Krueger JG. Human Langerhans cells induce distinct IL-22-producing CD4+ T cells lacking IL-17 production. Proc Natl Acad Sci U S A 2009; 106:21795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature biotechnology 2015; 33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dambacher J, Beigel F, Zitzmann K, De Toni E, Göke B, Diepolder HM, et al. The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut 2009; 58:1207–17. [DOI] [PubMed] [Google Scholar]
- 23.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 2012; 13:991–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu D, Notarbartolo S, Croonenborghs T, Patel B, Cialic R, Yang TH, et al. Transcriptional signature of human proinflammatory TH17 cells identifies reduced IL10 gene expression in multiple sclerosis. Nat Commun 2017; 8:1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aschenbrenner D, Foglierini M, Jarrossay D, Hu D, Weiner HL, Kuchroo VK, et al. An immunoregulatory and tissue-residency program modulated by c-MAF in human TH17 cells. Nat Immunol 2018; 19:1126–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Yang Y, Qiu G, Lal G, Wu Z, Levy DE, et al. c-Maf regulates IL-10 expression during Th17 polarization. The Journal of Immunology 2009; 182:6226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pot C, Jin H, Awasthi A, Liu SM, Lai CY, Madan R, et al. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J Immunol 2009; 183:797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobie JJ, Shah PR, Yang L, Rebhahn JA, Fowell DJ, Mosmann TR. T regulatory and primed uncommitted CD4 T cells express CD73, which suppresses effector CD4 T cells by converting 5′-adenosine monophosphate to adenosine. The Journal of Immunology 2006; 177:6780–6. [DOI] [PubMed] [Google Scholar]
- 29.Alam MS, Kurtz CC, Rowlett RM, Reuter BK, Wiznerowicz E, Das S, et al. CD73 is expressed by human regulatory T helper cells and suppresses proinflammatory cytokine production and Helicobacter felis-induced gastritis in mice. The Journal of infectious diseases 2009; 199:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chalmin F, Mignot G, Bruchard M, Chevriaux A, Végran F, Hichami A, et al. Stat3 and Gfi-1 transcription factors control Th17 cell immunosuppressive activity via the regulation of ectonucleotidase expression. Immunity 2012; 36:362–73. [DOI] [PubMed] [Google Scholar]
- 31.Harden JL, Lewis SM, Lish SR, Suarez-Farinas M, Gareau D, Lentini T, et al. The tryptophan metabolism enzyme L-kynureninase is a novel inflammatory factor in psoriasis and other inflammatory diseases. J Allergy Clin Immunol 2016; 137:1830–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson KJ, Allen RL. Regulation of T- cell immunity by leucocyte immunoglobulin- like receptors: innate immune receptors for self on antigen- presenting cells. Immunology 2009; 127:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe S, Wagatsuma K, Ichikawa E, Takahashi H. Abnormal distribution of epidermal protein antigens in psoriatic epidermis. The Journal of dermatology 1991; 18:143–51. [DOI] [PubMed] [Google Scholar]
- 34.Müller A, Hennig A, Lorscheid S, Grondona P, Schulze-Osthoff K, Hailfinger S, et al. IκBζ is a key transcriptional regulator of IL-36–driven psoriasis-related gene expression in keratinocytes. Proceedings of the National Academy of Sciences 2018; 115:10088–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riis JL, Johansen C, Vestergaard C, Bech R, Kragballe K, Iversen L. Kinetics and differential expression of the skin- related chemokines CCL27 and CCL17 in psoriasis, atopic dermatitis and allergic contact dermatitis. Experimental dermatology 2011; 20:789–94. [DOI] [PubMed] [Google Scholar]
- 36.Garza LA, Yang C-C, Zhao T, Blatt HB, Lee M, He H, et al. Bald scalp in men with androgenetic alopecia retains hair follicle stem cells but lacks CD200-rich and CD34-positive hair follicle progenitor cells. The Journal of clinical investigation 2011; 121:613–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia M, Hu S, Fu Y, Jin W, Yi Q, Matsui Y, et al. CCR10 regulates balanced maintenance and function of resident regulatory and effector T cells to promote immune homeostasis in the skin. J Allergy Clin Immunol 2014; 134:634–44 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C, Xu M, Coyne J, Wang WB, Davila ML, Wang Y, et al. Psoriasis-associated impairment of CCL27/CCR10-derived regulation leads to IL-17A/IL-22-producing skin T-cell overactivation. J Allergy Clin Immunol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den Brink SC, Sage F, Vértesy Á, Spanjaard B, Peterson-Maduro J, Baron CS, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nature methods 2017; 14:935. [DOI] [PubMed] [Google Scholar]
- 40.Autengruber A, Gereke M, Hansen G, Hennig C, Bruder D. Impact of enzymatic tissue disintegration on the level of surface molecule expression and immune cell function. European Journal of Microbiology and Immunology 2012; 2:112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nestle FO, Zheng XG, Thompson CB, Turka LA, Nickoloff BJ. Characterization of dermal dendritic cells obtained from normal human skin reveals phenotypic and functionally distinctive subsets. J Immunol 1993; 151:6535–45. [PubMed] [Google Scholar]
- 42.Lowes MA, Chamian F, Abello MV, Fuentes-Duculan J, Lin SL, Nussbaum R, et al. Increase in TNF-alpha and inducible nitric oxide synthase-expressing dendritic cells in psoriasis and reduction with efalizumab (anti-CD11a). Proc Natl Acad Sci U S A 2005; 102:19057–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klicznik MM, Morawski PA, Hollbacher B, Varkhande SR, Motley SJ, Kuri-Cervantes L, et al. Human CD4(+)CD103(+) cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci Immunol 2019; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe R, Gehad A, Yang C, Scott LL, Teague JE, Schlapbach C, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Science translational medicine 2015; 7:279ra39–ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burns LA, Maroof A, Marshall D, Steel KJA, Lalnunhlimi S, Cole S, et al. Presence, function, and regulation of IL-17F-expressing human CD4(+) T cells. Eur J Immunol 2020; 50:568–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, et al. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell 2015; 163:1400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, et al. The Aryl hydrocarbon Receptor (AhR) interacts with c-Maf to promote the differentiation of IL-27-induced regulatory type 1 (TR1) cells. Nature immunology 2010; 11:854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Q, Harden JL, Anderson CD, Egilmez NK. Tolerogenic Phenotype of IFN-gamma-Induced IDO+ Dendritic Cells Is Maintained via an Autocrine IDO-Kynurenine/AhR-IDO Loop. J Immunol 2016; 197:962–70. [DOI] [PubMed] [Google Scholar]
- 49.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol 2004; 4:762–74. [DOI] [PubMed] [Google Scholar]
- 50.Kulkarni A, Anderson AG, Merullo DP, Konopka G. Beyond bulk: a review of single cell transcriptomics methodologies and applications. Curr Opin Biotechnol 2019; 58:129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


