Abstract
The host-microbiota cross-talk represents an important factor contributing to innate immune response and host resistance during infection. It has been shown that probiotic lactobacilli exhibit the ability to modulate innate immunity and enhance pathogen elimination. Here we showed that heat-inactivated probiotic strain Lactobacillus curvatus BGMK2-41 stimulates immune response and resistance of the Caenorhabditis elegans against Staphylococcus aureus and Pseudomonas aeruginosa. By employing qRT-PCR and western blot analysis we showed that heat-inactivated BGMK2-41 activated PMK-1/p38 MAPK immunity pathway which prolongs the survival of C. elegans exposed to pathogenic bacteria in nematode killing assays. The C. elegans pmk-1 mutant was used to demonstrate a mechanistic basis for the antimicrobial potential of BGMK2-41, showing that BGMK2-41 upregulated PMK-1/p38 MAPK dependent transcription of C-type lectins, lysozymes and tight junction protein CLC-1. Overall, this study suggests that PMK-1/p38 MAPK‐dependent immune regulation by BGMK2-41 is essential for probiotic-mediated C. elegans protection against gram-positive and gram-negative bacteria and could be further explored for development of probiotics with the potential to increase resistance of the host towards pathogens.
Subject terms: Immunology, Microbiology
Introduction
Innate immunity provides the first line of defense against invading microbes with nonspecific detection of conserved features of pathogens1. In mammals, discrimination of these conserved molecules occurs through Pattern Recognition Receptors (PRRs), such as Toll-like Receptors (TLRs) resulting in activation of intracellular signaling cascades that rapidly induce the expression of distinct genes involved in the inflammatory and immune responses2. The molecular pathways initially triggered by pathogens are highly conserved in a large variety of organisms ranging from flies and nematodes to mammals3.
Caenorhabditis (C.) elegans is a small soil nematode naturally exposed to numerous microbes. From early adulthood, nematodes feed on bacteria which pass through the pharynx to the gut, colonize the intestinal lumen and establish their gut microbiota4. This close interaction of nematode and bacteria led C. elegans to evolve ways of avoiding dangerous pathogens and to adapt to hostile environment5. To defend against infection, C. elegans possesses certain evolutionarily conserved defense mechanisms including the PMK-1 pathway, an orthologue of p38 mitogen-activated protein kinase (MAPK)6, the transforming growth factor β (TGF-β) signaling denote as DBL-1 pathway7 and the ERK1/2 MAPK homolog MPK-1 that can be activated in response to specific pathogens5. The most studied PMK-1 pathway coordinates defense against a variety of ingested pathogens by regulating the expression of different antimicrobial effectors. Downstream targets of PMK-1 include genes that encode antimicrobial proteins, such as antimicrobial peptides, C-type lectins and lysozymes3. Like in mammals, pathogen recognition in worms could be mediated through TLR ortholog TOL-1, which is required for immunity of C. elegans to certain gram-negative bacteria8. As most bacterial species that are known to be pathogenic for C. elegans also cause infections in humans9, C. elegans has become a useful model system for innate immunity studies in terms of pathogen-host interactions10.
In laboratory conditions, C. elegans are reared on a single bacterial strain of Escherichia coli OP504. The composition of bacterial diet plays an important role in the control of C. elegans well-being by modulating animal physiology including metabolism, development and reproduction11. These bacterivore nematodes have been used to study dietary effects and changes in physiology and transcriptomic signatures after exposure to commensal bacteria and probiotics12. Probiotics are recognized as beneficial microorganisms, known to exert numerous positive effects on human health13,14. Probiotic-host cross-talk primarily results in the modulation of innate immunity which is important to determine the host resistance to infection15. Several studies already demonstrated that some Lactobacillus strains when applied alive activated immune and stress responses in the worms which increased resistance of the nematodes to certain bacterial infections16,17. Beside live probiotic bacteria, UV‐killed and heat-inactivated bacteria, also exhibited beneficial effects on the worms. It has been shown that lactobacilli applied unviable, as postbiotics, tune SKN-1/NRF2 or HLH-30/TFEB signaling in the nematodes which consequently extended C. elegans lifespan and improved general health markers18,19. However, the data about potential of heat-inactivated probiotic bacteria to enhance survival of the host towards different pathogens, especially Staphylococcus aureus and Pseudomonas aeruginosa, remain limited. Reports showed that S. aureus and P. aeruginosa infection causes death of the C. elegans by triggering intestinal epithelium distention and destruction followed by intracellular invasion and complete degradation of internal organs20. This means that epithelial barrier represents the major site of host defense system, beside above-mentioned canonical pathways21. In light of the growing antibiotic resistance of these two pathogens, the stimulation of innate immunity and maintenance of barrier integrity, as alternative strategy, is increasingly being explored.
Therefore, following the postbiotic trend of replacing live probiotic bacteria with bacterial biomolecules and/or inactivated bacteria in order to get stable and better controlled biological response22, here we report that heat-inactivated Lactobacillus curvatus BGMK2-41 strain activated PMK-1/p38 MAPK dependent immune response to control the production of hosts antimicrobials which increased survival of the worms exposed to both gram-positive and gram-negative pathogenic bacteria.
Results
Pre-conditioning of C. elegans with BGMK2-41 prolongs the survival of nematodes exposed to pathogenic bacteria
To evaluate the immunomodulatory potential of heat-inactivated BGMK2-41 we used C. elegans as an in vivo model system and schematic overview of experimental approach is presented in Fig. 1A. Results revealed that animals previously exposed to heat-inactivated BGMK2-41 exhibited enhanced survival upon infection with both S. aureus ATCC 25923 (Fig. 1B) and P. aeruginosa PA14 (Fig. 1C) pathogenic strains in comparation to the exposure to heat-inactivated OP50, used as a control. Furthermore, we were able to demonstrate that gram-negative bacteria exhibit more lethal infection which correlates with the increased intestinal accumulation of P. aeruginosa PA14 (9.4 × 103 CFUs), whereas only 0.58 × 103 CFUs accumulated in the case of S. aureus ATCC 25923. However, prolonged survival of nematodes previously exposed to BGMK2-41 was not a consequence of decreased intestinal load of pathogenic bacteria, as we detected no differences in CFUs between BGMK2-41 treated and control animals (Fig. 1D,E). These findings imply that BGMK2-41 enhances the survival of C. elegans without decreasing bacterial accumulation and colonization.
Figure 1.
Heat-inactivated Lactobacillus curvatus BGMK2-41 prolongs the survival of C. elegans exposed to pathogenic bacteria. (A) An overview of experimental approach for evaluation of BGMK2-41 effects on infected worms’ lifespan and gene/protein. Survival curve of WT animals exposed to (B) S. aureus ATCC 25923 and (C) P. aeruginosa PA14 after overnight feeding of L4 stage worms with heat-inactivated control (E. coli OP50) or BGMK2-41 bacteria (n = 25–30 worms per group, results are representative of 2 independent assays). CFUs level of (D) S. aureus ATCC 25923 and (E) P. aeruginosa PA14 present in the BGMK2-41 pre-treated worms after 1 day of exposure to pathogens (n = 10 worms per group, results are representative of 2 independent assays). All values are presented as mean ± SD and the student’s t-test was used to compare the treated group relative to control. The log-rank (Mantel-Cox) test was used to assess the p-value in nematode killing assays (***P < 0.001). The statistical analysis and graphs were done in GraphPad Prism version 8.0.0 for Mac, GraphPad Software, www.graphpad.com. (A) was designed by using free trial of Adobe Illustrator, www.adobe.com/products/illustrator.
Heat-inactivated BGMK2-41 activates a canonical PMK-1/p38 MAPK immune pathway
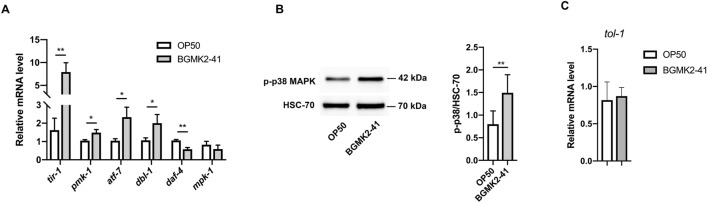
To address whether BGMK2-41 upregulates gene expression of the above-mentioned immune pathways, we performed qRT-PCR analysis in wild-type N2 worms. We have found that all genes involved in PMK-1/p38 MAPK signaling (tir-1, pmk-1, atf-7) were significantly upregulated (Fig. 2A). Remarkably, we detected elevated mRNA levels for dbl-1 gene, but when we analyzed further DBL-1 signaling, we detected decreased transcription of downstream daf-4 gene (Fig. 2A). Finally, mpk-1 expression was similar in BGMK2-41 treated and control worms which excluded ERK1/2 MAPK involvement in worms’ resistance phenomenon (Fig. 2A). Next, we confirmed the activation of PMK-1/p38 MAPK pathway by determining the level of the activated form of p38 MAPK and showed that its phosphorylation was upregulated in C. elegans treated with BGMK2-41 relative to OP50 control (Fig. 2B).
Figure 2.
Heat-inactivated Lactobacillus curvatus BGMK2-41 stimulates a canonical p38MAPK immune pathway in C. elegans. (A) Expression of immune-related genes measured by qRT-PCR in WT animals after 24 h of treatment with heat-inactivated BGMK2-41 relative to heat-inactivated E. coli OP50 control. (B) Representative western blot with densitometric analysis showing the levels of phospho-p38 MAPK isolated from WT animals after overnight BGMK2-41 treatment. HSC-70 was used as a loading control. (C) Levels of tol-1 mRNA measured in day 1 old WT worms treated with BGMK2-41. All results were obtained from three independent experiments and the data are presented as mean ± SD and student’s t-test was used to compare the treated group relative to control (*P < 0.05, **P < 0.01). The statistical analysis and graphs were done in GraphPad Prism version 8.0.0 for Mac, GraphPad Software, www.graphpad.com.
When we looked upstream of PMK-1/p38 MAPK pathway and examine evolutionarily conserved TOL-1/TLR role in immune response stimulated by BGMK2-41, we did not detect differences in tol-1 mRNA level between treated and control groups (Fig. 2C). Therefore, we concluded that BGMK2-41 triggers a canonical PMK-1/p38 MAPK immune pathway, most likely in TOL-1/TLR independent manner and that alternative mechanisms may exist for the detection of conserved BGMK2-41 molecules.
The PMK-1/p38 MAPK is essential for the enhanced resistance of the worms to pathogenic bacteria
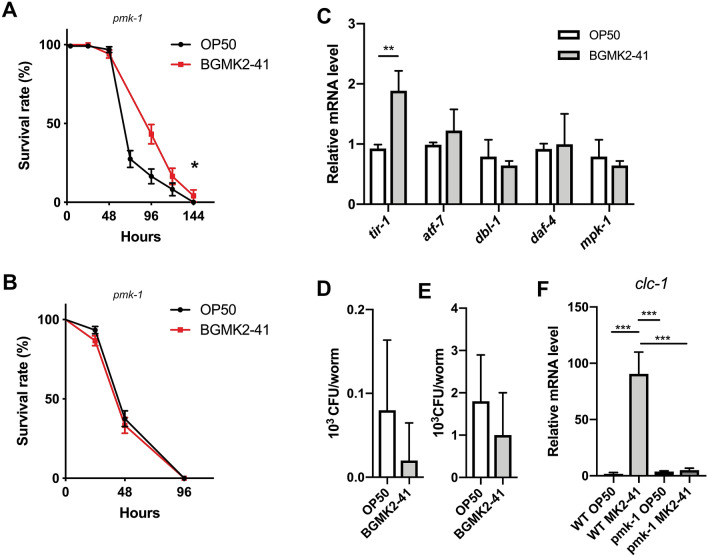
To evaluate whether PMK-1/p38 MAPK is functionally involved in host defense against pathogenic bacteria triggered by BGMK2-41, we infected C. elegans loss-of-function pmk-1 mutant and followed survival over time. Deletion of pmk-1 reduced enhanced survival induced by pre-conditioning of the worms with BGMK2-41 during exposure to S. aureus ATCC 25923 (Fig. 3A). Moreover, increased resistance of the worms previously exposed to BGMK2-41 was completely abrogated in pmk-1 mutant exposed to P. aeruginosa PA14 (Fig. 3B). Inactivation of pmk-1 did not substantially alter BGMK2-41 induced expression of tir-1 mRNA, a gene encoding a highly conserved Toll/IL-1 resistance (TIR) domain protein acting upstream of PMK-1, observed in pmk-1 mutant (Fig. 3C). On the other hand, mRNA levels of downstream atf-7 transcription factor responsible for the expression of secreted immune effectors, were not changed upon treatment with BGMK2-41 suggesting that BGMK2-41 activates TIR-1, but impaired PMK-1/p38 MAPK signaling did not elicit protective response against pathogens (Fig. 3C). The defective PMK-1/p38 MAPK signaling was confirmed by the absence of p38 MAPK phosphorylation in pmk-1 mutant (Supplementary Fig. 1). Finally, qRT-PCR analysis in pmk-1 mutant showed that genes involved in DBL-1 and MPK-1 immune pathways were unchanged by BGMK2-41 treatment excluding the possible activation of these two defense programs (Fig. 3C).
Figure 3.
The p38 MAPK deficiency abrogates protective effects of Lactobacillus curvatus BGMK2-41. Survival curve of pmk-1 loss-of-function animals exposed to (A) S. aureus ATCC 25923 and (B) P. aeruginosa PA14 after overnight feeding of L4 stage worms with heat-inactivated control (E. coli OP50) or BGMK2-41 bacteria (n = 25–30 worms per group, results are representative of 2 independent assays). (C) Expression of immune-related genes analyzed by qRT-PCR in the L4 stage of the pmk-1 mutant after overnight treatment with heat-inactivated BGMK2-41 (n = 3, three independent experiments). CFUs values of (D) S. aureus ATCC 25923 and (E) P. aeruginosa PA14 present in the pmk-1 deficient BGMK2-41 treated worms after 1 day of exposure to pathogens (n = 10 worms per group, results are representative of 2 independent assays). (F) Tight junction barrier assessment by determination of clc-1 mRNA levels in day 1 old WT and pmk-1 worms treated with heat-inactivated BGMK2-41 (results from three independent experiments). All values are presented as mean ± SD. Student’s t-test was used to compare the treated group relative to control. The log-rank (Mantel-Cox) test was used to assess the p-value in nematode killing assays (*P < 0.05, **P < 0.01, ***P < 0.001). The statistical analysis and graphs were done in GraphPad Prism version 8.0.0 for Mac, GraphPad Software, www.graphpad.com.
Gut colonization analysis for both pathogens in pmk-1 mutant, showed similar CFUs values between the experimental and control groups, as we previously determined in N2 worms (Fig. 3D,E). This observation excluded the impact of BGMK2-41 on pathogens gut colonization and confirmed that prolonged survival of BGMK2-41 treated worms was independent from pathogen intestinal burden.
As immune activation did not result in a decrease of pathogens in the intestines, we further looked in the integrity of the intestinal barrier. We tested the expression of mRNA encoding the tight junction protein claudin-like in Caenorhabditis (CLC-1), mainly expressed in the epithelial cells of digestive tubes23. The expression analysis revealed that BGMK2-41 strongly stimulated the expression of the claudin-related clc-1 gene in N2 worms, but this effect was completely abolished in pmk-1 mutant suggesting that BGMK2-41 strengthen gut epithelial barrier through upregulation of tight junction’s claudin in PMK-1/p38 MAPK dependent manner (Fig. 3F).
The p38 MAPK pathway triggered by heat-inactivated BGMK2-41 controls expression of various antimicrobial genes
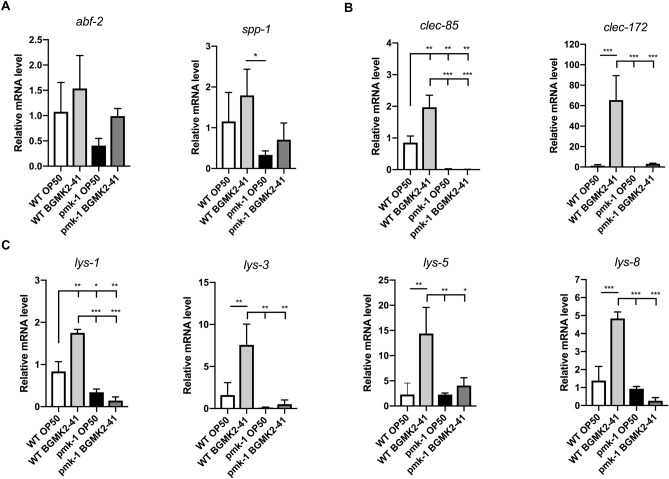
Next, we performed transcriptional profiling of the selected antimicrobial genes as markers of the wider host response, by comparing their expression in N2 animals and pmk-1 mutants after exposure to BGMK2-41. These genes include antimicrobial peptides (abf-2, spp-1), C-type lectins (clec-85, clec-172) and lysozymes (lys-1, lys-3, lys-5, lys-8). First, the BGMK2-41 failed to upregulate the antimicrobial peptides both in N2 and pmk-1 animals, excluding their role in BGMK2-41 orchestrated defense program (Fig. 4A). However, the BGMK2-41 treatment of N2 worms results in a major upregulation of the rest of the tested genes, with clec-172 and lys-5 as the most highly expressed (Fig. 4B,C). Remarkably, a drastic reduction of mRNAs of C-type lectins and lysozymes were detected in pmk-1 worms (Fig. 4B,C). As a result, we concluded that BGMK2-41 enhanced survival of the worms is mediated by the production of C-type lectins and lysozymes as antimicrobial effectors controlled by PMK-1/p38 MAPK activity.
Figure 4.
Heat-inactivated Lactobacillus curvatus BGMK2-41 controls the expression of antimicrobial proteins via p38 MAPK. Expression of (A) antimicrobial peptides, (B) C-type lectins and (C) lysozymes measured by qRT-PCR in day 1 old WT and pmk-1 worms treated with heat-inactivated BGMK2-41 (results from three independent experiments). All values are presented as mean ± SD. Student’s t-test was used to compare the treated group relative to control (*P < 0.05, **P < 0.01, ***P < 0.001). The statistical analysis and graphs were done in GraphPad Prism version 8.0.0 for Mac, GraphPad Software, www.graphpad.com.
Discussion
Probiotic lactobacilli can elicit distinct immunomodulatory effects in the host ranging from immune stimulation to suppression in a species-specific manner24,25. In the present study, we found that PMK-1/p38 MAPK‐dependent immune activation by heat-inactivated Lb. curvatus BGMK2-41 is essential for C. elegans protection against S. aureus ATCC 25923 and P. aeruginosa PA14, as loss of PMK-1/p38 MAPK almost completely abrogated survival of the worms exposed to pathogens. Further, we show that p38 MAPK orchestrated a specific transcriptional response which strengthened the gut epithelial barrier and stimulated the production of C-type lectins and lysozymes to improve the survival of infected worms.
Despite a growing body of evidence reporting the beneficial effects of different Lactobacillus species, only a few studies have attempted to elucidate the health-promoting potential of the Lb. curvatus strains. The literature data showed that different isolates of Lb. curvatus exhibit beneficial role in dyslipidemia and obesity26. Moreover, in the case of Lb. curvatus effects on the immune system, one report has shown that Lb. curvatus WiKim38 triggered the activation of dendritic cells and stimulated IL-10 production to suppress chemically-induced colitis in mice27. However, as the data about Lb. curvatus impact on gut epithelial barrier, mucosal and innate immunity is lacking, our study provides evidence of the immunostimulatory potential of Lb. curvatus.
The p38 MAPK has been established as a major regulator of the innate immune response, especially with regard to the production of proinflammatory cytokines28. Its modulation by probiotics, like Lb. acidophilus NCFM, has been described as a key factor in controlling the production of cytokines and chemokines of gut epithelial cells necessary for the restriction of pathogen invasion29. Like in mammals, PMK-1/p38 MAPK activation in C. elegans also mediates the expression of secreted immune response genes as a response to recognized microbe-associated molecular patterns (MAMPs)3. Beside pathogenic bacteria previously described to activate this pathway in nematodes, probiotic lactobacilli exhibit some of their beneficial effects through p38 MAPK. It has been described that Lb. fermentum JDFM216 effectively stimulated PMK-1/p38 MAPK pathway and increased longevity and immunity of C. elegans towards S. aureus and E. coli30. These results are in accordance with our findings in term of BGMK2-41 mediated protective effect against pathogens, but another study aimed to screen a library of different lactic acid bacteria (LAB) reported that the heat-inactivated LAB did not reduce the susceptibility of the worms to P. aeruginosa31. In this study, we detected the most prominent survival upon infection with gram-positive bacteria suggesting that BGMK2-41 and S. aureus share the same core network of host protective responses probably due to their structural similarities’ characteristic for gram-positive bacteria. To the best of our knowledge, this is the first study demonstrating the positive effect of heat-inactivated lactobacilli applied as postbiotic towards these two pathogens.
S. aureus and P. aeruginosa are frequent multiresistant nosocomial pathogens with increasing prevalence and ability to delay wound healing of hospitalized patients32. With growing resistance to antimicrobial agents, stimulation of the immune response by BGMK2-41 which enhanced resistance to S. aureus ATCC 25923 and P. aeruginosa PA14 infection, could be of great importance for patients affected by wound healing disorders and cutaneous infections33. Further, by applying the postbiotic concept, we succeeded to showed that heat-inactivated BGMK2-41 is sufficient to elicit an effective immune response against both pathogenic bacteria which excluded the potential adverse effect of live probiotics when it comes to their application to wounded tissue or intake by immunocompromise individuals34. Consistent with this, it has been shown that the gene expression profiles elicited by live or dead En. faecium, also used as probiotic35, are extremely similar and dependent on the immune regulator PMK-1/p38 MAPK36. Except on gene expression level, a protective effect of heat-inactivated Lb. plantarum 133 (LP133) and Lb. fermentum 21 (LP21) has been demonstrated against Salmonella and Yersinia infection in C. elegans37. These data pointed that conserved heat-stable moieties of Enterococcus and Lactobacillus mediated transcriptional response and worms’ survival to infection. It seems that BGMK2-41 with its heat-stable features evoked a protective response of the C. elegans against S. aureus ATCC 25923 and P. aeruginosa PA14. Additionally, failure of BGMK2-41 to activate TOL-1/TLR led to conclusion that potential other receptors are engaged in recognition of BGMK2-41 MAMPs, which is not surprise considering that a lot of studies pointed that the TOL-1/TLR signaling is not significantly involved in the primary response against gram-positive bacteria in the intestinal cells of C. elegans38. As TOL-1/TLR represents one of the major activators of antimicrobial peptide ABF-2, the unchanged expression of abf-2 gene further justifies the conclusion that BGMK2-41 activates PMK-1/p38 MAPK in TOL-1/TLR independent fashion8. On the other hand, strong activation of TIR-1 observed in N2 worms and pmk-1 mutant could control the innate immunity in C. elegans independently of TOL-1/TLR39. Although, the elevated mRNA levels for dbl-1 gene pointed to the possible activation of this pathway as well, we excluded this possibility by detecting decreased expression of daf-4 gene, which encodes receptor of TGF-beta signaling.
Probiotic lactobacilli and their secreted molecules have a well-described role in the reinforcement of the gut intestinal barrier by increasing expression of tight junction proteins40,41. As BGMK2-41-mediated C. elegans resistance to both tested pathogens was uncoupled from their CFUs number in the gut, we propose that upregulated expression of clc-1 consequently strengthened the tight junctions, which may be responsible for the observed longevity extension of infected worms. Moreover, the important way that hosts can defend against the pathogens is by tolerating them42. A mechanism of tolerance does not result in decrease of pathogen load, which is in accordance with the results we obtained for BGMK2-41, suggesting that reinforcement of the gut intestinal barrier could occur due to increasing tolerance to infection. When it comes to p38 MAPK role in tight junction dynamics, we provide evidence that clc-1 expression was dependent on p38 MAPK phosphorylation. It has been shown that p38 MAPK induced upregulation of claudin-1 during regeneration of rat hepatocytes43. On the contrary, p38 MAPK could mediate tight junction disruption in distal renal tubular cells induced by calcium oxalate crystals44. Therefore, having in mind the reported dual role of p38 MAPK, our study shows a significant positive correlation between p38 MAPK and claudin-like protein in in vivo model. Finally, lactobacilli could mediate worms’ resistance to methicillin-resistant S. aureus (MRSA) infection by employing DBL-1 immune signaling. This effect was independent from changes in MRSA gut colonization and CFUs45, which support our conclusion that BGMK2-41 does not have to affect the number of pathogenic bacteria to increase host survival during infection. Also, our CFUs result is in accordance with S. aureus and P. aeruginosa pathogenicity in the worms, which is reflected in pathogens accumulation in the intestinal lumen in the form of clumps surrounded by an extracellular matrix20. This characteristic phenotype of the infection could impact the CFUs numeration.
Taken together, our study offers a safer alternative besides the traditional use of antibiotics by providing molecular mechanism behind the immunostimulatory effect of heat-inactivated Lb. curvatus BGMK2-41. The results presented here indicate that modulation of the p38 MAPK pathway could be of particular interest in order to restrain infection via strengthening the gut epithelial barrier and innate immunity.
Materials and methods
C. elegans strains and maintenance
C. elegans was maintained on nematode growth medium (NGM) plates seeded with E. coli OP50 strain at 20 °C by using standard growing protocols46. To obtained synchronized population egg-bearing worms from mix population were collected from NGM plates by using M9 buffer. Eggs were extracted in the cleaning solution containing 0.5 M NaOH with 1% Na-hypochlorite and then washed with M9 buffer at least three times. Eggs were transferred to OP50 seeded NGM plates and incubated overnight at 20 °C to obtain synchronized L1 animals. In order to avoid progeny hatching and maintain synchronized population, NGM plates were supplemented with 20 μM 5-Fluorodeoxyuridine (FudR, Sigma-Aldrich) where was necessary. The wild‐type N2 (Bristol) and KU25 pmk-1 (km25) strains were used in the study. E. coli OP50 and worms’ strains originate from the collection of Caenorhabditis Genetics Center, University of Minnesota.
Bacterial strains and growth conditions
Strain Lactobacillus curvatus BGMK2-41 is a part of the IMGGE collection of microorganisms previously isolated from an artisanal Serbian fermented dairy product Kajmak47. The strain was cultivated in MRS broth (Merck) at 37 °C under anaerobic conditions using Anaerocult A (Merck). P. aeruginosa PA14, S. aureus ATCC 25923 and E. coli OP50 were cultivated in LB broth at 37 °C with shaking/aerobic conditions.
Preparation of heat-inactivated bacteria and treatments
Overnight grown culture of Lb. curvatus BGMK2-41 were pelleted by centrifugation at 5000 × g for 10 min at room temperature, washed twice in phosphate-buffered saline (PBS) and resuspended in the same volume of LB medium as OP50. Heat-inactivation for both BGMK2-41 and OP50 was performed at 60 °C for 60 min followed by inoculation of bacterial suspension (10 μl) on MRS/LB agar plates and incubation overnight at 37 °C in order to check the efficacy of inactivation treatment. The NGM plates were prepared by spreading the heat-inactivated bacterial suspensions on 9 cm plates and dried at room temperature. Age‐synchronous worms were grown to the L4 stage on live E. coli OP50 followed by transfer to NGM plates containing appropriate heat-inactivated treatment.
Nematode killing assays
In the nematode killing assays, we used S. aureus ATCC 25923 and P. aeruginosa PA14 found to elicit distinct immune responses in the infected nematodes20. The L4 stage worms were preconditioned with heat-inactivated BGMK2-41 or OP50 for 24 h before being transferred to pathogens containing plates.
P. aeruginosa killing assay was done as described by Troemel et al.3 Briefly, overnight culture of P. aeruginosa PA14 strain was spread on modified 3.5 mm NGM plates containing 0.35% peptone and incubated for 24 h at 37 °C followed by additional incubation of the plates for two days at 25 °C. A total of 25–30 one day old adult worms were transferred to each pathogen plate and a total of 100–120 worms (4 plates) were used per condition.
S. aureus killing assay was performed by spreading the overnight culture of S. aureus ATCC 25923 strain onto 3.5 mm NGM plates and incubate for 24 h at 37 °C followed by transfer of 100–120 one day old adult worms to 4 pathogen plates per condition. All killing assays were performed at 20 °C and death events scored every day by gentle prodding with a platinum wire. The worms that escaped were censored.
Colony forming unit assay (CFU)
The colonization of worms’ intestines with pathogenic bacteria was determined according to the method described by Yuen and Ausubel36, with slight modifications. Briefly, after the treatment with heat-inactivated BGMK2-41 or OP50, the worms were exposed to S. aureus ATCC 25923 and P. aeruginosa PA14 for 24 h. The infected nematodes were picked onto unseeded NGM agar plates in order to remove any external bacteria and then 10 worms were transferred to a 2 ml tube containing 25 mM tetramisole hydrochloride (Sigma-Aldrich) and 0.01% Triton X-100 (Sigma-Aldrich) in M9 wash buffer. The infected worms were washed 3 times with a total volume of 250 µl of the buffer. Afterwards, 200 µl of the buffer was removed and 100 µl of M9 buffer containing 20 mM tetramisole hydrochloride and 2% Triton X-100 and 0.5 mm silicon beads were added to the tubes. The tubes were vortexed in Disruptor Genie (Scientific Industries Inc, USA) at maximum speed for 5 min to release the bacteria from the worm intestine without impairing bacterial viability. The suspension was serially diluted and plated onto LB agar plates to enumerate the bacterial colonies. The results were expressed as CFU unit per worm.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Approximately 200 worms were collected from 9 cm treatment plates using M9 buffer and washed three times to remove the remaining bacteria. Total RNA was extracted using a Trizol reagent (Thermo Fisher Scientific) and treated with DNase I (DNA-free DNA Removal Kit) according to the manufacturer′s protocol (Thermo Fisher Scientific). RevertAid Reverse Transcriptase (Thermo Fisher Scientific) was used to transcribe 0.5 µg of isolated RNA as a template, with random hexamers (Thermo Fisher Scientific) and RiboLock RNase inhibitor (Thermo Fisher Scientific) also used in the reactions. Synthesized cDNA was then subjected to qRT-PCR analysis using SYBR Green PCR Master Mix (Thermo Fisher Scientific) in a 7500 real-time PCR machine (Applied Biosystems) under the following conditions: 10 min at 95 °C activation, 40 cycles of 15 s at 95 °C and 60 s at 60 °C. All results are normalized against the control act-1 gene19 and expressed as relative target abundance using the 2−ΔΔCt method48. Primers used in the study are listed in Table 1. All primers were purchased from Thermo Fisher Scientific. For each condition, three independent replicates were used.
Table 1.
The list of primers used in the study.
| Primer name | Primer sequence 5ʹ–3ʹ | Reference |
|---|---|---|
| tir-1 forward | CCGACCACCAAAGAAATGCC | This work |
| tir-1 reverse | CTTGGTCCACCGATGCTTCT | |
| pmk-1 forward | ACTTCATCCGACTCCACGAG | This work |
| pmk-1 reverse | CAGCAGCACAAACAGTTCCA | |
| atf-7 forward | AGAAATAAGGCTGCGGCTGT | This work |
| atf-7 reverse | TTTCAGCCTCCATTGCTTGA | |
| tol-1 forward | GCTCACCAACATCGAGCAATTC | This work |
| tol-1 reverse | GCGTTTCCAGCCAAATTCAC | |
| clc-1 forward | CCACTCACCCTCTTTGCAGT | This work |
| clc-1 reverse | CGAGTATCCAAGCTGCGAGT | |
| mpk-1 forward | TGGAGGGCAGAATCCTGTTT | This work |
| mpk-1 reverse | CAACAAGCTTTTCAGCGGGA | |
| dbl-1 forward | TTTTGCGGCGAACAAATCGT | This work |
| dbl-1 reverse | TTCGCTGTTGCCTGTTTGTG | |
| daf-4 forward | GCCAAGGACGATCATTTCGC | This work |
| daf-4 reverse | TGGGAACTTGTCCTTCTTCAA | |
| act-1 forward | TGCAGAAGGAAATCACCGCT | 19 |
| act-1 reverse | CGGACTCGTCGTATTCTTGC | |
| abf-2 forward | TTCCTTGCACTTCTCCTGGC | This work |
| abf-2 reverse | GACGACCGCTTCGTTTCTTG | |
| spp-1 forward | TCGTCGAGGGTGGAGAGAAG | This work |
| spp-1 reverse | ACGCCTTGTCTGGAGAATCC | |
| clec-85 forward | ACTATGTCGCTGAGAGCACG | This work |
| clec-85 reverse | TCCGGAGACTGGGTAAGACA | |
| clec-172 forward | AGTTTGGGAATTCGTGCTACA | This work |
| clec-172 reverse | CCGTGCTCCATTCCGAAGAT | |
| lys-1 forward | GGATCTGGAGCATTCGACACA | This work |
| lys-1 reverse | GCTGGGGAGGTAACCTGAATC | |
| lys-3 forward | TGCGAAGAATCTGGGCTTGT | This work |
| lys-3 reverse | GTGGCCTGTCTCCATGGTC | |
| lys-5 forward | TGCAGTGTTGTAAGTGCAGC | This work |
| lys-5 reverse | CATCGACATCAGTGAGGCCA | |
| lys-8 forward | TTGTCCGTGCATACAACCCA | This work |
| lys-8 reverse | TCCTTGCTTGCTTGAAGCCG |
Western blotting
After worm’s collection (approximately 200 animals) from treatment plates, proteins were isolated according to the protocol described by Herholz et al.49 Protein concentration was measured with the BCA Assay Kit (Thermo Fisher Scientific) and 20 μg of extracted proteins were separated on 12% SDS–PAGE and transferred to a 0.2 mm nitrocellulose membrane (GE Healthcare). Western blotting was performed overnight at 4 °C with antibodies against: HSC-70 (1:2000, Santa Cruz Biotechnology, #sc-7298) and phospho-p38 MAPK (1:1000, Cell Signaling, #9211). Chemiluminescence was detected by using ChemiDoc Touch Imaging System with Image Lab Touch Software (Bio-Rad). The Image Lab Touch software accurately estimates the shortest exposure time. The Image Resolution/Sensitivity scale was set to 4 × 4 pixel binning settings and pictures captured by using Rapid Auto Exposure mode. The intensity of the bands was quantified in ImageJ (National Institutes of Health, NIH) software by using plot lanes and wand tool. The results were normalized to HSC-70 loading control and values for OP50 control were adjusted to 1 in order to calculate fold of change. For each condition, three independent replicates were used.
Statistical analysis
All values are presented as mean ± standard deviation (SD). The differences between the control and experimental groups were compared using Student’s t-test, while one-way ANOVA followed by Tukey post hoc test was used for multiple comparisons. The differences between survival curves in nematode killing assays were analyzed using the log-rank (Mantel-Cox) test. A p value less than 0.05 was considered statistically significant. The statistical analysis and graphs were done in GraphPad Prism version 8.0.0 for Mac, GraphPad Software, San Diego, California USA, www.graphpad.com.
Supplementary Information
Acknowledgements
The authors are grateful to Aleksandra Trifunovic for providing the KU25 pmk-1 (km25) strain.
Author contributions
M.D., S.J., J.Đ., N.P., D.R. performed the experiments. M.D., J.Đ., N.P., I.S., N.G. interpreted findings. M.D. and D.R. prepared figures. M.D. wrote the manuscript. I.S. and N.G. supervised the work. M.D., I.S. and N.G. designed the study. All authors read and approved the manuscript.
Funding
This work was supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Grant No. 451–03-9/2021–14/200042). The study was further supported by the Science Fund of the Republic of Serbia, Diaspora Collaboration Program grant (PLASH, 6426409) and by the Center for Leadership Development, Start Up for Science grant.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-00698-5.
References
- 1.Paludan SR, Pradeu T, Masters SL, Mogensen TH. Constitutive immune mechanisms: mediators of host defence and immune regulation. Nat. Rev. Immunol. 2021;21:137–150. doi: 10.1038/s41577-020-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Troemel ER, et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:1725–1739. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han B, et al. Microbial genetic composition tunes host longevity. Cell. 2017;169:1249–1262.e13. doi: 10.1016/j.cell.2017.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ermolaeva MA, Schumacher B. Insights from the worm: the C. elegans model for innate immunity. Semin. Immunol. 2014;26:303–309. doi: 10.1016/j.smim.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim DH, et al. A conserved p38 MAP kinase pathway in C. elegans innate immunity. Science (80-) 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 7.Mallo GV, et al. Inducible antibacterial defense system in C. elegans. Curr. Biol. 2002;12:1209–1214. doi: 10.1016/S0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 8.Tenor JL, Aballay A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 2008;9:103–109. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darby, C. Interactions with microbial pathogens. WormBook 1–15 (2005) 10.1895/wormbook.1.21.1. [DOI] [PMC free article] [PubMed]
- 10.Gravato-Nobre MJ, Hodgkin J. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell. Microbiol. 2005;7:741–751. doi: 10.1111/j.1462-5822.2005.00523.x. [DOI] [PubMed] [Google Scholar]
- 11.Stuhr NL, Curran SP. Bacterial diets differentially alter lifespan and healthspan trajectories in C. elegans. Commun. Biol. 2020;3:653. doi: 10.1038/s42003-020-01379-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roselli M, et al. C. elegans and probiotics interactions from a prolongevity perspective. Int. J. Mol. Sci. 2019;20:1–14. doi: 10.3390/ijms20205020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bajić SS, et al. GABA potentiate the immunoregulatory effects of Lactobacillus brevis BGZLS10-17 via ATG5-dependent autophagy in vitro. Sci. Rep. 2020;10:130. doi: 10.1038/s41598-019-56889-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinić M, et al. Lactobacillus fermentum postbiotic-induced autophagy as potential approach for treatment of acetaminophen hepatotoxicity. Front. Microbiol. 2017;8:119. doi: 10.3389/fmicb.2017.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klaenhammer TR, Kleerebezem M, Kopp MV, Rescigno M. The impact of probiotics and prebiotics on the immune system. Nat. Rev. Immunol. 2012;12:728–734. doi: 10.1038/nri3312. [DOI] [PubMed] [Google Scholar]
- 16.Zhou M, Liu X, Yu H, Gong J. Lactobacillus regulates C. elegans cell signaling to combat Salmonella infection. Front. Immunol. 2021;12:1–10. doi: 10.3389/fimmu.2021.653205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou M, et al. Cell signaling of C. elegans in response to enterotoxigenic E. coli infection and Lactobacillus zeae protection. Front. Immunol. 2018;9:1–11. doi: 10.3389/fimmu.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa H, et al. Effects and mechanisms of prolongevity induced by Lactobacillus gasseri SBT2055 in C. elegans. Aging Cell. 2016;15:227–236. doi: 10.1111/acel.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinic M, et al. Host-commensal interaction promotes health and lifespan in Caenorhabditis elegans through the activation of HLH-30/TFEB-mediated autophagy. Aging (Albany. NY). 2021;13:8040–8054. doi: 10.18632/aging.202885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irazoqui JE, et al. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010;6:1–24. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allaire JM, et al. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol. 2018;39:677–696. doi: 10.1016/j.it.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Cuevas-González PF, Liceaga AM, Aguilar-Toalá JE. Postbiotics and paraprobiotics: From concepts to applications. Food Res. Int. 2020;136:109502. doi: 10.1016/j.foodres.2020.109502. [DOI] [PubMed] [Google Scholar]
- 23.Simske JS, Hardin J. Claudin family proteins in C. elegans. Methods Mol. Biol. 2011;762:147–169. doi: 10.1007/978-1-61779-185-7_11. [DOI] [PubMed] [Google Scholar]
- 24.Lukic J, et al. Interaction of Lactobacillus fermentum BGHI14 with rat colonic mucosa: implications for colitis induction. Appl. Environ. Microbiol. 2013;79:5735–5744. doi: 10.1128/AEM.01807-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dinic M, et al. Exopolysaccharide produced by probiotic strain Lactobacillus paraplantarum BGCG11 reduces inflammatory hyperalgesia in rats. Front. Pharmacol. 2018;9:1–12. doi: 10.3389/fphar.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, et al. Latilactobacillus curvatus: A candidate probiotic with excellent fermentation properties and health benefits. Foods. 2020;9:1–20. doi: 10.3390/foods9101366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jo SG, et al. Lactobacillus curvatus WiKim38 isolated from kimchi induces IL-10 production in dendritic cells and alleviates DSS-induced colitis in mice. J. Microbiol. 2016;54:503–509. doi: 10.1007/s12275-016-6160-2. [DOI] [PubMed] [Google Scholar]
- 28.Tten HB, van den Blink I, Pronk P, Drillenburg MP, van Peppelenbosch SJHD. Dichotomal role of inhibition of p38 MAPK with SB 203580 in experimental colitis. Gut. 2002;50:507–513. doi: 10.1136/gut.50.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Y, et al. Lactobacillus acidophilus induces cytokine and chemokine production via NF-κB and p38 mitogen-activated protein kinase signaling pathways in intestinal epithelial cells. Clin. Vaccine Immunol. 2012;19:603–608. doi: 10.1128/CVI.05617-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh S, et al. Probiotic Lactobacillus fermentum strain JDFM216 stimulates the longevity and immune response of C. elegans through a nuclear hormone receptor. Sci. Rep. 2018;8:1–10. doi: 10.1038/s41598-018-25333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chelliah R, et al. In vitro and in vivo defensive effect of probiotic LAB against Pseudomonas aeruginosa using C. elegans model. Virulence. 2018;9:1489–1507. doi: 10.1080/21505594.2018.1518088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pastar I, et al. Interactions of methicillin resistant Staphylococcus aureus USA300 and P. aeruginosa in polymicrobial wound infection. PLoS ONE. 2013;8:1–11. doi: 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukic J, et al. Probiotics or pro-healers: the role of beneficial bacteria in tissue repair. Wound Repair Regen. 2017;25:912–922. doi: 10.1111/wrr.12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsilingiri K, Rescigno M. Postbiotics: what else? Benef. Microbes. 2013;4:101–107. doi: 10.3920/BM2012.0046. [DOI] [PubMed] [Google Scholar]
- 35.Popović N, et al. The influence of heat-killed Enterococcus faecium BGPAS1–3 on the tight junction protein expression and immune function in differentiated Caco-2 cells infected with Listeria monocytogenes ATCC 19111. Front. Microbiol. 2019;10:1006. doi: 10.3389/fmicb.2019.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuen GJ, Ausubel FM. Both live and dead Enterococci activate C. elegans host defense via immune and stress pathways. Virulence. 2018;9:683–699. doi: 10.1080/21505594.2018.1438025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J, et al. Heat-killed Lactobacillus spp. cells enhance survivals of C. elegans against Salmonella and Yersinia infections. Lett. Appl. Microbiol. 2015;61:523–530. doi: 10.1111/lam.12478. [DOI] [PubMed] [Google Scholar]
- 38.Pujol N, et al. A reverse genetic analysis of components of the Toll signaling pathway in C. elegans. Curr. Biol. 2001;11:809–821. doi: 10.1016/S0960-9822(01)00241-X. [DOI] [PubMed] [Google Scholar]
- 39.Couillault C, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 40.Popović N, et al. Yogurt produced by novel natural starter cultures improves gut epithelial barrier in vitro. Microorganisms. 2020;8:165. doi: 10.3390/microorganisms8101586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bajic SS, et al. GABA-producing natural dairy isolate from artisanal zlatar cheese attenuates gut inflammation and strengthens gut epithelial barrier in vitro. Front. Microbiol. 2019;10:960. doi: 10.3389/fmicb.2019.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vorburger C, Perlman SJ. The role of defensive symbionts in host–parasite coevolution. Biol. Rev. 2018;93:1747–1764. doi: 10.1111/brv.12417. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto T, et al. p38 MAP-kinase regulates function of gap and tight junctions during regeneration of rat hepatocytes. J. Hepatol. 2005;42:707–718. doi: 10.1016/j.jhep.2004.12.033. [DOI] [PubMed] [Google Scholar]
- 44.Peerapen P, Thongboonkerd V. p38 MAPK mediates calcium oxalate crystal-induced tight junction disruption in distal renal tubular epithelial cells. Sci. Rep. 2013;3:1–8. doi: 10.1038/srep01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mørch MGM, et al. The TGF-β ligand DBL-1 is a key player in a multifaceted probiotic protection against MRSA in C. elegans. Sci. Rep. 2021;11:1–14. doi: 10.1038/s41598-021-89831-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brenner S. The genetics of C. elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jokovic N, et al. A survey of the lactic acid bacteria isolated from Serbian artisanal dairy product kajmak. Int. J. Food Microbiol. 2008;127:305–311. doi: 10.1016/j.ijfoodmicro.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Herholz M, et al. KLF-1 orchestrates a xenobiotic detoxification program essential for longevity of mitochondrial mutants. Nat. Commun. 2019;10:1–15. doi: 10.1038/s41467-019-11275-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.